- 1Computational Systems Biology Group, National Centre for Biotechnology, Spanish National Research Council, Madrid, Spain
- 2Plant Molecular Genetics Department, National Centre for Biotechnology, Spanish National Research Council, Madrid, Spain
To some extent contradicting the classical paradigm of the relationship between protein 3D structure and function, now it is clear that large portions of the proteomes, especially in higher organisms, lack a fixed structure and still perform very important functions. Proteins completely or partially unstructured in their native (functional) form are involved in key cellular processes underlain by complex networks of protein interactions. The intrinsic conformational flexibility of these disordered proteins allows them to bind multiple partners in transient interactions of high specificity and low affinity. In concordance, in plants this type of proteins has been found in processes requiring these complex and versatile interaction networks. These include transcription factor networks, where disordered proteins act as integrators of different signals or link different transcription factor subnetworks due to their ability to interact (in many cases simultaneously) with different partners. Similarly, they also serve as signal integrators in signaling cascades, such as those related to response to external stimuli. Disordered proteins have also been found in plants in many stress-response processes, acting as protein chaperones or protecting other cellular components and structures. In plants, it is especially important to have complex and versatile networks able to quickly and efficiently respond to changing environmental conditions since these organisms cannot escape and have no other choice than adapting to them. Consequently, protein disorder can play an especially important role in plants, providing them with a fast mechanism to obtain complex, interconnected and versatile molecular networks.
Protein Intrinsic Disorder
It is now recognized that a large fraction of the proteome, especially in eukaryotic organisms, lacks a fixed 3D structure in its native form. In these proteins, either the complete chain (“ intrinsically disordered/unstructured protein,” IDP/IUP) or part of it (“ intrinsically disordered/unstructured region,” IDR/IUR) do not adopt a folded structure in its functional form, but exist as a flexible mobile polypeptide (Dunker and Obradovic, 2001; Tompa, 2002; Uversky, 2013).
Intrinsically disordered proteins/intrinsically disordered region were detected from diverse experimental evidences of lack of fixed structure: e.g., missing segments in X-Ray-derived structures or lack of constraints to define a unique structure in NMR. At the sequence level, IDRs are characterized by long stretches of charged and polar residues almost lacking hydrophobic residues, which consequently do not allow the formation of hydrophobic cores to initiate folding (Romero et al., 2001; Dyson and Wright, 2005; Tompa, 2005). This particular (highly biased) amino-acid composition was the basis of the first approaches for detecting IDPs/IDRs from primary sequences. Later, as more examples of experimentally determined IDRs accumulated, specific predictors were trained with them, such as PONDR (Romero et al., 1997) or DISOPRED (Ward et al., 2004). These, together with the latest methodologies based on physical principles, e.g., FoldIndex (Prilusky et al., 2005) and IUPRED (Dosztanyi et al., 2005), constitute the current toolbox for predicting disorder from primary sequences.
Research in this type of proteins was delayed in part by the fact that they apparently contradicted the classic “ structure-function relationship” paradigm, which states that a protein has to be folded in a fixed 3D conformation in order to perform its function. In IDPs, it is actually their lack of structure what is instrumental to perform their particular functions. This is because in most cases the molecular function of these polypeptides is related to transient binding to multiple (different) partners. Such a particular way of interacting could not be achieved by “ fixed” surfaces, but only by those able to adapt to different conformations. Indeed, in many cases IDRs become structured upon binding to a partner, and in some cases the same IDR can adopt different bound structures depending on the partner (Tompa, 2005). This entropy reduction due to the structural gain associated to the binding is in part responsible for the special characteristics of the disorder-mediated interactions. Besides binding, IDRs also act as flexible linkers and “ springs” within the cell (Dunker et al., 2002; Tompa, 2002; Cozzetto and Jones, 2013).
