
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 06 May 2014
Sec. Plant Pathogen Interactions
Volume 5 - 2014 | https://doi.org/10.3389/fpls.2014.00178
This article is part of the Research Topic Plant cell wall in pathogenesis, parasitism and symbiosis View all 15 articles
The battle between plants and microbes is evolutionarily ancient, highly complex, and often co-dependent. A primary challenge for microbes is to breach the physical barrier of host cell walls whilst avoiding detection by the plant’s immune receptors. While some receptors sense conserved microbial features, others monitor physical changes caused by an infection attempt. Detection of microbes leads to activation of appropriate defense responses that then challenge the attack. Plant cell walls are formidable and dynamic barriers. They are constructed primarily of complex carbohydrates joined by numerous distinct connection types, and are subject to extensive post-synthetic modification to suit prevailing local requirements. Multiple changes can be triggered in cell walls in response to microbial attack. Some of these are well described, but many remain obscure. The study of the myriad of subtle processes underlying cell wall modification poses special challenges for plant glycobiology. In this review we describe the major molecular and cellular mechanisms that underlie the roles of cell walls in plant defense against pathogen attack. In so doing, we also highlight some of the challenges inherent in studying these interactions, and briefly describe the analytical potential of molecular probes used in conjunction with carbohydrate microarray technology.
The dynamic interplay between pathogen and plant host is the product of millions of years of co-evolution. This struggle is often described in terms of an “arms race,” a fitting term considering the investment required and the significance for both sides. The goal for the plant is to keep healthy and fertile, but for pathogens the strategy varies. Biotrophic and hemibiotrophic microorganisms rely on keeping their host alive and unaware of their presence, at least until later stages of infection. In contrast, necrotrophs live and feed on dead and dying cells and can therefore use a more forceful attack strategy (Davidsson et al., 2013). The frontline of the plant defense system consists of physical and chemical barriers such as the cell wall, waxes, hairs, antimicrobial enzymes, and secondary metabolites. If these obstacles are overcome, the pathogen is still confronted by elaborate surveillance systems in which molecular sentinels operate to activate resistance responses (Jones and Dangl, 2006).
Molecular components that serve essential functions for the fitness or survival of microbes are often highly conserved. For plants, detection of such microbial fingerprints also referred to as pathogen-associated molecular patterns (PAMPs), is a warning of impending attack (Medzhitov and Janeway, 1997). Consequently, a key aspect of plant innate immunity is the ability to recognize PAMPs such as bacterial flagellin, lipopolysaccharides, peptidoglycans, and fungal chitin (Boller and Felix, 2009). In addition to sensing PAMPs, the ability to sense a compromised “self” by detecting damage-associated molecular patterns (DAMPs) such as released plant cell wall fragments is a central part of plant defense (Boller and Felix, 2009). Both PAMP- and DAMP-recognition activates PAMP-triggered immunity (PTI) which in general prevents microbial colonization. To escape detection and PTI induction, a common strategy among adapted microorganisms is to secrete a range of effector proteins that can modulate PTI components (Jones and Dangl, 2006). The stealth afforded by the microbial effectors can in turn be counteracted in the plant by an intracellular surveillance system consisting of an array of nucleotide-binding leucine-rich-repeat proteins that seek to detect the presence of such effector proteins, and enable induction of effector-triggered immunity (ETI). ETI is often associated with a localized cell death termed the hypersensitive response which functions to restrict spread of this more progressive stage of microbial attack (Jones and Dangl, 2006; Dodds and Rathjen, 2010). Hence the important feature of PTI is the ability to sense infectious-self and non-self, whereas for ETI it is the ability to sense microbe-mediated modifications inferred on points of vulnerability in the host. By guarding these weak points or even setting up decoys to confuse invaders, ETI is an efficient safety net for more progressed infections (van der Hoorn and Jones, 2004; Jones and Dangl, 2006).
Pathogen-associated molecular pattern perception is mediated by ligand-binding surface-exposed transmembrane pattern-recognition receptors (PRRs) of either the receptor-like kinase (RLK) or receptor-like proteins (RLPs) families. Both types of modular proteins are single-pass transmembrane proteins with extracellular domains, but where RLKs have an intracellular kinase domain; RLPs lack this cytosolic signaling domain (Monaghan and Zipfel, 2012). The archetypical bacterial PRRs are elongation factor Tu (EF-Tu) receptor (EFR), a leucine-rich repeat RLK (LRR-RLK) that recognizes the abundant cytoplasmic protein EF-Tu; and the related flagellin sensing 2 (FLS2) that recognizes flagellin, the principal component of bacterial flagella. These two PAMPs are often characterized by their minimal requirement peptide epitopes, elf18 and flg22 (Gomez-Gomez et al., 1999; Kunze et al., 2004; Boller and Felix, 2009). Sensing of the elf18 peptide appears to be restricted to the Brassicaceae family, while in rice recognition of EF-Tu, occurs via the EFa50 region (Furukawa et al., 2013). Interestingly, transgenic expression of Arabidopsis (Arabidopsis thaliana) EFR in Solanaceae species that cannot perceive EF-Tu is sufficient to confer resistance to a broad range of phytopathogenic bacteria, suggesting high conservation of the responses downstream of PAMP recognition (Lacombe et al., 2010). PTI induction leads to a series of early and late responses. The early responses occur within minutes to hours, and consist of rapid ion fluxes across the plasma membrane, an oxidative burst, activation of mitogen-activated protein kinases (MAPKs) and calcium-dependent protein kinases (CDPKs), and induction of defense-related genes. Deposition of callose, inhibition of seedling growth and PAMP induced resistance are later responses that occur within days (Boller and Felix, 2009).
A good example of the intricate evolutionary arms race between a pathogen and its plant host concerns the polysaccharide chitin (Figure 1). Chitin, a homopolymer of β-(1→4)-linked N-acetylglucosamine (GlcNAc) units, is the major structural component of fungal cell walls, and is also a main constituent of insect exoskeletons, crustacean shells, and the eggs and gut linings of nematodes (Bueter et al., 2013; Hadwiger, 2013). Chitin is an obvious PAMP-candidate and an ideal point of attack during plant defense responses since glucosamine polymers are not found in plants. It is therefore not surprising that an evolutionarily conserved strategy toward fungi and insects in plants is based on secreting chitinases – hydrolytic enzymes, which can break down microbial chitin polymers (Figure 1; Hadwiger, 2013). However, as usual, countermeasures have also evolved, the biotrophic fungal pathogen Cladosporium fulvum combats the action of chitinases by secreting the apoplastic effector Avr4, a chitin-binding lectin that functions to protect the integrity of the fungal cell wall against chitinases (Figure 1; van den Burg et al., 2006). Heterologous expression of Avr4 in Arabidopsis or tomato camouflages the chitin thereby increasing the virulence of several fungal pathogens (van Esse et al., 2007). That is unless the host harbors Cf-4, an extracellular membrane-anchored LRR protein that mediates Avr4 perception (Figure 1) and activates the hypersensitive response (Thomas et al., 1997; Takken et al., 1999). In addition to this forceful degradation strategy, chitin is also recognized as a PAMP in rice (Oryza sativa) by a dual recognition system consisting of the lysine motif (LysM)–RLK CHITIN ELICITOR RECEPTOR KINASE-1 OsCERK1 and the LysM-RLP CHITIN OLIGOSACCHARIDE ELICITOR-BINDING PROTEIN OsCEBiP (Shimizu et al., 2010). In Arabidopsis there is no contribution to signaling from the major chitin-oligosaccharide binding CEBiP, and AtCERK1 seems to act alone as the chitin PRR (Figure 1; Iizasa et al., 2010; Petutschnig et al., 2010; Shinya et al., 2012). The biological activity of chitin oligomers depends on their size, as the highest PAMP-activity is found for heptamers and octamers. Higher oligomeric chitin fragments like octamers can bind two or more AtCERK1 and this ligand-induced dimerization activates the receptor (Liu et al., 2012). To avoid chitin-induced PTI, C. fulvum also secretes the evolutionally conserved LysM-containing effector extracellular protein 6 (Ecp6) during infection. Ecp6 is a scavenger of chitin fragments released by chitinases and out-competes chitin-PRR binding to avoid fungal detection (Figure 1; Bolton et al., 2008; de Jonge et al., 2010; Sanchez-Vallet et al., 2013). Whether Ecp6 is recognized in plants is still unknown. In all cases, this example demonstrates the continuous battle between pathogen and host and how one party’s armor is continuously being evolutionarily countered by the opponent.
