
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 20 August 2013
Sec. Plant Development and EvoDevo
Volume 4 - 2013 | https://doi.org/10.3389/fpls.2013.00302
This article is part of the Research TopicUnderstanding plant adaptation and diversification: evo-devo and beyondView all 16 articles
Malpighiaceae possess flowers with a unique bilateral symmetry (zygomorphy), which is a hypothesized adaptation associated with specialization on neotropical oil bee pollinators. Gene expression of two representatives of the CYC2 lineage of floral symmetry TCP genes, CYC2A and CYC2B, demarcate the adaxial (dorsal) region of the flower in the characteristic zygomorphic flowers of most Malpighiaceae. Several clades within the family, however, have independently lost their specialized oil bee pollinators and reverted to radial flowers (actinomorphy) like their ancestors. Here, we investigate CYC2 expression associated with four independent reversals to actinomorphy. We demonstrate that these reversals are always associated with alteration of the highly conserved CYC2 expression pattern observed in most New World (NW) Malpighiaceae. In NW Lasiocarpus and Old World (OW) Microsteria, the expression of CYC2-like genes has expanded to include the ventral region of the corolla. Thus, the pattern of gene expression in these species has become radialized, which is comparable to what has been reported in the radial flowered legume clade Cadia. In striking contrast, in NW Psychopterys and OW Sphedamnocarpus, CYC2-like expression is entirely absent or at barely detectable levels. This is more similar to the pattern of CYC2 expression observed in radial flowered Arabidopsis. These results collectively indicate that, regardless of geographic distribution, reversals to similar floral phenotypes in this large tropical angiosperm clade have evolved via different genetic changes from an otherwise highly conserved developmental program.
Convergence is an evolutionary process in which similar features reoccur independently across the Tree of Life (Donoghue, 2005; Protas et al., 2006; Conway Morris, 2008; McGhee, 2011; Wake et al., 2011). These similarities are commonly thought to have arisen as a result of adaptation to similar selective pressures, rather than due to inheritance from a common ancestor. Convergent evolution thus represents an important evolutionary process in shaping the design of organismal diversity. It has been a challenge to explain the origin of convergence, however, since the underlying genetic and developmental bases of this phenomenon remains largely unknown, with notable recent exceptions [reviewed in Gompel and Prud'homme (2009); Conte et al. (2012)]. Evolutionary developmental genetics, however, has facilitated the characterization of several types of convergence operating at the genetic level. In some cases, studies have revealed convergence in terms of genetic change (e.g., Sucena et al., 2003; Prud'homme et al., 2006; Rosenblum et al., 2010), but in others, distinct genetic mechanisms were uncovered (e.g., Hoekstra and Nachman, 2003; Wittkopp et al., 2003; Steiner et al., 2009). A special case of convergence is reversion, the reappearance of ancestral phenotypes. Here, we use the flowering plant family Malpighiaceae, which has been the subject of recent floral developmental genetic studies (Zhang et al., 2010, 2012), to investigate the genetic patterns associated with four independent examples of reversion to an ancestral floral phenotype. We also discuss the diverse evolutionary trajectories underlying these independent reversions.
Malpighiaceae are a family of tropical trees, shrubs, and vines of both the New World (NW) and Old World (OW) tropics and subtropics. The zygomorphic (bilaterally symmetrical) floral morphology of the more than 1000 NW species of this clade is very distinctive and highly conserved, especially with regard to symmetry and pollinator reward (Figure 1A). The NW Malpighiaceae flower has a single upright dorsal banner petal that is strongly differentiated from other petals in the corolla whorl, and appears to orient and attract an extremely limited suite of pollinators, principally female bees of the tribes Centridini, Tetrapedini, and Tapinotaspidini (Vogel, 1974). These bees use their mandibles to grasp the base of this distinct dorsal banner petal and then use their fore and mid legs to access the oil glands borne in pairs on the abaxial surface of the sepals. It is thought that the stereotypical floral morphology of NW Malpighiaceae is maintained primarily due to this specialization on oil-bee pollinators (Vogel, 1974; Anderson, 1979).
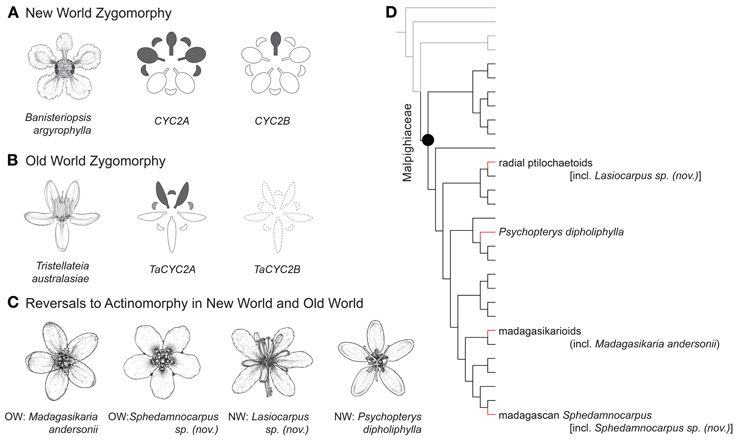
Figure 1. Phylogenetic positions, floral morphology, and CYC2 expression of Malpighiaceae. (A) Banisteriopsis argyrophylla illustrates the stereotypical NW floral morphology and pattern of CYC2 expression in NW Malpighiaceae (expression shown in gray). (B) Tristellateia australasiae represents the OW flowers that have departed from NW morphology with two dorsal petals (CYC2B is missing shown as dash line). (C) Madagasikaria andersonii, Lasiocarpus sp. (nov.), Psychopterys dipholiphylla and Sphedamnocarpus sp. (nov.) represent four OW floral phenotypes that have evolved independently from a similar NW type ancestor. (D) Phylogeny indicating relationships of the four focal clades: radial ptilochaetoids [including Lasiocarpus sp. (nov.)], Psychopterys dipholiphylla, madagasikarioids (including Madagasikaria andersonii) and Malagasy Sphedamnocarpus [including Sphedamnocarpus sp. (nov.)]. We examined the representatives of these four clades in this study. Gray lines highlight the radially symmetrical sister groups of Malpighiaceae, i.e., Centroplacaceae and Elatinaceae; black lines highlight Malpighiaceae species with the stereotypical NW floral morphology; red highlights the three OW clades with floral morphologies reversed to radially symmetry. The phylogenetic relationships inferred here are highly supported by previous studies (Davis et al., 2001; Davis and Chase, 2004; Wurdack and Davis, 2009; Davis and Anderson, 2010; Xi et al., 2012). Note: floral line drawings are not proportional to the sizes of flowers.
We recently established the likely genetic basis for the origin of this floral zygomorphy in NW Malpighiaceae, especially in regards to the unique banner petal morphology (Zhang et al., 2010, 2012). CYCLOIDEA2-like (CYC2) transcription factors of the ECE clade within the TCP gene family have been shown to be critical for establishing floral symmetry and to have been repeatedly recruited to regulate floral zygomorphy in diverse angiosperm lineages [reviewed in Howarth and Donoghue (2006); Preston and Hileman (2009); Citerne et al. (2010)]. The common finding in all of these studies, using model species with functional data and correlative patterns of gene expression from non-model species, is the persistent expression of CYC2 homologs in dorsal floral organs [reviewed in Preston and Hileman (2009); Citerne et al. (2010)]. Moreover, CYC2 loss-of-function mutants exhibit a fully actinomorphic (radially symmetrical) phenotype in which all floral organs gain ventral floral organ identity (Luo et al., 1996; Cubas et al., 1999b; Busch and Zachgo, 2007), or exhibit greatly reduced dorsal floral organ identity (Feng et al., 2006; Broholm et al., 2008). Conversely, ectopic expression of CYC, such as in the backpetals mutant of Antirrhinum, produces a radialized phenotype due to dorsalization of the corolla (Luo et al., 1999).
