- Phytopathology and Biochemistry, Biocenter Klein Flottbek, University of Hamburg, Hamburg, Germany
Plant diseases are one of the most studied subjects in the field of plant science due to their impact on crop yield and food security. Our increased understanding of plant–pathogen interactions was mainly driven by the development of new techniques that facilitated analyses on a subcellular and molecular level. The development of labeling technologies, which allowed the visualization and localization of cellular structures and proteins in live cell imaging, promoted the use of fluorescence and laser-scanning microscopy in the field of plant–pathogen interactions. Recent advances in new microscopic technologies opened their application in plant science and in the investigation of plant diseases. In this regard, in planta Förster/Fluorescence resonance energy transfer has demonstrated to facilitate the measurement of protein–protein interactions within the living tissue, supporting the analysis of regulatory pathways involved in plant immunity and putative host–pathogen interactions on a nanoscale level. Localization microscopy, an emerging, non-invasive microscopic technology, will allow investigations with a nanoscale resolution leading to new possibilities in the understanding of molecular processes.
Introduction
The plant cell wall and its outer cuticle represent the first line of defense to biotic and abiotic stress. Based on its crucial role in plant defense, the cell wall also constitutes a primary target of plant pathogen attack and is constantly subject to various extrinsic, physical forces. While pathogens try to enter the cell to establish infection structures for further colonization of the tissue, which can be associated with a reprogramming of the plant metabolism for its own purpose, as in the case of obligate biotrophic and hemibiotrophic pathogens with their release of effectors (Giraldo and Valent, 2013), the plant responds to the attack by a variety of cell wall-associated defense reactions.
Even though we already have a detailed insight into the processes and signaling pathways that follow recognition of pathogens by plasma membrane-bound receptors (Hamann, 2012; Wolf et al., 2012), we only have little knowledge about processes that occur directly at the cell wall. In recent years, growing evidence suggests that a mechanism for cell wall integrity may exists, which monitors and maintains functional integrity of the cell and includes restructuring and rebuilding of cell wall components (Knepper and Day, 2010; Steinbrenner et al., 2012). In this regard, the plant cell wall seems to be more dynamic as previously expected. In response to pathogen attack, the main cell wall polymer cellulose, a (1,4)-β-glucan, forms a three-dimensional network with the (1,3)-β-glucan callose (Eggert et al., 2014), a cell wall polymer that is directly associated with the plant’s innate immunity (Hardham et al., 2007). A prerequisite of these advances in cell wall visualization is the increasing number of advanced molecular dyes and techniques that have become available for high resolution imaging of cell wall integrity processes and for localization of individual cell wall components.
Here, we describe the role that Förster/Fluorescence resonance energy transfer (FRET) and localization microscopy has played and probably is going to play in investigating plant–pathogen interaction by highlighting processes occurring at the plant cell wall and being part of cell wall integrity mechanisms. In contrast to scanning electron microscopy, transmission electron microscopy, or atomic force microscopy, FRET and localization microscopy are both suitable to visualize processes and interactions between different components with a resolution in the submicron to nanometer scale in live cell imaging where the tissue is still intact and in its native state maintaining the full functionality of enzymes as well as arrangement of cell wall fibrils and other components.
This perspective focuses on the new and emerging possibilities in subcellular investigation of plant–pathogen interactions and the understanding of how individual molecules, such as callose and cellulose, and its regulating enzymes allow plants to perceive pathogens and pathogens to infect their hosts.
FRET Microscopy
Förster/Fluorescence resonance energy transfer is commonly used to image the interaction of fluorescent labeled molecules or proteins in living cells. The physical principle of FRET is a distance-dependent interaction between the electronic excited states of two dye molecules where excitation energy is transferred from a donor molecule to an acceptor molecule without emission of a photon. This transfer of energy only happens if (i) the absorption spectrum of the acceptor overlap with the fluorescence emission spectrum of the donor and (ii) donor and acceptor molecules are in close proximity (1–10 nm; Jares-Erijman and Jovin, 2003; Grecco and Verveer, 2011). Donor and acceptor molecules might be either fused to different putative interaction partners or linked with each other by a spacer. In the first case, energy transfer takes place as soon as both partners bind each other. In second case, changes in protein folding induced by shifts in the redox state, tension, or pH reduce the distance of the FRET pair partners to a level that allows energy transfer (Gjetting et al., 2012). The FRET pair can function as a FRET sensor that generally consists of a substrate-specific binding domain, which is flanked by a suitable donor on the one and acceptor on the other site. Substrate binding causes a change in distance or orientation of the two fluorophore that is translated into a measurable change in energy transfer. FRET sensors have been developed for a large spectrum of substrates and are used to analyze dynamic processes in mammals and plant cells (Okumoto et al., 2012; Jones et al., 2013; Okumoto, 2014). Interestingly, mammal sensors were successfully used in plant cells to identify new sugar transporters (Chen et al., 2010), highlighting the comparability of basic cellular mechanisms in different biological kingdoms.
