
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 13 February 2014
Sec. Plant Genetics and Genomics
Volume 5 - 2014 | https://doi.org/10.3389/fpls.2014.00042
This article is part of the Research Topic Reversible Ubiquitylation in Plant Biology View all 13 articles
 Vincent Duplan1,2
Vincent Duplan1,2 Susana Rivas1,2*
Susana Rivas1,2*Reversible protein ubiquitination plays a crucial role during the regulation of plant immune signaling. E3 ubiquitin (Ub)-ligase enzymes, which are classified into different families depending on their structural and functional features, confer the specificity of substrate and are the best characterized components of the ubiquitination cascade. E3 Ub-ligases of different families have been shown to be involved in all steps of plant immune responses. Indeed, they have been involved in the first steps of pathogen perception, as they appear to modulate perception of pathogen-associated molecular patterns by pattern-recognition receptors at the plasma membrane and to regulate the accumulation of nucleotide-binding leucine-rich repeat-type intracellular immune receptors. In addition, E3 Ub-ligase proteins are also involved in the regulation of the signaling responses downstream of pathogen perception through targeting vesicle trafficking components or nuclear transcription factors, for instance. Finally, we also discuss the case of microbial effector proteins that are able to target host E3 Ub-ligases, or to act themselves as E3 Ub-ligases, in their attempt to subvert the host proteasome to promote disease.
Reversible protein conjugation with ubiquitin (Ub), or ubiquitination, is a key regulatory mechanism that controls a variety of cellular processes in eukaryotic cells, including DNA repair, gene transcription, protein activation or receptor trafficking, although the best characterized function of Ub involves selective protein degradation through the 26S proteasome (Vierstra, 2009). Ub becomes covalently attached to lysine residues of intracellular targets via an ATP-dependent reaction cascade that involves the sequential action of three enzymes: E1 (Ub-activating), E2 (Ub-conjugating), and E3 (Ub-ligase). The importance of the Ub-related pathway is underlined by the finding that the Arabidopsis genome encodes more than 1600 genes (> 6% of the total genome) involved in Ub/26S proteasome system (UPS)-related functions. Most of these genes (> 1400) encode putative E3 Ub-ligases (Mazzucotelli et al., 2006). E3 proteins are classified into four main subfamilies depending on their structural features and mechanism of action: HECT (Homologous to E6-associated protein C-Terminus), RING (Really Interesting New Gene), U-Box and CRL (Cullin-RING Ligases; Vierstra, 2009). HECT proteins form an Ub-E3 intermediate before transfer of Ub to the substrate (Downes et al., 2003). RING and U-box proteins are structurally related single polypeptides that, respectively, use zinc chelation and hydrogen bonds/salt bridges to transfer Ub from the Ub-E2 intermediate to the target (Stone et al., 2005; Yee and Goring, 2009). Ub-ligases containing a RING domain can act independently or as part of a multisubunit CRL complex such as the SCF (Skp1, Cullin, F-box)-type ligase. In this complex, substrate recognition is provided by the F-box protein, whereas the RING protein binds to the E2 (Hua and Vierstra, 2011). By contrast, some RING-type Ub-ligases act independently and determine substrate specificity allowing the interaction between the E2 and the target protein by tethering them in close proximity (Vierstra, 2009).
Plants have developed a multi-layered defense system to ensure their survival in a microbe-rich environment. A first line of pathogen detection is activated after recognition of highly conserved PAMPs/MAMPs (pathogen-/microbe-associated molecular patterns) by specific plant PRRs (pattern-recognition receptors), leading to a form of basal resistance called PTI (PAMP-triggered immunity; Jones and Dangl, 2006). Thriving pathogens evolved to secrete virulence effectors that inactivate crucial PTI regulators thereby counteracting plant defenses. In turn, plants gained the ability to recognize these effectors through resistance (R) proteins, for the most part intracellular NB-LRR (nucleotide-binding-leucine-rich repeat) immune sensors, that lead to a more efficient form of resistance called ETI (effector-triggered immunity; Jones and Dangl, 2006). This specific resistance is frequently associated to development of the hypersensitive response (HR), a form of programmed cell death at the infection site that prevents pathogen spreading (Coll et al., 2011).
