
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 09 October 2013
Sec. Plant Physiology
Volume 4 - 2013 | https://doi.org/10.3389/fpls.2013.00398
This article is part of the Research Topic Nitric Oxide Signalling in Plants View all 22 articles
Nitric oxide (NO) is currently considered a ubiquitous signal in plant systems, playing significant roles in a wide range of responses to environmental and endogenous cues. During the signaling events leading to these plant responses, NO frequently interacts with plant hormones and other endogenous molecules, at times originating remarkably complex signaling cascades. Accumulating evidence indicates that virtually all major classes of plant hormones may influence, at least to some degree, the endogenous levels of NO. In addition, studies conducted during the induction of diverse plant responses have demonstrated that NO may also affect biosynthesis, catabolism/conjugation, transport, perception, and/or transduction of different phytohormones, such as auxins, gibberellins, cytokinins, abscisic acid, ethylene, salicylic acid, jasmonates, and brassinosteroids. Although still not completely elucidated, the mechanisms underlying the interaction between NO and plant hormones have recently been investigated in a number of species and plant responses. This review specifically focuses on the current knowledge of the mechanisms implicated in NO–phytohormone interactions during the regulation of developmental and metabolic plant events. The modifications triggered by NO on the transcription of genes encoding biosynthetic/degradative enzymes as well as proteins involved in the transport and signal transduction of distinct plant hormones will be contextualized during the control of developmental, metabolic, and defense responses in plants. Moreover, the direct post-translational modification of phytohormone biosynthetic enzymes and receptors through S-nitrosylation will also be discussed as a key mechanism for regulating plant physiological responses. Finally, some future perspectives toward a more complete understanding of NO–phytohormone interactions will also be presented and discussed.
As sessile organisms, plants must rely on highly sophisticated signaling mechanisms to adjust their growth, shape, and metabolism with the constant changes in their environment. Playing a key role in this process, plant hormones integrate a multitude of internal and external cues into coordinated metabolic and developmental responses, which, in turn, maximize plant fitness under diverse ontogenetic and environmental contexts. To effectively carry out such critical function, distinct plant hormones intensively interact among themselves and also with other endogenous signaling substances (Santner et al., 2009).
Among these hormone-interacting molecules, the gaseous free radical nitric oxide (NO) has recently gained special interest in the research community given its involvement in a number of signaling cascades controlling plant responses ranging from seed germination to plant senescence (Neill et al., 2003; Wilson et al., 2008; Mur et al., 2012a). Whereas great strides have been made in recent years in understanding the mechanistic relationship between NO and phytohormones in certain physiological responses (Leon and Lozano-Juste, 2011; Terrile et al., 2012; Feng et al., 2013), the exact nature of the interaction between these substances in many developmental, metabolic, and defense events still remains remarkably elusive. In some cases, for instance, it is known that both NO and plant hormones are able to influence a given response, but it is not clear whether they share a common signaling cascade or just modulate the same plant event via parallel, independent signaling pathways.
As mentioned by Hancock et al. (2011), characterizing the precise function of NO in a particular signaling event is more difficult than it might appear. Firstly, the particular chemical characteristics of NO inexorably imply peculiar mechanisms for “sensing” the presence and levels of this signaling molecule. Instead of a unique or very few receptors, NO likely interacts with a wide range of target proteins via direct modification of protein structure (Figure 1). Through these chemical modifications of target proteins, NO may trigger changes in their activities and cellular functions, ultimately leading to the transduction of the NO message into plant responses.
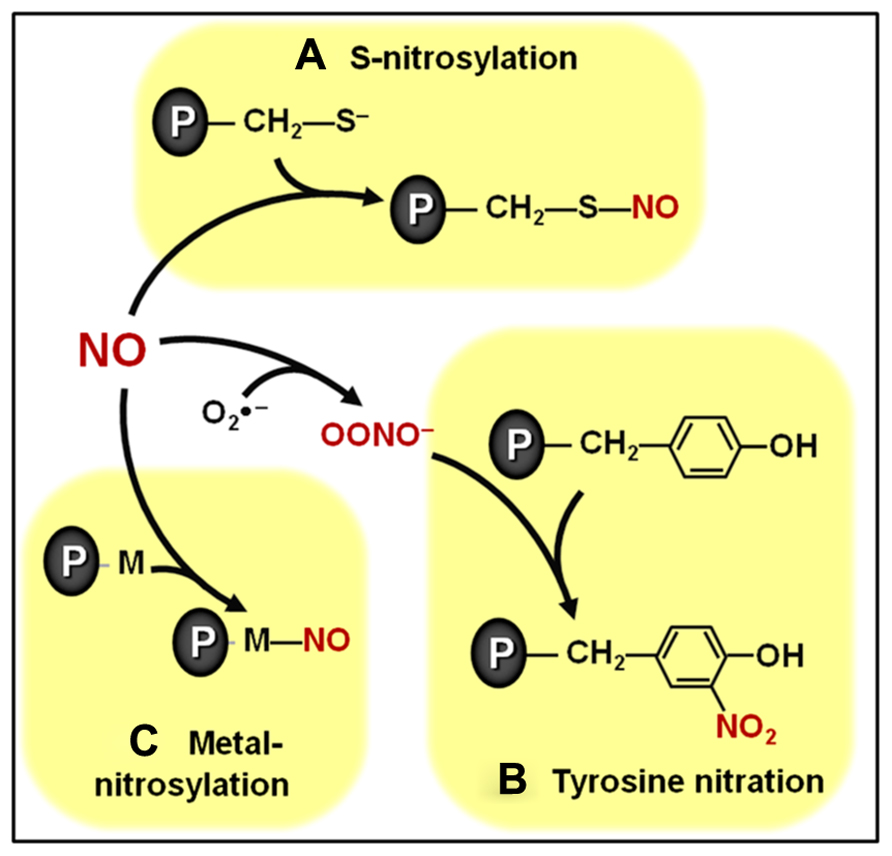
FIGURE 1. Overview of biologically relevant NO-dependent post-translational modifications (PTMs). (A) S-nitrosylation of cysteine residues. (B) Tyrosine nitration. (C) Metal nitrosylation. Proteins are represented with gray ovals and “P” letters.
Among the biologically relevant NO-dependent post-translational modifications (PTMs), the covalent modification of cysteine residues through a processes known as S-nitrosylation (Figure 1A) has been emerging as a critically important mechanism intermediating NO signal transduction in plants (Lindermayr et al., 2005; Astier et al., 2011, 2012). This specific, reversible and regulated NO-dependent PTM has been implicated as potentially controlling the function of components of plant processes as diverse as cellular architecture, photosynthesis, genetic information processing, protection against oxidative stress, defense responses to biotic and abiotic stresses, hormonal signaling, among others (Lindermayr et al., 2005; Romero-Puertas et al., 2008; Astier et al., 2011, 2012; Astier and Lindermayr, 2012). Currently, some of the best characterized examples of S-nitrosylation in plant systems include the modulation of phytohormone biosynthetic enzymes (Lindermayr et al., 2006), receptors (Terrile et al., 2012), and signal transduction proteins (Feng et al., 2013), which will be discussed in more detail later in this review. The specificity of this NO-triggered PTM is essentially based on the fact that only cysteine residues surrounded by particular neighboring amino acids seem to be the target of S-nitrosylation (Astier et al., 2011; Kovacs and Lindermayr, 2013).
A second physiologically relevant NO-dependent PTM depends on the reaction between NO and reactive oxygen species (ROS), such as superoxide (), resulting in the production of NO-derived species, such as peroxynitrite (ONOO-), which, in turn, can covalently modify tyrosine residues through a process known as tyrosine nitration (Figure 1B; Astier and Lindermayr, 2012). Initially considered an irreversible process, tyrosine denitration is now believed to occur either enzymatically or non-enzymatically (Abello et al., 2009; Vandelle and Delledonne, 2011; Astier and Lindermayr, 2012). Reinforcing such reversibility in tyrosine nitration, transient, rather than permanent, changes in the abundance of nitrated proteins have already been reported in the literature (Cecconi et al., 2009). More research is required to better define the biological relevance of this NO-dependent protein modification in plants, which apparently may target proteins involved in many basic cellular processes, such as photosynthesis, respiration and nitrogen metabolism (Cecconi et al., 2009; Chaki et al., 2009b; Lozano-Juste et al., 2011; Tanou et al., 2012)
In addition to S-nitrosylation and tyrosine nitration, a third important NO-dependent PTM involves the binding of NO to transition metal centers of metalloproteins in a process known as metal nitrosylation (Figure 1C). Currently, one of the best characterized examples of metal nitrosylation is the activation of soluble guanylate cyclase (sGC) in animal systems (Ignarro et al., 1999). In plants, although cyclic guanosine monophosphate (cGMP) has already been reported as an important intermediate in several NO-induced processes, including root development, mitochondrial respiration, nodule functioning, and defense responses (Durner et al., 1998; Pagnussat et al., 2003; Ederli et al., 2008; Keyster et al., 2010; Wang et al., 2010), more studies are still required to clarify whether metal nitrosylation also regulates plant sGC.
Regardless of the specific type of NO-triggered PTM considered, these chemical modifications may represent a central mechanism through which NO impacts signaling networks responsible for controlling plant development and metabolism. In responses regulated by plant hormones, for instance, these PTMs might facilitate the influence of NO on hormonal production and/or action via three distinct but non-exclusive mechanisms. The first mechanism implicates NO-dependent chemical modifications of proteins (e.g., transcription factors, regulatory proteins, and channels) whose functions may not be directly implicated in plant hormone metabolism, distribution, or signaling but, instead, may influence the abundance of other proteins more intimately implicated in such specific roles (Figure 2A). In contrast, a second and more direct way involves the NO-triggered PTM of proteins directly associated with the production, degradation, conjugation, transport, perception, or signaling transduction of plant hormones (Figure 2B). For example, in the first mechanism, NO may chemically modify a transcription factor that stimulates the production of a hypothetical enzyme responsible for hormone degradation, whereas in the second mechanism, NO would directly interact and modify the activity, stability, and/or cellular localization of this degradative enzyme (Figures 2A, B). A third possibility recently described in the literature involves the direct chemical reaction between NO-derivates (e.g., peroxynitrite) and certain hormonal species (e.g., zeatin), rendering products with altered biological activity (Figure 2C). Specific examples of all three of these mechanisms of NO–phytohormone interaction will be provided and discussed later in this review.
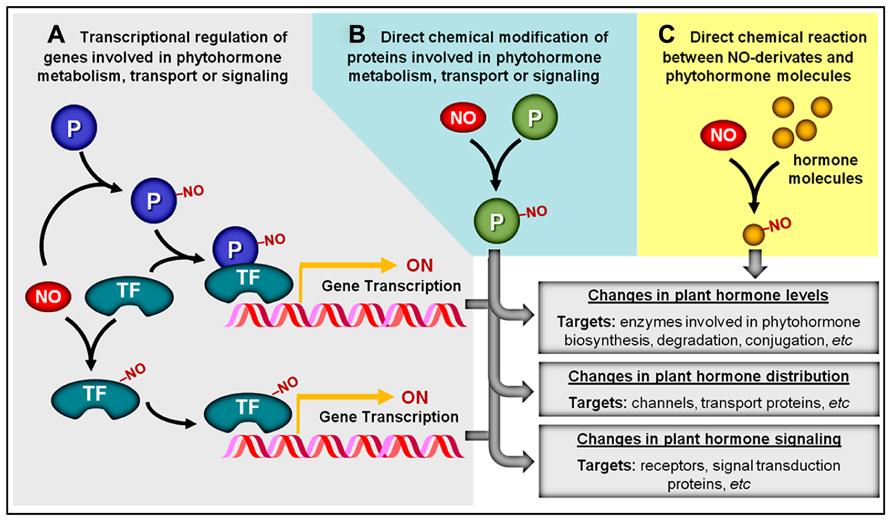
FIGURE 2. Overview of potential NO–phytohormone interaction mechanisms. (A) By chemically modifying transcription factors (TF) and other proteins (P), NO may influence the transcription level of genes involved in phytohormone metabolism, transport, or signal transduction. (B) NO may post-translationally modify proteins (P) directly involved in the production, distribution, or signaling of plant hormones. (C) NO or NO-derived reactive species might also chemically react with certain plant hormonal species, rendering products with altered biological activity. NO-dependent chemical modifications are represented by “–NO.”
Considering that a massive number of proteins, peptides, and other molecules may undergo changes in their structure and activity via direct NO-dependent chemical modifications (Astier et al., 2011, 2012; Astier and Lindermayr, 2012) and an equivalent amount of genes may have their transcription levels influenced by NO (Polverari et al., 2003; Parani et al., 2004; Grun et al., 2006; Besson-Bard et al., 2009), one pertinent question that arises is how NO signals can confer sufficient specificity to trigger coordinated downstream effects. Although answering this question involves a certain degree of speculation at this point in the research of NO signaling in plants, aspects such as spatial and temporal signaling compartmentation and a precise control of NO biosynthesis and removal might possibly be key to explaining how a molecule as small as NO might be responsible for controlling so many plant responses.
As is the case with cytosolic Ca2+, a strict temporal and spatial regulation of NO levels inside each plant cell might be essential for delivering sufficiently specific NO signals. The transient generation of “NO hot-spots,” in particular plant cell compartments, could lead to compartmentalized protein modifications (Neill et al., 2008b), and, consequently, the NO signals may be sensed by a specific group of proteins responsible for a particular set of cellular functions. A possible mechanism for assuring such localized action of NO could be the existence of macromolecular modules including all major NO signaling components (e.g., NO biosynthetic enzymes, NO removal enzymes, and targets of NO-dependent PTMs). Although such macromolecular complexes have not yet been described in plants, recent models for NO-mediated stress signaling in animal systems suggest, for instance, that the control of certain membrane calcium channels via reversible S-nitrosylation is facilitated by the close proximity of these channels to the NO-generating enzyme (Stamler and Meissner, 2001). Therefore, in this case, instead of a global change in cellular NO levels, the transient production of this signaling molecule at particular regions of the animal cell may control the activity of nearby target proteins via reversible S-nitrosylation (Martinez-Ruiz et al., 2013). As an ultimate consequence, such compartmentation and fine-tuned dynamics of NO production could minimize a certain spatial promiscuity in terms of concomitant occurrence of NO, NO-derivates, and their target proteins.
A relevant bottleneck for advances in the evaluation of the possible existence of such NO signaling macromolecular modules in plants is the still incipient characterization not only of the targets of NO-dependent PTMs but also of the biosynthetic and removal machinery responsible for controlling NO levels inside the plant cell compartments. Interestingly, though, compartmentalized production of NO has already been reported in plant cells. Foissner et al. (2000), for example, reported that after challenging epidermal tobacco cells with the elicitor cryptogein, NO accumulation first appeared in the plastids and subsequently in other cell compartments, such as the nucleus and the cytoplasm.
Placing NO as an element of a given signaling cascade necessarily implies that changes in its levels or cellular localization might occur during the course of the signaling event. Therefore, characterizing the specific changes in the NO biosynthetic and degradation mechanisms responsible for delivering adequate concentrations of this molecule at the right time and place seems a logical step in any research interested in discriminating the actual role of NO during the regulation of specific plant responses. However, the relevance of the different origins of NO in plants is still poorly understood; as a consequence, controversy and ambiguity are still frequently found in the current literature (Kaiser and Planchet, 2006; Gupta et al., 2011).
Besides the non-enzymatic NO production, which is believed to occur only under very specific conditions (Bethke et al., 2004), so far, seven potential enzymatic sources of NO have been identified in plants (Figure 3A; Gupta et al., 2011). Among them, nitrate reductase (NR) and NO synthase-like (NOS-like) activities are currently considered as the most likely candidates for the production of NO under physiologically relevant conditions (Neill et al., 2008b; Mur et al., 2012a). Since the discovery that plant NR could produce NO both under in vitro and in vivo conditions (Harper, 1981), a great deal of evidence has indicated this enzyme as one of the major plant biosynthetic sources of NO (Rockel et al., 2002; Meyer et al., 2005; Kaiser et al., 2010). Supporting this view, pharmacological and genetic approaches in different plant species, organs, tissues, and experimental conditions have revealed that NR inhibition frequently results in decreased NO production (Planchet and Kaiser, 2006; Oliveira et al., 2009; Freschi et al., 2010; Kolbert et al., 2010; Lombardo and Lamattina, 2012). On the other hand, the existence of NOS-like activity in plants is exclusively supported by biochemical and pharmacological evidence since a canonical NOS gene or a mutant deficient in NOS-like-dependent NO production has not been identified in higher plants yet (Corpas et al., 2006; Gupta et al., 2011; Mur et al., 2012a). Thus far, the organism more closely related to higher plants in which such a gene was described is the photosynthetic microalgae Ostreococcus tauri (Foresi et al., 2010; Correa-Aragunde et al., 2013), which belongs to a basal branch of the flowering plant evolutionary tree.
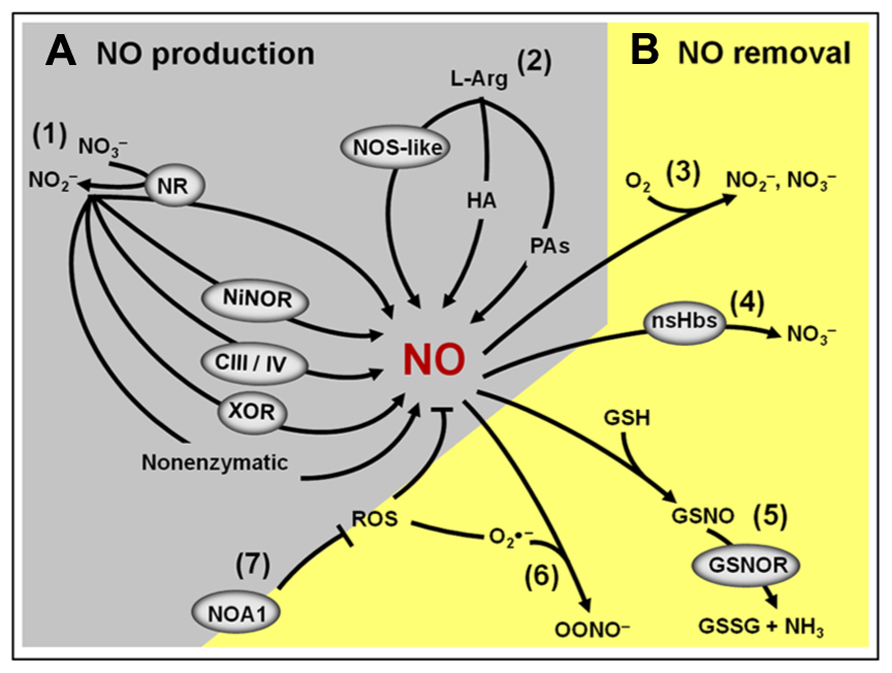
FIGURE 3. Overview of the NO production and removal mechanisms in plants. (A) Main components of the NO biosynthetic machinery: (1) Nitrite-dependent NO production in plants includes a non-enzymatic pathway and several enzymatic pathways involving the action of cytosolic and plasma membrane nitrate reductases (NR), nitrite–NO reductase (NiNOR), mitochondrial electron transport chain (CIII/IV) and xanthine oxidoreductase (XOR). (2) L-Arginine-dependent NO production pathway involves a non-identified nitric oxide synthase (NOS)-like enzyme and two still poorly characterized pathways using hydroxylamine (HA) or polyamines (PAs) as substrates. (B) Main components of the NO removal machinery: (3) the reaction of NO with molecular oxygen leads to the spontaneous production of nitrite and nitrate. (4) NO can react with non-symbiotic hemoglobins (nsHbs) resulting in nitrate formation. (5) Alternatively, NO may react with reduced glutathione (GSH) to form S-nitrosoglutathione (GSNO), which, in turn, can be converted into oxidized GSSG and ammonia by the action of GSNO reductase (GSNOR). (6) NO can also react with superoxide (), resulting in the formation of peroxynitrite (OONO-). (7) By influencing the production of reactive oxygen species (ROS), NO-ASSOCIATED 1 (NOA1) protein indirectly impacts NO levels in plants.