FRET in Analyzing Protein–Protein Interaction of Plant Penetration Resistance
Over 150 years ago, deBary (1863) discovered cell wall thickenings in plants, so called papillae, at sites where fungal pathogens penetrated through the cell wall. Chemical analyses of papillae have identified callose, a (1,3)-β-glucan with some (1,6)-branches (Aspinall and Kessler, 1957), as the most common constituent among others, which may also include protein (e.g., peroxidases, antimicrobial thionins), phenolics, and other constituents (Aist and Williams, 1971; Mercer et al., 1974; Aist, 1976; Mims et al., 2000). The formation as well as degradation of papilla requires a high spatial and temporal regulation of transport processes between the infection site, the plasma membrane, and the trans-Golgi network. Alteration or disruption of these regulatory processes cannot only result in increased susceptibility to pathogen attack, but also induce complete penetration resistance to powdery mildews, which are biotrophic fungal pathogens. This was illustrated in studies with Arabidopsis (Arabidopsis thaliana) where overexpression of GSL5 (GLUCAN SYNTHASE-LIKE 5, also known as POWDERY MILDEW RESISTENT 4), a gene encoding a stress-induced callose synthase (Jacobs et al., 2003; Nishimura et al., 2003), resulted in early and elevated callose biosynthesis at sites of attempted penetration by the adapted powdery mildew Golovinomyces cichoracearum and the non-adapted powdery mildew Blumeria graminis f.sp. hordei. These enhanced callose deposits prevented pathogen ingress (Ellinger et al., 2013). Penetration resistance to fungal pathogens was also observed after disruption of mildew resistance locus O (MLO) protein family members in Arabidopsis infected with G. cichoracearum and G. orontii as well as the necrotrophic fungus Botrytis cinerea (Consonni et al., 2006, 2010), in tomato (Solanum lycopersicum) after infection with the powdery mildew Oidium neolycopersici (Bai et al., 2008), and in barley (Hordeum vulgare) after B. graminis f.sp. hordei infection (Jørgensen, 1992; Piffanelli et al., 2004). MLO proteins have been characterized as a family of plasma membrane-localized MLO proteins that are required for successful entry of adapted powdery mildew species in leaf epidermal cells (Panstruga, 2005).
Förster/Fluorescence resonance energy transfer microscopy has been used to analyze the recruitment and interaction dynamics of components that contribute to plant penetration resistance, which strongly promoted this technique in the field of plant–pathogen interaction. Using FRET-acceptor photo bleaching (APB; Karpova et al., 2003) and FRET-Fluorescence lifetime imaging microscopy (FLIM; Becker, 2012), new spatiotemporal information about the interaction of MLO and calmodulin, a cytoplasmatic calcium sensor (Cheval et al., 2013), and a new function of MLO was obtained (Iwai and Uyeda, 2008), which had remained undetected before using these advanced microscopic technologies. A prominent field of FRET-APB application is to verify dynamic protein–protein interaction between cytosolic and membrane-bound proteins, like the interaction of the endoplasmic reticulum (ER)-resident BAX INHIBITOR-1 (BI-1) protein with the cytochrome oxidase CYP83A1 during inoculation with the adapted powdery mildew fungus Erysiphe cruciferarum (Weis et al., 2013). A further field of FRET-APB application could be the localization of membrane-bound enzymes or enzyme complexes that are involved in reorganization and reinforcement of the cell wall after pathogen attack. In this regard, the stress-induced callose synthase GSL5 from Arabidopsis could be a suitable target. Its involvement in pathogen-induced cell wall rearrangements was clearly shown (Jacobs et al., 2003; Nishimura et al., 2003; Eggert et al., 2014); and a successful fluorescence-tagging was also demonstrated (Ellinger et al., 2013; Naumann et al., 2013). Because quantitative proteomics of plasma membrane microdomains from poplar (Populus trichocarpa) cell suspension cultures suggested a localization of callose synthases in lipid rafts (Srivastava et al., 2013), a FRET analysis of tagged GSL5 and lipid raft-resident protein used as markers could reveal whether this specific localization would also occur in intact plant tissue. In addition, this analysis might provide information about the mechanisms of enzyme translocation that was already observed during infection (Ellinger et al., 2013). Because it has been suggested that a callose synthase complex is formed at sites of callose biosynthesis (Verma and Hong, 2001), FRET-ABP can be used as a microscopic tool to screen for possible interaction partners of the callose synthase GSL5 at sites of attempted fungal penetration. A promising target for a screening would be monomeric GTPases (Figure 1) that were already identified as putative interaction and complex forming partners in Saccharomyces cerevisiae where a GTPase from the Rho family might control the phosphorylation status of the callose synthase (Calonge et al., 2003), and in Arabidopsis where the Rho-like GTPase Rop1 might be involved in regulating callose biosynthesis of GSL6 at cell plate through interaction with the UDP-glucose transferase UGT1 (Hong et al., 2001).
