
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 13 November 2013
Sec. Plant Genetics and Genomics
Volume 4 - 2013 | https://doi.org/10.3389/fpls.2013.00454
This article is part of the Research TopicEpigenetic Mechanisms of Plant Responses to the EnvironmentView all 15 articles
Polycomb group (PcG) proteins act in an evolutionarily conserved epigenetic pathway that regulates chromatin structures in plants and animals, repressing many developmentally important genes by modifying histones. PcG proteins can form at least two multiprotein complexes: Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2, respectively). The functions of Arabidopsis thaliana PRCs have been characterized in multiple stages of development and have diverse roles in response to environmental stimuli. Recently, the mechanism that precisely regulates Arabidopsis PcG activity was extensively studied. In this review, we summarize recent discoveries in the regulations of PcG at the three different layers: the recruitment of PRCs to specific target loci, the polyubiquitination and degradation of PRC2, and the antagonism of PRC2 activity by the Trithorax group proteins. Current knowledge indicates that the powerful activity of the PcG pathway is strictly controlled for specific silencing of target genes during plant development and in response to environmental stimuli.
Multicellular eukaryotic organisms develop from a single cell called a zygote, which goes through cell division and differentiation to develop into multiple tissues and organs. Precise control of gene expression under the guidance of developmental cues/signals or environmental stimuli is strictly regulated to ensure proper development of organisms. Plants and animals have evolved to have multiple methods to regulate gene expression, among which epigenetic regulation is essential for correct genome-wide gene expression profiles (Russo et al., 1996; Liu et al., 2010a).
Polycomb group (PcG) proteins, one of the evolutionarily conserved epigenetic pathways, have critical roles in plant and animal development via regulation of gene expression levels (Whitcomb et al., 2007; Molitor and Shen, 2013). In the model plant Arabidopsis thaliana, PcG proteins are incorporated into two multi-protein complexes: Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2, respectively), which both have functions in the epigenetic repression of gene expression via histone modifications (Schatlowski et al., 2008; Molitor and Shen, 2013). The major function of PRC2 is to trimethylate lysine 27 on histone H3 (H3K27me3), while PRC1 recognizes the H3K27me3 marker and mono-ubiquitinates histone H2A (H2Aub; Schatlowski et al., 2008; Molitor and Shen, 2013).
The core component of Arabidopsis PRC2 is the SET-domain H3K27 methyltransferase CURLY LEAF (CLF; Goodrich et al., 1997; Schubert et al., 2006). In addition, SWINGER (SWN) has partially redundant functions with CLF (Chanvivattana et al., 2004), and MEDEA (MEA) functions as a methyltransferase in the endosperm (Grossniklaus et al., 1998). Some other assistant proteins are also required to form PRC2, such as EMBRYONIC FLOWER2 (EMF2; Yoshida et al., 2001). There are about 4,400 gene loci that are modified by H3K27me3 in Arabidopsis seedlings (Zhang et al., 2007a). Such modifications are dynamic and the modification patterns differ among organs and tissues. Genome-wide identification of H3K27me3-modificed loci revealed that this dynamic modification occurs during the shoot apical meristem (SAM)-to-leaf organ formation (Lafos et al., 2011), the embryo-to-seedling phase transition (Bouyer et al., 2011), in leaf-to-callus regeneration (He et al., 2012), and in the endosperm (Weinhofer et al., 2010). Therefore, PRC2-mediated H3K27me3 is precisely controlled during development.
PRC1 components have also been characterized in Arabidopsis. LIKE HETEROCHROMATIN PROTEIN1 (LHP1), also called TERMINAL FLOWER2 (TFL2), can recognize chromatin that is modified by H3K27me3, and its genome-wide binding sites show significant overlap with the H3K27me3 modification (Turck et al., 2007; Zhang et al., 2007b). There are two groups of RING-domain proteins in PRC1: two AtRING1 proteins and three AtBMI1 proteins (Xu and Shen, 2008; Bratzel et al., 2010; Chen et al., 2010). These RING-domain proteins function in the modification of H2Aub (Bratzel et al., 2010). Although the genome-wide identification of H2Aub loci has not been performed, it is possible that the role of PRC1 is also developmentally controlled (Kim et al., 2012).