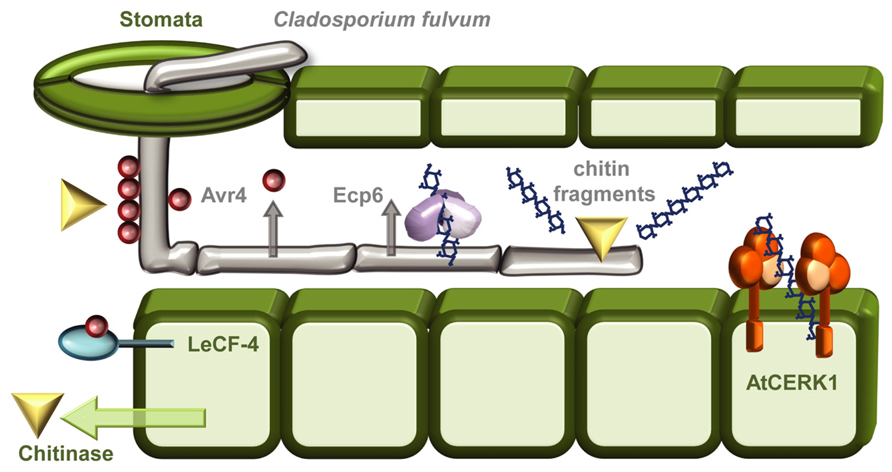
FIGURE 1. The “arms race” between Cladosporium fulvum and plant hosts. As a general protective measure against fungal infections plants respond to infection attempts by secreting chitinases into the apoplastic space. The tomato leaf mould fungus C. fulvum can enter its host through stomatal openings, and grows as extracellular hyphae. To shield against the action of these chitin-degrading enzymes the fungus camouflages its chitin-containing cell walls by cloaking them with the chitin-binding effector Avr4 (van den Burg et al., 2006; Hadwiger, 2013). The presence of Avr4 can be recognized by the Tomato RLP Cf-4, leading to induction of the hypersensitive response (Thomas et al., 1997; Takken et al., 1999). Chitin oligomers released by chitinases are recognized as PAMPs, in Arabidopsis by the RLK CERK1 (Iizasa et al., 2010; Petutschnig et al., 2010), and in rice by both OsCERK1 and the RLP OsCEBiP (Shimizu et al., 2010). To escape chitin-induced PTI C. fulvum can secrete Ecp6 an effector that functions as a chitin-scavenger removing the chitin oligomers released by the chitinases (de Jonge et al., 2010; Kombrink and Thomma, 2013; Sanchez-Vallet et al., 2013).
A vital feature of plant innate immunity is the early recognition of potential pathogens via perception of PAMPs and DAMPs. This recognition is mediated by designated surface-localized PRRs and sets in train a range of early and late responses that ensure a specialized and appropriate response is transmitted by a shared downstream pathway. Some of these steps are already mapped whereas others are yet to be discovered.
After ligand recognition, several PRRs rapidly form complexes with the regulatory RLK BRI1-ASSOCIATED KINASE1 (BAK1; Figure 2; Monaghan and Zipfel, 2012). Indeed, complex formation between the co-receptor and the flagellin receptor FLS2 occurs within seconds of flg22 treatment (Schulze et al., 2010). BAK1 was believed to function solely as a signal enhancer rather than being involved in ligand-binding. However, it has recently been demonstrated that BAK1 acts as a co-receptor in FLS2 mediated flg22 recognition by interacting with the bound ligand (Sun et al., 2013). BAK1 belongs to the somatic embryogenesis-related kinases; a small family of five LRR-RLKs (Heese et al., 2007; Roux et al., 2011). In addition to FLS2, BAK1 has also been shown to interact with EFR and the DAMP receptors PEPR1 and PEPR2 (Krol et al., 2010; Yamaguchi et al., 2010). Apart from its association with PRRs, BAK1 is also involved in the perception of brassinosteroids through its interaction with the RLK BRASSINOSTEROID INSENSITIVE 1 (BRI1; Li et al., 2002; Nam and Li, 2002). PAMP-binding and complex formation leads to auto- and trans-phosphorylation between PRR and co-receptor, as well as the plasma membrane localized receptor-like cytoplasmic kinase BOTRYTIS-INDUCED KINASE 1 (BIK1) and related PBS1-LIKE (PBL) kinases (Lu et al., 2010; Zhang et al., 2010). BIK1 plays a central role in conveying signals from several PRRs including EFR, FLS2, CERK1, and PEPR1/PEPR2. PAMP/DAMP induced phosphorylation of BIK1 prompts dissociation from the PRR complex which may permit BIK1 to move toward and activate downstream signaling targets (Lu et al., 2010; Zhang et al., 2010; Liu et al., 2013).
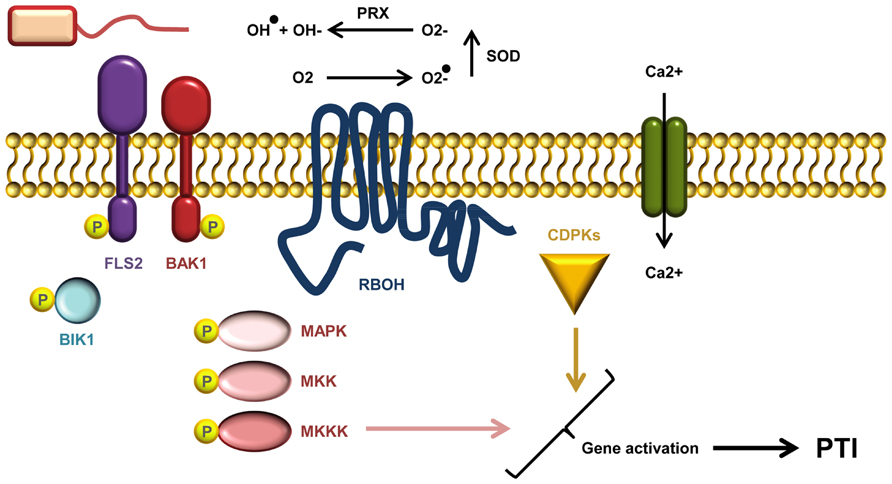
FIGURE 2. A current model of AtFLS2 signaling. Upon flg22 binding, a complex between FLS2, BIK1, BAK1 (and other SERKs) is formed. Complex formation triggers multiple rapid phosphorylation events resulting in BIK1 release. The signal transduction downstream of ligand perception includes a Ca2+ burst, activation CDPKs and AtRbohD required for the ROS burst, and induction MAPK cascades. Activation of CDPKs and MAPKs is required for full induction of defense genes.
Calcium ions serve as important second messengers in eukaryotes. Both abiotic and biotic stress responses lead to rapid transient fluctuations in the concentration of intracellular calcium ions (Figure 2). The low basal cytosolic concentration allows rapid spatial and temporal changes in the calcium flux. Such calcium signatures are sensed by stimulus-specific Ca2+-binding sensors that can mediate the downstream signal transduction. Deciphering the calcium signatures enables the affected cells to respond appropriately to diverse stimuli (DeFalco et al., 2010). During PTI, CDPKs function as Ca2+-sensors that are required to obtain complete transcriptional reprogramming (Figure 2). The four functionally redundant Arabidopsis CDPKs, CPK4, CPK5, CPK6, and CPK11 define a sub-clade required for flg22 induced PTI responses (Boudsocq et al., 2010). The transcriptional changes effected by the CDPKs are mostly independent of the MAPK pathway (see below), but the combination of CDPKs and MAPKs is needed to activate at least four early transcriptional regulatory programs induced by flg22 (Boudsocq et al., 2010; Wurzinger et al., 2011).
An oxidative burst of reactive oxygen species (ROS) such as ·O2-, ·OH, and H2O2 is one of the early measurable events in PTI (Figure 2). In Arabidopsis, the ROS burst is facilitated by the RESPIRATORY BURST OXIDASE HOMOLOGUE D (RbohD), an NADPH oxidase (Torres et al., 2002). The activity of RbohD is dependent on calcium and phosphorylation (Ogasawara et al., 2008; Kimura et al., 2012), and interestingly RbohD is phosphorylated by CDPK5 (Dubiella et al., 2013). In potato (Solanum tuberosum) StRbohB is phosphorylated by the plasma membrane-localized potato StCDPK5, and expression of constitutively active StCDPK5 in Nicotiana benthamiana leads to induction of ROS (Kobayashi et al., 2007).
Like the CDPKs, MAPK cascades also function to transmit signals within the cell via differential phosphorylation. In such cascades MAP kinase-kinase-kinases (MAPKKKs) phosphorylate MAP kinase-kinases (MAPKKs), which in turn phosphorylate MAP-kinases (Figure 2; Rasmussen et al., 2012). The Arabidopsis MAP kinases MPK3, MPK4, MPK6 as well as the recently documented MPK11 are all activated by PAMP treatment (Rasmussen et al., 2012). They designate at least two different pathways: one that triggers MPK4 activation by a module consisting of the two MAPKKs MKK1 and MKK2, and MEKK1; and another that leads to MPK3/MPK6 activation, via MKK4 and MKK5, and possibly MEKK1 (Rasmussen et al., 2012). Activation of MAPK cascades is, together with CDPK activation, vital for obtaining the transcriptional reprogramming needed to mount a full PTI response (Figure 2; Boudsocq et al., 2010). Even though some genes require both MAPK and CDPK activation to be induced there seems to be a branching of PTI signals on some level (Boudsocq et al., 2010; Xu et al., 2014). Loss of the flg22-induced ROS in Arabidopsis rbohD does not affect MPK3 and MPK6 activation, and activation of these MAP kinases is also not required for ROS burst (Xu et al., 2014).
