- Abteilung Pflanzenproteomik, Institut für Pflanzengenetik, Leibniz Universität Hannover, Hannover, Germany
The respiratory electron transport chain (ETC) couples electron transfer from organic substrates onto molecular oxygen with proton translocation across the inner mitochondrial membrane. The resulting proton gradient is used by the ATP synthase complex for ATP formation. In plants, the ETC is especially intricate. Besides the “classical” oxidoreductase complexes (complex I–IV) and the mobile electron transporters cytochrome c and ubiquinone, it comprises numerous “alternative oxidoreductases.” Furthermore, several dehydrogenases localized in the mitochondrial matrix and the mitochondrial intermembrane space directly or indirectly provide electrons for the ETC. Entry of electrons into the system occurs via numerous pathways which are dynamically regulated in response to the metabolic state of a plant cell as well as environmental factors. This mini review aims to summarize recent findings on respiratory electron transfer pathways in plants and on the involved components and supramolecular assemblies.
Introduction
During cellular respiration, organic compounds are oxidized to generate usable chemical energy in the form of ATP. The respiratory electron transport chain (ETC) of mitochondria is at the center of this process. Its core consists of four oxidoreductase complexes, the NADH dehydrogenase (complex I), the succinate dehydrogenase (complex II), the cytochrome c reductase (complex III) and the cytochrome c oxidase (complex IV), as well as of two mobile electron transporters, cytochrome c, and the lipid ubiquinone. Overall, electrons are transferred from the coenzymes NADH or FADH2 onto molecular oxygen which is reduced to water. Three of the four oxidoreductase complexes (complexes I, III and IV) couple their electron transfer reactions with proton translocation across the inner mitochondrial membrane. As a result, a proton gradient is formed which can be used by the ATP synthase complex (complex V) for the phosphorylation of ADP. In its classically described form, cellular respiration is based on a linear ETC (from NADH via complexes I, III, and IV to molecular oxygen). However, electrons can enter and leave the ETC at several alternative points. This is especially true for the plant ETC system, which is highly branched. In this review we aim to integrate current knowledge on the ETC system in plants with respect to its components, electron transport pathways and supramolecular structure.
Components of the Plant ETC System
The “classical” oxidoreductase complexes of the respiratory chain (given in dark blue in Figure 1) resemble their homologues in animal mitochondria but at the same time have some clear distinctive features (reviewed in Millar et al., 2008, 2011; Rasmusson and Moller, 2011; van Dongen et al., 2011; Jacoby et al., 2012). Complex I is especially large in plant mitochondria and includes nearly 50 different subunits (Braun et al., 2014). Compared to its homologs from bacteria and other eukaryotic lineages it has an extra domain which includes carbonic anhydrase-like proteins. The function of this additional domain is currently unclear but it has been suggested to be important in the context of an inner-cellular CO2 transfer mechanism to provide mitochondrial CO2 for carbon fixation in chloroplasts (Braun and Zabaleta, 2007; Zabaleta et al., 2012). Complex II is composed of four subunits in bacteria and mitochondria of animals and fungi. In plants complex II includes homologs of these subunits but additionally four extra proteins of unknown function (Millar et al., 2004; Huang and Millar, 2013). In contrast, the subunit composition of complex III from plants is highly similar to the ones in yeast and bovine mitochondria (Braun and Schmitz, 1995a). The two largest subunits of this protein complex, termed “core proteins” in animals and fungi, are homologous to the two subunits of the mitochondrial processing peptidase (MPP) which removes pre-sequences of nuclear-encoded mitochondrial proteins after their import into mitochondria. In animal mitochondria, the core proteins are proteolytically inactive. Instead, an active MPP is present within the mitochondrial matrix. In contrast, the core subunits of complex III from plants have intact active sites (Braun et al., 1992; Glaser et al., 1994). Indeed, complex III isolated from plant mitochondria efficiently removes pre-sequences of mitochondrial pre-proteins. The differing functional states of complex III in diverse eukaryotic lineages might reflect different evolutionary stages of this protein complex (Braun and Schmitz, 1995b). Also complex IV has some extra subunits in mitochondria of plants (Millar et al., 2004). Eight subunits are homologous to complex IV subunits from other groups of eukaryotes and another six putative subunits represent proteins of unknown functions.
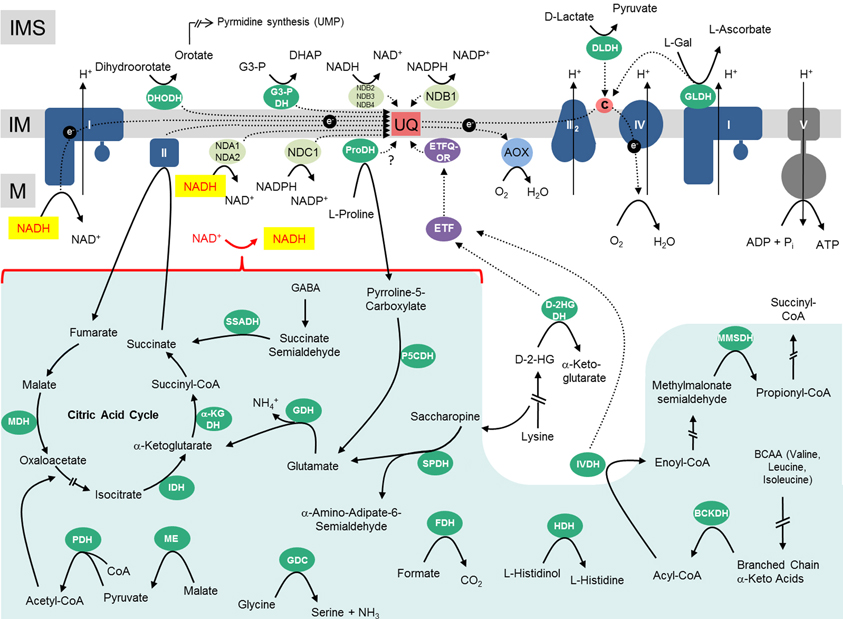
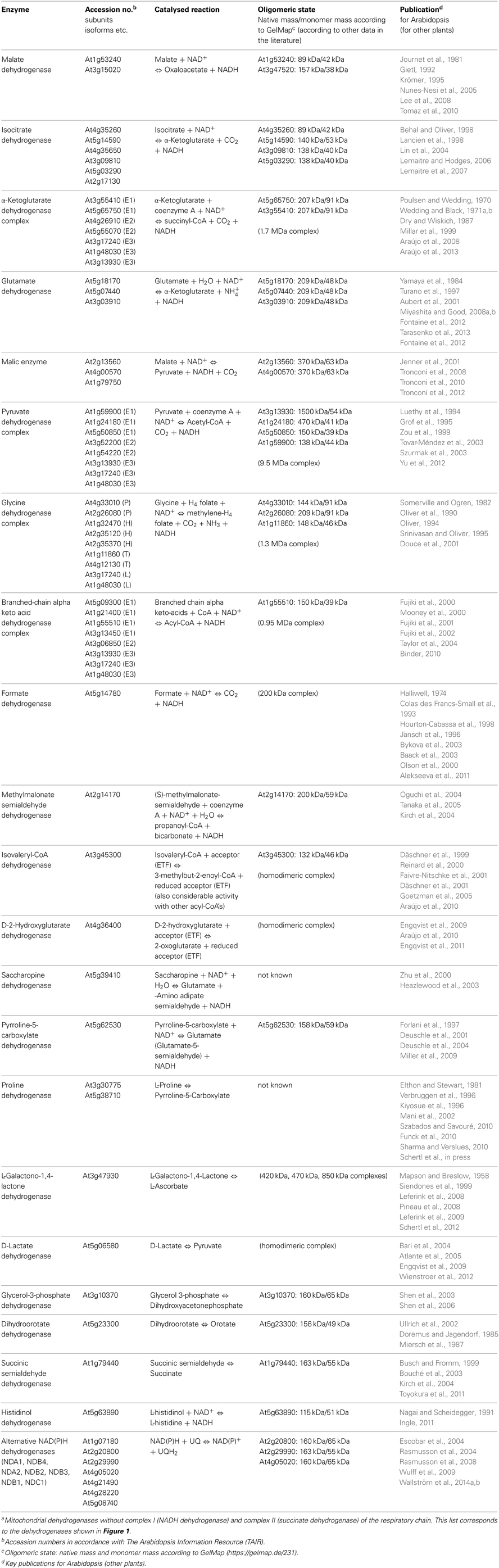
