
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 05 March 2014
Sec. Plant Physiology
Volume 5 - 2014 | https://doi.org/10.3389/fpls.2014.00079
This article is part of the Research Topic Biogenesis of the oxidative phosphorylation machinery in plants. From gene expression to complex assembly View all 11 articles
The ribosome filter hypothesis posits that ribosomes are not simple non-selective translation machines but may also function as regulatory elements in protein synthesis. Recent data supporting ribosomal filtering come from plant mitochondria where it has been shown that translation of mitochondrial transcripts encoding components of oxidative phosphorylation complexes (OXPHOS) and of mitoribosomes can be differentially affected by alterations in mitoribosomes. The biogenesis of mitoribosome was perturbed by silencing of a gene encoding a small-subunit protein of the mitoribosome in Arabidopsis thaliana. As a consequence, the mitochondrial OXPHOS and ribosomal transcripts were both upregulated, but only the ribosomal proteins were oversynthesized, while the OXPHOS subunits were actually depleted. This finding implies that the heterogeneity of plant mitoribosomes found in vivo could contribute to the functional selectivity of translation under distinct conditions. Furthermore, global analysis indicates that biogenesis of OXPHOS complexes in plants can be regulated at different levels of mitochondrial and nuclear gene expression, however, the ultimate coordination of genome expression occurs at the complex assembly level.
It is a widely held belief that post-transcriptional control dominates in the regulation of mitochondrial gene expression in plants, but the experimental evidence was until recently limited to events affecting RNA quality and quantity (Binder and Brennicke, 2003). Translational regulation in plant mitochondria was expected but, until now (Kwaśniak et al., 2013), only hypothetical. The basic process of translation involves the decoding of the mRNA-encoded information into proteins by ribosomes. Therefore, historically, ribosomes were considered to have a constitutive rather than a regulatory function, and the efficiency of translation was believed to be determined either by features of the mRNA itself or by mRNA-binding factors. However, a number of observations indicated that ribosomes themselves could also differentially affect the translation of particular mRNAs (Mauro and Edelman, 2007). This mini-review summarizes current knowledge on ribosomal regulation of OXPHOS biogenesis.
All ribosomes consist of the large subunit (LSU) and small subunit (SSU) composed of both proteins and rRNAs. Originally, ribosomes of an organism were viewed as homogeneous entities showing no variation in their rRNA or protein composition despite the observation of a growth rate-dependent ribosome heterogeneity in Escherichia coli made over 40 years ago (Deusser, 1972). The earliest evidence demonstrating heterogeneity of ribosomes in a eukaryotic cell emerged in 1981 during studies on the social amoeba Dictyostelium discoideum (Ramagopal and Ennis, 1981). The ribosomes from the different developmental stages of D. discoideum varied in their composition and covalent modifications (Ramagopal, 1992). Since then ample evidence has accumulated supporting the view that heterogeneous pools of ribosomes exhibiting variations in the RNA and or the protein components are present in bacteria and in eukaryotic cells (Brygazov et al., 2013; Filipovska and Rackham, 2013). Since the focus of this review is the translational regulation in plant mitochondria the heterogeneity of plant mitoribosomes will be presented in more detail.
The plant mitoribosome contains three rRNA molecules encoded in the mitochondrial DNA, 26S and 5S for the LSU, and 18S for the SSU. The rRNAs undergo several important post-transcriptional modifications (pseudouridylation and methylation; Bonen, 2004), and an rRNA methyltransferase required for the dimethylation of two conserved adenines in the mitochondrial 18S rRNA of Arabidopsis was characterized (Richter et al., 2010). In contrast to the rRNAs, the mitoribosomal proteins are encoded by both the mitochondrial and nuclear genomes. The sets of proteins encoded by these two genomes vary between plant species (Salinas et al., 2013), and the exact protein composition of plant mitoribosomes has not been fully determined yet. A recent bioinformatics analysis identified 71 genes encoding mitoribosomal proteins in A. thaliana, among them eight in the mitochondrial genome (Sormani et al., 2011). It should be emphasized that these numbers do not represent unique genes, since 16 of mitochondrial ribosomal proteins are encoded by more than one copy of gene. Potato and broad bean proteomics found 68–80 mitoribosomal proteins (Pinel et al., 1986; Maffey et al., 1997). Thus, compared with the bacterial ribosomes with only 54 proteins (Wittmann, 1982), the plant mitoribosome is apparently more complex. The plant mitochondrial ribosomal proteins are of two evolutionarily distinct types, those with a direct bacterial origin and those recruited during the evolution (Salinas et al., 2013). Additionally, in Arabidopsis two proteins have been identified to be associated with mitoribosomes, namely PNM1 and PPR336 (Uyttewaal et al., 2008; Hammani et al., 2011). They belong to the pentatricopeptide repeat (PPR) protein family that is particularly large in higher plants, but their role in mitochondrial translation is still unknown.