How PRCs specifically and dynamically recognize their targets during development and in different tissues or organs is a key question for understanding the mechanism that controls the PcG pathway. Currently, there are two types of recruiters found in Arabidopsis: transcription factors and non-coding RNAs (ncRNAs).
Transcription factors usually bind to certain specific cis elements, termed Polycomb response elements (PREs), and recruit PRCs to their specific targets via direct interaction with PRCs (Schwartz and Pirrotta, 2008). AGAMOUS (AG), a MADS-box transcription factor that is essential for establishing floral organ identity and termination of floral meristem (FM), represses WUSCHEL (WUS) expression in FM (Bowman et al., 1989; Lenhard et al., 2001; Lohmann et al., 2001). AG binds to the WUS locus at CArG boxes and then recruits PRC2-mediated H3K27me3 and LHP1 to repress WUS (Liu et al., 2011). Mutation in AG results in the decreased H3K27me3 level and loss of LHP1 binding at the WUS locus. In this case, CArG boxes may serve as the PRE in WUS repression. However, it is not clear whether AG directly or indirectly recruits PcG proteins.
BREVIPEDICELLUS (BP) and KNOTTED-LIKE FROM ARABIDOPSIS THALIANA2 (KNAT2) are members of the KNOX gene family and are expressed in the SAM but are silenced in leaves (Lincoln et al., 1994; Ori et al., 2000; Pautot et al., 2001). The MYB-domain transcription factor ASYMMETRIC LEAVES1 (AS1) and the LOB-domain transcription factor AS2 form a protein complex to repress BP and KNAT2 expression in leaves (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001; Iwakawa et al., 2002; Sun et al., 2002; Lin et al., 2003; Xu et al., 2003). The AS1-AS2 complex targets the cis elements in the promoters of BP and KNAT2 (Guo et al., 2008; Li et al., 2012). Recently, it was shown that AS1-AS2 interacts with PRC2 to recruit it to the BP and KNAT2 loci (Lodha et al., 2013). Mutations in AS1, AS2, and the AS1-AS2-binding sites in BP and KNAT2 promoters all result in decreased H3K27me3 levels at BP and KNAT2 loci and ectopic expression of the two KNOX genes in leaves. In this case, the AS1-AS2-binding sites are likely to serve as PREs in PcG-mediated gene silencing.
Another possible PRE was found in the promoter of LEAFY COTYLEDON2 (LEC2; Berger et al., 2011), a gene involved in embryo development, but which is silenced in the post-embryo stage (Stone et al., 2001). A cis element, called Repressive LEC2 Element (RLE), was identified to recruit PRC2 to trimethylate the LEC2 locus (Berger et al., 2011). RLE is essential for LEC2 repression in the post-embryo stage. PRC2 is unable to trimethylate H3K27 at the LEC2 locus once RLE is mutated, leading to ectopic expression of LEC2 in the post-embryo stage. An RLE-driven reporter gene could be repressed, accompanied by H3K27me3 modification at the transgene locus, suggesting that RLE is necessary and sufficient to recruit PRC2 for histone modification and gene silencing. It seems important in the future to identify the transcription factor that binds RLE and is able to interact with PRC2.
An analysis of cis-regulatory elements in the promoter of FLOWERING LOCUS T (FT), a key gene that controls flowering time (Kardailsky et al., 1999; Kobayashi et al., 1999), also indicated the existence of PRE within the promoter region (Adrian et al., 2010). However, the exact sequence that serves as the PRE and the transcription factor that binds the PRE of the FT promoter have not yet been identified.
The B3 domain proteins VP1/ABI3-LIKE1 (VAL1) and VAL2 are key regulators in the prevention of embryo traits in somatic tissues via repression of embryo specific genes. The loss-of-function double mutant val1 val2 results in somatic embryogenesis, with ectopic expressions of embryo genes at the post-embryo stage (Suzuki et al., 2007). Somatic embryogenesis and ectopic expression of embryo genes were also observed in mutants corresponding to PRC1 components (Bratzel et al., 2010; Chen et al., 2010). A recent study showed that VAL proteins interact with PRC1 and recruit PRC1-mediated H2Aub to initiate repression of the embryonic genes after germination (Yang et al., 2013). The H2Aub modification at the embryo-gene loci is lost in val1 val2 and Atbmi1a Atbmi1b mutants. Therefore, VALs may serve as a recruiter for PRC1.