Figure 1. Mitochondrial dehydrogenases and the respiratory chain. Within the mitochondrial matrix (M) numerous dehydrogenases generate NADH by oxidizing various carbon compounds. NADH subsequently is re-oxidized at the inner mitochondrial membrane (IM) by the respiratory electron transfer chain (ETC). The electrons of NADH can enter the ETC through complex I or at the ubiquinone level via alternative NAD(P)H-dehydrogenases. Besides, some dehydrogenases of the mitochondrial matrix transfer electrons to ubiquinone via the ETF/ETFQOR system. Proline dehydrogenase possibly directly transfers electrons onto ubiquinone. In the intermembrane space (IMS), electrons from NAD(P)H generated in the cytoplasm can be inserted into the ETC via alternative NAD(P)H dehydrogenases. Furthermore, some dehydrogenases of the IMS can directly transfer electrons onto ubiquinone or cytochrome c. Color code—dark blue, protein complexes of the ETC; blue, AOX; purple, ETF/ETFQQ system; light green, alternative NAD(P)H dehydrogenases of the ETC; green, dehydrogenases; red, ubiquinone and cytochrome c; yellow, NADH produced by dehydrogenases of the mitochondrial matrix/NADH re-oxidized by complex I or internal alternative NADH dehydrogenases; dark gray, ATP synthase complex; light green background, NADH producing dehydrogenases of the mitochondrial matrix. Abbreviations—alphabetically ordered. I, complex I; II, complex II; III, complex III; IV, complex IV; V, complex V; α-KGDH, α-ketoglutarate dehydrogenase; AOX, alternative oxidase; BCKDH, branched-chain α-ketoacid dehydrogenase complex; c, cytochrome c; D-2HGDH, D-2-hydroxyglutarate dehydrogenase; DHODH, dihydroorotate dehydrogenase; DLDH, D-lactate dehydrogenase; ETF, electron transfer flavoprotein; ETFQOR, electron transfer flavoprotein ubiquinone oxidoreductase; FDH, formate dehydrogenase; GDC, glycine dehydrogenase; GDH, glutamate dehydrogenase; GLDH, L-galactono-1,4-lactone dehydrogenase; G3-PDH, glyceraldehyde 3-phosphate dehydrogenase; HDH, histidinol dehydrogenase; IDH, isocitrate dehydrogenase; IVDH, isovaleryl-coenzyme A dehydrogenase; MDH, malate dehydrogenase; ME, malic enzyme; MMSDH, methylmalonate-semialdehyde dehydrogenase; NDA1/2, NDB2/3/4, alternative NADH dehydrogenase; NDC1, NDB1, alternative NADPH dehydrogenase; P5CDH, pyrroline-5-carboxylate dehydrogenase; PDH, pyruvate dehydrogenase; ProDH, proline dehydrogenase; SPDH, saccharopine dehydrogenase; SSADH, succinic semialdehyde dehydrogenase; UQ, ubiquinone. For further information of the enzymes see Table 1.
The ETC of plant mitochondria additionally includes several so-called “alternative oxidoreductases”: the alternative oxidase (AOX; light blue in Figure 1) and several functionally distinguishable alternative NAD(P)H dehydrogenases (alternative NDs, light green in Figure 1). Findings on their functional roles have been reviewed recently (Rasmusson et al., 2008; Rasmusson and Moller, 2011; Moore et al., 2013). AOX directly transfers electrons from ubiquinol to molecular oxygen and therefore constitutes an alternative electron exit point of the ETC. As a result, complexes III and IV are excluded from respiratory electron transport. The alternative NAD(P)H dehydrogenases serve as alternative electron entry points of the plant ETC and may substitute complex I. They differ with respect to co-factor requirement and localization at the outer or inner surface of the inner mitochondrial membrane (external alternative NDs, internal alternative NDs). Some of the genes encoding alternative NDs are activated by light (Rasmusson et al., 2008; Rasmusson and Moller, 2011). The latter enzymes are considered to be important during photorespiration and all alternative enzymes during various stress conditions. Since none of the alternative oxidoreductases couple electron transfer with proton translocation across the inner mitochondrial membrane, their enzymatic function is believed to be important in the context of an overflow protection mechanism for the ETC which is especially relevant during high-light conditions.
Finally, dehydrogenases (dark green in Figure 1; Table 1) can directly or indirectly insert electrons into the respiratory chain (Rasmusson et al., 2008; Rasmusson and Moller, 2011). Numerous dehydrogenases of the mitochondrial matrix generate NADH which is re-oxidized by complex I and the internal alternative NDs. However, some dehydrogenases directly transfer electrons onto ubiquinone [dihydroorotate dehydrogenase (DHODH), glyceraldehyde 3-phosphate dehydrogenase (G3-PDH) and possibly proline dehydrogenase (ProDH)] or onto cytochrome c [L-galactone-1,4-lactone dehydrogenase (GLDH) and D-lactate dehydrogenase (DLDH)]. Furthermore, at least two dehydrogenases [isovaleryl-coenzyme A dehydrogenase (IVDH) and D-2-hydroxyglutarate dehydrogenase (D-2HGDH)] transfer electrons onto ubiquinone via a short electron transfer chain composed of the “electron transfer flavoprotein” and the “electron transfer flavoprotein-ubiquinone oxidoreductase” (ETF and ETFQ-OR, purple in Figure 1) (Ishizaki et al., 2005, 2006; Araújo et al., 2010). IVDH is involved in the branched chain amino acid catabolism and D-2HGDH in the catabolism of lysine. In plants, degradation of amino acids for respiration was shown to be especially important during carbon starvation conditions, e.g., extended darkness (Araújo et al., 2011). In contrast to animal mitochondria, fatty acid oxidation does not take place in plant mitochondria and the involved dehydrogenases consequently are absent. Instead, additional metabolic pathways occur in plants, e.g., the final step of an ascorbic acid biosynthesis pathway, which is catalyzed by GLDH. Electrons of L-galactono-1,4-lactone (GL) oxidation are inserted into the ETC via cytochrome c (Bartoli et al., 2000). Proline, besides being a building block for protein biosynthesis, is used as an osmolyte in plant cells. Proline is catabolized in mitochondria by a two-step process involving pyrroline-5-carboxylate dehydrogenase (P5CDH) and ProDH (Szabados and Savouré, 2010). P5CDH produces NADH, whereas ProDH represents a flavoenzyme which is assumed to transfer electrons directly or indirectly onto ubiquinone. Some additional dehydrogenases occur in plant mitochondria in the mitochondrial matrix and the intermembrane space which also contribute electrons to the ETC (Figure 1, Table 1). However, in some cases their mitochondrial localization is not entirely certain and should be further investigated by future research.