The first indication of a possible heterogeneity of plant mitoribosomes comes from a study on four paralogs of ribosomal protein L12 in potato (Delage et al., 2007). At the RNA level these paralogs are expressed simultaneously and at similar abundance. All four L12 variants were present in the mitochondrial ribosome fraction, but they showed a divergent tendency to dissociate upon treatments that affects ribosomes integrity. The presence of the four paralogs of slightly different proportion suggests either occurrence of heterogeneous L12 composition among each mitoribosome and/or a heterogeneous population of mitochondrial ribosomes in the plant cell.
A heterogeneity of plant mitoribosomes is also implied by several developmental phenotypes of mutants defective in a mitoribosomal protein (Schippers and Mueller-Roeber, 2010). Slightly different phenotypes connected with defects in leaf morphology have been reported for three Arabidopsis mutants. In all three, the leaves are smaller, but an irregular leaf shape is characteristic for the mutant with reduced expression of both MRPS3 and MRPL16 (Sakamoto et al., 1996) as well as the mutant with RNAi-dependent silencing of MRPS10 (Majewski et al., 2009), but not for the mutant with a 90% reduction in MRPL11 transcript (Pesaresi et al., 2006). The alteration in leaf morphology was observed also in maize rps3-rpl16 mutants, which produces severely stunted plants with striations on the leaves (Hunt and Newton, 1991). The role of mitoribosomal proteins in other specific developmental processes is underlined by several specific Arabidopsis mutants with defects in genes: MRPL14, which is essential for ovule development (Schneitz et al., 1998; Skinner et al., 2001), MRPL21 and MRPS11 which are required during female gametophyte development (Portereiko et al., 2006), as well as MRPS16 gene, in which transposon-induced knockout causes an embryo-defective lethal phenotype (Tsugeki et al., 1996). Thus, specific mitoribosomal proteins appear to influence selectively different phases of the plant development, hinting on the existence of specialized ribosomes with the translational activity critical at specific developmental stages. This view is strengthened by the finding that the expression of individual A. thaliana genes encoding mitochondrial ribosomal proteins is highly variable during leaf development (Schippers and Mueller-Roeber, 2010). A functional heterogeneity of ribosomes could also be generated by interactions with accessory proteins as well as posttranslational modifications of ribosomal proteins, but the significance, if any of these factors for plant mitoribosomes is unknown.
The apparent ribosome heterogeneity formed a basis for the ribosome filter hypothesis in which the ribosome acts like a filter selecting specific mRNAs and consequently modulating translation (Mauro and Edelman, 2007). The different subpopulations of ribosomes would vary in their ability to translate specific subsets of mRNA and thus the ribosomes would selectively control translation and, consequently, gene expression. In accordance with the ribosome filter hypothesis, it has recently been shown that the biogenesis of OXPHOS complexes in A. thaliana mitochondria is subject to translational control by mitoribosomes (Kwaśniak et al., 2013).
Mitochondrial translation is indispensable for the biogenesis of the OXPHOS machinery and mitoribosomes simply because several protein subunits of the both types of complexes are synthesized in mitochondria. Spice has been added to that simple story by recent data indicating that plant mitoribosomes do not synthesize proteins encoded in the mitochondrial genome non-selectively but rather execute a transcript-specific translational control (Kwaśniak et al., 2013).
This finding is fully consistent with the ribosome filter theory. RNAi-mediated silencing leads to the generation of unique, heterogeneous population of ribosomes. Ribosomes lacking the S10 protein coexisted with wild-type ones and an excess of free LSU (Figure 1). It should be underlined that the rps10 mutant did not suffer from an insufficiency of wild-type mitoribosomes since the silencing provoked a compensatory response increasing the overall abundance of both small and LSUs. The polymorphic population of mitoribosomes turned out to translate two subsets of mRNAs, those encoding OXPHOS subunits and ribosomal proteins, with altered efficiency compared to the wild-type. The majority of the OXPHOS transcripts were translated less efficiently, whereas most of the mitoribosome protein transcripts were translated with an enhanced efficiency relative to the wild-type. It should be emphasized that in the RPS10-silenced plants the mitochondrial transcripts for both OXPHOS and ribosomal proteins were up-regulated. Thus, the altered translation was not correlated with the transcript level but was due to an altered efficiency of their binding by the mitoribosomes. As a consequence, the rate of synthesis of the OXPHOS proteins was below that observed in wild-type mitochondria despite the over-abundance of their mRNAs. Thus, the diversified mitochondrial translation efficiency modulated profoundly the outcome of the earlier steps of mitochondrial genome expression. The effect of the altered SSU and/or the excess of free LSU on the efficiency of protein synthesis in the rps10 mitochondria was postulated to have resulted from the formation of unique interactions between those subunits and other components of the translation apparatus. It is tempting to speculate that similar translational regulation could occur in wild-type plants, namely under conditions that lead to heterogeneity of mitoribosome population in response to environmental or developmental signals.
