- 1Centro de Biotecnologia, Universidade Federal do Rio Grande do Sul, Porto Alegre, Rio Grande do Sul, Brazil
- 2Departamento de Botânica, Universidade Federal do Rio Grande do Sul, Porto Alegre, Rio Grande do Sul, Brazil
- 3Centro de Ciências Biológicas e da Saúde, Programa de Pós-Graduação em Biotecnologia, Centro Universitário UNIVATES, Lajeado, Rio Grande do Sul, Brazil
Senescence is a coordinated process where a plant, or a part of it, engages in programmed cell death to salvage nutrients by remobilizing them to younger tissues or to developing seeds. As Fe and Zn deficiency are the two major nutritional disorders in humans, increased concentration of these nutrients through biofortification in cereal grains is a long-sought goal. Recent evidences point to a link between the onset of leaf senescence and increased Fe and Zn remobilization. In wheat, one member of the NAC (NAM, ATAF, and CUC) transcription factor (TF) family (NAM-B1) has a major role in the process, probably regulating key genes for the early onset of senescence, which results in higher Fe and Zn concentrations in grains. In rice, the most important staple food for nearly half of the world population, the NAM-B1 ortholog does not have the same function. However, other NAC proteins are related to senescence, and could be playing roles on the same remobilization pathway. Thus, these genes are potential tools for biofortification strategies in rice. Here we review the current knowledge on the relationship between senescence, Fe and Zn remobilization and the role of NAC TFs, with special attention to rice. We also propose a working model for OsNAC5, which would act on the regulation of nicotianamine (NA) synthesis and metal–NA remobilization.
Fe and Zn Biofortification
Iron (Fe) and zinc (Zn) are essential micronutrients for almost all living organisms and are two of the most versatile metals in biology. Fe participates as a catalytic cofactor in multiple metabolic pathways (photosynthesis, respiration, hormone synthesis, nitrogen fixation, DNA synthesis and repair) due to the ability to participate on reversible redox reactions as Fe2+ (ferrous) and Fe3+ (ferric) ions (Puig et al., 2007). Zn does not participate directly on redox reactions, since it occurs in a single oxidation state, but is a key structural component of around 300 enzymes and 2,000 transcription factors (TFs; Palmgren et al., 2008; Prasad, 2012). Both Fe and Zn are present in low quantities in most plant staple foods, leading to Fe and Zn deficiency in humans. Malnutrition of these micronutrients are leading risk factors for disability and death worldwide, especially to children eating cereal-based diets, with low intake of micronutrient-rich foods as meat, poultry, fish, fruits, legumes, and vegetables.
Strategies to alleviate micronutrient malnutrition include fortification (addition during food processing) and supplementation (ingestion of pills or sachets). Although somewhat successful, these approaches are not widely accessible due to logistic and economic issues. A very cost-effective alternative is biofortification, the increase of bioavailable concentrations of an element in edible portions of crops before harvesting (for comprehensive reviews see White and Broadley, 2005; Sperotto et al., 2012a; Carvalho and Vasconcelos, 2013).
Biofortification includes different approaches like soil fertilization or foliar application, conventional breeding and/or transgenic strategies. Mineral fertilization is an effective method to increase seed mineral concentrations, but can be problematic due to continuous cost and environmental carryover. Conventional breeding has been used for decades. Although there is genetic diversity available within existing germplasm collections, rice seems to have the narrowest range, making substantial increases in Fe and Zn concentrations more difficult compared to maize and wheat (Kennedy and Burlingame, 2003; Gómez-Galera et al., 2010; Sperotto et al., 2012a). Thus, it seems imperative that transgenic approaches be used to enable significant increases in Fe and Zn content and bioavailability.
Single or multiple-transgene insertions into the rice genome have successfully increased Fe concentration in grains. Independent over-expression of OsNAS genes produced the most promising results so far (Johnson et al., 2011), while other multi-transgene approaches also increased grain Fe concentration (Wirth et al., 2009; Masuda et al., 2012). However, except in the study from Johnson et al. (2011), the levels are still not effective enough to impact human nutrition. An unexplored avenue would be the controlled expression of regulatory genes involved in key processes to Fe and Zn seed allocation. This approach has already been performed to generate rice plants more resistant to Fe deficiency (Kobayashi et al., 2007; Ogo et al., 2011). In order to do that for mineral concentrations in grains, we still need to identify the molecular players relevant to their transport within the plant and during remobilization.
Senescence Processes: What, When, and How?
