- Department 1 (Willmitzer), Central Metabolism, Max Planck Institute for Plant Physiology, Potsdam, Germany
Whole genome sequencing and the relative ease of transcript profiling have facilitated the collection and data warehousing of immense quantities of expression data. However, a substantial proportion of genes are not yet functionally annotated a problem which is particularly acute for transport proteins. In Arabidopsis, for example, only a minor fraction of the estimated 700 intracellular transporters have been identified at the molecular genetic level. Furthermore it is only within the last couple of years that critical genes such as those encoding the final transport step required for the long distance transport of sucrose and the first transporter of the core photorespiratory pathway have been identified. Here we will describe how transcriptional coordination between genes of known function and non-annotated genes allows the identification of putative transporters on the premise that such co-expressed genes tend to be functionally related. We will additionally extend this to include the expansion of this approach to include phenotypic information from other levels of cellular organization such as proteomic and metabolomic data and provide case studies wherein this approach has successfully been used to fill knowledge gaps in important metabolic pathways and physiological processes.
Introduction
Over the last 15 years or so, co-expression analysis has emerged as a powerful statistical tool which is based on the guilt-by-association approach. This approach assumes that if transcript levels of a gene of unknown function correspond tightly with those of genes of known function then it is highly likely that the gene of unknown function plays a role in the same biological process as the known gene (Tohge and Fernie, 2012; Saito et al., 2013; Stitt, 2013). There are a number of caveats to this approach including the influence of the type of expression data used to construct the co-expression networks and the statistical methods used to evaluate them. However, these have been discussed in detail elsewhere [see reviews by (Usadel et al., 2009; Bordych et al., 2013; Stitt, 2013)] and when properly considered this strategy can prove very effective.
The earliest large scale use of this approach was performed in yeast in a two step approach. First, similarity scores were assigned to each possible gene pair on the comparison of their gene expression levels across a wide range of conditions. Secondly the resultant distance matrix, comprised of all possible similarity scores was organized (“clustered”), in a manner allowing the facile identification of genes showing the most similar expression patterns (Eisen et al., 1998). Given the simplicity of this approach it has also been rapidly adopted in microbial (Fribourg et al., 2001; Gasch and Eisen, 2002; Mao et al., 2005; Zhang et al., 2005), mammalian (Taniguchi et al., 2002; Voehringer et al., 2000; Altman and Raychaudhuri, 2001; Raychaudhuri et al., 2001; Lee et al., 2004; Li et al., 2004; Prieto et al., 2008), and plant research (Maleck et al., 2000; Schaffer et al., 2001; Goda et al., 2008). For plant research several web-based tools including ATTED-II (Obayashi et al., 2009; Obayashi et al., 2011), AraNet (Hwang et al., 2011), Expression Angler of the Bio-Array resource [BAR; (Toufighi et al., 2005], KappaViewer (Sakurai et al., 2011), GeneCAT (Mutwil et al., 2008), Genevestigator (Zimmermann et al., 2004), and VirtualPlant (Katari et al., 2010) simplify this task yet further. One of the strongest demonstrations of the power of this technique in plants comes from its early utility in the identification of further genes involved in secondary cell wall and hemicellulose synthesis in Arabidopsis (Brown et al., 2005; Persson et al., 2005; Cocuron et al., 2007). These studies used the three major cellulose synthase A (CESA) genes as a bait (i.e., expression profile query), to construct networks and thus isolate novel functional genes displaying similar expression patterns. Subsequent confirmation of the biological function of a large number of these genes has been achieved and documented in a number of publications (Bringmann et al., 2012; Ruprecht and Persson, 2012; Sanchez-Rodriguez et al., 2012). A second area of plant metabolism in which the approach has proven highly informative is secondary metabolism. This is perhaps not surprising since secondary metabolism is often regulated directly at the transcriptional level and known to be under the control of a wide range of transcription factors including the MYB transcription factors. The initial use of this approach was in constructing a flavonoid co-expression network to identify a flavonol-3′-O-methyltransferase (Tohge et al., 2007) and flavonol-7-O-rhamnosyltransferse (Yonekura-Sakakibara et al., 2007). However, it has subsequently been used to find other flavonoid genes (Yonekura-Sakakibara et al., 2008), glucosinolate MYB regulators (Hirai et al., 2007), anthocyanin glucosyltransferase (Yonekura-Sakakibara et al., 2012), phospholipid sugar transferase (Okazaki et al., 2009), and lignin biosynthetic genes (Ehlting et al., 2005; Vanholme et al., 2013). These studies have thus allowed us to make considerable advances in defining genes associated with metabolism per se. That said, even for primary metabolism, there remain many essential proteins for which the corresponding gene has not yet been identified. This is particularly problematic for transport proteins, of which estimates based on entirely different approaches suggest that a total of 6500 membrane transporters exist in Arabidopsis (Schwacke et al., 2003), while 700 intracellular transporters are required merely to maintain the primary metabolic network of the same species (Mintz-Oron et al., 2012). Recent articles concerning chloroplast-, peroxisomal-, vacuolar-, ER-, and plasma membrane-transport all indicate a gradual increase in the functional elucidation of all types of transport proteins (Liu and Bush, 2006; Palmieri et al., 2011; Rieder and Neuhaus, 2011; Weber and Linka, 2011; Martinoia et al., 2012; Hoffmann et al., 2013). However, the number of identified transporter proteins falls well short of either of the predicted numbers given above, i.e., there are a vast number of putative transport proteins but the vast majority have either only homology-based annotations or no functional characterization whatsoever. The recent review by Schroeder et al. (2013) reiterates the importance of transporters in metabolic engineering strategies and as such defines the identification of the permeome as of clear strategical importance in sustaining crop productivity. As one approach toward this goal, in this mini-review we detail (i) the potential of co-expression as a stand-alone approach for aiding in the definition of metabolite transporters and (ii) how other phenotypic data can be integrated with that of gene expression in order to enhance chances of successful gene annotation.