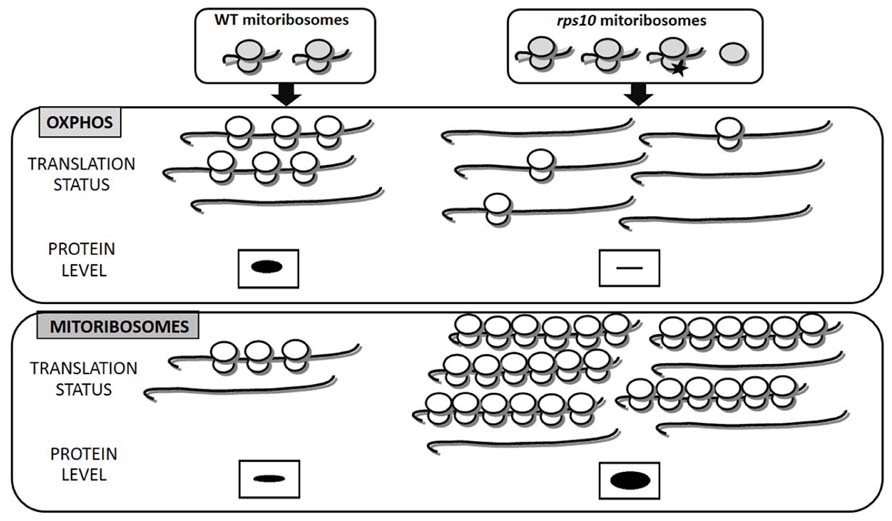
FIGURE 1. Heterogeneity of mitoribosomes differentially affects the translation of mitochondrial OXPHOS and ribosomal proteins (based on Kwaśniak et al., 2013). In rps10 protein synthesis is carried out by a heterogeneous population of mitoribosomes characterized by an excess of free LSU subunits and a portion of SSUs lacking the S10 protein (black asterisk). Decreased translation of OXPHOS in rps10 results mainly from the reduced number of ribosomes per mRNA, whereas increased translation of mitoribosomal proteins results from enhanced ribosomal loading and also increased ribosomal density. In consequence, OXPHOS proteins are depleted, whereas mitoribosomal proteins over-abundant in rps10 mitochondria compared to wild-type ones. The amount of proteins translated by mitoribosomes is represented by the size of the black bands.
According to existing reports plant mitochondrial RNAs have long 5′ untranslated regions (UTR), which could be involved in translational efficiency by interacting with altered SSU and/or the excess of free LSU. However, no obvious motifs have been found within the 5′ UTR sequences suggesting that these parts of mRNAs are involved in gene-specific translation regulation (Kazama et al., 2013), whereas in rps10 rather group-specific translational regulation was observed. Future studies are awaited to understand how differential translation of OXPHOS and mitoribosomal proteins is achieved in the rps10 mutant.
The importance of mitochondrial translational regulation for OXPHOS complex I biogenesis has recently been suggested basing on analysis of an A. thaliana mutant with a depletion of the mitochondrial matrix iron-sulfur protein required for NADH dehydrogenase (INDH)(Wydro et al., 2013). INDH was shown not to be a complex I-specific translational regulator but rather to have a more general role in mitochondrial translation. Depending on the A. thaliana growth stage and the extent of INDH depletion diverse alterations in the mitochondrial translation rate and/or protein pattern were observed in the indh mutants. A strong depletion of INDH decreased the rate of mitochondrial translation, whereas the steady state levels of almost all mitochondrial transcripts examined were unaltered or increased.