Senescence represents endogenously degenerative processes which ultimately lead to organ death. However, it is not a passive process (Yoshida, 2003), but rather a series of coordinated and controlled events. Decline of photosynthesis, chloroplast and chlorophyll degradation, dismantling of biomolecules and decrease in cellular metabolic activities take place, which result in available nutrients and metabolites that can be transported from source (the tissue that supplies nutrients, most commonly green tissues) to sink (the net importer of nutrients, younger or reproductive organs) through the vascular system (Thomas, 2013). Part of leaf senescence seems to be regulated by sugar levels (Rolland et al., 2006; Sperotto et al., 2007), since a senescence-related loss of chlorophyll or protein can be induced by increased sugar contents (Wingler et al., 1998). Hormones and nutrients also contribute to regulation of senescence in source tissues, especially cytokinins, which have a senescence-delaying effect (Sperotto et al., 2009; Davies and Gan, 2012).
The source–sink signaling is not fully understood, and it depends on the species and circumstances. It is already known that senescence in source leaves can be delayed by removal of strong sinks (Zavaleta-Mancera et al., 1999). Elevated levels of N alter sugar signaling in source leaves (Thomas, 2013) and have a significant impact on Fe and Zn acquisition and grain allocation in wheat (Kutman et al., 2011). According to Shi et al. (2012), a sufficient N supply inhibits Fe export from source leaves, but N deficiency enhances Fe pools in source leaves and stimulates Fe export from senescing leaves to sink tissues in barley, corroborating previous findings that high protein concentrations in N-fertilized leaves tend to immobilize Fe and delay senescence (Marschner, 1995).
Nitrogen remobilization from source tissues to seeds (Hortensteiner and Feller, 2002) has received much more attention than metal remobilization over the last decades, since grain yield has been treated as the most important trait to be improved. Part of the seed N is acquired by the roots, but N remobilization from almost all vegetative organs also contributes to the seed N loading, especially during senescence processes (Burstin et al., 2007). In this way, the senescence process partly satisfies grain N concentration, as well with other minerals (Himelblau and Amasino, 2001). However, it is already known that application of fertilizer N generally decreases whole-plant remobilization efficiency (Bahrani et al., 2011).
Previous work has shown that although seed minerals are partially supplied by continuous uptake and translocation during reproductive growth, remobilization of previous stored minerals in green source tissues is also important (Jiang et al., 2007; Waters and Grusak, 2008; Sperotto et al., 2012b), as commonly seen for N. In rice, it was shown that Fe remobilization is dependent on Fe status: under low or sufficient Fe supply, flag leaf Fe remobilization is observed; under high but non-toxic Fe concentrations, there is no Fe remobilization, presumably because of continuous root uptake (Sperotto et al., 2012b). It is also known that mineral remobilization from leaves to seeds can be enhanced by senescence (Zhang et al., 1995; Uauy et al., 2006; Distelfeld et al., 2007; Shi et al., 2012). As several proteins include Fe and Zn ions, a substantial level of metals can be released during leaf senescence due to the high level of protein degradation (Waters et al., 2009).
Recent molecular studies have shown that senescence processes are driven by TF networks that regulate the expression of several senescence-related genes (Guo et al., 2004; Lin and Wu, 2004). One of the most important families of genes described as associated with senescence, and also with nutrient remobilization from source organs to developing seeds, is the NAC (NAM, ATAF, and CUC) family of TFs (Guo et al., 2004; Uauy et al., 2006).
NAC Transcription Factors
Proteins of the NAC family are one of the largest classes of plant-specific TFs. Roles of many NAC TFs have been demonstrated in diverse plant developmental processes. The earliest reports include the NAM (non-apical meristem) protein from petunia (Petunia hybrida); nam mutants lack the shoot apical meristem (SAM) and die at the seedling stage (Souer et al., 1996). The CUC1/CUC2 (cup-shaped cotyledon) TF from Arabidopsis, which participates in the development of embryos and flowers (Aida et al., 1997), defines the boundary domain around organs in the meristem (Nikovics et al., 2006). Later, NAC proteins have been related to diverse processes such as auxin and ethylene signaling (He et al., 2005; Park et al., 2011), cell wall formation (Wang et al., 2011), biotic and abiotic stresses (Puranik et al., 2012), and senescence (Kou et al., 2012).
NAC proteins contain a highly conserved N-terminal domain known as the NAC domain, which has been implicated in DNA binding (Duval et al., 2002; Ernst et al., 2004) as well as protein–protein interactions, forming homodimers or heterodimers with TFs from the NAC family (Ernst et al., 2004; Jeong et al., 2009) or other families (Xie et al., 2000; Greve et al., 2003). The NAC domain reveals a fold consisting of a twisted beta-sheet surrounded by a few helical elements (Ernst et al., 2004). On the other hand, C-terminal regions of NAC proteins are highly divergent (Olsen et al., 2005; Fang et al., 2008), and are related to transcriptional regulation (Xie et al., 2000; Duval et al., 2002).