In 2003, studies revealed that NO Associated1 (AtNOA1), formerly described as AtNOS1 (Guo et al., 2003; Guo and Crawford, 2005; Zemojtel et al., 2006), also significantly influences NO generation in Arabidopsis. However, according with the latest consensus in the literature, AtNOA1 encodes a chloroplast-localized cGTPase probably involved in ribosome assembly and subsequent mRNA translation to proteins in this organelle (Flores-Perez et al., 2008; Moreau et al., 2008). Therefore, the reduced NO production observed in noa1 mutants is currently interpreted as an indirect outcome of disturbances in chloroplast metabolism due to the lack of AtNOA1 function (Zemojtel et al., 2006; Gas et al., 2009). More recently, this mutant was crossed with the NR-deficient nia1-nia2 mutant of Arabidopsis, generating a triple mutant (nia1,2noa1,2), which presented no detectable NO production and a range of physiological and developmental disturbances (Lozano-Juste and Leon, 2010a), thereby reinforcing the physiological importance of these pathways for determining the endogenous NO levels in plants.
Another important and frequently neglected aspect that may influence NO metabolism and signaling in plants is the presence of efficient mechanisms for removing the NO signal from a particular cell type or compartment as soon as it is no longer required. Besides the inherent chemical instability of NO in the presence of oxygen, this molecule might also be removed from plant tissues by several biochemical mechanisms (Figure 3B; Neill et al., 2008b; Mur et al., 2012a). Firstly, NO can be removed by reacting with ROS, such as superoxide anions, generating peroxynitrite. Secondly, NO may interact with plant proteins, such as non-symbiotic hemoglobins (nsHbs), which facilitates its oxidation to nitrate (Perazzolli et al., 2006). Finally, NO might also react with thiol proteins and peptides, resulting in the formation of S-nitrosothiols. In plant tissues, one of the most abundant low-molecular-mass S-nitrosothiols is the intracellular antioxidant glutathione, which may react with NO or with the NO-derivate N2O3, generating S-nitrosoglutathione (GSNO; Neill et al., 2008b; Mur et al., 2012a). The GSNO formed can spontaneously liberate NO or be metabolized by the enzyme S-nitrosoglutathione reductase (GSNOR), originating oxidized glutathione (GSSG) and NH3 (Barroso et al., 2006; Corpas et al., 2008b; Leterrier et al., 2011). Besides being an intracellular NO reservoir, GSNO may also be transported between cells, possibly playing a critical role as a vehicle of the NO signal throughout the plant body (Corpas et al., 2013).
Before exploring the general mechanisms underlying the interactions between NO and phytohormones, it is worth mentioning that a great diversity of methodological approaches, experimental designs, and plant models have been used in NO research, which sometimes makes it difficult to directly compare the literature data. In terms of methodological approaches, for instance, a considerable variety of analytical techniques have been employed to determine NO levels in plant systems, including the Griess and the hemoglobin assays, electron spin resonance, laser-based photoacoustic detection, ozone-based chemiluminescence, and various fluorescent probes (reviewed by Vitecek et al., 2008 and Mur et al., 2011). As expected, these different methods provide distinct information. For example, it is always a challenge to compare results obtained by gas-phase NO detection techniques (e.g., chemiluminescence or laser photoacoustic) with fluorescent methods for in situ NO detection since these two groups of techniques differ greatly in their specificity, spatial resolution, and capacity to indicate the actual concentration of NO inside the target cells (Mur et al., 2011). Furthermore, evaluations of NO levels under the same experimental conditions by two or more independent methods, although recommended (Mur et al., 2012a; Gupta and Igamberdiev, 2013), are rarely carried out (Besson-Bard et al., 2008).
Besides measuring NO itself, alternatives to access NO and other RNS signaling inside the cells, such as the abundance of NO-triggered chemical modifications on proteins and peptides, have also recently drawn increasing attention of the plant research community, providing, in some cases, spectacularly relevant information. A number of technical options are currently available for such proposes, including the determination of S-nitrosothiol levels in plant extracts by reductive gas-phase chemiluminescence (Valderrama et al., 2007; Corpas et al., 2008b; Lee et al., 2008; Chaki et al., 2009a), immunolocalization of particular S-nitrosylated thiols or proteins (Barroso et al., 2006; Valderrama et al., 2007; Corpas et al., 2008a; Chaki et al., 2009a; Leterrier et al., 2011) or even proteomic profiling of proteins chemically modified by NO or NO-derivates (Lindermayr et al., 2005; Chaki et al., 2009b; Astier et al., 2011; Kovacs and Lindermayr, 2013), which, among other aspects, may facilitate the identification of the specific targets of NO-dependent PTMs in distinct plant responses.
Another relevant aspect to be considered in NO–phytohormone interaction studies is that the simple observation of changes in NO levels triggered by exogenous plant hormones does not necessarily imply a straightforward relationship between NO and the hormonal stimulus. Firstly, the exogenous application of a signaling substance might potentially induce global, unspecific changes in plant biochemistry, metabolism, and development. Secondly, modifications in NO levels might sometimes result from excessive levels of exogenous hormones; therefore, whenever possible, the actual concentration of particular phytohormone species inside the plant cells and tissues should be determined following the supplementation with these substances. Finally, some plant hormones may affect the biosynthesis and signaling of others (Santner et al., 2009); consequently, the establishment of a direct correlation between the pharmacological effect of a specific plant hormone on a given cellular response is not always an easy task. To overcome such a lack of specificity and potentially artificial effects, the use of transgenic and mutant plants with altered production, degradation or signaling of particular hormonal classes as well as a detailed characterization of several elements involved in phytohormone and NO metabolisms and signaling transduction have proven to be a powerful strategy for accessing the mechanistic relationship between these substances (Desikan et al., 2002; Leon and Lozano-Juste, 2011; Terrile et al., 2012).
Despite these methodological disparities and the limited literature information currently available, there is virtually no doubt that NO and phytohormones interact at multiple, diversified levels. Depending on the signaling cascade, NO has been demonstrated to act either upstream or downstream of plant hormones (Hancock et al., 2011; Simontacchi et al., 2013). Obviously, placing NO downstream of the hormonal stimuli in a signaling route necessarily means that the NO biosynthetic, degradation, conjugation, or deconjugation machinery may be affected at certain point between the perception of hormonal stimulus and the induction of the plant response. Therefore, the time period between the hormonal message input and the detection of changes in endogenous NO levels represents valuable information. In some cases, lag phases compatible with changes in the transcripts level or protein abundance of NO-synthesizing or removal enzymes have been reported (Pagnussat et al., 2002; Freschi et al., 2010). However, under some particular circumstances, the lag phase observed between the application of plant hormones and the rise in NO endogenous levels has been shown to be of just few minutes (Tun et al., 2001; Garcia-Mata and Lamattina, 2002; Huang et al., 2004; Tun et al., 2006; Sun et al., 2010), which indicates that the post-translational regulation of proteins involved in NO metabolism rather than their de novo synthesis might sometimes be implicated.
When acting upstream of phytohormones, NO seems able to modulate elements controlling either the plant hormone levels (e.g., biosynthetic, degradation, and conjugation enzymes), distribution (e.g., transport proteins) or signaling (e.g., receptors and signal transduction proteins). This modulation has been shown to occur either at the transcriptional (Bethke et al., 2007; Liu et al., 2009; Manjunatha et al., 2010; Xu et al., 2010; Leon and Lozano-Juste, 2011) or post-translational levels (Lindermayr et al., 2006; Terrile et al., 2012; Feng et al., 2013); however, some post-transcriptional or even translational regulation of hormone-related proteins by NO, although not yet demonstrated, cannot be ruled out.
Based on the basic information provided thus far, the current state-of-the-art of the interplay between NO and each one of the major classes of plant hormones [i.e., auxins, cytokinins, gibberellins (GAs), abscisic acid (ABA), and ethylene] will now be discussed. Although discussed here in an isolated manner, it is important to keep in mind that very frequently, if not always, plant hormones intensively interact with each other during the induction and establishment of plant responses. However, future studies will still be required to mechanistically explain exactly how distinct plant hormones concomitantly interact with NO to regulate specific plant events.
Synergistic effects of auxin and NO have been observed during the regulation of a series of plant responses, including root organogenesis (Pagnussat et al., 2002, 2003, 2004; Lanteri et al., 2006), gravitropic responses (Hu et al., 2005), root nodule formation (Pii et al., 2007), root responses to iron deficiency (Chen et al., 2010), activation of cell division and embryogenic cell formation (Ötvös et al., 2005), NR activity stimulation (Du et al., 2008), among others. In virtually all of these cases, NO was identified to function downstream of auxins, apparently through linear signaling pathways. Increased NO production has frequently been observed after exogenous auxin application (Pagnussat et al., 2002; Correa-Aragunde et al., 2004; Hu et al., 2005; Lombardo et al., 2006) or in auxin overproducer mutants (Chen et al., 2010), being especially evident in plant tissues or cells undergoing auxin-dependent physiological responses. On the other hand, no or weak stimulation in NO production by auxins has been reported in some particular experimental conditions or cell types (Tun et al., 2001; Guo et al., 2003), suggesting that the auxin-dependent NO production may occur exclusively under specific temporal and spatial contexts (Hu et al., 2005).
Currently, most of the reports on NO and auxin interaction are focused on plant root responses, with relatively little information available on the crosstalk between these two signaling molecules in shoot or reproductive tissues. During the last decade, detailed information about the interaction between NO and auxin during root growth and development was provided by a series of studies conducted by Lamattina and colleagues, including the interplay between these molecules during adventitious roots formation (Pagnussat et al., 2002, 2003, 2004), lateral root development (Correa-Aragunde et al., 2004), and root hair initiation and elongation (Lombardo et al., 2006). In almost all of these studies, the removal of NO by scavengers significantly decreased typical auxin-dependent root responses, such as the activation of mitogen-activated protein kinases (MAPKs) during the adventitious root formation (Pagnussat et al., 2004) and induction of cell cycle genes during lateral root formation (Correa-Aragunde et al., 2006).
Also focusing on root tissues responses, Chen et al. (2010) identified a direct correlation between auxin availability, root NO levels and the expression of iron acquisition genes and other Fe deficiency-associated stress responses, providing further support for the action of NO as a downstream element in the auxin signaling pathway. Similarly, a clear spatial correlation was also observed between the asymmetric auxin distribution and the endogenous NO localization during the gravitropic bending in soybean roots (Hu et al., 2005) and during indeterminate nodule formation in roots of Medicago species infected by auxin-overproducing rhizobia (Pii et al., 2007).
A possible role for NR as the major biosynthetic source of the auxin-induced NO production during some plant root responses has been suggested (Kolbert and Erdei, 2008). Kolbert et al. (2008), for instance, reported that the NO production during the auxin-induced lateral root development in Arabidopsis requires NR activity since the NR-deficient double mutant nia1,nia2 failed to increase NO generation in response to exogenous auxin, whereas no evidence for an involvement of NOS in this response was observed. NR-dependent NO production was also shown to be crucially important for the adequate vesicle trafficking during root hair formation because exogenous NO application completely restored the abnormal vesicle formation and trafficking as well as root hair growth in the nia1,nia2 Arabidopsis mutant (Lombardo and Lamattina, 2012). In a few cases, however, such as during the auxin-regulated NO generation under Fe deficiency and during the gravitropic bending in soybean roots, evidence indicates the involvement of not only NR but also NOS and/or NOA1 in the auxin-induced NO generation (Hu et al., 2005; Chen et al., 2010).
Considering that many of these root responses, including root hair formation and lateral root development, respond to both auxins and nitrate supply, NR-dependent NO generation might be a key integrator of exogenous and endogenous cues leading to the control of plant root biology. Although the precise mechanism through which auxin trigger NR-dependent NO generation has still not been fully characterized, literature data indicate a promotive effect of this plant hormone on NR protein, activity and gene transcription (Vuylsteker et al., 1997; Du et al., 2008).
Besides these impacts of auxin on NO production, recent studies have demonstrated that NO might also modulate auxin metabolism, transport, and signaling. For example, NO has been demonstrated to enhance root indole-3-acetic acid (IAA) levels in cadmium-treated Medicago truncatula seedlings by reducing its degradation via IAA oxidase activity (Figure 4), thereby positively impacting auxin equilibrium and ameliorating cadmium toxicity (Xu et al., 2010). In addition, pharmacological treatments and NO-overproducing mutants indicated that, at high concentrations, NO inhibits acropetal auxin transport in Arabidopsis roots by reducing the abundance of the auxin efflux protein PIN-FORMED 1 (PIN1) via a proteasome-independent post-transcriptional mechanism (Fernández-Marcos et al., 2011). This NO-dependent decrease in PIN1 protein levels and consequent disturbance in root auxin transport resulted in severe reductions in root meristem size and activity in primary roots due to a reduction in cell division and a promotion in cell differentiation, compromising the root apical meristem maintenance and primary root growth (Fernández-Marcos et al., 2011).
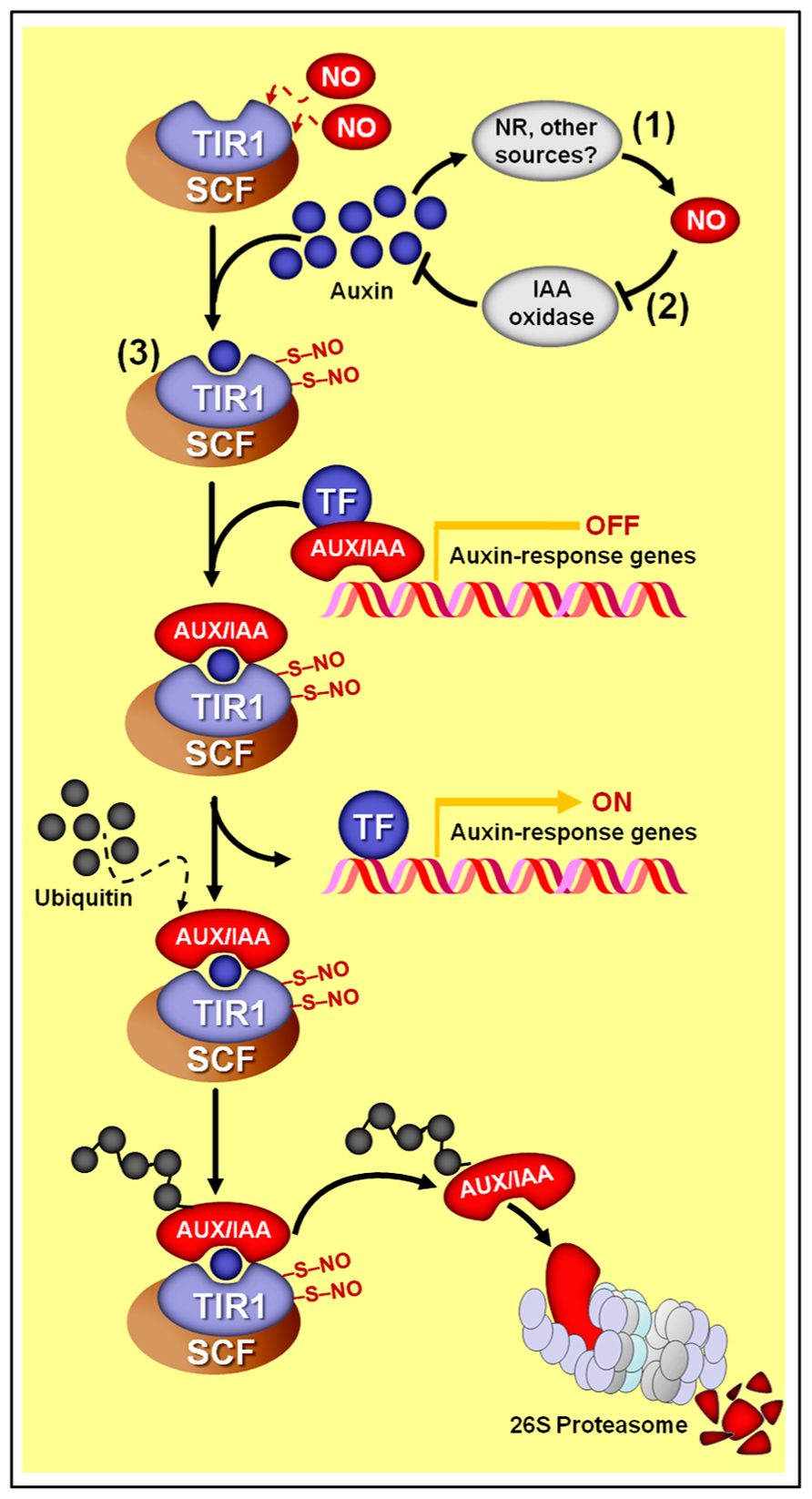
FIGURE 4. Schematic representation of NO–auxin synergist interactions. (1) Auxins stimulate NO production in several plant materials and experimental conditions. In most cases, nitrate reductase (NR) seems to be the main biosynthetic source of auxin-induced NO production. (2) In M. truncatula roots, NO promotes auxin accumulation by repressing its degradation via IAA-oxidase. (3) In Arabidopsis, NO might also positively impact auxin signaling since the auxin receptor TRANSPORT INHIBITOR RESPONSE 1 (TIR1) may undergo S-nitrosylation at cys-140 and cys-480, which promotes its interaction with AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) proteins. Subsequently, TIR1 marks AUX/IAA proteins to degradation through SCF-26S proteasome-mediated proteolysis, thereby de-repressing the transcription of auxin-regulated genes. Protein S-nitrosylation is represented by “–S–NO.”
Finally, a direct influence of NO on auxin perception and signal transduction has also been suggested based on the recent demonstration that the auxin receptor protein TIR1 (TRANSPORT INHIBITOR RESPONSE 1) undergoes S-nitrosylation at two particular cysteine residues (cys-140 and cys-480) (Terrile et al., 2012). This S-nitrosylation of TIR1 seems to promote its interaction with AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) proteins, which are transcriptional repressors of genes associated with auxin responses (Figure 4). Being part of an E3 ubiquitin ligase complex, TIR1 marks AUX/IAA proteins to proteasome degradation, de-repressing the expression of auxin-dependent genes. Therefore, as a result, the increased TIR1–AUX/IAA interaction caused by TIR1 S-nitrosylation may facilitate AUX/IAA degradation via proteasome and subsequently promote auxin-dependent gene expression (Terrile et al., 2012). A possible impact of S-nitrosylation on the capacity of TIR1 to bind auxin could also be a possible outcome of this NO-dependent PTM, but further investigations are still required on this subject.
Furthermore, evidence indicates that nsHbs might also influence and modify the auxin signaling and action site by modulating the endogenous NO levels. Hunt et al. (2002), for example, detected a drastic modification in auxin-regulated root morphology and development in transgenic lines of Arabidopsis overexpressing class 1 nsHb, which could be interpreted as the result of changes in the content and/or bioactivity of NO in these plants.