Electron Entry Pathways into the ETC
All electrons enter the ETC via NAD(P)H (generated by a variety of dehydrogenases in the mitochondrial matrix or the intermembrane space/the cytoplasm) or via flavine nucleotides (FADH2, FMNH2), which generally are bound to proteins termed flavoproteins. Consequently, the following electron entry pathways into the ETC can be defined: (i) the Matrix NAD(P)H pathway, (ii) the Matrix-FADH2 pathway, (iii) the Intermembrane-space-NAD(P)H pathway, and (iv) the Intermembrane-space-FADH2/FMDH2 pathway (Figure 2).
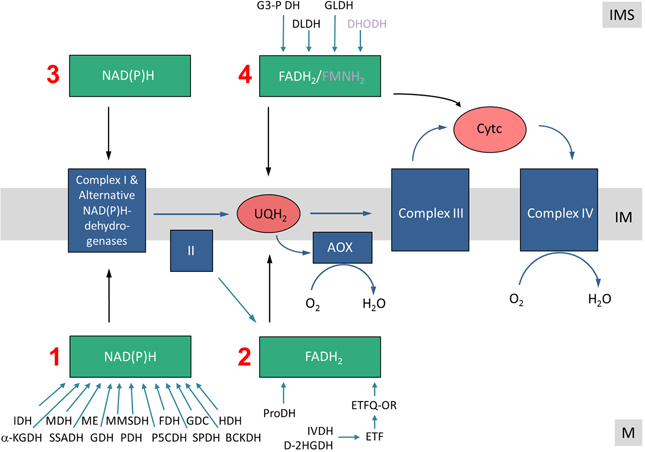
Figure 2. Electron entry pathways into the mitochondrial electron transport chain in plants. Electrons enter the respiratory chain via four different pathways. (1) The Matrix-NAD(P)H pathway. Various dehydrogenases oxidize carbon compounds in the mitochondrial matrix. Electrons are transferred in the form of NADH to the ETC. NADH is re-oxidized by complex I or the internal alternative NAD(P)H dehydrogenases. (2) The Matrix-FADH2 pathway. FAD-containing enzymes oxidize carbon compounds in the mitochondrial matrix and directly (ProDH?) or indirectly (via the ETF/ETFQQ system) transfer electrons to the ubiquinone pool. (3) The IMS-NAD(P)H pathway. Cytoplasmically formed NAD(P)H is re-oxidized via external alternative dehydrogenases. (4) The IMS-FADH2 pathway. FAD/FMN-containing enzymes oxidize carbon compounds in the mitochondrial intermembrane space. Electrons are transferred either to the ubiquinone or the cytochrome c. M, matrix; IM, inner membrane; IMS, intermembrane space. Abbreviations—alphabetically ordered. I, complex I; II, complex II; III, complex III; IV, complex IV; α-KGDH, α-ketoglutarte dehydrogenase; AOX, alternative oxidase; BCKDH, branched-chain α-ketoacid dehydrogenase complex; Cytc, cytochrome c; D-2HGDH, D-2-hydroxyglutarate dehydrogenase; DHODH, dihydroorotate dehydrogenase; DLDH, D-lactate dehydrogenase; ETF, electron transfer flavoprotein; ETFQOR, electron transfer flavoprotein ubiquinone oxidoreductase; FDH, formate dehydrogenase; GDC, glycine dehydrogenase; GDH, glutamate dehydrogenase; GLDH, L-galactono-1,4-lactone dehydrogenase; G3-PDH, glyceraldehyde 3-phosphate dehydrogenase; HDH, histidinol dehydrogenase; IDH, isocitrate dehydrogenase; IVDH, isovaleryl-coenzyme A dehydrogenase; MDH, malate dehydrogenase; ME, malic enzyme; MMSDH, methylmalonate-semialdehyde dehydrogenase; P5CDH, pyrroline-5-carboxylate dehydrogenase; PDH, pyruvate dehydrogenase; ProDH, proline dehydrogenase; SPDH, saccharopine dehydrogenase; SSADH, succinic semialdehyde dehydrogenase; UQH2, ubiquinol.
Different metabolic processes, which vary depending on the physiological state of the plant cell, contribute to the four electron entry pathways. During stable carbohydrate conditions, electrons for the respiratory chain can be supplied by NADH and FADH2 provided by the tricarboxylic acid (TCA) cycle. This is believed to be the standard mode of cellular respiration in non-green plant tissues or green tissues at night and resembles the basic situation in animal cells. However, during photosynthesis, NADH generation of the TCA cycle is reduced because some of its intermediates are used for anabolic reactions (reviewed in Sweetlove et al., 2010). Furthermore, the pyruvate dehydrogenase (PDH) complex is deactivated in plant mitochondria in the light by phosphorylation (Budde and Randall, 1990). At the same time photorespiration leads to an increase in NADH formation in the mitochondrial matrix by the activity of the glycine dehydrogenase complex (GDC). Indeed, at high-light conditions, NADH formed by GDC is believed to be the main substrate of the ETC, and not the NADH formed by the enzymes of the TCA cycle. At the same time, plant cells might become over-reduced in the presence of high-light. In this situation alternative oxidoreductases can insert excess electrons into the respiratory chain without contributing to the proton gradient. Upon carbon starvation conditions (e.g., extended darkness) electrons from the breakdown of amino acids are provided to the ETC (Araújo et al., 2011). Especially after release of salt stress the amino acid proline is used as an electron source (Szabados and Savouré, 2010). In summary, electron entry into the ETC is a highly flexible process in plants which much depends on light, the metabolic state of the cell as well as environmental stress factors.
Supramolecular Structure of the ETC System
The ETC is based on defined protein-protein interactions. Most stable interactions occur within the four “classical” oxidoreductase complexes of the respiratory chain. Indeed, complexes I to IV can be isolated in intact form by various biochemical and electrophoretic procedures. Furthermore, several lines of evidence indicate that complexes I, III and IV interact within the inner mitochondrial membrane forming respiratory supercomplexes (reviewed in Dudkina et al., 2008). Complex I as well as complex IV associate with dimeric complex III (I + III2 and IV2 + III2 supercomplexes). An even larger supercomplex includes complexes I, III2, and IV and was proposed to be called “respirasome” because it can autonomously catalyzes the overall ETC reaction in the presence of ubiquinone and cytochrome c. The alternative oxidoreductases of the plant ETC seem not to be part of the respiratory supercomplexes. However, alternative NDs were found to be part of other protein complexes of about 160 kDa (Klodmann et al., 2011) or 150–700 kDa (Rasmusson and Agius, 2001).
Experimental data also indicate that several of the mitochondrial dehydrogenases form protein complexes. TCA cycle enzymes can assemble forming multienzyme clusters (Barnes and Weitzman, 1986). In addition, some of the mitochondrial dehydrogenases interact with ETC complexes, e.g., malate dehydrogenase has been reported to interact with complex I in animal mitochondria (Fukushima et al., 1989; see Braun et al., 2014 for review). Information on the native state of mitochondrial dehydrogenases furthermore comes from the GelMap project (Klodmann et al., 2011). Using 2D Blue native/SDS PAGE and systematic protein identifications, various dehydrogenases were described (Figure 3, Table 1). Native molecular mass of the dehydrogenases in many cases much exceeds the molecular mass of the monomeric proteins (Table 1, column 3). This indicates that probably most mitochondrial dehydrogenases form part of defined higher order structures.