In Malpighiaceae, two clades of CYC2 were identified, CYC2A and CYC2B, which resulted from a duplication event coincident with the origin of the family (Zhang et al., 2010). In most NW species these loci are differentially expressed along the dorsoventral axis such that CYC2A is expressed in the dorsal banner petal and two adjacent lateral petals while CYC2B is restricted solely to the banner petal (Figure 1A). This pattern of CYC2 expression is conserved across three distantly related NW species [Janusia guaranitica A. Juss., Byrsonima crassifolia Kunth (Zhang et al., 2010), and Bunchosia glandulifera (Jacq.) Kunth (Zhang et al., 2012)], that span the origin of the family and its unique floral morphology (Davis and Anderson, 2010; Zhang et al., 2010, 2012). These results indicate that CYC2 genes likely play a key role in this plant pollinator mutualism and are thus good candidates for understanding the genetic basis of derived floral symmetries in Malpighiaceae.
The oil bees that pollinate NW Malpighiaceae are absent in the OW (Vogel, 1974; Michener, 2000), which is particularly interesting because most OW Malpighiaceae lack the oil glands and floral banner petals (Vogel, 1990; Davis, 2002; Davis et al., in review) that appear to be critical to the NW pollination syndrome (Anderson, 1979). These OW species possess either an altered form of floral zygomorphy with two dorsal petals or completely actinomorphic flowers (Figure 1) (Davis and Anderson, 2010). Recent phylogenetic studies suggest that these derived forms of zygomorphy and actinomorphy evolved three and eight times, respectively (Figure A1) (Davis and Anderson, 2010; Zhang et al., 2010, 2012). Two of the eight reversals to actinomorphy may have evolved from an ancestor with the altered form of floral zygomorphy (Figure A1) (Davis and Anderson, 2010; Zhang et al., 2010, 2012). The only obvious pollinator rewards of these atypical Malpighiaceae are pollen, which likely reflects their shifts to new pollinators (Lobreau-Callen, 1989; Davis, 2002). Malpighiaceae therefore represent a set of natural experiments involving replicate clades that initially possessed the ancestral zygomorphic NW floral morphology but diverged dramatically in the absence of their specialist oil-bee pollinators.
Recently, we built upon the genetic and developmental framework established for NW Malpighiaceae to examine parallel evolution of three OW Malpighiaceae lineages that evolved the altered form of zygomorphy described above (Figure 1B) (Zhang et al., 2012). In all three cases, the species exhibit a loss of CYC2B function, and a strikingly similar shift in the expression of CYC2A that is coincident with their shift in floral symmetry. These results indicate that similar floral phenotypes of OW Malpighiaceae have evolved via parallel genetic changes from an otherwise highly conserved developmental program. Interestingly, in two of these three zygomorphic OW clades, Sphedamnocarpus and Acridocarpus-Brachylophon, this altered form of zygomorphy is closely related to actinomorphic species (Figure A1) (Davis and Anderson, 2010; Zhang et al., 2010), which facilitates the extension of our studies into additional forms of symmetry.
Here, we continue with a similar line of inquiry, but turn our attention to investigate genetic modifications to this conserved developmental program that are associated with reversals to floral actinomorphy in four clades of Malpighiaceae. In addition to our analysis of two OW genera, two NW clades that have also independently lost the typical NW floral morphology and have become actinomorphic were also analyzed. These represent four phylogenetically independent contrasts to investigate reversals to actinomorphy from zygomorphic flowered ancestors that are also geographically distinct. We show that the conserved CYC2 program in the NW zygomorphic flowers is modified in each of the four actinomorphic flowered clades: Microsteira and Sphedamnocarpus from the OW, and Psychopterys and Lasiocarpus from the NW (Figures 1C,D). Our results indicate that these reversals in floral symmetry are the result of distinct modifications to the conserved NW CYC2 program, including the loss of CYC2 expression in Malagasy Sphedamnocarpus and NW Psychopterys, and the radialized expansion of expression in OW Microsteira and NW Lasiocarpus. Thus, CYC2-like genes seem to play diverse roles in the evolution of floral symmetry in Malpighiaceae. The loss of banner petal-associated CYC2B expression is particularly common, but has been combined in different ways with either shifts to radialized expression or complete loss of CYC2A expression, consistent with each type of symmetry. By analyzing these patterns of CYC expression in an evolutionary context, we gain a comprehensive understanding of the diversification of this genetic program. Within this single tropical flowering plant clade we find that essentially all potential mechanisms for facilitating shifts to actinomorphy are observed, highlighting the context-dependent and stochastic nature of developmental evolution.
We first identified CYC2-like homologs from eight species using degenerate primers and exhaustive screening of PCR clones as described in the Materials and Methods and in Zhang et al. (2010); Zhang et al. (2012) (Table A1). These species represent the four Malpighiaceae lineages that have reverted to actinomorphy: the radial flowered OW madagasikarioid clade (represented by Madagasikaria andersonii C. Davis, Microsteira sp., and Rhynchophora phillipsonii W. R. Anderson), the OW Malagasy Sphedamnocarpus clade [represented by Philgamia glabrifolia Arènes, and Sphedamnocarpus sp. (nov.)], the NW Psychopterys clade (represented by the only species Psychopterys dipholiphylla (Small) W. R. Anderson and S. Corso), and the NW ptilochaetoid clades (represented by Lasiocarpus sp. and Ptilochaeta nudipes Griseb.). The phylogenetic positions of these CYC2 homologs were inferred using our previously published CYC2 homologs, which sampled broadly across Malpighiaceae, including taxa that are closely related to the additions here (Zhang et al., 2010, 2012). Phylogenetic relationships inferred from CYC2 mirror our understanding of accepted species tree relationships (Davis et al., 2001, 2002; Davis and Anderson, 2010). Lasiocarpus, Ptilochaeta, and Psychopterys dipholiphylla maintain both copies of CYC2A and CYC2B, but in contrast, CYC2B was not detected in the madagasikarioid clade where we sampled Madagasikaria andersonii C. Davis, Microsteria sp., and Rhynchophora phillipsonii W. R. Anderson. (Figure 2, Table A1) (Zhang et al., 2010). These results were confirmed by Southern analyses in Microsteria sp. and Rhynchophora phillipsonii (Figure A2). Meanwhile, the CYC2A in two radialized species of Malagasy Sphedamnocarpus [i.e., Philgamia glabrifolia Arènes and Sphedamnocarpus sp. (nov.)], could not be amplified by PCR, but Southern analyses identified more than one copy of CYC2 (Figure A2). It is worth noting that in our previous paper (Zhang et al., 2012), we examined an African Sphedamnocarpus, a close relative of Malagasy Sphedamnocarpus, which exhibits a greatly altered zygomorphy from most NW species. CYC2A and CYC2B were both identified with no bias from this African species. These results suggest that CYC2A in species of Malagasy Sphedamnocarpus has diverged such that we cannot easily isolate it via degenerate PCR from genomic DNA or cDNA.