Disordered proteins/regions are associated with key cellular processes such as signaling cascades, transcription regulation, cell cycle control and chaperone activity (Iakoucheva et al., 2002; Uversky et al., 2005; Tompa et al., 2006; Xie et al., 2007). These processes require reversible transient interactions of high specificity and low affinity, eventually with different partners, exactly the type of interactions mediated by these unstructured polypeptides. Consequently, far from being “ rare” or anecdotic, disordered proteins are among the most important proteins in a given proteome, and their mutation is, in many cases, either lethal or leads to diseases (Iakoucheva et al., 2002; Midic et al., 2009). Indeed, the possibility of interacting with multiple partners makes IDPs being “ hubs” (highly connected nodes) in protein interaction networks (Haynes et al., 2006) which are themselves related to lethality (Jeong et al., 2001). For example, the highly studied human transcription factor (TF) p53 is disordered in half of its length and indeed uses these IDRs to interact with its more than hundred different known partners (Oldfield et al., 2008). Similarly, signaling networks are branched and interconnected, and they require transient interactions of high specificity with different partners, making unstructured proteins excellent candidates for them. Another prototypical example are the molecular chaperones, for which a growing body of evidence points to the involvement of disorder in the activity of many of them (Kovacs and Tompa, 2012). Many chaperones contain IDRs (which are involved in the regulation of the chaperone or in the interaction with the substrate itself) or are fully disordered (IDPs). Interacting through disordered segments allows these chaperones to help in the folding of a much broader range of substrates.
Within these long disordered segments, particular stretches of amino-acids, generally with increased evolutionary conservation, have been found to be important for determining the interaction specificity. They can be seen as a sort of “ functional sites” within disordered segments. These include “ molecular recognition features” (MoRFs), which have a tendency to form certain secondary structures (α-MoRFs, β-MoRFs, …) realized when they bind to a partner (Fuxreiter et al., 2004; Mohan et al., 2006), “eukaryotic linear motifs” (ELMs; Gould et al., 2010), and “short linear motifs” (SLiMs; Diella et al., 2008).
These proteins are not only involved in central cellular processes but they are also more abundant than previously anticipated. The development of specific predictors able to detect IDPs/IDRs from primary sequences, and their massive application to complete proteomes rendered surprising results. Almost 1/3 of eukaryotic proteins are mostly disordered and half of them contains at least one long IDR (>30 residues). This rises to 70% for proteins involved in signaling (Iakoucheva et al., 2002; Vucetic et al., 2003; Ward et al., 2004).
Moreover, there is a relationship between disorder content and what one intuitively regards as “ organism complexity.” Even if this is controversial mainly due to the imprecise definition and quantification of “ organismal complexity,” at least there is a clear difference between the relatively low disorder content of prokaryotic organisms and the high disorder found in eukarya (Ward et al., 2004; Schad et al., 2011). This can be related to the involvement of disorder in cellular processes that are apparently more complex and interconnected in higher and multicellular organisms (cell cycle control, signaling cascades, etc.).
Taking together all these observations point toward the involvement of disorder in the generation of the highly-connected and intricate molecular interaction networks which underlie the complex biological processes characteristic of higher organisms. Indeed, protein interactions mediated by IDRs are recognized as a way of introducing plasticity in protein interaction networks (Tompa et al., 2005; Uversky et al., 2005). Along the same line, it has also been shown that in many cases alternative splicing isoforms are characterized by the addition/deletion of IDRs so as to add/remove interacting regions and consequently tune the “ wiring” of the networks these isoforms are involved in (Romero et al., 2006; Buljan et al., 2013).
Protein Disorder in Plants
Large-Scale Quantifications of Disorder
In principle, protein disorder in plant proteomes follows the same trends reported for other species. A number of studies focused on plant model organisms showed that disorder is present in the typical processes involving transient interactions with multiple partners. For example, a genome-wide analysis of protein disorder in Arabidopsis thaliana (Pietrosemoli et al., 2013) showed that the biological processes more enriched in disordered proteins were related to cell cycle, signaling, DNA metabolism, RNA splicing, etc. In this study, disorder predictions were generated for all proteins in this model organism. These data, together with a functional classification of the proteins in biological processes, allowed to evaluate the degree of disorder of the different biological processes. Carrying out the same process for the Human proteome allows to perform comparative studies on the usage of disorder in both organisms. The proteome of A. thaliana follows the expected trend regarding whole disorder content: as an eukaryotic organism, it has much more disorder than bacterial proteomes and, leaving apart discussions on the definition of “ organism complexity” and its quantification, Arabidopsis is globally less disordered than Human, an organism intuitively regarded as of higher complexity (Schad et al., 2011; Pietrosemoli et al., 2013).