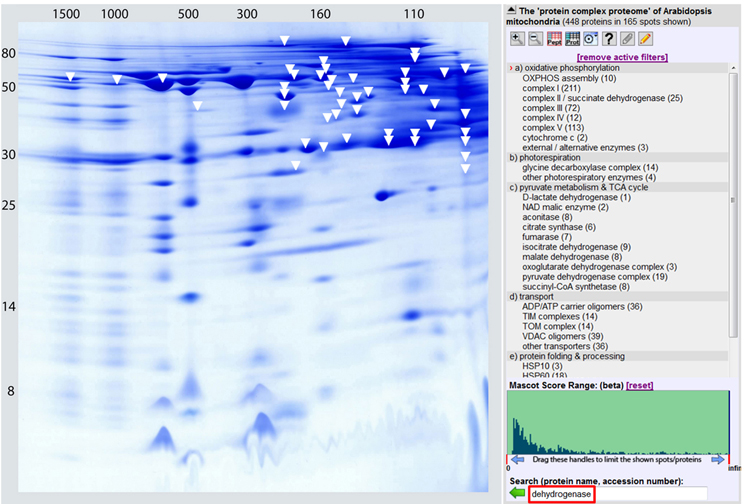
Figure 3. The dehydrogenase subproteome of plant mitochondria. Mitochondrial proteins from Arabidopsis thaliana were separated by 2D Blue native/SDS PAGE and displayed via GelMap (https://gelmap.de/231#). Protein separation under native condition was from left to right, protein separation in the presence of SDS from top to bottom. Molecular masses of standard proteins are given to the left/above the 2D gel. All proteins annotated as “dehydrogenase” are indicated by white arrows. Exception: The subunits of complex I (NADH dehydrogenase) and complex II (succinate dehydrogenase) are not indicated on the figure.
Conclusion and Outlook
Cellular respiration in plants is an especially dynamic system. The classical protein complexes of the ETC have extra functions and several alternative oxidoreductases occur. A network of mitochondrial dehydrogenases directly or indirectly supplies electrons for the respiratory chain. Insertion of electrons via various pathways is highly dependent on the metabolic state of the plant cell. The regulation of electron entry pathways into the respiratory chain is only partially understood and might, besides others, depend on the formation of supramolecular structures. Non-invasive experimental procedures will be necessary to physiologically investigate the function of these structures by future research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tatjana Hildebrandt and Holger Eubel, Leibniz University Hannover, for critically reading the manuscript. This research project was supported by the Deutsche Forschungs-gemeinschaft (DFG), grant Br 1829/10-2. Furthermore we acknowledge support by the Open Access Publishing Fund of Leibniz Universität Hannover, which is funded by the DFG.
References
Alekseeva, A. A., Savin, S. S., and Tishkov, V. I. (2011). NAD (+)-dependent formate dehydrogenase from plants. Acta Nat. 3, 38–54.
Araújo, W. L., Ishizaki, K., Nunes-Nesi, A., Larson, T. R., Tohge, T., Krahnert, I., et al. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22, 1549–1563. doi: 10.1105/tpc.110.075630
Araújo, W. L., Nunes-Nesi, A., Trenkamp, S., Bunik, V. I., and Fernie, A. R. (2008). Inhibition of 2-oxoglutarate dehydrogenase in potato tuber suggests the enzyme is limiting for respiration and confirms its importance in nitrogen assimilation. Plant Physiol. 148, 1782–1796. doi: 10.1104/pp.108.126219
Araújo, W. L., Tohge, T., Ishizaki, K., Leaver, C. J., and Fernie, A. R. (2011). Protein degradation - an alternative respiratory substrate for stressed plants. Trends Plant Sci. 16, 489–498. doi: 10.1016/j.tplants.2011.05.008
Araújo, W. L., Trofimova, L., Mkrtchyan, G., Steinhauser, D., Krall, L., Graf, A., et al. (2013). On the role of the mitochondrial 2-oxoglutarate dehydrogenase complex in amino acid metabolism. Amino Acids 44, 683–700. doi: 10.1007/s00726-012-1392-x
Atlante, A., de Bari, L., Valenti, D., Pizzuto, R., Paventi, G., and Passarella, S. (2005). Transport and metabolism of D-lactate in Jerusalem artichoke mitochondria. Biochim. Biophys. Acta 1708, 13–22. doi: 10.1016/j.bbabio.2005.03.003
Aubert, S., Bligny, R., Douce, R., Gout, E., Ratcliffe, R. G., and Roberts, J. K. (2001). Contribution of glutamate dehydrogenase to mitochondrial glutamate metabolism studied by (13)C and (31)P nuclear magnetic resonance. J. Exp. Bot. 52, 37–45. doi: 10.1093/jexbot/52.354.37
Baack, R. D., Markwell, J., Herman, P. L., and Osterman, J. C. (2003). Kinetic behavior of the Arabidopsis thaliana leaf formate dehydrogenase is thermally sensitive. J. Plant Physiol. 160, 445–450. doi: 10.1078/0176-1617-00995
Bari, R., Kebeish, R., Kalamajka, R., Rademacher, T., and Peterhänsel, C. (2004). A glycolate dehydrogenase in the mitochondria of Arabidopsis thaliana. J. Exp. Bot. 55, 623–630. doi: 10.1093/jxb/erh079
Barnes, S. J., and Weitzman, P. D. (1986) Organization of citric acid cycle enzymes into a multienzyme cluster. FEBS Lett. 201, 267–270. doi: 10.1016/0014-5793(86)80621-4
Bartoli, C. G., Pastori, G. M., and Foyer, C. H. (2000). Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol. 123, 335–344. doi: 10.1104/pp.123.1.335
Behal, R. H., and Oliver, D. J. (1998). NAD(+)-dependent isocitrate dehydrogenase from Arabidopsis thaliana. Characterization of two closely related subunits. Plant Mol. Biol. 36, 691–698. doi: 10.1023/A:1005923410940
Binder, S. (2010). Branched-chain amino acid metabolism in Arabidopsis thaliana. Arabidopsis Book 8, e0137. doi: 10.1199/tab.0137
Bouché, N., Fait, A., Bouchez, D., Møller, S. G., and Fromm, H. (2003). Mitochondrial succinic-semialdehyde dehydrogenase of the gamma-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc. Natl. Acad. Sci. U.S.A. 100, 6843–6848. doi: 10.1073/pnas.1037532100
Braun, H. P., Binder, S., Brennicke, A., Eubel, H., Fernie, A. R., Finkemeier, I., et al. (2014). The life of plant mitochondrial complex I. Mitochondrion. doi: 10.1016/j.mito.2014.02.006. [Epub ahead of print].
Braun, H. P., Emmermann, M., Kruft, V., and Schmitz, U. K. (1992). The general mitochondrial processing peptidase from potato is an integral part of cytochrome c reductase of the respiratory chain. EMBO J. 11, 3219–3227.
Braun, H. P., and Schmitz, U. K. (1995a). The bifunctional cytochrome c reductase/processing peptidase complex from plant mitochondria. J. Bioenerg. Biomembr. 27, 423–436. doi: 10.1007/BF02110005
Braun, H. P., and Schmitz, U. K. (1995b) Are the “core” proteins of the mitochondrial bc1 complex evolutionary relics of a processing peptidase? Trends Biochem. Sci. 20, 171–175. doi: 10.1016/S0968-0004(00)88999-9
Braun, H. P., and Zabaleta, E. (2007) Carbonic anhydrase subunits of the mitochondrial NADH dehydrogenase complex (complex I) in plants. Physiologia Plantarum 129, 114–122. doi: 10.1111/j.1399-3054.2006.00773.x
Budde, R. J., and Randall, D. D. (1990). Pea leaf mitochondrial pyruvate dehydrogenase complex is inactivated in vivo in a light-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 87, 673–676. doi: 10.1073/pnas.87.2.673
Busch, K. B., and Fromm, H. (1999). Plant succinic semialdehyde dehydrogenase. Cloning, purification, localization in mitochondria, and regulation by adenine nucleotides. Plant Physiol. 121, 589–597. doi: 10.1104/pp.121.2.589
Bykova, N. V., Stensballe, A., Egsgaard, H., Jensen, O. N., and Moller, I. M. (2003). Phosphorylation of formate dehydrogenase in potato tuber mitochondria. J. Biol. Chem. 278, 26021–26030. doi: 10.1074/jbc.M300245200
Colas des Francs-Small, C., Ambard-Bretteville, F., Small, I. D., and Rémy, R. (1993). Identification of a major soluble protein in mitochondria from nonphotosynthetic tissues as NAD-dependent formate dehydrogenase. Plant Physiol. 102, 1171–1177. doi: 10.1104/pp.102.4.1171
Däschner, K., Couée, I., and Binder, S. (2001). The mitochondrial isovaleryl-coenzyme a dehydrogenase of arabidopsis oxidizes intermediates of leucine and valine catabolism. Plant Physiol. 126, 601–612. doi: 10.1104/pp.126.2.601
Däschner, K., Thalheim, C., Guha, C., Brennicke, A., and Binder, S. (1999). In plants a putative isovaleryl-CoA-dehydrogenase is located in mitochondria. Plant Mol. Biol. 39, 1275–1282.