Ubiquitination is a key signal for endosomal sorting of membrane proteins (Raiborg and Stenmark, 2009). Upon ligand perception FLS2 is being ubiquitinated by two PLANT U-BOX (PUB) E3 ligases PUB12 and PUB13 (Lu et al., 2011). This may well be part of a strategy to terminate the signal initiated by PAMP recognition. FLS2 is endocytosed and targeted for endocytic degradation approximately 30–60 min post-elicitation, and the activated FLS2 travels through early, late and multi-vesicular endosomes toward its vacuolar destination (Robatzek et al., 2006; Chinchilla et al., 2007; Beck et al., 2012b; Smith et al., 2014). The removal of ligand-bound receptors de-sensitizes the system after initial PAMP-treatment. After two hours the de-sensitized cells begin to be re-sensitized through de novo synthesis mediated replenishment of the receptor, preparing them for a new round of ligand perception (Smith et al., 2014). FLS2 has recently been shown to interact with two subunits of ENDOSOMAL SORTING COMPLEXES REQUIRED FOR TRANSPORT-I (ESCRT-I; Spallek et al., 2013). ESCRT complexes are responsible for identifying ubiquitinated vesicular cargoes, sorting them for degradation, and depositing them in intraluminal vesicles to prevent their recycling (Raiborg and Stenmark, 2009). Loss of the E3 ligases or ESCRT-I components affects flg22-triggered immune reactions underlining the importance of correct endosomal trafficking of FLS2 in sustaining a sufficient level of immunity (Lu et al., 2011; Spallek et al., 2013). In addition removal of activated receptors, trafficking might also play an integrated part in signaling transduction as part of the signaling responses may happen from endosomal compartments (Geldner and Robatzek, 2008; Beck et al., 2012a).
The final consequence of PTI is the induction of resistance responses that will prevent microbial colonization. Other late responses are the inhibition of seedling growth inhibition occurring within days of continuous PAMP treatment, and deposition of callose about 16 h after PAMP treatment (Boller and Felix, 2009). Callose is found during attempted fungal infections in cell wall fortifications termed papillae, structures that are assembled at fungal penetration sites (Underwood, 2012). The linear β-(1→3)-D-Glcp homo-polymer is produced by callose synthases; among these is the Arabidopsis POWDERY MILDEW RESISTANT 4 (PMR4), the predominant synthase responsible for defense-induced callose deposition (Vogel and Somerville, 2000; Nishimura et al., 2003; Ham et al., 2007; Chen and Kim, 2009). All the callose deposited in response to flg22, and most of the chitosan-induced depositions are synthesized by PMR4 (Luna et al., 2011). In addition to differences in their dependency on PMR4, callose depositions induced by these two PAMPs also differ in their requirements for other signaling components as only the flg22-induced callose depends on RbohD (Luna et al., 2011). Callose depositions like the papillae are an important feature of immunity, and are thought to reinforce the cell wall at fungal penetration sites to impede infections (Underwood, 2012). In agreement with this, overexpression of PMR4 in Arabidopsis results in enlarged callose deposits, and gain of penetration resistance against both virulent and non-adapted powdery mildew strains (Ellinger et al., 2013; Naumann et al., 2013). Absence of callose depositions also leads to susceptibility toward the non-adapted hemibiotrophic bacteria Pseudomonas syringae pv. phaseolicola (Ham et al., 2007). Surprisingly the pmr4 mutant was identified by its increased powdery mildew resistance, a phenotype that is dependent on the defense phytohormone salicylic acid (Vogel and Somerville, 2000; Nishimura et al., 2003; Huibers et al., 2013). These observations are counterintuitive both with respect to the pattern of callose deposition during defense induction, and the recent evidence from overexpression lines, supporting a role for callose in the plants defensive strategy (Vogel and Somerville, 2000; Ellinger et al., 2013; Naumann et al., 2013). One explanation for such a discrepancy could be that PMR4 is guarded by a resistance gene. Accordingly, loss of PMR4 might be perceived as a breach on the plants defensive fences and thus set about induction of ETI via elevation of salicylic acid.
Plant cell walls not only provide structure to the plant body but also act as barriers against biotic and abiotic stresses. The cell wall, sometimes covered with a cuticle, is usually the first obstacle encountered by pathogens, and to penetrate this barrier microbes have evolved an arsenal of wall degrading enzymes which are key virulence factors (Figure 3; Nuhse, 2012; Davidsson et al., 2013).
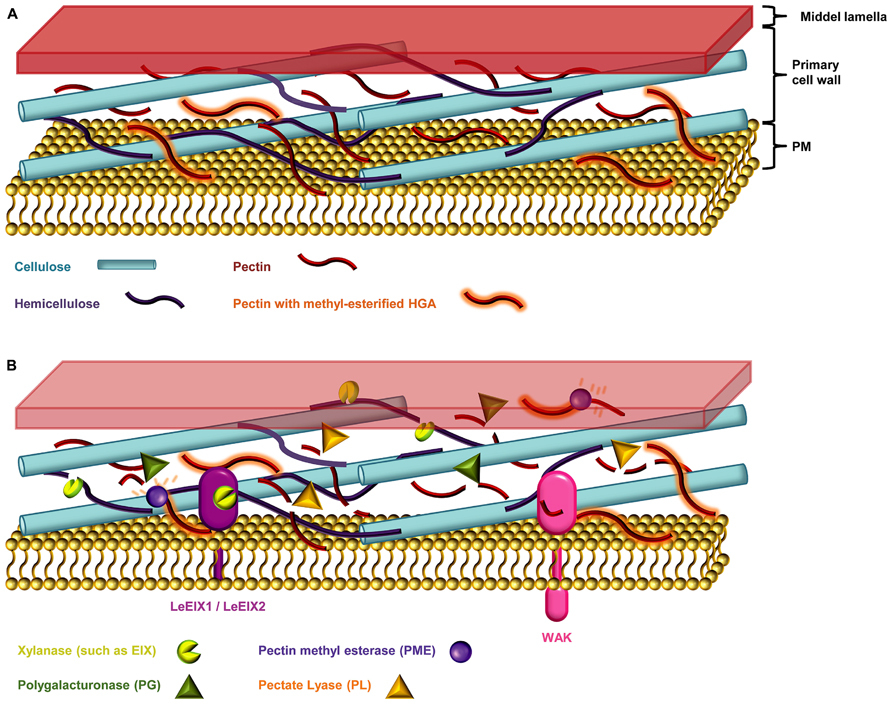
FIGURE 3. The plant cell wall. (A) The primary cell wall is constructed of a web of cellulose micro-fibrils, hemicellulose polysaccharides, and the hetero-polysaccharide pectin (Endler and Persson, 2011). (B) Necotrophic pathogens devour their hosts by degrading the plant cell wall, and secreting necrosis-inducing proteins and toxins (Davidsson et al., 2013). The host entry step of such microbes is thought to happen mainly via secretion of cell wall degrading enzymes such as cellulases, pectinases, and hemicellulases (Choquer et al., 2007; Davidsson et al., 2013). Xylanase, degrades the linear backbone of the predominant hemicellulose xylan into xylose residues (Belien et al., 2006; Scheller and Ulvskov, 2010). Pectinases such as polygalacturonases (PGs) and pectate lyases (PLs) degrades the pectic homogalacturonan (HGA) backbone. The HGA in newly synthesized pectin is methyl-esterified, which protects it from degradation by pectinases. De-esterification of HGA by the action of microbial pectin methyl esterases (PMEs) enables access PGs and PLs (Annis and Goodwin, 1997; Wolf et al., 2009; Lionetti et al., 2012). Endo-PGs cleave un-esterified regions while concomitantly releasing the oligogalacturonides (OGAs), oligomeric fragments of the HGA backbone (Annis and Goodwin, 1997; Ferrari et al., 2013). The wall-associated kinases (WAKs) are DAMP sensors that monitor the integrity of pectin by sensing the presence of OGAs (Ferrari et al., 2013). Plants can counter the cocktail of cell wall degrading enzymes by producing proteinaceous inhibitors such as the xylanase inhibitors and PG inhibitor proteins (PGIPs; Belien et al., 2006).
Most plant cell walls are based on a co-extensive load-bearing network of cellulose micro-fibrils cross-linked with hemicelluloses (Figure 3; Fry, 2004; Scheller and Ulvskov, 2010; Pauly et al., 2013). In the primary walls of the growing parts of plants, this network is embedded in a matrix of pectic polysaccharides. In the secondary cell walls of mature, non-growing tissues pectin is less abundant but walls are reinforced with lignin (Endler and Persson, 2011). Although most cell walls are based on these main components they differ considerably in their fine structures and three dimensional architectures. This heterogeneity is reflected in the diversity of strategies that pathogens have evolved to breach them, including the secretion of numerous glycosyl hydrolases (Annis and Goodwin, 1997). In response to an attack, plants may deposit certain reinforcing polymers, notably callose, phenolic complexes, and employ toxic compounds (Huckelhoven, 2007). However, these physical and chemical responses are only one part of the cell walls role in defense and the extensive ability of pathogens to degrade cell wall component has a cost. Disturbance of cell wall integrity (which may also cause deformation of adjacent portion of the plasma membrane) and the release of degradation fragments are monitored by plants and changed cell wall status is an important trigger for defense mechanisms (Hematy et al., 2009). Despite the undoubted importance of cell walls in plant defense there are many aspects that are poorly understood. For example, most work has focused on a limited set of cell wall polymers (notably callose, extensins, and lignin) and the potential roles of many other cell wall components are obscure. Another largely unexplored aspect of cell walls and defense is the evolution of and co-evolution of defense strategies (Sørensen et al., 2010). Discussed below are the major cell wall components in the context of pathogenic responses to them, and the prospects for advancing our understanding using emerging microarray technologies.
