- Laboratory of Plant Pathology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan
Many plant viruses have positive-strand RNA [(+)RNA] as their genome. Therefore, it is not surprising that RNA-binding proteins (RBPs) play important roles during (+)RNA virus infection in host plants. Increasing evidence demonstrates that viral and host RBPs play critical roles in multiple steps of the viral life cycle, including translation and replication of viral genomic RNAs, and their intra- and intercellular movement. Although studies focusing on the RNA-binding activities of viral and host proteins, and their associations with membrane targeting, and intercellular movement of viral genomes have been limited to a few viruses, these studies have provided important insights into the molecular mechanisms underlying the replication and movement of viral genomic RNAs. In this review, we briefly overview the currently defined roles of viral and host RBPs whose RNA-binding activity have been confirmed experimentally in association with their membrane targeting, and intercellular movement of plant RNA virus genomes.
Introduction
Positive-strand RNA [(+)RNA] plant viruses are the most abundant in plant viruses, and cause diverse diseases in host plants. Because of the RNA nature of their genomes, it is not surprising that RNA-binding proteins (RBPs) of viral and host origin affect multiple steps of virus infection. It is known that viral RNA (vRNA) has critical non-template functions in addition to the essential function as a replication template (Pathak et al., 2011). After entry into the host cell, the genomic RNA of the (+)RNA virus serves as mRNA in the production of the replication proteins. The viral replication proteins recognize the viral genomic RNAs rapidly and specifically from within a pool of abundant cellular RNAs (e.g., rRNA, tRNA, and mRNA) before vRNAs are degraded by antiviral mechanisms. Plant (+)RNA viruses use organelle membranes, such as those of endoplasmic reticulum (ER), peroxisome, chloroloplast, mitochondrion, and tonoplast, by remodeling their structures for vRNA replication (Miller and Krijnse-Locker, 2008; Laliberté and Sanfaçon, 2010; Nagy and Pogany, 2011; Hyodo and Okuno, 2014). Thus, one of the critical steps in early replication is the selective recruitment of the (+)RNA template to the membrane and establishment of the viral replication complexes (VRCs).
Following the translation and replication processes, (+)RNA plant viruses spread their infection into neighboring uninfected cells through plasmodesmata (PD) by using their encoded movement protein (MP). Some plant viruses move through PD in the form of virus particles and others are thought to move in ribonucleoprotein (RNP) complexes that contain MP (Scholthof, 2005). MPs from a wide range of genera of plant viruses have been reported to bind nucleic acids in a sequence-non-specific manner (Waigmann et al., 2004; Lucas, 2006). The interaction between MPs and viral genomic RNAs is required for efficient virus cell-to-cell movement.
In this review, we overview the currently defined roles of viral- and host-derived RBPs in virus infection by focusing on the proteins whose RNA-binding activity has been confirmed experimentally in association with their membrane targeting, and intercellular movement of plant RNA virus genomes. We recommend readers consult several current reviews that discuss other topics relating to the roles of host RBPs during RNA virus infection (Li and Nagy, 2011; Huh and Paek, 2013).
Viral RBPs Involved in Membrane Targeting of Viral Genomic RNAs and their Replication
Red clover necrotic mosaic virus (RCNMV) has a bipartite genome, RNA1 and RNA2 (Okuno and Hiruki, 2013). An interesting feature of RCNMV is that RNA1 and RNA2 use different mechanisms for template selection during replication. RNA1 encodes an auxiliary replication protein p27 and the RNA-dependent RNA polymerase (RdRp) p88pol. p27 and p88pol localize to the ER membrane, where vRNA replication occurs (Turner et al., 2004; Kusumanegara et al., 2012; Hyodo et al., 2013). The membrane association of p27 is mediated by a stretch of 20 amino acids located in its N-terminal region that forms an amphipathic α-helix (Kusumanegara et al., 2012). p27 interacts directly with many partners such as p27 itself, p88pol, viral genomic RNAs, and host heat shock proteins, Hsp70 and Hsp90, and ADP ribosylation factor 1, and has multiple functions during virus infection (Mine et al., 2010, 2012; Iwakawa et al., 2011; Hyodo et al., 2013). RNA2 encodes no replication proteins, and its replication depends entirely on p27 and p88pol supplied by RNA1. To recruit the replication proteins, RNA2 has a Y-shaped RNA element (YRE) in its 3′-untranslated region (UTR; An et al., 2010). An in vitro RNA-aptamer (Strepto-Tag) affinity and immunoprecipitation assay showed that the YRE is necessary and sufficient to interact with p27 and with the 480 kDa replication complex (Iwakawa et al., 2011). The RNA-binding activity of p27 is required for recruiting RNA2 to the membrane (Hyodo et al., 2011).
p27–p27/p88pol interactions are not required for specific RNA-binding, suggesting that a monomeric form of p27 can recognize the YRE as the hallmark of RCNMV RNA2 and avoid the recruitment of unrelated RNAs from the pool of heterogeneous RNAs (Figure 1, Step 1). Interestingly, the p27–YRE interaction is modulated by Hsp90 (Mine et al., 2012). The p27–YRE interaction is disrupted by pharmacological inhibition of Hsp90, which also compromises the interaction between p27 and Hsp90, suggesting that Hsp90 modulates the conformation of p27 to make it suitable for recognizing YRE. This is consistent with the observations that purified recombinant p27 expressed in Escherichia coli has no affinity for the YRE in vitro, whereas p27 directly and specifically recognizes this RNA element in plant-derived cell-free extracts (Hyodo et al., 2011; Iwakawa et al., 2011; Mine et al., 2012).