Many NAC genes have been involved in responses to various environmental stresses like drought, cold, salinity, pathogen attack, and wounding. For recent reviews on NAC TFs in stress response, see Nakashima et al. (2012) and Puranik et al. (2012). In rice, the stress-responsive NAC group (SNAC) includes some already characterized members. OsNAC5, OsNAC6, and OsNAC10 are induced by abiotic stresses, abscisic acid (ABA), and methyl jasmonic acid, a plant hormone that activates defense responses against herbivores and pathogens (Ohnishi et al., 2005; Sperotto et al., 2009; Jeong et al., 2010; Takasaki et al., 2010; Song et al., 2011); OsNAC6 is also induced by biotic stresses (such as wounding and blast disease; Nakashima et al., 2007). Rice plants over-expressing either OsNAC5, OsNAC9, or OsNAC10 under the control of the root-specific RCc3 promoter improved tolerance to abiotic stresses during the vegetative stage of growth and, most importantly, at the reproductive stage, with a concomitant increase in grain yield (Jeong et al., 2010, 2013; Redillas et al., 2012). Another characterized SNAC from rice, OsNAC4, was proposed to lead to hypersensitive response in plants after an appropriate pathogen recognition signal is encountered (Kaneda et al., 2009). In Arabidopsis, the SNAC genes ANAC019, ANAC055, and ANAC072 are induced by pathogen attack and wounding, and transgenic plants over-expressing either one showed a significant increase in drought tolerance (Tran et al., 2004). Considering that these proteins group with the rice paralogs SNAC1, OsNAC3, OsNAC4, OsNAC5, and OsNAC6, their function seems to be conserved.
Several members of the NAC family have been functionally characterized as playing a prominent role in leaf senescence. In Arabidopsis, almost one-fifth of the predicted 109 NAC members are in the database of senescence leaf expression sequence tags (dbEST; Guo et al., 2004). Characterization of NAC TFs involved in senescence processes is also available for other plant species, like rice OsNAC5 (Sperotto et al., 2009); wheat NAM-B1 (Uauy et al., 2006); and bamboo BeNAC1 (Chen et al., 2011). The relation between NAC TFs, senescence and nutrient remobilization will be discussed in the next section.
Senescence and Metal Remobilization: are NAC Transcription Factors Bridging the Gap?
In monocarpic plants such as wheat and rice, whole-plant senescence is a coordinated process where catabolic activity provides nutrients that are exported and remobilized to developing grains (Matile et al., 1996; Hortensteiner and Feller, 2002). The Gpc-B1 quantitative trait loci (QTL) from wild emmer wheat (Triticum turgidum ssp. dicoccoides) was first described as conferring high grain protein content in wheat across diverse environments (Chee et al., 2001; Olmos et al., 2003), an important trait for improving bread and pasta quality. Although already mapped and used in breeding programs, Gpc-B1 locus was cloned later by Uauy et al. (2006), which found the causal gene to be a NAC TF, NAM-B1. NAM-B1 expression is up-regulated after anthesis in flag leaves and accelerates senescence. In modern wheat varieties, a 1-bp frame-shift insertion at the NAM-B1 coding sequence results in a truncated version of the protein, while the wild relative has an intact, fully functional NAM-B1. Silencing of NAM-B1 and other NAM paralogs mimicked the insertion effect, resulting in delayed senescence, decreased grain protein, and lower Fe and Zn concentrations due to reduced nutrient remobilization from vegetative tissues (Uauy et al., 2006; Waters et al., 2009). Moreover, a transcriptomic study showed enrichment of sequences related to transport in wild-type (WT) compared to NAM-B1 RNA interference (RNAi) lines during senescence (Cantu et al., 2011). Taken together, these results established NAM-B1 as a positive regulator of senescence and nutrient remobilization during grain maturation, suggesting that an early senescence onset could lead to increased Fe and Zn concentrations in grains (Uauy et al., 2006).
To describe genes with similar functions in other crops, an obvious avenue would be looking at orthologous proteins. The closest homolog of NAM-B1 in the rice genome, named ONAC010 (LOC_Os07g3792; Uauy et al., 2006), has a function in flower development but not in senescence, as neither ONAC010 loss-of-function nor over-expression have the expected effects on senescence timing (Distelfeld et al., 2012). Thus, another paralog with lower sequence similarity could play the NAM-B1 role in rice plants. In this context, OsNAC5 was demonstrated to be a senescence associated gene that is up-regulated during grain maturation in rice flag leaves (Sperotto et al., 2009). OsNAC5 is regulated by ABA, a hormone with a known central role in senescence processes (Lim et al., 2007). A comparison of diverse cultivars showed a positive correlation of OsNAC5 expression in flag leaves before and during anthesis with final Fe, Zn, and protein concentrations in mature grains (Sperotto et al., 2009, 2010). These results showed that OsNAC5 expression pattern resembles that of NAM-B1, and suggested that OsNAC5 could act during senescence-associated nutrient remobilization to rice grains, probably downstream on the senescence onset pathway.