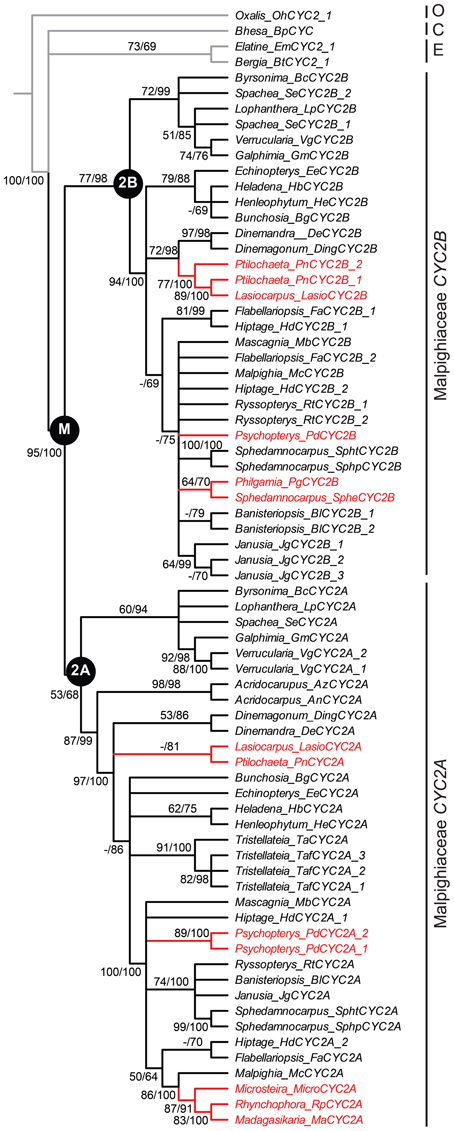
Figure 2. Phylogeny of CYC2-like genes for Malpighiaceae with the four reversal lineages highlighted. Bayesian majority rule consensus topology shown; clades with ≥50% maximum likelihood (ML) bootstrap support and ≥60% Bayesian posterior probabilities depicted above lines, respectively. ML bootstrap support <50% indicated with a hyphen. Inferred gene tree is reflective of accepted species tree relationships (Davis and Anderson, 2010). Accessions highlighted in red include the four Malpighiaceae clades examined here that exhibit reversal floral phenotypes—madagasikarioids and Malagasy Sphedamnocarpus in OW, and radial ptilochaetoids and Psychopterys dipholiphylla in NW. See Materials and Methods and Table A1 for species identities and voucher information. C, Centroplacaceae; E, Elatinaceae; M, Malpighiaceae; O, Oxalidaceae.
To examine whether these CYC2 homologs are involved in floral development, locus-specific reverse transcription (RT)-PCR was carried out at several stages of floral development (Figure 3). Most NW Malpighiaceae express both CYC2A and CYC2B consistently throughout floral development (Figure 3A) (Zhang et al., 2010, 2012). The late stage expression of CYC2 like genes has been shown to be critical for developing zygomorphic flowers in many lineages (Luo et al., 1996, 1999; Cubas et al., 1999a; Feng et al., 2006; Busch and Zachgo, 2007; Broholm et al., 2008; Wang et al., 2008). In sharp contrast, RT-PCR revealed that CYC2 homologs are not expressed in the OW Sphedamnocarpus sp. (nov.) floral buds and show exceptionally low activity in the NW Psychopterys dipholiphylla, especially at late stages of floral development (Figures 3B,C). We used quantitative RT-PCR to further examine the spatial pattern of CYC2 expression in actinomorphic flowered Psychopterys dipholiphylla. This experiment revealed that the low expression levels of CYC2A and CYC2B that are present have become more radialized by expanding into the ventral region of the corolla (Figure 4A). However, both of these actinomorphic flowered lineages show greatly down regulated CYC2 expression: CYC2 expression is more radialized in the corolla of Psychopterys dipholiphylla (Figure 4A), but at barely detectable levels (Figure 3C); while, Sphedamnocarpus sp. expresses no CYC2 during floral development (Figure 3B).
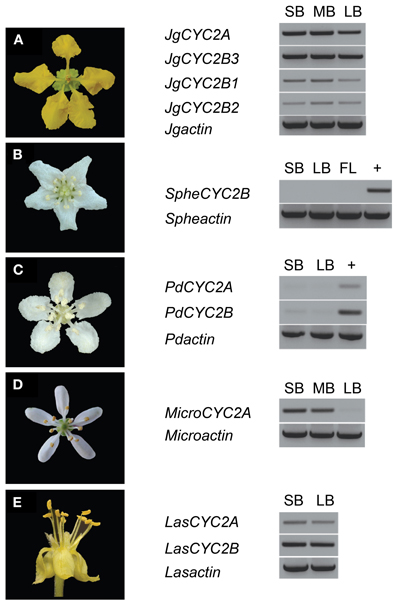
Figure 3. Locus-specific RT-PCR for CYC2-like gene expression in Malpighiaceae. (A) Janusia guaranitica shows the consistent CYC2A and CYC2B expression in NW Malpighiaceae. (B–E) The temporal pattern of CYC2 expression in the derived actinomorphic Malpighiaceae Sphedamnocarpus sp. (B) Psychopterys dipholiphylla (C) Microsteria sp. (D) Lasiocarpus sp. (E) +, genomic DNA was used as a control for PCR efficiency. MicroCYC2B and SpheCYC2A were not included in the analyses due to failure to recover these loci using exhaustive PCR and clone screening described in the Materials and Methods. ACTIN-specific primers were used as a positive control (Zhang et al., 2010). Abbreviations are as follows: SB, small buds ~10–20% of full size buds; MB, medium buds ~40–60% of full size buds; LB, large buds ~70–90% of full size buds; FL, open flowers.
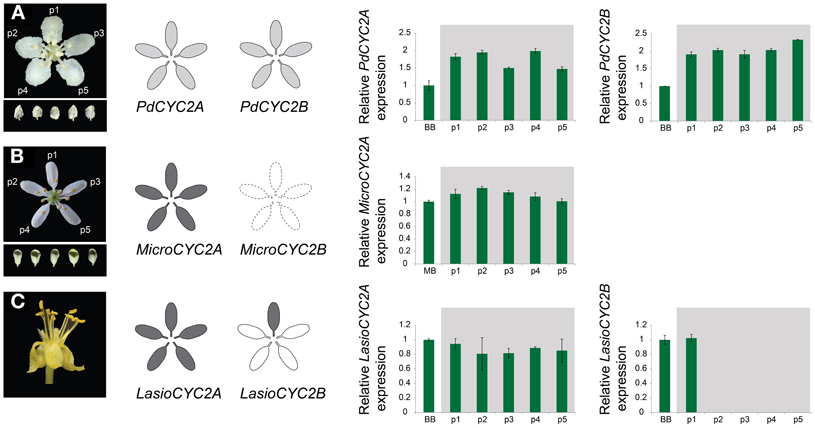
Figure 4. Quantitative RT-PCR (qRT-PCR) expression of CYC2-like genes for the reversed floral morphologies in the Malpighiaceae Psychopterys dipholiphylla (A), Microsteria sp. (B), and Lasiocarpus sp. (C). MicroCYC2B was not included in this analysis due to inability to recover it from Microsteria sp. Grayscale shading on floral diagrams summarizes the relative strength of the spatial pattern of CYC2 expression in the corolla. qRT-PCR expression data was determined for dissected floral organs at mid to late stage (also see Materials and Methods). Expression levels are relative to the control β-tubulin. Error bars represent standard errors. p1–p5, five dissected petals from a single flower. Levels of gene expression among p1-p5 are not significantly different (P > 0.05) for PdCYC2A, PdCYC2B, MicroCYC2A, and LasioCYC2A.
In the OW Microsteria sp. and the NW Lasiocarpus sp., CYC2 is expressed at a high level throughout floral development (Figures 3D,E). Moreover, quantitative RT-PCR revealed that CYC2A expression is expanded to include the ventral region for Microsteria sp. and Lasiocarpus sp. (Figures 4B,C). While MicroCYC2A and LasCYC2A expression is similarly high across the entire corolla, LasCYC2B is expressed only in the innermost single petal in Lasiocarpus sp. Species of Malpighiaceae have very conserved floral aestivation, in which the innermost petal in the corolla always forms the conspicuous dorsal banner petal in the NW species (Eichler, 1878; Zhang et al., 2010). This suggests that LasCYC2B is expressed in the petal that is homologous to the banner petal of most NW Malpighiaceae. The OW madagasikarioids and the NW radial ptilochaetoids therefore share very similar changes in the pattern of CYC2A expression by expanding gene expression such that it is equally distributed in all five petals.