In spite of this lower overall disorder content, there are some biological processes that are more enriched in disorder in A. thaliana than in Human. Many of these processes are related to the detection and response to external (environmental) stimuli (Pietrosemoli et al., 2013). These include processes related to the perception of light, response to abiotic stress, protein folding (chaperones) and secondary metabolism (mediating plant response to stress). A hypothesis to explain that these processes related to the perception and response to stimuli are more disordered in plants than in organisms of higher complexity involves that plants might have evolved very complex, versatile and intricate systems for interacting with the environment since, being sessile organisms, they cannot escape from environmental hazards and changes, as animals do, and have no other option than responding to them (Pietrosemoli et al., 2013). Protein disorder is a possible way for increasing the “ wiring” (connectivity) of the molecular networks underlying a given biological system. As a consequence, such system becomes more intricate and complex. This relationship between disorder in plants and their increased ability to respond to changing conditions has also been noted by other authors (Sun et al., 2013).
Examples of Involvement of Disorder in Plants
The involvement of protein intrinsic disorder in a number of plant molecular systems has been studied in detail. Again, in all the cases protein disorder allows the proteins in these systems to interact transiently with multiple partners with high specificity and low affinity. Moreover, in general these systems follow the trend commented above regarding the relationship between disorder in plants and versatility/complexity in the response to stimuli and changing conditions.
Maybe the most studied example of disordered plant systems are the dehydrins (Mouillon et al., 2006; Kovacs et al., 2008; Sun et al., 2013). This large and diverse family of proteins, involved in the response to drought and other environmental stresses, is almost completely disordered: their content of “ standard” secondary structure elements (α-helix and β-strand) is low, and they present a significant content of poly-Pro helices (Mouillon et al., 2006). This family includes protein chaperones such as ERD10 and ERD14 (Kovacs et al., 2008; Tompa and Kovacs, 2010) as well as proteins involved in the binding of metal ions, protection of membranes, and global protection of the cell during the highly compact dry state characteristic of plant seeds (Sun et al., 2013). It looks like dehydrins have evolved for maintaining this disorder state and avoid forming compact folded structures (Mouillon et al., 2006). The conformational flexibility associated to the disordered state allows them to sequester water, ions, proteins (as chaperones), and perform all the other molecular functions associated to their roles in responding to water-related stresses.
Another plant-specific family of proteins heavily relying on disorder for functioning is the GRAS family (Sun et al., 2011; Sun et al., 2012). These proteins play an important role in plant development and are involved in signal transduction cascades, such as those related to hormone response. Within these cascades, they act as integrators of signals (i.e., from different hormones or environmental inputs). It is indeed their disordered region (present in the N-terminal) that allows them to interact with multiple partners through different binding sites (MoRFs, see Introduction) and consequently integrate the signals they represent. The most conserved C-terminal domain of this family (which is actually what characterizes it) is structurally ordered and contains also motifs involved in protein interaction. Among them, there are Leucine-rich regions probably involved in interactions with TFs so as to transduce the integrated signals downstream.
In some cases, this integration of signals occurs at the level of the TF itself, due to the presence of disordered domains (besides the ordered DNA binding domain), which allows the TF to be influenced by multiple partners. For example the NAC family of plant TF is involved in a variety of processes such as plant defense, stress response or development. These proteins present a conserved (structured) N-terminal DNA-binding domain and a more variable intrinsically disordered C-terminal region (Jensen et al., 2010; Sun et al., 2013; Figure 1). This region acquires local structure (α-helix) when binding to the multiple partners of these proteins. This mechanism by which a TF is influenced by multiple partners through disordered regions, which also happens with other plant TFs such as the basic leucine zipper domain (bZIP) family (Yoon et al., 2006), is similar to that of the human p53 commented.