Transgenic plants bearing constructs with OsNAC5 under the control of a constitutive or a root-specific promoter were generated in a recent work. When OsNAC5 was expressed only in roots, plants increased root diameter and improved recovery after drought stress (Jeong et al., 2013). Interestingly, transcriptomic analysis of roots from both transgenic lines showed commonly up-regulated genes, indicating potential targets of OsNAC5, but not necessarily linked to changes in root morphology (Jeong et al., 2013). Among them, Nicotianamine Synthase 2 (OsNAS2) and Yellow Stripe-Like 2 (OsYSL2), two genes related to metal homeostasis, were up-regulated. OsNAS2 is a key enzyme in nicotianamine (NA) synthesis, a low-molecular weight compound that chelates metals and a precursor for phytosiderophore synthesis (Higuchi et al., 1999), while OsYSL2 is an Fe–NA transporter expressed in phloem cells (Koike et al., 2004). Both NA and OsYSL2 are involved in Fe seed loading and metal long distance transport through the phloem (Koike et al., 2004; Curie et al., 2009; Klatte et al., 2009; Ishimaru et al., 2010; Schuler et al., 2012).
Different studies point that regulation of NA synthesis and metal–NA complex transporters could be involved in remobilization. Constitutive over-expression or activation-tagging of OsNAS1, OsNAS2, or OsNAS3 genes in rice increased concentrations of Fe, Zn, or both in grains (Lee et al., 2009, 2011, 2012; Johnson et al., 2011). Concomitant insertion of constructs driving Hordeum vulgare NAS1 constitutive expression, OsYSL2 expression in phloem cells and endosperm and Ferritin in endosperm, led to increased Fe concentrations (and Zn to a lower extent) in grains (Masuda et al., 2012). In H. vulgare, both dark and N deficiency-induced senescence up-regulated HvNAS2 expression in leaves, resulting in increased phytosiderophore concentration rather than NA (Shi et al., 2012). In A. thaliana, loss-of-function of OsYSL2 homologs, AtYSL1 and AtYSL3, resulted in reduced remobilization of metals to seeds during senescence (Waters et al., 2006).
Therefore, we speculate that OsNAC5 has a role in senescence and metal movement to grains by controlling, either directly or indirectly, the biosynthesis of NA and metal transport through the phloem. Our proposed model, based on data from the literature, is shown in Figure 1: (A) a senescence signal is sensed by the cell, activating signaling molecules that regulate the onset of senescence; (B) OsNAC5 transcription is up-regulated as part of the senescence-induced nutrient remobilization process; (C) OsNAC5 protein up-regulates, either directly or indirectly, OsNAS2 and OsYSL2 transcription, as well as other targets (not necessarily related to metal remobilization); (D) OsNAS2 increases NA production, which binds free Fe coming from cellular degradation; (E) after efflux from the cell, OsYSL2 acquires the Fe–NA complex into phloem cells for long distance transport. It is important to note that, while OsYSL2 was not demonstrated to transport Zn–NA complexes, NA is able to bind Zn2+, and Zn–NA complexes are the major Zn form found in the rice phloem sap (Nishiyama et al., 2012). Thus, other transporters could be playing a similar role to OsYSL2 to load Zn–NA into the phloem.