We observed that the conserved CYC2 pathway of the stereotypical NW Malpighiaceae changes dramatically in four clades that have each experienced reversals to floral actinomorphy. Interestingly, this occurs against two strikingly different biogeographical backgrounds. Two actinomorphic clades on opposite sides of the Atlantic Ocean, the OW Microsteira sp. and NW Lasiocarpus sp., shift their expression to be broad, whereas the similarly distributed OW Malagasy Sphedamnocarpus and NW Psychopterys lack late stage CYC2-like gene expression (Figure 5). These contrasting patterns seem to reflect the two general CYC2-dependent mechanisms that may be responsible for the transition to actinomorphy from a zygomorphic ancestor: (i) ectopic CYC2 expression resulting in phenotypic dorsalization of the entire flower, or (ii) loss of function resulting in phenotypic ventralization. However, we cannot exclude the possibility that CYC2 downstream factors may also play a role for floral radialization. The presence of both patterns underscores the fact that each of these four clades independently transitioned to actinomorphy in response to the loss of their specialist NW oil bee pollinators, traversing distinct genetic trajectories in the process.
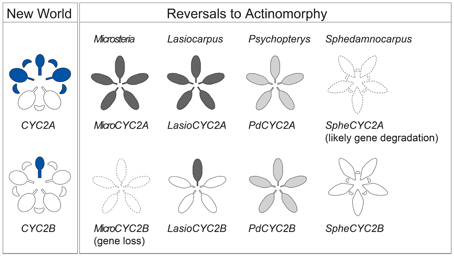
Figure 5. Summary of CYC2-like gene expression. Expression of CYC2-like genes in NW Malpighiaceae and in reversals to actinomorphy in the four Malpighiaceae clades represented, from left to right, by Microsteria sp., Lasiocarpus sp., Psychopterys dipholiphylla, and Sphedamnocarpus sp. The blue shading of the NW Species indicates late stage CYC2 gene expression (Zhang et al., 2010, 2012). The gradient shading in the derived actinomorphic species, from white to dark gray, indicates increasing intensity of CYC2 expression, respectively. The dotted outlines indicate the loss of gene copy, or inability to recover this gene due to its likely degradation.
The evolutionary potential for such modification of a pre-existing CYC-dependent program was immediately recognized in early studies of CYC mutants in Antirrhinum (Coen and Nugent, 1994; Coen et al., 1995). However, while loss-of-function represents an attractively simple model for reversals to actinomorphy, Donoghue et al. (1998) pointed out that a gain of function model in which expression is expanded into the ventral petals seems more likely from a morphological perspective. Diverse studies have, in fact, revealed both patterns. Ventral expansion of CYC2 expression has been observed in the actinomorphic flowers of the legume Cadia (Citerne et al., 2006) and in Tengia, a member of the Gesneriaceae (Pang et al., 2010). Conversely, a different actinomorphic Gesneriaceae, Bournea leiophylla, exhibits rapid down-regulation of CYC2 expression at later developmental stages (Zhou et al., 2008), suggesting loss-of-function. Similarly, the actinomorphic flowers of Plantago, a member of the Plantaginaceae, are associated with degeneration of both the CYC-based dorsal identity and MYB-based ventral identity programs, although one CYC2 homolog is still broadly expressed (Preston et al., 2011).
Currently, we have data on CYC2 homolog expression patterns from ten major clades within Malpighiaceae: three with the stereotypical NW zygomorphy, three with the altered form of OW zygomorphy, and four that are completely actinomorphic. Analyzing these results in a phylogenetic context provides insight into the major evolutionary events associated with diversification of floral morphology across the family (Figure 6). The origin of zygomorphy in the NW ancestors of the family appears to be closely associated with a duplication event that gave rise to the CYC2A and 2B lineages. Analyses of divergent taxa with the distinct NW zygomorphy reveal a pattern in which CYC2A is expressed in the banner petal and two lateral petals while CYC2B is narrowly confined to the banner petal alone (Figure 6) (Zhang et al., 2010, 2012). Similar to what has been observed in many other systems [reviewed in Citerne et al. (2010)], the expression of these CYC2 paralogs in partially overlapping domains may create a dosage-based mechanism whereby the banner petal is distinguished by the highest concentration of CYC2A+B, followed by the lateral petals with CYC2A alone, and the ventral petals expressing no CYC2 homologs. The origin of this program in the Malpighiaceae, however, is clouded by the presence of two contrasting patterns in their closest actinomorphic flowered relatives (Figure 6) (Zhang et al., 2010). Reconstruction of the ancestral condition is equivocal as to whether absence of expression or broad expression is ancestral (Zhang et al., 2010). Although the close radial flowered relative of Malpighiaceae, Bergia (Elatinaceae), may seem atypical in having broad CYC2 expression, a strikingly similar pattern has been observed in the asterid clade Dipsacales (Howarth et al., 2011). In this case, broad CYC2 expression in actinomorphic Viburnum appears to be the ancestral condition predating evolution of dorsal-restricted expression in the zygomorphic Caprifoliaceae. However, these two examples in the Malpighiales and Dipsacales highlight the general lack of both expression and functional data from actinomorphic flowers, and raise the possibility that the Arabidopsis pattern, which has been assumed to represent the ancestral actinomorphic condition (Cubas et al., 2001), may not actually be the best representative. More research is needed on CYCLOIDEA function and expression in radial flowered angiosperm clades.
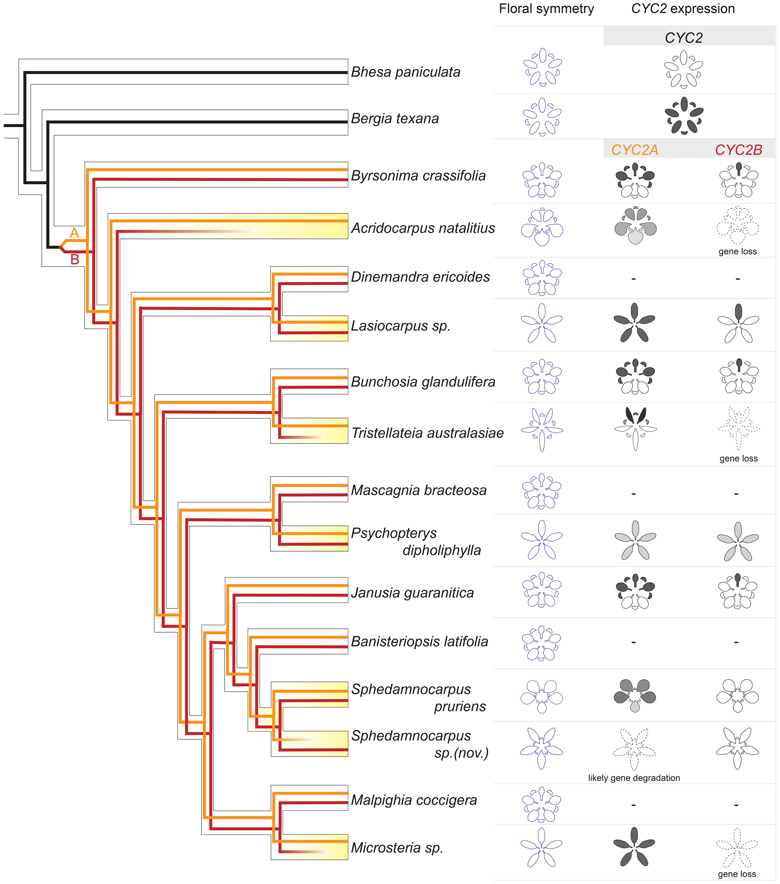
Figure 6. A synopsis of CYC2 evolution underlying floral symmetry evolution in Malpighiaceae and close relatives. The hollow-branch phylogeny indicates species tree relationships based on Davis and Anderson (2010). The solid-line phylogeny indicates CYC2 gene tree, which was embedded within the species tree. Black lines: the CYC2 gene lineage in close relatives, Centroplacaceae (e.g., Bhesa paniculata) and Elatinaceae (e.g., Bergia texana); dark red and orange lines: CYC2A and CYC2B, respectively. CYC2 paralogs originated through a gene duplication that occurred in the common ancestor of all Malpighiaceae. Most species of NW Malpighiaceae have maintained both CYC2A and CYC2B copies, while many of their OW counterparts (e.g., Acridocarpus natalitius, Tristellateia australasiae, Sphedamnocarpus sp. (nov.), and Microsteria sp.) lost one of the two CYC2 copies indicating as fainting out lines. The expression patterns of CYC2 genes at late stages of floral development is summarized for all of the sampled outgroup and Malpighiaceae taxa: the radial outgroup genera Bhesa paniculata and Bergia texana; three zygomorphic species of NW Malpighiaceae that are phylogenetically distant related and span the origin of Malpighiaceae, Bunchosia glandulifera, Byrsonima crassifolia, and Janusia guaranitica; and several species that lost the NW zygomorphy, which are highlighted in yellow. OW species: Acridocarpus natalitius, Sphedamnocarpus pruriens, Sphedamnocarpus sp.(nov.), and Tristellateia australasiae; NW species: Lasiocarpus sp., and Psychopterys dipholiphylla. A dash indicates NW counterparts for which CYC2 genes were cloned but the expression data is not available.