During the last few years, accumulating evidence has indicated complex and multilevel interactions between NO and cytokinins. Both synergistic and antagonistic interactions between NO and cytokinins have been described depending on the physiological response, plant species and experimental approach. Evidence implying a possible participation of NO in cytokinin signal transduction was first obtained during the accumulation of the red pigment betalaine in Amaranthus caudatus seedlings, which was shown to positively respond not only to cytokinins but also to NO gas or donors (Scherer and Holk, 2000). Since then, a number of studies have reported rapid and dose-dependent increases in NO production triggered by μM concentrations of cytokinins in both plant cell cultures (Tun et al., 2001; Carimi et al., 2005) and intact seedlings (Tun et al., 2008; Shen et al., 2012). In Arabidopsis seedlings, for instance, zeatin triggered increases in NO production within 3 min via a biosynthetic mechanism sensitive to arginine analogs and apparently independent of NR activity (Tun et al., 2008). However, other evidence revealed unchanged or even lower NO levels after cytokinin treatments or in mutant or transgenic plants with increased cytokinin production (Xiao-Ping and Xi-Gui, 2006; Romanov et al., 2008; Liu et al., 2013). Moreover, no obvious influence of exogenous application or depletion of NO has been observed on some early signaling events leading to the induction of primary cytokinin responses, such as the activation of cytokinin-responsive Arabidopsis response regulator (ARR)5 promoter in seedlings (Romanov et al., 2008).
Examples of synergistic interaction between cytokinins and NO include the control of leaf senescence (Mishina et al., 2007), programmed cell death (PCD; Carimi et al., 2005), photosynthesis adaptability to drought stress (Shao et al., 2010), cell division, and differentiation (Shen et al., 2012), among others. Studies of the integrated influence of NO and cytokinins on plant senescence program have demonstrated that natural, dark- or dehydration-induced leaf senescence can be minimized by exogenous NO application (Cheng et al., 2002; Mishina et al., 2007). In addition, mutant or transgenic plants exhibiting decreased NO levels usually display precocious senescence in detached leaves and intact plants (Guo and Crawford, 2005; Mishina et al., 2007), which can sometimes be alleviated by exogenous cytokinin supplementation (Mishina et al., 2007). Although still limited in terms of current commercial application, this antisenescence trait of NO and cytokinins has been proven to extend post-harvest life of agronomically relevant fruits and vegetables (Leshem and Wills, 1998; Leshem et al., 1998; Leshem et al., 2001).
Further indicating a protective and antisenescence role of NO and cytokinins, Shao et al. (2010) reported increased NO levels during the cytokinin-induced photosynthetic adaptability to drought stress and described a good correlation between NO production and NR activity during this adaptive plant response to water limitation. In contrast, however, NOS-like-dependent increases in NO generation have been suggested to act as an intermediate during the acceleration of cell apoptosis induced by high cytokinin dosages since cell death was alleviated when cytokinins were supplied along with NOS inhibitors or NO scavengers to Arabidopsis cell cultures (Carimi et al., 2005).
The interaction between cytokinins and NO during the regulation of plant cell division has also been recently studied in more detail. Among other evidence, NO deficiency caused either by loss of the gene NOA1 or due to NO scavenger treatments was demonstrated to result in severe inhibition of cytokinin-induced transcriptional activation of the cell cycle gene CYCD3;1 (CYCLIN-D3;1) and the subsequent callus initiation from somatic plant tissues, implying that NO may act downstream of cytokinins in the control of plant cell mitotic cycles (Shen et al., 2012). In this study, roots of Atnoa1 mutant were described as severely impaired in cytokinin-induced NO production and less sensitive to cytokinins than wild-type (WT) ones (Shen et al., 2012).
Contrary to the above described synergistic relationships between NO and cytokinins, literature data have also suggested an opposite interaction between these signaling molecules in some plant responses (Xiao-Ping and Xi-Gui, 2006). Studies conducted on epidermal strips of Vicia faba indicated that exogenous cytokinins efficiently reduced NO generation in guard cells exposed to the NO donor sodium nitroprusside (SNP) as well as promoted stomata reopening under dark condition due to the abolishment of the dark-induced increases in endogenous NO, which was interpreted as evidence of a potential scavenging action of cytokinins on the NO produced under these situations (Xiao-Ping and Xi-Gui, 2006).
Consistent with these results, Wilhelmová et al. (2006) also observed a negative correlation between endogenous cytokinin and NO levels in transgenic tobacco plants with either increased or decreased cytokinin levels. More recently, Liu et al. (2013) reported that cytokinins might intimately participate in NO catabolism since some cytokinin species, such as zeatin, can chemically react with peroxynitrite, leading to the production of cytokinin derivates with virtually no biological activity (Figure 5). Moreover, these authors verified that exogenous zeatin alleviates the severity of the phenotypes attributed to excessive NO levels in the Arabidopsis NO-overproducer nox1 (nitric oxide overexpression 1) mutant, and this same ameliorative effect was observed when nox1 plants were crossed with a cytokinin-overproducing mutant (Liu et al., 2013). Based on these biological and chemical data, Liu et al. (2013) postulated that these two signaling molecules (NO and cytokinins) might interact by modulating each other’s homeostatic levels and bioactivity (Figure 5). Such peculiar mechanism of interaction between cytokinins and NO, in which one of the substances directly interferes with the levels of another simply by a chemical combination of two molecules (Figure 2C), is quite different from the interaction at biosynthetic or signaling levels usually observed for other NO–phytohormone crosstalks (Figures 2A, B) and certainly deserves further attention.
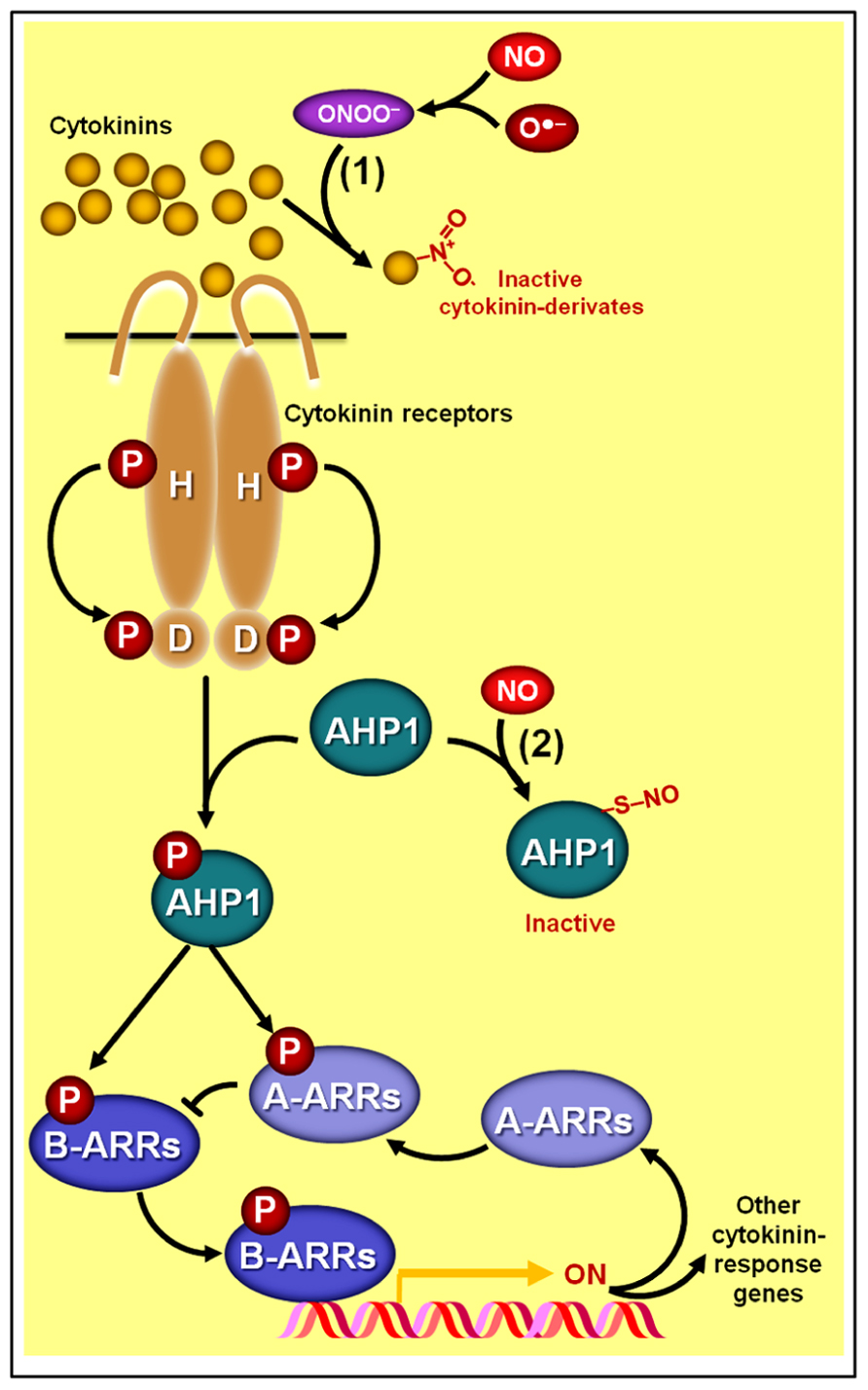
FIGURE 5. Schematic representation of NO–cytokinin antagonistic interactions. (1) Certain cytokinin species such as zeatin may chemically react with peroxynitrite (ONOO-), producing derivates with virtually no biological activity. (2) NO might also negatively impact cytokinin signaling since the protein HISTIDINE PHOSPHOTRANSFER PROTEIN 1 (AHP1), a key element in the phosphorelay mechanism involved in cytokinin transduction in Arabidopsis, may undergo S-nitrosylation at cys-115, rendering this protein incapable of transferring phosphoryl groups from the cytokinin receptors to the ARABIDOPSIS RESPONSE REGULATORs (ARRs). Protein S-nitrosylation and phosphorylation are represented by “–S–NO” and “P,” respectively.
Additionally, strong evidence indicating a direct impact of NO on the cytokinin signaling pathway has recently been uncovered (Feng et al., 2013). Besides corroborating previous observations that Arabidopsis mutant lines with excessive NO levels display more limited responsiveness to cytokinins, Feng et al. (2013) revealed that the phosphorelay mechanism central to the signaling transduction of this hormonal class can be severely impaired by the S-nitrosylation of a particular cysteine residue (cys 115) of the HISTIDINE PHOSPHOTRANSFER PROTEIN 1 (AHP1), hindering the transfer of phosphoryl groups from cytokinin receptors to AHP1 and subsequently to response regulators (ARRs; Figure 5). Confirming the importance of this NO-dependent post-translational protein modification for the cytokinin signal transduction, these authors have demonstrated that non-nitrosylatable mutation of AHP1 consistently relieved the inhibitory effect of NO on cytokinin responses whereas a nitrosomimetic mutation of this protein severely compromised cytokinin responses (Feng et al., 2013).
An additional, less direct way through which cytokinins might modulate NO levels in plant systems seems to rely on the regulatory effect of these hormones on the expression of nsHbs (Hunt et al., 2001; Ross et al., 2004; Bustos-Sanmamed et al., 2011). Cytokinin-triggered changes in the expression of certain nsHbs have been described for several plant models (Ross et al., 2004; Bustos-Sanmamed et al., 2011). Moreover, transgenic and mutant plants with altered levels of particular nsHb classes have frequently displayed alterations in plant responses typically controlled by cytokinins (Hunt et al., 2001; Wang et al., 2011). For instance, marked changes in shoot organogenesis and altered expression of genes associated with cytokinin perception and signaling have been observed in Arabidopsis lines silencing or overexpressing class 1 or class 2 nsHbs (Wang et al., 2011). In the transgenic lines overexpressing nsHbs, cytokinin feedback repressors (Type-A ARRs) were repressed, whereas cytokinin activators (Type-B ARRs) and receptors were stimulated (Figure 3), culminating in a higher sensitivity of the tissues to the cytokinin-induced shoot organogenesis (Wang et al., 2011). Unfortunately, NO content was not measured in these transgenic lines; therefore, a direct correlation between the higher responsiveness to cytokinins observed in nsHb overexpressing lines and their possibly lower NO levels could not be established.
Both important “stress-related” molecules, NO and ABA intensively crosstalk during certain signaling cascades triggered by environmental challenges, such as water limitation and UV-B radiation, which ultimately leads to the induction of plant adaptive responses, such as stomatal closure and antioxidant defenses (Neill et al., 2008a; Tossi et al., 2009; Hancock et al., 2011). During the induction of these plant stress responses, NO mainly acts as a downstream element in the ABA signaling pathway since the impairment in NO production or its removal from tissues usually decreases or even eliminates ABA responses while the inhibition of ABA production typically does not affect the induction of these responses by exogenous NO application. On the other hand, during the regulation of certain developmental events not directly linked to plant stress responses, such as seed dormancy breaking, NO seems to counteract ABA effects (Bethke et al., 2006; Lozano-Juste and Leon, 2010a, b), suggesting a certain level of specificity in the NO–ABA interaction mechanisms, which may depend on the physiological events under analysis (e.g., stomatal closure versus seed dormancy release) or even the type of plant cell, tissue, or organ considered (e.g., guard cell versus seed tissues).
In some cases, such as during the induction of stomatal closure (Neill et al., 2002; Desikan et al., 2004; Bright et al., 2006) and during the up-regulation of the gene transcription and activities of antioxidant enzymes (Zhang et al., 2007; Lu et al., 2009; Zhang et al., 2009), ABA-induced NO generation seems to depend on H2O2 synthesis, suggesting this ROS as a mediator in NO-dependent ABA responses (Figure 6). In addition, the calcium/calmodulin system and MAPKs have also being identified as downstream elements of NO signaling during the regulation of plant antioxidant defenses induced either by ABA or H2O2 (Zhang et al., 2007; Sang et al., 2008). Moreover, cGMP has also been demonstrated to participate in NO-dependent ABA signaling, apparently acting downstream of NO and upstream of cytosolic Ca2+ (Figure 6; Dubovskaya et al., 2011). Similarly, type 2C protein phosphatases (PP2Cs), which acts as negative regulators of ABA signaling, have also been suggested to play a role as putative crosstalk elements between ABA receptors and NO-mediated ABA signal transduction, possibly acting downstream of NO in the complex networks controlling ABA-triggered stomatal closure (Desikan et al., 2002).
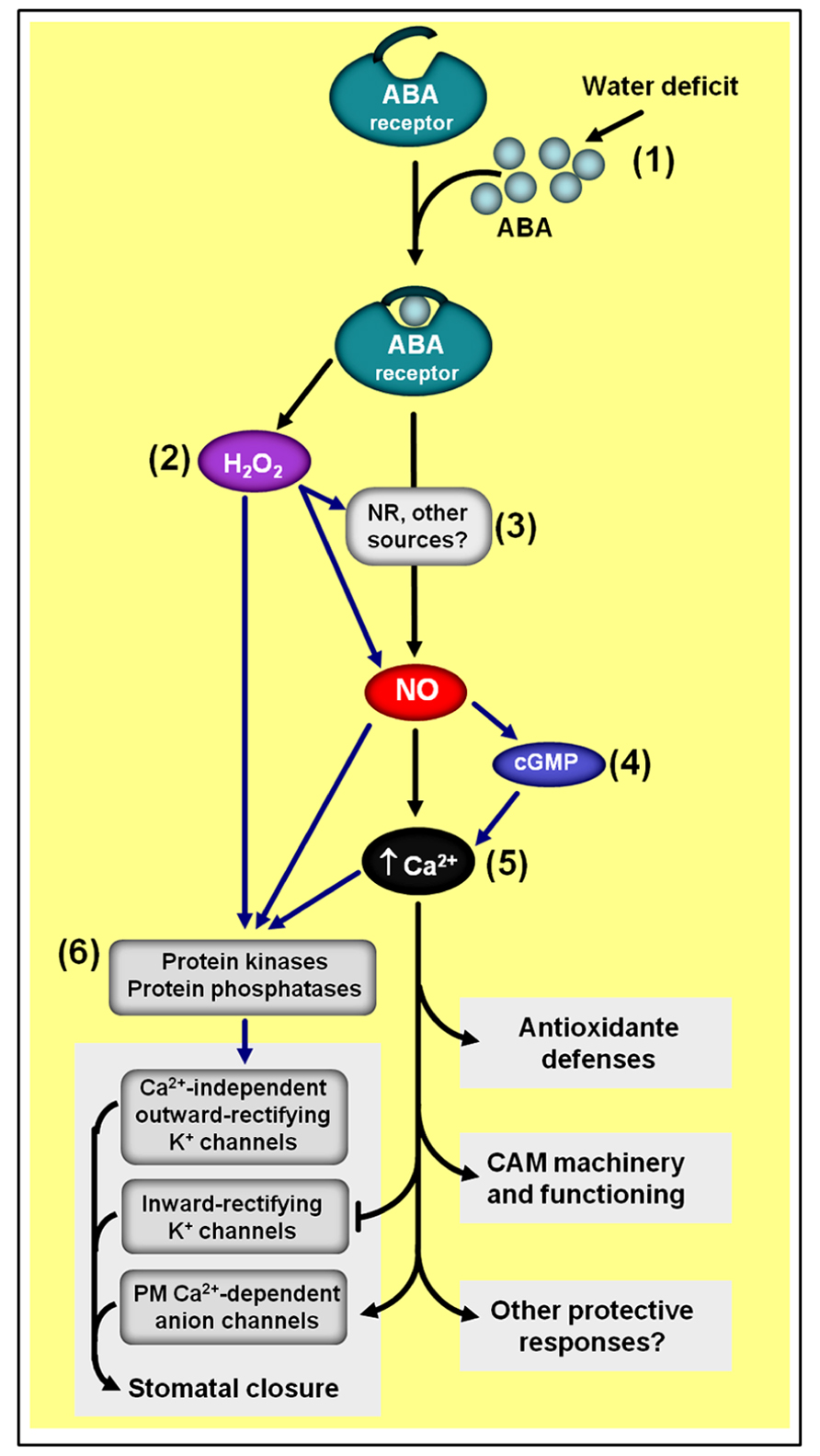
FIGURE 6. Simplified schematic representation of NO–ABA interactions during defense responses to water shortage. (1) Water deficiency usually increases endogenous ABA levels. (2) ABA-induced NO generation depends on hydrogen peroxide (H2O2) synthesis. (3) NR seems to be one of the main sources of ABA-induced NO production. (4) NO-triggered changes in cytosolic calcium (Ca2+) seem to involve cyclic guanosine monophosphate (cGMP). (5) The calcium/calmodulin system is a key downstream element of NO/ABA signaling. (6) Protein kinases and phosphatases are typical targets of H2O2, NO, and Ca2+/calmodulin during ABA-induced responses. Black arrows indicate signaling steps shared by all three drought responses considered in the scheme (i.e., stomatal closure, antioxidant defenses, and Crassulacean acid metabolism induction). Blue arrows indicate some steps currently described only for the regulation of stomatal closure and/or antioxidant defenses. ABA- and NO-independent signaling pathways are not represented in this schematic representation.
Since the discovery that NO scavengers could reduce ABA-induced stomata closure in turgid leaves of different plant species (Garcia-Mata and Lamattina, 2002; Neill et al., 2002), intensive research has been dedicated to characterize the mechanisms underlying the interplay between these two molecules in guard cell signaling networks (reviewed in Neill et al., 2008a; Hancock et al., 2011; Simontacchi et al., 2013), leading to the identification of several NO targets during the ABA-induced guard cell responses. Among these targets, plasma membrane calcium-dependent anion channels and inward-rectifying K+ channels have been demonstrated to be activated and deactivated, respectively, by NO as a consequence of increases in guard cell cytoplasmatic Ca2+ levels (Figure 6) due to NO-triggered release of this anion from intercellular stores (Garcia-Mata et al., 2003).