In plants, the UPS pathway, and more particularly E3 Ub-ligase proteins, have been shown to be involved in responses to a variety of stimuli (Vierstra, 2009; Robert-Seilaniantz et al., 2011). Since the finding that the SCF complex-interacting protein SGT1 (Suppressor of G2 allele of skp1) is an essential component of R gene-triggered disease resistance provided a first connection between the UPS and plant immune signaling (Azevedo et al., 2002), evidence that E3 Ub-ligase proteins act as regulators of plant immunity has increasingly accumulated (Trujillo and Shirasu, 2010; Marino et al., 2012). Indeed, modulation of the expression of E3 Ub-ligase-encoding genes has been reported following elicitor treatment or inoculation with different pathogens. Moreover, misregulation of E3 Ub-ligase gene expression, using overexpressing, RNA interference (RNAi) and/or mutant lines, results in modulation of plant defense responses following pathogen inoculation [reviewed in (Marino et al., 2012)]. Therefore, it has become increasingly evident that plant E3 Ub-ligase proteins play important roles in the regulation of immune signaling, although the proteins targeted by Ub-ligases are only known in a limited number of cases, and our current knowledge of the involved molecular mechanisms is thus only partial. Here we review positive and negative roles played by E3 Ub-ligases during the regulation of various steps of plant immunity, from pathogen recognition to downstream signaling during both PTI and ETI responses. Due to space limitations, we focus on recent reports about E3 Ub-ligases for which a target protein has been identified during the plant response to bacterial or fungal pathogens, since these particular examples provide insight into the cellular processes involved in regulation of immune signaling. For an overview on UPS-related pathways in response to viral infection we refer the reader to a recent review (Alcaide-Loridan and Jupin, 2012). We also discuss the case of microbial effectors that, to promote disease, either target host E3 Ub-ligases or act as Ub-ligases inside plant cells (Figure 1).
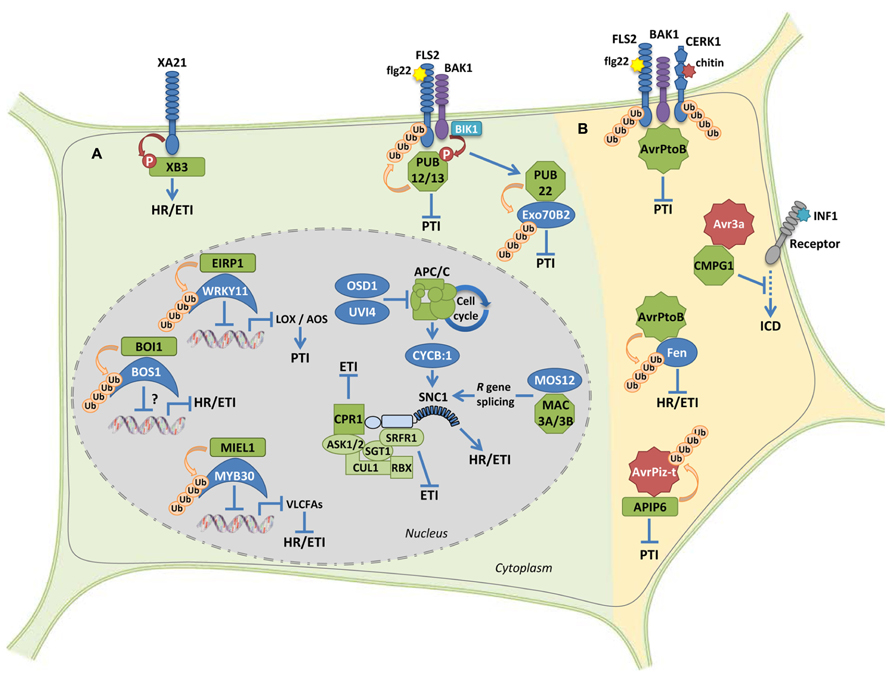
FIGURE 1. Ub-ligase proteins during the regulation of plant immune signaling. (A) Examples of positive and negative regulatory roles on plant immunity by host E3 Ub-ligases. The RING-type protein XB3 is required for the accumulation of the receptor protein XA21 and promotes downstream HR and defense responses. U-box proteins PUB12 and PUB13 attenuate flg22-mediated PTI signaling. The U-box protein PUB22 targets the exocyst complex subunit Exo70B2, which leads to attenuation of PTI signaling. Accumulation of the intracellular immune receptor SNC1 appears to be controlled by the SCF complex through the interaction with both the F-box CPR1 and the SGT1-interacting protein SRFR1. Modulation of SNC1 levels by the multisubunit cell cycle-related E3 ligase APC/C or the U-box proteins MAC3A and MAC3B provide additional molecular mechanisms to control immunity. The interaction between the RING-type E3 Ub-ligase EIRP1, and the TF WRK11 results in suppressed transcriptional activation of WRKY11 target genes AOS and LOX1, thereby enhancing PTI. The RING E3 ligase MIEL1 interacts with the MYB TF MYB30, which results in weakened transcriptional activation of VLCFA-related genes and therefore suppressed HR and defense responses. The RING protein BOI1 interacts with and ubiquitinates the MYB TF BOS1 that is required for pathogen resistance. However, the effect of BOI1 on BOS1 accumulation and transcriptional activation remains unknown. (B) Microbial effector proteins acting as or interacting with E3 ligase proteins in the host. The bacterial effector AvrPtoB is able to suppress both PTI and ETI signaling though its respective interaction with FLS2/CERK1 PRRs and the cytoplasmic kinase Fen. The oomycete effector AVR3a suppresses INF1-induced cell death by interacting with and stabilizing the host U-box E3 ligase CMPG1. The fungal effector protein AvrPiz-t targets the RING E3 ligase APIP6, which in turn is able to ubiquitinate AvrPiz-t, resulting in suppressed PTI. RING-, F-box- and U-box-type E3 Ub-ligases are, respectively, represented by rectangles, squares, and octagons. All proteins displaying E3 ligase activity are represented in green. Effectors are represented by stars. See the text for details.