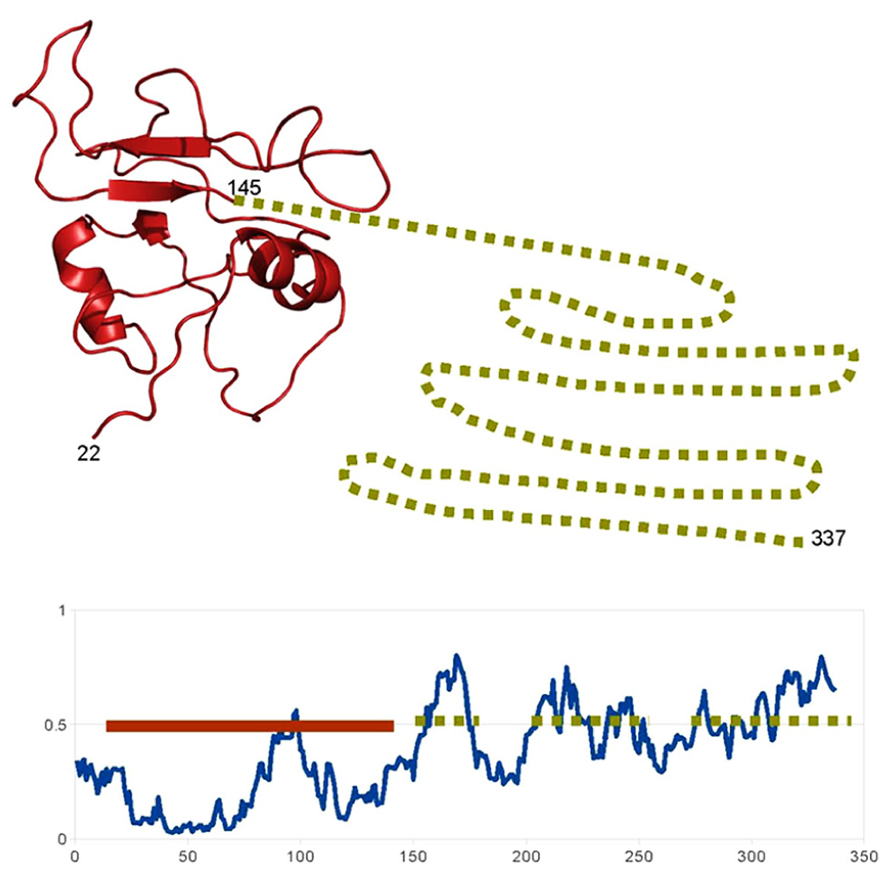
FIGURE 1. Example of a highly disordered protein in A. thaliana. Schematic representation of the structural features of the “ putative NAC domain-containing protein 94” (Uniprot: NAC94_ARATH). The disorder prediction of IUPRED (Dosztanyi et al., 2005) show that, according with the standard 0.5 threshold, most of the C-terminal part of the protein (from 150 to 337) is probably unstructured (dotted lines). The N-terminal DNA binding domain is probably structured and, indeed, a structural model can be generated based on the structure of a homolog (PDB:4dul_B, 62% sequence identity; solid line). So probably this protein “ looks like” the representation above: a short structured DNA-binding domain followed by a long flexible disordered region, involved in the binding of different partners.
It was also proposed that many chloroplast proteins whose genes were originally encoded by the chloroplast genome acquire disordered regions as they are transferred to the nucleus (Yruela and Contreras-Moreira, 2012). In concordance with its prokaryotic origin, proteins coded in the chloroplast genome almost lack disordered regions. Nevertheless, it looks like the “ eukaryotic machinery” of the nucleus adds disorder to them once they become coded there. This reinforces the idea of the relationship between disorder and the emergence of the complex molecular machineries associated to eukaryotic organisms.
Conclusions
In summary, recent research is showing that, in contrast to the classical dogma, intrinsic disorder is an important feature for many proteins to function. In general, protein disorder allows interaction versatility and adds complexity to the interactomes. It is likely a way in which evolution can increase the complexity of biological networks without increasing excessively the size of the genomes. In plants, the predominance of intrinsic disorder in proteins involved in responses to environmental conditions could be explained as a requirement of these processes to be more complex due to the special characteristics of these sessile organisms.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the members of the Computational System Biology Group (CNB-CSIC) for interesting discussions. This work was partially supported by project BIO2010-22109 from the Spanish Ministry of Science and Innovation.