Tightly packed crystalline cellulose micro-fibrils serve a primary structural function in the cell wall (Endler and Persson, 2011; Nuhse, 2012). The fibrils are composed of hydrogen-bonded β-(1→4)-D-Glcp chains, and have a condensed nature that makes them generally resistant to degradation via glycosyl hydrolases. However, recent work has highlighted the role of another class of oxidative enzymes, the lytic polysaccharide monoxygenases that have the capacity to degrade recalcitrant crystalline cell wall components, including cellulose (Vaaje-Kolstad et al., 2010; Hemsworth et al., 2014). Cellulose micro-fibrils are synthesized by large multimeric complexes containing cellulose synthase catalytic subunits (CESAs; Endler and Persson, 2011). It is generally considered that in Arabidopsis, and probably other angiosperms, there are two CESA subfamilies with distinct roles. Whilst CESAs 1, 3, and 6 are responsible for the formation of cellulose in primary walls, CESAs 4, 7, and 8 synthesize cellulose in secondary walls (Endler and Persson, 2011). However, recent work has demonstrated that some functional cross over exists across these groups in vitro although it is not clear if these new CESA pairings are significant in planta (Carroll et al., 2012).
As in many cases where cell wall integrity is perturbed, compensatory mechanisms have evolved to limit the effects of disturbance to cellulose structure or deposition. Cellulose deficient mutants typically exhibit increased lignification (Cano-Delgado et al., 2003; Hamann, 2012). Interestingly, these changes appear to have effects beyond the purely structural. Such mutants also display enhanced defense responses (Hamann, 2012) as mutants in the primary wall CESA3 are more resistant toward powdery mildew (Ellis and Turner, 2001; Cano-Delgado et al., 2003). Additionally defects in the secondary wall CESAs; CESA4, CESA7, and CESA8, also lead to elevated resistance against broad-host necrotrophic pathogens like the fungus Plectosphaerella cucumerina and the soil-borne bacterium Ralstonia solanacearum (Hernandez-Blanco et al., 2007). Consistent with the genetic evidence, similar results have also been described for treatment with the cellulose biosynthesis inhibitor isoxaben (Hamann et al., 2009; Hamann, 2012). In Arabidopsis, isoxaben-mediated wall damage and lignin production is induced via an RbohD-dependent mechanism, and fine-tuned through a jasmonic-acid-dependent negative feedback loop (Hamann et al., 2009; Denness et al., 2011). The defense associated cellulose deprivation phenotypes suggest that the cell wall damage triggers defense responses, and suggest the presence of a tight cell wall integrity monitoring system. The RLK THESEUS1 (THE1) seems to serve a role in this as it has been implicated in the response to cell wall damage. The the1 mutant was identified as a suppressor of the cesA6 null allele procuste1-1. Mutations in THE1 reduce the ectopic lignification of several cellulose deficient mutants (Hematy et al., 2007).
Hemicelluloses are a diverse group of polysaccharides usually characterized by having a β-(1→4)-linked backbone of mannose, glucose, or xylose. The central role of hemicelluloses is to fortify the cell wall by interaction with cellulose and sometimes lignin (Scheller and Ulvskov, 2010; Endler and Persson, 2011). Xylans are the predominant hemicelluloses in secondary plant cell walls. The common characteristic of xylans is a backbone of repeating β-(1→4)-D-Xylp residues most often substituted by Araf and GlcpA (Scheller and Ulvskov, 2010). The arabinosyl residues can in addition contain ferulic acid groups esterified to the O-5 position of the carboxyl group (Smith and Hartley, 1983). These ferulate esters can be oxidatively cross-linked to lignin possibly incorporating xylan into the lignin complex to advance strengthening of the network (Tanner and Morrison, 1983; Mcneil et al., 1984; Harris and Stone, 2009). Some phytopathogenic microbes secrete xylanases, which are enzymes that can degrade the linear xylan backbone into xylose units (Belien et al., 2006). Hemicellulose breakdown by xylanases weakens the cell wall and enables the microbe to breach it (Belien et al., 2006). Fungi of the Trichoderma spp. produce the ETHYLENE-INDUCING XYLANASE (EIX) that is recognized as a PAMP (Furman-Matarasso et al., 1999; Belien et al., 2006). In tomato (Lycopersicum esculentum), EIX is sensed by the cell wall-derived RLPs LeEix1 and LeEix2 (Figure 3; Ron and Avni, 2004).
In addition to feruloylation cell wall glycans can also be subjected to methylation and acetylation. REDUCED WALL ACETYLATION 2 (RWA2) has been shown to be responsible for the acetylation of numerous pectic and non-pectic polymers in Arabidopsis, and rwa2 knock-out mutants have (Mcneil et al., 1984) increased resistance toward Botrytis cinerea (Manabe et al., 2011). Furthermore de-acetylation of xyloglucan and pectin in transgenic plants exhibit increased accessibility to degrading enzymes which could be part of a defense strategy to release oligosaccharides which can act as defense elicitors (Pogorelko et al., 2013).
The complex hetero-polysaccharide pectin is a major cell wall matrix component that is abundant in primary cell walls. It is composed of a series of structurally distinct domains acting as backbones or side chains to the pectic polysaccharides (Wolf et al., 2009). Back bones are comprised either of contiguous α-(1→4)-D-GalpA residues (homogalacturonan or HGA) or repeating dimers of α-(1→4)-D-GalpA-α-(1→2)-L-Rhap residues (rhamogalacturonan). Within the pectin complex especially HGA appears to have special significance in the context of defense responses. HGA can be methyl esterified at C6 and acetylated at C2–C3 and regulation of these substitutions enable plants to fine-tune the functionality of HGA to suit prevailing local requirements. For example non-esterified HGA carrying negatively charged free carboxyl groups may be subject to cross-linking via calcium leading to the formation of gels that are required for cell adhesion and other structural roles (Cabrera et al., 2008).
Some of the first enzymes that phytopathogenic fungi secrete during infections are endo-polygalacturonases (PGs) which cleave HGA (Figure 3), thereby degrading cell wall integrity and aiding pathogen access (Annis and Goodwin, 1997). Moreover, HGA degradation releases oligogalacturonide (OGA) fragments from the backbone, which act as potent defense response elicitors (Galletti et al., 2009). The WALL-ASSOCIATED KINASES (WAKs) are thought to be DAMP sensors that monitor the integrity of pectin by detecting the presence of OGAs with a degree of polymerization between 10 and 15 (Ferrari et al., 2013). In the context of degradation by microbial enzymes the level of HGA methyl esterification is critically important. Endo-PGs and pectate lyases (PLs) preferentially cleave non-esterified HGA and these enzymes frequently act in concert with pectin methyl esterases (PMEs) that de-methyl esterify HGA to create endo-PG and PL cleavage sites (Figure 3). Moreover, another class of microbial enzymes – the pectin lyases act preferentially on highly methyl esterified HGA.
Homogalacturonan and the microbial enzymes that degrade it are good examples of how the high complexity of cell wall components is countered by sets of highly specific microbial enzymes. Nevertheless, the role of pectin in plant defense cannot only be viewed in straightforward terms of polysaccharide integrity versus enzymatic breakdown. An example is provided by the elevated activity of certain endogenous Arabidopsis PMEs in response to infection with the necrotrophic fungus Alternaria brassicicola, the hemibiotrophic bacterial pathogen P. syringae pv. maculicola ES4326, or by treatment with PAMPs (Bethke et al., 2014). The increase in PME activity in such infected plants leads to a concomitant decrease in methyl esterification of HGA, which doubtlessly renders the HGA more susceptible toward endo-PGs (Bethke et al., 2014). It seems counterintuitive to increase the activity of an endogenous wall degrading enzyme during a pathogen attack as this could aid microbial entry. One explanation is that the enhanced production of OGAs – and thereby an associated potential activation of DAMP signaling – is worth the price of a decrease in HGA integrity. It is also important to consider the broader picture of the 66 PMEs in Arabidopsis in terms of redundancy and spatiotemporal regulation. For example, in the case of A. brassicicola it is worth noting that the observed increase in PME activity occurred rather late after pathogen challenge. With this in mind it may be the case that the purpose of enhanced PME activity is localized HGA de-methyl esterification and the concomitant formation of gel structures to bolster damaged walls (Bethke et al., 2014).