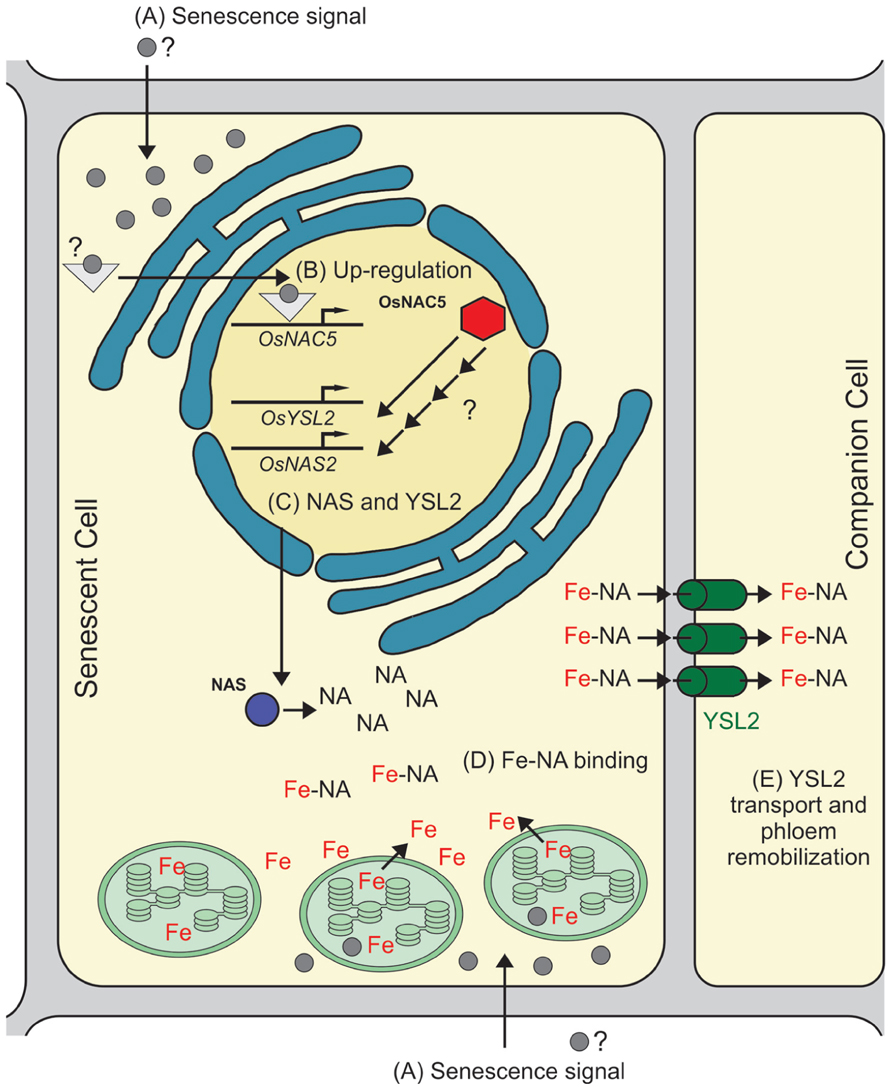
FIGURE 1. Proposed model for OsNAC5 role in senescence and metal remobilization. The model is based on indirect evidence provided by several studies, especially on Sperotto et al. (2009) and Jeong et al. (2013), and shows only Fe remobilization, although a similar pathway is likely to be involved in Zn and other metals remobilization. (A) A signal is sensed by the cell, triggering the senescence-associated cellular components degradation, including chloroplasts, the main site of Fe concentration in the cell. (B) OsNAC5 transcription is up-regulated by the senescence downstream signaling pathway. (C) OsNAC5 protein is produced and triggers the up-regulation of OsNAS2 and OsYSL2 transcription (based on microarray data presented in the work performed by Jeong et al., 2013). The increased transcription observed is either directly or indirectly regulated by OsNAC5. (D) OsNAS2 protein increases intracellular NA concentration, which in turn chelates free Fe coming from chloroplast and other cellular components degradation. (E) The Fe–NA complex is transported across the plasma membrane of the senescent cell, and then transported into phloem by OsYSL2, which allows Fe–NA complex long distance translocation. It is important to point the possibility that Fe and NA are exported independently from the cell, interacting in the apoplast and then transported into the phloem by OsYSL2.
However, work analyzing distinct OsNAC5 over-expressing lines did not show up-regulation of OsNAS2 or OsYSL2 (Takasaki et al., 2010). We should consider that the increased expression observed by Jeong et al. (2013) was a result from roots transcriptomic analyses, which is not the tissue where remobilization takes place. Moreover, up-regulation of these genes could be an indirect effect rather than a result of OsNAC5 binding to OsNAS2 or OsYSL2 promoters.
Transcriptomic analysis of NAM-B1 RNAi wheat lines did not reveal down-regulation of homologous sequences to NAS2 or YSL2 compared to WT, but rather putative ZIP (zinc-regulated/iron-regulated transporter) and NRAMP (natural resistance associated macrophage protein) metal transporters (Cantu et al., 2011), indicating that NAS and YSL homologs are not regulated by NAM-B1. This could be due to the fact that NAM-B1 and OsNAC5 are not necessarily regulating the same set of genes. NAM-B1 silencing leads to late senescence, while the functional version from wheat wild relative accelerates it. On the other hand, OsNAC5 over-expressing plants were not reported as senescing earlier than WT (Takasaki et al., 2010; Song et al., 2011; Jeong et al., 2013), indicating that OsNAC5 increased expression cannot trigger senescence alone. Silencing of OsNAC5 did not lead to late senescence as well (Song et al., 2011). OsNAC5 is known to homo- and heterodimerize with other TFs (Jeong et al., 2009), which could be necessary for OsNAC5-mediated response. Thus, it seems that OsNAC5 is acting more downstream on the senescence pathway than NAM-B1, or even in a distinct parallel regulatory network, regulating a different set of senescence-related processes.