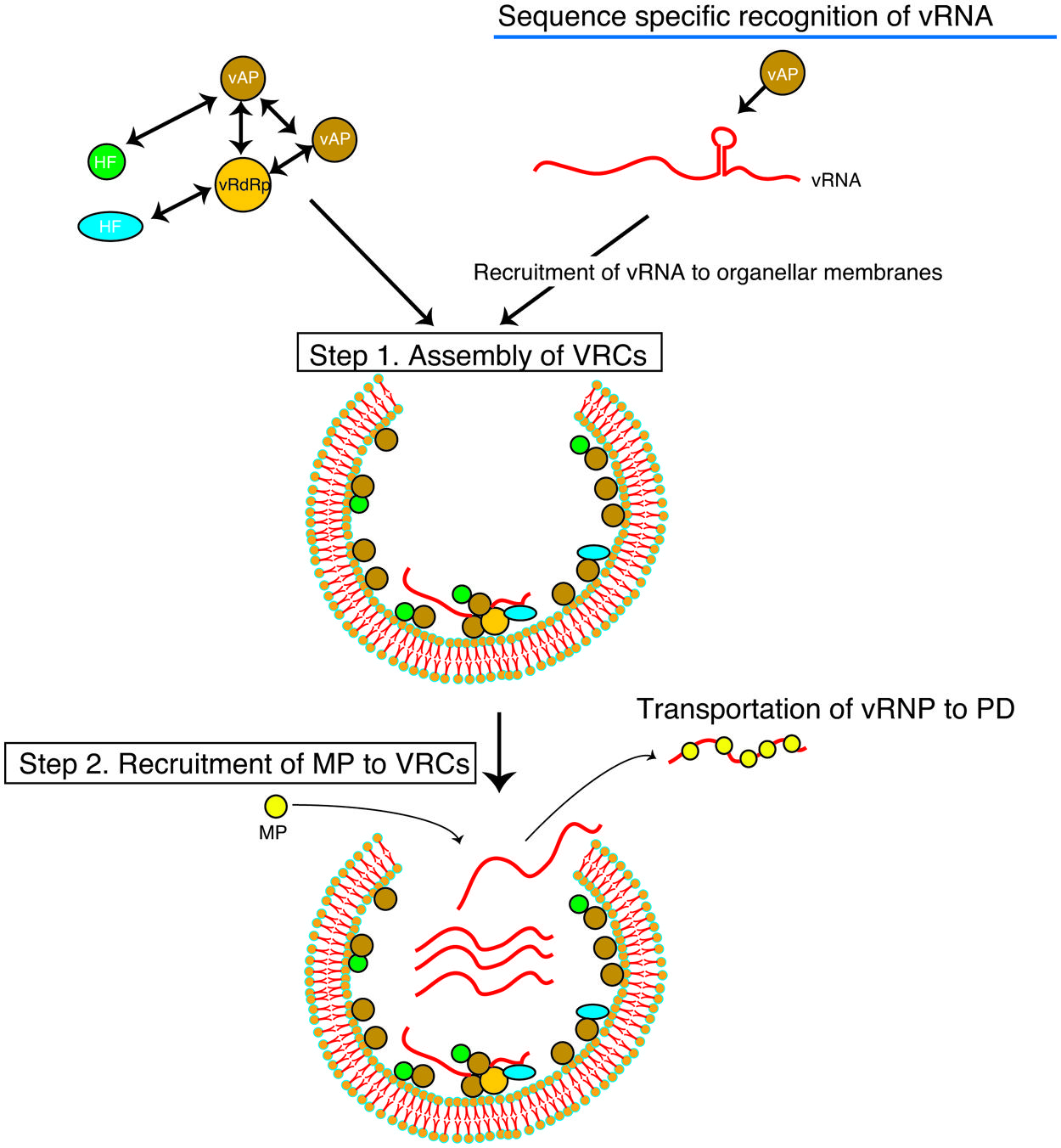
FIGURE 1. Schematic representation of the multiplication steps of a (+)RNA virus. Viral genomic RNA serves as the template for translation of the replicase proteins (vRdRp, viral RNA dependent RNA polymerase; vAP, auxiliary replicase protein). vAP recognizes vRNA in a sequence-specific manner and recruits to a selected membrane. vAP and vRdRp proteins interact with each other to form viral replication complexes (VRCs) with host proteins (HF) and vRNA (Step 1). Viral movement protein (MP) that is translated near the VRCs binds to vRNA. In the case of RCNMV, the MP expressed ectopically is recruited to the VRCs (Step 2; Kaido et al., 2009). MP binds to vRNA in or near the VRCs in a sequence non-specific manner and is transported to plasmodesmata (PD).
RNA1 does not have YRE-like structures and is not recognized by the replication proteins supplied in trans (Okamoto et al., 2008; Iwakawa et al., 2011). However, both p27 and p88pol interact with their ongoing translation templates in cis (Iwakawa et al., 2011). The RNA-binding activity of p27 is also important for the replication of RNA1 (Hyodo et al., 2011).
Replication of Tomato bushy stunt virus (TBSV) and related Cucumber necrosis virus (CNV) takes place at the peroxisomal membrane (McCartney et al., 2005; Panavas et al., 2005). TBSV and CNV infections cause the formation of peroxisomal multivesicular bodies, which may be derived from ER membranes (Barajas et al., 2009; Rochon et al., 2014). Auxiliary replication protein p33 of TBSV that has multiple functions during infection (Nagy et al., 2012) localizes to peroxisomes and plays a role in recruiting the vRNA to the peroxisome membrane. p33 has both non-specific and specific RNA-binding activities (Rajendran and Nagy, 2003; Pogany et al., 2005; Stork et al., 2011). In the specific binding mode, p33 binds to an internal replication element (IRE) located within the p92pol RdRp coding region of the viral genome (Pogany et al., 2005). The absence of the p33–p33/p92 interaction domain in p33 prevents specific RNA binding but allows non-specific RNA binding, suggesting that a multimeric (possibly dimeric) form of this protein is involved in the specific recognition of the viral replication template. The C-C mismatch within a conserved RNA helix of IRE is likely to be the hallmark of the TBSV replication template because mutations that disrupt the C-C mismatch completely abolish p33 binding to the IRE and p33-mediated recruitment to replication sites (Monkewich et al., 2005; Pogany et al., 2005; Pathak et al., 2012).
RNA replication of Brome mosaic virus (BMV) takes place in ER membrane-associated invaginations or spherules induced by replication protein 1a (Schwartz et al., 2002). In these sites, vRNAs are strongly protected from nucleases and presumably other antiviral factors (Schwartz et al., 2002). Although direct binding between 1a and vRNAs has not been demonstrated, 1a recruits template RNAs to the ER (Chen and Ahlquist, 2000; Chen et al., 2001; Schwartz et al., 2002). The intergenic region of RNA3 contains a box B motif that is conserved with the TΨC loop of tRNAs. The same box B motif is found in the 5′-UTRs of RNAs 1 and 2. These box B motifs and flanking sequences are likely to be required for the recruitment of the BMV RNAs into the VRCs, because mutations in box B motifs disrupt 1a-mediated in vivo stabilization of RNA2 and RNA3 (Janda and Ahlquist, 1998; Sullivan and Ahlquist, 1999; Chen et al., 2001).
A recent publication by Kawamura-Nagaya et al. (2014) showed that a 126-kDa replication protein of Tobacco mosaic virus (TMV) binds the 5′-UTR of the genomic RNA cotranslationally, and that this binding plays an important role in regulation of vRNA translation and replication.