Several E3 ligase proteins have been identified as modulators of the first steps of pathogen recognition by plant cells, as they appear to be able to target both PRR and NB-LRR proteins in order to prevent unnecessary activation of defense signaling. In rice (Oryza sativa), the RING-type E3 Ub-ligase XB3 interacts with the receptor-like kinase (RLK) protein XA21, which confers resistance to bacterial blight caused by Xanthomonas oryzae pv. oryzae (Xoo; Wang et al., 2006). XB3 has been shown to be required for XA21 accumulation and XA21-mediated resistance to Xoo, suggesting that it most likely targets a protein that modulates accumulation of XA21 (Wang et al., 2006). XA21 is able to phosphorylate XB3 but the molecular mechanism underlining activation of defense responses by XB3 remains to be elucidated. Overexpression of members of the XB3 family from rice, Arabidopsis and citrus in Nicotiana benthamiana induces cell death and this effect is dependent on XB3 catalytic activity, suggesting an evolutionarily conserved role for the XB3 protein family in regulating plant programmed cell death (Huang et al., 2013).
In Arabidopsis, the U-box E3 ligases Plant U-Box12 (PUB12) and PUB13 have been involved in attenuation of PTI responses triggered by perception of flagellin, or its active peptide derivative flg22, by the PRRs Flagellin Sensing2 (FLS2) and its co-receptor BAK1. In response to flg22, PUB12 and PUB13 form a BAK1-dependent complex with FLS2 and are able to polyubiquitinate FLS2, but not BAK1 (Lu et al., 2011). BAK1 phosphorylates PUB12 and PUB13 and this phosphorylation is enhanced by flg22 and by the FLS2/BAK1-associated kinase BIK1 (Lu et al., 2011). flg22-dependent signaling is enhanced in pub12 or pub13 mutant plants and, in agreement with PUB12 and PUB13 promoting FLS2 degradation, pub12 pub13 double mutant plants displayed increased resistance to bacterial infection. These data are consistent with the fact that FLS2 undergoes flg22-induced endocytosis and subsequent degradation (Robatzek et al., 2006; Gohre et al., 2008; Beck et al., 2012; Choi et al., 2013). However, whether stabilization of FLS2 in pub12pub13 plants reflects FLS2 accumulation at the plasma membrane or within an endosomal compartment remains to be determined. A recent report showed that FLS2 degradation occurs in a flg22 time- and dose-dependent manner, which may play a significant role in turning over ligand-occupied FLS2, but the role of PUB12 and PUB13 in this process was not determined in this study (Smith et al., 2013).