Deuschle, K., Funck, D., Forlani, G., Stransky, H., Biehl, A., Leister, D., et al. (2004). The role of [Delta]1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell 16, 3413–3425. doi: 10.1105/tpc.104.023622
Deuschle, K., Funck, D., Hellmann, H., Däschner, K., Binder, S., and Frommer, W. B. (2001). A nuclear gene encoding mitochondrial Delta-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J. 27, 345–356. doi: 10.1046/j.1365-313X.2001.01101.x
Doremus, H. D., and Jagendorf, A. T. (1985). Subcellular localization of the pathway of de novo pyrimidine nucleotide biosynthesis in pea leaves. Plant Physiol. 79, 856–861. doi: 10.1104/pp.79.3.856
Douce, R., Bourguignon, J., Neuburger, M., and Rébeillé, F. (2001). The glycine decarboxylase system: a fascinating complex. Trends Plant Sci. 6, 167–176. doi: 10.1016/S1360-1385(01)01892-1
Dry, I. B., and Wiskich, J. T. (1987). 2-Oxoglutarate dehydrogenase and pyruvate dehydrogenase activities in plant mitochondria: interaction via a common coenzyme a pool. Arch. Biochem. Biophys. 257, 92–99. doi: 10.1016/0003-9861(87)90546-7
Dudkina, N. V., Sunderhaus, S., Boekema, E. J., and Braun, H.-P. (2008). The higher level of organization of the oxidative phosphorylation system: mitochondrial supercomplexes. J. Bioenerg. Biomembr. 40, 419–424. doi: 10.1007/s10863-008-9167-5
Elthon, T. E., and Stewart, C. R. (1981). Submitochondrial location and electron transport characteristics of enzymes involved in proline oxidation. Plant Physiol. 67, 780–784. doi: 10.1104/pp.67.4.780
Engqvist, M., Drincovich, M. F., Flügge, U.-I., and Maurino, V. G. (2009). Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and beta-oxidation pathways. J. Biol. Chem. 284, 25026–25037. doi: 10.1074/jbc.M109.021253
Engqvist, M. K. M., Kuhn, A., Wienstroer, J., Weber, K., Jansen, E. E. W., Jakobs, C., et al. (2011). Plant D-2-hydroxyglutarate dehydrogenase participates in the catabolism of lysine especially during senescence. J. Biol. Chem. 286, 11382–11390. doi: 10.1074/jbc.M110.194175
Escobar, M. A., Franklin, K. A., Svensson, A. S., Salter, M. G., Whitelam, G. C., and Rasmusson, A. G. (2004). Light regulation of the Arabidopsis respiratory chain. Multiple discrete photoreceptor responses contribute to induction of type II NAD(P)H dehydrogenase genes. Plant Physiol. 136, 2710–2721. doi: 10.1104/pp.104.046698
Faivre-Nitschke, S. E., Couée, I., Vermel, M., Grienenberger, J. M., and Gualberto, J. M. (2001). Purification, characterization and cloning of isovaleryl-CoA dehydrogenase from higher plant mitochondria. Eur. J. Biochem. 268, 1332–1339. doi: 10.1046/j.1432-1327.2001.01999.x
Fontaine, J.-X., Tercé-Laforgue, T., Armengaud, P., Clément, G., Renou, J.-P., Pelletier, S., et al. (2012). Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell 24, 4044–4065. doi: 10.1105/tpc.112.103689
Fontaine, J.-X., Tercé-Laforgue, T., Bouton, S., Pageau, K., Lea, P. J., Dubois, F., et al. (2013). Further insights into the isoenzyme composition and activity of glutamate dehydrogenase in Arabidopsis thaliana. Plant Signal. Behav. 8, e23329. doi: 10.4161/psb.23329
Forlani, G., Scainelli, D., and Nielsen, E. (1997). [delta]1-pyrroline-5-carboxylate dehydrogenase from cultured cells of potato (purification and properties). Plant Physiol. 113, 1413–1418.
Fujiki, Y., Ito, M., Itoh, T., Nishida, I., and Watanabe, A. (2002). Activation of the promoters of Arabidopsis genes for the branched-chain alpha-keto acid dehydrogenase complex in transgenic tobacco BY-2 cells under sugar starvation. Plant Cell Physiol. 43, 275–280. doi: 10.1093/pcp/pcf032
Fujiki, Y., Ito, M., Nishida, I., and Watanabe, A. (2001). Leucine and its keto acid enhance the coordinated expression of genes for branched-chain amino acid catabolism in Arabidopsis under sugar starvation. FEBS Lett. 499, 161–165. doi: 10.1016/S0014-5793(01)02536-4
Fujiki, Y., Sato, T., Ito, M., and Watanabe, A. (2000). Isolation and characterization of cDNA clones for the e1beta and E2 subunits of the branched-chain alpha-ketoacid dehydrogenase complex in Arabidopsis. J. Biol. Chem. 275, 6007–6013. doi: 10.1074/jbc.275.8.6007
Fukushima, T., Decker, R. V., Anderson, W. M., and Spivey, H. O. (1989). Substrate channeling of NADH in mitochondrial redox processes. J. Biol. Chem. 264, 16483–16488.
Funck, D., Eckard, S., and Müller, G. (2010). Non-redundant functions of two proline dehydrogenase isoforms in Arabidopsis. BMC Plant Biol. 10:70. doi: 10.1186/1471-2229-10-70
Gietl, C. (1992). Malate dehydrogenase isoenzymes: cellular locations and role in the flow of metabolites between the cytoplasm and cell organelles. Biochim. Biophys. Acta 1100, 217–234. doi: 10.1016/0167-4838(92)90476-T
Glaser, E., Eriksson, A., and Sjöling, S. (1994). Bifunctional role of the bc1 complex in plants. Mitochondrial bc1 complex catalyses both electron transport and protein processing. FEBS Lett. 346, 83–87. doi: 10.1016/0014-5793(94)00312-2
Goetzman, E. S., Mohsen, A.-W. A., Prasad, K., and Vockley, J. (2005). Convergent evolution of a 2-methylbutyryl-CoA dehydrogenase from isovaleryl-CoA dehydrogenase in Solanum tuberosum. J. Biol. Chem. 280, 4873–4879. doi: 10.1074/jbc.M412640200
Grof, C. P., Winning, B. M., Scaysbrook, T. P., Hill, S. A., and Leaver, C. J. (1995). Mitochondrial pyruvate dehydrogenase. Molecular cloning of the E1 alpha subunit and expression analysis. Plant Physiol. 108, 1623–1629. doi: 10.1104/pp.108.4.1623
Halliwell, B. (1974). Oxidation of formate by peroxisomes and mitochondria from spinach leaves. Biochem. J. 138, 77–85.