Host RBPs Involved in Subcellular Localization of Viral and Subviral RNAs and Their Replication
Eukaryotic translation elongation factor 1A (eEF1A) is an abundant cellular protein that functions in protein degradation, apoptosis, nucleocytoplasmic trafficking, heat shock, and organization of the actin cytoskeleton in addition to its canonical role in delivering aminoacyl-tRNA to the elongating ribosome (Mateyak and Kinzy, 2010). Several RNA viruses have been reported to use eEF1A for RNA replication by a variety of mechanisms (Li et al., 2013). Studies of TBSV have provided information about the eEF1A function in viral template recruitment. A replication silencer element (RSE) located within the 3′-UTR of TBSV (+)RNA is the binding site of eEF1A (Li et al., 2009). The RSE is a cis-acting RNA element essential for TBSV replicase complex assembly and (–)RNA synthesis (Pogany et al., 2003; Panaviene et al., 2005). Pharmacological inhibitors of eEF1A, such as didemnin B (DB) and gamendazole (GM), block eEF1A–vRNA interaction, inhibit the membrane association of the TBSV-derived RNA replicon, and repress vRNA synthesis in vitro (Li et al., 2010). These findings suggest that eEF1A stimulates the recruitment of the (+)RNA template to the site of replication. Consistent with this, neither DB nor GM inhibits the RNA synthesis on the pre-assembled viral replicase complex (Li et al., 2010).
Bamboo mosaic virus (BaMV) RNA has been detected in chloroplasts, a putative replication site of BaMV (Lin et al., 1993). Chloroplast phosphoglycerate kinase (chl-PGK) of Nicotiana benthamiana can bind to the 3′-UTR of BaMV (+) RNA (Lin et al., 2007). PGK is an ATP-generating enzyme that acts in the glycolytic, gluconeogenesis, and photosynthetic pathways. chl-PGK is a nuclear-encoded protein that possesses an N-terminal targeting signal, which mediates posttranslational protein transport into the chloroplast (Jarvis and López-Juez, 2013). The role of chl-PGK during BaMV infection was reported recently (Cheng et al., 2013). In protoplasts derived from chl-PGK-silenced N. benthamiana, the accumulation of BaMV coat protein (CP) is inhibited, suggesting that PGK is required for the replication of BaMV. Interestingly, overexpression of cytosolic- or nuclear-localized PGK mutants has detrimental effects on BaMV infection. This is consistent with the author’s prediction that these PGK mutants mislead BaMV RNA away from chloroplasts to the cytoplasm or the nucleus that are unsuitable sites for viral replication. BaMV seems to use the trafficking system of chl-PGK to target vRNA to the chloroplasts for replication. In support of this, artificial chloroplast-targeting EF1a, which binds to BaMV (+)RNA, can partially complement BaMV accumulation in chl-PGK-knockdown N. benthamiana (Lin et al., 2007; Cheng et al., 2013). BaMV RNA localization is detected throughout the chloroplasts, suggesting that BaMV does not simply associate with the chloroplast outer membrane but that it enters the stroma (Lin et al., 1993; Cheng et al., 2013). It will be interesting to investigate the possible involvement of the chloroplast translocation machinery in BaMV localization.
A recent publication by Chaturvedi et al. (2014) showed that a tomato bromodomain-containing host protein (BRP1, also known as VIRP1) binds a satellite RNA of Cucumber mosaic virus (CMV), and that BRP1 plays an important role in the nuclear importation and multiplication of the satellite RNA. BRP1 also binds Potato spindle tuber viroid RNA both in vitro and in vivo, and is involved in the nuclear transportation and multiplication of the viroid (Martínez de Alba et al., 2003; Kalantidis et al., 2007). Further studies on host RBPs are awaited to elucidate the replication and movement mechanism of these subviral pathogens.
Viral RBPs Involved in the Intercellular Movement of vRNAs
Plant RNA viruses encode MPs that play a central role in the transport of viral genomes into neighboring uninfected cells. CPs also play ancillary roles in the virus cell-to-cell movement of some viruses (see reviews by Waigmann et al., 2004; Lucas, 2006; Harries and Nelson, 2008; Harries et al., 2010; Peña and Heinlein, 2012). Viruses that do not require virion formation for cell-to-cell movement are thought to form viral RNPs (vRNP) that contain viral genomic RNA and MP for the transport to and through PD. TMV MP, the first MP shown to bind nucleic acids, can bind both single-stranded RNA and DNA in a sequence-non-specific and cooperative manner, but cannot bind double-stranded DNA (Citovsky et al., 1990, 1992). After the first report of TMV MP binding to nucleic acids, the binding of other MPs to nucleic acids has been reported for 26 other viruses, including (+)RNA viruses, (-) RNA viruses, and DNA viruses (reviewed by Waigmann et al., 2004). In the past decade, at least nine MPs have been added to the list of nucleic acid-binding MPs: Cowpea mosaic virus (CPMV; Carvalho et al., 2004); Prunus necrotic ringspot virus (PNRSV; Herranz and Pallás, 2004); Rice yellow stunt virus (Huang et al., 2005); Apple chlorotic leaf spot virus (Isogai and Yoshikawa, 2005); Rice dwarf virus (Ji et al., 2011); Parietaria mottle virus (Martínez et al., 2014); Pelargonium flower break virus (Martínez-Turiño and Hernández, 2011); Melon necrotic spot virus (Navarro et al., 2006); Rice stripe virus (Xiong et al., 2008). Most MPs can bind single-stranded nucleic acids, and only three viral MPs (one geminivirus and two hordeiviruses) have been shown to bind double-stranded nucleic acids (Bleykasten et al., 1996; Donald et al., 1997; Rojas et al., 1998).