Another noteworthy insight into the roles of pectin in defense is provided by the loss of function mutants of the endogenous plant enzyme POWDERY MILDEW RESISTANT 6 MUTANT (PMR6; Vogel et al., 2002). PMR6 has similarity to PLs, and loss of function leads to enhanced levels of intact pectin and but also lower levels of potential OGA release. One explanation for the enhanced resistance to powdery mildew in pmr6 is that increased levels of inaccessible HGA per se provide more protection. But, the fact that pmr6 only exhibits this elevated resistance against certain powdery mildew species (Erysiphe cichoracearum and E. orontii) implies a less generic and more subtle effect. For example, it is possible that alterations in pmr6 cell wall composition make these mutants poor hosts specifically for Erysiphe spp. It certainly seems highly unlikely that a plant gene would have evolved to serve the needs of a pathogen, and the pleotropic effects observed in pmr6 are consistent with a role as a susceptibility factor. It is plausible that pmr6 resistance is a specialized form of disease resistance, possibly based on the loss of a host susceptibility gene required by the pathogen for growth and development (Vogel et al., 2002).
Lignin is a phenolic polymer mainly deposited in secondary cell wall during the last stages of cell differentiation. It displaces the aqueous phase of the cell wall, encasing cellulose and matrix polysaccharides and providing enhanced mechanical strength and a water-impermeable barrier (Albersheim et al., 2011). Lignin is also required for reinforcing vascular cells that transport water under negative pressure as a result of transpiration. The importance of lignin in these tissues is demonstrated by vascular collapse in lignin deficient plants (Piquemal et al., 1998; Jones et al., 2001). Lignin is built from monolignols, with up to three different types in higher plants which appear to be incorporated into the lignin polymer in a non-predictable fashion (Martone et al., 2009; Vanholme et al., 2010). This apparently random pattern of synthesis may be significant in relation to microbial enzymes which have typically evolved to break polymers with structurally consistent cleavage sites (Sarkar et al., 2009).
Lignin has multiple roles in plant defense and lignin, or lignin-like phenolic polymers are often rapidly deposited in response to both biotic and abiotic stresses (Sattler and Funnell-Harris, 2013). Lignin not only acts as physical barrier to pathogen invasion, but the phenylpropanoid pathway responsible for lignin biosynthesis may also be recruited for defense purposes. For example, this pathway underpins the synthesis of other phenolic compounds including phytoalexins, stilbenes, coumarins, and flavonoids – some of which have been implicated in plant defense (Weiergang et al., 1996; Dicko et al., 2005; Lozovaya et al., 2007). Indeed, there is evidence that in some plants, salicylic acid which is known to be a key component of some defense pathways, may also be produced by the phenylpropanoid pathway (Ruuhola et al., 2003; Pan et al., 2006). Understanding the role of lignin in plant defense has received considerable attention in the context of engineering plants for use as bioenergy feedstocks. Lignin contributes substantially to the recalcitrance of cell walls to deconstruction into fermentable sugars and for this reason reducing lignin content has become an important goal for feedstock biotechnology research. However, since lignin is also required for plant defense there is probably a lower limit for lignin reduction beyond which plant would become unacceptably vulnerable to pathogens (Sattler and Funnell-Harris, 2013). The roles of lignin in defense and the importance of this for bioenergy feedstocks are reviewed in (Sattler and Funnell-Harris, 2013).
Often, an infection involves a multifaceted strategy. In the case of B. cinerea the host entry step is thought to happen mainly via secretion of cell wall degrading enzymes such as a PMEs, endo-PGs, and endo-β-1,4-xylanase often encoded by multi-genic families (Choquer et al., 2007). Cellulases, pectinases, and hemicellulases are also secreted from bacteria like the Pectobacterium and Dickeya genera necrotrophic soft rot enterobacteria (Davidsson et al., 2013). At first glance, plants would seem defenseless against such savage enzyme cocktails, however, they can counter by producing proteinaceous inhibitors of cell wall degrading enzymes such as the xylanase inhibitors and PG inhibitor proteins (PGIPs; Belien et al., 2006). The cunning strategy of making PGIPs is not to inhibit pectin degradation entirely, but to shift it toward producing longer OGAs that can be sensed as DAMPs (Federici et al., 2006).
Even from a concise overview such as provided above, it is clear that analyzing the subtle, complex and often rapid changes that affect cell walls in response to pathogens is a formidable technical challenge. In contrast to proteins and nucleotides, the polysaccharides from which cell walls are primarily made cannot readily be sequenced, synthesized, or expressed. Although cell walls are usually physically tough, they are also typically highly plastic and can undergo rapid modifications in response to specific local conditions, even within cell wall micro-domains. The analytical difficulties are compounded by the sheer complexity and multilayered nature of host/pathogen interactions, and the differing nature of these interactions depending on developmental stage, organ, or tissue. Conventional biochemical techniques for carbohydrate analysis, for example methylation analysis and ion exchange chromatography, are powerful but low throughput and usually require large amounts of material. They also involve the complete or partial destruction of cell walls into their component parts – which inevitably means that information about three dimensional cell wall architectures is lost. Molecular probes, for example monoclonal antibodies (mAbs) and carbohydrate binding modules (CBMs) have been invaluable for advancing our understanding of plant cell walls per se, and have immense potential for providing insight into host pathogen interactions at the cellular and subcellular levels. There is an extensive repertoire of probes directed against plant cell wall components (Lee et al., 2011), but this is not matched by probes for pathogens and their associated effectors. For example, our understanding of the precise role of chitosan (the de-acetylated form of chitin) during pathogenesis is hampered by a lack of suitable probes (Sorlier et al., 2003; Hoell et al., 2010; Baker et al., 2011).
One technique that appears to offer considerable promise for plant/pathogen interaction research is based on carbohydrate microarrays (Moller et al., 2007). This technique combines the specificity of mAbs and CBMs with the multiplexed analysis capacity of robotically produced microarrays, and simultaneously provides information about the relative abundance of multiple epitopes present in large sample sets (Tyler et al., 2014). This approach has been extensively used for cell wall analysis in numerous contexts (Tronchet et al., 2010; Sørensen et al., 2011; Fangel et al., 2012; Moore et al., 2014; Zhang et al., 2014), but has only been used in a very limited way for plant defense research (Nguema-Ona et al., 2013). Typically, at least 20 mAbs or CBMs are used in a single analysis, and several 100 samples can be processed in 2 days. Analysis starts with the preparation of cell wall material (although native plant material can also be used) which is then treated with a series of solvents to release cell wall components. For example, extraction with 1,2-diaminocyclohexanetetraacetic acid followed by sodium hydroxide is expected to release pectins and hemicellulose respectively. These extractions are then printed onto a suitable surface such as nitrocellulose membrane or modified glass slides using a microarray robot, and the resultant arrays probed with mAbs and/or CBMs and then quantified. The fact that relatively small amounts of cell wall material are needed (0.5–10 mg) and the ability to rapidly analyze large numbers of samples are attractive attributes in the context of plant/pathogen interaction research. Currently, the analysis of cell walls in the context of plant defense is often limited to polymers with well characterized roles, for example extensins and callose. The described technology can potentially provide the means to greatly increase the scope of cell wall analysis, both in terms of the number of cell wall components analyzed and the number of variables in an experimental set up. Our initial experiments indicate that glycan arrays are powerful tools for monitoring the progression of cell wall changes during defense responses and are well suited to the parallel analysis of, for example, multiple mutants, elicitors, or conditions. Furthermore we anticipate that analysis of cell wall glycomes will be integrated with transcriptomic analysis of defense related genes using the same material. Moreover, although the technique has mostly been applied to carbohydrate analysis, the same approach could be applied to investigate proteins and their conjugates of relevance to pathogenic interactions. So long as analytes can be extracted and immobilized, and providing that probes are available the scope of analysis can be extended.
Our current knowledge of the interactions between plants and pathogens at the plant cell wall level, and how this affects downstream signaling is still limited. To obtain more insight we need to expand our understanding of the full repertoire of cell wall modifications that occurs during microbial interactions. Carbohydrate microarrays could be used to monitor the complex interplay between microbes and plants, and may reveal additional, perhaps more subtle, cell wall modifications to those documented so far. This technique enables rapid and multiplexed analysis of multiple changes in wall composition and could be used as a tool to provide new insight into the dynamic nature of host/pathogen interactions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Milena Roux and Dr. Jacqueline Monaghan for critically reading and commenting on the manuscript.
Albersheim, P., Darvill, A., Roberts, K., Sederoff, R., and Staehelin, A. (ed.). (2011). Plant Cell Walls. New York, NY: Garland Science, Taylor and Francis Publishing Group, LLC, 52–61.