Identification of Transporters in the Model Plant Arabidopsis
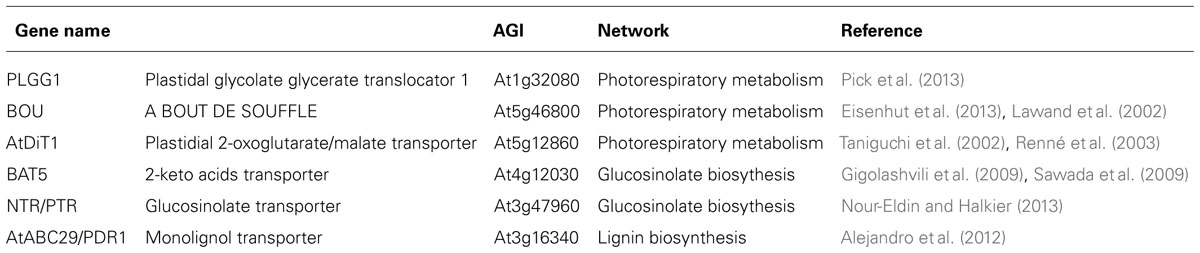
As mentioned above bioinformatics strategies based either on features in protein amino acid sequences or on transport steps required to allow a functional subcellular metabolism have enable us to set an upper limit to the number of metabolite transporters in the plant cell. Despite this considerable research effort is warranted to elucidate the function of these carriers. That said, amongst many important breakthroughs in transporter identification, critical advances have been made both in the cloning of the first transporters of the core photorespiratory pathway (Bordych et al., 2013; Pick et al., 2013) and amino acid metabolism (Liu and Bush, 2006) as well as the initial characterization of a glucosinolate transporter (Gigolashvili et al., 2009; Sawada et al., 2009), epicatechin conjugates transporter (Marinova et al., 2007), and a lignol transporter (Alejandro et al., 2012). In this section we will detail the role of co-expression studies in these discoveries (Table 1).
Transporters involved in photorespiration
The use of co-expression analysis with regard to the identification of photorespiratory transporters has recently been expertly reviewed (Bordych et al., 2013) so we will only cover it briefly here. In their analysis the gene PLGG1 (At1g32080) was ranked as a highly promising candidate transporter and plgg1-1 knockout plants develop chlorotic regions along the leaf lamina when grown under ambient air (NC) conditions while the transporter was recently characterized as the plastidic glycerate/glycolate transporter (Pick et al., 2013). Similarly, the A BOUT DE SOUFFLE (BOU) protein was successfully identified as a transporter involved in shuttling intermediates in the photorespiratory C2 cycle (Lawand et al., 2002; Eisenhut et al., 2013). Bou knockout plants were demonstrated to suffer in ambient air, but grow much like the wild-type when kept under high CO2 conditions. Moreover, the glycine level was greatly increased in comparison to that of wild-type plants, while mitochondrial glycine degradation is strongly reduced in the mutant. Although the specific substrate transported via BOU has not been identified, results collated to date seem to suggest it is likely to be a glycine decarboxylase co-factor (Bordych et al., 2013). A third candidate, the plastidial 2-oxoglutarate (2-OG)/malate transporter (AtDiT1) was found in a co-expression analysis approach and its sequence homology with DiT2.1 (AtpDCT1; Taniguchi et al., 2002; Renné et al., 2003). The function of this gene was subsequently confirmed by phenotypic analysis of the gene knockout mutant (dit1 mutant) which was shown to suffer under normal growth conditions, and displayed retarded development, small leaf size, frequently emerging shoots, and a decrease in chlorophyll content (Kinoshita et al., 2011). AtDiT1 provides the chloroplast with the 2-OG utilized by Ferredoxin-dependent glutamate synthase (FD-GOGAT) in the chloroplast and constructs a double-transporter system together with the AtpDCT1 protein. Thus this protein participates, albeit one step removed, in the export of synthesized glutamate and re-fixation of ammonium ions as the result of the photorespiratory cycle (Schneidereit et al., 2006; Kinoshita et al., 2010). Despite the success of these three examples the function of the other genes highlighted in this photorespiratory co-expression study remain to be confirmed.