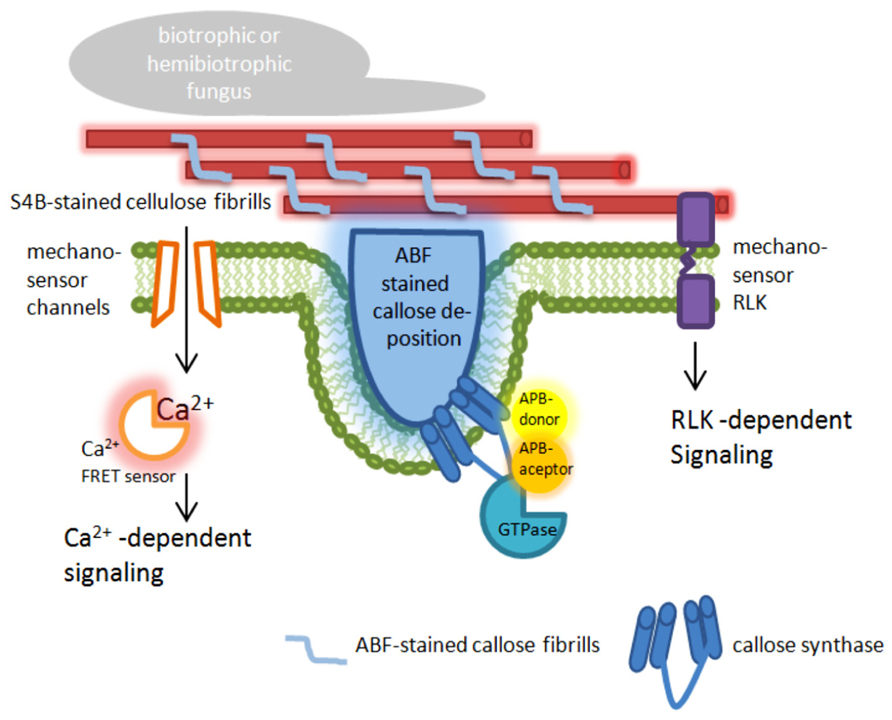
FIGURE 1. Targets for nanoscale microcopy tools to analyze plant immunity-related cell wall modifications. The presented model highlights possible targets at the plasma membrane and the cell wall in epidermal leaf cells of plants attacked by pathogens. ABF, aniline blue fluorochrome [fluorescent dye specific for the (1,3)-β-glucan callose]; S4B, pontamine fast scarlet 4B [fluorescent dye specific for the (1,4)-β-glucan cellulose]; APB, acceptor photo bleaching; FRET, Förster/Fluorescence resonance energy transfer; RLK, receptor-like kinase.
FRET for Sensing Ion Influx and Mechanical Stress
During entry of pathogenic fungi into plant tissue, a localized deformation of the cell surface occurs. The perception of those punctual mechanical signals induces a dynamic protein interaction (as described above) and a very rapid reorganization of actin microfilaments, ER, and peroxisoms (Koh et al., 2005). This is also associated with ion flux within the plant cell (Meng and Sachs, 2011). Using the FRET based Ca2+ sensor Yellow Cameleon 3.6 (Nagai et al., 2004), it was demonstrated in root hairs that localized cell wall deformation induced a monophasic Ca2+ increase starting from the site of stress and spreading through the cytoplasm, which finally activated extracellular production of reactive oxygen species at the cell wall (Monshausen et al., 2008, 2009). At the site of fungal ingress, also callose is deposited in the apoplastic space (Figure 1). The amount of deposited callose depended on the cytoplasmatic Ca2+ level because the presence of chelators or inhibitors of Ca2+ channels reduced callose biosynthesis (Mercer et al., 1974; Mims et al., 2000). It has been speculated that possible stress-activated Ca2+-permeable channels might be gated by changes in membrane tension (Bhat et al., 2005).
