- Department of Crop and Soil Sciences, Cornell University, Ithaca, NY, USA
Copper (Cu) is an essential micronutrient that performs a remarkable array of functions in plants including photosynthesis, cell wall remodeling, flowering, and seed set. Of the world's major cereal crops, wheat, barley, and oat are the most sensitive to Cu deficiency. Cu deficient soils include alkaline soils, which occupy approximately 30% of the world's arable lands, and organic soils that occupy an estimated 19% of arable land in Europe. We used Brachypodium distachyon (brachypodium) as a proxy for wheat and other grain cereals to initiate analyses of the molecular mechanisms underlying their increased susceptibility to Cu deficiency. In this report, we focus on members of the CTR/COPT family of Cu transporters because their homologs in A. thaliana are transcriptionally upregulated in Cu-limited conditions and are involved either in Cu uptake from soils into epidermal cells in the root, or long-distance transport and distribution of Cu in photosynthetic tissues. We found that of five COPT proteins in brachypodium, BdCOPT3, and BdCOPT4 localize to the plasma membrane and are transcriptionally upregulated in roots and leaves by Cu deficiency. We also found that BdCOPT3, BdCOPT4, and BdCOPT5 confer low affinity Cu transport, in contrast to their counterparts in A. thaliana that confer high affinity Cu transport. These data suggest that increased sensitivity to Cu deficiency in some grass species may arise from lower efficiency and, possibly, other properties of components of Cu uptake and tissue partitioning systems and reinforce the importance of using brachypodium as a model for the comprehensive analyses of Cu homeostasis in cereal crops.
Introduction
Copper (Cu) is an essential micronutrient for all organisms because it acts as a cofactor for enzymes participating in important biological processes such as respiration, photosynthesis, and scavenging of oxidative stress (Burkhead et al., 2009; Merchant, 2010; Ravet and Pilon, 2013). In addition to these functions, plants also employ Cu for the perception of ethylene, nitrogen metabolism, molybdenum cofactor synthesis, cell wall remodeling, response to pathogens, flowering, and seed set (Shorrocks and Alloway, 1988; Marschner, 1995; Burkhead et al., 2009; Mendel and Kruse, 2012; Ravet and Pilon, 2013). This remarkable array of functions is attributed to the ability of Cu to change its oxidation state (Cu2+ ↔ Cu+). However, the same property imposes toxicity when free Cu ions accumulate in cells in excess because of their ability to promote oxidative stress (Valko et al., 2005). Copper availability, and thus crop productivity on agricultural soils depend on soil type and agricultural practices (Shorrocks and Alloway, 1988; Marschner, 1995; Solberg et al., 1999). For example, Cu deficiency develops on alkaline soils due to low solubility of Cu at high pH, and on organic soils due to Cu binding to organic matter (Shorrocks and Alloway, 1988; Marschner, 1995; Solberg et al., 1999). While Cu deficiency can be remedied by the application of Cu-based fertilizers, this strategy is not environmentally friendly, and the repeated use of fertilizers, as well as Cu-based pesticides, has led to the build-up of toxic levels of Cu in soils (Shorrocks and Alloway, 1988; Marschner, 1995). In this regard, organic farming has emerged as a preferred production system that relies on using natural fertilizers; however natural fertilizers increase soil organic matter and thus, further reduce Cu bioavailability. Sensitivity to Cu bioavailability in soils varies among crops species. Of the major cereal crops, wheat is regarded as the most sensitive to Cu deficiency (Shorrocks and Alloway, 1988; Solberg et al., 1999). In contrast, crops like canola have not shown Cu deficiency symptoms or responded to Cu fertilizer when grown on soils that would cause Cu deficiency in wheat or barley (Solberg et al., 1999). Higher sensitivity of some cereal crops to Cu deficiency compared to other crop species has been recognized in farm reports and fact sheets from some states in the United States and different countries in the world. Among the major cereal crops, sensitivity to Cu deficiency is reported to be in the following rank order, from higher to lower sensitivity: winter wheat > spring wheat > flax > barley > oats > triticale > rye (Shorrocks and Alloway, 1988; Solberg et al., 1999). Remarkably, the molecular mechanisms that underlie this tendency are unknown.
Plants tightly regulate Cu homeostasis to prevent deficiency while avoiding toxicity. This regulation includes transcriptional control of genes encoding proteins that are involved in Cu uptake, trafficking, tissue partitioning, and reallocation among Cu requiring enzymes. Of known Cu transporters, members of the CTR/COPT family are among the main contributors to initial Cu uptake in plants, the green alga Chlamydomonas reinhardtii, yeast, Drosophila, and humans (Merchant, 2010; Nevitt et al., 2012; Ravet and Pilon, 2013). The plant CTR/COPT family is best characterized in Arabidopsis thaliana and Oryza sativa, where it is represented by six and seven members, respectively (Peñarrubia et al., 2010; Yuan et al., 2011). A. thaliana COPT1, COPT2, and COPT6 are transcriptionally regulated by Cu deficiency, localize to the plasma membrane, mediate Cu uptake, and complement the growth defect of the S. cerevisiae Cu uptake mutant lacking functional Cu transporters, Ctr1p, Ctr2p, and Ctr3p (ctr1Δctr3Δ or ctr1Δctr2Δctr3Δ) (Sancenon et al., 2004; Andres-Colas et al., 2010; Jung et al., 2012; Garcia-Molina et al., 2013; Gayomba et al., 2013; Perea-García et al., 2013). COPT1 and COPT2 function primarily in Cu uptake into the root, while COPT6 also contributes to Cu partitioning in photosynthetic tissues (Jung et al., 2012; Garcia-Molina et al., 2013). In contrast, COPT5 localizes to the tonoplast and the pre-vacuolar compartment and functions by remobilizing Cu from these organelles during Cu deficiency (Garcia-Molina et al., 2011; Klaumann et al., 2011). CTR/COPT proteins homotrimerize to form a pore within the membrane to transport Cu across the lipid bilayer, but can also form heterocomplexes with other CTR/COPT family members and/or other proteins (Lee et al., 2002; De Feo et al., 2009; Yuan et al., 2010, 2011). Studies in A. thaliana have shown that while COPT6 interacts with COPT1, this interaction is not required for the ability of COPT6 or COPT1 to complement the Cu uptake deficiency phenotype of the S. cerevisiae ctr1Δctr2Δctr3Δ mutant strain (Jung et al., 2012). Unlike CTR/COPT transporters from A. thaliana, CTR/COPT transporters from O. sativa complement the Cu uptake mutant of yeast only when are co-expressed with another OsCOPT member, and OsCOPT7 is the only high-affinity Cu transporter in the O. sativa CTR/COPT family (Yuan et al., 2010, 2011). These findings suggest that some properties of CTR/COPT transporters are distinct between grass and non-grass species. These differences are not entirely surprising, given that A. thaliana is only distantly related to the Poaceae family and lacks many biological features of monocotyledonous grass crops (Brkljacic et al., 2011).
A member of the grass species, Brachypodium distachyon (from then on referred to as brachypodium) has emerged as a valuable experimental model for the study of small-grain cereals due to its less complex and fully sequenced genome, the continued development of numerous genetic resources, including efficient protocols for Agrobacterium-mediated transformation (Vogel and Hill, 2008), whole genome TILLING mutant alleles (Brkljacic et al., 2011; Thole et al., 2012), a T-DNA insertion mutant collection (http://brachypodium.pw.usda.gov/TDNA/; Bragg et al., 2012), as well as other attributes such as short life cycle (8–12 weeks), small stature (15–20 cm), diploid accessions, self-fertility, and simple growth requirements (Brkljacic et al., 2011). Because brachypodium and wheat grains have similar structure, brachypodium is an attractive model for molecular, genetic, and genomic studies of Cu homeostasis of wheat (Brkljacic et al., 2011; Mochida et al., 2011; Mur et al., 2011). Furthermore, root anatomy of brachypodium and wheat is similar and is distinct from the root anatomy of another Poaceae family member, O. sativa, which is specialized for overcoming anaerobic conditions associated with submerged roots, and thus, it is suggested that brachypodium and wheat may have similar root-related genes, including those that are involved in mineral ion uptake (Chochois et al., 2012). Hence, brachypodium becomes a preferred model for studies of ion homeostasis, as was recently demonstrated by the analyses of brachypodium Yellow Stripe-Like (YSL) proteins, which are involved in uptake and internal translocation of iron (Fe) (Yordem et al., 2011).
Members of the CTR/COPT family of Cu transporters are the best characterized in both, A. thaliana and rice, but corresponding members in brachypodium have not yet been characterized, and are not fully annotated, according to database collections such as the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/). In contrast, a specialized database for A. thaliana membrane proteins, ARAMEMNON 7.0 (http://aramemnon.botanik.uni-koeln.de/; Schwacke et al., 2003) has a more complete annotation of putative membrane proteins of A. thaliana and provides their homologs from other species, including brachypodium, based on amino acid similarity and motif organization. According to in silico predictions using ARAMEMNON 7.0, we have identified five putative CTR/COPT family members in brachypodium and provided the initial characterization of their function in Cu homeostasis. We have also developed growth conditions for studies of Cu homeostasis in brachypodium as a model for small grain cereals and established protocols for the isolation and transfection of protoplasts isolated from brachypodium mesophyll cells. We used these procedures, along with functional complementation assays in the S. cerevisiae ctr1Δctr2Δctr3Δ mutant strain, gene expression analyses, and yeast-two-hybrid assays of protein-protein interactions to study members of the CTR/COPT family of Cu transporters in brachypodium.