Bile acid transporter family
The plastidic bile acid transporter 5 (BAT5) was associated with glucosinolate metabolism on the basis of its co-expression with known genes of glucosinolate metabolism (Gigolashvili et al., 2009; Sawada et al., 2009). This was importantly confirmed by the fact that loss of function and reduced expression of BAT5 resulted in considerably decreased glucosinolate levels (Gigolashvili et al., 2009; Sawada et al., 2009). However, sodium-coupled transport activity of recombinant BAT5 has yet to be demonstrated. Recently, glucosinolate transport to seeds was characterized as being carried out by At3g47960, a member of the nitrate/peptide (NTR/PTR) transporter family, in an approach independent of co-expression analysis (Nour-Eldin and Halkier, 2013). Returning to the BAT, using a targeted variation on the co-expression theme, another member of this family – namely BAT1 – was putatively identified (and subsequently confirmed), as a plastidial sodium-dependent pyruvate transporter (Furumoto et al., 2011). In this study the authors used comparative transcriptome analyses between a C3 plant species, Flaveria pringlei, and the closely related C4 plant species F. trinervia and F. bidentis to identify three novel C4 species abundant genes predicted to encode chloroplast membrane proteins. Unlike C3 plant species, which only contain a sodium-dependent pyruvate transporter, both sodium-dependent and sodium-independent pyruvate transport have been reported in a range of C4 species (Aoki et al., 1992). Given this fact Furumoto et al. (2011) used their cross-species analyses to search for the gene encoding the sodium-dependent pyruvate transporter using the following criteria; (i) given its essential role in C4 photosynthesis it should be expressed at considerably higher levels in C4 than C3 plants and (ii) that its expression should be low in plants of the proton-dependent C4 plant species but equivalent in plants displaying sodium-dependent pyruvate transport. Wider comparative transcriptomics allowed the exclusion of one of the three candidate genes. Crucially functional analysis revealed BAT1, on the basis of its efficient import of pyruvate and physiological characterization of Arabidopsis mutant, to be the plastid sodium-dependent pyruvate carrier (Furumoto et al., 2011). In this study the authors were further able to pinpoint BAT1 as functioning in C4 and in the methyl erythritol phosphate pathway in C3 plants. The search for the mitochondrial pyruvate transporter is, however, ongoing.
Lignin transporters
In order to identify genes involved in monolignol transport, Alejandro et al. (2012) performed a co-expression network analysis with the ABCG transporter subfamily (previously called WBCs and PDRs) of Arabidopsis using the ATTED-II database (http://atted.jp/). Given that members of the ABCG subfamily have been shown to transport a broad range of fatty acids and terpenoids they wondered whether this class could also be implicated in the transport of phenolic compounds. The results revealed that AtABCG29/PDR1, a member of the full-size ABCG subfamily, exhibited a high co-expression ratio with three genes of the phenylpropanoid biosynthesis pathway, which is involved in the synthesis of lignin and flavonoids. The well-correlated genes correspond to two 4-coumarate coenzyme A (CoA) ligases (4CL2 and 4CL5), which convert hydroxycinnamic acids into hydroxycinnamoyl CoA esters, and one caffeoyl CoA-O-methyltransferase catalyzing the conversion of caffeoyl CoA into feruloyl CoA. Moreover, seven further genes related to phenylpropanoid biosynthesis are co-expressed with AtABCG29, albeit with lower co-expression ratios. In concordance with these results Ehlting et al. (2005) also reported that AtABCG29 showed an expression pattern in primary stems consistent with that of monolignol biosynthetic genes and increased lignin content. Subsequent characterization of AtABCG29 revealed that yeasts expressing this transporter exhibited increased tolerance to p-coumaryl by means of excreting this monolignol whilst AtABCG29 deficient mutants revealed that they contained less lignin as well as modifications to secondary metabolites underlining the importance of p-coumaryl alcohol levels in the cytosol (Alejandro et al., 2012). Similarly, a targeted co-expression analyses looking for transporters which were specifically highly expressed in the phloem was used alongside metabolome analyses to uncover that ABCG9, ABCG11, and ABCG14 are involved in lipid/sterol homeostasis regulation (Le Hir et al., 2013).