Evidence for the involvement of protein phosphorylation upstream of intracellular calcium release has also been obtained, implicating protein kinases as additional targets of NO action within ABA-regulated guard cell signaling (Sokolovski et al., 2005). Moreover, NO has also been reported to directly modulate calcium-independent outward-rectifying K+ channels possibly by post-translationally modifying these channels or closely associated regulatory proteins (Sokolovski and Blatt, 2004). As a final consequence, this NO-dependent modulation of both Ca2+-dependent and Ca2+-independent ion channels at the plasma membrane of guard cells facilitates osmotic solute loss, thereby reducing guard cell turgor and promoting stomatal closure.
It is worth mentioning that NO has been suggested to play a role as a second messenger shared by multiple hormonal signaling cascades involved in the intricate guard cell network responsible for coordinating stomatal movement in higher plants, mediating not only the ABA signal but also ethylene (Liu et al., 2010), salicylic acid (SA; Hao et al., 2010), methyl jasmonate (Saito et al., 2009), auxin, and cytokinins (Xiao-Ping and Xi-Gui, 2006). Curiously, though, NO apparently is not an absolute requirement during the ABA signaling cascades leading to stomatal closure (Ribeiro et al., 2009) or the inhibition of light-induced stomatal opening (Yan et al., 2007; Yang et al., 2008); therefore, the existence of both NO-dependent and NO-independent pathways in ABA-induced guard cell responses is currently being suggested. Of course, more studies are clearly needed to better characterize a possible integrative, but apparently non-essential, role of NO during the regulation of stomatal movements by distinct environmental and hormonal stimuli.
At least in bromeliads, NO and ABA also seem to intensively interact to control Crassulacean acid metabolism (CAM) expression (Freschi et al., 2010; Mioto and Mercier, 2013), which, in turn, facilitates the survival of these plants under water- and nutrient-limited environments. As during the regulation of stomatal movements, NO apparently acts downstream of ABA and upstream of cytosolic calcium in the ABA-dependent signaling cascade leading to the up-regulation of the CAM machinery (Figure 6), and does not participate in the ABA-independent pathway also responsible for the regulation of this plant stress response (Freschi et al., 2010). The regulation of CAM expression in bromeliads as well as the control of stomata movements in Arabidopsis seem to have NR activity as the main source of the ABA-induced NO production (Desikan et al., 2002; Freschi et al., 2010).
While a number of pharmacological and genetic studies have reported higher endogenous NO levels following increases in plant tissue ABA concentration (i.e., NO action downstream of ABA; Zhang et al., 2009), NO-triggered changes in ABA biosynthesis and catabolism (i.e., NO action upstream of ABA) have rarely been described. In one of the few examples, Liu et al. (2009) reported that during the seed dormancy breaking in Arabidopsis, a rapid accumulation of NO in the endosperm layer preceded a decrease in ABA concentration, which was associated with a pronounced rise in the transcript and protein levels of the ABA 8′-hydroxylase CYP707A2, a key enzyme in ABA catabolism. Moreover, exogenous NO and the NO scavenger carboxy-PTIO (cPTIO), respectively, induced and impaired CYP707A2 transcript accumulation during the imbibition period (Liu et al., 2009), further suggesting that the promotive effect of NO on seed dormancy break might indeed be associated with a stimulation of ABA catabolism.
In addition to modulating ABA catabolism, NO has also been described to affect the sensitivity of plant cells to ABA (Bethke et al., 2006; Lozano-Juste and Leon, 2010a, b). Bethke et al. (2006) reported that the NO donor SNP enhanced germination of dormant Arabidopsis seeds by decreasing the seed sensitivity to exogenous ABA. More recently, genetic evidence supporting this inhibitory effect of NO on ABA sensitivity was obtained by Lozano-Juste and Leon (2010a), b, who observed that the depletion of endogenous NO levels resulting from the generation of the nia1,2noa1-2 Arabidopsis triple mutant clearly led to ABA hypersensitivity. Among other features, this triple mutant displayed enhanced seed dormancy, decreased seed germination, and reduced seedling establishment in the presence of exogenous ABA, reinforcing the hypothesis that NO production during seed germination and initial seedling development counteracts the ABA inhibitory effects on these events. Interestingly, this ABA hypersensitivity continued through the post-germinative vegetative development of this triple mutant, as evidenced by the presence of increased expression of ABA-responsive genes, extreme drought resistance phenotype as well as higher responsiveness to ABA during stomatal closure (Lozano-Juste and Leon, 2010a, b). Curiously, dehydration- and ABA-dependent stomatal closure normally occurred in the presence of undetectable NO production in guard cells, corroborating the existence of a NO-independent pathway in this guard cell response (Ribeiro et al., 2009). Whether NO exerts its effects directly on ABA receptors or on some downstream element of ABA signaling cascade is obviously an important question that remains to be answered.
Nitric oxide has also been reported to influence several plant developmental events in which GAs play crucial roles, such as seed germination, hypocotyl elongation, acquisition of photomorphogenic traits, primary root growth, reorientation, and growth of pollen tubes, among others (Beligni and Lamattina, 2000; Prado et al., 2008; Tonón et al., 2010; Leon and Lozano-Juste, 2011); however, thus far, the actual interaction between NO and GAs has been described for only a limited number of these physiological events. In fact, most of our current knowledge of the mechanisms underlying the interplay between GAs and NO is restricted to the regulation of seed germination (Beligni et al., 2002; Bethke et al., 2007) and the inhibition of hypocotyl elongation during seedling de-etiolation (Leon and Lozano-Juste, 2011). During the control of these responses, NO has been described to act upstream of GA (Bethke et al., 2007), regulating both GA biosynthesis and perception/transduction (Leon and Lozano-Juste, 2011).
A certain level of antagonism between NO and GAs has been observed for most of the physiological processes in which both of these signaling compounds participate. A mounting body of evidence has indicated that DELLA proteins apparently represent a key crosstalk component between GA and NO signaling interactions (Figure 7; Leon and Lozano-Juste, 2011). DELLA proteins are a relatively small family of transcriptional regulators notably important for the integration of diverse hormonal signals, such as GAs, ethylene, jasmonate (JA), and ABA (Achard et al., 2003; Gao et al., 2011; Ross et al., 2011). During GA signaling transduction, for instance, the hormonal molecules interact with GA INSENSITIVE DWARF1 (GID1) receptors, which, in turn, binds a DELLA protein and subsequently directs the GA–GID1–DELLA complex to the E3 ubiquitin ligase SLEEPY1 (SLY1), thereby promoting DELLA degradation at the proteasome (Figure 7). Given that DELLAs mainly act by repressing the transcription of GA-regulated genes, the perception and transduction of the GA signal leads, as a final result, to a decrease in DELLA concentration into the cell and a consequent induction of GA-responsive genes.
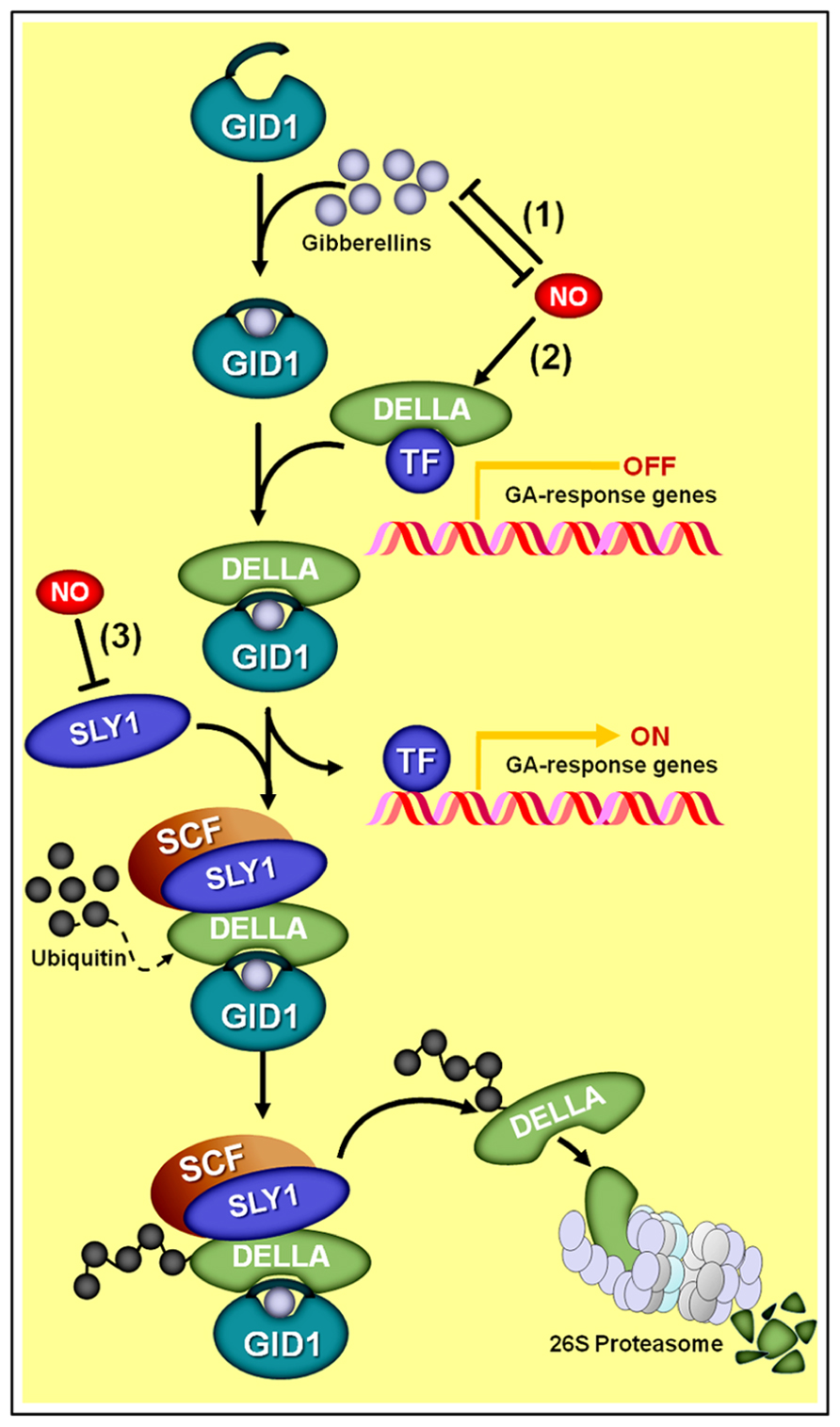
FIGURE 7. Schematic representation of NO–gibberellin antagonistic interactions. (1) A mutual antagonism controls the endogenous levels of NO and gibberellins in Arabidopsis seedlings. (2) Additionally, NO negatively influences GA signaling by promoting the accumulation of DELLA proteins, whose presence represses the transcription of GA-regulated genes. Since the degradation of DELLAs through SCF-26S proteasome-mediated proteolysis depends on the interaction of these proteins with the complex formed by active gibberellin molecules associated with the receptor GA INSENSITIVE DWARF1 (GID1) and the E3 ubiquitin ligase SLEEPY1 (SLY1), the NO-driven increase in DELLAs and reduction in SLY1 abundance (3) negatively impacts the transduction of the GA signal.
Interestingly, recent studies have indicated that NO triggers the opposite effect on cellular DELLA concentration, promoting the accumulation of this protein and a consequent negative impact on GA signal transduction (Figure 7). Essentially, this NO-driven DELLA accumulation can be interpreted as a reduction in tissue sensitivity to GA since a larger number of GA–GID1–DELLA complexes will need to be formed in order to mark an adequate quantity of DELLA proteins for proteasome degradation, thereby leading to a satisfactory level of transcriptional de-repression of GA-regulated genes. This differential effect of NO and GAs on DELLA regulation might account, at least in part, for the antagonism observed between these two signaling compounds during the regulation of physiological processes, such as hypocotyl elongation (Leon and Lozano-Juste, 2011) and primary root growth (Fernández-Marcos et al., 2012) in Arabidopsis.
In addition, studies performed on nia1,2noa1-2 seedlings revealed that this NO-deficient mutant presents defective DELLA accumulation associated with an up-regulation of the E3 ubiquitin ligase SLY1 (Figure 7), resulting in increased GA sensitivity and deficient de-etiolation under red light (Leon and Lozano-Juste, 2011). Further emphasizing the potential role for DELLAs in the GA–NO antagonistic interactions, exogenous NO was also demonstrated to induce the accumulation of GA-regulated DELLA proteins (Leon and Lozano-Juste, 2011), very likely by negatively regulating the GID1–SLY1 system of DELLA tagging for degradation (Figure 7). However, as pointed out by Leon and Lozano-Juste (2011), the regulation of DELLA turnover and activity may represent the main but not the only target for NO action in regulating plant growth and other GA-mediated developmental responses since DELLA-independent mechanisms might also be implicated.
Besides the negative action of NO on GA signaling network, a mutual antagonism controlling the endogenous levels of these two signaling molecules has also recently been proposed (Figure 7) (Leon and Lozano-Juste, 2011). Supporting this suggestion, etiolated seedlings of the GA-deficient Arabidopsis mutant ga1-3 have been shown to exhibit NO levels significantly higher than those observed in the WT genotype. Moreover, both ga1-3 mutant and WT seedlings showed reduced NO levels after GA3 supplementation, thereby suggesting that GAs negatively modulates NO production (Leon and Lozano-Juste, 2011). On the other hand, WT Arabidopsis seedlings treated with SNP presented a significant reduction in endogenous GA levels (Leon and Lozano-Juste, 2011). Based on a detailed analysis of the expression of Arabidopsis genes involved in GA biosynthesis (GA20oxidase and GA3oxidase) and catabolism (GA2oxidase), GA20ox3 was identified as the only gene significantly up-regulated in the NO-deficient nia1,2noa1-2 mutant and down-regulated in NO-treated WT seedlings (Leon and Lozano-Juste, 2011).
Under certain circumstances, however, NO seems to play a stimulatory rather than inhibitory role in the GA biosynthetic machinery (Bethke et al., 2007). Exemplifying such a synergist relationship, Bethke et al. (2007) reported that NO generation was required for the transcription of two GA3oxidase genes (GA3ox1 and GA3ox2) during the Arabidopsis seed dormancy breaking. Another indication of the positive interaction between GA and NO has recently been reported in wheat roots, for which the SNP-induced apical growth was associated with increased GA3 levels (He et al., 2012).
Apart from the above-mentioned evidence of NO acting upstream of GA, a certain level of uncertainty remains as to whether NO and GA actually share a common signaling route or just act through parallel, independent cascades during the regulation of some plant responses. During seed dormancy breaking, for instance, although there is virtually no doubt that both of these signal molecules promote germination in a number of species (Giba et al., 1998; Beligni and Lamattina, 2000; Kopyra and Gwozdz, 2003), whether and how NO and GA interact during this process still needs further characterization.
In fact, whereas a mounting body of evidence indicates that NO selectively interferes in some specific GA-induced events associated with the seed germination process, such as the longevity of cereal aleurone cells (Beligni et al., 2002), transcription of Myb transcription factor (GAMYB), and amylase synthesis (Wu et al., 2013), for some other responses associated with the germination process, no indications of additive or antagonistic responses have been found when both GA and NO were exogenously applied (Zhang et al., 2005). In addition, a rapid burst in NO production has been detected during early seed germination (Simontacchi et al., 2004), which has been speculated to be temporally dissociated from the action of GAs at later stages of seed germination (Zhang et al., 2005).
Regardless of whether or not NO and GA share a common signaling cascade during seed dormancy breaking, the stimulation of seed germination by either of these substances can be blocked by sufficiently high concentrations of ABA (Bethke et al., 2004, 2006; Sarath et al., 2006; Dong et al., 2012). Considering that NO may stimulate germination not only by breaking seed dormancy but also by alleviating the influence of environmental factors inhibitory to the germination process (Bethke et al., 2007), a NO-hormonal network much more complex than the interaction between NO, GA, and ABA might possibly be involved in the regulation of this critically important step in the plant life cycle.
A significant number of the currently available reports on the interaction between NO and ethylene suggest an antagonistic relationship between these two gaseous molecules (Leshem et al., 1998; Lamattina et al., 2003; Manjunatha et al., 2010). The first and presently most explored plant phenomenon in which NO was demonstrated to counteract ethylene production and action is the control of fruit ripening and the regulation of leaf and flower senescence (Leshem et al., 1998; Manjunatha et al., 2010). For these responses, ethylene has long been identified as a key promotive signal, and a large number of reports indicate that the production and perception mechanisms of this plant hormone are under strict regulation, depending not only on the plant developmental program but also on a number of environmental factors (Grbić and Bleecker, 1995; Fischer, 2012). Additional studies revealed that exogenous application of NO, either by direct fumigation or by means of NO-releasing chemicals, delays senescence of both vegetative and reproductive organs by negatively regulating a number of elements involved in ethylene production (Leshem and Haramaty, 1996; Leshem et al., 1998; Wills et al., 2000; Zhu et al., 2006; Liu et al., 2007; Manjunatha et al., 2010, 2012). Corroborating this pharmacological evidence, measurements of ethylene and NO emission during either fruit ripening (Leshem et al., 1998; Leshem and Pinchasov, 2000) or plant senescence (Magalh a, 2000; Corpas et al., 2004) revealed an opposite trend between these gases, in which ethylene production increases, whereas NO levels decrease during the induction and establishment of these processes.
Recent studies have revealed that the inhibition of fruit ethylene production by NO may be attributed to a reduction in the transcript level and/or activity of key ethylene biosynthetic enzymes (Manjunatha et al., 2010). In vegetative and reproductive plant tissues, ethylene production depends on the conversion of the S-adenosyl methionine (SAM), derived from “Yang cycle,” into the immediate ethylene precursor 1-aminocyclopropane 1-carboxylic acid (ACC) through ACC synthase (ACS) activity (Figure 8). The ACC formed may be subsequently converted to ethylene due to the activity of a second enzyme, the ACC oxidase (ACO; Yang and Hoffman, 1984). Since the abundance of ACC, ACS, and ACO in plant tissues represents a critical aspect for determining ethylene production rates (Barry et al., 1996; Barry et al., 2000), an inhibitory effect of NO on any of these elements can be expected to be an efficient mechanism for down-regulating ethylene synthesis.