If we turn our attention back to the clades in which the NW zygomorphy is lost, we see that these transitions are consistently associated with extreme reduction or loss of CYC2B expression. The only exception in this regard is Lasiocarpus, where it seems likely that the radial, strong expression of CYC2A negates the remaining banner petal-specific expression of CYC2B. Interestingly, this pattern directly parallels what has been observed in the actinomorphic legume Cadia, where expression of one CYC2 copy has been maintained at weak to moderate expression levels in dorsal petals, while the second copy is broadly expressed at very high levels (Citerne et al., 2006). Considering CYC2A expression patterns, there is a much greater diversity in patterns of expression across clades, but a highly consistent correlation with floral morphology. When the NW-type zygomorphy is altered via a shift to two dorsal petals instead of one, the axis of CYC2A expression shifts in concert, although this new expression domain can be arrayed in a differential gradient across the corolla (i.e., Acridocarpus and Sphedamnocarpus puriens) or tightly restricted to the two dorsal petals (i.e., Tristellateia) (Figure 6; Zhang et al., 2012). When actinomorphy evolves, the pattern of expression is much more variable, but still completely consistent with morphology. Although we might be tempted to conclude that Lasiocarpus and Microsteria flowers are essentially dorsalized while those of Psychopterys and Sphedamnocarpus sp.(nov.) are ventralized, this is difficult to determine without information on other aspects of radial organ identity. In the Plantago case described above, data on the CYC2 loci alone might lead to a conclusion that the flowers are dorsalized, but the complete absence of downstream components of both the dorsal and ventral identity pathways suggests that they are actually lateralized (Preston et al., 2011). Unfortunately, there is no current data to suggest that the functions of these downstream factors are conserved outside the Lamiales, so we have no candidates for effectors of CYC2 function in Malpighiales. What is clear is that despite three distinct floral forms, the expression of CYC2 homologs correlates incredibly well with floral morphology across Malpighiaceae, providing further evidence that these genes play crucial roles in the development and evolution of floral symmetry in this clade.
In addition to illuminating broader mechanisms of morphological evolution, our molecular studies have helped us better understand the small scale character state transitions that gave rise to divergent floral forms across Malpighiaceae, particularly in Sphedamnocarpus. This OW genus is comprised of two major clades: an African clade represented by S. puriens, which bears yellow or cream-colored flowers with the altered NW zygomorphy, and a Malagasy clade represented by S. sp. (nov.), which bears white actinomorphic flowers (Davis and Anderson, 2010). The closest sister genus of Sphedamnocarpus in the NW is Banisteriopsis C. B. Rob, whose yellow flowers have the typical banner petal zygomorphy (Davis and Anderson, 2010). Simple ancestral character state reconstruction of floral symmetry cannot resolve whether the Sphedamnocarpus ancestral state was NW zygomorphy, the altered OW zygomorphy, or actinomorphy (Figure A1), but a much clearer evolutionary scenario becomes apparent with the addition of our molecular data. Both Sphedamnocarpus species lack CYC2B expression, indicating that this was most likely the case in their common ancestor. Given that loss of CYC2B expression is closely associated with loss of the NW zygomorphic pattern, we would hypothesize that the common ancestor of all Sphedamnocarpus did not possess the banner petal morphology. Furthermore, since the actinomorphy of S. sp. (nov.) is associated with complete loss of CYC2A, it seems unlikely that this state represents the ancestral condition (i.e., a scenario that would invoke the re-evolution of the zygomorphic pattern of CYC2 gene expression in zygomorphic African Sphedamnocarpus). Thus, we can propose a model for the stepwise reversion to actinomorphy in this lineage. First, CYC2B expression was lost in the OW ancestor, most likely in concert with the evolution of the altered form of zygomorphy. This ancestor then gave rise to the African clade with its shifted axis of graded differential CYC2A expression, as well as to the Malagasy clade in which loss of CYC2A facilitated the transition to complete actinomorphy. This is the most parsimonious explanation of the current data, but further sampling of this fascinating lineage will allow our hypothesis to be further tested and refined as needed. Thus, the Sphedamnocarpus clade represents a very nice example of the way in which phylogenetically focused studies of floral gene expression can illuminate otherwise uncertain ancestral character state reconstructions involving important morphological innovations.
Taken together, our study illustrates both the constrained and contingent nature of evolution. While certain patterns are fairly predictable, such as loss of CYC2B function in association with loss of the stereotypical NW zygomorphy, nothing is absolutely certain, as evidenced by the retention of CYC2B in Lasiocarpus. There are alternative genetic solutions to the problem of shifting pollinator availability, even when the morphological outcomes appear convergent. In this way, the Malpighiaceae offer a fascinating context in which to study the intersection between biogeography, pollinator interactions, floral morphology, and genetic evolution. Further studies will continue to build on and expand these models, hopefully providing a clearer picture of the complex evolutionary processes at work during the diversification of the family.
Specimens of Microsteria sp. and Rhynchophora phillipsonii W. R. Anderson are from Toliara, Madagascar; Philgamia glabrifolia Arènes from Mountain Ibity, Fianarantsoa, Madagascar; Sphedamnocarpus sp.(nov.) from Isalo National Park, Fianarantsoa, Madagascar; Lasiocarpus sp. from the municipio de Huitzuco de los Figueroa, Guerrero, Mexico; and Psychopterys dipholiphylla (Small) W. R. Anderson and S. Corso from the municipio de Taxco, Guerrero, Mexico (also see Table A1).
We used the 77 degenerate primer pair combinations followed by exhaustive clone screening as described in Zhang et al. (2010, 2012) to isolate CYC2-like genes from our target species. These primers amplify CYC-like genes from many major Malpighiales clades, including especially the sister families of Malpighiaceae. CYC2-like amplicons spanning the TCP and R domains were obtained following our previous methods [described in Zhang et al. (2010)]. The PCR products were cloned, and more than 200 clones were screened for each sample to identify CYC2 homologs.