Methods and Materials
Plant Material and Growth Conditions
Seeds of the 21-3 inbred line of brachypodium (Vogel and Hill, 2008) were sterilized in 100% ethanol for 1 min and rinsed three times with sterile water. The lamella and palea were softened by further incubation in sterile water for 2 h and removed with forceps, taking care not to damage the seed. Prepared seeds were then spread onto rinsed perlite irrigated with a standard hydroponic solution, described below. After stratification at 4°C for 24 h, plants were grown for 7 days at 22°C; 12-h light/12-h dark photoperiod at photosynthetic photon flux density of 150 μmol photons m−2s−1 before transferring to a hydroponic solution prepared as described (Arteca and Arteca, 2000) except that Cu (as CuSO4) was added at a higher concentration of 0.25 μM. For Cu limitation and sensitivity assays, 7-day-old seedlings were transferred from perlite to hydroponic solution supplemented with the indicated concentrations of CuSO4, or without CuSO4 but with the specific Cu chelator, bathocuproine disulfonate (BCS) (Rapisarda et al., 2002). Plants were grown for 18 days before subsequent analyses. For all experiments, the hydroponic solution was changed every 7 days.
RNA Extraction
Root and leaf tissues were separated from 25-day-old plants grown under the indicated conditions and flash-frozen in liquid nitrogen. Samples were homogenized in liquid nitrogen using a mortar and a pestle, and total RNA was isolated using the Plant RNA Kit (Omega Bio-Tek), according to the manufacturer's instructions. Genomic DNA in RNA samples was digested with DNAse I (Omega Bio-Tek) prior to cDNA synthesis using the iScript cDNA Synthesis kit (BioRad).
Quantitative Real-Time (qRT)-PCR Analysis
Prior to qRT-PCR analysis, primers (Supplemental Table 1) and cDNA concentrations were optimized to reach a qRT-PCR amplification efficiency of 100 ± 10%. Two microliters of 10-fold diluted cDNA was used as a template in a total reaction volume of 10 μl containing 500 nM of each PCR primer, 50 mM KCl, 20 mM Tris-HCI, pH 8.4, 0.2 mM dNTPs, and 1.25 units of iTaq DNA polymerase in iQ SYBR Green Supermix (BioRad), containing 3 mM MgCl2, SYBR Green I, 20 nM fluoresceine, and stabilizers. PCR was carried out using the CFX96 Real-Time PCR system (BioRad). The thermal cycling parameters were as follows: denaturation at 95°C for 3 min, followed by 39 cycles of 95°C for 10 s then 55°C for 30 s. Amplicon dissociation curves, i.e., melting curves, were recorded after cycle 39 by heating from 60 to 95°C with 0.5°C increments and an average ramp speed of 3.3°C s−1. Data were analyzed using the CFX Manager Software, version 1.5 (BioRad). A brachypodium gene encoding ACTIN2 was used as a reference for normalizing gene expression. qRT-PCR experiments were conducted using three independent biological samples, each consisting of three technical replicates, unless indicated otherwise. Statistical analysis was performed using the Relative Expression Software Tool (REST, Qiagen; Pfaffl et al., 2002).
Plasmid Construction
Total RNA isolated from brachypodium leaves and oligonucleotides pairs that are indicated in Supplemental Table 1 were used for RT-PCR amplification of BdCOPT1, BdCOPT3, BdCOPT4, and BdCOPT5 cDNAs with or without the stop codon. Primers were designed to contain attB sites on resulting PCR products for subsequent Gateway cloning (Invitrogen) into the DONR-Zeo entry vector (Invitrogen) and appropriate destination vectors described below.
Functional Complementation of the S. cerevisiae Copper Uptake Deficient ctr1Δctr2Δctr3Δ Mutant Strain
S. cerevisiae SEY6210 (MATa ura3-52 leu2-3,-112 his3Δ200 trp1Δ901 lys2-801 suc2Δ9) wild-type and the ctr1Δctr2Δctr3Δ triple mutant (MATa ura3-52 his3Δ200 trp1-901 ctr1::ura3::Knr ctr2::HIS3 ctr3::TRP1) used for functional complementation assays were the generous gift of Dr. Dennis Thiele (Duke University). Yeast cells were transformed with YES3-Gate-BdCOPT1, YES3-Gate-BdCOPT3, YES3-Gate-BdCOPT4, YES3-Gate-BdCOPT5 constructs or an empty YES3-Gate vector using the Frozen-EZ yeast Transformation II Kit (Zymo Research). Transformants were selected for uracil prototrophy on YNB medium (YNB-Ura) containing 0.67% (w/v) Yeast Nitrogen Base without amino acids (Difco), 0.077% (w/v) CSM-Ura, 0.05% (w/v) NaCl, 2% dextrose, 2% (w/v) agar. Respiration competence was evaluated by testing the ability of transformants to grow on the non-fermentable carbon sources, glycerol and ethanol (YPEG) as described (Dancis et al., 1994a; Puig et al., 2002). Briefly, transformants were grown in liquid YNB-Ura to an OD600 nm = 1.0, serially-10-fold diluted and spotted onto YPEG medium containing 1% (w/v) yeast extract, 2% (w/v) bacto-peptone, 3% (v/v) glycerol 2% (v/v) ethanol, and 2% (w/v) agar and the indicated concentration of CuSO4 or onto YNB-Ura for controls. Plates were incubated for 3 days at 30°C.
Isolation of Protoplasts from Brachypodium Leaves
Protoplasts were isolated from leaves of 25-day-old brachypodium grown as described above. The protoplast isolation procedure was based on (Zhai et al., 2009). Briefly, 0.2 g of young leaf tissue was immersed in 15 ml of filter-sterilized TVL solution (Supplemental Table 2), finely chopped with a fresh razor blade and transferred to a 200 ml beaker containing 20 ml of Enzyme solution (Supplemental Table 2). The beaker was wrapped in aluminum foil to protect samples from light and samples were vacuum infiltrated for 30 min before incubation at 30°C for 60 min. The mixture was then agitated at 35 rpm at room temperature for 18–20 h. Released protoplasts were collected into 50-ml Falcon centrifuge tubes by carefully sieving the mixture through eight layers of cheesecloth, pre-wetted with W5 solution (Supplemental Table 2). To increase the protoplast yield, the cheesecloth was rinsed with an additional 10 ml of W5 solution. Sieved protoplasts were then carefully overlaid with 5 ml of W5 solution and left at room temperature for 1 h to allow protoplasts to float to the interface of Enzyme solution and W5 solution. Fifteen milliliters of protoplasts were collected from the interface and transferred into a new 50 ml Falcon tube containing 20 ml of W5 solution. Protoplasts were collected by centrifugation for 7 min at 100 × g. The residual Enzyme solution was removed by two rounds of rinsing protoplasts with 10 ml W5 solution and centrifuged for 5 min at 60 × g. Purified protoplasts were resuspended in 3–5 ml W5 solution and the protoplast yield was evaluated by cell counting using a hemocytometer. Protoplast viability was evaluated using the Plant Cell Viability Assay Kit (Sigma), according to manufacturer's recommendations.
Transfection of Protoplasts with Plasmid DNA
Protoplasts were transfected using a procedure adopted from (Jung et al., 2011). Briefly, purified protoplasts were incubated on ice for 30 min and centrifuged for 5 min at 60 × g and W5 solution was removed. Protoplasts were then re-suspended in MMG solution (Supplemental Table 2) and 100 μl aliquots were transferred into a 2-ml round-bottom microcentrifuge tube. Plasmid DNA (5–10 μg) was added to protoplasts and mixed gently. For controls, DNA was omitted and replaced with equivalent volumes of sterile water (mock transfection). Transfection was initiated by the addition of 110 μl of PEG-calcium solution (Supplemental Table 2). Protoplasts were gently mixed with PEG-calcium solution by tapping the tube followed by incubation for 7 min at room temperature. Transfection was terminated by diluting the mixture by an addition of 700 μl of W5 solution. Transfected protoplasts were collected by centrifugation for 2 min at 100 × g, and supernatant was removed to leave 50–100 μl of protoplast suspension. Each sample was then brought up to a volume of 1 ml with W5 solution and incubated in the dark at room temperature for 18 h.
Subcellular Localization and Fluorescent Microscopy
For studies of the subcellular localization in brachypodium protoplasts, BdCOPT3, and BdCOPT4 cDNAs lacking the stop codon were fused at the C-terminus to the enhanced green fluorescent protein (EGFP) of the SAT6-N1-EGFP-Gate vector and expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The resulting 35Spro-BdCOPT-EGFP constructs or SAT6-N1-EGFP-Gate lacking cDNA inserts were transfected into brachypodium protoplasts as described above. Plasma membranes were stained with 50 μM FM4-64 as described (Ueda et al., 2001).
For studies of subcellular localization in S. cerevisiae, entry clones containing BdCOPT1, BdCOPT3, BdCOPT4, or BdCOPT5 cDNAs without the stop codon were fused at C-terminus with EGFP in the YES3-EGFP-Gate vector (Jung et al., 2012). The resulting constructs and the empty YES3-EGFP-Gate vector were transformed into S. cerevisiae ctr1Δctr2Δctr3Δ triple mutant using the Frozen-EZ yeast Transformation II Kit (Zymo Research) and transformants were selected on YNB-Ura medium, as described above. Subcellular localization was assessed in cells grown overnight in liquid YNB-Ura.
EGFP- and FM4-64- mediated fluorescence, and chlorophyll autofluorescence were visualized using FITC (for EGFP) or rhodamine (FM4-64 and chlorophyll) filter sets of the Axio Imager M2 microscope equipped with the motorized Z-drive (Zeiss). Z-stack (1.3 μm-thick) images were collected with the high-resolution AxioCam MR Camera and then 3D deconvoluted using an inverse filter algorithm of the Zeiss AxioVision 4.8 software. Images were processed using the Adobe Photoshop software package, version 12.0.