Similar to PRR proteins, intracellular NB-LRR immune receptors are also targeted by E3 Ub-ligases, which appear to control R protein accumulation at multiple levels. First, in Arabidopsis, the F-box motif E3 Ub-ligase CPR1 interacts with and down-regulates the accumulation of the NB-LRR R proteins Suppressor of npr1-1 Constitutive1 (SNC1) and Resistant to P. syringae2 (RPS2), resulting in attenuation of immune signaling (Cheng et al., 2011; Gou et al., 2012). Second, accumulation of SNC1 and RPS4, an additional NB-LRR immune receptor, are also negatively regulated by Suppressor of rps4-RLD1 (SRFR1), a tetratricopeptide repeat protein (Kim et al., 2010; Li et al., 2010). Since (i) SNC1 levels increased in CPR1-overexpressing plants treated with the proteasome inhibitor MG132 (Gou et al., 2012); (ii) SRFR1 interacts with SGT1; and (iii) increased SNC1 and RPS4 accumulation was also observed in sgt1 mutant plants (Li et al., 2010), stability of SNC1 and RPS4 is likely regulated by SRFR1 through SGT1 interaction with the SCF complex, revealing an additional molecular mechanism to prevent autoimmnunity. Third, mutation of MOS12 (modifier of snc1-12), that encodes an Arabidopsis Arg-rich protein homologous to human cyclin L, resulted in altered SNC1 and RPS4 splicing patterns and protein levels (Xu et al., 2012). Interestingly, MOS12 interacts with the nuclear U-Box E3 ligases MAC3A and MAC3B, which are required for full R protein-mediated resistance, suggesting that MOS12, MAC3A, and MAC3B contribute to the fine-tuning of R gene expression, in a process that appears to be critical for directing appropriate defense outputs (Monaghan et al., 2009; Xu et al., 2012). Finally, a recent report showed an intriguing link between cell cycle regulation and defense signaling (Bao et al., 2013). Omission of the Second Division (OSD1) and its homolog UV-B-Insensitive 4 (UVI4) are two negative regulators of the multisubunit E3 Ub-ligase APC/C (anaphase-promoting complex/cyclosome) that regulates cell cycle progression in Arabidopsis. Overexpression of either OSD1 or UVI4 leads to downregulation of APC/C activity, overaccumulation of the APC/C degradation target CYCB1;1, upregulation of several R genes, including SNC1, and spontaneous cell death and enhanced disease resistance to virulent bacteria (Bao et al., 2013). These data provide further evidence of the intricate control exerted on immune receptor levels in order to regulate defense activation.
Pathogen perception by PRRs stimulates a cascade of signaling events including changes in ion fluxes across the plasma membrane, production of reactive oxygen species (ROS), induction of mitogen-activated protein kinases (MAPKs), modulation of host gene transcription and callose deposition at the plant cell wall. Amplitude and duration of these signaling responses must be tightly regulated to ensure an appropriate response. In addition to their role in internalization or degradation of receptors to attenuate downstream signaling, E3 Ub-ligase proteins also regulate the accumulation of plant components involved in defense-related signaling.
Similar to PUB12 and PUB13, PUB22 acts in concert with PUB23 and PUB24 to negatively regulate PTI responses in Arabidopsis (Trujillo et al., 2008). Following elicitation with various PAMPs, pub22/pub23/pub24 triple mutants displayed enhanced early signaling responses, indicating that these three PUB proteins target components involved in defense signaling triggered by different PRRs (Trujillo et al., 2008). Indeed, the exocyst complex subunit Exo70B2 that is involved in vesicle tethering during exocytosis, has been identified as a cellular target of PUB22 (Stegmann et al., 2012). PUB22 is stabilized in response to flg22 treatment (potentially by inhibition of its autocatalytic ubiquitination activity), leading to Exo70B2 ubiquitination and proteasomal degradation. Exo70B2 is required for both immediate (ROS production, MAPK activation) and later responses (PTI marker gene expression, root growth inhibition) triggered by several PAMPs, indicative of a role in signaling. Indeed, exo70B2 mutant plants displayed enhanced susceptibility to pathogens (Stegmann et al., 2012). Together, these data suggest a mechanism by which Exo70B2 levels are regulated by quick changes in PUB22 turnover in response to PAMPs and identify a first component of vesicle trafficking required for regulation of plant PTI signaling. In view of these data, it has been proposed that Exo70B2 may contribute to recycling of plasma membrane proteins involved in PAMP-triggered signaling, including NADPH oxidases, ion channels or RLKs such as FLS2. Exo70B2 degradation by PUB22 would thus attenuate the recycling pathway redirecting positive signaling components into the vacuolar degradation pathway and downregulating signaling (Stegmann et al., 2012). As previously discussed, PUB12/PUB13-mediated ubiquitination of FLS2 is expected to modulate its intracellular trafficking (Lu et al., 2011). Since degradation of integral membrane proteins is mediated by the vacuole, signal attenuation is probably simultaneously regulated at various levels of vesicle trafficking.