Annis, S. L., and Goodwin, P. H. (1997). Recent advances in the molecular genetics of plant cell wall-degrading enzymes produced by plant pathogenic fungi. Eur. J. Plant Pathol. 103, 1–14. doi: 10.1023/A:1008656013255
Baker, L. G., Specht, C. A., and Lodge, J. K. (2011). Cell wall chitosan is necessary for virulence in the opportunistic pathogen Cryptococcus neoformans. Eukaryot. Cell 10, 1264–1268. doi: 10.1128/Ec.05138-11
Beck, M., Heard, W., Mbengue, M., and Robatzek, S. (2012a). The INs and OUTs of pattern recognition receptors at the cell surface. Curr. Opin. Plant Biol. 15, 367–374. doi: 10.1016/j.pbi.2012.05.004
Beck, M., Zhou, J., Faulkner, C., Maclean, D., and Robatzek, S. (2012b). Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24, 4205–4219. doi: 10.1105/tpc.112.100263
Belien, T., Van Campenhout, S., Robben, J., and Volckaert, G. (2006). Microbial endoxylanases: effective weapons to breach the plant cell-wall barrier or, rather, triggers of plant defense systems? Mol. Plant Microbe Interact. 19, 1072–1081. doi: 10.1094/Mpmi-19-1072
Bethke, G., Grundman, R. E., Sreekanta, S., Truman, W., Katagiri, F., and Glazebrook, J. (2014). Arabidopsis PECTIN METHYLESTERASEs contribute to immunity against Pseudomonas syringae. Plant Physiol. 164, 1093–1107. doi: 10.1104/pp.113.227637
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Bolton, M. D., Van Esse, H. P., Vossen, J. H., De Jonge, R., Stergiopoulos, I., Stulemeijer, I. J. E., et al. (2008). The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 69, 119–136. doi: 10.1111/j.1365-2958.2008.06270.x
Boudsocq, M., Willmann, M. R., Mccormack, M., Lee, H., Shan, L., He, P., et al. (2010). Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464, 418–422. doi: 10.1038/nature08794
Bueter, C. L., Specht, C. A., and Levitz, S. M. (2013). Innate sensing of chitin and chitosan. PLoS Pathog. 9:e1003080. doi: 10.1371/journal.ppat.1003080
Cabrera, J. C., Boland, A., Messiaen, J., Cambier, P., and Van Cutsem, P. (2008). Egg box conformation of oligogalacturonides: the time-dependent stabilization of the elicitor-active conformation increases its biological activity. Glycobiology 18, 473–482. doi: 10.1093/glycob/cwn027
Cano-Delgado, A., Penfield, S., Smith, C., Catley, M., and Bevan, M. (2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34, 351–362. doi: 10.1046/j.1365-313X.2003.01729.x
Carroll, A., Mansoori, N., Li, S., Lei, L., Vernhettes, S., Visser, R. G., et al. (2012). Complexes with mixed primary and secondary cellulose synthases are functional in Arabidopsis plants. Plant Physiol. 160, 726–737. doi: 10.1104/pp.112.199208
Chen, X. Y., and Kim, J. Y. (2009). Callose synthesis in higher plants. Plant Signal. Behav. 4, 489–492. doi: 10.4161/psb.4.6.8359
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nurnberger, T., Jones, J. D., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. doi: 10.1038/nature05999
Choquer, M., Fournier, E., Kunz, C., Levis, C., Pradier, J. M., Simon, A., et al. (2007). Botrytis cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 277, 1–10. doi: 10.1111/j.1574-6968.2007.00930.x
Davidsson, P. R., Kariola, T., Niemi, O., and Palva, E. T. (2013). Pathogenicity of and plant immunity to soft rot pectobacteria. Front. Plant Sci. 4:191. doi: 10.3389/fpls.2013.00191
DeFalco, T. A., Bender, K. W., and Snedden, W. A. (2010). Breaking the code: Ca2+ sensors in plant signalling. Biochem. J. 425, 27–40. doi: 10.1042/BJ20091147
de Jonge, R., Van Esse, H. P., Kombrink, A., Shinya, T., Desaki, Y., Bours, R., et al. (2010). Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329, 953–955. doi: 10.1126/science.1190859
Denness, L., Mckenna, J. F., Segonzac, C., Wormit, A., Madhou, P., Bennett, M., et al. (2011). Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 156, 1364–1374. doi: 10.1104/pp.111.175737
Dicko, M. H., Gruppen, H., Barro, C., Traore, A. S., Van Berkel, W. J., and Voragen, A. G. (2005). Impact of phenolic compounds and related enzymes in sorghum varieties for resistance and susceptibility to biotic and abiotic stresses. J. Chem. Ecol. 31, 2671–2688. doi: 10.1007/s10886-005-7619-5
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812x
Dubiella, U., Seybold, H., Durian, G., Komander, E., Lassig, R., Witte, C. P., et al. (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. U.S.A. 110, 8744–8749. doi: 10.1073/pnas.1221294110
Ellinger, D., Naumann, M., Falter, C., Zwikowics, C., Jamrow, T., Manisseri, C., et al. (2013). Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol. 161, 1433–1444. doi: 10.1104/pp.112.211011
Ellis, C., and Turner, J. G. (2001). The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13, 1025–1033. doi: 10.1105/tpc.13.5.1025
Endler, A., and Persson, S. (2011). Cellulose synthases and synthesis in Arabidopsis. Mol. plant 4, 199–211. doi: 10.1093/mp/ssq079
Fangel, J. U., Ulvskov, P., Knox, J. P., Mikkelsen, M. D., Harholt, J., Popper, Z. A., et al. (2012). Cell wall evolution and diversity. Front. Plant Sci. 3:152. doi: 10.3389/fpls.2012.00152
Federici, L., Di Matteo, A., Fernandez-Recio, J., Tsernoglou, D., and Cervone, F. (2006). Polygalacturonase inhibiting proteins: players in plant innate immunity? Trends Plant Sci. 11, 65–70. doi: 10.1016/j.tplants.2005.12.005
Ferrari, S., Savatin, D. V., Sicilia, F., Gramegna, G., Cervone, F., and Lorenzo, G. D. (2013). Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 4:49. doi: 10.3389/fpls.2013.00049
Fry, S. C. (2004). Primary cell wall metabolism: tracking the careers of wall polymers in living plant cells. New Phytol. 161, 641–675. doi: 10.1111/j.1469-8137.2003.00980.x
Furman-Matarasso, N., Cohen, E., Du, Q., Chejanovsky, N., Hanania, U., and Avni, A. (1999). A point mutation in the ethylene-inducing xylanase elicitor inhibits the beta-1-4-endoxylanase activity but not the elicitation activity. Plant Physiol. 121, 345–351. doi: 10.1104/pp.121.2.345
Furukawa, T., Inagaki, H., Takai, R., Hirai, H., and Che, F. S. (2013). Two distinct EF-Tu epitopes induce immune responses in rice and Arabidopsis. Mol. Plant Microbe Interact. 27, 113–124. doi: 10.1094/MPMI-10-13-0304-R
Galletti, R., De Lorenzo, G., and Ferrari, S. (2009). Host-derived signals activate plant innate immunity. Plant Signal. Behav. 4, 33–34. doi: 10.4161/psb.4.1.7224
Geldner, N., and Robatzek, S. (2008). Plant receptors go endosomal: a moving view on signal transduction. Plant Physiol. 147, 1565–1574. doi: 10.1104/pp.108.120287
Gomez-Gomez, L., Felix, G., and Boller, T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18, 277–284. doi: 10.1046/j.1365-313X.1999.00451.x
Hadwiger, L. A. (2013). Multiple effects of chitosan on plant systems: solid science or hype. Plant Sci. 208, 42–49. doi: 10.1016/j.plantsci.2013.03.007
Ham, J. H., Kim, M. G., Lee, S. Y., and Mackey, D. (2007). Layered basal defenses underlie non-host resistance of Arabidopsis to Pseudomonas syringae pv. phaseolicola. Plant J. 51, 604–616. doi: 10.1111/j.1365-313X.2007.03165.x
Hamann, T. (2012). Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front. Plant Sci. 3:77. doi: 10.3389/fpls.2012.00077
Hamann, T., Bennett, M., Mansfield, J., and Somerville, C. (2009). Identification of cell-wall stress as a hexose-dependent and osmosensitive regulator of plant responses. Plant J. 57, 1015–1026. doi: 10.1111/j.1365-313X.2008.03744.x
Harris, P. J., and Stone, B. A. (eds). (2009). Chemistry and Molecular Organization of Plant Cell Walls, Chap. 4. Oxford: Blackwell Publishing Ltd.