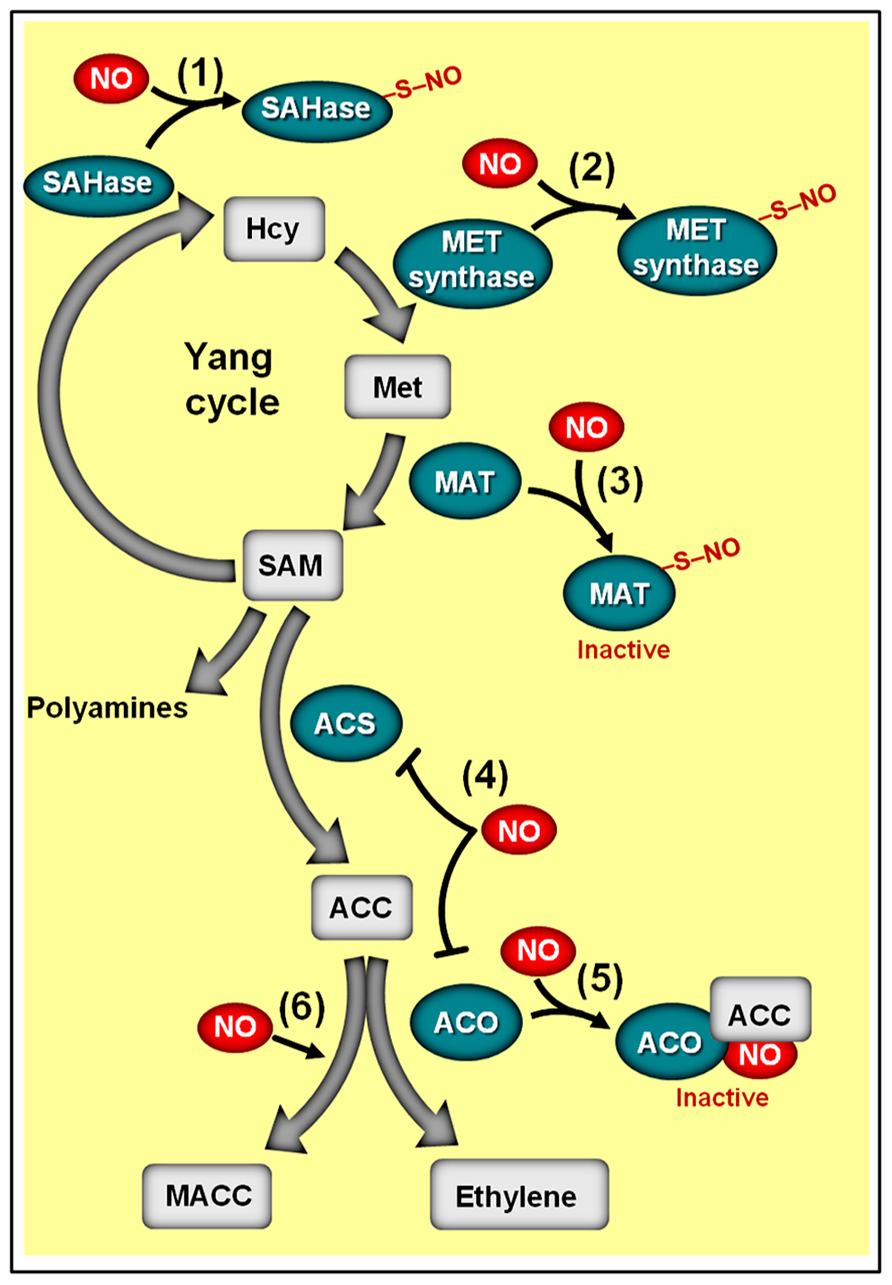
FIGURE 8. Schematic representation of NO-ethylene antagonistic interactions. The methylmethionine cycle enzymes adenosyl homocysteinase (SAHase) (1) and methionine synthase (MET synthase) (2), whose activities are responsible for the production of homocysteinase (Hcy) and methionine (Met), respectively, may undergo S-nitrosylation. (3) Additionally, the activity of the Arabidopsis methionine adenosyltransferase 1 (MAT1) can be suppressed by S-nitrosylation, thereby repressing the conversion of methionine (Met) to S-adenosyl methionine (SAM). (4) In ripening climacteric fruits, NO has been shown to inhibit the transcript levels of 1-aminocyclopropane 1-carboxylic acid (ACC) synthase (ACS) and/or ACC oxidase (ACO). (5) NO can also inhibit ACO activity by directly binding this enzyme, resulting in the ACO–NO binary complex, which subsequently originates a ternary stable complex ACO–NO–ACC. (6) NO-driven accumulation of non-volatile ACC metabolite 1-malonyl aminocyclopropane-1-carboxylic acid (MACC) has also been reported. Biosynthetic enzymes are represented with green ovals and metabolic substrates and products with gray rectangles. Protein S-nitrosylation is represented by “–S–NO.” Note that the impact of S-nitrosylation on the activities of SAHase, MET synthase, and ACS remains to be determined.
As revealed by a series of studies on climacteric fruits, exogenous NO indeed has the capacity to modulate both the transcription and the activity of both ACS and ACO (Figure 8), consequently impacting not only the levels of ethylene production but also the accumulation of ACC (Manjunatha et al., 2010). In tomato fruits, for instance, although the expression of all ACS homologs remained virtually unchanged following NO fumigation, the transcript abundance of ACO genes, such as LeACO1, LeACOH2, and LeACO4, and the levels of ethylene emission were reduced and/or delayed when NO was applied before the start of the ripening process (Eum et al., 2009). In banana fruits, on the other hand, NO negatively impacted the expression of both ACS and ACO homologs, leading to a reduction in ACO activity and ethylene emission as well as an accumulation of ACC (Cheng et al., 2009).
Apart from controlling the transcript levels of ACS and ACO, NO may also regulate ACS activity via S-nitrosylation (Abat and Deswal, 2009) and influence ACO activity by a mechanism involving the direct binding of NO to the enzyme, resulting in the ACO–NO binary complex, which is then chelated by ACC to produce the ternary stable complex ACO–NO–ACC (Figure 8) (Tierney et al., 2005; Zhu et al., 2006; Manjunatha et al., 2010). Currently, the impacts of S-nitrosylation on ACS activity remain uncharacterized, and the occurrence of the ACO–NO–ACC ternary complex is exclusively described during in vitro studies conducted on recombinant ACO (Tierney et al., 2005); therefore, the actual in vivo implications of such regulatory mechanisms still need further elucidation. Nevertheless, the hypothetical formation of an ACO–NO–ACC complex has already been inferred as possibly responsible for the reduction of ACO activity in climacteric peach (Prunus persica) fruits subjected to NO fumigation, which resulted in a concomitant decrease in ethylene emission and accumulation of ACC (Zhu et al., 2006). In this specific case, the NO-induced reduction of ACO activity was accompanied by an increment in the accumulation of the non-volatile ACC metabolite 1-malonyl aminocyclopropane-1-carboxylic acid (MACC; Figure 8), which was interpreted as a secondary effect of NO during the ripening of these fruits (Zhu et al., 2006).
Besides stimulating the irreversible conversion of ACC into MACC, NO may also negatively impact the turnover of SAM, which is the main precursor molecule for ACC synthesis. Supporting this assumption, proteomic analysis of Arabidopsis plants revealed that the methylmethionine cycle enzymes adenosyl homocysteinase (SAHase), methionine synthase (MET synthase) and methionine adenosyltransferase (MAT, also known as SAM synthase), whose activities are responsible for the production of homocysteinase (HCY), methionine (Met), and SAM, respectively, may undergo S-nitrosylation (Figure 8). In addition, similar analyses conducted on GSNO-treated protein extracts of Kalanchoe pinnata (Abat et al., 2008) and Brassica juncea (Abat and Deswal, 2009) also identified cobalamin-independent MET synthases as a common target of S-nitrosylation. Whereas the influence of S-nitrosylation on the activities of SAHase and MET synthase has yet to be determined, a detailed study conducted by Lindermayr et al. (2006) revealed that the activity of MAT1, one of the three Arabidopsis MAT isoforms, is indeed suppressed via S-nitrosylation at cys-114, having as a logical consequence the depletion of the SAM pool and a reduction in ethylene production. Curiously, the study conducted by Lindermayr et al. (2006) was the first detailed characterization of S-nitrosylation in plant systems, opening up a new window of opportunities for accessing the actual relevance of this NO-dependent post-translational regulatory mechanism in plant signaling.
In contrast to the above-mentioned evidence of an antagonistic relationship between NO and ethylene during the maturation, senescence, and abscission of plant organs, a number of reports have also indicated that NO donors, such as SNP, might sometimes stimulate, rather than negate, ethylene production in certain plant materials, such as non-senescent leaf tissues of Arabidopsis, tobacco, and maize (Magalhães et al., 2000; Ederli et al., 2006; Wang et al., 2006; Mur et al., 2008; Ahlfors et al., 2009) and apple embryos (Gniazdowska et al., 2007). In tobacco leaves, for instance, SNP infiltration has been show to stimulate ACS expression (Ederli et al., 2006; Mur et al., 2008), whereas in Arabidopsis roots the application of GSNO positively impacted the transcript levels of not only ACS but also other key ethylene biosynthetic enzymes, such as SAM synthetases, ACOs, and 5-methylthioribose kinase (MTK; Garcia et al., 2011). Further emphasizing a stimulatory influence of NO on ethylene biosynthesis, ethylene production is usually elevated when the NO accumulation is promoted via suppression of nsHbs gene expression (Manac’h-Little et al., 2005; Hebelstrup et al., 2012). Similarly, the increased NO production observed in transgenic tobacco lines expressing mammalian NOS were accompanied by a higher expression of ACO and some other ethylene-related genes (Chun et al., 2012). Moreover, a concomitant increase in both ethylene and NO emission has been consistently observed both in tobacco leaves undergoing bacterially triggered hypersensitive response (Mur et al., 2012b) and in Arabidopsis and cucumber (Cucumis sativus) roots subjected to Fe deficiency (Garcia et al., 2011).
Besides these indications of a positive influence of NO on ethylene production, some data also seem to support a stimulatory role of ethylene on NO production under certain circumstances (Garcia et al., 2011). Earlier in the research of NO–ethylene interaction in plants, Leshem and Haramaty (1996) reported that exogenous ACC induced significant increases in both ethylene and NO emission in pea (Pisum sativum) leaves. More recently, Garcia et al. (2011) have also detected increased NO levels in the root subapical region of Arabidopsis and cucumber plants exposed to ACC. In addition, these authors reported that inhibitors of ethylene biosynthesis and action completely abolished the increases in NO levels in roots of plants subjected to Fe deficiency. In contrast, ethylene supplementation or depletion, respectively, repressed and promoted NO production during the abscission of mature olive fruits (Parra-Lobato and Gomez-Jimenez, 2011), which apparently indicates that under certain circumstances ethylene may negatively, rather than positively, impact the endogenous NO levels. In agreement with this, ethylene has sometimes been shown to induce class 1 nsHbs (Qu et al., 2006; Bustos-Sanmamed et al., 2011), which in turn may lead to reductions in tissue concentration of NO.
Surprisingly, the possible influence of NO on ethylene signal transduction elements has remained virtually unexplored, both during antagonistic (e.g., fruit ripening and leaf senescence) and synergistic (e.g., plant defense to biotic stresses and Fe deficiency) interactions between these signaling substances. Therefore, it is currently unknown whether NO might regulate the transcripts levels or activities of receptors, signal transduction proteins and/or transcription factors involved in ethylene signaling, which would very likely impact the sensitivity of the plant tissues to this plant hormone. In one of the few studies on this line, Niu and Guo (2012) demonstrated that the dark-induced early senescence phenotype of the Arabidopsis NO-deficient mutant noa1 was suppressed by mutation in ETHYLENE INSENSITIVE 2 (EIN2) and indicated that this protein might act downstream of NO signaling, possibly playing a key role as a crosstalk point between ethylene and NO signaling cascades.
Besides interacting with the five “classical” phytohormone classes, NO has also been reported to crosstalk with other plant hormones, including JAs, SA, polyamines, and brassinosteroids. Some of these interactions, such as the interplay between NO, SA, and JA in plant defense responses, have been investigated in great detail, uncovering impressively complex NO–phytohormone interaction networks. A detailed discussion about these interactions is beyond the scope of the present work; instead, just some brief, general comments, and examples of these NO–phytohormone crosstalks will be provided below.
As recently reviewed by Yu et al. (2012) and Mur et al. (2013), during the induction of plant defense responses against biotic challenges, NO positively impacts the production of both SA and JA (Feechan et al., 2005; Chun et al., 2012; Mur et al., 2012b) and, at the same time, NO modulates SA signaling by controlling the oligomerization status of the translational activator NON-EXPRESSER OF PATHOGENESIS-RELATED GENE1 (NPR1) via S-nitrosylation at cys156 (Tada et al., 2008; Figure 9). S-nitrosylation of NPR1 facilitates its oligomerization (Tada et al., 2008) and permanence in the cytosol (Fu et al., 2012), where it may interact with SA receptors (NPR3/4). Following such interaction with NPR3/4, the S-nitrosylated cys156 of NPR1 is reduced (Tada et al., 2008), promoting NPR1 monomer formation and its consequent migration to the nucleus, where this protein may interact with several TGA-class transcription factors that subsequently activate promoters of SA-responsive genes (Mur et al., 2012b; Figure 9). In contrast, the presence of the S-nitrosylated, oligomeric form of NPR1 in the cytosol facilitates the repression of JA-triggered responses (Spoel et al., 2003). Consequently, this NO-dependent PTM of NPR1 seems to play a key integrative role during the hormonal signaling cascades leading to coordinated plant immunity responses (Yu et al., 2012; Mur et al., 2013). In parallel, S-nitrosylation of SA-BINDING PROTEIN 3 (SABP3) at cys280, which takes place during late stages of bacterial infection, represses its capacity to bind SA, and antagonizes the expression of plant immunity responses (Wang et al., 2009), thereby representing a negative feedback loop apparently essential for the correct regulation of SA-modulated plant defense against biotic challenges (Figure 9).
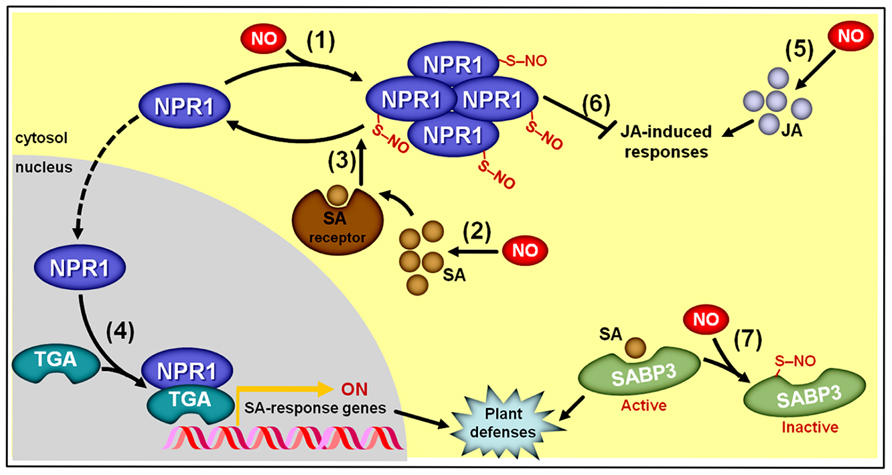
FIGURE 9. Simplified schematic representation of NO, salicylic acid (SA), and jasmonic acid (JA) interactions during plant responses to biotic challenges. (1) S-nitrosylation of NON-EXPRESSER OF PATHOGENESIS-RELATED GENE1 (NPR1) at cys156 promotes its oligomerization and permanence in the cytosol. (2) NO stimulates SA biosynthesis. (3) Oligomeric NPR1 is denitrosylated following its interaction with SA receptors, which promotes the formation of monomeric NPR1. (4) Monomeric NPR1 translocates to the nucleus, where this protein binds TGA-class transcription factors, which subsequently activate promoters of SA-responsive genes. (5) NO also stimulates JA biosynthesis. (6) Cytosolic, oligomeric NPR1 represses JA-triggered responses. (7) At late stages of bacterial infection, S-nitrosylation of SA-binding protein 3 (SABP3) at cys280 represses its SA binding capacity, thereby promoting a negative feedback loop during the defense signaling pathway. Protein S-nitrosylation is represented by “–S–NO.”
Accumulating evidence indicates that NO might also mediate both developmental and stress responses induced by polyamines (Wimalasekera et al., 2011). Briefly, very rapid NO production has been observed in plant tissues exposed to mM concentration of polyamines (Tun et al., 2006), which has sometimes been interpreted as an indication of a potential NO biosynthetic pathway involving the catabolism of these plant hormones (Wimalasekera et al., 2011). Given the absence of a lag phase between the application of polyamines and the rise in NO endogenous levels (Tun et al., 2006), it is currently assumed that these hormones might be directly converted to NO by the action of one or more enzymes, whose identities are yet to be determined (Wimalasekera et al., 2011). So far, it is only known that the polyamine-induced NO production can be quenched by mammalian NOS inhibitors and is not affected in Arabidopsis NR-deficient mutants (Tun et al., 2006; Wimalasekera et al., 2011). Whether polyamines act as substrates, cofactors, or signals for promoting NO synthesis also needs to be better determined; therefore, monitoring the formation of 15NO from isotopic-labeled polyamines in plant tissues or extracts seems an important experiment in future studies. A possible influence of NO on polyamine metabolism has been demonstrated in some studies (Fan et al., 2013) but not in others (Arasimowicz-Jelonek et al., 2009); consequently, this topic still deserves further investigation.
Considering that polyamines and ethylene share SAM as a common precursor, all the basic NO-dependent mechanisms controlling the SAM pools discussed earlier in this review (Figure 8) might also indirectly affect polyamine synthesis in plants. In addition, we also need to keep in mind that L-arginine is substrate for the production of polyamines, via arginase and arginine decarboxylase activities, as well as NO, via NOS-like activities; therefore, the availability of this particular amino acid might also influence NO/polyamine connections in plants and other organisms. In mammals, for instance, the occurrence of an arginine switch, in which NOS and arginase compete for arginine, seems to be supported by a great deal of experimental evidence (Satriano, 2004). In parallel, literature data in the animal field also indicates that polyamines such as spermidine and spermine influence NO production via NOS activity during diverse physiological responses (Guerra et al., 2006), which may represent an important source of information to guide current and future research on the NO and polyamine interactions in plants. The polyamine precursor agmatine, for example, has been demonstrated to act either as an alternative substrate or a competitive inhibitor of mammalian NOS, depending on the isoform or physiological process taken under consideration (Satriano, 2003; Raghavan and Dikshit, 2004), thereby indicating a possible role for this compound as an endogenous regulator of NO generation in mammals. In plants, the ameliorative effects of both polyamines and NO under stressful conditions (Arasimowicz-Jelonek et al., 2009; Wimalasekera et al., 2011; Gupta et al., 2013) might represent an important driving force to stimulate further studies on the interaction between these critically important signaling compounds.
Generation of information about whether and how NO and brassinosteroids interact has only recently begun (Zhang et al., 2011; Tossi et al., 2013). In one of these studies, Zhang et al. (2011) demonstrated that nM concentrations of brassinosteroids promoted rapid increases in the NO levels of leaf mesophyll cells, which together with some other evidence allowed the authors to place NO as a possible intermediate in the brassinosteroid-induced ABA biosynthesis in maize leaves. More recently, Tossi et al. (2013) reported that NR and NOS-like activities are probably involved in the brassinosteroid-induced NO production in Arabidopsis and that NO very likely mediates brassinosteroid-triggered modifications in plant root architecture.
Despite the methodological difficulties and conceptual complexity intrinsically involved in the elucidation of the exact mechanisms responsible for interconnecting plant hormones and NO signaling during the coordination of plant metabolism and development, some cutting edge insights into the NO–phytohormone crosstalks have recently been achieved.
Many downstream and upstream components of the NO signaling cascades have been identified, and NO-dependent PTMs, notably S-nitrosylation, have emerged as critical mechanisms controlling key elements involved in plant hormone production and signaling. As highlighted in the course of this review, by chemically modifying these hormone-related proteins, NO may modify plant hormone metabolism and signaling at multiple, diversified levels. The identification and functional analysis of the protein targets of NO-dependent PTMs and whose action determines the delicate hormonal homeostasis in plants has been, and will probably continue to be, an approach of upmost relevance in NO–phytohormone studies.