As stated above a vast number of transport proteins remain uncharacterized and physiologically important transporters such as the mitochondrial pyruvate and folate transporters as well as practically all amino acid transporters remain to be molecularly characterized. The examples presented here suggest that the co-expression approach will have utility in identifying genes encoding transporters for specific metabolites.
Use of co-expression analysis in pinpointing process-related transporters
With the exception of the identification of the photorespiratory transporters described above most uses of co-expression we have described thus far have related to the identification of (metabolite) specific transporters, however, the utility of the approach goes far beyond this application. The photorespiratory transporters are the best example to date of taking a broader approach, however, several further studies have followed this route albeit not to such a conclusive end. Three examples of this come from our own work wherein we looked at (i) genes co-expressed on dark induced senescence (Araújo et al., 2010; Araújo et al., 2011), (ii) genes co-expressed following exposure to high levels of several light species including UV-B irradiance (Tohge et al., 2011), and (iii) genes co-expressed with barley tonoplast proteins (Tohge et al., 2011). The first two approaches identified nine transporters and nine transport related proteins as putative membrane transporters involved in senescence and the UV-B responsive phenolic secondary metabolism, respectively. The former study exhibited considerable overlap in targets to co-expression and cis associated regulatory element analysis of mitochondrially associated proteins following imposition of a broad range of mitochondrial stresses (Holt et al., 2006), providing further support for the correctness of the putative functional assignment which we suggested. The latter study was, however, slightly more complicated in that it formed clusters on the basis of already identified tonoplast proteins but gave suggested functions including transport of phenylpropanoids (Multidrug resistant type transporter and H+ dependent transporter) and mugineic acid (ABC transporter and transport related protein which is in the gene family of glutathione S-transferase). It is important to note, however, that these candidate genes are yet to be validated by functional analysis.
Conclusion and Outlook
Recent years have seen impressive advances in our understanding of transport protein function, however, many gaps remain (Weber and Linka, 2011; Rolland et al., 2012; Sweetlove and Fernie, 2013). While the co-expression approach has been used effectively for transport function predictions being, at least partially, responsible for many, of the discoveries reviewed in this article it probably remains an underexploited tool. The layering of datasets beyond those at the transcriptional level (Tohge and Fernie, 2012), alongside more sophisticated cross-species comparisons such as that illustrated in the Furomoto study will ultimately likely be more tractable in asking specific pathway or process based questions. That said the recent characterization of the plant ammonium transceptor De Michele et al. (2013) suggests, at least in theory, that such approaches may ultimately also prove powerful in linking transporters to signal transduction cascades and the processes which they control.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research activity of Takayuki Tohge is supported by the European Commission’s Directorate-General for Research within the 7th Framework Program (FP7/2007-2013) under grant agreements 270089 (MULTIBIOPRO), and supported by the Alexander von Humboldt Foundation. Funding from the Max-Planck-Society (to Takayuki Tohge and Alisdair R. Fernie) is also gratefully acknowledged.
References
Alejandro, S., Lee, Y., Tohge, T., Sudre, D., Osorio, S., Park, J., et al. (2012). AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 10, 1207–1212. doi: 10.1016/j.cub.2012.04.064
Altman, R. B., and Raychaudhuri, S. (2001). Whole-genome expression analysis: challenges beyond clustering. Curr. Opin. Struct. Biol. 11, 340–347. doi: 10.1016/s0959-440x(00)00212-8
Aoki, N., Ohnishi, J., and Kanai, R. (1992). Two different mechanisms for transport of pyruvate into mesophyll chloroplasts of c-4 plants-a comparative-study. Plant Cell Physiol. 33, 805–809.