Although we should be cautious in the analysis of the available evidence, our model seems to be holding as the first attempt to point out the mechanism of NAC proteins in mineral remobilization in crops. Further work will be necessary to clearly elucidate the role of OsNAC5 in senescence and metal remobilization, as well to functionally demonstrate which genes are controlled by this TF. If true, the regulation of metal remobilization by OsNAC5 could be an interesting avenue for biofortification strategies.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Janette Palma Fett and Marta Wilton Vasconcelos for critical reading of the manuscript.
References
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. doi: 10.1105/tpc.9.6.841
Bahrani, A., Heidari, H., Abad, S., and Aynehband, A. (2011). Nitrogen remobilization in wheat as influenced by nitrogen application and post-anthesis water deficit during grain filling. Afr. J. Biotechnol. 10, 10585–10594. doi: 10.5897/AJB11.013
Burstin, J., Marget, P., Huart, M., Moessner, A., Mangin, B., Duchene, C., et al. (2007). Developmental genes have pleiotropic effects on plant morphology and source capacity, eventually impacting on seed protein content and productivity in pea. Plant Physiol. 144, 768–781. doi: 10.1104/pp.107.096966
Cantu, D., Pearce, S. P., Distelfeld, A., Christiansen, M. W., Uauy, C., Akhunov, E., et al. (2011). Effect of the down-regulation of the high Grain Protein Content (GPC) genes on the wheat transcriptome during monocarpic senescence. BMC Genomics 12:492. doi: 10.1186/1471-2164-12-492
Carvalho, S. M. P., and Vasconcelos, M. W. (2013). Producing more with less: strategies and novel technologies for plant-based food biofortification. Food Res. Int. (in press). doi: 10.1016/j.foodres.2012.12.021
Chee, P. W., Elias, E. M., Anderson, J. A., and Kianian, S. F. (2001). Evaluation of a high grain protein QTL from Triticum turgidum L. var. dicoccoides in an adapted durum wheat background. Crop Sci. 41, 295–301. doi: 10.2135/cropsci2001.412295x
Chen, Y., Qiu, K., Kuai, B., and Ding, Y. (2011). Identification of an NAP-like transcription factor BeNAC1 regulating leaf senescence in bamboo (Bambusa emeiensis ‘Viridiflavus’). Physiol. Plant. 42, 361–371. doi: 10.1111/j.1399-3054.2011.01472.x
Curie, C., Cassin, G., Couch, D., Divol, F., Higuchi, K., Le Jean, M., et al. (2009). Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann. Bot. 103, 1–11. doi: 10.1093/aob/mcn207
Davies, P. J., and Gan, S. (2012). Towards an integrated view of monocarpic plant senescence. Russian J. Plant Physiol. 59, 467–478. doi: 10.1134/S102144371204005X
Distelfeld, A., Cakmak, I., Peleg, Z., Ozturk, L., Yazici, A. M., Budak, H., et al. (2007). Multiple QTL-effects of wheat Gpc-B1 locus on grain protein and micronutrient concentrations. Physiol. Plant. 129, 635–643. doi: 10.1111/j.1399-3054.2006.00841.x
Distelfeld, A., Pearce, S. P., Avni, R., Scherer, B., Uauy, C., Piston, F., et al. (2012). Divergent functions of orthologous NAC transcription factors in wheat and rice. Plant Mol. Biol. 78, 515–524. doi: 10.1007/s11103-012-9881-6
Duval, M., Hsieh, T. F., Kim, S. Y., and Thomas, T. L. (2002). Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 50, 237–248. doi: 10.1023/A:1016028530943
Ernst, H. A., Olsen, A. N., Larsen, S., and Lo Leggio, L. (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5, 297–303. doi: 10.1038/sj.embor.7400093
Fang, Y., You, J., Xie, K., Xie, W., and Xiong, L. (2008). Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics 280, 547–563. doi: 10.1007/s00438-008-0386-6
Gómez-Galera, S., Rojas, E., Sudhakar, D., Zhu, C., Pelacho, A. M., Capell, T., et al. (2010). Critical evaluation of strategies for mineral fortification of staple food crops. Transgenic Res. 19, 165–180. doi: 10.1007/s11248-009-9311-y
Greve, K., La Cour, T., Jensen, M. K., Poulsen, F. M., and Skriver, K. (2003). Interactions between plant RING-H2 and plant-specific NAC (NAM/ATAF1/2/CUC2) proteins: RING-H2 molecular specificity and cellular localization. Biochem. J. 371, 97–108. doi: 10.1042/BJ20021123
Guo, Y., Cai, Z., and Gan, S. (2004). Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27, 521–549. doi: 10.1111/j.1365-3040.2003.01158.x
He, X. J., Mu, R. L., Cao, W. H., Zhang, Z. G., Zhang, J. S., and Chen, S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916. doi: 10.1111/j.1365-313X.2005.02575.x