In Arabidopsis, two ncRNAs, COLD INDUCED LONG ANTISENSE INTRAGENIC RNA (COOLAIR) and COLD ASSISTED INTRONIC NONCODING RNA (COLDAIR; Swiezewski et al., 2009; Heo and Sung, 2011; Ietswaart et al., 2012), regulate FLOWERING LOCUS C (FLC) expression. FLC is a flowering repressor that is essential for vernalization in response to cold treatment (Michaels and Amasino, 1999; Sheldon et al., 1999). COOLAIR is an antisense RNA that is transcribed in response to cold treatment (Swiezewski et al., 2009). COOLAIR is alternatively polyadenylated at the 3′ end, resulting in a proximal poly(A) site or a distal ploy(A) site (Liu et al., 2010b). The proximal poly(A) site stimulates the activity of FLD, a homolog of the human LYSINE SPECIFIC DEMETHYLASE1 (LSD1; Sanda and Amasino, 1996; Liu et al., 2007), to reduce the H3K4me2 level at the FLC locus, leading to a transition from an active chromatin state to a repressive state (Liu et al., 2010b). The reduction of H3K4me2 might benefit the H3K27me3 modification; thus, COOLAIR acts as an indirect recruiter of PRC2. However, how FLD is activated using the proximal site of COOLAIR remains unknown.
COLDAIR is a sense ncRNA that contains a 5′ cap, but no poly(A) tail (Heo and Sung, 2011). COLDAIR is induced by low temperature, and its transcription reaches a maximum level after 20 days of cold treatment, which is about 10 days later than COOLAIR. COLDAIR can directly interact with the CXC domain of the core PRC2 components CLF and SWN. In COLDAIR knockdown plants, CLF is not properly recruited to FLC, resulting in insufficient H3K27me3 modification at the FLC locus, consistent with the late flowering phenotype of these plants. Therefore, COLDAIR serves as a direct recruiter for PRC2 in Arabidopsis.
The Arabidopsis PRC2 is post-translationally regulated by the F-box protein UPWARD CURLY LEAF1 (UCL1; Jeong et al., 2011). UCL1 is a component of the SCF E3 ubiquitin ligase complex, which has a role in the degradation of proteins via polyubiquitination and the 26S proteasome pathway (Vierstra, 2003). Both activation tagging dominant mutant ucl1-D, and plants overexpressing UCL1 under the control of the CaMV 35S promoter resulted in phenotypes that were similar to the clf mutant, with ectopic expression of some typical PRC2 targets, whose loci exhibit decreased H3K27me3 levels (Jeong et al., 2011). These results indicated that UCL1 inhibits PRC2 activity. Additionally, UCL1 directly interacts with CLF, but not with MEA, and overexpression of UCL1 causes a reduced CLF protein level. Therefore, UCL1 was thought to polyubiquitinate CLF and to degrade CLF through the 26S proteasome pathway.
Interestingly, UCL1 is expressed in the endosperm, where CLF and MEA transcripts are detectable. However, overexpression of CLF in the endosperm causes mea-like phenotypes, suggesting that the CLF protein level is strictly controlled in the endosperm. Jeong et al. suggested that UCL1 functions in the endosperm to specifically degrade CLF, and therefore to prevent CLF from competing with MEA during the formation of PRC2 (Jeong et al., 2011).
Trithorax group (TrxG) proteins were first identified in Drosophila and function in antagonism of PcG (Schuettengruber et al., 2011). The first identified plant TrxG protein was Arabidopsis HOMOLOG OF TRITHORAX1 (ATX1), which encodes a SET-domain protein that specifically trimethylates H3K4 (Alvarez-Venegas et al., 2003; Pien et al., 2008; Saleh et al., 2008). Two other SET-domain proteins were shown to participate in genome-wide control of histone methylation: SDG8 is responsible for H3K36me2/3 (Zhao et al., 2005; Xu et al., 2008), and SDG2 acts for H3K4me2/3 (Berr et al., 2010; Guo et al., 2010). The histone modifications H3K4me2/3 and H3K36me2/3 function in activation of gene expression, showing the opposite role of PcG-mediated H3K27me3 (Zhang et al., 2007a, 2009; Roudier et al., 2011). However, the molecular mechanism whereby H3K4me2/3 and H3K36me2/3 antagonize H3K27me3 is not clear in Arabidopsis. A recent study showed that the 35S enhancer decreases the H3K27me3 level but increases the H3K4me3 level at the insert locus (Chen et al., 2013). This suggests that cis enhancer sequences may play a role to recruit TrxG proteins to restrict the PRC2-mediated H3K27me3.