In Chinese wild grapevine (Vitis pseudoreticulata), EIRP1 is an active E3 Ub-ligase whose RING domain is necessary for its activity and also mediates the interaction with the WRKY nuclear transcription factor (TF) VpWRKY11 (Yu et al., 2013). Similar to EIRP1, expression of VpWRKY11 was rapidly induced following fungal infection. VpWRKY11 activated the expression of AOS (Allene Oxide Synthase) and LOX2 (Lipoxygenase2), two JA-responsive genes that function as negative regulators of basal resistance in Arabidopsis (Journot-Catalino et al., 2006). Additionally, in agreement with the observation that co-expression with EIRP1 results in proteasomal degradation of VpWRKY11, AOS and LOX2 expression was, respectively, repressed and induced in EIRP1-overexpressing and RNAi plants. Moreover, EIRP1 overexpression in Arabidopsis conferred enhanced resistance to fungal and bacterial pathogens, which correlated with reduced expression of WRKY11, AOS, and LOX2 (Yu et al., 2013). Together, these data identify a RING-type Ub-ligase that plays a positive role in activation of resistance by targeting a TF that acts as a negative regulator of plant defenses.
In contrast, the Arabidopsis RING-type Ub-ligase MIEL1 acts as a negative regulator of plant resistance (Marino et al., 2013). Indeed, MIEL1 interacts with the MYB TF MYB30, which activates plant defense responses by up-regulating the expression of genes involved in the production of very long chain fatty acids (VLCFAs; Raffaele et al., 2008). The MYB30-MIEL1 nuclear interaction leads to MYB30 proteasomal degradation, reduced expression of VLCFA-related MYB30 target gene expression and, therefore, attenuation of plant immune responses (Marino et al., 2013). MIEL1 expression is rapidly repressed in inoculated cells, suggesting that (i) in the absence of the pathogen, MIEL1 may negatively regulate plant defense activation through degradation of MYB30 and that (ii) after pathogen inoculation, repression of MIEL1 expression may release MYB30 negative regulation, triggering defense (Marino et al., 2013).
BOI1 is an additional nuclear RING-type Ub-ligase that interacts with and ubiquitinates the MYB TF BOS1, which confers resistance to several pathogens in Arabidopsis (Mengiste et al., 2003; Luo et al., 2010). This finding suggests that BOS1 may be a target of BOI1. However, no effect of BOI1 on BOS1 transcriptional activity has been reported and both bos1 mutant and BOI1 RNAi Arabidopsis plants (in which the BOS1 protein is expected to accumulate) display enhanced susceptibility to fungal infection (Luo et al., 2010). Therefore, whether BOI1 is able to directly regulate BOS1 protein accumulation remains to be determined.
The important role played by E3 Ub-ligases during the establishment of plant immune responses to pathogen attack is highlighted by the discovery of microbial effector proteins that evolved the ability to interfere with these host UPS components to promote disease. For example, the AvrPiz-t effector from the rice blast fungus Magnaporthe oryzae is translocated into rice cells, where it is able to mediate suppression of PAMP-induced ROS production, inducing susceptibility to M. oryzae (Park et al., 2012). AvrPiz-t appears to inhibit the Ub-ligase activity of APIP6, a rice RING-type Ub-ligase that is also able to ubiquitinate AvrPiz-t in vitro (Park et al., 2012). Interestingly, AvrPiz-t and APIP6 are both degraded when transiently coexpressed in N. benthamiana. Since APIP6 positively regulates flg22-induced ROS generation, induction of defense-related gene expression, and rice resistance to M. oryzae, targeting of APIP6 by AvrPiz-t results in suppression of rice PTI responses (Park et al., 2012).
The effector AVR3a from the oomycete Phytophthora infestans prevents development of cell death induced by P. infestans elicitin INF1. The finding that AVR3a targets and stabilizes the U-box-type Ub-ligase CMPG1 revealed the molecular mechanism behind AVR3a negative regulation of ICD (INF1-triggered Cell Death; Bos et al., 2010). CMPG1 Ub-ligase activity is required for ICD as well as for cell death following elicitor perception at the plasma membrane (Gonzalez-Lamothe et al., 2006; Gilroy et al., 2011). Considering that AVR3a is essential for P. infestans virulence, stabilization of CMPG1 by AVR3a suggests that this effector is able to suppress ICD during the biotrophic phase of infection by modifying CMPG1 activity, impeding normal proteasomal degradation of both CMPG1 and its host targets (Bos et al., 2010).