One significant function of RNA-binding by MP is to protect vRNA from the degradation by RNAse. For example, the binding of CMV MP to vRNA is rather weak and is RNAse-sensitive (Li and Palukaitis, 1996). CMV uses its CP to strengthen the weak binding to form RNAse-resistant vRNP complexes used in cell-to-cell movement (Nagano et al., 2001; Andreev et al., 2004; Kim et al., 2004). Triple gene block protein 1 (TGBp1) of Poa semilatent virus (PSLV) forms a high concentration salt-resistant vRNP (Kalinina et al., 2001), which may confer the ability for cell-to-cell and systemic movement in a CP-independent manner. The importance of the RNA-binding ability of MPs in viral cell-to-cell movement was confirmed by mutational analysis of the RNA-binding domains of RCNMV and PNRSV MPs (Giesman-Cookmeyer and Lommel, 1993; Herranz et al., 2005). The RNA-binding domains of PSLV MP (TGBp1) are unnecessary for cell-to-cell movement but are required for the systemic movement of the virus (Kalinina et al., 2001). Interestingly, the salt-resistant tight binding of Turnip crinkle virus (TCV) MP is a restricting factor for systemic infection by the virus (Wobbe et al., 1998). TCV strain B has a greater ability to spread in Arabidopsis thaliana ecotype Di-17 compared with TCV strain M. The two MPs differ only by one amino acid, and the binding to RNA by the MP of strain M has a greater tolerance to high salt concentration compared with that by the MP of strain B. It is possible that improper dissociation of the MP of strain M from vRNA can block the viral proliferation steps in newly infected cells.
It is interesting that CPMV MP can bind to the single-stranded nucleic acids despite the fact that CPMV moves through PD in the form of virions (van Lent et al., 1990; Wellink et al., 1993). The formation of a MP-vRNA complex of CPMV may be required for systemic infection, or CPMV MP may have distinct roles other than the movement of virions through PD.
All MPs that bind to nucleic acids reported so far do not exhibit sequence specificity. This raises the question of how MPs find vRNAs in infected cells. It is noteworthy that MPs of several RNA viruses, including TMV (Heinlein et al., 1998; Asurmendi et al., 2004; Kawakami et al., 2004), Potato virus X (PVX; Bamunusinghe et al., 2009; Tilsner et al., 2012, 2013), BMV (Dohi et al., 2001), and RCNMV (Kaido et al., 2009), colocalize with their VRCs. This may enable MPs to attach to viral genomes in the vicinity (Figure 1, Step 2). RCNMV MP is recruited to the VRC through the interaction with a host protein contained in the VRC (Kaido et al., 2009, 2011; Kaido and Okuno, unpublished results).
The only reported example of RNA cis-element that is required for virus cell-to-cell movement is the 5′-terminal stem–loop structure (SL1) of PVX genomic RNA. The green fluorescent protein gene containing the 107 nt terminal sequence of the PVX 5′-UTR was transported effectively to neighbor cells by cobombarded PVX (Lough et al., 2006). Immunoelectron microscopy and atomic-force microscopy revealed that the 5′-proximal region of PVX RNA is recognized by its CP, and that PVX MP (TGBp1) interacts with the CP (Karpova et al., 2006; Zayakina et al., 2008). Because PVX does not require virion formation for its cell-to-cell movement (Lough et al., 2000), PVX seems to move between cells through the CP-mediated TGBp1–vRNA complex.
Host RBPs Involved in the Intercellular Movement of vRNAs
In addition to MPs, plant viruses are thought to use host proteins for the cell-to-cell and systemic movement of viral genomes. To date, more than 40 cellular proteins have been reported to interact with MPs and some of these have been identified as a host factor involved in a variety of processes related to virus movement (Harries and Ding, 2011; Niehl and Heinlein, 2011). However, the RNA-binding activity of these host proteins remains to be defined.
Using two-dimensional electrophoresis, northwestern blot analysis, and mass spectrometry, 24 proteins of N. benthamiana were identified as being able to interact with the PVX 5′-terminal SL1 sequence (Cho et al., 2012b). Of these, two RBPs have been reported to be involved in the movement of a plant virus. NbDnaJ protein was shown by an electro-mobility shift assay to interact specifically with the minus-strand SL1 in vitro. NbDnaJ also interacts with PVX CP in planta, and negatively affects the systemic movement of the virus (Cho et al., 2012c). NbMPB2Cb, another SL1-interacting protein, interacts with PVX CP, TGBp1, and TGBp2 in planta, and negatively regulates the systemic movement of PVX (Cho et al., 2012a). NbMPB2Cb changed its subcellular localization from microtubules to ER in association with PVX infection. NbMPB2Cb may disturb the formation of TGBp2-induced ER-derived vesicles and this may negatively affect the functions of the vesicles by RNA–protein or protein–protein interactions.
Perspectives
Viral and host RBPs play diverse critical roles during infection. Some RBPs recruit vRNAs from the cytosol to the membranous replication sites, and others pick up and deliver RNAs to neighboring cells. These RBPs may require partners to accomplish their roles, and these partners may differ, depending on the infection stage; translation, replication, intra-, and inter-cellular movement. Viral replication proteins, such as RCNMV p27 and TBSV p33, are multifunctional proteins that interact with viral and host proteins in addition to vRNAs and play central roles in virus infection. It is also noteworthy that, during infection by (+)RNA viruses, translation, replication, and intra- and intercellular movements are often closely linked (Tilsner and Oparka, 2012). To investigate the regulatory mechanisms underlying these processes, we need to identify more RBPs and their partners. The recently developed interactome approach comprising in vivo UV cross-linking, capture of polyadenylated RNA on oligo(dT)-coated beads, and release of bound proteins by nuclease digestion has successfully identified a number of RBPs that bind specifically to cellular mRNAs (Baltz et al., 2012; Castello et al., 2012; Mitchell et al., 2013). Application of this approach combined with the RNA aptamer pulldown assay will be useful for identifying candidate RBPs involved in membrane targeting, intracellular trafficking and intercellular movement of plant RNA virus genomes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by a Grant-in Aid for Scientific Research (A) (22248002) from the Japan Society for the Promotion of Science.