ULTRAPETALA1 (ULT1), a plant-specific SAND domain protein, is another TrxG protein (Carles and Fletcher, 2009). Overexpression of ULT1 resulted in phenotypes similar to those of clf, and caused derepression of PcG target genes with decreased H3K27me3 levels. ULT1 is able to directly bind to ATX1 and guides it to the target genes.
RELATIVE OF EARLY FLOWERING6 (REF6) is a JmjC-domain protein (Noh et al., 2004) that specifically demethylates H3K27me2/3 in vivo and in vitro (Lu et al., 2011). Overexpression of REF6 caused an lhp1-like phenotype (Lu et al., 2011). In addition, mutation of REF6 could partially rescue the clf phenotype, and resulted in hypermethylation of H3K27me3 for hundreds of genes. Characterizationof REF6 revealed that the H3K27me3 modification is a reversible process, and such regulation is critical to balance PcG activity (Lu et al., 2011).
The ATP-dependent chromatin remodeling factor PICKLE (PKL) is involved in antagonism of PcG (Aichinger et al., 2009, 2011), and the pkl mutant partially rescued developmental defects of the clf mutant, including the up-curled leaves and early flowering. Conversely, pkl enhanced the defects of the clf swn double mutant in the root. In roots, PKL activates the expression of EMF2, CLF, andSWN, and this could explain why pkl enhances clf swn. In addition, PKL is very important for the activity of the root apical meristem (RAM). In the pkl mutant, a number of genes that respond to the activity of RAM were silenced because of the increased H3K27me3 level on these genes (Aichinger et al., 2011). However, several other studies suggested that PKL has a role in the promotion of PRC2-mediated H3K27me3 and that the PKL protein associates with H3K27me3-enriched loci (Zhang et al., 2008, 2012). Further studies to clarify the genome-wide function of PKL will improve our understanding of how the chromatin remodeling pathway coordinates with the PcG pathway to regulate their downstream targets. Overall, these studies revealed that chromatin structure, which is organized by chromatin remodeling factors using energy from ATP hydrolysis, is essential for regulation of histone modification states.
Controls for the PcG action at different levels are essential to ensure its specificity and activity (see the model in Figure 1). However, many questions still remain to be answered. First, although several candidates that recruit PRC1 and PRC2 to the target genes have been characterized, the common rule of the recruitment remains unclear. For example, how do these transcription factors bind PRCs? If PRCs are recruited generally by transcription factors, common domains or motifs might exist in the PcG proteins and transcription factors, similar to the binding of the TOPLESS corepressor and transcription factors (Causier et al., 2012). On the other hand, if ncRNAs are the common factors that associate with PRCs, the RNA-binding domain of PcG proteins may contain various ncRNAs. Detection of these ncRNAs might be helpful in studying the specificity of PcG. In addition, how the ncRNAs are transcribed in response to developmental or environmental cues is not clear. Recently, it was proposed that the chromatin structure and its modification status also affect the recruitment of PRCs to target loci in the “chromatin sampling” model (Klose et al., 2013), and it was also shown that PRC2 binding sites contain GAGA factor binding sequences (Deng et al., 2013).
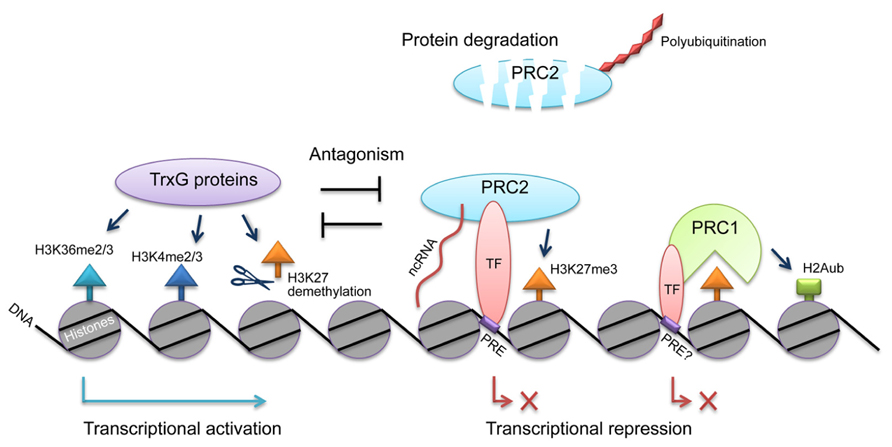
FIGURE 1. Regulation of PcG actions by transcription factors, ncRNAs, polyubiquitination, and TrxG proteins. PcG complexes PRC1 and PRC2 can be recruited to specific loci by transcription factors (TF) and ncRNAs. The PRC2 component CLF is post-translationally regulated by polyubiquitination, leading to protein degradation. PcG and TrxG antagonize each other to reach a dynamic regulation of gene expression.