In addition to microbial effectors that are able to target host E3 Ub-ligase proteins, examples of effectors that present E3 Ub-ligase-related domains have also been reported in a diversity of pathogenic microbes including bacteria, fungi, oomycetes, viruses and nematodes (Marino et al., 2012). The best characterized example of microbial E3 ligases is the AvrPtoB effector from Pseudomonas syringae that presents a C-terminal domain with remarkable structural homology with RING- and U-box-type Ub-ligases (Janjusevic et al., 2006). AvrPtoB is a modular effector able to suppress PTI signaling. Indeed, AvrPtoB N-terminal domain is able to interact with the PRRs FLS2 and the chitin receptor CERK1, whereas its C-terminal Ub-ligase domain mediates PRR proteasomal degradation (Gohre et al., 2008; Gimenez-Ibanez et al., 2009). AvrPtoB is additionally able to target the co-receptor protein BAK1 (Shan et al., 2008) and to interfere with MAPK activation downstream of FLS2 (He et al., 2006), although independently of AvrPtoB Ub-ligase activity. Remarkably, AvrPtoB is also able to suppress ETI signaling. Indeed, AvrPtoB interacts with and mediates proteasomal degradation of Fen, a tomato protein kinase that activates plant immunity in response to P. syringae carrying Ub-ligase deficient forms of AvrPtoB, as well as cell death responses when overexpressed in N. benthamiana (Rosebrock et al., 2007).
Evidence of the involvement of the UPS pathway in the regulation of plant immunity is rapidly mounting. E3 Ub-ligase proteins are the best characterized UPS components playing a role in immune signaling at multiple levels. Nevertheless, based on induction of their expression following elicitation, E1 and E2 enzymes have also been suggested to contribute to plant disease resistance although the molecular mechanisms by which they regulate this process remain poorly characterized (Marino et al., 2012). Interestingly, a recent report provides a first example of the direct involvement of Ub-conjugating proteins in the regulation of plant immunity. In tomato, Fen-interacting protein 3 (Fni3) is a homolog of the Arabidopsis E2 enzyme Ubc13. Through interaction with its cofactor S. lycoperiscum Uev (Suv), which is an inactive Ub-conjugating enzyme variant, Fni3 catalyzes Lys-63-linked ubiquitination, a non-proteolytic regulatory signal (Mural et al., 2013). Fni3 interacts with Fen but does not affect Fen stability. Fni3 Ub-conjugating activity and interaction with Fen are required for cell death triggered by Fen overexpression in N. benthamiana and by several R protein/effector pairs (Mural et al., 2013). Together these results suggest that, in addition to conventional Lys-48-linked ubiquitination that mainly serves as a signal for proteasomal degradation of substrate proteins, other ubiquitination forms are important regulators of plant immune signaling. Consistent with this idea, Lys-63-linked ubiquitination has also been shown to be a target of manipulation by plant pathogens, as the PthA effector from the bacterial pathogen Xanthomonas axonopodis pv. citri is able to target Ubc13 and Uev in citrus (Domingues et al., 2010).
In conclusion, further characterization of additional UPS components as well as of the distinct fates of ubiquitination targets should contribute to dissecting the complex regulation of plant immune signaling by the UPS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Our work is supported by the French Laboratory of Excellence project “TULIP” (ANR-10-LABX-41; ANR-11-IDEX-0002-02).
Alcaide-Loridan, C., and Jupin, I. (2012). Ubiquitin and plant viruses, let’s play together! Plant Physiol. 160, 72–82. doi: 10.1104/pp.112.201905
Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. doi: 10.1126/science.1067554
Bao, Z., Yang, H., and Hua, J. (2013). Perturbation of cell cycle regulation triggers plant immune response via activation of disease resistance genes. Proc. Natl. Acad. Sci. U.S.A. 110, 2407–2412. doi: 10.1073/pnas.1217024110
Beck, M., Zhou, J., Faulkner, C., MacLean, D., and Robatzek, S. (2012). Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24, 4205–4219. doi: 10.1105/tpc.112.100263
Bos, J. I., Armstrong, M. R., Gilroy, E. M., Boevink, P. C., Hein, I., Taylor, R. M., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. U.S.A. 107, 9909–9914. doi: 10.1073/pnas.0914408107