The newly acquired sequences of CYC2-like genes from Microsteria sp., Rhynchophora phillipsonii, Philgamia glabrifolia, Sphedamnocarpus sp.(nov.), Lasiocarpus sp., and Psychopterys dipholiphylla were aligned with a previously available matrix including several ingroup accessions of Malpighiaceae, and the outgroup families, Elatinaceae, Centroplacaceae, and Oxalidaceae, by eye with reference to the translated amino acid sequences using MacClade 4.06 (Maddison and Maddison, 2003). The phylogeny in Figure 2 was based on amino acid sequence analyses. We applied the WAG + G model of amino acid evolution to the aligned CYC2 data set as determined by the AIC criterion in ProtTEST (Abascal et al., 2005). One thousand maximum likelihood bootstrap replicates were conducted using RAxML-VI-HPC (Stamatakis, 2006). Bayesian analyses were implemented in MrBayes ver. 3.1.2 (Huelsenbeck and Ronquist, 2001) under the mixed amino acid model. Analyses using nucleotide sequence data with third codon positions excluded under the best-fit model (“GTR + I + Γ”) for these data as determined using the Akaike Information Criterion (Akaike, 1973) in MODELTEST 3.06 (Posada and Crandall, 1998), yielded a topology nearly identical to the amino acid sequence data (not shown). One hundred ML bootstrap replicates were conducted with the optimal model of sequence evolution. Bayesian analyses were also conducted using the same model and default priors for the rate matrix, branch lengths, and gamma shape parameter. A Dirichlet distribution was used for the base frequency parameters and an uninformative prior was used for the starting tree topology. Four chains were initiated with a random starting tree and run for two million generations sampled every 1000 generations. Stationarity was determined using Tracer v1.4.1. (http://tree.bio.ed.ac.uk/software/tracer/). We sampled from the posterior distribution to calculate clade posterior probabilities following a burn-in of 1000 trees. DNA sequences of the newly acquired CYC2-like genes have been deposited in GenBank, under accession numbers KF514885-KF514893.
Ten μg of genomic DNA was digested from Microsteria sp., Rhynchophora phillipsonii, Philgamia glabrifolia, and Sphedamnocarpus sp. (nov.) with restriction enzyme EcoRI, fractionated on 0.8% agarose gels, and blotted onto a positively charged nylon membrane (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) following the protocol in Zhang et al. (2010). A fragment containing the 3′ end of the TCP domain and the variable region between the TCP and R domains was used as a template to synthesize probes for detecting CYC2-like genes. A mixture of CYC2 sequences of SphpCYC2A and SphpCYC2B of Sphedamnocarpus pruriens, SpheCYC2B of Sphedamnocarpus sp. (nov.), and PgCYC2B of Philgamia glabrifolia in equal molar concentration was used as a template to synthesize our 32P labeled probe to examine Philgamia glabrifolia, and Sphedamnocarpus sp. (nov.); and CYC2 sequences of TmCYC2A and TmCYC2B of Triaspis macropteron, a member of the madagasikarioids and MicroCYC2A of Microsteria sp. to examine Microsteria sp., and Rhynchophora phillipsonii. We previously showed that the number of bands in the EcoRI digest is a reliable indicator of CYC2 copy number (Zhang et al., 2010). Here, we identified a single band in the EcoRI digest for both Microsteria sp., and Rhynchophora phillipsonii, while two bands in the EcoRI digest for both Philgamia glabrifolia, and Sphedamnocarpus sp. (nov.) (Figure A2).
Floral buds from different developmental stages were prepared in liquid nitrogen in the field. They were grouped as small buds (~10–20% of full size buds); medium buds (~40–60% of full size buds); large buds (~70–90% of full size buds); and open flowers. All materials were preserved in cryogenic containers, and were processed in the lab using the RNAqueous kit (Ambion, Austin, TX, USA). Floral organs from medium to late stage samples, ~50–80% of flower bud size just before anthesis, were dissected in the lab from a single bud. These buds were dissected using the RNAlater® -ICE Kit (Ambion-Applied Biosystems, Austin, TX, USA) The micro-dissected samples were processed using the RNAqueous Micro kit (Ambion, Austin, TX, USA). The details about this method described in Zhang et al. (2012).
RT-PCR was performed using locus specific primers (Table A2) to examine the expression of CYC2. The sequence identity of RT-PCR fragments was further confirmed by sequencing.
Samples from a single flower were analyzed in three separate qRT-PCRs. Three biological replicates, from three separate flowers, were conducted for each species. qRT-PCR reactions were conducted using PerfeCTa® SYBR® Green FastMix®, Low ROX™ (Quanta BioSciences, Inc., Gathersburg, MD) using the Stratagene Mx3005P QPCR System. Class I β-tubulin was used as a control to normalize the qRT-PCR (Oakley et al., 2007). CYC2 expression levels were calculated relative to β-tubulin using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Details about the method and statistical analyses were described in Zhang et al. (2012).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank W. Anderson, D. Boufford, and members of the Davis and Kramer laboratories for comments on our manuscript; D. Boufford, J. Andriantiana, M. Petru, and A. Arnold for help with fieldwork. This work supports by National Science Foundation Grants DEB-0544039 and AToL EF 04-31242 (to Charles C. Davis).
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. doi: 10.1093/bioinformatics/bti263
Akaike, H. (1973). “Information theory and an extension of the maximum likelihood principle,” in Second International Symposium in Information Theory, eds B. N. Petrov and F. Csaki (Budapest: Akademiai Kiado), 267–281.
Anderson, W. R. (1979). Floral conservatism in neotropical Malpighiaceae. Biotropica 11, 219–223. doi: 10.2307/2388042
Broholm, S. K., Tähtiharju, S., Laitinen, R.A., Albert, V. A., Teeri, T. H., and Elomaa, P. (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. U.S.A. 105, 9117–9122. doi: 10.1073/pnas.0801359105
Busch, A., and Zachgo, S. (2007). Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proc. Natl. Acad. Sci. U.S.A. 104, 16714–16719. doi: 10.1073/pnas.0705338104
Citerne, H., Jabbour, F., Nadot, S., and Damerval, C. (2010). “The evolution of floral symmetry,” in Advances in Botanical Research, eds J. C. Kader and M. Delseny (London: Elsevier), 85–137.
Citerne, H. L., Pennington, R. T., and Cronk, Q. C. B. (2006). An apparent reversal in floral symmetry in the legume Cadia is a homeotic transformation. Proc. Natl. Acad. Sci. U.S.A. 103, 12017–12020. doi: 10.1073/pnas.0600986103
Coen, E. S., and Nugent, J. M. (1994). Evolution of flowers and inflorescences. Development 1994(Suppl.), 107–116.
Coen, E. S., Nugent, J. M., Lou, D. A., Bradley, D., Cubas, P., Chadwick, M., et al. (1995). Evolution of floral symmetry. Philos. Trans. R. Soc. Lond. B Biol. Sci. 350, 35–38. doi: 10.1098/rstb.1995.0134
Conte, G. L., Arnegard, M. E., Peichel, C. L., and Schluter, D. (2012). The probability of genetic parallelism and convergence in natural populations. Proc. R. Soc. B Biol. Sci. 279, 5039–5047. doi: 10.1098/rspb.2012.2146
Conway Morris, S. (2008). “Evolution and convergence: some wider considerations,” in The Deep Structure of Biology: is Convergence Sufficiently Ubiquitous to Give a Directional Signal? ed S. Conway Morris (West Conshohocken, PA: Templeton Foundation Press), 46–67.