Second, the mechanism guiding the balance between PcG and TrxG activities in regulating developmental processes in plants is currently unknown. In animals, several transcription factors can regulate both PcG and TrxG activity, providing a dynamic and reversible epigenetic state (Schwartz and Pirrotta, 2008). Therefore, comparison of the regulation mechanisms of PcG and TrxG may be helpful to understand how a balance is established between PcG-mediated gene silencing and TrxG-mediated gene activation.
Third, some novel proteins were identified in plant PRCs, for example, the CLF interacting protein BLISTER (BLI; Schatlowski et al., 2010) and LHP1-INTERACTING FACTOR2 (LIF2; Latrasse et al., 2011). It will be of interest to test whether these proteins act as regulators of PRCs. LIF2 is an RNA-binding protein, suggesting that PRC1 may also be subject to regulation from RNAs.
Finally, recent studies revealed that PRC1 and PRC2 physically interact in Arabidopsis (Derkacheva et al., 2013), indicating the possibility that the two PcG complexes have a crosstalk in silencing common targets. Further studies on mechanisms that regulate PcG activity would be helpful to understand the epigenetic regulation of plant development.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors apologize for references not cited because of space limitations. This work was supported by grants from the National Basic Research Program of China (973 Program, 2012CB910503).
Adrian, J., Farrona, S., Reimer, J. J., Albani, M. C., Coupland, G., and Turck, F. (2010). cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22, 1425–1440. doi: 10.1105/tpc.110.074682
Aichinger, E., Villar, C. B., Di Mambro, R., Sabatini, S., and Kohler, C. (2011). The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23, 1047–1060. doi: 10.1105/tpc.111.083352
Aichinger, E., Villar, C. B., Farrona, S., Reyes, J. C., Hennig, L., and Kohler, C. (2009). CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 5:e1000605. doi: 10.1371/journal.pgen.1000605
Alvarez-Venegas, R., Pien, S., Sadder, M., Witmer, X., Grossniklaus, U., and Avramova, Z. (2003). ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 13, 627–637. doi: 10.1016/S0960-9822(03)00243-4
Berger, N., Dubreucq, B., Roudier, F., Dubos, C., and Lepiniec, L. (2011). Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell 23, 4065–4078. doi: 10.1105/tpc.111.087866
Berr, A., Mccallum, E. J., Menard, R., Meyer, D., Fuchs, J., Dong, A., et al. (2010). Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell 22, 3232–3248. doi: 10.1105/tpc.110.079962
Bouyer, D., Roudier, F., Heese, M., Andersen, E. D., Gey, D., Nowack, M. K., et al. (2011). Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 7:e1002014. doi: 10.1371/journal.pgen.1002014
Bowman, J. L., Smyth, D. R., and Meyerowitz, E. M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. doi: 10.1105/tpc.1.1.37
Bratzel, F., Lopez-Torrejon, G., Koch, M., Del Pozo, J. C., and Calonje, M. (2010). Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20, 1853–1859. doi: 10.1016/j.cub.2010.09.046
Byrne, M. E., Barley, R., Curtis, M., Arroyo, J. M., Dunham, M., Hudson, A., et al. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. doi: 10.1038/35050091
Carles, C. C., and Fletcher, J. C. (2009). The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev. 23, 2723–2728. doi: 10.1101/gad.1812609
Causier, B., Ashworth, M., Guo, W., and Davies, B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158, 423–438. doi: 10.1104/pp.111.186999
Chanvivattana, Y., Bishopp, A., Schubert, D., Stock, C., Moon, Y. H., Sung, Z. R., et al. (2004). Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131, 5263–5276. doi: 10.1242/dev.01400
Chen, X., Huang, H., and Xu, L. (2013). The CaMV 35S enhancer has a function to change the histone modification state at insertion loci in Arabidopsis thaliana. J. Plant Res. In press. doi: 10.1007/s10265-013-0580-4