The data indicating a heterogeneity of plant mitoribosomes combined with the finding that translation could be the overriding regulatory stage of mitochondrial gene expression regulation suggest that in plants the status of the mitoribosomal population could be an essential regulator of expression of mitochondrially encoded proteins in response to developmental or environmental signals. The postulated ribosome-mediated selectivity of mitochondrial translation should be reflected by a poor correlation between the steady-state abundance of mitochondrial transcripts and the encoded proteins. However, despite the recent intense studies of the plant mitochondrial transcriptome and proteome no enough quantitative data exist to substantiate such a claim. Relations between the transcript and the protein level are mainly known for the nuclear-encoded mitochondrial proteins (Law et al., 2012; Tan et al., 2012).
Biogenesis of OXPHOS complexes requires balanced production of nuclear- and mitochondrially encoded subunits. Some reports indicate that in plants coordination of the expression of OXPHOS genes between the nuclear and mitochondrial genomes may occur at the transcript level (Topping and Leaver, 1990; Smart et al., 1994; Ribichich et al., 2001; Howell et al., 2006), but recent global analyses of expression of the mitochondrial and nuclear genomes in plants underscore the crucial role of the post-transcriptional coordination (Giegé et al., 2005; Leister et al., 2011; Law et al., 2012; Kwaśniak et al., 2013). Furthermore, Giegé et al. (2005) and Kwaśniak et al. (2013) studying biogenesis of OXPHOS complexes under sugar starvation conditions and in the rps10 mutant, respectively, reached the same conclusion that the ultimate coordination of expression of the two genomes occurs at the level of protein complex assembly. The authors of both those papers assumed that an excess of unassembled subunits were degraded by nuclear-encoded ATP-dependent proteases. Apart from this similarity, there were also differences between the two experimental systems, as the RPS10 silencing mainly down-regulated the biosynthesis of the mitochondrially encoded subunits whereas under sugar starvation the biogenesis of OXPHOS complexes was regulated mostly by curbing the expression of the respective nuclear genes. As a consequence, the nuclear-encoded proteins in rps10 and the mitochondrially encoded proteins under sugar starvation had to be degraded to bring their abundance down to the level of their partners. One is tempted to suggest that depending on the nature or the duration of the stress the direct response of the two collaborating genomes is in fact limited to one or the other and the expression of the second genome is adjusted ultimately at the complex assembly level.
In line with the above predictions, an up-regulation of mitochondrial ATP-dependent proteases was indeed observed in the rps10 mutant. An induction of nuclear-encoded mitochondrial proteases and chaperones upon accumulation of unfolded or misfolded proteins in the mitochondrial matrix is a hallmark of the mitochondrial unfolded-protein response (UPRmt) that has been reported for mammals and Caernohabditis elegans (Haynes and Ron, 2010; Haynes et al., 2013). Conceivably, also in plants UPRmt is activated by the stoichiometric imbalance between the nDNA- and mtDNA-encoded OXPHOS proteins caused by the silencing of RPS10 expression and restores the balance between the two sets of the subunits by selectively degrading the ones in excess.
In contrast to plants, where translational regulation has just emerged as an important step of the biogenesis of OXPHOS complexes, a diversity of mitochondrial translation control mechanisms have been reported in other organisms, particularly in Saccharomyces cerevisiae. One such mechanism is associated with translational activators which interact with mitochondrial mRNAs, in most cases at their 5′ UTR, to facilitate their translation (Smits et al., 2010). It is unclear whether all translational activators bind the mRNA directly or whether some of them interact with other components of the translation machinery (Herrmann et al., 2013). Ample data show that the translational activators can be part of a feedback mechanism coupling the rate of synthesis of mitochondrial proteins to the rate of their assembly into complexes, known as control by epistasy of synthesis (CES). This mechanism was first reported for chloroplasts (Choquet and Wollman, 2002). In the yeast mitochondria the best known such feedback loop is the one that controls the synthesis of Cox1. This regulation is achieved by the translational activator Mss51 which interacts with the Cox1 mRNA, newly synthesized unassembled Cox1 protein, and other Cox assembly factors during Cox1 maturation/assembly (Fontanesi et al., 2010). Assembly defects lead to a sequestration of Mss51 and consequent, stoppage of Cox1 synthesis. A link between mitochondrial translation and complex assembly is also supported by another regulatory mechanism found in yeast, in which generation of assembly competent proteins during mitochondrial translation requires the mitoribosomal protein MPRL36 (Prestele et al., 2009). Mitoribosomes with a C-terminal domain-deficient MRPL36 protein still carry out translation, but the resulting polypeptides fail to be properly assembled into OXPHOS complexes and are rapidly degraded. The authors suggested that during translation, MRPL36 interacts with the ribosome at an as-yet unidentified step, which could be a critical for the production of assembly competent products.