Heese, A., Hann, D. R., Gimenez-Ibanez, S., Jones, A. M., He, K., Li, J., et al. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. U.S.A. 104, 12217–12222. doi: 10.1073/pnas.0705306104
Hematy, K., Cherk, C., and Somerville, S. (2009). Host-pathogen warfare at the plant cell wall. Curr. Opin. Plant Biol. 12, 406–413. doi: 10.1016/j.pbi.2009.06.007
Hematy, K., Sado, P. E., Van Tuinen, A., Rochange, S., Desnos, T., Balzergue, S., et al. (2007). A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17, 922–931. doi: 10.1016/j.cub.2007.05.018
Hemsworth, G. R., Henrissat, B., Davies, G. J., and Walton, P. H. (2014). Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat. Chem. Biol. 10, 122–126. doi: 10.1038/nchembio.1417
Hernandez-Blanco, C., Feng, D. X., Hu, J., Sanchez-Vallet, A., Deslandes, L., Llorente, F., et al. (2007). Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19, 890–903. doi: 10.1105/tpc.106.048058
Hoell, I. A., Vaaje-Kolstad, G., and Eijsink, V. G. H. (2010). Structure and function of enzymes acting on chitin and chitosan. Biotechnol. Genet. Eng. Rev. 27, 331–366. doi: 10.1080/02648725.2010.10648156
Huckelhoven, R. (2007). Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45, 101–127. doi: 10.1146/annurev.phyto.45.062806.094325
Huibers, R. P., Loonen, A. E., Gao, D., Van Den Ackerveken, G., Visser, R. G., and Bai, Y. (2013). Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS ONE 8:e67467. doi: 10.1371/journal.pone.0067467
Iizasa, E., Mitsutomi, M., and Nagano, Y. (2010). Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J. Biol. Chem. 285, 2996–3004. doi: 10.1074/jbc.M109.027540
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jones, L., Ennos, A. R., and Turner, S. R. (2001). Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J. 26, 205–216. doi: 10.1046/j.1365-313x.2001.01021.x
Kimura, S., Kaya, H., Kawarazaki, T., Hiraoka, G., Senzaki, E., Michikawa, M., et al. (2012). Protein phosphorylation is a prerequisite for the Ca2+-dependent activation of Arabidopsis NADPH oxidases and may function as a trigger for the positive feedback regulation of Ca2+ and reactive oxygen species. Biochim. Biophys. Acta 1823, 398–405. doi: 10.1016/j.bbamcr.2011.09.011
Kobayashi, M., Ohura, I., Kawakita, K., Yokota, N., Fujiwara, M., Shimamoto, K., et al. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19, 1065–1080. doi: 10.1105/tpc.106.048884
Kombrink, A., and Thomma, B. P. (2013). LysM effectors: secreted proteins supporting fungal life. PLoS Pathog. 9:e1003769. doi: 10.1371/journal.ppat.1003769
Krol, E., Mentzel, T., Chinchilla, D., Boller, T., Felix, G., Kemmerling, B., et al. (2010). Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 285, 13471–13479. doi: 10.1074/jbc.M109.097394
Kunze, G., Zipfel, C., Robatzek, S., Niehaus, K., Boller, T., and Felix, G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16, 3496–3507. doi: 10.1105/tpc.104.026765
Lacombe, S., Rougon-Cardoso, A., Sherwood, E., Peeters, N., Dahlbeck, D., Van Esse, H. P., et al. (2010). Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369. doi: 10.1038/nbt.1613
Lee, K. J. D., Marcus, S. E., and Knox, J. P. (2011). Cell wall biology: perspectives from cell wall imaging. Mol. Plant 4, 212–219. doi: 10.1093/Mp/Ssq075
Li, J., Wen, J., Lease, K. A., Doke, J. T., Tax, F. E., and Walker, J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. doi: 10.1016/S0092-8674(02)00812-7
Lionetti, V., Cervone, F., and Bellincampi, D. (2012). Methyl esterification of pectin plays a role during plant-pathogen interactions and affects plant resistance to diseases. Plant Physiol. 169, 1623–1630. doi: 10.1016/j.jplph.2012.05.006
Liu, T., Liu, Z., Song, C., Hu, Y., Han, Z., She, J., et al. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164. doi: 10.1126/science.1218867
Liu, Z., Wu, Y., Yang, F., Zhang, Y., Chen, S., Xie, Q., et al. (2013). BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. U.S.A. 110, 6205–6210. doi: 10.1073/pnas.1215543110
Lozovaya, V. V., Lygin, A. V., Zernova, O. V., Ulanov, A. V., Li, S. X., Hartman, G. L., et al. (2007). Modification of phenolic metabolism in soybean hairy roots through down regulation of chalcone synthase or isoflavone synthase. Planta 225, 665–679. doi: 10.1007/s00425-006-0368-z
Lu, D., Lin, W., Gao, X., Wu, S., Cheng, C., Avila, J., et al. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332, 1439–1442. doi: 10.1126/science.1204903
Lu, D., Wu, S., Gao, X., Zhang, Y., Shan, L., and He, P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 496–501. doi: 10.1073/pnas.0909705107
Luna, E., Pastor, V., Robert, J., Flors, V., Mauch-Mani, B., and Ton, J. (2011). Callose deposition: a multifaceted plant defense response. Mol. Plant Microbe Interact. 24, 183–193. doi: 10.1094/MPMI-07-10-0149
Manabe, Y., Nafisi, M., Verhertbruggen, Y., Orfila, C., Gille, S., Rautengarten, C., et al. (2011). Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol. 155, 1068–1078. doi: 10.1104/pp.110.168989
Martone, P. T., Estevez, J. M., Lu, F. C., Ruel, K., Denny, M. W., Somerville, C., et al. (2009). Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr. Biol. 19, 169–175. doi: 10.1016/j.cub.2008.12.031
Medzhitov, R., and Janeway, C. A. Jr. (1997). Innate immunity: impact on the adaptive immune response. Curr. Opin. Immunol 9, 4–9. doi: 10.1016/S0952-7915(97)80152-5
Moller, I., Sorensen, I., Bernal, A. J., Blaukopf, C., Lee, K., Obro, J., et al. (2007). High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J. 50, 1118–1128. doi: 10.1111/j.1365-313X.2007.03114.x
Monaghan, J., and Zipfel, C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. doi: 10.1016/j.pbi.2012.05.006
Moore, J. P., Nguema-Ona, E., Fangel, J. U., Willats, W. G. T., Hugo, A., and Vivier, M. A. (2014). Profiling the main cell wall polysaccharides of grapevine leaves using high-throughput and fractionation methods. Carbohydr. Polym. 99, 190–198. doi: 10.1016/j.carbpol.2013.08.013
Nam, K. H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. doi: 10.1016/S0092-8674(02)00814-0
Naumann, M., Somerville, S., and Voigt, C. (2013). Differences in early callose deposition during adapted and non-adapted powdery mildew infection of resistant Arabidopsis lines. Plant Signal. Behav. 8:e24408. doi: 10.4161/psb.24408
Nguema-Ona, E., Moore, J. P., Fagerstrom, A. D., Fangel, J. U., Willats, W. G. T., Hugo, A., et al. (2013). Overexpression of the grapevine PGIP1 in tobacco results in compositional changes in the leaf arabinoxyloglucan network in the absence of fungal infection. BMC Plant Biol. 13:46. doi: 10.1186/1471-2229-13-46
Mcneil, M., Darvill, A. G., Fry, S. C., and Albersheim, P. (1984). Structure and function of the primary-cell walls of plants. Annu. Rev. Biochem. 53, 625–663. doi: 10.1146/annurev.bi.53.070184.003205
Nishimura, M. T., Stein, M., Hou, B. H., Vogel, J. P., Edwards, H., and Somerville, S. C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301, 969–972. doi: 10.1126/science.1086716
Nuhse, T. S. (2012). Cell wall integrity signaling and innate immunity in plants. Front. Plant Sci. 3:280. doi: 10.3389/fpls.2012.00280
Ogasawara, Y., Kaya, H., Hiraoka, G., Yumoto, F., Kimura, S., Kadota, Y., et al. (2008). Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J. Biol. Chem. 283, 8885–8892. doi: 10.1074/jbc.M708106200
Pan, Q. H., Zhan, J. C., Liu, H. T., Zhang, J. H., Chen, J. Y., Wen, P. F., et al. (2006). Salicylic acid synthesized by benzoic acid 2-hydroxylase participates in the development of thermotolerance in pea plants. Plant Sci. 171, 226–233. doi: 10.1016/j.plantsci.2006.03.012
Pauly, M., Gille, S., Liu, L. F., Mansoori, N., De Souza, A., Schultink, A., et al. (2013). Hemicellulose biosynthesis. Planta 238, 627–642. doi: 10.1007/s00425-013-1921-1
Petutschnig, E. K., Jones, A. M. E., Serazetdinova, L., Lipka, U., and Lipka, V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285, 28902–28911. doi: 10.1074/jbc.M110.116657
Piquemal, J., Lapierre, C., Myton, K., O’Connell, A., Schuch, W., Grima-Pettenati, J., et al. (1998). Down-regulation of cinnamoyl-CoA reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J. 13, 71–83. doi: 10.1046/j.1365-313X.1998.00014.x
Pogorelko, G., Lionetti, V., Fursova, O., Sundaram, R. M., Qi, M. S., Whitham, S. A., et al. (2013). Arabidopsis and Brachypodium distachyon transgenic plants expressing Aspergillus nidulans acetylesterases have decreased degree of polysaccharide acetylation and increased resistance to pathogens. Plant Physiol. 162, 9–23. doi: 10.1104/pp.113.214460
Raiborg, C., and Stenmark, H. (2009). The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452. doi: 10.1038/nature07961
Rasmussen, M. W., Roux, M., Petersen, M., and Mundy, J. (2012). MAP kinase cascades in Arabidopsis innate immunity. Front. Plant Sci. 3:169. doi: 10.3389/fpls.2012.00169
Robatzek, S., Chinchilla, D., and Boller, T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20, 537–542. doi: 10.1101/gad.366506
Ron, M., and Avni, A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16, 1604–1615. doi: 10.1105/Tpc.022475
Roux, M., Schwessinger, B., Albrecht, C., Chinchilla, D., Jones, A., Holton, N., et al. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2440–2455. doi: 10.1105/tpc.111.084301
Ruuhola, T., Julkunen-Tiitto, R., and Vainiotalo, P. (2003). In vitro degradation of willow salicylates. J. chem. ecol. 29, 1083–1097.