Split-Ubiquitin Membrane Yeast Two-Hybrid System (MYTH)
Vectors and S. cerevisiae strains THY.AP4 (MATa leu2-3,112 ura3-52 trp1-289 lexA::HIS3 lexA::ADE2 lexA::lacZ) and THY.AP5 (MATα URA3 leu2-3,112 trp1-289 his3-Δ1 ade2Δ::loxP) for MYTH were obtained from the Frommer lab (Stanford University) depository at Arabidopsis Biological Resource Center (ABRC) http://www.arabidopsis.org/abrc/index.jsp. AtCOPT6-Cub-PLV and AtCOPT6-NubG fusions were previously described (Jung et al., 2012). Full-length BdCOPT1, BdCOPT3, BdCOPT4, and BdCOPT5 cDNAs without stop codons were introduced into the MetYC-dest and pXN-dest22-3HA destination vectors by Gateway cloning (Invitrogen) to generate bait, BdCOPT-CubPLV, and prey, BdCOPT-NubG, constructs in THY.AP4 and THY.AP5 strains, respectively. In all cases, C- and N terminal fragments of ubiquitin were placed at the C-terminus of the BdCOPT proteins. Protein interactions were selected in diploid cells after 2 days of growth on SC medium lacking adenine and histidine. Interactions were verified using β-galactosidase assays, as detailed in Kittanakom et al. (2009).
Construction of Phylogenetic Tree
The phylogenetic tree was built using the Neighbor-Joining method (Saitou and Nei, 1987). The bootstrap consensus tree inferred from 500 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the number of differences method (Nei and Kumar, 2000) and are in units of the number of amino acid differences per sequence. The analysis involved 22 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 75 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011) with A. thaliana IRT1 as an outgroup.
Accession Numbers
Accession numbers for genes used in this study were according to nomenclature from ARAMEMNON 7.0 (http://aramemnon.botanik.uni-koeln.de/) and MIPS (http://mips.helmholtz-muenchen.de/plant/brachypodium/): Bradi1g24180 (BdCOPT1), Bradi1g24190 (BdCOPT2), Bradi2g51210 (BdCOPT3), Bradi4g31330 (BdCOPT4), Bradi5g09580 (BdCOPT5), Bradi1g10630.1 (BdACTIN), and At2g26975 (AtCOPT6).
Results
The Predicted COPT Transporters of Brachypodium Share Conserved Features of the CTR/COPT Family
To identify putative members of the CTR/COPT family in brachypodium, we used the Plant Membrane Protein database, ARAMEMNON 7.0 (http://aramemnon.botanik.uni-koeln.de/; Schwacke et al., 2003), which has the most complete annotation of putative CTR/COPT transporters based on amino acid similarity and motif organization of CTR/COPT proteins in different species. We found that the brachypodium genome possesses five genes that encode putative CTR/COPT transporters Bradi1g24180, Bradi1g24190, Bradi2g51210, Bradi4g31330, Bradi5g09580, designated COPT1 through COPT5 (alias BdCOPT1-BdCOPT5) (Table 1). BdCOPT1 and BdCOPT2 are located on chromosome 1, while BdCOPT3, BdCOPT4, and BdCOPT5 are found on chromosome 2, 4, and 5, respectively (Table 1). Based on the BdCOPT gene structures in the ARAMEMNON 7.0 database, BdCOPT1, BdCOPT3, BdCOPT4, and BdCOPT5 lack introns, similar to COPT genes in rice (Yuan et al., 2011) and A. thaliana (http://arabidopsis.org). In contrast, BdCOPT2 possesses two introns (Figure 1A).
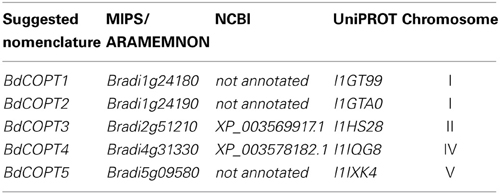
Table 1. The proposed nomenclature and accession numbers of putative brachypodium COPT transporters annotated at different databases.
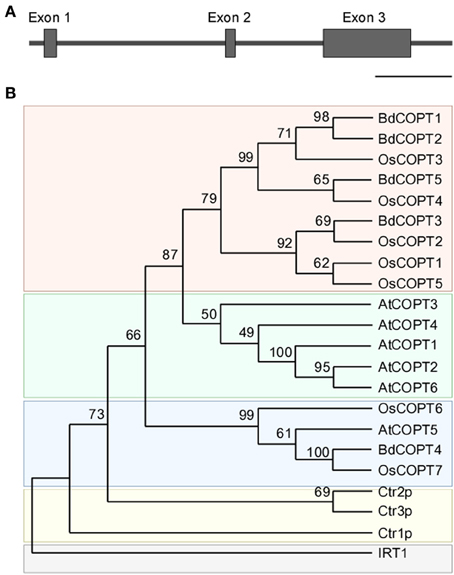
Figure 1. Gene structure of BdCOPT2 and phylogenetic analysis of the brachypodium COPT family. (A) Gene structure of BdCOPT2. Note the presence of introns, which are absent in other plant CTR/COPT members. Scale bar = 500 bp. (B) A phylogenetic tree of CTR/COPT members of A. thaliana (designated as AtCOPT1-6), O. sativa (designated as OsCOPT1-7), brachypodium (designated as BdCOPT1-5), and S. cerevisiae (designated as Ctr1p-CTR3p). The A. thaliana protein, IRT1, is included as an outgroup.
BdCOPTs share 16–69% amino acid sequence identity and 26–73% sequence similarity to each other (Table 2). Phylogenetic analysis revealed that a majority of CTR/COPT proteins from brachypodium and O. sativa cluster together and are separate from CTR/COPTs from A. thaliana (Figure 1B). One exception is BdCOPT4, which forms a separate cluster along with AtCOPT5, OsCOPT7, and OsCOPT6 (Figure 1B). BdCOPT4 is highly similar (65%) to AtCOPT5, whereas other BdCOPT proteins share 38–54% similarity to their counterparts in A. thaliana (Table 3). Similar to OsCOPT members (Yuan et al., 2011), the nucleotide sequence of BdCOPT genes is GC-rich, ranging from GC content of 67.9% (BdCOPT2) to 73.2% (BdCOPT3), in contrast to an average GC content of 50% in A. thaliana COPTs.
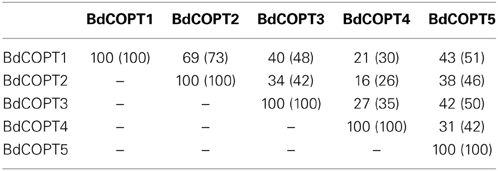
Table 2. Percentage of amino acid identity (similarity) among putative COPT proteins in brachypodium.
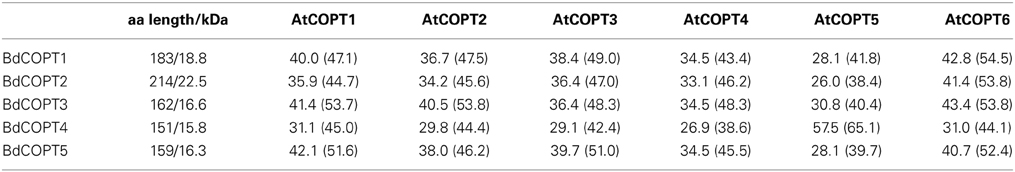
Table 3. Amino acid length (aa length), molecular mass (shown in kDa), and percentage of amino acid identity (similarity) of putative brachypodium COPT (BdCOPT1-5) transporters in comparison to their homologs in A. thaliana (AtCOPT1-6).
CTR/COPT family members possess conserved structural features that include three putative transmembrane helices (TMs), the N- and C-termini located toward the extracellular space and cytosol respectively, N-terminally-located methionine-rich motifs (Mets motifs), and MXXXM-X12-GXXXG motifs located within TM2 and TM3, respectively (Puig et al., 2002; De Feo et al., 2007, 2009; Peñarrubia et al., 2010). Importantly, MXXXM motifs of TM2 in Ctr1p of S. cerevisiae, and COPT2 and COPT6 of A. thaliana contain a positionally conserved Met residue that is essential for the ability of these proteins to transport Cu (Puig et al., 2002; Jung et al., 2012; Gayomba et al., 2013). Some CTR/COPT proteins also contain the C-terminal cysteine-rich CXC motif, which is suggested to bind Cu ions for transfer to cytosolic Cu chaperones, or to downregulate Ctr1p activity in response to toxic Cu levels (Puig et al., 2002; Xiao et al., 2004; Wu et al., 2009). This motif is present in A. thaliana COPT1 and COPT2, but is absent in COPT6; nevertheless, COPT6 is a functional Cu transporter (Jung et al., 2012).
Computer algorithm-assisted analysis of membrane topology and motif organization in brachypodium CTR/COPT proteins revealed that all BdCOPTs, except for BdCOPT3, are predicted to contain the classical three TMs (Figure 2) with N-terminal domains oriented toward the extracellular space, while C-terminal domains are predicted to be located in the cytosol. In contrast, BdCOPT3 is predicted to have two transmembrane domains, with both N- and C-termini located in the cytosol. While polypeptides of all BdCOPT included the N-terminal Mets motifs, the distribution of MXXXM-X12-GXXXG motifs varied within different BdCOPT polypeptides (Figure 2). For example, MXXXM motifs and thus, positionally conserved essential Met residues, were located within TM2 in BdCOPT3, BdCOPT4, and BdCOPT5, but outside of TM2 in BdCOPT1 and BdCOPT2. Although the predicted membrane topology of BdCOPT proteins has to be validated experimentally, the location of essential positionally conserved Met residues of the MXXXM motif outside of the predicted membrane domain suggest that BdCOPT1 and BdCOPT2 might not mediate Cu transport. The C-terminal CXC motif is present in BdCOPT3 as a CC motif and in BdCOPT4 as a CXC motif, but is absent in BdCOPT1, BdCOPT2, and BdCOPT5 (Figure 2).