A negative feedback mechanism for the regulation of mitochondrial translation based on an interplay between the translation and m-AAA protease, the key component of the mitochondrial quality control system has also been proposed for yeast (Nolden et al., 2005). The m-AAA protease not only degrades misfolded or damaged proteins, but also processes the mitoribosomal protein MRPL32. This maturation seems to be required for mitochondrial translation since the synthesis of mitochondrially encoded proteins was substantially impaired in cells lacking the m-AAA protease. Under certain conditions, misfolded, non-native respiratory subunits can compete with MRPL32 for binding to the m-AAA protease and thereby hamper the MRPL32 processing, ribosome assembly, and mitochondrial translation. MRPL32 processing and mitochondrial translation resume when the level of the respiratory subunits decreases as a consequence of their m-AAA-mediated degradation. It is worth mentioning in this context that plant m-AAA is able to process the yeast ribosomal protein MRPL32 (Kolodziejczak et al., 2002; Piechota et al., 2010). However, no direct evidence has been provided that plant m-AAA proteases can process a plant homolog of this mitoribosomal protein.
Given the mammalian mitochondrial transcripts contain no or only a short untranslated regulatory 5′ region (Koc and Spremulli, 2003), it seems unlikely that in mammals the synthesis of mitochondrial OXPHOS subunits is subject to CES, as it is in yeast. Instead, some reports indicate that members of the PPR protein family function as translational regulators of OXPHOS biogenesis in mammalian cells. One of them is leucine-rich PPR containing (LRPPRC) protein reported not only to coordinate mitochondrial translation but also to stabilize mitochondrial mRNAs and promote their polyadenylation (Sasarman et al., 2010). In the absence of LRPPRC the translation pattern is misregulated, with excessive translation of some transcripts and no translation of others. Interestingly, the two homologs of LRPPRC protein in fission yeast, Ppr4, and Ppr5, that also affect translation, play opposite roles in the expression of the cox1 transcript (Kühl et al., 2011). Ppr4 specifically activates the translation of the cox1 mRNA, whereas Ppr5 is a general negative regulator of mitochondrial translation. Furthermore, in humans another PPR protein – PTCD1 has been proposed to inhibit mitochondrial translation probably by decreasing the stability of the mitochondrial leucine tRNAs (Rackham et al., 2009). Other noteworthy human PPR proteins implicated in translation include PTCD2 involved in the maturation of cytochrome b transcripts (Xu et al., 2008) and PTCD3 associated with the SSU of mitochondrial ribosomes by binding 12S rRNA and necessary for efficient mitochondrial translation (Davies et al., 2009).
In bacteria mRNA abundance is the primary factor controlling the rate of protein synthesis. Recent studies have shown that in plant mitochondria the synthesis of OXPHOS subunits is surprisingly insensitive to the accumulation of respective transcripts (Kwaśniak et al., 2013; Wydro et al., 2013). It seems that, similarly to the chloroplast system (Marin-Navarro et al., 2007), the expression of mitochondrial genes in plants is mainly regulated at the level of translation. Compared with transcriptional regulation, translational regulation combined with selective degradation allows the organelles to modulate protein abundance more rapidly.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by grant N N301 784940 from the Ministry of Science and Higher Education.