Araújo, W. L., Ishizaki, K., Nunes-Nesi, A., Larson, T. R., Tohge, T., Krahnert, I., et al. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22, 1549–1563. doi: 10.1105/tpc.110.075630
Araújo, W. L., Tohge, T., Ishizaki, K., Leaver, C. J., and Fernie, A. R. (2011). Protein degradation – an alternative respiratory substrate for stressed plants. Trends Plant Sci. 16, 489–498. doi: 10.1016/j.tplants.2011.05.008
Bordych, C., Eisenhut, M., Pick, T. R., Kuelahoglu, C., and Weber, A. P. M. (2013). Co-expression analysis as tool for the discovery of transport proteins in photorespiration. Plant Biol. 15, 686–693. doi: 10.1111/plb.12027
Bringmann, M., Li, E., Sampathkumar, A., Kocabek, T., Hauser, M.-T., and Persson, S. (2012). POM-POM2/cellulose synthase interacting1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 24, 163–177. doi: 10.1105/tpc.111.093575
Brown, D. M., Zeef, L. A. H., Ellis, J., Goodacre, R., and Turner, S. R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281–2295. doi: 10.1105/tpc.105.031542
Cocuron, J.-C., Lerouxel, O., Drakakaki, G., Alonso, A. P., Liepman, A. H., Keegstra, K., et al. (2007). A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc. Natl. Acad. Sci. U.S.A. 104, 8550–8555. doi: 10.1073/pnas.0703133104
De Michele, R., Ast, C., Loqué, D., Ho, C.-H., Andrade, S. L., Lanquar, V., et al. (2013). Fluorescent sensors reporting the activity of ammonium transceptors in live cells. eLIFE 2:e00800. doi: 10.7554/eLife.00800
Ehlting, J., Mattheus, N., Aeschliman, D. S., Li, E. Y., Hamberger, B., Cullis, I. F., et al. (2005). Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 42, 618–640. doi: 10.1111/j.1365-313X.2005.02403.x
Eisen, M. B., Spellman, P. T., Brown, P. O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95, 14863–14868. doi: 10.1073/pnas.95.25.14863
Eisenhut, M., Planchais, S., Cabassa, C., Guivarc’h, A., Justin, A.-M., Taconnat, L., et al. (2013). Arabidopsis A BOUT DE SOUFFLE is a putative mitochondrial transporter involved in photorespiratory metabolism and is required for meristem growth at ambient CO2 levels. Plant J. 73, 836–849. doi: 10.1111/tpj.12082
Fribourg, S., Romier, C., Werten, S., Gangloff, Y. G., Poterszman, A., and Moras, D. (2001). Dissecting the interaction network of multiprotein complexes by pairwise coexpression of subunits in E. coli. J. Mol. Biol. 306, 363–373. doi: 10.1006/jmbi.2000.4376
Furumoto, T., Yamaguchi, T., Ohshima-Ichie, Y., Nakamura, M., Tsuchida-Iwata, Y., Shimamura, M., et al. (2011). A plastidial sodium-dependent pyruvate transporter. Nature 476, 472–475. doi: 10.1038/nature10250
Gasch, A. P., and Eisen, M. B. (2002). Exploring the conditional coregulation of yeast gene expression through fuzzy k-means clustering. Genome Biol. 3, 1–22. doi: 10.1186/gb-2002-3-11-research0059
Getz, G., Levine, E., and Domany, E. (2000). Coupled two-way clustering analysis of gene microarray data. Proc. Natl. Acad. Sci. U.S.A. 97, 12079–12084. doi: 10.1073/pnas.210134797
Gigolashvili, T., Yatusevich, R., Rollwitz, I., Humphry, M., Gershenzon, J., and Flugge, U. I. (2009). The Plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. Plant Cell 21 1813–1829. doi: 10.1105/tpc.109.066399
Goda, H., Sasaki, E., Akiyama, K., Maruyama-Nakashita, A., Nakabayashi, K., Li, W., et al. (2008). The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 55, 526–542. doi: 10.1111/j.1365-313X.2008.03510.x
Hirai, M. Y., Sugiyama, K., Sawada, Y., Tohge, T., Obayashi, T., Suzuki, A., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 6478–6483. doi: 10.1073/pnas.0611629104
Hoffmann, C., Plocharski, B., Haferkamp, I., Leroch, M., Ewald, R., Bauwe, H., et al. (2013). From endoplasmic reticulum to mitochondria: absence of the Arabidopsis ATP antiporter endoplasmic reticulum adenylate transporter1 perturbs photorespiration. Plant Cell 25, 2647–2660. doi: 10.1105/tpc.113.113605
Holt K. E., Millar A. H., and Whelan J. (2006) ModuleFinder and CoReg: alternative tools for linking gene expression modules with promoter sequences motifs to uncover gene regulation mechanisms in plants. Plant Methods. 2:8. doi: 10.1186/1746-4811-2-8