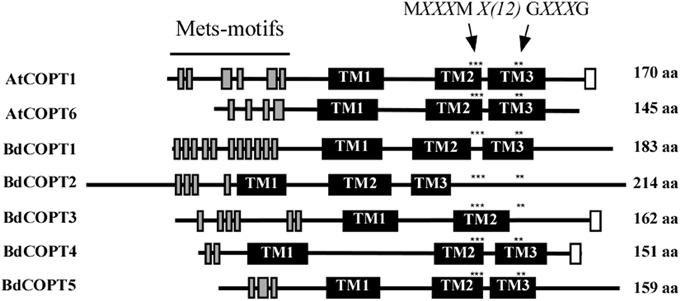
Figure 2. A diagram showing conserved motifs within the primary sequence of BdCOPT proteins and their homologs in A. thaliana, AtCOPT1 and AtCOPT6. Topology predictions are based on the TMHMM software, version 1.0. Shown are methionine-rich motifs within the predicted N-terminal domain (Mets-motifs, gray bars), transmembrane domains (TM1, TM2, and TM3; black bars) and cysteine-rich motifs within the predicted C-terminal domain (white bars). Asterisks “***” and “**” indicate the location of conserved MXXXM and GXXXG motifs. Note that BdCOPT3 has a unique structure of two transmembrane domains and that BdCOPT3 and BdCOPT4 have cysteine-rich domains in the C-terminal domain.
BdCOPT3 and BdCOPT4 Respond Transcriptionally to Cu Status
To determine whether BdCOPT genes respond transcriptionally to Cu status of the plant like their counterparts in A. thaliana and O. sativa, we first developed growth conditions under which Cu limitation would minimally affect the growth and development of brachypodium. Seeds were germinated in perlite irrigated with a modified hydroponic medium supplemented with 0.25 μM CuSO4 (Supplemental Figure 1 and Methods and Materials) and grown for 7 days before transferring to hydroponic medium to grow further for 18 days under the following conditions: (1) control conditions (0.25 μM CuSO4); (2) Cu limited conditions (0 μM CuSO4); (3) Cu deficiency (0 μM CuSO4 and supplemented with 500 μM of the Cu chelator, bathocuprione disulfonate (BCS); and (4) Cu excess (3 μM CuSO4). While there were no signs of chlorosis in leaves of plants grown under Cu deficiency or Cu excess (Supplemental Figure 2A), plants grown under Cu deficiency had decreased height, and shoot and root dry weight compared to plants grown under control conditions (Supplemental Figure 2B). Plants grown under Cu excess had decreased shoot and root dry weight, but were of the same height as control plants. Cu limitation did not significantly alter plant growth or shoot and root biomass (Supplemental Figure 2B).
We then analyzed the steady-state levels of BdCOPT mRNAs in different plant tissues of brachypodium grown under the Cu regimes described above. Expression studies in roots (Figure 3A) revealed that Cu limitation increased mRNA expression of BdCOPT3 and BdCOPT4. Cu deficiency increased BdCOPT3 and BdCOPT4 expression even further and, in addition, increased expression of BdCOPT1. There were no statistically significant differences in mRNA expression levels of BdCOPT2 and BdCOPT5 in root tissue of plants grown under these conditions (Figure 3A). In roots of plants grown under Cu toxicity, BdCOPT4 was the only gene whose expression was responsive to this treatment (Figure 3A). In young leaves (Figure 3B), Cu limitation elevated the abundance of BdCOPT3 and BdCOPT4 transcripts, which increased even further under Cu deficiency. In young leaves of plants grown under Cu toxicity, expression of BdCOPT4 decreased, but expression of BdCOPT2 increased. Expression studies in old leaves (Figure 3C) of plants grown in Cu limited or Cu deficient conditions showed an increase of the abundance of BdCOPT3 and BdCOPT4 transcripts. In contrast, both, Cu toxic and Cu limited conditions, increased expression of BdCOPT1, while expression of BdCOPT2 and BdCOPT5 were downregulated under Cu deficiency.
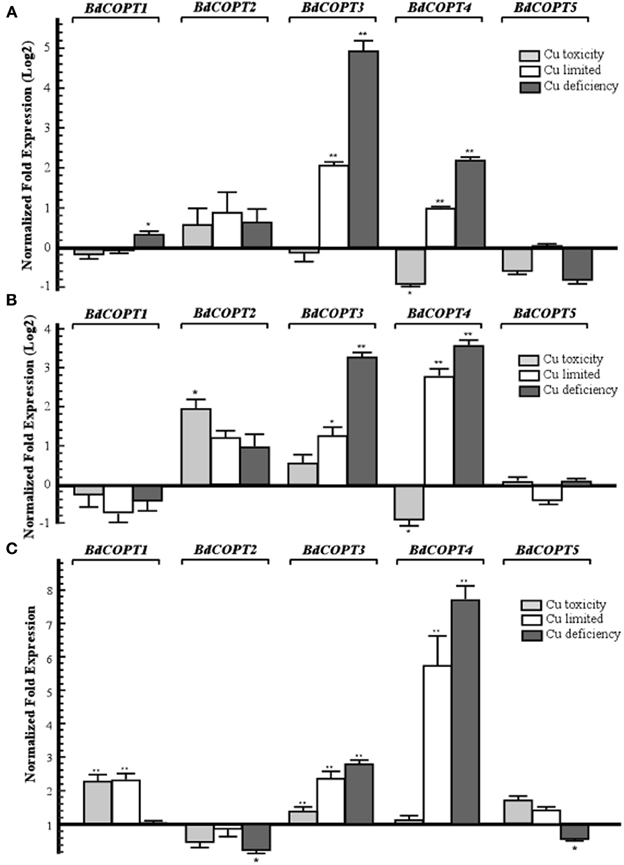
Figure 3. Quantitative real-time (qRT)-PCR analysis of the effect of Cu on expression COPT1 through 5 in roots (A), young leaves (B), and older leaves (C) of brachypodium. For all treatments, 7-day-old wild-type seedlings were transferred to hydroponic solution and were grown for 18 days in medium that, in addition to macro- and micronutrients, contained either 0.25 μM CuSO4 (control conditions), 0 μM CuSO4 (Cu limited), 0 μM CuSO4 + 500 μM BCS (Cu deficiency), or 3 μM CuSO4 (Cu toxicity). Error bars show SE (n = 9). Differences of the mean values between control and treated plants are indicated as *p ≤ 0.05 or **p ≤ 0.001. Results are presented relative to the expression of genes under control conditions.
To summarize, of five BdCOPT genes, BdCOPT3, and BdCOPT4, are the most responsive to fluctuations in Cu availability, and changes in their expression are most prominent in roots and young leaves. BdCOPT2 is significantly upregulated under Cu toxicity only in young leaves, while BdCOPT1 is regulated by Cu mainly in old leaves. Expression of BdCOPT5 was the least responsive to Cu availability and was downregulated only by Cu deficiency and only in old leaves.
Heterologously Expressed BdCOPT3, BdCOPT4, and BdCOPT5 Partially Rescue Growth Defects of the S. cerevisiae ctr1Δctr2Δctr3ΔCu Uptake Mutant on Non-Fermentable Growth Medium
We next tested whether BdCOPT proteins are involved in Cu transport. In this regard, the S. cerevisiae ctr1Δctr2Δctr3Δ mutant, which lacks high-affinity plasma membrane-localized Cu uptake transporters Ctr1p and Ctr3p, and the vacuolar membrane-localized Ctr2p, is deficient in Cu uptake and release of Cu from the vacuole, and has been successfully used to identify Cu transport capabilities of CTR/COPT transporters from higher plants (Kampfenkel et al., 1995; Barhoom et al., 2008; Yuan et al., 2010, 2011; Garcia-Molina et al., 2011; Klaumann et al., 2011; Jung et al., 2012; Gayomba et al., 2013). Analyses of Cu transport capabilities using yeast as a heterologous system is based on the fact that cells lacking functional Cu uptake systems are unable to deliver Cu to cytochrome c oxidase in the mitochondrial respiratory chain, preventing cell growth on non-fermentable carbon sources such as glycerol and ethanol (YPEG medium), unless high concentrations of exogenous Cu are added to the growth medium (Dancis et al., 1994b; Glerum et al., 1996). Therefore, we expected that if BdCOPT proteins act as high-affinity Cu transporters, then their expression in the S. cerevisiae ctr1Δctr2Δctr3Δ mutant would promote Cu uptake and suppress growth defects of the mutant on non-fermentable medium.
Here, we focused on analyses of BdCOPT1, BdCOPT3, BdCOPT4, and BdCOPT5 due to difficulty in cloning of BdCOPT2. As a positive control, we used the previously characterized CTR/COPT transporter from A. thaliana, AtCOPT6 (Jung et al., 2012). We found that all yeast lines grew at the same rate on standard medium (YNB-Ura) supplied with glucose as a carbon source or on YPEG medium supplemented with 100 μM CuSO4 (Figure 4). In contrast, only the empty vector-expressing wild-type cells and ctr1Δctr2Δctr3Δ cells expressing AtCOPT6 grew well on YPEG medium (Figure 4), suggesting that none of BdCOPTs tested were able to rescue the growth defect of the S. cerevisiae mutant on YPEG medium. However, addition of a low concentration of Cu (10 μM CuSO4) to YPEG medium allowed BdCOPT3 (Figure 4B), BdCOPT4 (Figure 4C), and BdCOPT5 (Figure 4D) but not BdCOPT1 (Figure 4A) to complement partially the growth defect of ctr1Δctr2Δctr3Δ cells. These results suggest that transport capabilities of brachypodium COPT transporters differ from their counterparts in A. thaliana.