Binder, S., and Brennicke, A. (2003). Gene expression in plant mitochondria: transcriptional and post-transcriptional control. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 181–189. doi: 10.1098/rstb.2002.1179
Bonen, L. (2004). “Translational machinery in plant organelles,” in Molecular Biology and Biotechnology of Plant Organelles, eds H. Daniell and C. Chase (Netherlands: Springer) 323–345. doi: 10.1007/978-1-4020-3166-3_12
Brygazov, K., Vesper, O., and Moll, I. (2013). Ribosome heterogeneity: another level of complexity in bacterial translation regulation. Curr. Opin. Microbiol. 16, 133–139. doi: 10.1016/j.mib.2013.01.009
Choquet, Y., and Wollman, F. A. (2002). Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 529, 39–42. doi: 10.1016/S0014-5793(02)03260-X
Davies, S. M., Rackham, O., Shearwood, A. M., Hamilton, K. L., Narsai, R., Whelan, J., et al. (2009). Pentatricopeptide repeat domain protein 3 associates with the mitochondrial small ribosomal subunit and regulates translation. FEBS Lett. 583, 1853–1858. doi: 10.1016/j.febslet.2009.04.048
Delage, L., Giegé, P., Sakamoto, M., and Maréchal-Drouard, L. (2007). Four paralogues of RPL12 are differentially associated to ribosome in plant mitochondria. Biochimie 89, 658–668. doi: 10.1016/j.biochi.2007.02.002
Deusser, E. (1972). Heterogeneity of ribosomal populations in Escherichia coli cells grown in different media. Mol. Gen. Genet. 119, 249–258. doi: 10.1007/BF00333862
Filipovska, A., and Rackham, O. (2013). Specialization from synthesis: how ribosome diversity can customize protein function. FEBS Lett. 587, 1189–1197. doi: 10.1016/j.febslet.2013.02.032
Fontanesi, F., Soto, I. C., Horn, D., and Barrientos, A. (2010). Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol. Cell. Biol. 30, 245–259. doi: 10.1128/MCB.00983-09
Giegé, P., Sweetlove, L. J., Cognat, V., and Leaver, C. J. (2005). Coordination of nuclear and mitochondrial genome expression during mitochondrial biogenesis in Arabidopsis. Plant Cell 17, 1497–1512. doi: 10.1105/tpc.104.030254
Hammani, K., Gobert, A., Hleibieh, K., Choulier, L., Small, I., and Giegé, P. (2011). An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell 23, 730–740. doi: 10.1105/tpc.110.081638
Haynes, C. M., Fiorese, C. J., and Lin, Y. F. (2013). Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends Cell Biol. 23, 311–318. doi: 10.1016/j.tcb.2013.02.002
Haynes, C. M., and Ron, D. (2010). The mitochondrial UPR – protecting organelle protein homeostasis. J. Cell Sci. 123, 3849–3855. doi: 10.1242/jcs.075119
Herrmann, J. M., Woellhaf, M. W., and Bonnefoy, N. (2013). Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim. Biophys. Acta 1833, 286–294. doi: 10.1016/j.bbamcr.2012.03.007
Howell, K. A., Narsai, R., Carroll, A., Ivanova, A., Lohse, M., Usadel, B., et al. (2006). Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol. 149, 961–980. doi: 10.1104/pp.108.129874
Hunt, M. D., and Newton, K. J. (1991). The NCS3 mutation: genetic evidence for the expression of ribosomal protein genes in Zea mays mitochondria. EMBO J. 10, 1045–1052.
Kazama, T., Yagi, Y., Toriyama, K., and Nakamura, T. (2013). Heterogeneity of the 5’-end in plant mRNA may be involved in mitochondrial translation. Front. Plant Sci. 4:517. doi: 10.3389/fpls.2013.00517
Koc, E. C., and Spremulli, L. L. (2003). RNA-binding proteins of mammalian mitochondria. Mitochondrion 2, 277–291. doi: 10.1016/S1567-7249(03)00005-9
Kolodziejczak, M., Kolaczkowska, A., Szczesny, B., Urantowka, A., Knorpp, C., Kieleczawa, J., et al. (2002). A higher plant mitochondrial homologue of the yeast m-AAA protease. Molecular cloning, localization, and putative function. J. Biol. Chem. 277, 43792–43798. doi: 10.1074/jbc.M203831200
Kühl, I., Dujeancourt, L., Gaisne, M., Herbert, C. J., and Bonnefoy, N. (2011). A genome wide study in fission yeast reveals nine PPR proteins that regulate mitochondrial gene expression. Nucleic Acids Res. 39, 8029–8041. doi: 10.1093/nar/gkr511
Kwaśniak, M., Majewski, P., Skibior, R., Adamowicz, A., Czarna, M., and Janska, H. (2013). Silencing of nuclear RPS10 gene encoding mitochondrial ribosomal protein alters translation in Arabidopsis mitochondria. Plant Cell 25, 1855–1867. doi: 10.1105/tpc.113.111294
Law, S. R., Narsai, R., Taylor, N. L., Delannoy, E., Carrie, C., Giraud, E., et al. (2012). Nucleotide and RNA metabolism prime translational initiation in the earliest events of mitochondrial biogenesis during Arabidopsis germination. Plant Physiol. 158, 1610–1627. doi: 10.1104/pp.111.192351