Cheng, Y. T., Li, Y., Huang, S., Huang, Y., Dong, X., Zhang, Y., et al. (2011). Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc. Natl. Acad. Sci. U.S.A. 108, 14694–14699. doi: 10.1073/pnas.1105685108
Choi, S. W., Tamaki, T., Ebine, K., Uemura, T., Ueda, T., and Nakano, A. (2013). RABA members act in distinct steps of subcellular trafficking of the Flagellin Sensing2 receptor. Plant Cell 25, 1174–1187. doi: 10.1105/tpc.112.108803
Coll, N. S., Epple, P., and Dangl, J. L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. doi: 10.1038/cdd.2011.37
Domingues, M. N., De Souza, T. A., Cernadas, R. A., de Oliveira, M. L., Docena, C., Farah, C. S., et al. (2010). The Xanthomonas citri effector protein PthA interacts with citrus proteins involved in nuclear transport, protein folding and ubiquitination associated with DNA repair. Mol. Plant Pathol. 11, 663–675. doi: 10.1111/j.1364-3703.2010.00636.x
Downes, B. P., Stupar, R. M., Gingerich, D. J., and Vierstra, R. D. (2003). The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 35, 729–742. doi: 10.1046/j.1365-313X.2003.01844.x
Gilroy, E. M., Taylor, R. M., Hein, I., Boevink, P., Sadanandom, A., and Birch, P. R. (2011). CMPG1-dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytol. 190, 653–666. doi: 10.1111/j.1469-8137.2011.03643.x
Gimenez-Ibanez, S., Hann, D. R., Ntoukakis, V., Petutschnig, E., Lipka, V., and Rathjen, J. P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423–429. doi: 10.1016/j.cub.2009.01.054
Gohre, V., Spallek, T., Haweker, H., Mersmann, S., Mentzel, T., Boller, T., et al. (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18, 1824–1832. doi: 10.1016/j.cub.2008.10.063
Gonzalez-Lamothe, R., Tsitsigiannis, D. I., Ludwig, A. A., Panicot, M., Shirasu, K., and Jones, J. D. (2006). The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18, 1067–1083. doi: 10.1105/tpc.106.040998
Gou, M., Shi, Z., Zhu, Y., Bao, Z., Wang, G., and Hua, J. (2012). The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J. 69, 411–420. doi: 10.1111/j.1365-313X.2011.04799.x
He, P., Shan, L., Lin, N. C., Martin, G. B., Kemmerling, B., Nurnberger, T., et al. (2006). Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125, 563–575. doi: 10.1016/j.cell.2006.02.047
Hua, Z., and Vierstra, R. D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62, 299–334. doi: 10.1146/annurev-arplant-042809-112256
Huang, X., Liu, X., Chen, X., Snyder, A., and Song, W. Y. (2013). Members of the XB3 family from diverse plant species induce programmed cell death in Nicotiana benthamiana. PLoS ONE 8:e63868. doi: 10.1371/journal.pone.0063868
Janjusevic, R., Abramovitch, R. B., Martin, G. B., and Stebbins, C. E. (2006). A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311, 222–226. doi: 10.1126/science.1120131
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Journot-Catalino, N., Somssich, I. E., Roby, D., and Kroj, T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18, 3289–3302. doi: 10.1105/tpc.106.044149
Kim, S. H., Gao, F., Bhattacharjee, S., Adiasor, J. A., Nam, J. C., and Gassmann, W. (2010). The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 6:e1001172. doi: 10.1371/journal.ppat.1001172
Li, Y., Li, S., Bi, D., Cheng, Y. T., Li, X., and Zhang, Y. (2010). SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog. 6:e1001111. doi: 10.1371/journal.ppat.1001111
Lu, D., Lin, W., Gao, X., Wu, S., Cheng, C., Avila, J., et al. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332, 1439–1442. doi: 10.1126/science.1204903
Luo, H., Laluk, K., Lai, Z., Veronese, P., Song, F., and Mengiste, T. (2010). The Arabidopsis Botrytis Susceptible1 Interactor defines a subclass of RING E3 ligases that regulate pathogen and stress responses. Plant Physiol. 154, 1766–1782. doi: 10.1104/pp.110.163915
Marino, D., Froidure, S., Canonne, J., Ben Khaled, S., Khafif, M., Pouzet, C., et al. (2013). Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat. Commun. 4, 1476. doi: 10.1038/ncomms2479
Marino, D., Peeters, N., and Rivas, S. (2012). Ubiquitination during plant immune signaling. Plant Physiol. 160, 15–27. doi: 10.1104/pp.112.199281
Mazzucotelli, E., Belloni, S., Marone, D., De Leonardis, A., Guerra, D., Di Fonzo, N., et al. (2006). The E3 ubiquitin ligase gene family in plants: regulation by degradation. Curr. Genomics 7, 509–522. doi: 10.2174/138920206779315728