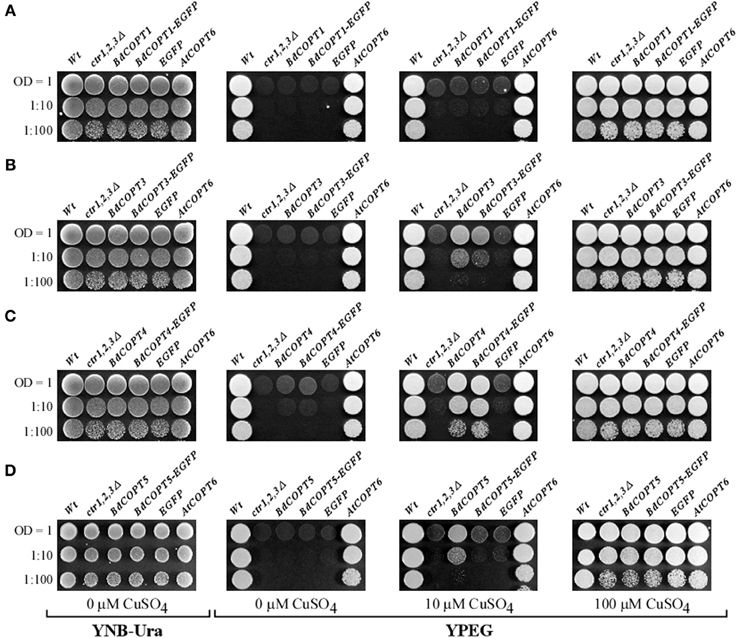
Figure 4. BdCOPT3, BdCOPT4, and BdCOPT5 rescue the growth defect of the S. cerevisiae ctr1Δctr2Δctr3Δ triple mutant on ethanol/glycerol medium (YPEG). The ctr1Δctr2Δctr3Δ mutant was transformed with the YES3-Gate vector harboring BdCOPT1 (A), BdCOPT3 (B), BdCOPT4 (C), and BdCOPT5 (D) along with corresponding EGFP-fusions and spotted onto YPEG plates supplemented with the indicated concentrations of CuSO4. As negative controls, the ctr1Δctr2Δctr3Δ mutant strain was transformed with the empty YES3-Gate (ctr1,2,3Δ) or empty YES3-EGFP-Gate vector (EGFP). The isogenic wild-type, SEY6210, transformed with the empty YES3-Gate vector (Wt) and ctr1Δctr2Δctr3Δ cells transformed with YES3-Gate harboring the A. thaliana COPT6 (AtCOPT6) cDNA insert were used as positive controls.
BdCOPT3 and BdCOPT4 Localize to the Plasma Membrane in S. cerevisiae Cells and Brachypodium Protoplasts
S. cerevisiae Ctr1p and Ctr3p localize to the plasma membrane and contribute to Cu uptake into the cell (Dancis et al., 1994a; Peña et al., 2000), while Ctr2p localizes to the vacuolar membrane and remobilizes Cu from this internal store upon Cu deficiency (Rees et al., 2004). To determine whether BdCOPTs rescue growth defects of the S. cerevisiae ctr1Δctr2Δctr3Δ mutant by facilitating Cu uptake into the cell from the external medium or by vacuolar remobilization, we determined their subcellular localization in yeast cells as well as in brachypodium protoplasts.
For studies in S. cerevisiae, we inserted BdCOPT3, BdCOPT4, and BdCOPT5 cDNAs without the stop codon into the YES3-EGFP-Gate vector to generate translational C-terminal EGFP fusions. We then verified whether the BdCOPT-EGFP constructs were functional by expressing them in the ctr1Δctr2Δctr3Δ mutant and assessing the growth of transformed cells on YPEG media. We found that expression of BdCOPT3-, and BdCOPT4-EGFP fusions in ctr1Δctr2Δctr3Δ cells resulted in growth phenotypes mirroring results of cells expressing un-tagged proteins (Figures 4B,C). However, the BdCOPT5-EGFP construct was unable to rescue growth of ctr1Δctr2Δctr3Δ cells, suggesting that fusing EGFP with BdCOPT5 resulted in the loss of its activity (Figure 4D). Since unfunctional BdCOPT5-EGFP might mislocalize in yeast cells as well as in brachypodium protoplasts, this construct was omitted from subsequent study. We did not analyze the subcellular localization of BdCOPT1 for the same reason. Fluorescent microscopy revealed that EGFP-mediated fluorescence in BdCOPT3-EGFP and BdCOPT4-EGFP expressing cells localized mainly to the cell periphery and the distribution of the EGFP signal was distinct in S. cerevisiae cells expressing the EGFP-only vector (Figure 5). These data suggest that BdCOPT3 and BdCOPT4 localize to the plasma membrane in this heterologous system.
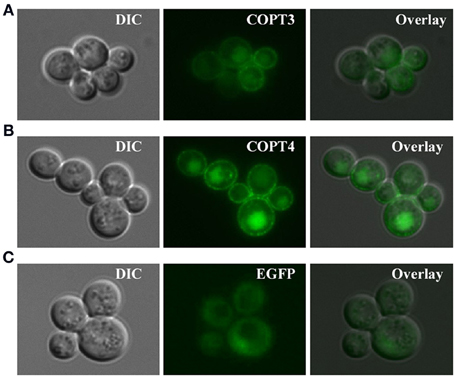
Figure 5. Subcellular localization of BdCOPT3-EGFP (A) or BdCOPT4-EGFP (B) fusions or EGFP (C) in S. cerevisiae ctr1Δctr2Δctr3Δ cells. Superimposed images (Overlay) from differential interference contrast microscopy (DIC) and EGFP-mediated fluorescence (EGFP) show that BdCOPT3 and BdCOPT4 localize to the plasma membrane and that the pattern of fluorescence of EGFP-fused proteins is distinct from EGFP.
We next analyzed the subcellular localization of BdCOPT3 and BdCOPT4 in brachypodium using transient expression in protoplasts. After establishing a procedure for the isolation of viable protoplasts from brachypodium (Figure 6), we transfected protoplasts with BdCOPT3-EGFP or BdCOPT4-EGFP constructs with C-terminal EGFP fusions expressed from the CaMV promoter of the SAT-N1-EGFP-Gate vector, or with the empty SAT-N1-EGFP-Gate vector. We found that EGFP-mediated fluorescence originating from BdCOPT3-EGFP or BdCOPT4-EGFP constructs was located at the periphery of transfected protoplasts and did not overlap with chlorophyll-mediated autofluorescence (Figures 7A,B and Supplemental Figures 3A,B). Furthermore, fluorescence from BdCOPT3- and BdCOPT4-EGFP constructs was distinct from the fluorescence pattern exhibited by protoplasts transfected with the empty EGFP vector (Figure 7C and Supplemental Figure 3C). We note that internally-localized EGFP-mediated fluorescence is likely an artifact of the degradation of the BdCOPT-EGFPs. We then co-stained protoplasts expressing BdCOPT3- or BdCOPT4-EGFP with the lipophylic dye, FM4-64, which selectively labels the plasma membrane under low-temperature conditions (Vida and Emr, 1995). We found strict co-localization of BdCOPT3- or BdCOPT4-EGFP and FM4-64-mediated fluorescence (Figures 7A,B), suggesting that BdCOPT3 and BdCOPT4 are located at the plasma membrane.
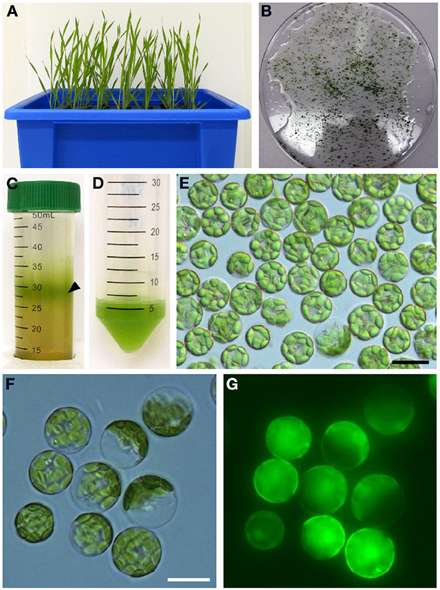
Figure 6. Isolation of protoplasts from brachypodium. Hydroponically grown 25-day-old plants (A) were used for the isolation of protoplasts from leaf tissue. Chopped brachypodium leaves in filter-sterilized TVL solution are shown in (B). Enzymatic digestion of the cell wall and fractionation by sucrose density gradient yielded protoplasts at the interface of the enzyme solution and W5 solution (C, black arrow). Protoplasts were collected and purified from sucrose density gradient solution (D) and visualized under microscopy using bright-field filter sets (E). In our method, 0.2 g of leaf tissue from 25-day-old seedlings yields 5 × 106–107 protoplasts. Close-up of brachypodium protoplasts through bright-field (F) and FITC (G) filter sets to assess protoplast viability after staining with the membrane-permeable non-fluorescent dye, fluorescein diacetate. After diffusion into viable protoplasts fluorescein diacetate is hydrolyzed into a polar compound, causing the cytosoplasm of the cell to fluoresce under the FITC filter set. Scale bar = 20 μm.