Currently, the physiological relevance of NO-dependent chemical modifications of phytohormone-related proteins has been poorly investigated in planta; therefore, this remains a rich area for future investigation. Clarifying how these NO-triggered PTMs, particularly S-nitrosylation and tyrosine nitration, actually control protein activity, subcellular localization as well as protein–protein, protein–DNA, protein–cofactors, or even protein–hormone binding capacity will inexorably involve the use of a wide range of experimental strategies and methodological approaches, some of which are currently available (e.g., overexpression of modified proteins in mutant genetic backgrounds) and others yet to be developed. Since some proteins are targets of multiple NO-dependent PTMs, sometimes even involving different types of these chemical modifications (e.g., both S-nitrosylation and nitration; Lozano-Juste et al., 2011; Astier et al., 2012), it would be enlightening to determine the impacts of concomitant NO-triggered modifications on the same protein.
Moreover, characterizing how these target proteins are chemically modified by NO and NO-derivates at the right time and place seems to be another promising area of progress in NO–phytohormone interactions. Addressing this question inevitably implies dealing with several critical aspects of the NO physiology that still require further elucidation. Firstly, the basic mechanisms responsible for NO production, removal, and transport in plants continues to represent a critical impediment for advances in the clarification of how NO levels are temporally and spatially controlled by plant hormones and other stimuli. A fine-tuned equilibrium between NO production and removal (e.g., biosynthesis versus degradation, conjugation versus deconjugation) might possibly exist to determine both the localization and the concentration of NO and NO-derivates within the plant cells. Secondly, given the impressive diversity of target proteins, which are ubiquitously distributed within the plant cells, the existence of a certain subcellular compartmentation in NO production and action is an assumption that urgently needs to be investigated in greater detail. Moreover, a concentration-dependent action mode for NO has also been proposed (Mur et al., 2013), in which distinct responses may be triggered depending on the abundance of this free radical. Obviously, the development of more sensitive and specific means to determine the subcellular localization and concentration of NO and NO-derivates is critical for further advances in this area. Thirdly, considering that both S-nitrosylation and tyrosine nitration are apparently reversible events, more conclusive studies on the denitrosylation and denitration systems as well as the general turnover of S-nitrosylated and nitrated proteins in plant cells also seems a logical requirement for a deeper understanding of the dynamics of these regulatory processes. Similar to the action of protein phosphatases during the regulation of protein phosphorylation, denitrosylases and denitrases may possibly play an important role in defining the kinetics of the NO impacts on plant signaling cascades.
Another aspect that also deserves further attention is the potential existence of feed-forward cycles, in which NO modulates the production and/or signaling of specific plant hormones and these same hormonal species influence the machinery responsible for controlling NO endogenous levels. As described in the course of this review, accumulating pharmacological and genetic evidence demonstrates that representatives of virtually all classes of plant hormones may impact, at least at a certain degree, the endogenous concentration and/or distribution of NO and, also very frequently, literature data seems to indicate that changes in NO levels might trigger alterations on the metabolism and/or signaling of many, if not all, hormonal classes. It is not clear, however, whether these processes occur at the same place and time, which is critical for generating authentic feed-forward cycles involving these signaling substances. Naturally, a more complete characterization of the actual impacts of specific plant hormones on the NO biosynthetic and removal machinery (e.g., NR, NOA1, GSNOR, nsHbs) seems a key step in such research topic.
Additionally, as also discussed earlier in this review, NO might affect the signaling transduction of certain hormones, such as auxins and GAs, by modulating signaling elements (e.g., receptor, signaling transduction molecules) that impact the general dynamics of ubiquitination and proteasome-dependent degradation of repressor proteins (e.g., AUX/IAA and DELLA proteins). Interestingly, in animal systems, NO has consistently been shown to influence protein stability via regulation of ubiquitination and proteasome-dependent proteolysis (Hess et al., 2005) and, at least in humans, ubiquitin ligases themselves are targets of S-nitrosylation (Chung et al., 2004). Considering that several plant hormone signaling transduction mechanisms are based on the ubiquitination and subsequent proteasome-dependent degradation of repressor proteins, investigating whether NO might also directly affect this protein degradation labeling system in plants seems a promising venue for uncovering additional mechanisms possibly involved in NO signaling in plant systems.
Another intriguing question that remains to be answered is how plants can distinguish endogenously produced NO signals from the NO naturally present in the environment (e.g., atmosphere, rhizosphere). Whereas the gaseous and highly diffusible nature of NO may promote certain movement of this molecule inside the plant tissue and at the plant–environment interface, the high reactivity and inherent instability of NO may possibly limit the diffusion of this free radical through biological tissues. In several aspects, this seems a relevant and challenging question to be answered in the future.
Finally, we must remain open-minded to conceive increasingly complex NO–phytohormone interconnection nodes since new targets of NO-dependent PTMs and other upstream and downstream elements of NO signaling cascades will likely be identified in the future. At the same time, more complete pictures mechanistically explaining how multiple plant hormones may simultaneously interact with NO to control specific plant responses might also emerge, very likely leading to exciting new models of NO–phytohormone interaction networks. Moreover, this whole scenario will be further complicated when the intensive research conducted today in a restricted number of plant models (e.g., Arabidopsis, tomato, rice) is extended to a broader range of plant species and environmental contexts. Altogether, this knowledge will improve our ability to define the actual roles of NO during the regulation of the distinct plant responses controlled by this multipurpose signaling molecule and may also lead to new opportunities to manipulate NO–phytohormone interactions and, thus, regulate plant growth, development, and metabolism.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author wishes to thank members of his lab for helpful comments on this manuscript. Work in the author’s laboratory on NO signaling has been supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Abat, J. K., and Deswal, R. (2009). Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: change in S-nitrosylation of Rubisco is responsible for the inactivation of its carboxylase activity. Proteomics 9, 4368–4380. doi: 10.1002/pmic.200800985
Abat, J. K., Mattoo, A. K., and Deswal, R. (2008). S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata - ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J. 275, 2862–2872. doi: 10.1111/j.1742-4658.2008.06425.x
Abello, N., Kerstjens, H. A., Postma, D. S., and Bischoff, R. (2009). Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J. Proteome Res. 8, 3222–3238. doi: 10.1021/pr900039c
Achard, P., Vriezen, W. H., Van Der Straeten, D., and Harberd, N. P. (2003). Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15, 2816–2825. doi: 10.1105/tpc.015685
Ahlfors, R., Brosche, M., and Kangasjarvi, J. (2009). Ozone and nitric oxide interaction in Arabidopsis thaliana: a role for ethylene? Plant Signal. Behav. 4, 878–879. doi: 10.4161/psb.4.9.9428
Arasimowicz-Jelonek, M., Floryszak-Wieczorek, J., and Kubis, J. (2009). Interaction between polyamine and nitric oxide signaling in adaptive responses to drought in cucumber. J. Plant Growth Regul. 28, 177–186. doi: 10.1007/s00344-009-9086-7
Astier, J., Kulik, A., Koen, E., Besson-Bard, A., Bourque, S., Jeandroz, S., et al. (2012). Protein S-nitrosylation: what’s going on in plants? Free Radic. Biol. Med. 53, 1101–1110. doi: 10.1016/j.freeradbiomed.2012.06.032
Astier, J., and Lindermayr, C. (2012). Nitric oxide-dependent posttranslational modification in plants: an update. Int. J. Mol. Sci. 13, 15193–15208. doi: 10.3390/ijms131115193
Astier, J., Rasul, S., Koen, E., Manzoor, H., Besson-Bard, A., Lamotte, O., et al. (2011). S-nitrosylation: an emerging post-translational protein modification in plants. Plant Sci. 181, 527–533. doi: 10.1016/j.plantsci.2011.02.011
Barroso, J. B., Corpas, F. J., Carreras, A., Rodriguez-Serrano, M., Esteban, F. J., Fernandez-Ocana, A., et al. (2006). Localization of S-nitrosoglutathione and expression of S-nitrosoglutathione reductase in pea plants under cadmium stress. J. Exp. Bot. 57, 1785–1793. doi: 10.1093/jxb/erj175
Barry, C. S., Blume, B., Bouzayen, M., Cooper, W., Hamilton, A. J., and Grierson, D. (1996). Differential expression of the 1-amino-cyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 9, 525–535. doi: 10.1046/j.1365-313X.1996.09040525.x
Barry, C. S., Llop-Tous, M. I., and Grierson, D. (2000). The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 123, 979–986. doi: 10.1104/pp.123.3.979
Beligni, M. V., Fath, A., Bethke, P. C., Lamattina, L., and Jones, R. L. (2002). Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol. 129, 1642–1650. doi: 10.1104/pp.002337
Beligni, M. V., and Lamattina, L. (2000). Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 210, 215–221. doi: 10.1007/PL00008128
Besson-Bard, A., Astier, J., Rasul, S., Wawer, I., Dubreuil-Maurizi, C., Jeandroz, S., et al. (2009). Current view of nitric oxide-responsive genes in plants. Plant Sci. 177, 302–309. doi: 10.1016/j.plantsci.2009.06.006
Besson-Bard, A., Griveau, S., Bedioui, F., and Wendehenne, D. (2008). Real-time electrochemical detection of extracellular nitric oxide in tobacco cells exposed to cryptogein, an elicitor of defence responses. J. Exp. Bot. 59, 3407–3414. doi: 10.1093/jxb/ern189
Bethke, P. C., Gubler, F., Jacobsen, J. V., and Jones, R. L. (2004). Dormancy of Arabidopsis seeds and barley grains can be broken by nitric oxide. Planta 219, 847–855. doi: 10.1007/s00425-004-1282-x
Bethke, P. C., Libourel, I. G. L., Aoyama, N., Chung, Y. Y., Still, D. W., and Jones, R. L. (2007). The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 143, 1173–1188. doi: 10.1104/pp.106.093435
Bethke, P. C., Libourel, I. G. L., and Jones, R. L. (2006). Nitric oxide reduces seed dormancy in Arabidopsis. J. Exp. Bot. 57, 517–526. doi: 10.1093/jxb/erj060
Bright, J., Desikan, R., Hancock, J. T., Weir, I. S., and Neill, S. J. (2006). ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 45, 113–122. doi: 10.1111/j.1365-313X.2005.02615.x
Bustos-Sanmamed, P., Tovar-Mendez, A., Crespi, M., Sato, S., Tabata, S., and Becana, M. (2011). Regulation of nonsymbiotic and truncated hemoglobin genes of Lotus japonicus in plant organs and in response to nitric oxide and hormones. New Phytol. 189, 765–776. doi: 10.1111/j.1469-8137.2010.03527.x
Carimi, F., Zottini, M., Costa, A., Cattelan, I., De Michele, R., Terzi, M., et al. (2005). NO signalling in cytokinin-induced programmed cell death. Plant Cell Environ. 28, 1171–1178. doi: 10.1111/j.1365-3040.2005.01355.x
Cecconi, D., Orzetti, S., Vandelle, E., Rinalducci, S., Zolla, L., and Delledonne, M. (2009). Protein nitration during defense response in Arabidopsis thaliana. Electrophoresis 30, 2460–2468. doi: 10.1002/elps.200800826
Chaki, M., Fernandez-Ocana, A. M., Valderrama, R., Carreras, A., Esteban, F. J., Luque, F., et al. (2009a). Involvement of reactive nitrogen and oxygen species (RNS and ROS) in sunflower-mildew interaction. Plant Cell Physiol. 50, 265–279. doi: 10.1093/pcp/pcn196
Chaki, M., Valderrama, R., Fernandez-Ocana, A. M., Carreras, A., Lopez-Jaramillo, J., Luque, F., et al. (2009b). Protein targets of tyrosine nitration in sunflower (Helianthus annuus L.) hypocotyls. J. Exp. Bot. 60, 4221–4234. doi: 10.1093/jxb/erp263
Chen, W. W., Yang, J. L., Qin, C., Jin, C. W., Mo, J. H., Ye, T., et al. (2010). Nitric oxide acts downstream of auxin to trigger root ferric-chelate reductase activity in response to iron deficiency in Arabidopsis. Plant Physiol. 154, 810–819. doi: 10.1104/pp.110.161109
Cheng, F. Y., Hsu, S. Y., and Kao, C. H. (2002). Nitric oxide counteracts the senescence of detached rice leaves induced by dehydration and polyethylene glycol but not by sorbitol. Plant Growth Regul. 38, 265–272. doi: 10.1023/A:1021529204978
Cheng, G., Yang, E., Lu, W., Jia, Y., Jiang, Y., and Duan, X. (2009). Effect of nitric oxide on ethylene synthesis and softening of banana fruit slice during ripening. J. Agric. Food Chem. 57, 5799–5804. doi: 10.1021/jf901173n
Chun, H. J., Park, H. C., Koo, S. C., Lee, J. H., Park, C. Y., Choi, M. S., et al. (2012). Constitutive expression of mammalian nitric oxide synthase in tobacco plants triggers disease resistance to pathogens. Mol. Cells 34, 463–471. doi: 10.1007/s10059-012-0213-0
Chung, K. K. K., Thomas, B., Li, X., Pletnikova, O., Troncoso, J. C., Marsh, L., et al. (2004). S-Nitrosylation of parkin regulates ubiquitination and compromises Parkin’s protective function. Science 304, 1328–1331. doi: 10.1126/science.1093891
Corpas, F. J., Alché, J. D. D., and Barroso, J. B. (2013). Current overview of S-nitrosoglutathione (GSNO) in higher plants. Front. Plant Sci. 4:126. doi: 10.3389/fpls.2013.00126
Corpas, F. J., Barroso, J. B., Carreras, A., Quiros, M., Leon, A. M., Romero-Puertas, M. C., et al. (2004). Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiol. 136, 2722–2733. doi: 10.1104/pp.104.042812
Corpas, F. J., Barroso, J. B., Carreras, A., Valderrama, R., Palma, J. M., Leon, A. M., et al. (2006). Constitutive arginine-dependent nitric oxide synthase activity in different organs of pea seedlings during plant development. Planta 224, 246–254. doi: 10.1007/s00425-005-0205-9
Corpas, F. J., Carreras, A., Esteban, F. J., Chaki, M., Valderrama, R., Del Río, L. A., et al. (2008a). Localization of S-nitrosothiols and assay of nitric oxide synthase and S-nitrosoglutathione reductase activity in plants. Method Enzymol. 437, 561–574. doi: 10.1016/S0076-6879(07)37028-6
Corpas, F. J., Chaki, M., Fernandez-Ocana, A., Valderrama, R., Palma, J. M., Carreras, A., et al. (2008b). Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant Cell Physiol. 49, 1711–1722. doi: 10.1093/pcp/pcn144
Correa-Aragunde, N., Foresi, N., and Lamattina, L. (2013). Structure diversity of nitric oxide synthases (NOS): the emergence of new forms in photosynthetic organisms. Front. Plant Sci. 4:232. doi: 10.3389/fpls.2013.00232
Correa-Aragunde, N., Graziano, M., Chevalier, C., and Lamattina, L. (2006). Nitric oxide modulates the expression of cell cycle regulatory genes during lateral root formation in tomato. J. Exp. Bot. 57, 581–588. doi: 10.1093/jxb/erj045
Correa-Aragunde, N., Graziano, M., and Lamattina, L. (2004). Nitric oxide plays a central role in determining lateral root development in tomato. Planta 218, 900–905. doi: 10.1007/s00425-003-1172-7
Desikan, R., Cheung, M. K., Bright, J., Henson, D., Hancock, J. T., and Neill, S. J. (2004). ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J. Exp. Bot. 55, 205–212. doi: 10.1093/jxb/erh033
Desikan, R., Griffiths, R., Hancock, J., and Neill, S. (2002). A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99, 16314–16318. doi: 10.1073/pnas.252461999
Dong, T. T., Tong, J. H., Xiao, L. T., Cheng, H. Y., and Song, S. Q. (2012). Nitrate, abscisic acid and gibberellin interactions on the thermoinhibition of lettuce seed germination. Plant Growth Regul. 66, 191–202. doi: 10.1007/s10725-011-9643-5