The sensing of mechanical stress and the resulting induction of callose biosynthesis or cellulose remodeling likely occurs via cell wall sensors. These mechanosensory proteins with their putative carbohydrate-binding domains, like lectin receptor kinases (Vaid et al., 2013), cell wall-associated kinases (Kohorn and Kohorn, 2012), or the THESEUS 1 receptor-like kinase (Cheung and Wu, 2011), might be linked to the cytoskeleton or to glycosylated proteins and polysaccharides of the cell wall. Here, they could transmit information about deformation of the cell wall via kinase-dependent phosphorylation of target proteins. Although neither phosphorylation of cell wall integrity target proteins nor tension-depended activation of cell wall sensors was shown so far in plant–fungus interactions, functional FRET sensors for phosphorylation (Sato and Umezawa, 2004; Mirabet et al., 2011) as well as tension (Nagai et al., 2004; Monshausen et al., 2009; Ellinger et al., 2013; Monshausen and Haswell, 2013) already exist. As stated by Ehrhardt and Frommer (2012), these tools have to make useable to the plant science community.
However, a major challenge in investigating cell wall integrity during plant–pathogen interaction using FRET technology is the autofluorescence in photosynthetic active tissue and especially of cell wall material. Therefore, fluorophores for these applications have to be carefully selected. An alternative would be the use of bioluminescent proteins, like the Ca2+-sensitive aequorin (Kunkel et al., 2007) where excitation is caused by a chemical reaction instead of light. In general, it has to be considered that FRET efficiency strongly depends on the distance that separates the FRET pair and the spatial orientation of the fluorophores. In most applications that we referred to in this article, fluorophores were fused to the proteins of interest. However, this protein modification may change its conformation, activity, or even stoichiometry in a protein-protein-interaction. Therefore, a lack of FRET efficiency cannot only indicate a non-interaction of proteins but also an inappropriate protein modification. As a consequence, various constructs with alternative fusion sites or linker usage would have to be tested to eventually distinguish between inappropriate modification and actual protein non-interaction. Nevertheless, for both, negative and positive FRET signals, appropriate controls have to be included; and results of FRET experiments should be verified by other methods like protein co-precipitation experiments in case of protein-protein interactions.
Localization Microscopy
A great advantage of FRET microscopy is the possibility to resolve molecule or protein interaction at nanoscale in live cell imaging. However, the resolution of the imaging system itself is not increased in FRET application. As a consequence, it is possible to determine that a specific interaction between the partners of FRET pair occurs in the cell, but it is not possible to determine on a nanoscale level where exactly the interaction takes place in the cell.
To overcome this limitation, new imaging technologies are now available, which allow the localization of molecules and proteins below the diffraction limit and are commonly referred to as super-resolution microscopy (Hell, 2007; Agrawal et al., 2013; Requejo-Isidro, 2013). These new microscopic techniques include stimulated emission depletion fluorescence (STED) microscopy with a possible resolution of 35 nm in the far field (Hell and Wichmann, 1994), but without reports of successful application on intact plant tissue due to the relatively high laser energy that destroys the tissue. Successful application of a super-resolution microscopy technique on intact plant cells was reported for structured illumination microscopy (SIM). This technique allowed a super-resolution time-lapse imaging of microtubule dynamics and organization in Arabidopsis (Komis et al., 2014) and, recently, the visualization and localization of specific domains and effector proteins at the extrahaustorial membrane of the pathogenic oomycete Phytophthora infestans (Bozkurt et al., 2014). In SIM applications, it is possible to achieve a lateral resolution that exceeds the classical diffraction limit by a factor of two, resulting in a possible axially resolution of 400 nm and up to 100 nm in x–y direction (Gustafsson, 2000; Schermelleh et al., 2008). This relatively high resolution can be exceeded by a factor of five in localization microscopy, which is one of the most dynamic and evolving fields of nanoscale imaging. In localization microscopy techniques such as photoactivated localization microscopy (PALM; Betzig et al., 2006) or stochastic optical reconstruction microscopy (STORM; Rust et al., 2006; Heilemann et al., 2008), a lateral resolution as high as 20 and 50 nm in the axial direction can be achieved (Bates et al., 2008). Like SIM, localization microscopy has already been successfully applied on intact plant tissue to visualize and localize cell wall polymers with a nanoscale resolution of below 50 nm (Liesche et al., 2013; Eggert et al., 2014). It turned out that both, the cellulose-specific fluorescent dye pontamine fast scarlet 4B (S4B; Anderson et al., 2010) and the callose-specific fluorescent dye aniline blue fluorochrome (ABF; Evans et al., 1984) revealed stochastic intensity fluctuations and photoblinking in stained cell walls (Eggert et al., 2014), which is a prerequisite for an application in localization microscopy. Besides a confirmation of the previously known orientation and size of the S4B-stained cellulose fibrils, new information about the interaction