Higuchi, K., Suzuki, K., Nakanishi, H., Yamaguchi, H., Nishizawa, N. K., and Mori, S. (1999). Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol. 119, 471–480. doi: 10.1104/pp.119.2.471
Himelblau, E., and Amasino, R. M. (2001). Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J. Plant Physiol. 158, 1317–1323. doi: 10.1078/0176-1617-00608
Hortensteiner, S., and Feller, U. (2002). Nitrogen metabolism and remobilization during senescence. J. Exp. Bot. 53, 927–937. doi: 10.1093/jexbot/53.370.927
Ishimaru, Y., Masuda, H., Bashir, K., Inoue, H., Tsukamoto, T., Takahashi, M., et al. (2010). Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 62, 379–390. doi: 10.1111/j.1365-313X.2010.04158.x
Jeong, J. S., Kim, Y. S., Baek, K. H., Jung, H., Ha, S. H., Choi, Y. D., et al. (2010). Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–197. doi: 10.1104/pp.110.154773
Jeong, J. S., Kim, Y. S., Redillas, M. C., Jang, G., Jung, H., Bang, S. W., et al. (2013). OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11, 101–114. doi: 10.1111/pbi.12011
Jeong, J. S., Park, Y. T., Jung, H., Park, S. H., and Kim, J. K. (2009). Rice NAC proteins act as homodimers and heterodimers. Plant Biotechnol. Rep. 3, 127–134. doi: 10.1007/s11816-009-0081-z
Jiang, W., Struik, P. C., Lingna, J., van Keulen, H., Ming, Z., and Stomph, T. J. (2007). Uptake and distribution of root-applied or foliar-applied 65Zn after flowering in aerobic rice. Ann. Appl. Biol. 150, 383–391. doi: 10.1111/j.1744-7348.2007.00138.x
Johnson, A. A., Kyriacou, B., Callahan, D. L., Carruthers, L., Stangoulis, J., Lombi, E., et al. (2011). Constitutive overexpression of the OsNAS gene family reveals single-gene strategies for effective iron- and zinc-biofortification of rice endosperm. PLoS ONE 6:e24476. doi: 10.1371/journal.pone.0024476
Kaneda, T., Taga, Y., Takai, R., Iwano, M., Matsui, H., Takayama, S., et al. (2009). The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J. 28, 926–936. doi: 10.1038/emboj.2009.39
Kennedy, G., and Burlingame, B. (2003). Analysis of food composition data on rice from a plant genetic resources perspective. Food Chem. 80, 589–596. doi: 10.1016/S0308-8146(02)00507-1
Klatte, M., Schuler, M., Wirtz, M., Fink-Straube, C., Hell, R., and Bauer, P. (2009). The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol. 150, 257–271. doi: 10.1104/pp.109.136374
Kobayashi, T., Ogo, Y., Itai, R. N., Nakanishi, H., Takahashi, M., Mori, S., et al. (2007). The transcription factor IDEF1 regulates the response to and tolerance of iron deficiency in plants. Proc. Natl. Acad. Sci. U.S.A. 104, 19150–19155. doi: 10.1073/pnas.0707010104
Koike, S., Inoue, H., Mizuno, D., Takahashi, M., Nakanishi, H., Mori, S., et al. (2004). OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 39, 415–424. doi: 10.1111/j.1365-313X.2004.02146.x
Kou, X., Watkins, C. B., and Gan, S. S. (2012). Arabidopsis AtNAP regulates fruit senescence. J. Exp. Bot. 63, 6139–6147. doi: 10.1093/jxb/ers266
Kutman, U. B., Yildiz, B., and Cakmak, I. (2011). Effect of nitrogen on uptake, remobilization and partitioning of zinc and iron throughout the development of durum wheat. Plant Soil 342, 149–164. doi: 10.1007/s11104-010-0679-5
Lee, S., Jeon, U. S., Lee, S. J., Kim, Y. K., Persson, D. P., Husted, S., et al. (2009). Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc. Natl. Acad. Sci. U.S.A. 106, 22014–22019. doi: 10.1073/pnas.0910950106
Lee, S., Kim, Y. S., Jeon, U. S., Kim, Y. K., Schjoerring, J. K., and An, G. (2012). Activation of rice nicotianamine synthase 2 (OsNAS2) enhances iron availability for biofortification. Mol. Cells 33, 269–275. doi: 10.1007/s10059-012-2231-3
Lee, S., Persson, D. P., Hansen, T. H., Husted, S., Schjoerring, J. K., Kim, Y. S., et al. (2011). Bio-available zinc in rice seeds is increased by activation tagging of nicotianamine synthase. Plant Biotechnol. J. 9, 865–873. doi: 10.1111/j.1467-7652.2011.00606.x
Lim, P. O., Kim, H. J., and Nam, H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Lin, J. F., and Wu, S. H. (2004). Molecular events in senescing Arabidopsis leaves. Plant J. 39, 612–628. doi: 10.1111/j.1365-313X.2004.02160.x
Masuda, H., Ishimaru, Y., Aung, M. S., Kobayashi, T., Kakei, Y., Takahashi, M., et al. (2012). Iron biofortification in rice by the introduction of multiple genes involved in iron nutrition. Sci. Rep. 2, 543. doi: 10.1038/srep00543
Matile, P., Hortensteiner, S., Thomas, H., and Krautler, B. (1996). Chlorophyll breakdown in senescent leaves. Plant Physiol. 112, 1403–1409.