Cubas, P., Coen, E., and Zapater, J. M. M. (2001). Ancient asymmetries in the evolution of flowers. Curr. Biol. 11, 1050–1052. doi: 10.1016/S0960-9822(01)00295-0
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999a). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
Cubas, P., Vincent, C., and Coen, E. (1999b). An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401, 157. doi: 10.1038/43657
Davis, C. C. (2002). Madagasikaria (Malpighiaceae): a new genus from Madagascar with implications for floral evolution in Malpighiaceae. Am. J. Bot. 89, 699–706. doi: 10.3732/ajb.89.4.699
Davis, C. C., and Anderson, W. R. (2010). A complete generic phylogeny of Malpighiaceae inferred from nucleotide sequence data and morphology. Am. J. Bot. 97, 2031–2048. doi: 10.3732/ajb.1000146
Davis, C. C., Anderson, W. R., and Donoghue, M. J. (2001). Phylogeny of Malpighiaceae: evidence from chloroplast ndhF and trnL-F nucleotide sequences. Am. J. Bot. 88, 1830–1846. doi: 10.2307/3558360
Davis, C. C., Bell, C. D., Mathews, S., and Donoghue, M. J. (2002). Laurasian migration explains Gondwanan disjunctions: evidence from Malpighiaceae. Proc. Natl. Acad. Sci. U.S.A. 99, 6833–6837. doi: 10.1073/pnas.102175899
Davis, C. C., and Chase, M. W. (2004). Elatinaceae are sister to Malpighiaceae; Peridiscaceae belong to Saxifragales. Am. J. Bot. 91, 262–273. doi: 10.3732/ajb.91.2.262
Donoghue, M. J. (2005). Key innovations, convergence, and success: macroevolutionary lessons from plant phylogeny. Paleobiology 31, 77–93. doi: 10.1666/0094-8373(2005)031[0077:KICASM]2.0.CO;2
Donoghue, M. J., Ree, R. H., and Baum, D. A. (1998). Phylogeny and the evolution of flower symmetry in the Asteridae. Trends Plant Sci. 3, 311–317. doi: 10.1016/S1360-1385(98)01278-3
Feng, X., Zhao, Z., Tian, Z., Xu, S., Luo, Y., Cai, Z., et al. (2006). Control of petal shape and floral zygomorphy in Lotus japonicus. Proc. Natl. Acad. Sci. U.S.A. 103, 4970–4975. doi: 10.1073/pnas.0600681103
Gompel, N., and Prud'homme, B. (2009). The causes of repeated genetic evolution. Dev. Biol. 332, 36–47. doi: 10.1016/j.ydbio.2009.04.040
Hoekstra, H. E., and Nachman, M. W. (2003). Different genes underlie adaptive melanism in different populations of rock pocket mice. Mol. Ecol. 12, 1185–1194. doi: 10.1046/j.1365-294X.2003.01788.x
Howarth, D. G., and Donoghue, M. J. (2006). Phylogenetic analysis of the "ECE" (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. U.S.A. 103, 9101–9106. doi: 10.1073/pnas.0602827103
Howarth, D. G., Martins, T., Chimney, E., and Donoghue, M. J. (2011). Diversification of CYCLOIDEA expression in the evolution of bilateral flower symmetry in Caprifoliaceae and Lonicera (Dipsacales). Ann. Bot. 107, 1521–1532. doi: 10.1093/aob/mcr049
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lobreau-Callen, D. (1989). Les Malpighiaceae et leurs pollinisateurs. Coadaptation ou coévolution. Bulletin du Museum National d'Histoire Naturelle. Section B, Adansonia: Botanique Phytochimie 11, 79–94.
Luo, D., Carpenter, R., Copsey, L., Vincent, C., Clark, J., and Coen, E. (1999). Control of organ asymmetry in flowers of Antirrhinum. Cell 99, 367–376. doi: 10.1016/S0092-8674(00)81523-8
Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799. doi: 10.1038/383794a0
Maddison, D. R., and Maddison, W. P. (2003). MacClade 4: Analysis of Phylogeny and Character Evolution. Version 4.0.6 Edn. Sunderland: Sinauer Associates.
Oakley, R. V., Wang, Y. S., Ramakrishna, W., Harding, S. A., and Tsai, C. J. (2007). Differential expansion and expression of alpha- and beta-tubulin gene families in Populus. Plant Physiol. 145, 961–973. doi: 10.1104/pp.107.107086
Pang, H. B., Sun, Q. W., He, S. Z., and Wang, Y. Z. (2010). Expression pattern of CYC-like genes relating to a dorsalized actinomorphic flower in Tengia (Gesneriaceae). J. Syst. Evol. 48, 309–317. doi: 10.1111/j.1759-6831.2010.00091.x
Posada, D., and Crandall, K. A. (1998). MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Preston, J. C., and Hileman, L. C. (2009). Developmental genetics of floral symmetry evolution. Trends Plant Sci. 14, 147–154. doi: 10.1016/j.tplants.2008.12.005
Preston, J. C., Martinez, C. C., and Hileman, L. C. (2011). Gradual disintegration of the floral symmetry gene network is implicated in the evolution of a wind-pollination syndrome. Proc. Natl. Acad. Sci. U.S.A. 108, 2343–2348. doi: 10.1073/pnas.1011361108
Protas, M. E., Hersey, C., Kochanek, D., Zhou, Y., Wilkens, H., Jeffery, W. R., et al. (2006). Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat. Genet. 38, 107–111. doi: 10.1038/ng1700
Prud'homme, B., Gompel, N., Rokas, A., Kassner, V. A., Williams, T. M., Yeh, S.-D., et al. (2006). Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440, 1050–1053. doi: 10.1038/nature04597
Rosenblum, E. B., Rompler, H., Schoneberg, T., and Hoekstra, H. E. (2010). Molecular and functional basis of phenotypic convergence in white lizards at White Sands. Proc. Natl. Acad. Sci. U.S.A. 107, 2113–2117. doi: 10.1073/pnas.0911042107