of cellulose and callose during pathogen attack was provided. Callose fibrils migrated into and penetrated through the preexisting cellulosic cell wall, which resulted in the formation of a three-dimensional polymer network (Eggert et al., 2014). This is a first example where localization microscopy helped to uncover previously unknown, plant immunity-related alterations and rearrangements of cell wall precisely at the site of attempted fungal penetration. Hence, localization microscopy can be used to examine localized cell wall changes induced either by stress or processes related to maintain cell wall integrity. In this regard, it would be useful to test additional fluorescent dyes that specifically label cell wall components or polymers other than cellulose and callose to receive a complete, three-dimensional overview of the cell wall and its changes in response to different types of stress. An alternative for fluorescent dyes would be the use of antibodies that are specific for different polymers or oligosaccharides of the plant cell wall. These antibodies are already known and have been tested for their specificity in intact plant tissue (Pattathil et al., 2010). For their application in localization microscopy, the primary antibodies could be either directly labeled, for example with fluorophores like CAGE552 that belongs to a class of caged rhodamines (Belov et al., 2010), or detected by an appropriate secondary antibody, which was successfully tested for the pathogen-induced callose deposition (Eggert et al., 2014). However, the use of primary and secondary antibody can decrease the maximum resolution compared to direct labeling (Ries et al., 2012; Eggert et al., 2014). In a next step, localization microscopy of cell wall polymers and components could be combined with the detection of tagged proteins within the cell wall, the apoplast, or the plasma membrane. The plasma membrane currently represents the z-direction limit in localization microscopy of intact, uncut plant tissue because this imaging technique is usually combined with total internal reflection microcopy resulting in a restriction of imaging to approximately 100–200 nm in z-direction (Cleemann et al., 1997; Martin-Fernandez et al., 2013). However, this limitation of imaging in localization microscopy would be sufficient to analyze important processes related to cell wall rearrangement as indicated in Figure 1.
The opportunities that localization microscopy already offers for nanoscale imaging of intact plant tissue, and that will likely be expended in the future, raises the question whether this new imaging tool would replace FRET due to the possibility that tagged proteins and labeled molecules could be directly imaged. At the current technical status of localization microscopy, this is not very likely because this imaging technique is limited in practice to resolutions of tens of nanometers, which is still far above the distance of FRET interactions with 1–10 nm and would not be sufficient to conclusively prove direct molecular interactions. In a future perspective, the implementation of super-resolution FRET microscopy as described by Grecco and Verveer (2011) could be a strategy to overcome limitations of both techniques and combine their superior benefits.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agrawal, U., Reilly, D. T., and Schroeder, C. M. (2013). Zooming in on biological processes with fluorescence nanoscopy. Curr. Opin. Biotechnol. 24, 646–653. doi: 10.1016/j.copbio.2013.02.016
Aist, J. R. (1976). “Papillae and related wound plugs of plant cells,” in Annual Review of Phytopathology, eds K. F. Baker, G. A. Zentmeyer, and E. B. Cowling (Palo Alto: Annual Reviews Inc.), 145–163.
Aist, J. R., and Williams, P. H. (1971). The cytology and kinetics of cabbage root hair penetration by Plasmodiophora brassicae. Can. J. Bot. 49, 2023–2034. doi: 10.1139/b71-284
Anderson, C. T., Carroll, A., Akhmetova, L., and Somerville, C. (2010). Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 152, 787–796. doi: 10.1104/pp.109.150128
Aspinall, G. O., and Kessler, G. (1957). The Structure of Callose from the Grape Vine. London: Chemistry and Industry.
Bai, Y., Pavan, S., Zheng, Z., Zappel, N. F., Reinstadler, A., Lotti, C., et al. (2008). Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of mlo function. Mol. Plant Microbe Interact. 21, 30–39. doi: 10.1094/MPMI-21-1-0030
Bates, M., Huang, B., and Zhuang, X. (2008). Super-resolution microscopy by nanoscale localization of photo-switchable fluorescent probes. Curr. Opin. Chem. Biol. 12, 505–514. doi: 10.1016/j.cbpa.2008.08.008
Becker, W. (2012). Fluorescence lifetime imaging – techniques and applications. J. Microsc. 247, 119–136. doi: 10.1111/j.1365-2818.2012.03618.x
Belov, V. N., Wurm, C. A., Boyarskiy, V. P., Jakobs, S., and Hell, S. W. (2010). Rhodamines NN: a novel class of caged fluorescent dyes. Angew. Chem. Int. Ed. Engl. 49, 3520–3523. doi: 10.1002/anie.201000150
Betzig, E., Patterson, G. H., Sougrat, R., Lindwasser, O. W., Olenych, S., Bonifacino, J. S., et al. (2006). Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645. doi: 10.1126/science.1127344
Bhat, R. A., Miklis, M., Schmelzer, E., Schulze-Lefert, P., and Panstruga, R. (2005). Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. U.S.A. 102, 3135–3140. doi: 10.1073/pnas.0500012102
Bozkurt, T. O., Richardson, A., Dagdas, Y. F., Mongrand, S., Kamoun, S., and Raffaele, S. (2014). The plant membrane-associated REM1.3 remorin accumulates in discrete perihaustorial domains and enhances susceptibility to Phytophthora infestans. Plant Physiol. doi: 10.1104/pp.114.235804 [Epub ahead of print].