References
Buljan, M., Chalancon, G., Dunker, A. K., Bateman, A., Balaji, S., Fuxreiter, M., et al. (2013). Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr. Opin. Struct. Biol. 23, 443–450. doi: 10.1016/j.sbi.2013.03.006
Cozzetto, D., and Jones, D. T. (2013). The contribution of intrinsic disorder prediction to the elicidation of protein function. Curr. Opin. Struct. Biol. 23, 467–472. doi: 10.1016/j.sbi.2013.02.001
Diella, F., Haslam, N., Chica, C., Budd, A., Michael, S., Brown, N. P., et al. (2008). Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 13:6580–6603. doi: 10.2741/3175
Dosztanyi, Z., Csizmok, V., Tompa, P., and Simon, I. (2005). IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434. doi: 10.1093/bioinformatics/bti541
Dunker, A. K., Brown, C. J., Lawson, J. D., Iakoucheva, L. M., and Obradovic, Z. (2002). Intrinsic disorder and protein function. Biochemistry 41, 6573–6582. doi: 10.1021/bi012159+
Dunker, A. K., and Obradovic, Z. (2001). The protein trinity – linking function and disorder. Nat. Biotechnol. 19, 805–806. doi: 10.1038/nbt0901-805
Dyson, H. J., and Wright, P. E. (2005). Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208. doi: 10.1038/nrm1589
Fuxreiter, M., Simon, I., Friedrich, P., and Tompa, P. (2004). Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 338, 1015–1026. doi: 10.1016/j.jmb.2004.03.017
Gould, C. M., Diella, F., Via, A., Puntervoll, P., Gemund, C., Chabanis-Davidson, S., et al. (2010). ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 38, D167–D180. doi: 10.1093/nar/gkp1016
Haynes, C., Oldfield, C. J., Ji, F., Klitgord, N., Cusick, M. E., Radivojac, P., et al. (2006). Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comp. Biol. 2:e100. doi: 10.1371/journal.pcbi.0020100
Iakoucheva, L. M., Brown, C. J., Lawson, J. D., Obradovic, Z., and Dunker, A. K. (2002). Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 323, 573–584. doi: 10.1016/S0022-2836(02)00969-5
Jensen, M. K., Kjaersgaard, T., Nielsen, M. M., Galberg, P., Petersen, K., O’ Shea, C., et al. (2010). The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426, 183–196. doi: 10.1042/BJ20091234
Jeong, H., Mason, S. P., Barabási, A. L., and Oltvai, Z. N. (2001). Lethality and centrality in protein networks. Nature 411, 41–42. doi: 10.1038/35075138
Kovacs, B., and Tompa, P. (2012). Diverse functional manifestations of intrinsic structural disorder in molecular chaperones. Biochem. Soc. Trans. 4, 963–968. doi: 10.1042/BST20120108
Kovacs, D., Kalmar, E., Torok, Z., and Tompa, P. (2008). Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 147, 381–390. doi: 10.1104/pp.108.118208
Midic, U., Oldfield, C. J., Dunker, A. K., Obradovic, Z., and Uversky, V. N. (2009). Protein disorder in the human diseasome: unfoldomics of human genetic diseases. BMC Genomics 10:S12. doi: 10.1186/1471-2164-10-S1-S12
Mohan, A., Oldfield, C. J., Radivojac, P., Vacic, V., Cortese, M. S., Dunker, A. K., et al. (2006). Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 362, 1043–1059. doi: 10.1016/j.jmb.2006.07.087
Mouillon, J.-M., Gustafsson, P., and Harryson, P. (2006). Structural investigation of disordered stress proteins. Comparison of full-length dehydrins with isolated peptides of their conserved segments. Plant Physiol. 141, 638–650. doi: 10.1104/pp.106.079848
Oldfield, C. J., Meng, J., Yang, J. Y., Yang, M. Q., Uversky, V. N., and Dunker, A. K. (2008). Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics 9:S1. doi: 10.1186/1471-2164-9-S1-S1
Pietrosemoli, N., García-Martín, J. A., Solano, R., and Pazos, F. (2013). Genome-wide analysis of protein disorder in Arabidopsis thaliana: implications for plant environmental adaptation. PLoS ONE 8:e55524. doi: 10.1371/journal.pone.0055524
Prilusky, J., Felder, C. E., Zeev-Ben-Mordehai, T., Rydberg, E. H., Man, O., Beckmann, J. S., et al. (2005). FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21, 3435–3438. doi: 10.1093/bioinformatics/bti537
Romero, P., Obradovic, Z., Kissinger, C., Villafranca, J. E., and Dunker, A. K. (1997). Identifying disordered regions in proteins from amino acid sequence. Proc. Int. Conf. Neural Netw. 1, 90–95.