Stamatakis, A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Steiner, C. C., Rompler, H., Boettger, L. M., Schoneberg, T., and Hoekstra, H. E. (2009). The genetic basis of phenotypic convergence in beach mice: similar pigment patterns but different genes. Mol. Biol. Evol. 26, 35–45. doi: 10.1093/molbev/msn218
Sucena, E., Delon, I., Jones, I., Payre, F., and Stern, D. L. (2003). Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 424, 935–938. doi: 10.1038/nature01768
Vogel, S. (1974). Ölblumen und ölsammelnde Bienen [Oil flowers and oil-collecting bees]. Tropische und Subtropische Pflanzenwelt 7, 283–547.
Vogel, S. (1990). History of the Malpighiaceae in the light of pollination ecology. Mem. N.Y. Bot. Gard. 55, 130–142.
Wake, D. B., Wake, M. H., and Specht, C. D. (2011). Homoplasy: from detecting pattern to determining process and mechanism of evolution. Science 331, 1032–1035. doi: 10.1126/science.1188545
Wang, Z., Luo, Y. H., Li, X., Wang, L. P., Xu, S. L., Yang, J., et al. (2008). Genetic control of floral zygomorphy in pea (Pisum sativum L.). Proc. Natl. Acad. Sci. U.S.A. 105, 10414–10419. doi: 10.1073/pnas.0803291105
Wittkopp, P. J., Williams, B. L., Selegue, J. E., and Carroll, S. B. (2003). Drosophila pigmentation evolution: divergent genotypes underlying convergent phenotypes. Proc. Natl. Acad. Sci. U.S.A. 100, 1808–1813. doi: 10.1073/pnas.0336368100
Wurdack, K. J., and Davis, C. C. (2009). Malpighiales phylogenetics: gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am. J. Bot. 96, 1551–1570. doi: 10.3732/ajb.0800207
Xi, Z. X., Ruhfel, B. R., Schaefer, H., Amorim, A. M., Sugumaran, M., Wurdack, K. J., et al. (2012). Phylogenomics and a posteriori data partitioning resolve the cretaceous angiosperm radiation Malpighiales. Proc. Natl. Acad. Sci. U.S.A. 109, 17519–17524. doi: 10.1073/pnas.1205818109
Zhang, W., Kramer, E. M., and Davis, C. C. (2010). Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. Proc. Natl. Acad. Sci. U.S.A. 107, 6388–6393. doi: 10.1073/pnas.0910155107
Zhang, W., Kramer, E. M., and Davis, C. C. (2012). Similar genetic mechanisms underlie the parallel evolution of floral phenotypes. PLoS ONE 7:e36033. doi: 10.1371/journal.pone.0036033
Zhou, X.-R., Wang, Y.-Z., Smith, J. F., and Chen, R. (2008). Altered expression patterns of TCP and MYB genes relating to the floral developmental transition from initial zygomorphy to actinomorphy in Bournea (Gesneriaceae). New Phytol. 178, 532–543. doi: 10.1111/j.1469-8137.2008.02384.x
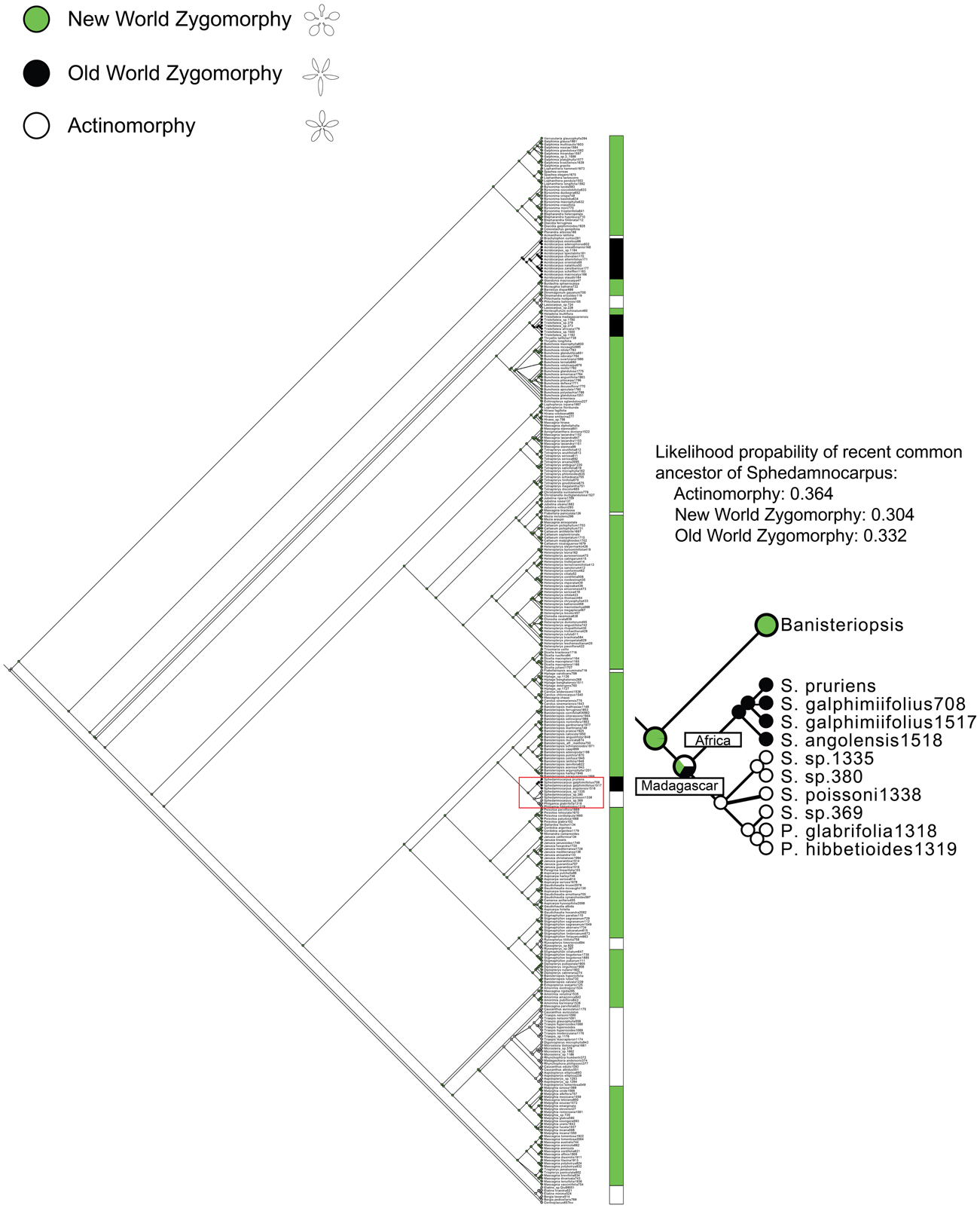
Figure A1. Ancestral character state reconstruction of floral symmetry. ML analysis indicates the NW zygomorphy has been lost nine times in Malpighiaceae. OW zygomorphy has evolved from an NW ancestor three times in parallel; the NW zygomorphy has also been reversed to actinomorphy eight times in the family. The part of tree is enlarged to show the evolutionary transition of floral symmetry in Sphedamnocarpus. These results indicate that the ancestral character state reconstruction suggests the common ancestor of the Sphedamnocarpus is equivocal among NW zygomorphy, OW zygomorphy, or actinomorphy. The comparative expression data help to distinguish between these scenarios (see Discussion).
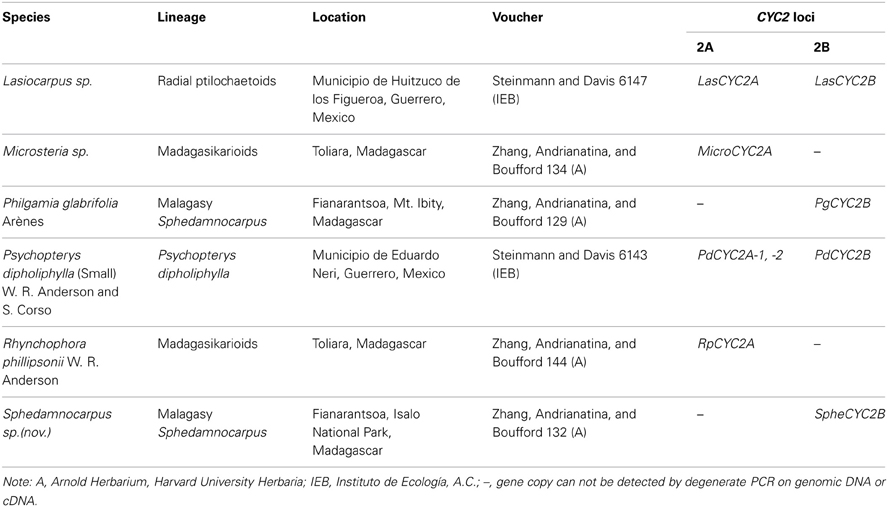
Table A1. Species sampled in this study, with collection locations, voucher information, and CYC2 loci.
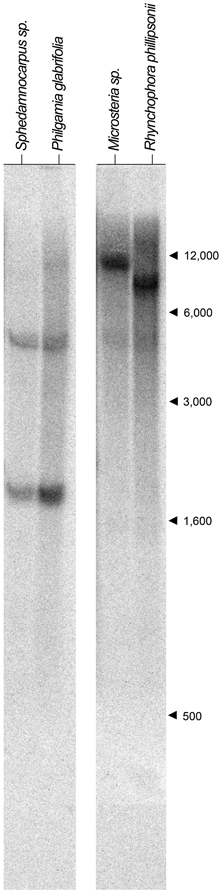
Figure A2. CYC2-like gene Southern hybridization results for Sphedamnocarpus sp.,Philgamia glabrifolia,Microsteria sp.,and Rhynchophora phillipsonii. Restriction digests using EcoRI are shown for Sphedamnocarpus sp.,Philgamia glabrifolia,Microsteria sp., and Rhynchophora phillipsonii. Arrows and numbers indicate molecular size markers (in base pairs).
Keywords: CYC2-like genes, development, floral symmetry, Malpighiaceae, reversals
Citation: Zhang W, Steinmann VW, Nikolov L, Kramer EM and Davis CC (2013) Divergent genetic mechanisms underlie reversals to radial floral symmetry from diverse zygomorphic flowered ancestors. Front. Plant Sci. 4:302. doi: 10.3389/fpls.2013.00302
Received: 26 April 2013; Paper pending published: 02 June 2013;
Accepted: 20 July 2013; Published online: 20 August 2013.
Edited by:
Jill Christine Preston, University of Vermont, USAReviewed by:
Lena Hileman, University of Kansas, USACopyright © 2013 Zhang, Steinmann, Nikolov, Kramer and Davis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenheng Zhang, Department of Biology, Virginia Commonwealth University, 1000 West Cary Street, Richmond, VA 23284, USA e-mail:d3poYW5nNUB2Y3UuZWR1;
Charles C. Davis, Department of Organismic and Evolutionary Biology, Harvard University Herbaria, 22 Divinity Avenue, Cambridge, MA 02138, USA e-mail:Y2RhdmlzQG9lYi5oYXJ2YXJkLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.