Du, S. T., Zhang, Y. S., Lin, X. Y., Wang, Y., and Tang, C. X. (2008). Regulation of nitrate reductase by nitric oxide in Chinese cabbage pakchoi (Brassica chinensis L.). Plant Cell Environ. 31, 195–204. doi: 10.1111/j.1365-3040.2007.01750.x
Dubovskaya, L. V., Bakakina, Y. S., Kolesneva, E. V., Sodel, D. L., Mcainsh, M. R., Hetherington, A. M., et al. (2011). cGMP-dependent ABA-induced stomatal closure in the ABA-insensitive Arabidopsis mutant abi1–1. New Phytol. 191, 57–69. doi: 10.1111/j.1469-8137.2011.03661.x
Durner, J., Wendehenne, D., and Klessig, D. F. (1998). Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. U.S.A 95, 10328–10333. doi: 10.1073/pnas.95.17.10328
Ederli, L., Meier, S., Borgogni, A., Reale, L., Ferranti, F., Gehring, C., et al. (2008). cGMP in ozone and NO dependent responses. Plant Signal. Behav. 3, 36–37. doi: 10.4161/psb.3.1.4818
Ederli, L., Morettini, R., Borgogni, A., Wasternack, C., Miersch, O., Reale, L., et al. (2006). Interaction between nitric oxide and ethylene in the induction of alternative oxidase in ozone-treated tobacco plants. Plant Physiol. 142, 595–608. doi: 10.1104/pp.106.085472
Eum, H. L., Kim, H. B., Choi, S. B., and Lee, S. K. (2009). Regulation of ethylene biosynthesis by nitric oxide in tomato (Solanum lycopersicum L.) fruit harvested at different ripening stages. Eur. Food Res. Technol. 228, 331–338. doi: 10.1007/s00217-008-0938-3
Fan, H. F., Du, C. X., and Guo, S. R. (2013). Nitric oxide enhances salt tolerance in cucumber seedlings by regulating free polyamine content. Environ. Exp. Bot. 86, 52–59. doi: 10.1016/j.envexpbot.2010.09.007
Feechan, A., Kwon, E., Yun, B.-W., Wang, Y., Pallas, J. A., and Loake, G. J. (2005). A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U.S.A. 102, 8054–8059. doi: 10.1073/pnas.0501456102
Feng, J., Wang, C., Chen, Q., Chen, H., Ren, B., Li, X., et al. (2013). S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nature 4, 1529–1529. doi: 10.1038/ncomms2541
Fernández-Marcos, M., Sanz, L., Lewis, D. R., Muday, G. K., and Lorenzo, O. (2011). Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-dependent acropetal auxin transport. Proc. Natl. Acad. Sci. U.S.A. 108, 18506–18511. doi: 10.1073/pnas.1108644108
Fernández-Marcos, M., Sanz, L., and Lorenzo, O. (2012). Nitric oxide: an emerging regulator of cell elongation during primary root growth. Plant Signal. Behav. 7, 196–200. doi: 10.4161/psb.18895
Fischer, A. M. (2012). The complex regulation of senescence. Crit. Rev. Biotechn. 31, 124–147. doi: 10.1080/07352689.2011.616065
Flores-Perez, U., Sauret-Gueto, S., Gas, E., Jarvis, P., and Rodriguez-Concepcion, M. (2008). A mutant impaired in the production of plastome-encoded proteins uncovers a mechanism for the homeostasis of isoprenoid biosynthetic enzymes in Arabidopsis plastids. Plant Cell 20, 1303–1315. doi: 10.1105/tpc.108.058768
Foissner, I., Wendehenne, D., Langebartels, C., and Durner, J. (2000). In vivo imaging of an elicitor-induced nitric oxide burst in tobacco. Plant J. 23, 817–824. doi: 10.1046/j.1365-313X.2000.00835.x
Foresi, N., Correa-Aragunde, N., Parisi, G., Calo, G., Salerno, G., and Lamattina, L. (2010). Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 22, 3816–3830. doi: 10.1105/tpc.109.073510
Freschi, L., Rodrigues, M. A., Domingues, D. S., Purgatto, E., Van Sluys, M. A., Magalhaes, J. R., et al. (2010). Nitric oxide mediates the hormonal control of Crassulacean acid metabolism expression in young pineapple plants. Plant Physiol. 152, 1971–1985. doi: 10.1104/pp.109.151613
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
Gao, X. H., Xiao, S. L., Yao, Q. F., Wang, Y. J., and Fu, X. D. (2011). An updated GA signaling ‘relief of repression’ regulatory model. Mol. Plant 4, 601–606. doi: 10.1093/mp/ssr046
Garcia-Mata, C., Gay, R., Sokolovski, S., Hills, A., Lamattina, L., and Blatt, M. R. (2003). Nitric oxide regulates K+ and Cl- channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 100, 11116–11121. doi: 10.1073/pnas.1434381100
Garcia, M. J., Suarez, V., Romera, F. J., Alcantara, E., and Perez-Vicente, R. (2011). A new model involving ethylene, nitric oxide and Fe to explain the regulation of Fe-acquisition genes in Strategy I plants. Plant Physiol. Biochem. 49, 537–544. doi: 10.1016/j.plaphy.2011.01.019
Garcia-Mata, C., and Lamattina, L. (2002). Nitric oxide and abscisic acid cross talk in guard cells. Plant Physiol. 128, 790–792. doi: 10.1104/pp.011020
Gas, E., Flores-Perez, U., Sauret-Gueto, S., and Rodriguez-Concepcion, M. (2009). Hunting for plant nitric oxide synthase provides new evidence of a central role for plastids in nitric oxide metabolism. Plant Cell 21, 18–23. doi: 10.1105/tpc.108.065243
Giba, Z., Grubisic, D., Todorovic, S., Sajc, L., Stojakovic, D., and Konjevic, R. (1998). Effect of nitric oxide - releasing compounds on phytochrome - controlled germination of Empress tree seeds. Plant Growth Regul. 26, 175–181. doi: 10.1023/A:1006131215297
Gniazdowska, A., Dobrzynska, U., Babanczyk, T., and Bogatek, R. (2007). Breaking the apple embryo dormancy by nitric oxide involves the stimulation of ethylene production. Planta 225, 1051–1057. doi: 10.1007/s00425-006-0384-z
Grbić, V., and Bleecker, A. B. (1995). Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 8, 595–602. doi: 10.1046/j.1365-313X.1995.8040595.x
Grun, S., Lindermayr, C., Sell, S., and Durner, J. (2006). Nitric oxide and gene regulation in plants. J. Exp. Bot. 57, 507–516. doi: 10.1093/jxb/erj053
Guerra, G., Mello, C., Sauzem, P., Berlese, D., Furian, A., Tabarelli, Z., et al. (2006). Nitric oxide is involved in the memory facilitation induced by spermidine in rats. Psychopharmacology 186, 150–158. doi: 10.1007/s00213-006-0376-5
Guo, F. Q., and Crawford, N. M. (2005). Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell 17, 3436–3450. doi: 10.1105/tpc.105.037770
Guo, F. Q., Okamoto, M., and Crawford, N. M. (2003). Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302, 100–103. doi: 10.1126/science.1086770
Gupta, K., Dey, A., and Gupta, B. (2013). Plant polyamines in abiotic stress responses. Acta Physiol. Plant. 35, 2015–2036. doi: 10.1007/s11738-013-1239-4
Gupta, K. J., Fernie, A. R., Kaiser, W. M., and Van Dongen, J. T. (2011). On the origins of nitric oxide. Trends Plant Sci. 16, 160–168. doi: 10.1016/j.tplants.2010.11.007
Gupta, K. J., and Igamberdiev, A. U. (2013). Recommendations of using at least two different methods for measuring NO. Front. Plant Sci. 4:58. doi: 10.3389/fpls.2013.00058
Hancock, J. T., Neill, S. J., and Wilson, I. D. (2011). Nitric oxide and ABA in the control of plant function. Plant Sci. 181, 555–559. doi: 10.1016/j.plantsci.2011.03.017
Hao, F., Zhao, S., Dong, H., Zhang, H., Sun, L., and Miao, C. (2010). Nia1 and Nia2 are involved in exogenous salicylic acid-induced nitric oxide generation and stomatal closure in Arabidopsis. J. Integr. Plant Biol. 52, 298–307. doi: 10.1111/j.1744-7909.2010.00920.x
Harper, J. E. (1981). Evolution of nitrogen oxide(s) during in vivo nitrate reductase assay of soybean leaves. Plant Physiol. 68, 1488–1493. doi: 10.1104/pp.68.6.1488
He, H. Y., He, L. F., Gu, M. H., and Li, X. F. (2012). Nitric oxide improves aluminum tolerance by regulating hormonal equilibrium in the root apices of rye and wheat. Plant Sci. 183, 123–130. doi: 10.1016/j.plantsci.2011.07.012
Hebelstrup, K. H., Van Zanten, M., Mandon, J., Voesenek, L. A. C. J., Harren, F. J. M., Cristescu, S. M., et al. (2012). Haemoglobin modulates NO emission and hyponasty under hypoxia-related stress in Arabidopsis thaliana. J. Exp. Bot. 63, 5581–5591. doi: 10.1093/jxb/ers210
Hess, D. T., Matsumoto, A., Kim, S. O., Marshall, H. E., and Stamler, J. S. (2005). Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 6, 150–166. doi: 10.1038/nrm1569
Hu, X. Y., Neill, S. J., Tang, Z. C., and Cai, W. M. (2005). Nitric oxide mediates gravitropic bending in soybean roots. Plant Physiol. 137, 663–670. doi: 10.1104/pp.104.054494
Huang, X., Stettmaier, K., Michel, C., Hutzler, P., Mueller, M. J., and Durner, J. (2004). Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana. Planta 218, 938–946. doi: 10.1007/s00425-003-1178-1
Hunt, P. W., Klok, E. J., Trevaskis, B., Watts, R. A., Ellis, M. H., Peacock, W. J., et al. (2002). Increased level of hemoglobin 1 enhances survival of hypoxic stress and promotes early growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99, 17197–17202. doi: 10.1073/pnas.212648799
Hunt, P. W., Watts, R. A., Trevaskis, B., Llewelyn, D. J., Burnell, J., Dennis, E. S., et al. (2001). Expression and evolution of functionally distinct haemoglobin genes in plants. Plant Mol. Biol. 47, 677–692. doi: 10.1023/A:1012440926982
Ignarro, L. J., Cirino, G., Casini, A., and Napoli, C. (1999). Nitric oxide as a signaling molecule in the vascular system: an overview. J. Cardiovasc. Pharmacol. 34, 879–886. doi: 10.1097/00005344-199912000-00016
Kaiser, W. M., and Planchet, E. (2006). Nitric oxide production in plants. Facts and Fictions. Plant Signal. Behav. 1, 46–51. doi: 10.4161/psb.1.2.2435
Kaiser, W. M., Planchet, E., and Rümer, S. (2010). “Nitrate reductase and nitric oxide,” in Annual Plant Reviews, Vol. 42, Nitrogen Metabolism in Plants in the Post-Genomic Era, eds C. H. Foyer and H. Zhang (Oxford: Wiley-Blackwell), 127–145.
Keyster, M., Klein, A., and Ludidi, N. (2010). Endogenous NO levels regulate nodule functioning: potential role of cGMP in nodule functioning? Plant Signal. Behav. 5, 1679–1681. doi: 10.4161/psb.5.12.14041
Kolbert, Z., Bartha, B., and Erdei, L. (2008). Exogenous auxin-induced NO synthesis is nitrate reductase-associated in Arabidopsis thaliana root primordia. J. Plant Physiol. 165, 967–975. doi: 10.1016/j.jplph.2007.07.019
Kolbert, Z., and Erdei, L. (2008). Involvement of nitrate reductase in auxin-induced NO synthesis. Plant Signal. Behav. 3, 972–973.
Kolbert, Z., Ortega, L., and Erdei, L. (2010). Involvement of nitrate reductase (NR) in osmotic stress-induced NO generation of Arabidopsis thaliana L. roots. J. Plant Physiol. 167, 77–80. doi: 10.1016/j.jplph.2009.08.013
Kopyra, M., and Gwozdz, E. A. (2003). Nitric oxide stimulates seed germination and counteracts the inhibitory effect of heavy metals and salinity on root growth of Lupinus luteus. Plant Physiol. Biochem. 41, 1011–1017. doi: 10.1016/j.plaphy.2003.09.003
Kovacs, I., and Lindermayr, C. (2013). Nitric oxide-based protein modification: formation and site-specificity of protein S-nitrosylation. Front. Plant Sci. 4:137. doi: 10.3389/fpls.2013.00137
Lamattina, L., Garcia-Mata, C., Graziano, M., and Pagnussat, G. (2003). Nitric oxide: the versatility of an extensive signal molecule. Annu. Rev. Plant Biol. 54, 109–136. doi: 10.1146/annurev.arplant.54.031902.134752
Lanteri, M. L., Pagnussat, G. C., and Lamattina, L. (2006). Calcium and calcium-dependent protein kinases are involved in nitric oxide- and auxin-induced adventitious root formation in cucumber. J. Exp. Bot. 57, 1341–1351. doi: 10.1093/jxb/erj109
Lee, U., Wie, C., Fernandez, B. O., Feelisch, M., and Vierling, E. (2008). Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20, 786–802. doi: 10.1105/tpc.107.052647
Leon, J., and Lozano-Juste, J. (2011). Nitric oxide regulates DELLA content and PIF expression to promote photomorphogenesis in Arabidopsis. Plant Physiol. 156, 1410–1423. doi: 10.1104/pp.111.177741
Leshem, Y. Y., and Haramaty, E. (1996). The characterization and contrasting effects of the nitric oxide free radical in vegetative stress and senescence of Pisum sativum Linn foliage. J. Plant Physiol. 148, 258–263. doi: 10.1016/S0176-1617(96)80251-3
Leshem, Y. Y., and Pinchasov, Y. (2000). Non-invasive photoacoustic spectroscopic determination of relative endogenous nitric oxide and ethylene content stoichiometry during the ripening of strawberries Fragaria anannasa (Duch.) and avocados Persea americana (Mill.). J. Exp. Bot. 51, 1471–1473. doi: 10.1093/jexbot/51.349.1471
Leshem, Y. Y., Wills, R., and Ku, V. (2001). Applications of nitric oxide (NO) for postharvest control. Acta Hortic. 553, 571–575.
Leshem, Y. Y., and Wills, R. B. H. (1998). Harnessing senescence delaying gases nitric oxide and nitrous oxide: a novel approach to postharvest control of fresh horticultural produce. Biol. Plant. 41, 1–10. doi: 10.1023/A:1001779227767
Leshem, Y. Y., Wills, R. B. H., and Ku, V. V. V. (1998). Evidence for the function of the free radical gas - nitric oxide (NO) - as an endogenous maturation and senescence regulating factor in higher plants. Plant Physiol. Biochem. 36, 825–833. doi: 10.1016/S0981-9428(99)80020-5
Leterrier, M., Chaki, M., Airaki, M., Valderrama, R., Palma, J. M., Barroso, J. B., et al. (2011). Function of S-nitrosoglutathione reductase (GSNOR) in plant development and under biotic/abiotic stress. Plant Signal. Behav. 6, 789–793. doi: 10.4161/psb.6.6.15161
Lindermayr, C., Saalbach, G., Bahnweg, G., and Durner, J. (2006). Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J. Biol. Chem. 281, 4285–4291. doi: 10.1074/jbc.M511635200
Lindermayr, C., Saalbach, G., and Durner, J. (2005). Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 137, 921–930. doi: 10.1104/pp.104.058719
Liu, J., Liu, G. H., Hou, L. X., and Liu, X. (2010). Ethylene-induced nitric oxide production and stomatal closure in Arabidopsis thaliana depending on changes in cytosolic pH. Chin. Sci. Bull. 55, 2403–2409. doi: 10.1007/s11434-010-4033-3
Liu, M.-C., Song, W.-H., Zhu, S.-H., and Zhou, J. (2007). Effects of nitric oxide and exogenous ethylene treatments on ethylene biosynthesis in Feicheng peach. Agric. Sci. China 6, 290–295. doi: 10.1016/S1671-2927(07)60047-9
Liu, W.-Z., Kong, D.-D., Gu, X.-X., Gao, H.-B., Wang, J.-Z., Xia, M., et al. (2013). Cytokinins can act as suppressors of nitric oxide in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 1548–1553. doi: 10.1073/pnas.1213235110
Liu, Y. G., Shi, L., Ye, N. H., Liu, R., Jia, W. S., and Zhang, J. H. (2009). Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis. New Phytol. 183, 1030–1042. doi: 10.1111/j.1469-8137.2009.02899.x
Lombardo, M. C., Graziano, M., Polacco, J. C., and Lamattina, L. (2006). Nitric oxide functions as a positive regulator of root hair development. Plant Signal. Behav. 1, 28–33. doi: 10.4161/psb.1.1.2398
Lombardo, M. C., and Lamattina, L. (2012). Nitric oxide is essential for vesicle formation and trafficking in Arabidopsis root hairgrowth. J. Exp. Bot. 63, 4875–4885. doi: 10.1093/jxb/ers166
Lozano-Juste, J., Colom-Moreno, R., and Leon, J. (2011). In vivo protein tyrosine nitration in Arabidopsis thaliana. J. Exp. Bot. 62, 3501–3517. doi: 10.1093/jxb/err042
Lozano-Juste, J., and Leon, J. (2010a). Enhanced abscisic acid-mediated responses in nia1nia2noa1–2 triple mutant impaired in NIA/NR-and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiol. 152, 891–903. doi: 10.1104/pp.109.148023
Lozano-Juste, J., and Leon, J. (2010b). Nitric oxide modulates sensitivity to ABA. Plant Signal. Behav. 5, 314–316. doi: 10.4161/psb.5.3.11235
Lu, S. Y., Su, W., Li, H. H., and Guo, Z. F. (2009). Abscisic acid improves drought tolerance of triploid bermudagrass and involves H2O2- and NO-induced antioxidant enzyme activities. Plant Physiol. Biochem. 47, 132–138. doi: 10.1016/j.plaphy.2008.10.006
Magalhães, J., Monte, D., and Durzan, D. (2000). Nitric oxide and ethylene emission in Arabidopsis thaliana. Physiol Mol Biol Plants 6, 117–127.
Manac’h-Little, N., Igamberdiev, A. U., and Hill, R. D. (2005). Hemoglobin expression affects ethylene production in maize cell cultures. Plant Physiol. Biochem. 43, 485–489. doi: 10.1016/j.plaphy.2005.03.012
Manjunatha, G., Gupta, K. J., Lokesh, V., Mur, L. A., and Neelwarne, B. (2012). Nitric oxide counters ethylene effects on ripening fruits. Plant Signal. Behav. 7, 476–483. doi: 10.4161/psb.19523
Manjunatha, G., Lokesh, V., and Neelwarne, B. (2010). Nitric oxide in fruit ripening: trends and opportunities. Biotechnol. Adv. 28, 489–499. doi: 10.1016/j.biotechadv.2010.03.001
Martinez-Ruiz, A., Araujo, I. M., Izquierdo-Alvarez, A., Hernansanz-Agustin, P., Lamas, S., and Serrador, J. M. (2013). Specificity in S-nitrosylation: a short-range mechanism for NO signaling? Antioxid. Redox Signal. doi: 10.1089/ars.2012.5066 [Epub ahead of print].