Hwang, S., Rhee, S. Y., Marcotte, E. M., and Lee, I. (2011). Systematic prediction of gene function in Arabidopsis thaliana using a probabilistic functional gene network. Nat. Protoc. 6, 1429–1442. doi: 10.1038/nprot.2011.372
Katari, M. S., Nowicki, S. D., Aceituno, F. F., Nero, D., Kelfer, J., Thompson, L. P., et al. (2010). VirtualPlant: a software platform to support systems biology research. Plant Physiol. 152, 500–515. doi: 10.1104/pp.109.147025
Kinoshita, H., Nagasaki, J., Yoshikawa, N., Yamamoto, A., Takito, S., Kawasaki, M., et al. (2010). The chloroplastic 2-oxoglutarate/malate transporter has dual function as the malate valve and in carbon/nitrogen metabolism. Plant J. 65, 15–26. doi: 10.1111/j.1365-313X.2010.04397.x
Kinoshita H., Nagasaki J., Yoshikawa N., Yamamoto A., Takito S., Kawasaki M., et al. (2011). The chloroplastic 2-oxoglutarate/malate transporter has dual function as the malate valve and in carbon/nitrogen metabolism. Plant J. 65. 15–26. doi: 10.1111/j.1365-313X.2010.04397.x
Lawand, S., Dorne, A. J., Long, D., Coupland, G., Mache, R., and Carol, P. (2002). Arabidopsis a bout de souffle, which is homologous with mammalian carnitine acyl carrier, is required for postembryonic growth in the light. Plant Cell 14, 2161–2173. doi: 10.1105/Tpc.002485
Le Hir, R., Sorin, C., Chakraborti, D., Moritz, T., Schaller, H., Tellier, F., et al. (2013). ABCG9, ABCG11 and ABCG14 ABC transporters are required for vascular development in Arabidopsis. Plant J. 76, 811–24. doi: 10.1111/tpj.12334
Lee, H. K., Hsu, A. K., Sajdak, J., Qin, J., and Pavlidis, P. (2004). Coexpression analysis of human genes across many microarray data sets. Genome Res. 14, 1085–1094. doi: 10.1101/gr.1910904
Li, K. C., Liu, C. T., Sun, W., Yuan, S. S., and Yu, T. W. (2004). A system for enhancing genome-wide coexpression dynamics study. Proc. Natl. Acad. Sci. U.S.A. 101, 15561–15566. doi: 10.1073/pnas.0402962101
Liu, X., and Bush, D. R. (2006). Expression and transcriptional regulation of amino acid transporters in plants. Amino Acids 30, 113–120. doi: 10.1007/s00726-005-0248-z
Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K. A., et al. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. doi: 10.1038/82521
Mao, D. Q., Luo, Y., Zhang, J. H., and Zhu, J. (2005). A new strategy of cooperativity of biclustering and hierarchical clustering: A case of analyzing yeast genomic microarray datasets. Front. Biosci. 10:1619–1627.
Marinova, K., Pourcel, L., Weder, B., Schwarz, M., Barron, D., Routaboul, J. M., et al. (2007). The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19, 2023–2038. doi: 10.1105/tpc.106.046029
Martinoia, E., Meyer, S., De Angeli, A., and Nagy, R. (2012). Vacuolar transporters in their physiological context. Annu. Rev. Plant Biol. 63, 183–213. doi: 10.1146/annurev-arplant-042811-105608
Mintz-Oron, S., Meir, S., Malitsky, S., Ruppin, E., Aharoni, A., and Shlomi, T. (2012). Reconstruction of Arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc. Natl. Acad. Sci. U.S.A. 109, 339–344. doi: 10.1073/pnas.1100358109
Mutwil, M., Obro, J., Willats, W. G. T., and Persson, S. (2008). GeneCAT - novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res. 36, W320–W326. doi: 10.1093/nar/gkn292
Nour-Eldin, H. H., and Halkier, B. A. (2013). The emerging field of transport engineering of plant specialized metabolites. Curr. Opin. Biotechnol. 24, 263–270. doi: 10.1016/j.copbio.2012.09.006
Obayashi, T., Hayashi, S., Saeki, M., Ohta, H., and Kinoshita, K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37, D987–D991. doi: 10.1093/nar/gkn807
Obayashi, T., Nishida, K., Kasahara, K., and Kinoshita, K. (2011). ATTED-II Updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 52, 213–219. doi: 10.1093/pcp/pcq203
Okazaki, Y., Shimojima, M., Sawada, Y., Toyooka, K., Narisawa, T., Mochida, K., et al. (2009). A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 21, 892–909. doi: 10.1105/tpc.108.063925