Chen, D., Molitor, A., Liu, C., and Shen, W. H. (2010). The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 20, 1332–1344. doi: 10.1038/cr.2010.151
Deng, W., Buzas, D. M., Ying, H., Robertson, M., Taylor, J., Peacock, W. J., et al. (2013). Arabidopsis Polycomb Repressive Complex 2 binding sites contain putative GAGA factor binding motifs within coding regions of genes. BMC Genomics 14:593. doi: 10.1186/1471-2164-14-593
Derkacheva, M., Steinbach, Y., Wildhaber, T., Mozgova, I., Mahrez, W., Nanni, P., et al. (2013). Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 32, 2073–2085. doi: 10.1038/emboj.2013.145
Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E. M., and Coupland, G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51. doi: 10.1038/386044a0
Grossniklaus, U., Vielle-Calzada, J. P., Hoeppner, M. A., and Gagliano, W. B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280, 446–450. doi: 10.1126/science.280.5362.446
Guo, M., Thomas, J., Collins, G., and Timmermans, M. C. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20, 48–58. doi: 10.1105/tpc.107.056127
Guo, L., Yu, Y., Law, J. A., and Zhang, X. (2010). SET DOMAIN GROUP2 is the major histone H3 lysine 4 trimethyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 18557–18562. doi: 10.1073/pnas.1010478107
He, C., Chen, X., Huang, H., and Xu, L. (2012). Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet. 8:e1002911. doi: 10.1371/journal.pgen.1002911
Heo, J. B., and Sung, S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. doi: 10.1126/science.1197349
Ietswaart, R., Wu, Z., and Dean, C. (2012). Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 28, 445–453. doi: 10.1016/j.tig.2012.06.002
Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., et al. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43, 467–478. doi: 10.1093/pcp/pcf077
Jeong, C. W., Roh, H., Dang, T. V., Choi, Y. D., Fischer, R. L., Lee, J. S., et al. (2011). An E3 ligase complex regulates SET-domain polycomb group protein activity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108, 8036–8041. doi: 10.1073/pnas.1104232108
Kardailsky, I., Shukla, V. K., Ahn, J. H., Dagenais, N., Christensen, S. K., Nguyen, J. T., et al. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. doi: 10.1126/science.286.5446.1962
Kim, S. Y., Lee, J., Eshed-Williams, L., Zilberman, D., and Sung, Z. R. (2012). EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet. 8:e1002512. doi: 10.1371/journal.pgen.1002512
Klose, R. J., Cooper, S., Farcas, A. M., Blackledge, N. P., and Brockdorff, N. (2013). Chromatin sampling-an emerging perspective on targeting polycomb repressor proteins. PLoS Genet. 9:e1003717. doi: 10.1371/journal.pgen.1003717
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. doi: 10.1126/science.286.5446.1960
Lafos, M., Kroll, P., Hohenstatt, M. L., Thorpe, F. L., Clarenz, O., and Schubert, D. (2011). Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 7:e1002040. doi: 10.1371/journal.pgen.1002040
Latrasse, D., Germann, S., Houba-Herin, N., Dubois, E., Bui-Prodhomme, D., Hourcade, D., et al. (2011). Control of flowering and cell fate by LIF2, an RNA binding partner of the polycomb complex component LHP1. PLoS ONE 6:e16592. doi: 10.1371/journal.pone.0016592
Lenhard, M., Bohnert, A., Jurgens, G., and Laux, T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between Wuschel and Agamous. Cell 105, 805–814. doi: 10.1016/S0092-8674(01)00390-7
Li, Z., Li, B., Shen, W.-H., Huang, H., and Dong, A. (2012). TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J. 71, 99–107. doi: 10.1111/j.1365-313X.2012.04973.x
Lin, W. C., Shuai, B., and Springer, P. S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15, 2241–2252. doi: 10.1105/tpc.014969
Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6, 1859–1876. doi: 10.1105/tpc.6.12.1859