References
An, M., Iwakawa, H.-O., Mine, A., Kaido, M., Mise, K., and Okuno, T. (2010). A Y-shaped RNA structure in the 3’ untranslated region together with the trans-activator and core promoter of Red clover necrotic mosaic virus RNA2 is required for its negative-strand RNA synthesis. Virology 405, 100–109. doi: 10.1016/j.virol.2010.05.022
Andreev, I. G., Kim, S. H., Kajinina, N. O., Rakitina, D. V., Fitzgerald, A. G., Palukaitis, P.,et al. (2004). Molecular interactions between a plant virus movement protein and RNA: force spectroscopy investigation. J. Mol. Biol. 339, 1041–1047. doi: 10.1016/j.jmb.2004.04.013
Asurmendi, S., Berg, R. H., Koo, J. C., and Beachy, R. N. (2004). Coat protein regulates formation of replication complexes during Tobacco mosaic virus infection. Proc. Natl. Acad. Sci. U.S.A. 101, 1415–1420. doi: 10.1073/pnas.0307778101
Baltz, A. G., Munschauer, M., Schwanhäusser, B., Vasile, A., Murakawa, Y., Schueler, M.,et al. (2012). The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690. doi: 10.1016/j.molcel.2012.05.021
Bamunusinghe, D., Hemenway, C. L., Nelson, R. S., Sanderfoot, A. A., Ye, C. M., Silva, M. A. T.,et al. (2009). Analysis of potato virus X replicase and TGBp3 subcellular locations. Virology 393, 272–285. doi: 10.1016/j.virol.2009.08.002
Barajas, D., Jiang, Y., and Nagy, P. D. (2009). A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 5:e1000705. doi: 10.1371/journal.ppat.1000705
Bleykasten, C., Gilmer, D., Guilley, H., Richards, K. E., and Jonard, G. (1996). Beet necrotic yellow vein virus 42 kDa triple gene block protein binds nucleic acid in vitro. J. Gen. Virol. 77, 889–897. doi: 10.1099/0022-1317-77-5-889
Carvalho, C. M., Pouwels, J., van Lent, J. W. M., Bisseling, T., Goldbach, R. W., and Wellink, J. (2004). The movement protein of Cowpea mosaic virus binds GTP and single-stranded nucleic acid in vitro. J. Virol. 78, 1591–1594. doi: 10.1128/JVI.78.3.1591-1594.2004
Castello, A., Fischer, B., Eichelbaum, K., Horos, R., Beckmann, B. M., Strein, C.,et al. (2012). Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406. doi: 10.1016/j.cell.2012.04.031
Chaturvedi, S., Kalantidis, K., and Rao, A. L. N. (2014). A bromodomain-containing host protein mediates the nuclear importation of a satellite RNA of Cucumber mosaic virus. J. Virol. 88, 1890–1896. doi: 10.1128/JVI.03082-13
Chen, J., and Ahlquist, P. (2000). Brome mosaic virus polymerase-like protein 2a is recruited to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74, 4310–4318. doi: 10.1128/JVI.74.9.4318.2000
Chen, J., Noueiry, A., and Ahlquist, P. (2001). Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5’ proximal RNA2 signal. J. Virol. 75, 3207–3219. doi: 10.1128/JVI.75.7.3207-3219.2001
Cheng, S.-F., Huang, Y.-P., Chen, L.-H., Hsu, Y.-H., and Tsai, C.-H. (2013). Chloroplast phosphoglycerate kinase is involved in the targeting of Bamboo mosaic virus to chloroplasts in Nicotiana benthamiana plants. Plant Physiol. 163, 1598–1608. doi: 10.1104/pp.113.229666
Citovsky, V., Knorr, D., Schuster, G., and Zambryski, P. C. (1990). The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60, 637–647. doi: 10.1016/0092-8674(90)90667-4
Citovsky, V., Wong, M. L., Shaw, A., Prasad, B. V. V., and Zambryski, P. C. (1992). Visualization and characterization of tobacco mosaic virus movement protein binding to single-strand nucleic acids. Plant Cell 4, 397–411. doi: 10.1105/tpc.4.4.397
Cho, S.-Y., Cho, W. K., Choi, H.-S., and Kim, K.-H. (2012a). Cis-acting element (SL1) of Potato virus X controls viral movement by interacting with the NbMPB2Cb and viral proteins. Virology 427, 166–176. doi: 10.1016/j.virol.2012.02.005
Cho, S.-Y., Cho, W. K., and Kim, K.-H. (2012b). Identification of tobacco proteins associated with the stem-loop 1 RNAs of Potato virus X. Mol. Cells 33, 379–384. doi: 10.1007/s10059-012-2298-x
Cho, S.-Y., Cho, W. K., Sohn, S.-H., and Kim, K.-H. (2012c). Interaction of the host protein NbDnaJ with Potato virus X minus-strand stem-loop 1 RNA and capsid protein affects viral replication and movement. Biochem. Biophys. Res. Commun. 417, 451–456. doi: 10.1016/j.bbrc.2011.11.137
Dohi, K., Mori, M., Furusawa, I., Mise, K., and Okuno, T. (2001). Brome mosaic virus replicase proteins localize with the movement protein at infection-specific cytoplasmic inclusions in infected barley leaf cells. Arch. Virol. 146, 1607–1615.
Donald, R. G., Lawrence, D. M., and Jackson, A. O. (1997). The barley stripe mosaic virus 58-kilodalton βb protein is a multifunctional RNA binding protein. J. Virol. 71, 1538–1546.
Giesman-Cookmeyer, D., and Lommel, S. A. (1993). Alanine scanning mutagenesis of a plant virus movement protein identifies three functional domains. Plant Cell 5, 973–982. doi: 10.1105/tpc.5.8.973
Harries, P., and Ding, B. (2011). Cellular factors in plant virus movement: at the leading edge of macromolecular trafficking in plants. Virology 411, 237–243. doi: 10.1016/j.virol.2010.12.021
Harries, P. A., and Nelson, R. S. (2008). “Movement of viruses in plants,” in Encyclopedia of Virology, 3rd Edn, Vol. 3, eds B. W. J. Mahy and M. H. V. Van Regenmortel (Oxford: Elsevier Academic press), 348–355.