Palmieri, F., Pierri, C. L., De Grassi, A., Nunes-Nesi, A., and Fernie, A. R. (2011). Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J. 66, 161–181. doi: 10.1111/j.1365-313X.2011.04516.x
Persson, S., Wei, H. R., Milne, J., Page, G. P., and Somerville, C. R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. U.S.A. 102, 8633–8638. doi: 10.1073/pnas.0503392102
Pick, T. R., Braeutigam, A., Schulz, M. A., Obata, T., Fernie, A. R., and Weber, A. P. M. (2013). PLGG1, a plastidic glycolate glycerate transporter, is required for photorespiration and defines a unique class of metabolite transporters. Proc. Natl. Acad. Sci. U.S.A. 110, 3185–3190. doi: 10.1073/pnas.1215142110
Prieto, C., Risueno, A., Fontanillo, C., and De Las Rivas, J. (2008). Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. PLoS ONE 3:e3911. doi: 10.1371/journal.pone.0003911
Raychaudhuri, S., Sutphin, P. D., Chang, J. T., and Altman, R. B. (2001). Basic microarray analysis: grouping and feature reduction. Trends Biotechnol. 19, 189–193. doi: 10.1016/s0167-7799(01)01599-2
Renné, P., Dressen, U., Hebbeker, U., Hille, D., Flügge, U. I., Westhoff, P. et al. (2003). The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. Plant J. 35, 316–331. doi: 10.1046/j.1365-313X.2003.01806.x
Rieder, B., and Neuhaus, H. E. (2011). Identification of an Arabidopsis plasma membrane-located ATP transporter important for anther development. Plant Cell 23, 1932–1944. doi: 10.1105/tpc.111.084574
Rolland, N., Curien, G., Finazzi, G., Kuntz, M., Marechal, E., Matringe, M., et al. (2012). The Biosynthetic capacities of the plastids and integration between cytoplasmic and chloroplast processes. Annu. Rev. Genet. 46, 233–264. doi: 10.1146/annurev-genet-110410-132544
Ruprecht, C., and Persson, S. (2012). Co-expression of cell-wall related genes: new tools and insights. Front. Plant Sci. 3:1–7. doi: 10.3389/fpls.2012.00083
Saito, K., Yonekura-Sakakibara, K., Nakabayashi, R., Higashi, Y., Yamazaki, M., Tohge, T., et al. (2013). The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol. Biochem. 72, 21–34. doi: 10.1016/j.plaphy.2013.02.001
Sakurai, N., Ara, T., Ogata, Y., Sano, R., Ohno, T., Sugiyama, K., et al. (2011). KaPPA-View4: a metabolic pathway database for representation and analysis of correlation networks of gene co-expression and metabolite co-accumulation and omics data. Nucleic Acids Res. 39, D677-D684. doi: 10.1093/nar/gkq989
Sanchez-Rodriguez, C., Bauer, S., Hematy, K., Saxe, F., Ibanez, A. B., Vodermaier, V., et al. (2012). Chitinase-like1/pom-pom1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 24, 589–607. doi: 10.1105/tpc.111.094672
Sawada, Y., Toyooka, K., Kuwahara, A., Sakata, A., Nagano, M., Saito, K., et al. (2009). Arabidopsis bile acid: sodium symporter family protein 5 is involved in methionine-derived glucosinolate biosynthesis. Plant Cell Physiol. 50, 1579–1586. doi: 10.1093/pcp/pcp110
Schaffer, R., Landgraf, J., Accerbi, M., Simon, V., Larson, M., and Wisman, E. (2001). Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13, 113–123. doi: 10.1105/tpc.13.1.113
Schneidereit, J., Hausler, R. E., Fiene, G., Kaiser, W. M., and Weber, A. P. M. (2006). Antisense repression reveals a crucial role of the plastidic 2-oxoglutarate/malate translocator DiT1 at the interface between carbon and nitrogen metabolism. Plant J. 45, 206–224. doi: 10.1111/j.1365-313X.2005.02594.x
Schroeder, J. I., Delhaize, E., Frommer, W. B., Guerinot, M. L., Harrison, M. J., Herrera-Estrella, L., et al. (2013). Using membrane transporters to improve crops for sustainable food production. Nature 497, 60–66. doi: 10.1038/nature11909
Schwacke, R., Schneider, A., van der Graaff, E., Fischer, K., Catoni, E., Desimone, M., et al. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26. doi: 10.1104/pp.011577
Stitt, M. (2013). Systems-integration of plant metabolism: means, motive and opportunity. Curr. Opin. Plant Biol. 16, 381–388. doi: 10.1016/j.pbi.2013.02.012
Sweetlove, L. J., and Fernie, A. R. (2013). Spatial organization of metabolism within the plant cell. Annu. Rev. Plant Biol. 64, 723–746. doi: 10.1146/annurev-arplant-050312-120233