Calonge, T. M., Arellano, M., Coll, P. M., and Perez, P. (2003). Rga5p is a specific Rho1p GTPase-activating protein that regulates cell integrity in Schizosaccharomyces pombe. Mol. Microbiol. 47, 507–518. doi: 10.1046/j.1365-2958.2003.03312.x
Chen, L. Q., Hou, B. H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. doi: 10.1038/nature09606
Cheung, A. Y., and Wu, H. M. (2011). THESEUS 1, FERONIA and relatives: a family of cell wall-sensing receptor kinases? Curr. Opin. Plant Biol. 14, 632–641. doi: 10.1016/j.pbi.2011.09.001
Cheval, C., Aldon, D., Galaud, J. P., and Ranty, B. (2013). Calcium/calmodulin-mediated regulation of plant immunity. Biochim. Biophys. Acta 1833, 1766–1771. doi: 10.1016/j.bbamcr.2013.01.031
Cleemann, L., Dimassa, G., and Morad, M. (1997). Ca2+ sparks within 200 nm of the sarcolemma of rat ventricular cells: evidence from total internal reflection fluorescence microscopy. Adv. Exp. Med. Biol. 430, 57–65. doi: 10.1007/978-1-4615-5959-7_5
Consonni, C., Bednarek, P., Humphry, M., Francocci, F., Ferrari, S., Harzen, A., et al. (2010). Tryptophan-derived metabolites are required for antifungal defense in the Arabidopsis mlo2 mutant. Plant Physiol. 152, 1544–1561. doi: 10.1104/pp.109.147660
Consonni, C., Humphry, M. E., Hartmann, H. A., Livaja, M., Durner, J., Westphal, L., et al. (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38, 716–720. doi: 10.1038/ng1806
deBary, A. (1863). Recherches sur le développement de quelques champignons parasites. Ann. Sci. Nat. Bot. Biol. Vég. 20, 5–148.
Eggert, D., Naumann, M., Reimer, R., and Voigt, C. A. (2014). Nanoscale glucan polymer network causes pathogen resistance. Sci. Rep. 4:4159. doi: 10.1038/srep04159
Ehrhardt, D. W., and Frommer, W. B. (2012). New technologies for 21st century plant science. Plant Cell 24, 374–394. doi: 10.1105/tpc.111.093302
Ellinger, D., Naumann, M., Falter, C., Zwikowics, C., Jamrow, T., Manisseri, C., et al. (2013). Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol. 161, 1433–1444. doi: 10.1104/pp.112.211011
Evans, N. A., Hoyne, P. A., and Stone, B. A. (1984). Characteristics and specificity of the interaction of a fluorochrome from aniline blue (sirofluor) with polysaccharides. Carbohyd. Polym. 4, 215–230. doi: 10.1016/0144-8617(84)90012-2
Giraldo, M. C., and Valent, B. (2013). Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814. doi: 10.1038/nrmicro3119
Gjetting, K. S., Ytting, C. K., Schulz, A., and Fuglsang, A. T. (2012). Live imaging of intra- and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J. Exp. Bot. 63, 3207–3218. doi: 10.1093/jxb/ers040
Grecco, H. E., and Verveer, P. J. (2011). FRET in cell biology: still shining in the age of super-resolution? Chemphyschem 12, 484–490. doi: 10.1002/cphc.201000795
Gustafsson, M. G. (2000). Suprassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82–87. doi: 10.1046/j.1365-2818.2000.00710.x
Hamann, T. (2012). Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front. Plant Sci. 3:77. doi: 10.3389/fpls.2012.00077
Hardham, A. R., Jones, D. A., and Takemoto, D. (2007). Cytoskeleton and cell wall function in penetration resistance. Curr. Opin. Plant Biol. 10, 342–348. doi: 10.1016/j.pbi.2007.05.001
Heilemann, M., Van De Linde, S., Schuttpelz, M., Kasper, R., Seefeldt, B., Mukherjee, A., et al. (2008). Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47, 6172–6176. doi: 10.1002/anie.200802376
Hell, S. W. (2007). Far-field optical nanoscopy. Science 316, 1153–1158. doi: 10.1126/science.1137395
Hell, S. W., and Wichmann, J. (1994). Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780–782. doi: 10.1364/OL.19.000780
Hong, Z., Zhang, Z., Olson, J. M., and Verma, D. P. (2001). A novel UDP-glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell 13, 769–779. doi: 10.1105/tpc.13.4.769
Iwai, S., and Uyeda, T. Q. (2008). Visualizing myosin-actin interaction with a genetically-encoded fluorescent strain sensor. Proc. Natl. Acad. Sci. U.S.A. 105, 16882–16887. doi: 10.1073/pnas.0805513105