Heazlewood, J. L., Howell, K. A., Whelan, J., and Millar, A. H. (2003). Towards an analysis of the rice mitochondrial proteome. Plant Physiol. 132, 230–242. doi: 10.1104/pp.102.018986
Hourton-Cabassa, C., Ambard-Bretteville, F., Moreau, F, Davy de Virville, J., Rémy, R., and Francs-Small, C. C. (1998). Stress induction of mitochondrial formate dehydrogenase in potato leaves. Plant Physiol. 116, 627–635. doi: 10.1104/pp.116.2.627
Huang, S., and Millar, A. H. (2013). Succinate dehydrogenase: the complex roles of a simple enzyme. Curr. Opin. Plant Biol. 16, 344–349. doi: 10.1016/j.pbi.2013.02.007
Ishizaki, K., Larson, T. R., Schauer, N., Fernie, A. R., Graham, I. A., and Leaver, C. J. (2005). The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17, 2587–2600. doi: 10.1105/tpc.105.035162
Ishizaki, K., Schauer, N., Larson, T. R., Graham, I. A., Fernie, A. R., and Leaver, C. J. (2006). The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J. 47, 751–760. doi: 10.1111/j.1365-313X.2006.02826.x
Jacoby, R. P., Li, L., Huang, S., Pong Lee, C., Millar, A. H., and Taylor, N. L. (2012). Mitochondrial composition, function and stress response in plants. J. Integr. Plant Biol. 54, 887–906. doi: 10.1111/j.1744-7909.2012.01177.x
Jänsch, L., Kruft, V., Schmitz, U. K., and Braun, H. P. (1996). New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J. 9, 357–368. doi: 10.1046/j.1365-313X.1996.09030357.x
Jenner, H. L., Winning, B. M., Millar, A. H., Tomlinson, K. L., Leaver, C. J., and Hill, S. A. (2001). NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol. 126, 1139–1149. doi: 10.1104/pp.126.3.1139
Journet, E. P., Neuburger, M., and Douce, R. (1981). Role of glutamate-oxaloacetate transaminase and malate dehydrogenase in the regeneration of NAD for glycine oxidation by spinach leaf mitochondria. Plant Physiol. 67, 467–469. doi: 10.1104/pp.67.3.467
Kirch, H.-H., Bartels, D., Wei, Y., Schnable, P. S., and Wood, A. J. (2004). The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 9, 371–377. doi: 10.1016/j.tplants.2004.06.004
Kiyosue, T., Yoshiba, Y., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1996). A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell 8, 1323–1335.
Klodmann, J., Senkler, M., Rode, C., and Braun, H.-P. (2011). Defining the protein complex proteome of plant mitochondria. Plant Physiol. 157, 587–598. doi: 10.1104/pp.111.182352
Krömer, S. (1995) Respiration during photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 47–70. doi: 10.1146/annurev.pp.46.060195.000401
Lancien, M., Gadal, P., and Hodges, M. (1998). Molecular characterization of higher plant NAD-dependent isocitrate dehydrogenase: evidence for a heteromeric structure by the complementation of yeast mutants. Plant J. 16, 325–333. doi: 10.1046/j.1365-313x.1998.00305.x
Lee, C. P., Eubel, H., O'Toole, N., and Millar, A. H. (2008). Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol. Cell Proteomics 7, 1297–1316. doi: 10.1074/mcp.M700535-MCP200
Leferink, N. G. H., van den Berg, W. A. M., and van Berkel, W. J. H. (2008). l-Galactono-gamma-lactone dehydrogenase from Arabidopsis thaliana, a flavoprotein involved in vitamin C biosynthesis. FEBS J. 275, 713–726. doi: 10.1111/j.1742-4658.2007.06233.x
Leferink, N. G. H., van Duijn, E., Barendregt, A., Heck, A. J. R., and van Berkel, W. J. H. (2009). Galactonolactone dehydrogenase requires a redox-sensitive thiol for optimal production of vitamin C. Plant Physiol. 150, 596–605. doi: 10.1104/pp.109.136929
Lemaitre, T., and Hodges, M. (2006). Expression analysis of Arabidopsis thaliana NAD-dependent isocitrate dehydrogenase genes shows the presence of a functional subunit that is mainly expressed in the pollen and absent from vegetative organs. Plant Cell Physiol. 47, 634–643. doi: 10.1093/pcp/pcj030
Lemaitre, T., Urbanczyk-Wochniak, E., Flesch, V., Bismuth, E., Fernie, A. R., and Hodges, M. (2007). NAD-dependent isocitrate dehydrogenase mutants of Arabidopsis suggest the enzyme is not limiting for nitrogen assimilation. Plant Physiol. 144, 1546–1558. doi: 10.1104/pp.107.100677
Lin, M., Behal, R. H., and Oliver, D. J. (2004) Characterization of a mutation in the IDH-II subunit of the NAD1-dependent isocitrate dehydrogenase from Arabidopsis thaliana. Plant Sci. 166, 983–988. doi: 10.1016/j.plantsci.2003.12.012
Luethy, M. H., Miernyk, J. A., and Randall, D. D. (1994). The nucleotide and deduced amino acid sequences of a cDNA encoding the E1 beta-subunit of the Arabidopsis thaliana mitochondrial pyruvate dehydrogenase complex. Biochim. Biophys. Acta 1187, 95–98. doi: 10.1016/0005-2728(94)90171-6
Mani, S., van de Cotte, B., van Montagu, M., and Verbruggen, N. (2002). Altered levels of proline dehydrogenase cause hypersensitivity to proline and its analogs in Arabidopsis. Plant Physiol. 128, 73–83. doi: 10.1104/pp.010572
Mapson, L. W., and Breslow, E. (1958). Biological synthesis of ascorbic acid: L-galactono-gamma-lactone dehydrogenase. Biochem. J. 68, 395–406.
Miersch, J., Grancharov, K., Krauss, G. J., Spassovska, N., Karamanov, G., Maneva, L., et al. (1987). Biological activity and mode of action of some dihydroorotic and dihydroazaorotic acid derivatives. Biomed. Biochim. Acta 46, 307–315.