Taniguchi, M., Taniguchi, Y., Kawasaki, M., Takeda, S., Kato, T., Sato, S., et al. (2002). Identifying and characterizing plastidic 2-oxoglutarate/malate and dicarboxylate transporters in Arabidopsis thaliana. Plant Cell Physiol. 43, 706–717. doi: 10.1093/pcp/pcf109
Tohge, T., and Fernie, A. R. (2012). Co-expression and co-responses: within and beyond transcription. Front. Plant Sci. 3:248–248. doi: 10.3389/fpls.2012.00248
Tohge, T., Kusano, M., Fukushima, A., Saito, K., and Fernie, A. (2011). Transcriptional and metabolic programs following exposure of plants to UV-B irradiation. Plant Signal. Behav. 6, 1987–1992. doi: 10.4161/psb.6.12.18240
Tohge, T., Ramos, M. S., Nunes-Nesi, A., Mutwil, M., Giavalisco, P., Steinhauser, D., et al. (2011). Toward the storage metabolome: profiling the barley vacuole. Plant Physiol. 157, 1469–1482. doi: 10.1104/pp.111.185710
Tohge, T., Yonekura-Sakakibara, K., Niida, R., Watanabe-Takahashi, A., and Saito, K. (2007). Phytochemical genomics in Arabidopsis thaliana: A case study for functional identification of flavonoid biosynthesis genes. Pure Appl. Chem. 79, 811–823. doi: 10.1351/pac200779040811
Toufighi, K., Brady, S. M., Austin, R., Ly, E., and Provart, N. J. (2005). The botany array resource: e-Northerns, expression angling, and promoter analyses. Plant J. 43, 153–163. doi: 10.1111/j.1365-313X.2005.02437.x
Usadel, B., Obayashi, T., Mutwil, M., Giorgi, F. M., Bassel, G. W., Tanimoto, M., et al. (2009). Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ. 32, 1633–1651. doi: 10.1111/j.1365-3040.2009.02040.x
Vanholme, R., Cesarino, I., Rataj, K., Xiao, Y., Sundin, L., Goeminne, G., et al. (2013). Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341, 1103–1106. doi: 10.1126/science.1241602
Voehringer, D. W., Hirschberg, D. L., Xiao, J., Lu, Q., Roederer, M., Lock, C., et al. (2000). Gene microarray identification of redox and mitochondrial elements that control resistance or sensitivity to apoptosis. Proc. Natl. Acad. Sci. U.S.A. 97, 2680–2685. doi: 10.1073/pnas.97.6.2680
Weber, A. P. M., and Linka, N. (2011). Connecting the plastid: transporters of the plastid envelope and their role in linking plastidial with cytosolic metabolism. Annu. Rev. Plant Biol. 62, 53–77. doi: 10.1146/annurev-arplant-042110-103903
Yonekura-Sakakibara, K., Fukushima, A., Nakabayashi, R., Hanada, K., Matsuda, F., Sugawara, S., et al. (2012). Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 69, 154–167. doi: 10.1111/j.1365-313X.2011.04779.x
Yonekura-Sakakibara, K., Tohge, T., Matsuda, F., Nakabayashi, R., Takayama, H., Niida, R., et al. (2008). Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20, 2160–2176. doi: 10.1105/tpc.108.058040
Yonekura-Sakakibara, K., Tohge, T., Niida, R., and Saito, K. (2007). Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 282, 14932–14941. doi: 10.1074/jbc.M611498200
Zhang, L., King, O., Wong, S., Goldberg, D., Tong, A., Lesage, G., et al. (2005). Motifs, themes and thematic maps of an integrated Saccharomyces cerevisiae interaction network. J. Biol. 4, 6. doi: 10.1186/jbiol23
Keywords: informatic identification, transporters, plant metabolism, co-expression network
Citation: Tohge T and Fernie AR (2014) Lignin, mitochondrial family, and photorespiratory transporter classification as case studies in using co-expression, co-response, and protein locations to aid in identifying transport functions. Front. Plant Sci. 5:75. doi: 10.3389/fpls.2014.00075
Received: 03 November 2013; Accepted: 17 February 2014;
Published online: 17 March 2014.
Edited by:
Ilka Haferkamp, Technische Universität Kaiserslautern, GermanyReviewed by:
Norbert Rolland, Centre National de la Recherche Scientifique, FranceKate Warpeha, University of Illinois at Chicago, USA
Copyright © 2014 Tohge and Fernie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takayuki Tohge, Department 1 (Willmitzer), Central Metabolism, Max Planck Institute for Plant Physiology, Am Mühlenberg 1, 14476 Potsdam, Germany e-mail:dG9oZ2VAbXBpbXAtZ29sbS5tcGcuZGU=
 Takayuki Tohge
Takayuki Tohge Alisdair R. Fernie
Alisdair R. Fernie