Liu, X., Kim, Y. J., Muller, R., Yumul, R. E., Liu, C., Pan, Y., et al. (2011). AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing Wuschel through recruitment of polycomb group proteins. Plant Cell 23, 3654–3670. doi: 10.1105/tpc.111.091538
Liu, C., Lu, F., Cui, X., and Cao, X. (2010a). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61, 395–420. doi: 10.1146/annurev.arplant.043008.091939
Liu, F., Marquardt, S., Lister, C., Swiezewski, S., and Dean, C. (2010b). Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327, 94–97. doi: 10.1126/science.1180278
Liu, F., Quesada, V., Crevillen, P., Baurle, I., Swiezewski, S., and Dean, C. (2007). The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol. Cell 28, 398–407. doi: 10.1016/j.molcel.2007.10.018
Lodha, M., Marco, C. F., and Timmermans, M. C. (2013). The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev. 27, 596–601. doi: 10.1101/gad.211425.112
Lohmann, J. U., Hong, R. L., Hobe, M., Busch, M. A., Parcy, F., Simon, R., et al. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803. doi: 10.1016/S0092-8674(01)00384-1
Lu, F., Cui, X., Zhang, S., Jenuwein, T., and Cao, X. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43, 715–719. doi: 10.1038/ng.854
Michaels, S. D., and Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. doi: 10.1105/tpc.11.5.949
Molitor, A., and Shen, W. H. (2013). The polycomb complex PRC1: composition and function in plants. J. Genet. Genomics 40, 231–238. doi: 10.1016/j.jgg.2012.12.005
Noh, B., Lee, S. H., Kim, H. J., Yi, G., Shin, E. A., Lee, M., et al. (2004). Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16, 2601–2613. doi: 10.1105/tpc.104.025353
Ori, N., Eshed, Y., Chuck, G., Bowman, J. L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532.
Pautot, V., Dockx, J., Hamant, O., Kronenberger, J., Grandjean, O., Jublot, D., et al. (2001). KNAT2: evidence for a link between knotted-like genes and carpel development. Plant Cell 13, 1719–1734.
Pien, S., Fleury, D., Mylne, J. S., Crevillen, P., Inze, D., Avramova, Z., et al. (2008). ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20, 580–588. doi: 10.1105/tpc.108.058172
Roudier, F., Ahmed, I., Berard, C., Sarazin, A., Mary-Huard, T., Cortijo, S., et al. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30, 1928–1938. doi: 10.1038/emboj. 2011.103
Russo, V. E. A., Martienssen, R. A., and Riggs, A. D. (1996). Epigenetic Mechanisms of Gene Regulation. Woodbury: Cold Spring Harbor Laboratory Press.
Saleh, A., Alvarez-Venegas, R., Yilmaz, M., Le, O., Hou, G., Sadder, M., et al. (2008). The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell 20, 568–579. doi: 10.1105/tpc.107.056614
Sanda, S. L., and Amasino, R. M. (1996). Ecotype-specific expression of a flowering mutant phenotype in Arabidopsis thaliana. Plant Physiol. 111, 641–644.
Schatlowski, N., Creasey, K., Goodrich, J., and Schubert, D. (2008). Keeping plants in shape: polycomb-group genes and histone methylation. Semin. Cell Dev. Biol. 19, 547–553. doi: 10.1016/j.semcdb.2008.07.019
Schatlowski, N., Stahl, Y., Hohenstatt, M. L., Goodrich, J., and Schubert, D. (2010). The CURLY LEAF interacting protein BLISTER controls expression of polycomb-group target genes and cellular differentiation of Arabidopsis thaliana. Plant Cell 22, 2291–2305. doi: 10.1105/tpc.109.073403
Schubert, D., Primavesi, L., Bishopp, A., Roberts, G., Doonan, J., Jenuwein, T., et al. (2006). Silencing by plant polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25, 4638–4649. doi: 10.1038/sj.emboj.7601311
Schuettengruber, B., Martinez, A. M., Iovino, N., and Cavalli, G. (2011). Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12, 799–814. doi: 10.1038/nrm3230
Schwartz, Y. B., and Pirrotta, V. (2008). Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 20, 266–273. doi: 10.1016/j.ceb.2008.03.002
Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128, 1771–1783.
Sheldon, C. C., Burn, J. E., Perez, P. P., Metzger, J., Edwards, J. A., Peacock, W. J., et al. (1999). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458.