Harries, P. A., Schoelz, J. E., and Nelson, R. S. (2010). Intracellular transport of viruses and their components: utilizing the cytoskeleton and membrane highways. Mol. Plant Microbe Interact. 23, 1381–1393. doi: 10.1094/MPMI-05-10-0121
Heinlein, M., Padgett, H. S., Gens, J. S., Pickard, B. G., Casper, S. J., Epel, B. L.,et al. (1998). Changing patterns of localization of the Tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10, 1107–1120. doi: 10.1105/tpc.10.7.1107
Herranz, M. C., and Pallás, V. (2004). RNA-binding properties and mapping of the RNA-binding domain from the movement protein of Prunus necrotic ringspot virus. J. Gen. Virol. 85, 761–768. doi: 10.1099/vir.0.19534-0
Herranz, M. C., Sanchez-Navarro, J.-A., Saurí, A., Mingarro, I., and Pallás, V. (2005). Mutational analysis of the RNA-binding domain of the Prunus necrotic ringspot virus (PNRSV) movement protein reveals its requirement for cell-to-cell movement. Virology 339, 31–41. doi: 10.1016/j.virol.2005.05.020
Huang, Y.-W., Geng, Y.-F., Ying, X.-B., Chen, X.-Y., and Fang, R.-X. (2005). Identification of a movement protein of Rice yellow stunt rhabdovirus. J. Virol. 79, 2108–2114. doi: 10.1128/JVI.79.4.2108-2114.2005
Huh, S. U., and Paek, K.-H. (2013). Plant RNA binding proteins for control of RNA virus infection. Front. Physiol. 4:397. doi: 10.3389/fphys.2013.00397
Hyodo, K., Mine, A., Iwakawa, H.-O., Kaido, M., Mise, K., and Okuno, T. (2011). Identification of amino acids in auxiliary replicase protein p27 critical for its RNA-binding activity and the assembly of the replicase complex in Red clover necrotic mosaic virus. Virology 413, 300–309. doi: 10.1016/j.virol.2011.02.017
Hyodo, K., Mine, A., Taniguchi, T., Kaido, M., Mise, K., Taniguchi, H.,et al. (2013). ADP ribosylation factor 1 plays an essential role in the replication of a plant RNA virus. J. Virol. 87, 163–176. doi: 10.1128/JVI.02383-12
Hyodo, K., and Okuno, T. (2014). Host factors used by positive-strand RNA plant viruses for genome replication. J. Gen. Plant Pathol. 80, 123–135. doi: 10.1007/s10327-014-0505-7
Isogai, M., and Yoshikawa, N. (2005). Mapping of RNA-binding domain of the Apple chlorotic leaf spot virus movement protein. J. Gen. Virol. 86, 225–229. doi: 10.1099/vir.0.80493-0
Iwakawa, H.-O., Mine, A., Hyodo, K., An, M., Kaido, M., Mise, K.,et al. (2011). Template recognition mechanism by replicase proteins differ between bipartite positive-strand genomic RNAs of a plant virus. J. Virol. 85, 497–509. doi: 10.1128/JVI.01754-10
Janda, M., and Ahlquist, P. (1998). Brome mosaic virus RNA replication protein 1a dramatically increase in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. U.S.A. 95, 2227–2232.
Jarvis, P., and López-Juez, E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. doi: 10.1038/nrm3702
Ji, X., Qian, D., Wei, C., Ye, G., Zhang, Z., Wu, Z.,et al. (2011). Movement protein Pns6 of Rice dwarf phytoreovirus has both ATPase and RNA binding activities. PLoS ONE 6:e24986. doi: 10.1371/journal.pone.0024986
Kaido, M., Funatsu, N., Tsuno, Y., Mise, K., and Okuno, T. (2011). Viral cell-to-cell movement requires formation of cortical punctate structures containing Red clover necrotic mosaic virus movement protein. Virology 413, 205–215. doi: 10.1016/j.virol.2011.02.008
Kaido, M., Tsuno, Y., Mise, K., and Okuno, T. (2009). Endoplasmic reticulum targeting of the Red clover necrotic mosaic virus movement protein is associated with the replication of viral RNA1 but not that of RNA2. Virology 395, 232–242. doi: 10.1016/j.virol.2009.09.022
Kalantidis, K., Denti, M. A., Tzortzakaki, S., Marinou, E., Tabler, M., and Tsagris, M. (2007). Virp1 is a host protein with a major role in Potato spindle tuber viroid infection in Nicotiana plants. J. Virol. 81, 12872–12880. doi: 10.1128/JVI.00974-07
Kalinina, N. O., Rakitina, D. A., Yelina, N. E., Zamayatnin, A. A. Jr., Stroganova, T. A., Klinov, D. V.,et al. (2001). RNA-binding properties of the 63 kDa protein encoded by the triple gene block of poa semilatent hodeivirus. J. Gen. Virol. 82, 2569–2578.
Karpova, O. V., Zayakina, O. V., Arkhipenko, M. V., Sheval, E. V., Kiselyova, O. I., Poljakov, O. I.,et al. (2006). Potato virus X RNA-mediated assembly of single-tailed ternary ‘coat protein-RNA-movement protein’ complexes. J. Gen. Virol. 87, 2731–2740. doi: 10.1099/vir.0.81993-0
Kawakami, S., Watanabe, Y., and Beachy, R. N. (2004). Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. U.S.A. 101, 6291–6296. doi: 10.1073/pnas.0401221101
Kawamura-Nagaya, K., Ishibashi, K., Huang, Y. P., Miyashita, S., and Ishikawa, M. (2014). Replication protein of tobacco mosaic virus cotranslationally binds the 5’ untranslated region of genomic RNA to enable viral replication. Proc. Natl. Acad. Sci. U.S.A. 111, 1620–1628. doi: 10.1073/pnas.1321660111
Kim, S. H., Kalinina, N. O., Andreev, I. G., Ryabov, E. V., Fitzgerald, A. G., Taliansky, M. E.,et al. (2004). The C-terminal 33 amino acids of the cucumber mosaic virus 3a protein affect virus movement, RNA binding and inhibition of infection and translation. J. Gen. Virol. 85, 221–230. doi: 10.1099/vir.0.19583-0
Kusumanegara, K., Mine, A., Hyodo, K., Kaido, M., Mise, K., and Okuno, T. (2012). Identification of domains in p27 auxiliary replicase protein essential for its association with the endoplasmic reticulum membranes in Red clover necrotic mosaic virus. Virology 433, 131–141. doi: 10.1016/j.virol.2012.07.017
Laliberté, J.-F., and Sanfaçon, H. (2010). Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 48, 69–91. doi: 10.1146/annurev-phyto-073009-114239
Li, D., Wei, T., Abbott, C. M., and Harrich, D. (2013). The unexpected roles of eukaryotic translation elongation factors in RNA virus replication and pathogenesis. Microbiol. Mol. Biol. Rev. 77, 253–266. doi: 10.1128/MMBR.00059-12
Li, Q., and Palukaitis, P. (1996). Comparison of the nucleic acid- and NTP-binding properties of the movement protein of cucumber mosaic virus and tobacco mosaic virus. Virology 216, 71–79. doi: 10.1006/viro.1996.0035
Li, Z., and Nagy, P. D. (2011). Diverse roles of host RNA-binding proteins in RNA virus replication. RNA Biol. 8, 305–315. doi: 10.4161/rna.8.2.15391
Li, Z., Pogany, J., Panavas, T., Xu, K., Esposito, A. M., Kinzy, T. G.,et al. (2009). Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology 385, 245–260. doi: 10.1016/j.virol.2008.11.041
Li, Z., Pogany, J., Tupman, S., Esposito, A. M., Kinzy, T. G., and Nagy, P. D. (2010). Translation elongation factor 1A facilitates the assembly of the Tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 6:e1001175. doi: 10.1371/journal.ppat.1001175
Lin, J.-W., Ding, M.-P., Hsu, Y.-H., and Tsai, C.-H. (2007). Chloroplast phophoglycerate kinase, a gluconeogenetic enzyme, is required for efficient accumulation of Bamboo mosaic virus. Nucleic Acids Res. 35, 424–432. doi: 10.1093/nar/gkl1061
Lin, N.-S., Chen, C.-C., and Hsu, Y.-H. (1993). Post-embedding in situ hybridization for localization of viral nucleic acid in ultra-thin sections. J. Histochem. Cytochem. 41, 1513–1519.