Meyer, C., Lea, U. S., Provan, F., Kaiser, W. M., and Lillo, C. (2005). Is nitrate reductase a major player in the plant NO (nitric oxide) game? Photosynth. Res. 83, 181–189. doi: 10.1007/s11120-004-3548-3
Mioto, P. T., and Mercier, H. (2013). Abscisic acid and nitric oxide signaling in two different portions of detached leaves of Guzmania monostachia with CAM up-regulated by drought. J. Plant Physiol. 170, 996–1002. doi: 10.1016/j.jplph.2013.02.004
Mishina, T. E., Lamb, C., and Zeier, J. (2007). Expression of a nitric oxide degrading enzyme induces a senescence programme in Arabidopsis. Plant Cell Environ. 30, 39–52. doi: 10.1111/j.1365-3040.2006.01604.x
Moreau, M., Lee, G. I., Wang, Y., Crane, B. R., and Klessig, D. F. (2008). AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J. Biol. Chem. 283, 32957–32967. doi: 10.1074/jbc.M804838200
Mur, L. A., Prats, E., Pierre, S., Hall, M. A., and Hebelstrup, K. H. (2013). Integrating nitric oxide into salicylic acid and jasmonic acid/ ethylene plant defense pathways. Front. Plant Sci. 4:215. doi: 10.3389/fpls.2013.00215
Mur, L. A. J., Laarhoven, L. J. J., Harren, F. J. M., Hall, M. A., and Smith, A. R. (2008). Nitric oxide interacts with salicylate to regulate biphasic ethylene production during the hypersensitive response. Plant Physiol. 148, 1537–1546. doi: 10.1104/pp.108.124404 PMCid:PMC2577248
Mur, L. A. J., Mandon, J., Cristescu, S. M., Harren, F. J. M., and Prats, E. (2011). Methods of nitric oxide detection in plants: a commentary. Plant Sci. 181, 509–519. doi: 10.1016/j.plantsci.2011.04.003
Mur, L. A. J., Mandon, J., Persijn, S., Cristescu, S. M., Moshkov, I. E., Novikova, G. V., et al. (2012a). Nitric oxide in plants: an assessment of the current state of knowledge. AoB Plants 5, pls052. doi: 10.1093/aobpla/pls052
Mur, L. A. J., Sivakumaran, A., Mandon, J., Cristescu, S. M., Harren, F. J. M., and Hebelstrup, K. H. (2012b). Haemoglobin modulates salicylate and jasmonate/ethylene-mediated resistance mechanisms against pathogens. J. Exp. Bot. 63, 4375–4387. doi: 10.1093/jxb/ers116
Neill, S., Barros, R., Bright, J., Desikan, R., Hancock, J., Harrison, J., et al. (2008a). Nitric oxide, stomatal closure, and abiotic stress. J. Exp. Bot. 59, 165–176. doi: 10.1093/jxb/erm293
Neill, S., Bright, J., Desikan, R., Hancock, J., Harrison, J., and Wilson, I. (2008b). Nitric oxide evolution and perception. J. Exp. Bot. 59, 25–35. doi: 10.1093/jxb/erm218
Neill, S. J., Desikan, R., Clarke, A., Hurst, R. D., and Hancock, J. T. (2002). Hydrogen peroxide and nitric oxide as signalling molecules in plants. J. Exp. Bot. 53, 1237–1247. doi: 10.1093/jexbot/53.372.1237
Neill, S. J., Desikan, R., and Hancock, J. T. (2003). Nitric oxide signalling in plants. New Phytol. 159, 11–35. doi: 10.1046/j.1469-8137.2003.00804.x
Niu, Y.-H., and Guo, F.-Q. (2012). Nitric oxide regulates dark-induced leaf senescence through EIN2 in Arabidopsis. J. Integr. Plant Biol. 54, 516–525. doi: 10.1111/j.1744-7909.2012.01140.x
Oliveira, H. C., Justino, G. C., Sodek, L., and Salgado, I. (2009). Amino acid recovery does not prevent susceptibility to Pseudomonas syringae in nitrate reductase double-deficient Arabidopsis thaliana plants. Plant Sci. 176, 105–111. doi: 10.1016/j.plantsci.2008.09.017
Ötvös, K., Pasternak, T. P., Miskolczi, P., Domoki, M., Dorjgotov, D., Szucs, A., et al. (2005). Nitric oxide is required for, and promotes auxin-mediated activation of, cell division and embryogenic cell formation but does not influence cell cycle progression in alfalfa cell cultures. Plant J. 43, 849–860. doi: 10.1111/j.1365-313X.2005.02494.x
Pagnussat, G. C., Lanteri, M. L., and Lamattina, L. (2003). Nitric oxide and cyclic GMP are messengers in the indole acetic acid-induced adventitious rooting process. Plant Physiol. 132, 1241–1248. doi: 10.1104/pp.103.022228
Pagnussat, G. C., Lanteri, M. L., Lombardo, M. C., and Lamattina, L. (2004). Nitric oxide mediates the indole acetic acid induction activation of a mitogen-activated protein kinase cascade involved in adventitious root development. Plant Physiol. 135, 279–286. doi: 10.1104/pp.103.038554
Pagnussat, G. C., Simontacchi, M., Puntarulo, S., and Lamattina, L. (2002). Nitric oxide is required for root organogenesis. Plant Physiol. 129, 954–956. doi: 10.1104/pp.004036
Parani, M., Rudrabhatla, S., Myers, R., Weirich, H., Smith, B., Leaman, D. W., et al. (2004). Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnol. J. 2, 359–366. doi: 10.1111/j.1467-7652.2004.00085.x
Parra-Lobato, M. C., and Gomez-Jimenez, M. C. (2011). Polyamine-induced modulation of genes involved in ethylene biosynthesis and signalling pathways and nitric oxide production during olive mature fruit abscission. J. Exp. Bot. 62, 4447–4465. doi: 10.1093/jxb/err124
Perazzolli, M., Romero-Puertas, M. C., and Delledonne, M. (2006). Modulation of nitric oxide bioactivity by plant haemoglobins. J. Exp. Bot. 57, 479–488. doi: 10.1093/jxb/erj051
Pii, Y., Crimi, M., Cremonese, G., Spena, A., and Pandolfini, T. (2007). Auxin and nitric oxide control indeterminate nodule formation. BMC Plant Biol. 7:21. doi: 10.1186/1471-2229-7-21
Planchet, E., and Kaiser, W. M. (2006). Nitric oxide (NO) detection by DAF fluorescence and chemiluminescence: a comparison using abiotic and biotic NO sources. J. Exp. Bot. 57, 3043–3055. doi: 10.1093/jxb/erl070
Polverari, A., Molesini, B., Pezzotti, M., Buonaurio, R., Marte, M., and Delledonne, M. (2003). Nitric oxide-mediated transcriptional changes in Arabidopsis thaliana. Mol. Plant Microbe Interact. 16, 1094–1105. doi: 10.1094/MPMI.2003.16.12.1094
Prado, A. M., Colaco, R., Moreno, N., Silva, A. C., and Feijo, J. A. (2008). Targeting of pollen tubes to ovules is dependent on nitric oxide (NO) signaling. Mol. Plant 1, 703–714. doi: 10.1093/mp/ssn034
Qu, Z. L., Zhong, N. Q., Wang, H. Y., Chen, A. P., Jian, G. L., and Xia, G. X. (2006). Ectopic expression of the cotton non-symbiotic hemoglobin gene GhHbd1 triggers defense responses and increases disease tolerance in Arabidopsis. Plant. Cell Physiol. 47, 1058–1068. doi: 10.1093/pcp/pcj076
Raghavan, S. A., and Dikshit, M. (2004). Vascular regulation by the L-arginine metabolites, nitric oxide and agmatine. Pharmacol. Res. 49, 397–414. doi: 10.1016/j.phrs.2003.10.008
Ribeiro, D. M., Desikan, R., Bright, J., Confraria, A., Harrison, J., Hancock, J. T., et al. (2009). Differential requirement for NO during ABA-induced stomatal closure in turgid and wilted leaves. Plant Cell Environ. 32, 46–57. doi: 10.1111/j.1365-3040.2008.01906.x
Rockel, P., Strube, F., Rockel, A., Wildt, J., and Kaiser, W. M. (2002). Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 53, 103–110. doi: 10.1093/jexbot/53.366.103
Romanov, G. A., Lomin, S. N., Rakova, N. Y., Heyl, A., and Schmulling, T. (2008). Does NO play a role in cytokinin signal transduction? FEBS Lett. 582, 874–880. doi: 10.1016/j.febslet.2008.02.016
Romero-Puertas, M. C., Campostrini, N., Matte, A., Righetti, P. G., Perazzolli, M., Zolla, L., et al. (2008). Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics 8, 1459–1469. doi: 10.1002/pmic.200700536
Ross, E. J. H., Stone, J. M., Elowsky, C. G., Arredondo-Peter, R., Klucas, R. V., and Sarath, G. (2004). Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J. Exp. Bot. 55, 1721–1731. doi: 10.1093/jxb/erh211
Ross, J. J., Weston, D. E., Davidson, S. E., and Reid, J. B. (2011). Plant hormone interactions: how complex are they? Physiol. Plant. 141, 299–309. doi: 10.1111/j.1399-3054.2011.01444.x
Saito, N., Nakamura, Y., Mori, I. C., and Murata, Y. (2009). Nitric oxide functions in both methyl jasmonate signaling and abscisic acid signaling in Arabidopsis guard cells. Plant Signal. Behav. 4, 119–120. doi: 10.4161/psb.4.2.7537
Sang, J. R., Zhang, A., Lin, F., Tan, M. P., and Jiang, M. Y. (2008). Cross-talk between calcium-calmodulin and nitric oxide in abscisic acid signaling in leaves of maize plants. Cell. Res 18, 577–588. doi: 10.1038/cr.2008.39
Santner, A., Calderon-Villalobos, L. I., and Estelle, M. (2009). Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307. doi: 10.1038/nchembio.165
Sarath, G., Bethke, P. C., Jones, R., Baird, L. M., Hou, G. C., and Mitchell, R. B. (2006). Nitric oxide accelerates seed germination in warm-season grasses. Planta 223, 1154–1164. doi: 10.1007/s00425-005-0162-3
Satriano, J. (2003). Agmatine: at the crossroads of the arginine pathways. Ann. N. Y. Acad. Sci. 1009, 34–43. doi: 10.1196/annals.1304.004
Satriano, J. (2004). Arginine pathways and the inflammatory response: interregulation of nitric oxide and polyamines: review article. Amino Acids 26, 321–329. doi: 10.1007/s00726-004-0078-4
Scherer, G. F. E., and Holk, A. (2000). NO donors mimic and NO inhibitors inhibit cytokinin action in betalaine accumulation in Amaranthus caudatus. Plant Growth Regul. 32, 345–350. doi: 10.1023/A:1010750111550
Shao, R. X., Wang, K. B., and Shangguan, Z. P. (2010). Cytokinin-induced photosynthetic adaptability of Zea mays L. to drought stress associated with nitric oxide signal: probed by ESR spectroscopy and fast OJIP fluorescence rise. J. Plant Physiol. 167, 472–479. doi: 10.1016/j.jplph.2009.10.020
Shen, Q., Wang, Y.-T., Tian, H., and Guo, F.-Q. (2012). Nitric oxide mediates cytokinin functions in cell proliferation and meristem maintenance in Arabidopsis. Mol. Plant doi: 10.1093/mp/sss148 doi: 10.1093/mp/sss148
Simontacchi, M., Garcia-Mata, C., Bartoli, C. G., Santa-Maria, G. E., and Lamattina, L. (2013). Nitric oxide as a key component in hormone-regulated processes. Plant Cell Rep. 32, 853–866. doi: 10.1007/s00299-013-1434-1
Simontacchi, M., Jasid, S., and Puntarulo, S. (2004). Nitric oxide generation during early germination of sorghum seeds. Plant Sci. 167, 839–847. doi: 10.1016/j.plantsci.2004.05.028
Sokolovski, S., and Blatt, M. R. (2004). Nitric oxide block of outward-rectifying K+ channels indicates direct control by protein nitrosylation in guard cells. Plant Physiol. 136, 4275–4284. doi: 10.1104/pp.104.050344
Sokolovski, S., Hills, A., Gay, R., Garcia-Mata, C., Lamattina, L., and Blatt, M. R. (2005). Protein phosphorylation is a prerequisite for intracellular Ca2+ release and ion channel control by nitric oxide and abscisic acid in guard cells. Plant J. 43, 520–529. doi: 10.1111/j.1365-313X.2005.02471.x
Spoel, S. H., Koornneef, A., Claessens, S. M. C., Korzelius, J. P., Van Pelt, J. A., Mueller, M. J., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15, 760–770. doi: 10.1105/tpc.009159
Stamler, J. S., and Meissner, G. (2001). Physiology of nitric oxide in skeletal muscle. Physiol. Rev. 81, 209–237.
Sun, L. R., Hao, F. S., Lu, B. S., and Ma, L. Y. (2010). AtNOA1 modulates nitric oxide accumulation and stomatal closure induced by salicylic acid in Arabidopsis. Plant Signal. Behav. 5, 1022–1024. doi: 10.4161/psb.5.8.12293
Tada, Y., Spoel, S. H., Pajerowska-Mukhtar, K., Mou, Z. L., Song, J. Q., Wang, C., et al. (2008). Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. doi: 10.1126/science.1156970
Tanou, G., Filippou, P., Belghazi, M., Job, D., Diamantidis, G., Fotopoulos, V., et al. (2012). Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J. 72, 585–599. doi: 10.1111/j.1365-313X.2012.05100.x
Terrile, M. C., París, R., Calderón-Villalobos, L. I. A., Iglesias, M. J., Lamattina, L., Estelle, M., et al. (2012). Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 70, 492–500. doi: 10.1111/j.1365-313X.2011.04885.x
Tierney, D. L., Rocklin, A. M., Lipscomb, J. D., Que, L. Jr., and Hoffman, B. M. (2005). ENDOR studies of the ligation and structure of the non-heme iron site in ACC oxidase. J. Am. Chem. Soc. 127, 7005–7013. doi: 10.1021/ja0500862
Tonón, C., Terrile, M. C., Iglesias, M. J., Lamattina, L., and Casalongue, C. (2010). Extracellular ATP, nitric oxide and superoxide act coordinately to regulate hypocotyl growth in etiolated Arabidopsis seedlings. J. Plant Physiol. 167, 540–546. doi: 10.1016/j.jplph.2009.11.002
Tossi, V., Lamattina, L., and Cassia, R. (2009). An increase in the concentration of abscisic acid is critical for nitric oxide-mediated plant adaptive responses to UV-B irradiation. New Phytol. 181, 871–879. doi: 10.1111/j.1469-8137.2008.02722.x
Tossi, V., Lamattina, L., and Cassia, R. (2013). Pharmacological and genetical evidence supporting nitric oxide requirement for 2,4-epibrassinolide regulation of root architecture in Arabidopsis thaliana. Plant Signal. Behav. 8, pii: e24712. doi: 10.4161/psb.24712
Tun, N. N., Holk, A., and Scherer, G. F. E. (2001). Rapid increase of NO release in plant cell cultures induced by cytokinin. FEBS Lett. 509, 174–176. doi: 10.1016/S0014-5793(01)03164-7
Tun, N. N., Livaja, M., Kieber, J. J., and Scherer, G. F. E. (2008). Zeatin-induced nitric oxide (NO) biosynthesis in Arabidopsis thaliana mutants of NO biosynthesis and of two-component signaling genes. New Phytol. 178, 515–531. doi: 10.1111/j.1469-8137.2008.02383.x
Tun, N. N., Santa-Catarina, C., Begum, T., Silveira, V., Handro, W., Floh, E. I. S., et al. (2006). Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant Cell Physiol. 47, 346–354. doi: 10.1093/pcp/pci252
Valderrama, R., Corpas, F. J., Carreras, A., Fernandez-Ocana, A., Chaki, M., Luque, F., et al. (2007). Nitrosative stress in plants. FEBS Lett. 581, 453–461. doi: 10.1016/j.febslet.2007.01.006
Vandelle, E., and Delledonne, M. (2011). Peroxynitrite formation and function in plants. Plant Sci. 181, 534–539. doi: 10.1016/j.plantsci.2011.05.002
Vitecek, J., Reinohl, V., and Jones, R. L. (2008). Measuring NO production by plant tissues and suspension cultured cells. Mol. Plant 1, 270–284. doi: 10.1093/mp/ssm020
Vuylsteker, C., Leleu, O., and Rambour, S. (1997). Influence of BAP and NAA on the expression of nitrate reductase in excised chicory roots. J. Exp. Bot. 48, 1079–1085. doi: 10.1093/jxb/48.5.1079
Wang, X., Li, J., Liu, J., He, W., and Bi, Y. (2010). Nitric oxide increases mitochondrial respiration in a cGMP-dependent manner in the callus from Arabidopsis thaliana. Nitric Oxide 23, 242–250. doi: 10.1016/j.niox.2010.07.004
Wang, Y., Feng, H., Qu, Y., Cheng, J., Zhao, Z., Zhang, M., et al. (2006). The relationship between reactive oxygen species and nitric oxide in ultraviolet-B-induced ethylene production in leaves of maize seedlings. Environ. Exper. Bot. 57, 51–61. doi: 10.1016/j.envexpbot.2005.04.009
Wang, Y. P., Elhiti, M., Hebelstrup, K. H., Hill, R. D., and Stasolla, C. (2011). Manipulation of hemoglobin expression affects Arabidopsis shoot organogenesis. Plant Physiol. Biochem. 49, 1108–1116. doi: 10.1016/j.plaphy.2011.06.005
Wang, Y. Q., Feechan, A., Yun, B. W., Shafiei, R., Hofmann, A., Taylor, P., et al. (2009). S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J. Biol. Chem. 284, 2131–2137. doi: 10.1074/jbc.M806782200
Wilhelmová, N., Fuksova, H., Srbova, M., Mikova, D., Mytinova, Z., Prochazkova, D., et al. (2006). The effect of plant cytokinin hormones on the production of ethylene, nitric oxide, and protein nitrotyrosine in ageing tobacco leaves. Biofactors 27, 203–211. doi: 10.1002/biof.5520270118
Wills, R. B. H., Ku, V. V. V., and Leshem, Y. Y. (2000). Fumigation with nitric oxide to extend the postharvest life of strawberries. Postharvest Biol. Technol. 18, 75–79. doi: 10.1016/S0925-5214(99)00061-7
Wilson, I. D., Neill, S. J., and Hancock, J. T. (2008). Nitric oxide synthesis and signalling in plants. Plant Cell Environ. 31, 622–631. doi: 10.1111/j.1365-3040.2007.01761.x
Wimalasekera, R., Tebartz, F., and Scherer, G. F. E. (2011). Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Sci. 181, 593–603. doi: 10.1016/j.plantsci.2011.04.002
Wu, M. Z., Wang, F. Q., Zhang, C., Xie, Y. J., Han, B., Huang, J. J., et al. (2013). Heme oxygenase-1 is involved in nitric oxide- and cGMP-induced a-Amy2/54 gene expression in GA-treated wheat aleurone layers. Plant Mol. Biol. 81, 27–40. doi: 10.1007/s11103-012-9979-x
Xiao-Ping, S., and Xi-Gui, S. (2006). Cytokinin- and auxin-induced stomatal opening is related to the change of nitric oxide levels in guard cells in broad bean. Physiol. Plant. 128, 569–579. doi: 10.1111/j.1399-3054.2006.00782.x
Xu, J., Wang, W. Y., Yin, H. X., Liu, X. J., Sun, H., and Mi, Q. (2010). Exogenous nitric oxide improves antioxidative capacity and reduces auxin degradation in roots of Medicago truncatula seedlings under cadmium stress. Plant Soil 326, 321–330. doi: 10.1007/s11104-009-0011-4
Yan, J., Tsuichihara, N., Etoh, T., and Iwai, S. (2007). Reactive oxygen species and nitric oxide are involved in ABA inhibition of stomatal opening. Plant Cell Environ. 30, 1320–1325. doi: 10.1111/j.1365-3040.2007.01711.x
Yang, J., Matsumoto, Y., Etoh, T., and Iwai, S. (2008). Nitric oxide (NO)-dependent and NO-independent signaling pathways act in ABA-inhibition of stomatal opening. Plant Signal. Behav. 3, 131–132. doi: 10.4161/psb.3.2.5040
Yang, S. F., and Hoffman, N. E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35, 155–189. doi: 10.1146/annurev.pp.35.060184.001103
Yu, M., Yun, B. W., Spoel, S. H., and Loake, G. J. (2012). A sleigh ride through the SNO: regulation of plant immune function by protein S-nitrosylation. Curr. Opin. Plant Biol. 15, 424–430. doi: 10.1016/j.pbi.2012.03.005
Zemojtel, T., Frohlich, A., Palmieri, M. C., Kolanczyk, M., Mikula, I., Wyrwicz, L. S., et al. (2006). Plant nitric oxide synthase: a never-ending story? Trends Plant Sci. 11, 524–525. doi: 10.1016/j.tplants.2006.09.008
Zhang, A., Zhang, J., Zhang, J., Ye, N., Zhang, H., Tan, M., et al. (2011). Nitric oxide mediates brassinosteroid-induced ABA biosynthesis involved in oxidative stress tolerance in maize leaves. Plant Cell Physiol. 52, 181–192. doi: 10.1093/pcp/pcq187
Zhang, A. Y., Jiang, M. Y., Zhang, J. H., Ding, H. D., Xu, S. C., Hu, X. L., et al. (2007). Nitric oxide induced by hydrogen peroxide mediates abscisic acid-induced activation of the mitogen-activated protein kinase cascade involved in antioxidant defense in maize leaves. New Phytol. 175, 36–50. doi: 10.1111/j.1469-8137.2007.02071.x
Zhang, H., Shen, W. B., Zhang, W., and Xu, L. L. (2005). A rapid response of beta-amylase to nitric oxide but not gibberellin in wheat seeds during the early stage of germination. Planta 220, 708–716. doi: 10.1007/s00425-004-1390-7
Zhang, Y. M., Tan, J. L., Guo, Z. F., Lu, S. Y., He, S. J., Shu, W., et al. (2009). Increased abscisic acid levels in transgenic tobacco over-expressing 9 cis-epoxycarotenoid dioxygenase influence H2O2 and NO production and antioxidant defences. Plant Cell Environ. 32, 509–519. doi: 10.1111/j.1365-3040.2009.01945.x
Keywords: nitric oxide, plant hormones, auxin, cytokinin, gibberellin, abscisic acid, ethylene, S-nitrosylation
Citation: Freschi L (2013) Nitric oxide and phytohormone interactions: current status and perspectives. Front. Plant Sci. 4:398. doi: 10.3389/fpls.2013.00398
Received: 23 July 2013; Accepted: 19 September 2013;
Published online: 09 October 2013.
Edited by:
John Hancock, University of the West of England, UKReviewed by:
Xuelu Wang, Fudan University, ChinaCopyright © 2013 Freschi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luciano Freschi, Laboratory of Plant Physiology and Biochemistry, Department of Botany, University of Sao Paulo, Rua do Matao, 277, Sao Paulo 05508-090, Brazil e-mail:ZnJlc2NoaUB1c3AuYnI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.