Millar, A. H., Eubel, H., Jänsch, L., Kruft, V., Heazlewood, J. L., and Braun, H.-P. (2004). Mitochondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol. Biol. 56, 77–90. doi: 10.1007/s11103-004-2316-2
Millar, A. H., Hill, S. A., and Leaver, C. J. (1999). Plant mitochondrial 2-oxoglutarate dehydrogenase complex: purification and characterization in potato. Biochem. J. 343(Pt 2), 327–334. doi: 10.1042/0264-6021:3430327
Millar, A. H., Small, I. D., Day, D. A., and Whelan, J. (2008). Mitochondrial biogenesis and function in Arabidopsis. Arabidopsis Book 6, e0111. doi: 10.1199/tab.0111
Millar, A. H., Whelan, J., Soole, K. L., and Day, D. A. (2011). Organization and regulation of 1822 mitochondrial respiration in plants. Annu. Rev. Plant Biol. 62, 79–104. doi: 10.1146/annurev-arplant-042110-103857
Miller, G., Honig, A., Stein, H., Suzuki, N., Mittler, R., and Zilberstein, A. (2009). Unraveling delta1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J. Biol. Chem. 284, 26482–26492. doi: 10.1074/jbc.M109.009340
Miyashita, Y., and Good, A. G. (2008a). Glutamate deamination by glutamate dehydrogenase plays a central role in amino acid catabolism in plants. Plant Signal. Behav. 3, 842–843. doi: 10.4161/psb.3.10.5936
Miyashita, Y., and Good, A. G. (2008b). NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J. Exp. Bot. 59, 667–680. doi: 10.1093/jxb/erm340
Mooney, B. P., Henzl, M. T., Miernyk, J. A., and Randall, D. D. (2000). The dihydrolipoyl acyltransferase (BCE2) subunit of the plant branched-chain alpha-ketoacid dehydrogenase complex forms a 24-mer core with octagonal symmetry. Protein Sci. 9, 1334–1339. doi: 10.1110/ps.9.7.1334
Moore, A. L., Shiba, T., Young, L., Harada, S., Kita, K., and Ito, K. (2013). Unraveling the heater: new insights into the structure of the alternative oxidase. Annu. Rev. Plant Biol. 64, 637–663. doi: 10.1146/annurev-arplant-042811-105432
Nagai, A., and Scheidegger, A. (1991). Purification and characterization of histidinol dehydrogenase from cabbage. Arch. Biochem. Biophys. 284, 127–132. doi: 10.1016/0003-9861(91)90274-M
Nunes-Nesi, A., Carrari, F., Lytovchenko, A., Smith, A. M. O., Loureiro, M. E., Ratcliffe, R. G., et al. (2005). Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol. 137, 611–622. doi: 10.1104/pp.104.055566
Oguchi, K., Tanaka, N., Komatsu, S., and Akao, S. (2004). Methylmalonate-semialdehyde dehydrogenase is induced in auxin-stimulated and zinc-stimulated root formation in rice. Plant Cell Rep. 22, 848–858. doi: 10.1007/s00299-004-0778-y
Oliver, D. J. (1994) The glycine decarboxylase complex from plant mitochondria. Annu. Rev.Plant Physiol. Plant Mol. Biol. 45, 323–338. doi: 10.1146/annurev.pp.45.060194.001543
Oliver, D. J., Neuburger, M., Bourguignon, J., and Douce, R. (1990). Interaction between the component enzymes of the glycine decarboxylase multienzyme complex. Plant Physiol. 94, 833–839. doi: 10.1104/pp.94.2.833
Olson, B. J., Skavdahl, M., Ramberg, H., Osterman, J. C., and Markwell, J. (2000). Formate dehydrogenase in Arabidopsis thaliana: characterization and possible targeting to the chloroplast. Plant Sci. 159, 205–212. doi: 10.1016/S0168-9452(00)00337-X
Pineau, B., Layoune, O., Danon, A., and de Paepe, R. (2008). L-galactono-1,4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. J. Biol. Chem. 283, 32500–32505. doi: 10.1074/jbc.M805320200
Poulsen, L. L., and Wedding, R. T. (1970). Purification and properties of the alpha-ketoglutarate dehydrogenase complex of cauliflower mitochondria. J. Biol. Chem. 245, 5709–5717.
Rasmusson, A. G., and Agius, S. C. (2001) Rotenone-insensitive NAD(P)H dehydrogenases in plants: immunodetection and distribution of native proteins in mitochondria. Plant Physiol. Biochem. 39, 1057–1066. doi: 10.1016/S0981-9428(01)01334-1
Rasmusson, A. G., Geisler, D. A., and Møller, I. M. (2008). The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 8, 47–60. doi: 10.1016/j.mito.2007.10.004
Rasmusson, A. G., and Moller, I. M. (2011). “Mitochondrial electron transport and plant stress,” in Plant Mitochondria, ed F. Kempken (New York, NY: Springer), 357–381.
Rasmusson, A. G., Soole, K. L., and Elthon, T. E. (2004). Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu. Rev. Plant Biol. 55, 23–39. doi: 10.1146/annurev.arplant.55.031903.141720
Reinard, T., Janke, V., Willard, J., Buck, F., Jacobsen, H. J., and Vockley, J. (2000). Cloning of a gene for an acyl-CoA dehydrogenase from Pisum sativum L. and purification and characterization of its product as an isovaleryl-CoA dehydrogenase. J. Biol. Chem. 275, 33738–33743. doi: 10.1074/jbc.M004178200
Schertl, P., Cabassa, C., Saadallah, K., Bordenave, M., Savouré, A., and Braun, H.P. (in press). Biochemical characterization of ProDH activity in Arabidopsis mitochondria. FEBS J. doi: 10.1111/febs.12821
Schertl, P., Sunderhaus, S., Klodmann, J., Grozeff, G. E. G., Bartoli, C. G., and Braun, H.-P. (2012). L-galactono-1,4-lactone dehydrogenase (GLDH) forms part of three subcomplexes of mitochondrial complex I in Arabidopsis thaliana. J. Biol. Chem. 287, 14412–14419. doi: 10.1074/jbc.M111.305144
Sharma, S., and Verslues, P. E. (2010). Mechanisms independent of abscisic acid (ABA) or proline feedback have a predominant role in transcriptional regulation of proline metabolism during low water potential and stress recovery. Plant Cell Environ. 33, 1838–1851. doi: 10.1111/j.1365-3040.2010.02188.x
Shen, W., Wei, Y., Dauk, M., Tan, Y., Taylor, D. C., Selvaraj, G., et al. (2006). Involvement of a glycerol-3-phosphate dehydrogenase in modulating the NADH/NAD+ ratio provides evidence of a mitochondrial glycerol-3-phosphate shuttle in Arabidopsis. Plant Cell 18, 422–441. doi: 10.1105/tpc.105.039750
Shen, W., Wei, Y., Dauk, M., Zheng, Z., and Zou, J. (2003). Identification of a mitochondrial glycerol-3-phosphate dehydrogenase from Arabidopsis thaliana: evidence for a mitochondrial glycerol-3-phosphate shuttle in plants. FEBS Lett. 536, 92–96. doi: 10.1016/S0014-5793(03)00033-4
Siendones, E., Gonzalez-Reyes, J. A., Santos-Ocana, C., Navas, P., and C rdoba, F. (1999). Biosynthesis of ascorbic acid in kidney bean. L-galactono-gamma-lactone dehydrogenase is an intrinsic protein located at the mitochondrial inner membrane. Plant Physiol. 120, 907–912. doi: 10.1104/pp.120.3.907
Somerville, C. R., and Ogren, W. L. (1982). Mutants of the cruciferous plant Arabidopsis thaliana lacking glycine decarboxylase activity. Biochem. J. 202, 373–380.