Lough, T. J., Lee, R. H., Emerson, S. J., Forster, R. L. S., and Lucas, W. J. (2006). Functional analysis of the 5’ untranslated region of potexvirus RNA reveals a role in viral replication and cell-to-cell movement. Virology 351, 455–465. doi: 10.1016/j.virol.2006.03.043
Lough, T. J., Netzler, N. E., Emerson, S. J., Sutherland, P., Carr, F., Beck, D. L.,et al. (2000). Cell-to-cell movement of Potexviruses: evidence for ribonucleoprotein complex involving the coat protein and first triple gene block protein. Mol. Plant Microbe Interact. 13, 962–974. doi: 10.1094/MPMI.2000.13.9.962
Lucas, W. J. (2006). Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology 344, 169–184. doi: 10.1016/j.virol.2005.09.026
Martínez, C., Coll-Bonfill, N., Aramburu, J., Pallás, V., Aparicio, F., and Galipienso, L. (2014). Two basic (hydrophilic) regions in the movement protein of Parietaria mottle virus have RNA binding activity and are required for cell-to-cell transport. Virus Res. 184, 54–61. doi: 10.1016/j.virusres.2014.02.008
Martínez de Alba, A. E., Sägesser, R., Tabler, M., and Tsagris, M. (2003). A bromodomain-containing protein from tomato specifically binds potato spindle tuber viroid RNA in vitro and in vivo. J. Virol. 77, 9685–9694. doi: 10.1128/JVI.77.17.9685-9694.2003
Martínez-Turiño, S., and Hernández, C. (2011). A membrane-associated movement protein of Pelargonium flower break virus shows RNA-binding activity and contains a biologically relevant leucine zipper-like motif. Virology 413, 310–319. doi: 10.1016/j.virol.2011.03.001
Mateyak, M. K., and Kinzy, T. G. (2010). eEF1A: thinking outside the ribosome. J. Biol. Chem. 285, 21209–21213. doi: 10.1074/jbc.R110.113795
McCartney, A. W., Greenwood, J. S., Fabian, M. R., White, K. A., and Mullen, R. T. (2005). Localization of the Tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17, 3513–3531. doi: 10.1105/tpc.105.036350
Miller, S., and Krijnse-Locker, J. (2008). Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6, 363–374. doi: 10.1038/nrmicro1890
Mine, A., Hyodo, K., Tajima, Y., Kusumanegara, K., Taniguchi, T., Kaido, M.,et al. (2012). Differential roles of Hsp70 and Hsp90 in the assembly of the replicase complex of a positive-strand RNA plant virus. J. Virol. 86, 12091–12104. doi: 10.1128/JVI.01659-12
Mine, A., Hyodo, K., Takeda, A., Kaido, M., Mise, K., and Okuno, T. (2010). Interactions between p27 and p88 replicase proteins of Red clover necrotic mosaic virus play an essential role in viral RNA replication and suppression of RNA silencing via the 480-kDa viral replicase complex assembly. Virology 407, 213–224. doi: 10.1016/j.virol.2011.02.017
Mitchell, S. F., Jain, S., She, M., and Parker, R. (2013). Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 20, 127–133. doi: 10.1038/nsmb.2468
Monkewich, S., Lin, H.-X., Fabian, M. R., Xu, W., Na, H., Ray, D.,et al. (2005). The p92 polymerase coding region contains an internal RNA element required at an early step in Tombusvirus genome replication. J. Virol. 79, 4848–4858. doi: 10.1128/JVI.79.8.4848-4858.2005
Nagano, H., Mise, K., Furusawa, I., and Okuno, T. (2001). Conversion in the requirement of coat protein in cell-to-cell movement mediated by the Cucumber mosaic virus movement protein. J. Virol. 75, 8045–8053. doi: 10.1128/JVI.75.17.8045-8053.2001
Nagy, P. D., Barajas, D., and Pogany, J. (2012). Host factors with regulatory roles in tombusvirus replication. Curr. Opin. Virol. 2, 685–692. doi: 10.1016/j.coviro.2012.10.004
Nagy, P. D., and Pogany, J. (2011). “Replication of plant RNA viruses,” in Recent Advances in Plant Virology, eds C. Caranta, M. A. Aranda, M. Tepfer, and J. J. Lopez-Moya (Norfolk: Caister Academic Press), 19–46.