Jacobs, A. K., Lipka, V., Burton, R. A., Panstruga, R., Strizhov, N., Schulze-Lefert, P., et al. (2003). An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15, 2503–2513. doi: 10.1105/tpc.016097
Jares-Erijman, E. A., and Jovin, T. M. (2003). FRET imaging. Nat. Biotechnol. 21, 1387–1395. doi: 10.1038/nbt896
Jones, A. M., Grossmann, G., Danielson, J. A., Sosso, D., Chen, L. Q., Ho, C. H., et al. (2013). In vivo biochemistry: applications for small molecule biosensors in plant biology. Curr. Opin. Plant Biol. 16, 389–395. doi: 10.1016/j.pbi.2013.02.010
Jørgensen, J. H. (1992). Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63, 141–152. doi: 10.1007/BF00023919
Karpova, T. S., Baumann, C. T., He, L., Wu, X., Grammer, A., Lipsky, P., et al. (2003). Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microsc. 209, 56–70. doi: 10.1046/j.1365-2818.2003.01100.x
Knepper, C., and Day, B. (2010). From perception to activation: the molecular-genetic and biochemical landscape of disease resistance signaling in plants. Arabidopsis Book 8:e012. doi: 10.1199/tab.0124
Koh, S., Andre, A., Edwards, H., Ehrhardt, D., and Somerville, S. (2005). Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J. 44, 516–529. doi: 10.1111/j.1365-313X.2005.02545.x
Kohorn, B. D., and Kohorn, S. L. (2012). The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 3:88. doi: 10.3389/fpls.2012.00088
Komis, G., Mistrik, M., Samajova, O., Doskocilova, A., Ovecka, M., Illes, P., et al. (2014). Dynamics and organization of cortical microtubules as revealed by superresolution structured illumination microscopy. Plant Physiol. 165, 129–148. doi: 10.1104/pp.114.238477
Kunkel, M. T., Toker, A., Tsien, R. Y., and Newton, A. C. (2007). Calcium-dependent regulation of protein kinase D revealed by a genetically encoded kinase activity reporter. J. Biol. Chem. 282, 6733–6742. doi: 10.1074/jbc.M608086200
Liesche, J., Ziomkiewicz, I., and Schulz, A. (2013). Super-resolution imaging with Pontamine Fast Scarlet 4BS enables direct visualization of cellulose orientation and cell connection architecture in onion epidermis cells. BMC Plant Biol. 13:226. doi: 10.1186/1471-2229-13-226
Martin-Fernandez, M. L., Tynan, C. J., and Webb, S. E. (2013). A ‘pocket guide’ to total internal reflection fluorescence. J. Microsc. 252, 16–22. doi: 10.1111/jmi.12070
Meng, F., and Sachs, F. (2011). Visualizing dynamic cytoplasmic forces with a compliance-matched FRET sensor. J. Cell Sci. 124, 261–269. doi: 10.1242/jcs.071928
Mercer, P. C., Wood, R. K. S., and Greenwood, A. D. (1974). Resistance to anthracnose of French bean. Physiol. Plant Pathol. 4, 291–306. doi: 10.1016/0048-4059(74)90016-2
Mims, C. W., Sewall, T. C., and Richardson, E. A. (2000). Ultrastructure of the host-pathogen relationship in Entomosporium leaf spot disease of Photinia. Int. J. Plant Sci. 161, 291–295. doi: 10.1086/314246
Mirabet, V., Das, P., Boudaoud, A., and Hamant, O. (2011). The role of mechanical forces in plant morphogenesis. Annu. Rev. Plant Biol. 62, 365–385. doi: 10.1146/annurev-arplant-042110-103852
Monshausen, G. B., Bibikova, T. N., Weisenseel, M. H., and Gilroy, S. (2009). Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21, 2341–2356. doi: 10.1105/tpc.109.068395
Monshausen, G. B., and Haswell, E. S. (2013). A force of nature: molecular mechanisms of mechanoperception in plants. J. Exp. Bot. 64, 4663–4680. doi: 10.1093/jxb/ert204
Monshausen, G. B., Messerli, M. A., and Gilroy, S. (2008). Imaging of the Yellow Cameleon 3.6 indicator reveals that elevations in cytosolic Ca2+ follow oscillating increases in growth in root hairs of Arabidopsis. Plant Physiol. 147, 1690–1698. doi: 10.1104/pp.108.123638
Nagai, T., Yamada, S., Tominaga, T., Ichikawa, M., and Miyawaki, A. (2004). Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 10554–10559. doi: 10.1073/pnas.0400417101
Naumann, M., Somerville, S., and Voigt, C. A. (2013). Differences in early callose deposition during adapted and non-adapted powdery mildew infection of resistant Arabidopsis lines. Plant Signal. Behav. 8:e24408. doi: 10.4161/psb.24408