Srinivasan, R., and Oliver, D. J. (1995). Light-dependent and tissue-specific expression of the H-protein of the glycine decarboxylase complex. Plant Physiol. 109, 161–168. doi: 10.1104/pp.109.1.161
Sweetlove, L. J., Beard, K. F. M., Nunes-Nesi, A., Fernie, A. R., and Ratcliffe, R. G. (2010). Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci. 15, 462–470. doi: 10.1016/j.tplants.2010.05.006
Szabados, L., and Savouré, A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97. doi: 10.1016/j.tplants.2009.11.009
Szurmak, B., Strokovskaya, L., Mooney, B. P., Randall, D. D., and Miernyk, J. A. (2003). Expression and assembly of Arabidopsis thaliana pyruvate dehydrogenase in insect cell cytoplasm. Protein Expr. Purif. 28, 357–361. doi: 10.1016/S1046-5928(02)00712-X
Tanaka, N., Takahashi, H., Kitano, H., Matsuoka, M., Akao, S., Uchimiya, H., et al. (2005). Proteome approach to characterize the methylmalonate-semialdehyde dehydrogenase that is regulated by gibberellin. J. Proteome Res. 4, 1575–1582. doi: 10.1021/pr050114f
Tarasenko, V. I., Garnik, E. Y., and Konstantinov, Y. M. (2013). Rate of alternative electron transport in arabidopsis mitochondria affects the expression of the glutamate dehydrogenase gene gdh2. Dokl. Biochem. Biophys. 452, 234–236. doi: 10.1134/S1607672913050037
Taylor, N. L., Heazlewood, J. L., Day, D. A., and Millar, A. H. (2004). Lipoic acid-dependent oxidative catabolism of alpha-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol. 134, 838–848. doi: 10.1104/pp.103.035675
Tomaz, T., Bagard, M., Pracharoenwattana, I., Lindén, P., Lee, C. P., Carroll, A. J., et al. (2010). Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis. Plant Physiol. 154, 1143–1157. doi: 10.1104/pp.110.161612
Tovar-Méndez, A., Miernyk, J. A., and Randall, D. D. (2003) Regulation of pyruvate dehydrogenase complex activity in plant cells. Eur. J. Biochem. 270, 1043–1049. doi: 10.1046/j.1432-1033.2003.03469.x
Toyokura, K., Watanabe, K., Oiwaka, A., Kusano, M., Tameshige, T., Tatematsu, K., et al. (2011). Succinic semialdehyde dehydrogenase is involved in the robust patterning of Arabidopsis leaves along the adaxial-abaxial axis. Plant Cell Physiol. 52, 1340–1353. doi: 10.1093/pcp/pcr079
Tronconi, M. A., Fahnenstich, H., Gerrard Weehler, M. C., Andreo, C. S., Flügge, U.-I., Drincovich, M. F., et al. (2008). Arabidopsis NAD-malic enzyme functions as a homodimer and heterodimer and has a major impact on nocturnal metabolism. Plant Physiol. 146, 1540–1552. doi: 10.1104/pp.107.114975
Tronconi, M. A., Gerrard Wheeler, M. C., Drincovich, M. F., and Andreo, C. S. (2012). Differential fumarate binding to Arabidopsis NAD+-malic enzymes 1 and -2 produces an opposite activity modulation. Biochimie 94, 1421–1430. doi: 10.1016/j.biochi.2012.03.017
Tronconi, M. A., Maurino, V. G., Andreo, C. S., and Drincovich, M. F. (2010). Three different and tissue-specific NAD-malic enzymes generated by alternative subunit association in Arabidopsis thaliana. J. Biol. Chem. 285, 11870–11879. doi: 10.1074/jbc.M109.097477
Turano, F. J., Thakkar, S. S., Fang, T., and Weisemann, J. M. (1997). Characterization and expression of NAD(H)-dependent glutamate dehydrogenase genes in Arabidopsis. Plant Physiol. 113, 1329–1341. doi: 10.1104/pp.113.4.1329
Ullrich, A., Knecht, W., Piskur, J., and Löffler, M. (2002). Plant dihydroorotate dehydrogenase differs significantly in substrate specificity and inhibition from the animal enzymes. FEBS Lett. 529, 346–350. doi: 10.1016/S0014-5793(02)03425-7
van Dongen, J. T., Gupta, K. J., Ramírez-Aguilar, S. J., Araújo, W. L., Nunes-Nesi, A., and Fernie, A. R. (2011). Regulation of respiration in plants: a role for alternative metabolic pathways. J. Plant Physiol. 168, 1434–1443. doi: 10.1016/j.jplph.2010.11.004
Verbruggen, N., Hua, X. J., May, M., and van Montagu, M. (1996). Environmental and developmental signals modulate proline homeostasis: evidence for a negative transcriptional regulator. Proc. Natl. Acad. Sci. U.S.A. 93, 8787–8791. doi: 10.1073/pnas.93.16.8787
Wallström, S. V., Florez-Sarasa, I., Araújo, W. L., Aidemark, M., Fernández-Fernández, M., Fernie, A. R., et al. (2014a). Suppression of the external mitochondrial NADPH dehydrogenase, NDB1, in Arabidopsis thaliana affects central metabolism and vegetative growth. Mol. Plant 7, 356–368. doi: 10.1093/mp/sst115
Wallström, S. V., Florez-Sarasa, I., Araújo, W. L., Escobar, M. A., Geisler, D. A., Aidemark, M., et al. (2014b). Suppression of NDA-type alternative mitochondrial NAD(P)H dehydrogenases in Arabidopsis thaliana modifies growth and metabolism, but not high light stimulation of mitochondrial electron transport. Plant Cell Physiol. doi: 10.1093/pcp/pcu021. [Epub ahead of print].
Wedding, R. T., and Black, M. K. (1971a). Evidence for tighter binding of magnesium-thiamine pyrophosphate to -ketoglutarate dehydrogenase when activated by adenosine monophosphate. J. Biol. Chem. 246, 4097–4099.
Wedding, R. T., and Black, M. K. (1971b). Nucleotide activation of cauliflower alpha-ketoglutarate dehydrogenase. J. Biol. Chem. 246, 1638–1643.
Wienstroer, J., Engqvist, M. K. M., Kunz, H.-H., Flügge, U.-I., and Maurino, V. G. (2012). D-Lactate dehydrogenase as a marker gene allows positive selection of transgenic plants. FEBS Lett. 586, 36–40. doi: 10.1016/j.febslet.2011.11.020
Wulff, A., Oliveira, H. C., Saviani, E. E., and Salgado, I. (2009) Nitrite reduction and superoxide-dependent nitric oxide degradation by Arabidopsis mitochondria: influence of external NAD(P)H dehydrogenases and alternative oxidase in the control of nitric oxide levels. Nitric Oxide 21, 132–139. doi: 10.1016/j.niox.2009.06.003
Yamaya, T., Oaks, A., and Matsumoto, H. (1984). Characteristics of glutamate dehydrogenase in mitochondria prepared from corn shoots. Plant Physiol. 76, 1009–1013. doi: 10.1104/pp.76.4.1009
Yu, H., Du, X., Zhang, F., Zhang, F., Hu, Y., Liu, S., et al. (2012). A mutation in the E2 subunit of the mitochondrial pyruvate dehydrogenase complex in Arabidopsis reduces plant organ size and enhances the accumulation of amino acids and intermediate products of the TCA cycle. Planta 236, 387–399. doi: 10.1007/s00425-012-1620-3
Zabaleta, E., Martin, M. V., and Braun, H.-P. (2012). A basal carbon concentrating mechanism in plants? Plant Sci. 187, 97–104. doi: 10.1016/j.plantsci.2012.02.001
Zhu, X., Tang, G., and Galili, G. (2000) Characterization of the two saccharopine dehydrogenase isozymes of lysine catabolism encoded by the single composite AtLKR=SDH locus of Arabidopsis. Plant Physiol. 124, 1363–1372. doi: 10.1104/pp.124.3.1363
Keywords: plant mitochondria, electron transport chain, dehydrogenase, alternative oxidase, respiratory supercomplex
Citation: Schertl P and Braun H-P (2014) Respiratory electron transfer pathways in plant mitochondria. Front. Plant Sci. 5:163. doi: 10.3389/fpls.2014.00163
Received: 28 February 2014; Accepted: 07 April 2014;
Published online: 29 April 2014.
Edited by:
Daniel H. Gonzalez, Universidad Nacional del Litoral, ArgentinaReviewed by:
Joshua L. Heazlewood, Lawrence Berkeley National Laboratory, USANicolas L. Taylor, The University of Western Australia, Australia
Copyright © 2014 Schertl and Braun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hans-Peter Braun, Institut für Pflanzengenetik, Abteilung Pflanzenproteomik, Leibniz Universität Hannover, Herrenhäuser Str. 2, 30419 Hannover, Germany e-mail:YnJhdW5AZ2VuZXRpay51bmktaGFubm92ZXIuZGU=
 Peter Schertl
Peter Schertl Hans-Peter Braun
Hans-Peter Braun