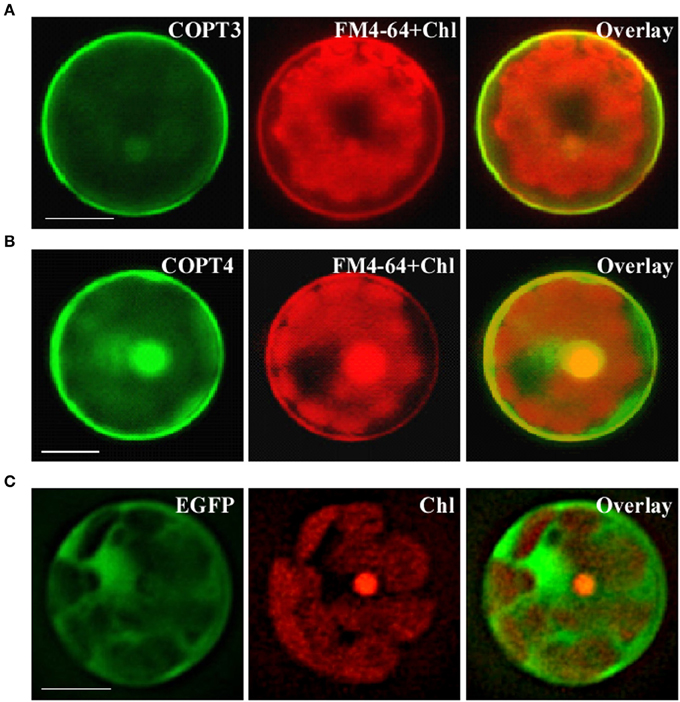
Figure 7. Subcellular localization of BdCOPT3 and BdCOPT4 in brachypodium protoplasts. Protoplasts isolated from leaves of 25-day-old plants were transfected with BdCOPT3-EGFP (A) or BdCOPT4-EGFP (B) constructs or the empty SAT6-N1-EGFP vector (C) and co-stained with the plasma-membrane dye, FM4-64. EGFP-mediated fluorescence derived from BdCOPT3-EGFP (COPT3) or BdCOPT4-EGFP, (COPT4), or from EGFP of the SAT6-N1-EGFP vector (EGFP) was detected using the FITC filter set while FM4-64 (FM4-64) and chlorophyll autofluorescence (Chl) were visualized using the Rhodamine filter set of an Axio Imager M2 microscope equipped with a motorized Z-drive (Zeiss). Images collected from FITC and Rhodamine filter sets were overlaid (Overlay) to show the plasma membrane subcellular localization of Cu transporters. Scale bar = 10 μm.
BdCOPT Proteins Interact with Each Other in Yeast-Two-Hybrid Assays
Homo- and heterodimerization of CTR/COPT proteins have been demonstrated in eukaryotes (Lee et al., 2002; De Feo et al., 2007, 2009; Yuan et al., 2010, 2011). Furthermore, although interaction of A. thaliana COPT6 with COPT1 is not required for the ability of these proteins to transport Cu (Jung et al., 2012), the activity of O. sativa CTR/COPT proteins seem to depend on their interactions with each other (Yuan et al., 2011). Therefore, we tested if the CTR/COPT family members in brachypodium would also interact with either themselves or/and with other family members. For this purpose we used the split-ubiquitin-based membrane yeast-two-hybrid (MYTH) approach (Kittanakom et al., 2009). In this system, a modified ubiquitin protein is split into its C- and N-terminal halves (Cub and NubG, respectively), which are fused to membrane-localized bait or prey proteins, respectively. The C-terminus of ubiquitin is attached to an artifical transcription factor, PLV (protein A-LexA-VP16). If bait and prey proteins are oriented in the cytosol and interactions occur, the modified ubiquitin is reconstituted and recognized by ubiquitin-specific proteases, which release PLV from Cub. PLV enters the nucleus to induce expression of lexA-controlled reporter genes ADE2, HIS3, and lacZ (Kittanakom et al., 2009), allowing protein interactions to be assessed by adenine and histidine prototrophy and by the β-galactosidase assay.
We fused Cub-PLV and NubG at the C-terminus of BdCOPT1, BdCOPT3, BdCOPT4, and BdCOPT5 since their predicted topology is consistent with a cytosolic orientation of their C-termini. We then co-expressed BdCOPT proteins in different combinations with themselves or with the empty Cub-PLV vector as a negative control (Figure 8). We included S. cerevisiae co-expressing AtCOPT6-Cub-PLV and AtCOPT6-NubG in our assays as a positive control since AtCOPT6 interacts with itself in the MYTH assay (Jung et al., 2012). To control for false positives, the empty Cub-PLV vector was co-expressed with BdCOPTs fused to NubG. These studies showed that all of the tested BdCOPT proteins interacted in both homo- and heterodimer combinations, regardless of whether interactions were assayed by growth on selective medium or with β-galactosidase (Figure 8). Whether these interactions are required for their ability to transport Cu remains to be elucidated.
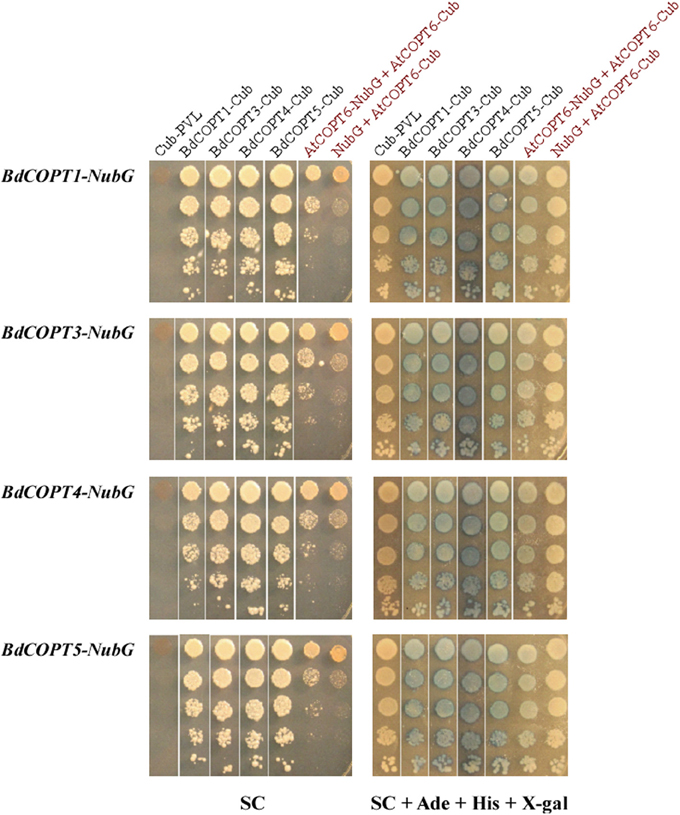
Figure 8. Analyses of protein-protein interactions of brachypodium CTR/COPT transporters using the split-ubiquitin membrane yeast two-hybrid system (MYTH). Shown are yeast cells co-expressing NubG constructs fused with BdCOPT1, BdCOPT3, BdCOPT4, or BdCOPT5 cDNA (BdCOPT1-, BdCOPT3-, BdCOPT4-, BdCOPT5-NubG) and a Cub-PLV construct lacking a cDNA insert (Cub-PLV) or Cub-PLV fused to BdCOPT1-, BdCOPT3-, BdCOPT4-, BdCOPT5 (BdCOPT1-, BdCOPT3-, BdCOPT4-, BdCOPT5-Cub). Interactions were visualized by colony formation on selective media SC medium (SC) or as blue colonies in a ß-galactosidase assay (SC + Ade + His + X-gal). Growth was monitored for 2 days under conditions indicated below each panel. Interactions of AtCOPT6 with itself (AtCOPT6-NubG + AtCOPT6-Cub), or lack of interactions between AtCOPT6-Cub-PLV and the empty NubG vector (NubG + AtCOPT6-Cub) were used as controls and are indicated in red. Shown are representative results of at least three biological replicates. SC, synthetic complete medium; Ade, adenine; His, histidine; X-gal, bromo-chloro-indolylgalactopyranoside.
Discussion
Biological attributes of brachypodium, such as root architecture, grain structure, and the continued development of molecular and genetic resources prompted us to establish this plant as a preferred model for Cu homeostasis studies in grain cereals (e.g., wheat, barley, and oat) which are reported to be more sensitive to Cu availability in agricultural soils in comparison to other crops (Shorrocks and Alloway, 1988; Solberg et al., 1999). To begin investigations of the underlying molecular basis of this phenomenon, we initiated studies of the brachypodium CTR/COPT Cu transporters since members of this family in A. thaliana provide an entry point for Cu into the root and are suggested to contribute to subsequent Cu partitioning in photosynthetic tissues (Burkhead et al., 2009; Merchant, 2010; Ravet and Pilon, 2013). Toward this goal, we have identified five putative CTR/COPT family members in brachypodium based on amino acid similarity and motifs organization of CTR/COPT proteins in different species and we classified them as BdCOPT1 through 5. Phylogenetic analyses of the predicted CTR/COPT members in brachypodium and their counterparts from a dicot, A. thaliana, and a monocot, O. sativa, show that, with the exception of BdCOPT4, brachypodium and O. sativa CTR/COPT proteins are more closely related to each other than to A. thaliana (Figure 1B), reflecting a closer evolutionary relationship of brachypodium to O. sativa than to A. thaliana. Furthermore, analysis of membrane topology and the motif organization in brachypodium CTR/COPT proteins revealed that while polypeptides of all BdCOPT included the N-terminal Mets motifs, the location of MXXXM-X12-GXXXG motifs varies within different BdCOPT polypeptides, affecting the location of the positionally conserved Met residue, which is essential for the translocation of Cu across lipid bilayer (Figure 2). For example, MXXXM motifs are predicted to be located within TM2 only in BdCOPT3, BdCOPT4, and BdCOPT5, similar to CTR/COPT family members in other species. In contrast, MXXXM motifs are predicted to be located outside of TM2 in BdCOPT1 and BdCOPT2, suggesting that these proteins might not be able to mediate Cu transport due to a shift in the position of the essential Mets motifs. Although the topology predictions have to be validated experimentally, the ability of BdCOPT3, BdCOPT4, and BdCOPT5, but not BdCOPT1, to rescue partially the Cu-deficiency associated respiratory defects of the S. cerevisiae ctr1Δctr2Δctr3Δ mutant (Figure 4) was consistent with this suggestion. We also note that BdCOPT3, BdCOPT4, and BdCOPT5 conferred growth to the yeast mutant only when a low concentration of Cu was added to the growth medium (Figure 4). This result might be interpreted using at least two not mutually exclusive scenarios: (1) BdCOPT3, BdCOPT4, and BdCOPT5 are low affinity Cu transporters, unlike their high-affinity counterparts from A. thaliana and/or (2) in order to confer high affinity transport, BdCOPT3, BdCOPT4, and/or BdCOPT5 must interact with other CTR/COPTs and/or other transporters. In this regard it has been shown that most of CTR/COPTs in O. sativa form dimers/trimers with other CTR/COPTs or other proteins to function as high-affinity transporters (Yuan et al., 2010, 2011). In contrast, the plasma membrane-localized CTR/COPT family members from A. thaliana are high-affinity transporters by themselves even though can form hetero-complexes (Nakagawa et al., 2010; Jung et al., 2012; Gayomba et al., 2013). We found that brachypodium CTR/COPT transporters homo- and heterooligomerize in the MYTH system (Figure 8); whether these interactions are required for transport capabilities of brachypodium CTR/COPTs has yet to be determined.