Stone, S. L., Kwong, L. W., Yee, K. M., Pelletier, J., Lepiniec, L., Fischer, R. L., et al. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U.S.A. 98, 11806–11811. doi: 10.1073/pnas.201413498
Sun, Y., Zhou, Q., Zhang, W., Fu, Y., and Huang, H. (2002). ASYMMETRIC LEAVES1, an Arabidopsis gene that is involved in the control of cell differentiation in leaves. Planta 214, 694–702. doi: 10.1007/s004250100673
Suzuki, M., Wang, H. H., and McCarty, D. R. (2007). Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 143, 902–911. doi: 10.1104/pp.106.092320
Swiezewski, S., Liu, F., Magusin, A., and Dean, C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799–802. doi: 10.1038/nature08618
Turck, F., Roudier, F., Farrona, S., Martin-Magniette, M. L., Guillaume, E., Buisine, N., et al. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3:e86. doi: 10.1371/journal.pgen.0030086
Vierstra, R. D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8, 135–142. doi: 10.1016/S1360-1385(03)00014-1
Weinhofer, I., Hehenberger, E., Roszak, P., Hennig, L., and Kohler, C. (2010). H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 6:e1001152. doi: 10.1371/journal.pgen.1001152
Whitcomb, S. J., Basu, A., Allis, C. D., and Bernstein, E. (2007). Polycomb Group proteins: an evolutionary perspective. Trends Genet. 23, 494–502. doi: 10.1016/j.tig.2007.08.006
Xu, L., and Shen, W.-H. (2008). Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr. Biol. 18, 1966–1971. doi: 10.1016/j.cub.2008.11.019
Xu, L., Xu, Y., Dong, A., Sun, Y., Pi, L., Xu, Y., et al. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130, 4097–4107. doi: 10.1242/dev.00622
Xu, L., Zhao, Z., Dong, A., Soubigou-Taconnat, L., Renou, J. P., Steinmetz, A., et al. (2008). Di- and tri-but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell Biol. 28, 1348–1360. doi: 10.1128/MCB.01607-07
Yang, C., Bratzel, F., Hohmann, N., Koch, M., Turck, F., and Calonje, M. (2013). VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 23, 1324–1329. doi: 10.1016/j.cub.2013.05.050
Yoshida, N., Yanai, Y., Chen, L., Kato, Y., Hiratsuka, J., Miwa, T., et al. (2001). EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13, 2471–2481.
Zhang, H., Bishop, B., Ringenberg, W., Muir, W. M., and Ogas, J. (2012). The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 159, 418–432. doi: 10.1104/pp.112.194878
Zhang, X., Bernatavichute, Y. V., Cokus, S., Pellegrini, M., and Jacobsen, S. E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10, R62. doi: 10.1186/gb-2009-10-6-r62
Zhang, X., Clarenz, O., Cokus, S., Bernatavichute, Y. V., Pellegrini, M., Goodrich, J., et al. (2007a). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5:e129. doi: 10.1371/journal.pbio. 0050129
Zhang, X., Germann, S., Blus, B. J., Khorasanizadeh, S., Gaudin, V., and Jacobsen, S. E. (2007b). The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 14, 869–871. doi: 10.1038/ nsmb1283
Zhang, H., Rider, S. D., Henderson, J. T., Fountain, M., Chuang, K., Kandachar, V., et al. (2008). The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J. Biol. Chem. 283, 22637–22648. doi: 10.1074/jbc.M802129200
Keywords: Arabidopsis, epigenetics, histone modification, Polycomb group, Polycomb Repressive Complex, Trithorax group
Citation: He C, Huang H and Xu L (2013) Mechanisms guiding Polycomb activities during gene silencing in Arabidopsis thaliana. Front. Plant Sci. 4:454. doi: 10.3389/fpls.2013.00454
Received: 01 October 2013; Paper pending published: 17 October 2013;
Accepted: 23 October 2013; Published online: 13 November 2013.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanReviewed by:
Paula Casati, Centro de Estudios Fotosinteticos-CONICET, ArgentinaCopyright © 2013 He, Huang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Xu, National Laboratory of Plant Molecular Genetics, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 300 Fenglin Road, Shanghai 200032, China e-mail:eHVsaW4wMUBzaWJzLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.