Leister, D., Wang, X., Haberer, G., Mayer, K. F., and Kleine, T. (2011). Intracompartmental and intercompartmental transcriptional networks coordinate the expression of genes for organellar functions. Plant Physiol. 157, 386–404. doi: 10.1104/pp.111.177691
Maffey, L., Degand, H., and Boutry, M. (1997). Partial purification of mitochondrial ribosomes from broad bean and identification of proteins encoded by the mitochondrial genome. Mol. Gen. Genet. 254, 365–371. doi: 10.1007/s004380050428
Majewski, P., Woloszynska, M., and Janska, H. (2009). Developmentally early and late onset of Rps10 silencing in Arabidopsis thaliana: genetic and environmental regulation. J. Exp. Bot. 60, 1163–1178. doi: 10.1093/jxb/ern362
Marin-Navarro, J., Manuell, A. L., Wu, J., and Mayfield, S. (2007). Chloroplast translation regulation. Photosynth. Res. 94, 359–374. doi: 10.1007/s11120-007-9183-z
Mauro, V. P., and Edelman, G. M. (2007). The ribosome filter redux. Cell Cycle 6, 2246–2251. doi: 10.4161/cc.6.18.4739
Nolden, M., Ehses, S., Koppen, M., Bernacchia, A., Rugarli, E. I., and Langer, T. (2005). The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123, 277–289. doi: 10.1016/j.cell.2005.08.003
Pesaresi, P., Masiero, S., Eubel, H., Braun, H. P., Bhushan, S., Glaser, E., et al. (2006). Nuclear photosynthetic gene expression is synergistically modulated by rates of protein synthesis in chloroplasts and mitochondria. Plant Cell 18, 970–991. doi: 10.1105/tpc.105.039073
Piechota, J., Kolodziejczak, M., Juszczak, I., Sakamoto, W., and Janska, H. (2010). Identification and characterization of high molecular weight complexes formed by matrix AAA proteases and prohibitins in mitochondria of Arabidopsis thaliana. J. Biol. Chem. 285, 12512–12521. doi: 10.1074/jbc.M109.063644
Pinel, C., Douce, R., and Mache, R. (1986). A study of mitochondrial ribosomes from the higher plant Solanum tuberosum L. Mol. Biol. Rep. 11, 93–97. doi: 10.1007/BF00364820
Portereiko, M. F., Sandaklie-Nikolova, L., Lloyd, A., Dever, C. A., Otsuga, D., and Drews, G. N. (2006). Nuclear fusion defective 1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant Physiol. 141, 957–965. doi: 10.1104/pp.106.079319
Prestele, M., Vogel, F., Reichert, A. S., Herrmann, J. M., and Ott, M. (2009). Mrpl36 is important for generation of assembly competent proteins during mitochondrial translation. Mol. Biol. Cell 20, 2615–2625. doi: 10.1091/mbc.E08-12-1162
Rackham, O., Davies, S. M., Shearwood, A. M., Hamilton, K. L., Whelan, J., and Filipovska, A. (2009). Pentatricopeptide repeat domain protein 1 lowers the levels of mitochondrial leucine tRNAs in cells. Nucleic Acids Res. 37, 5859–5867. doi: 10.1093/nar/gkp627
Ramagopal, S. (1992). The Dictyostelium ribosome: biochemistry, molecular biology, and developmental regulation. Biochem. Cell Biol. 70, 738–750. doi: 10.1139/o92-113
Ramagopal, S., and Ennis, H. L. (1981). Regulation of synthesis of cell-specific ribosomal proteins during differentiation of Dictyostelium discoideum. Proc. Natl. Acad. Sci. U.S.A. 78, 3083–3087. doi: 10.1073/pnas.78.5.3083
Ribichich, K. F., Tioni, M. F., Chan, R. L., and Gonzalez, D. H. (2001). Cell-type-specific expression of plant cytochrome c mRNA in developing flowers and roots. Plant Physiol. 125, 1603–1610. doi: 10.1104/pp.125.4.1603
Richter, U., Kühn, K., Okada, S., Brennicke, A., Weihe, A., and Börner, T. (2010). A mitochondrial rRNA dimethyladenosine methyltransferase in Arabidopsis. Plant J. 61, 558–569. doi: 10.1111/j.1365-313X.2009.04079.x
Sakamoto, W., Kondo, H., Murata, M., and Motoyoshi, F. (1996). Altered mitochondrial gene expression in a maternal distorted leaf mutant of Arabidopsis induced by chloroplast mutator. Plant Cell 8, 1377–1390. doi: 10.1105/tpc.8.8.1377
Salinas, T., Remacle, C., and Maréchal-Drouard, L. (2013). “Mitochondrial translation in green algae and higher plant,” in Translation in Mitochondria and Other Organelles, ed. A. M. Duchêne (Berlin: Springer Berlin-Heidelberg), 181–205. doi: 10.1007/978-3-642-39426-3_8
Sasarman, F., Brunel-Guitton, C., Antonicka, H., Wai, T., Shoubridge, E. A., and LSFC Consortium. (2010). LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell 21, 1315–1323. doi: 10.1091/mbc.E10-01-0047
Schippers, J. H. M., and Mueller-Roeber, B. (2010). Ribosomal composition and control of leaf development. Plant Sci. 179, 307–315. doi: 10.1016/j.plantsci.2010.06.012
Schneitz, K., Baker, S. C., Gasser, C. S., and Redweik, A. (1998). Pattern formation and growth during floral organogenesis: HUELLENLOS and AINTEGUMENTA are required for the formation of the proximal region of the ovule primordium in Arabidopsis thaliana. Development 125, 2555–2563.