We have also examined the subcellular localization of BdCOPTs by analyzing the localization pattern of functional BdCOPT3-EGFP and BdCOPT4-EGFP constructs by heterologous expression in yeast or transient expression in brachypodium protoplasts. Toward this goal we have established procedures for preparing viable protoplasts from brachypodium mesophyll cells and for protoplast transfection (Figure 6). These studies showed that both, BdCOPT3-EGFP and BdCOPT4-EGFP, localize to the plasma membrane, regardless of whether assays were done in yeast or protoplasts (Figures 5, 7), further suggesting that these two CTR/COPT proteins may function in Cu uptake. Finally, BdCOPT3 and BdCOPT4 genes were highly expressed in roots, and old and young leaves, and their expression was tightly regulated by Cu availability (Figure 3), as was shown for other CTR/COPT proteins from different species, including plants.
To conclude, this manuscript shows that brachypodium can be used for analyses of the molecular mechanisms underlying the increased susceptibility of small grain cereals to Cu deficiency and of Cu homeostasis overall. Analyses of the phylogenetic relationship between CTR/COPT proteins of brachypodium, O. sativa, and A. thaliana, and the clear divergence of A. thaliana CTR/COPT members, suggest that differences may exist in characteristics of the CTR/COPT proteins between monocots and dicots. This is further validated by studies of Cu transport capabilities of brachypodium CTR/COPTs. Heterologous expression of brachypodium CTR/COPTs in the S. cerevisiae Cu uptake mutant ctr1Δctr2Δctr3Δ suggest that increased sensitivity to Cu deficiency in some grass species may arise from lower efficiency of Cu uptake and, possibly, other properties of components of Cu uptake and tissue partitioning systems. It is also possible that the CTR/COPT family members in brachypodium require homo- or heterooligomerization for high-affinity Cu uptake, unlike corresponding family members in A. thaliana, reinforcing the importance of using brachypodium as a model for the comprehensive analyses of Cu homeostasis in cereal crops.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Dennis Thiele (Duke University, USA) for S. cerevisiae strains; Dr. Jian Hua (Cornell University, USA) for the SAT6-N1-EGFP plasmid. This work was supported by the Cornell University Agricultural Experiment Station USDA-CSREES NYC-FFF-Hatch #125433, USDA-CSREES NYC-MRF S1041 #125853 awarded to Olena K. Vatamaniuk.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00236/abstract
Supplemental Figure 1. Establishing growth conditions for brachypodium. Plants were grown on standard nutrient solution at 22°C and 12-h light/12-h dark photoperiod at photosynthetic photon flux density of 150 μmol photons m−2 s−1. The glume and lemma of seeds were removed and seeds were sown on rinsed perlite (A). Seven-day-old seedlings grown on perlite irrigated with standard nutrient solution (B). Seven-day-old seedlings were transferred from perlite to hydroponic medium and shown from above (C) and from the side view (D). Scale bar = 5 cm.
Supplemental Figure 2. Phenotypes of brachypodium grown under different copper conditions. (A) Twenty five-day-old plants grown in hydroponic solution without CuSO4 and supplemented with 500 μM BCS (500 BCS), or indicated concentrations of CuSO4. (B) Plants from (A) were used to measure height (Plant height), shoot dry weight (Shoot dry weight), and root dry weight (Root dry weight). Asterisks (**) indicate statistically significant differences (p ≤ 0.01, ANOVA) of the mean values of treatments compared to control conditions (0.25 μM CuSO4). Error bars show SD (n = 3).
Supplemental Figure 3. Subcellular localization of BdCOPT3 and BdCOPT4 in brachypodium protoplasts. Protoplasts were transfected with BdCOPT3-EGFP (A) or BdCOPT4-EGFP (B) constructs or empty SAT6-N1-EGFP vector (C) and co-stained with the plasma-membrane dye, FM4-64. EGFP-mediated fluorescence derived from BdCOPT3-EGFP (COPT3) or BdCOPT4-EGFP, (COPT4), or from EGFP of the SAT6-N1-EGFP vector (EGFP) was detected using the FITC filter set while FM4-64 (FM4-64) and chlorophyll autofluorescence (Chl) were visualized using the Rhodamine filter set of an Axio Imager M2 microscope equipped with the motorized Z-drive (Zeiss). Images collected from FITC and Rhodamine filter sets were overlaid (Overlay) to show the plasma membrane subcellular localization of the Cu transporters.
Supplemental Table 1. Primers used in this study. Sequences in italics indicate att sites used for Gateway cloning and “*” indicate sequences without a stop codon.
Supplemental Table 2. Solutions used in protoplast isolation and transfection. All solutions were filter sterilized before use.
References
Andres-Colas, N., Perea-Garcia, A., Puig, S., and Peñarrubia, L. (2010). Deregulated copper transport affects Arabidopsis development especially in the absence of environmental cycles. Plant Physiol. 153, 170–184. doi: 10.1104/pp.110.153676
Arteca, R. N., and Arteca, J. M. (2000). A novel method for growing Arabidopsis thaliana plants hydroponically. Physiol. Plantarum 108, 188–193. doi: 10.1034/j.1399-3054.2000.108002188.x
Barhoom, S., Kupiec, M., Zhao, X., Xu, J., and Sharon, A. (2008). Functional characterization of CgCTR2, a putative vacuole copper transporter that is involved in germination and pathogenicity in Colletotrichum gloeosporioides. Eukaryot. Cell 7, 1098–1108. doi: 10.1128/EC.00109-07
Bragg, J. N., Wu, J., Gordon, S. P., Guttman, M. E., Thilmony, R., Lazo, G. R., et al. (2012). Generation and characterization of the western regional research center brachypodium T-DNA insertional mutant collection. PLoS ONE 7:e41916. doi: 10.1371/journal.pone.0041916
Brkljacic, J., Grotewold, E., Scholl, R., Mockler, T., Garvin, D. F., Vain, P., et al. (2011). Brachypodium as a model for the grasses: today and the future. Plant Physiol. 157, 3–13. doi: 10.1104/pp.111.179531
Burkhead, J., Reynolds, K., Abdel-Ghany, S., Cohu, C., and Pilon, M. (2009). Copper homeostasis. New Phytol. 182, 799–816. doi: 10.1111/j.1469-8137.2009.02846.x
Chochois, V., Vogel, J. P., and Watt, M. (2012). Application of Brachypodium to the genetic improvement of wheat roots. J. Exp. Bot. 63, 3467–3474. doi: 10.1093/jxb/ers044
Dancis, A., Haile, D., Yuan, D. S., and Klausner, R. D. (1994a). The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J. Biol. Chem. 269, 25660–25667.