Nishimura, M. T., Stein, M., Hou, B. H., Vogel, J. P., Edwards, H., and Somerville, S. C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301, 969–972. doi: 10.1126/science.1086716
Okumoto, S. (2014). Quantitative imaging approaches for small-molecule measurements using FRET sensors in plants. Methods Mol. Biol. 1083, 55–64. doi: 10.1007/978-1-62703-661-0_5
Okumoto, S., Jones, A., and Frommer, W. B. (2012). Quantitative imaging with fluorescent biosensors. Annu. Rev. Plant Biol. 63, 663–706. doi: 10.1146/annurev-arplant-042110-103745
Panstruga, R. (2005). Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem. Soc. Trans. 33, 389–392. doi: 10.1042/BST0330389
Pattathil, S., Avci, U., Baldwin, D., Swennes, A. G., Mcgill, J. A., Popper, Z., et al. (2010). A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 153, 514–525. doi: 10.1104/pp.109.151985
Piffanelli, P., Ramsay, L., Waugh, R., Benabdelmouna, A., D’Hont, A., Hollricher, K., et al. (2004). A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature 430, 887–891. doi: 10.1038/nature02781
Requejo-Isidro, J. (2013). Fluorescence nanoscopy. Methods and applications. J. Chem. Biol. 6, 97–120. doi: 10.1007/s12154-013-0096-3
Ries, J., Kaplan, C., Platonova, E., Eghlidi, H., and Ewers, H. (2012). A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 9, 582–584. doi: 10.1038/nmeth.1991
Rust, M. J., Bates, M., and Zhuang, X. (2006). Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795. doi: 10.1038/nmeth929
Sato, M., and Umezawa, Y. (2004). Imaging protein phosphorylation by fluorescence in single living cells. Methods 32, 451–455. doi: 10.1016/j.ymeth.2003.10.013
Schermelleh, L., Carlton, P. M., Hasse, S., Shao, L., Winoto, L., Kner, P., et al. (2008). Subdiffraction multicolor imaginng of te nuclear periphery with 3D structured illumination microscopy. Science 320, 1332–1336. doi: 10.1126/science.1156947
Srivastava, V., Malm, E., Sundqvist, G., and Bulone, V. (2013). Quantitative proteomics reveals that plasma membrane microdomains from poplar cell suspension cultures are enriched in markers of signal transduction, molecular transport, and callose biosynthesis. Mol. Cell. Proteomics 12, 3874–3885. doi: 10.1074/mcp.M113.029033
Steinbrenner, A. D., Goritschnig, S., Krasileva, K. V., Schreiber, K. J., and Staskawicz, B. J. (2012). Effector recognition and activation of the Arabidopsis thaliana NLR innate immune receptors. Cold. Spring Harb. Symp. Quant. Biol. 77, 249–257. doi: 10.1101/sqb.2012.77.014860
Vaid, N., Macovei, A., and Tuteja, N. (2013). Knights in action: lectin receptor-like kinases in plant development and stress responses. Mol. Plant 6, 1405–1418. doi: 10.1093/mp/sst033
Verma, D. P., and Hong, Z. (2001). Plant callose synthase complexes. Plant Mol. Biol. 47, 693–701. doi: 10.1023/A:1013679111111
Weis, C., Huckelhoven, R., and Eichmann, R. (2013). LIFEGUARD proteins support plant colonization by biotrophic powdery mildew fungi. J. Exp. Bot. 64, 3855–3867. doi: 10.1093/jxb/ert217
Keywords: callose, cell wall integrity, FRET, innate immunity, localization microscopy, powdery mildew, plant defense, super-resolution
Citation: Ellinger D and Voigt CA (2014) The use of nanoscale fluorescence microscopic to decipher cell wall modifications during fungal penetration. Front. Plant Sci. 5:270. doi: 10.3389/fpls.2014.00270
Received: 27 April 2014; Accepted: 25 May 2014;
Published online: 18 June 2014.
Edited by:
Nuria Sanchez Coll, Centre for Research in Agricultural Genomics, SpainReviewed by:
Vincenzo Lionetti, “Sapienza” Università di Roma, ItalyTolga Osman Bozkurt, Imperial College London, UK
Copyright © 2014 Ellinger and Voigt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian A. Voigt, Phytopathology and Biochemistry, Biocenter Klein Flottbek, University of Hamburg, Ohnhorststrasse 18, 22609 Hamburg, Germany e-mail:Y2hyaXN0aWFuLnZvaWd0QHVuaS1oYW1idXJnLmRl
 Dorothea Ellinger
Dorothea Ellinger Christian A. Voigt
Christian A. Voigt