Navarro, J. A., Genovés, A., Climent, J., Saurí, A., Martínez-Gil, L., Mingarro, I.,et al. (2006). RNA-binding properties and membrane insertion of Melon necrotic spot virus (MNSV) double gene block movement proteins. Virology 356, 57–67. doi: 10.1016/j.virol.2006.07.040
Niehl, A., and Heinlein, M. (2011). Cellular pathways for viral transport through plasmodesmata. Protoplasma 248, 75–99. doi: 10.1007/s00709-010-0246-1
Okamoto, K., Nagano, H., Iwakawa, H. O., Mizumoto, H., Takeda, A., Kaido, M.,et al. (2008). cis-Preferential requirement of a -1 frameshift product p88 for the replication of Red clover necrotic mosaic virus RNA1. Virology 375, 205–212. doi: 10.1016/j.virol.2008.02.004
Okuno, T., and Hiruki, C. (2013). Molecular biology and epidemiology of dianthoviruses. Adv. Virus Res. 87, 37–74. doi: 10.1016/B978-0-12-407698-3.00002-8
Panavas, T., Hawkins, C. M., Panaviene, Z., and Nagy, P. D. (2005). The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology 338, 81–95. doi: 10.1016/j.virol.2005.04.025
Panaviene, Z., Panavas, T., and Nagy, P. D. (2005). Role of an internal and two 3’-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 79, 10608–10618. doi: 10.1128/JVI.79.16.10608-10618.2005
Pathak, K. B., Pogany, J., and Nagy, P. D. (2011). Non-template functions of the viral RNA in plant RNA virus replication. Curr. Opin. Virol. 1, 332–338. doi: 10.1016/j.coviro.2011.09.011
Pathak, K. B., Pogany, J., Xu, K., White, K. A., and Nagy, P. D. (2012). Defining the roles of cis-acting elements in Tombusvirus replicase assembly in vitro. J. Virol. 86, 156–171. doi: 10.1128/JVI.00404-11
Peña, E. J., and Heinlein, M. (2012). RNA transport during TMV cell-to-cell movement. Front. Plant Sci. 3:193. doi: 10.3389/fpls.2012.00193
Pogany, J., Fabian, M. R., White, K. A., and Nagy, P. D. (2003). A replication silencer element in a plus-strand RNA virus. EMBO J. 22, 5602–5611. doi: 10.1093/emboj/cdg523
Pogany, J., White, K. A., and Nagy, P. D. (2005). Specific binding of Tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 79, 4859–4869. doi: 10.1128/JVI.79.8.4859-4869.2005
Rajendran, K. S., and Nagy, P. D. (2003). Characterization of the RNA-binding domains in the replicase proteins of Tomato bushy stunt virus. J. Virol. 77, 9244–9258. doi: 10.1128/JVI.77.17.9244-9258.2003
Rojas, M. R., Noueiry, A. O., Lucas, W. J., and Gilbertson, R. L. (1998). Bean dwarf mosaic geminivirus movement proteins recognize DNA in a form- and size-specific manner. Cell 95, 105–113. doi: 10.1016/S0092-8674(00)81786-9
Rochon, D., Singh, B., Reade, R., Theilmann, J., Ghoshal, K., Alam, S. B.,et al. (2014). The p33 auxiliary replicase protein of Cucumber necrosis virus targets peroxisomes and infection induces de novo peroxisome formation from the endoplasmic reticulum. Virology 452–453, 133–142. doi: 10.1016/j.virol.2013.12.035
Scholthof, H. B. (2005). Plant virus transport: motions of functional equivalence. Trends Plant Sci. 10, 376–382. doi: 10.1016/j.tplants.2005.07.002
Schwartz, M., Chen, J., Janda, M., Sullivan, M., den Boon, J., and Ahlquist, P. (2002). A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9, 505–514. doi: 10.1016/S1097-2765(02)00474-4
Stork, J., Kovalev, N., Sasvari, Z., and Nagy, P. D. (2011). RNA chaperone activity of the tombusviral p33 replication protein facilitates initiation of RNA synthesis by the viral RdRp in vitro. Virology 409, 338–347. doi: 10.1016/j.virol.2010.10.015
Sullivan, M., and Ahlquist, P. (1999). A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73, 2622–2632.
Tilsner, J., Linnik, O., Louveaux, M., Roberts, I. M., Chapman, S. N., and Oparka, K. J. (2013). Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 201, 981–995. doi: 10.1083/jcb.201304003
Tilsner, J., Linnik, O., Wright, K. M., Bell, K., Roberts, A. G., Lacomme, C.,et al. (2012). The TGB1 movement protein of Potato virus X reorganizes actin and endomembranes into the X-body, a viral replication factory. Plant Physiol. 158, 1359–1370. doi: 10.1104/pp.111.189605
Tilsner, J., and Oparka, K. J. (2012). Missing links? – the connection between replication and movement of plant RNA viruses. Curr. Opin. Virol. 2, 705–711. doi: 10.1016/j.coviro.2012.09.007
Turner, K. A., Sit, T. L., Callaway, A. S., Allen, N. S., and Lommel, S. A. (2004). Red clover necrotic mosaic virus replication proteins accumulate at the endoplasmic reticulum. Virology 320, 276–290. doi: 10.1016/j.virol.2003.12.006
van Lent, J., Wellink, J., and Goldbach, R. (1990). Evidence for the involvement of the 58K and 48K proteins in the intercellular movement of cowpea mosaic virus. J. Gen. Virol. 71, 219–223. doi: 10.1099/0022-1317-71-1-219
Waigmann, E., Ueki, S., Trutnyeva, K., and Citovsky, V. (2004). The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. Crit. Rev. Plant Sci. 23, 195–250. doi: 10.1080/07352680490452807
Wellink, J., van Lent, J. W., Verver, J., Sijen, T., Goldbach, R. W., and van Kammen, A. (1993). The cowpea mosaic virus M RNA-encoded 48-kilodalton protein is responsible for induction of tubular structures in protoplasts. J. Virol. 67, 3660–3664.
Wobbe, K. K., Akgoz, M., Dempsey, D. A., and Klessig, D. F. (1998). A single amino acid change in turnip crinkle virus movement protein p8 affects RNA binding and virulence on Arabidopsis thaliana. J. Virol. 72, 6247–6250.
Xiong, R., Wu, J., Zhou, Y., and Zhou, X. (2008). Identification of a movement protein of the Tenuivirus Rice stripe virus. J. Virol. 82, 12304–12311. doi: 10.1128/JVI.01696-08
Zayakina, O., Arkhipenko, M., Kozlovsky, S., Nikitin, N., Smirnov, A., Susi, P.,et al. (2008). Mutagenic analysis of Potato virus X movement protein (TGBp1) and the coat protein (CP): in vitro TGBp1-CP binding and viral RNA translation activation. Mol. Plant Pathol. 9, 37–44. doi: 10.1111/j.1364-3703.2007.00445.x
Keywords: RNA-binding protein, RNA virus, cellular membrane, cell-to-cell movement, RNA replication
Citation: Hyodo K, Kaido M and Okuno T (2014) Host and viral RNA-binding proteins involved in membrane targeting, replication and intercellular movement of plant RNA virus genomes. Front. Plant Sci. 5:321. doi: 10.3389/fpls.2014.00321
Received: 12 May 2014; Accepted: 18 June 2014;
Published online: 07 July 2014.
Edited by:
Sergey Morozov, Moscow State University, RussiaReviewed by:
Helene Sanfacon, Pacific Agri-Food Research Centre – Agriculture and Agri-Food Canada, CanadaJosé-Antonio Daròs, Consejo Superior de Investigaciones Científicas, Spain
Copyright © 2014 Hyodo, Kaido and Okuno. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tetsuro Okuno, Laboratory of Plant Pathology, Graduate School of Agriculture, Kyoto University, Kitashirakawa, Sakyo-ku, Kyoto 606-8502, Japan e-mail:b2t1bm9Aa2Fpcy5reW90by11LmFjLmpw
 Kiwamu Hyodo
Kiwamu Hyodo Masanori Kaido
Masanori Kaido Tetsuro Okuno
Tetsuro Okuno