Dancis, A., Yuan, D., Haile, D., Askwith, C., Eide, D., Moehle, C., et al. (1994b). Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell 76, 393–402. doi: 10.1016/0092-8674(94)90345-X
De Feo, C., Aller, S., and Unger, V. (2007). A structural perspective on copper uptake in eukaryotes. Biometals 20, 705–716. doi: 10.1007/s10534-006-9054-7
De Feo, C. J., Aller, S. G., Siluvai, G. S., Blackburn, N. J., and Unger, V. M. (2009). Three-dimensional structure of the human copper transporter hCTR1. Proc. Natl. Acad. Sci. U.S.A. 106, 4237–4242. doi: 10.1073/pnas.0810286106
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Garcia-Molina, A., Andrés-Colás, N., Perea-García, A., del Valle-Tascón, S., Peñarrubia, L., and Puig, S. (2011). The intracellular Arabidopsis COPT5 transport protein is required for photosynthetic electron transport under severe copper deficiency. Plant J. 65, 848–860. doi: 10.1111/j.1365-313X.2010.04472.x
Garcia-Molina, A., Andres-Colas, N., Perea-Garcia, A., Neumann, U., Dodani, S. C., Huijser, P., et al. (2013). The Arabidopsis COPT6 transport protein functions in copper distribution under copper-deficient conditions. Plant Cell Physiol. 54, 1378–1390. doi: 10.1093/pcp/pct088
Gayomba, S. R., Jung, H. I., Yan, J., Danku, J., Rutzke, M. A., Bernal, M., et al. (2013). The CTR/COPT-dependent copper uptake and SPL7-dependent copper deficiency responses are required for basal cadmium tolerance in A. thaliana. Metallomics 5, 1262–1275. doi: 10.1039/c3mt00111c
Glerum, D. M., Shtanko, A., and Tzagoloff, A. (1996). Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 271, 14504–14509. doi: 10.1074/jbc.271.24.14504
Jung, H. I., Gayomba, S. R., Rutzke, M. A., Craft, E., Kochian, L. V., and Vatamaniuk, O. K. (2012). COPT6 is a plasma membrane transporter that functions in copper homeostasis in Arabidopsis and is a novel target of SQUAMOSA promoter-binding protein-like 7. J. Biol. Chem. 287, 33252–33267. doi: 10.1074/jbc.M112.397810
Jung, H. I., Zhai, Z., and Vatamaniuk, O. K. (2011). Direct transfer of synthetic double-stranded RNA into protoplasts of Arabidopsis thaliana. Methods Mol. Biol. 744, 109–127. doi: 10.1007/978-1-61779-123-9_8
Kampfenkel, K., Kushnir, S., Babiychuk, E., Inze, D., and Van Montagu, M. (1995). Molecular characterization of a putative Arabidopsis thaliana copper transporter and its yeast homologue. J. Biol. Chem. 270, 28479–28486. doi: 10.1074/jbc.270.47.28479
Kittanakom, S., Chuk, M., Wong, V., Snyder, J., Edmonds, D., Lydakis, A., et al. (2009). Analysis of membrane protein complexes using the split-ubiquitin membrane yeast two-hybrid (MYTH) system. Methods Mol. Biol. 548, 247–271. doi: 10.1007/978-1-59745-540-4_14
Klaumann, S., Nickolaus, S. D., Fürst, S. H., Starck, S., Schneider, S., Ekkehard Neuhaus, H., et al. (2011). The tonoplast copper transporter COPT5 acts as an exporter and is required for interorgan allocation of copper in Arabidopsis thaliana. New Phytol. 192, 393–404. doi: 10.1111/j.1469-8137.2011.03798.x
Lee, J., Peña, M. M., Nose, Y., and Thiele, D. J. (2002). Biochemical characterization of the human copper transporter ctr1. J. Biol. Chem. 277, 4380–4387. doi: 10.1074/jbc.M104728200
Mendel, R. R., and Kruse, T. (2012). Cell biology of molybdenum in plants and humans. Biochim. Biophys. Acta 1823, 1568–1579. doi: 10.1016/j.bbamcr.2012.02.007
Merchant, S. S. (2010). The elements of plant micronutrients. Plant Physiol. 154, 512–515. doi: 10.1104/pp.110.161810
Mochida, K., Yoshida, T., Sakurai, T., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L.-S. P. (2011). In silico analysis of transcription factor repertoires and prediction of stress-responsive transcription factors from six major Gramineae plants. DNA Res. 18, 321–332. doi: 10.1093/dnares/dsr019
Mur, L. A., Allainguillaume, J., Catalan, P., Hasterok, R., Jenkins, G., Lesniewska, K., et al. (2011). Exploiting the Brachypodium tool box in cereal and grass research. New Phytol. 191, 334–347. doi: 10.1111/j.1469-8137.2011.03748.x
Nakagawa, Y., Kikuchi, S., Sakamoto, Y., and Yano, A. (2010). Identification and characterization of CcCTR1, a copper uptake transporter-like gene, in Coprinopsis cinerea. Microbiol. Res. 165, 276–287. doi: 10.1016/j.micres.2009.05.004
Nei, M., and Kumar, S. (2000). Molecular Evolution and Phylogenetics. New York, NY: Oxford University Press.
Nevitt, T., Öhrvik, H., and Thiele, D. J. (2012). Charting the travels of copper in eukaryotes from yeast to mammals. Biochim. Biophys. Acta 1823, 1580–1593. doi: 10.1016/j.bbamcr.2012.02.011
Peña, M. M., Puig, S., and Thiele, D. J. (2000). Characterization of the Saccharomyces cerevisiae high affinity copper transporter Ctr3. J. Biol. Chem. 275, 33244–33251. doi: 10.1074/jbc.M005392200
Peñarrubia, L., Andres-Colas, N., Moreno, J., and Puig, S. (2010). Regulation of copper transport in Arabidopsis thaliana: a biochemical oscillator? J. Biol. Inorg. Chem. 15, 29–36. doi: 10.1007/s00775-009-0591-8
Perea-García, A., Garcia-Molina, A., Andrés-Colás, N., Vera-Sirera, F., Pérez-Amador, M. A., Puig, S., et al. (2013). Arabidopsis copper transport protein COPT2 participates in the cross talk between iron deficiency responses and low-phosphate signaling. Plant Physiol. 162, 180–194. doi: 10.1104/pp.112.212407
Pfaffl, M. W., Horgan, G. W., and Dempfle, L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. doi: 10.1093/nar/30.9.e36
Puig, S., Lee, J., Lau, M., and Thiele, D. (2002). Biochemical and genetic analysis of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J. Biol. Chem. 277, 26021–26030. doi: 10.1074/jbc.M202547200
Rapisarda, V. A., Volentini, S. I., Farias, R. N., and Massa, E. M. (2002). Quenching of bathocuproine disulfonate fluorescence by Cu(I) as a basis for copper quantification. Anal. Biochem. 307, 105–109. doi: 10.1016/S0003-2697(02)00031-3
Ravet, K., and Pilon, M. (2013). Copper and iron homeostasis in plants: the challenges of oxidative stress. Antioxid. Redox Signal. 19, 919–932. doi: 10.1089/ars.2012.5084
Rees, E. M., Lee, J., and Thiele, D. J. (2004). Mobilization of intracellular copper stores by the Ctr2 vacuolar copper transporter. J. Biol. Chem. 279, 54221–54229. doi: 10.1074/jbc.M411669200
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sancenon, V., Puig, S., Mateu-Andres, I., Dorcey, E., Thiele, D., and Peñarrubia, L. (2004). The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. J. Biol. Chem. 279, 15348–15355. doi: 10.1074/jbc.M313321200
Schwacke, R., Schneider, A., van der Graaff, E., Fischer, K., Catoni, E., Desimone, M., et al. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26. doi: 10.1104/pp.011577
Shorrocks, V. M., and Alloway, B. J. (1988). Copper in Plant, Animal and Human Nutrition Potters Bar. Hertfordshire: Copper Development Association.
Solberg, E., Evans, I., and Penny, D. (1999). Copper deficiency: diagnosis and correction. Agri-facts. Soil fertility/crop nutrition. alberta agriculture, food and rural development, Agdex 532–533, 1–9.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thole, V., Peraldi, A., Worland, B., Nicholson, P., Doonan, J. H., and Vain, P. (2012). T-DNA mutagenesis in Brachypodium distachyon. J. Exp. Bot. 63, 567–576. doi: 10.1093/jxb/err333
Ueda, T., Yamaguchi, M., Uchimiya, H., and Nakano, A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20, 4730–4741. doi: 10.1093/emboj/20.17.4730
Valko, M., Morris, H., and Cronin, M. T. (2005). Metals, toxicity and oxidative stress. Curr. Med. Chem. 12, 1161–1208. doi: 10.2174/0929867053764635
Vida, T. A., and Emr, S. D. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792. doi: 10.1083/jcb.128.5.779
Vogel, J., and Hill, T. (2008). High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep. 27, 471–478. doi: 10.1007/s00299-007-0472-y
Wu, X., Sinani, D., Kim, H., and Lee, J. (2009). Copper transport activity of yeast Ctr1 is down-regulated via its C terminus in response to excess copper. J. Biol. Chem. 284, 4112–4122. doi: 10.1074/jbc.M807909200
Xiao, Z., Loughlin, F., George, G., Howlett, G., and Wedd, A. (2004). C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) ions as a cuprous-thiolate polynuclear cluster: sub-femtomolar Cu(I) affinity of three proteins involved in copper trafficking. J. Am. Chem. Soc. 126, 3081–3090. doi: 10.1021/ja0390350
Yordem, B. K., Conte, S. S., Ma, J. F., Yokosho, K., Vasques, K. A., Gopalsamy, S. N., et al. (2011). Brachypodium distachyon as a new model system for understanding iron homeostasis in grasses: phylogenetic and expression analysis of Yellow Stripe-Like (YSL) transporters. Ann. Bot. 108, 821–833. doi: 10.1093/aob/mcr200
Yuan, M., Chu, Z., Li, X., Xu, C., and Wang, S. (2010). The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell 22, 3164–3176. doi: 10.1105/tpc.110.078022
Yuan, M., Li, X., Xiao, J., and Wang, S. (2011). Molecular and functional analyses of COPT/Ctr-type copper transporter-like gene family in rice. BMC Plant Biol. 11:69. doi: 10.1186/1471-2229-11-69
Keywords: Brachypodium, CTR/COPT transporters, copper transport, copper homeostasis, wheat
Citation: Jung H, Gayomba SR, Yan J and Vatamaniuk OK (2014) Brachypodium distachyon as a model system for studies of copper transport in cereal crops. Front. Plant Sci. 5:236. doi: 10.3389/fpls.2014.00236
Received: 06 February 2014; Accepted: 11 May 2014;
Published online: 30 May 2014.
Edited by:
Paloma Koprovski Menguer, Jonh Innes Centre, UKReviewed by:
Irene Murgia, Università degli Studi di Milano, ItalyElizabeth Pilon-Smits, Colorado State University, USA
Copyright © 2014 Jung, Gayomba, Yan and Vatamaniuk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olena K. Vatamaniuk, Department of Crop and Soil Sciences, Cornell University, 306 Tower Road, Bradfield Hall, Rm. 608, Ithaca, NY 14853, USA e-mail:b2t2MkBjb3JuZWxsLmVkdQ==
 Ha-il Jung
Ha-il Jung Sheena R. Gayomba
Sheena R. Gayomba Jiapei Yan
Jiapei Yan Olena K. Vatamaniuk
Olena K. Vatamaniuk