Mengiste, T., Chen, X., Salmeron, J., and Dietrich, R. (2003). The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15, 2551–2565. doi: 10.1105/tpc.014167
Monaghan, J., Xu, F., Gao, M., Zhao, Q., Palma, K., Long, C., et al. (2009). Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog 5:e1000526. doi: 10.1371/journal.ppat.1000526
Mural, R. V., Liu, Y., Rosebrock, T. R., Brady, J. J., Hamera, S., Connor, R. A., et al. (2013). The tomato Fni3 lysine-63-specific ubiquitin-conjugating enzyme and suv ubiquitin e2 variant positively regulate plant immunity. Plant Cell 25, 3615–3631. doi: 10.1105/tpc.113.117093
Park, C. H., Chen, S., Shirsekar, G., Zhou, B., Khang, C. H., Songkumarn, P., et al. (2012). The Magnaporthe oryzae Effector AvrPiz-t Targets the RING E3 Ubiquitin Ligase APIP6 to Suppress Pathogen-Associated Molecular Pattern-Triggered Immunity in Rice. Plant Cell 24, 4748–4762. doi: 10.1105/tpc.112.105429
Raffaele, S., Vailleau, F., Leger, A., Joubes, J., Miersch, O., Huard, C., et al. (2008). A MYB transcription factor regulates Very-Long-Chain Fatty Acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20, 752–767. doi: 10.1105/tpc.107.054858
Robatzek, S., Chinchilla, D., and Boller, T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20, 537–542. doi: 10.1101/gad.366506
Robert-Seilaniantz, A., Grant, M., and Jones, J. D. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Rosebrock, T. R., Zeng, L., Brady, J. J., Abramovitch, R. B., Xiao, F., and Martin, G. B. (2007). A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448, 370–374. doi: 10.1038/nature05966
Shan, L., He, P., Li, J., Heese, A., Peck, S. C., Nurnberger, T., et al. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4, 17–27. doi: 10.1016/j.chom.2008.05.017
Smith, J. M., Salamango, D. J., Leslie, M. E., Collins, C. A., and Heese, A. (2013). Sensitivity to flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLS2. Plant Physiol. 164, 440–454. doi: 10.1104/pp.113.229179
Stegmann, M., Anderson, R. G., Ichimura, K., Pecenkova, T., Reuter, P., Zarsky, V., et al. (2012). The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24, 4703–4716. doi: 10.1105/tpc.112.104463
Stone, S. L., Hauksdottir, H., Troy, A., Herschleb, J., Kraft, E., and Callis, J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137, 13–30. doi: 10.1104/pp.104.052423
Trujillo, M., Ichimura, K., Casais, C., and Shirasu, K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18, 1396–1401. doi: 10.1016/j.cub.2008.07.085
Trujillo, M., and Shirasu, K. (2010). Ubiquitination in plant immunity. Curr. Opin. Plant Biol. 13, 402–408. doi: 10.1016/j.pbi.2010.04.002
Vierstra, R. D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397. doi: 10.1038/nrm2688
Wang, Y. S., Pi, L. Y., Chen, X., Chakrabarty, P. K., Jiang, J., De Leon, A. L., et al. (2006). Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18, 3635–3646. doi: 10.1105/tpc.106.046730
Xu, F., Xu, S., Wiermer, M., Zhang, Y., and Li, X. (2012). The cyclin L homolog MOS12 and the MOS4-associated complex are required for proper splicing of plant resistance genes. Plant J. 70, 916–928. doi: 10.1111/j.1365-313X.2012.04906.x
Yee, D., and Goring, D. R. (2009). The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J. Exp. Bot. 60, 1109–1121. doi: 10.1093/jxb/ern369
Yu, Y., Xu, W., Wang, J., Wang, L., Yao, W., Yang, Y., et al. (2013). The Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase Erysiphe necator-induced RING finger protein 1 (EIRP1) activates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol. 200, 834–846. doi: 10.1111/nph.12418
Keywords: E3 ubiquitin-ligase, microbial effector, plant immunity, 26S proteasome, ubiquitination
Citation: Duplan V and Rivas S (2014) E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 5:42. doi: 10.3389/fpls.2014.00042
Received: 13 December 2013; Paper pending published: 14 January 2014;
Accepted: 28 January 2014; Published online: 13 February 2014.
Edited by:
Pascal Genschik, Centre National de la Recherche Scientifique, FranceReviewed by:
Xun Xu, Beijing Genomics Institute, ChinaCopyright © 2014 Duplan and Rivas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susana Rivas, Laboratoire des Interactions Plantes-Microorganismes (LIPM), UMR CNRS/INRA 2594/441, 24 Chemin de Borde Rouge-Auzeville, CS 52627, 31326 Castanet-Tolosan cedex, France e-mail:c3VzYW5hLnJpdmFzQHRvdWxvdXNlLmlucmEuZnI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.