Romero, P., Obradovic, Z., Li, X., Garner, E. C., Brown, C. J., and Dunker, A. K. (2001). Sequence complexity of disordered protein. Proteins 42, 38–48. doi: 10.1002/1097-0134(20010101)42:1
Romero, P. R., Zaidi, S., Fang, Y. Y., Uversky, V. N., Radivojac, P., Oldfield, C. J., et al. (2006). Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl. Acad. Sci. U.S.A. 103, 8390–8395. doi: 10.1073/pnas.0507916103
Schad, E., Tompa, P., and Hegyi, H. (2011). The relationship between proteome size, structural disorder and organism complexity. Genome Biol. 12, R120. doi: 10.1186/gb-2011-12-12-r120
Sun, X., Jones, W. T., and Rikkerink, E. H. A. (2012). GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling. Biochem. J. 442, 1–12. doi: 10.1042/BJ20111766
Sun, X., Rikkerink, E. H. A., Jones, W. T., and Uversky, V. N. (2013). Multifarious roles of intrinsic disorder in proteins illustrate its broad impact on plant biology. Plant Cell 25, 38–55. doi: 10.1105/tpc.112.106062
Sun, X., Xue, B., Jones, W. T., Rikkerink, E., Dunker, A. K., and Uversky, V. N. (2011). A functionally required unfoldome from the plant kingdom: intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 77, 205–223. doi: 10.1007/s11103-011-9803-z
Tompa, P. (2002). Intrinsically unstructured proteins. Trends Biochem. Sci. 27, 527–533. doi: 10.1016/S0968-0004(02)02169-2
Tompa, P. (2005). The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 579, 3346–3354. doi: 10.1016/j.febslet.2005.03.072
Tompa, P., Dosztanyi, Z., and Simon, I. (2006). Prevalent structural disorder in E. coli and S. cerevisiae proteomes. J. Proteome Res. 5, 1996–2000. doi: 10.1021/pr0600881
Tompa, P., and Kovacs, D. (2010). Intrinsically disordered chaperones in plants and animals. Biochem. Cell Biol. 88, 167–174. doi: 10.1139/O09-163
Tompa, P., Szász, C., and Buday, L. (2005). Structural disorder throws new light on moonlighting. Trends Biochem. Sci. 30, 484–489. doi: 10.1016/j.tibs.2005.07.008
Uversky, V. N. (2013). A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci. 6, 693–724. doi: 10.1002/pro.2261
Uversky, V. N., Oldfield, C. J., and Dunker, A. K. (2005). Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 18, 343–384. doi: 10.1002/jmr.747
Vucetic, S., Brown, C. J., Dunker, A. K., and Obradovic, Z. (2003). Flavors of protein disorder. Proteins 52, 573–584. doi: 10.1002/prot.10437
Ward, J. J., Sodhi, J. S., McGuffin, L. J., Buxton, B. F., and Jones, D. T. (2004). Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337, 635–645. doi: 10.1016/j.jmb.2004.02.002
Xie, H., Vucetic, S., Iakoucheva, L. M., Oldfield, C. J., Dunker, A. K., Uversky, V. N., et al. (2007). Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J. Proteome Res. 6, 1882–1898. doi: 10.1021/pr060392u
Yoon, M. K., Shin, J., Choi, G., and Choi, B. S. (2006). Intrinsically unstructured N-terminal domain of bZIP transcription factor HY5. Proteins 65, 856–866. doi: 10.1002/prot.21089
Keywords: protein function, protein structure, protein interactions, protein intrinsic disorder, biological networks, plant environmental responses
Citation: Pazos F, Pietrosemoli N, García-Martín JA and Solano R (2013) Protein intrinsic disorder in plants. Front. Plant Sci. 4:363. doi: 10.3389/fpls.2013.00363
Received: 28 June 2013; Accepted: 27 August 2013;
Published online: 12 September 2013.
Edited by:
Wolfgang Schmidt, Academia Sinica, TaiwanReviewed by:
Katja Baerenfaller, Swiss Federal Institute of Technology Zurich, SwitzerlandSandra Karin Tanz, The University of Western Australia, Australia
Copyright © 2013 Pazos, Pietrosemoli, García-Martín and Solano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Florencio Pazos, Computational Systems Biology Group, National Centre for Biotechnology, Spanish National Research Council, c/Darwin 3, Madrid 28049, Spain e-mail:cGF6b3NAY25iLmNzaWMuZXM=
†Present address: Juan A. García-Martín, Biology Department, Boston College, Boston, MA, USA