Skinner, D. J., Baker, S. C., Meister, R. J., Broadhvest, J., Schneitz, K., and Gasser, C. S. (2001). The Arabidopsis HUELLENLOS gene, which is essential for normal ovule development, encodes a mitochondrial ribosomal protein. Plant Cell 13, 2719–2730. doi: 10.1105/tpc.010323
Smart, C. J., Monéger, F., and Leaver, C. J. (1994). Cell-specific regulation of gene expression in mitochondria during anther development in sunflower. Plant Cell 6, 811–825.
Smits, P., Smeitink, J., and van den Heuvel, L. (2010). Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J. Biomed. Biotechnol. 2010, 24. doi: 10.1155/2010/737385
Sormani, R., Masclaux-Daubresse, C., Daniele-Vedele, F., and Chardon, F. (2011). Transcriptional regulation of ribosome components are determined by stress according to cellular compartments in Arabidopsis thaliana. PLoS ONE 6:e28070. doi: 10.1371/journal.pone.0028070
Tan, Y. F., Millar, A. H., and Taylor, N. L. (2012). Components of mitochondrial oxidative phosphorylation vary in abundance following exposure to cold and chemical stresses. J. Proteome Res. 11, 3860–3879. doi: 10.1021/pr3003535
Topping, J. F., and Leaver, C. J. (1990). Mitochondrial gene expression during wheat leaf development. Planta 182, 399–407. doi: 10.1007/BF02411391
Tsugeki, R., Kochieva, E. Z., and Federoff, N. V. (1996). A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J. 10, 479–489. doi: 10.1046/j.1365-313X.1996.10030479.x
Uyttewaal, M., Mireau, H., Rurek, M., Hammani, K., Arnal, N., Quadrado, M., et al. (2008). PPR336 is associated with polysomes in plant mitochondria. J. Mol. Biol. 375, 626–636. doi: 10.1016/j.jmb.2007.11.011
Wittmann, H. G. (1982). Components of bacterial ribosomes. Annu. Rev. Biochem. 51, 155–183. doi: 10.1146/annurev.bi.51.070182.001103
Wydro, M. M., Sharma, P., Foster, J. M., Bych, K., Meyer, E. H., and Balk, J. (2013). The evolutionarily conserved iron-sulfur protein INDH is required for complex I assembly and mitochondrial translation in Arabidopsis. Plant Cell. 25, 4014–4027. doi: 10.1105/tpc.113.117283
Xu, F., Ackerley, C., Maj, M. C., Addis, J. B., Levandovskiy, V., Lee, J., et al. (2008). Disruption of a mitochondrial RNA-binding protein gene results in decreased cytochrome b expression and a marked reduction in ubiquinol-cytochrome c reductase activity in mouse heart mitochondria. Biochem. J. 416, 15–26. doi: 10.1042/BJ20080847
Keywords: OXPHOS, mitoribosome, mitochondrial translation, ribosome filter hypothesis, ribosome heterogeneity, coordination of gene expression
Citation: Janska H and Kwasniak M (2014) Mitoribosomal regulation of OXPHOS biogenesis in plants. Front. Plant Sci. 5:79. doi: 10.3389/fpls.2014.00079
Received: 26 November 2013; Paper pending published: 13 January 2014;
Accepted: 19 February 2014; Published online: 05 March 2014.
Edited by:
Philippe Giegé, Centre National de la Recherche Scientifique, FranceReviewed by:
Linda Bonen, University of Ottawa, CanadaCopyright © 2014 Janska and Kwasniak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanna Janska, Molecular Biology of the Cell Department, Faculty of Biotechnology, University of Wroclaw, F. Joliot-Curie 14A, 50-383 Wroclaw, Poland e-mail:amFuc2thQGlibWIudW5pLndyb2MucGw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.