Sanchez-Vallet, A., Saleem-Batcha, R., Kombrink, A., Hansen, G., Valkenburg, D. J., Thomma, B. P., et al. (2013). Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. Elife 2:e00790. doi: 10.7554/eLife.00790
Sarkar, P., Bosneaga, E., and Auer, M. (2009). Plant cell walls throughout evolution: towards a molecular understanding of their design principles. J. Exp. Bot. 60, 3615–3635. doi: 10.1093/Jxb/Erp245
Sattler, S. E., and Funnell-Harris, D. L. (2013). Modifying lignin to improve bioenergy feedstocks: strengthening the barrier against pathogens? Front. Plant Sci. 4:70. doi: 10.3389/Fpls.2013.00070
Scheller, H. V., and Ulvskov, P. (2010). Hemicelluloses. Annu. Rev. Plant Biol. 61, 263–289. doi: 10.1146/annurev-arplant-042809-112315
Schulze, B., Mentzel, T., Jehle, A. K., Mueller, K., Beeler, S., Boller, T., et al. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285, 9444–9451. doi: 10.1074/jbc.M109.096842
Shimizu, T., Nakano, T., Takamizawa, D., Desaki, Y., Ishii-Minami, N., Nishizawa, Y., et al. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. doi: 10.1111/j.1365-313X.2010.04324.x
Shinya, T., Motoyama, N., Ikeda, A., Wada, M., Kamiya, K., Hayafune, M., et al. (2012). Functional characterization of CEBiP and CERK1 homologs in Arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 53, 1696–1706. doi: 10.1093/pcp/pcs113
Smith, J. M., Salamango, D. J., Leslie, M. E., Collins, C. A., and Heese, A. (2014). Sensitivity to Flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol. 164, 440–454. doi: 10.1104/pp.113.229179
Smith, M. M., and Hartley, R. D. (1983). Occurrence and nature of ferulic acid substitution of cell-wall polysaccharides in graminaceous plants. Carbohydr. Res. 118, 65–80. doi: 10.1016/0008-6215(83)88036-7
Sørensen, I., Domozych, D., and Willats, W. G. T. (2010). How have plant cell walls evolved? Plant Physiol. 153, 366–372. doi: 10.1104/pp.110.154427
Sørensen, I., Pettolino, F. A., Bacic, A., Ralph, J., Lu, F. C., O’Neill, M. A., et al. (2011). The charophycean green algae provide insights into the early origins of plant cell walls. Plant J. 68, 201–211. doi: 10.1111/j.1365-313X.2011.04686.x
Sorlier, P., Hartmann, D. J., Denuziere, A., Viton, C., and Domard, A. (2003). Preparation and development of anti-chitosan antibodies. J. Biomed. Mater. Res. A 67, 766–774. doi: 10.1002/Jbm.A.10132
Spallek, T., Beck, M., Ben Khaled, S., Salomon, S., Bourdais, G., Schellmann, S., et al. (2013). ESCRT-I mediates FLS2 endosomal sorting and plant immunity. PLoS Genet. 9:e1004035. doi: 10.1371/journal.pgen.1004035
Sun, Y., Li, L., Macho, A. P., Han, Z., Hu, Z., Zipfel, C., et al. (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628. doi: 10.1126/science.1243825
Takken, F. L., Thomas, C. M., Joosten, M. H., Golstein, C., Westerink, N., Hille, J., et al. (1999). A second gene at the tomato Cf-4 locus confers resistance to Cladosporium fulvum through recognition of a novel avirulence determinant. Plant J. 20, 279–288. doi: 10.1046/j.1365-313X.1999.00601.x
Tanner, G. R., and Morrison, I. M. (1983). Phenolic-carbohydrate complexes in the cell-walls of Lolium perenne. Phytochemistry 22, 1433–1439. doi: 10.1016/S0031-9422(00)84030-3
Thomas, C. M., Jones, D. A., Parniske, M., Harrison, K., Balint-Kurti, P. J., Hatzixanthis, K., et al. (1997). Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9, 2209–2224. doi: 10.1105/tpc.9.12.2209
Torres, M. A., Dangl, J. L., and Jones, J. D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. U.S.A. 99, 517–522. doi: 10.1073/pnas.012452499
Tronchet, M., Balague, C., Kroj, T., Jouanin, L., and Roby, D. (2010). Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 11, 83–92. doi: 10.1111/j.1364-3703.2009.00578.X
Tyler, L., Fangel, J. U., Fagerstrom, A. D., Steinwand, M. A., Raab, T. K., Willats, W. G., et al. (2014). Selection and phenotypic characterization of a core collection of Brachypodium distachyon inbred lines. BMC Plant Biol. 14:25. doi: 10.1186/1471-2229-14-25
Underwood, W. (2012). The plant cell wall: a dynamic barrier against pathogen invasion. Front. Plant Sci. 3:85. doi: 10.3389/fpls.2012.00085
Vaaje-Kolstad, G., Westereng, B., Horn, S. J., Liu, Z., Zhai, H., Sorlie, M., et al. (2010). An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222. doi: 10.1126/science.1192231
van den Burg, H. A., Harrison, S. J., Joosten, M. H., Vervoort, J., and De Wit, P. J. (2006). Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant Microbe Interact. 19, 1420–1430. doi: 10.1094/MPMI-19-1420
van der Hoorn, R. A., and Jones, J. D. (2004). The plant proteolytic machinery and its role in defence. Curr. Opin. Plant Biol. 7, 400–407. doi: 10.1016/j.pbi.2004.04.003
van Esse, H. P., Bolton, M. D., Stergiopoulos, I., de Wit, P. J., and Thomma, B. P. (2007). The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant Microbe Interact. 20, 1092–1101. doi: 10.1094/Mpmi-20-9-1092
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., and Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153, 895–905. doi: 10.1104/pp.110.155119
Vogel, J. P., Raab, T. K., Schiff, C., and Somerville, S. C. (2002). PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14, 2095–2106. doi: 10.1105/tpc.003509
Vogel, J., and Somerville, S. (2000). Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. U.S.A. 97, 1897–1902. doi: 10.1073/pnas.030531997
Weiergang, I., Hipskind, J. D., and Nicholson, R. L. (1996). Synthesis of 3-deoxyanthocyanidin phytoalexins in sorghum occurs independent of light. Physiol. Mol. Plant Pathol. 49, 377–388. doi: 10.1006/pmpp.1996.0060
Wolf, S., Mouille, G., and Pelloux, J. (2009). Homogalacturonan methyl-esterification and plant development. Mol. Plant. 2, 851–860. doi: 10.1093/mp/ssp066
Wurzinger, B., Mair, A., Pfister, B., and Teige, M. (2011). Cross-talk of calcium-dependent protein kinase and MAP kinase signaling. Plant Signal. Behav. 6, 8–12. doi: 10.4161/psb.6.1.14012
Xu, J., Xie, J., Yan, C., Zou, X., Ren, D., and Zhang, S. (2014). A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J. 77, 222–234. doi: 10.1111/tpj.12382
Yamaguchi, Y., Huffaker, A., Bryan, A. C., Tax, F. E., and Ryan, C. A. (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22, 508–522. doi: 10.1105/tpc.109.068874
Zhang, H., Fangel, J. U., Willats, W. G. T., Selig, M. J., Lindedam, J., Jorgensen, H., et al. (2014). Assessment of leaf/stem ratio in wheat straw feedstock and impact on enzymatic conversion. Glob. Change Biol. Bioenergy 6, 90–96. doi: 10.1111/Gcbb.12060
Keywords: plant cell wall, defense, PTI, PAMP, DAMP, callose, chitin, immunity
Citation: Malinovsky FG, Fangel JU and Willats WGT (2014) The role of the cell wall in plant immunity. Front. Plant Sci. 5:178. doi: 10.3389/fpls.2014.00178
Received: 14 February 2014; Accepted: 14 April 2014;
Published online: 06 May 2014.
Edited by:
Vincenzo Lionetti, Sapienza Università di Roma, ItalyReviewed by:
Thorsten Hamann, Norwegian University of Science and Technology, NorwayCopyright © 2014 Malinovsky, Fangel and Willats. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Frederikke G. Malinovsky, DNRF Center DynaMo and Copenhagen Plant Science Center, Department of Plant and Environmental Sciences, Faculty of Science, University of Copenhagen, Thorvaldsensvej 40, 1871 Frederiksberg Centret, Copenhagen, Denmark e-mail:ZmdtQFBMRU4ua3UuZGs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.