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nakashima, K., Tran, L. S., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., et al. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18, 2929–2945. doi: 10.1105/tpc.106.045617
Nishiyama, R., Kato, M., Nagata, S., Yanagisawa, S., and Yoneyama, T. (2012). Identification of Zn-nicotianamine and Fe-2′-Deoxymugineic acid in the phloem sap from rice plants (Oryza sativa L.). Plant Cell Physiol. 53, 381–390. doi: 10.1093/pcp/pcr188
Ogo, Y., Itai, R. N., Kobayashi, T., Aung, M. S., Nakanishi, H., and Nishizawa, N. K. (2011). OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol. Biol. 75, 593–605. doi: 10.1007/s11103-011-9752-6
Ohnishi, T., Sugahara, S., Yamada, T., Kikuchi, K., Yoshiba, Y., Hirano, H. Y., et al. (2005). OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes Genet. Syst. 80, 135–139. doi: 10.1266/ggs.80.135
Olmos, S., Distelfeld, A., Chicaiza, O., Schlatter, A. R., Fahima, T., Echenique, V., et al. (2003). Precise mapping of a locus affecting grain protein content in durum wheat. Theor. Appl. Genet. 107, 1243–1251. doi: 10.1007/s00122-003-1377-y
Olsen, A. N., Ernst, H. A., Leggio, L. L., and Skriver, K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. doi: 10.1016/j.tplants.2004.12.010
Palmgren, M. G., Clemens, S., Williams, L. E., Kramer, U., Borg, S., Schjorring, J. K., et al. (2008). Zinc biofortification of cereals: problems and solutions. Trends Plant Sci. 13, 464–473. doi: 10.1016/j.tplants.2008.06.005
Park, J., Kim, Y. S., Kim, S. G., Jung, J. H., Woo, J. C., and Park, C. M. (2011). Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol. 156, 537–549. doi: 10.1104/pp.111.177071
Prasad, A. S. (2012). Discovery of human zinc deficiency: 50 years later. J. Trace Elem. Med. Biol. 26, 66-69. doi: 10.1016/j.jtemb.2012.04.004
Puig, S., Andrés-Colás, N., García-Molina, A., and Peñarrubia, L. (2007). Copper and iron homeostasis in Arabidopsis: responses to metal deficiencies, interactions and biotechnological applications. Plant Cell Environ. 30, 271–290. doi: 10.1111/j.1365-3040.2007.01642.x
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Redillas, M. C. F. R., Jeong, J. S., Kim, Y. S., Jung, H., Bang, S. W., Choi, Y. D., et al. (2012). The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 10, 792–805. doi: 10.1111/j.1467-7652.2012.00697.x
Rolland, F., Baena-Gonzalez, E., and Sheen, J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709. doi: 10.1146/annurev.arplant.57.032905.105441
Schuler, M., Rellán-Álvarez, R., Fink-Straube, C., Abadía, J., and Bauer, P. (2012). Nicotianamine functions in the phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell 24, 2380–2400. doi: 10.1105/tpc.112.099077
Shi, R., Weber, G., Köster, J., Reza-Hajirezaei, M., Zou, C., Zhang, F., et al. (2012). Senescence-induced iron mobilization in source leaves of barley (Hordeum vulgare) plants. New Phytol. 195, 372–383. doi: 10.1111/j.1469-8137.2012.04165.x.
Song, S. Y., Chen, Y., Chen, J., Dai, X. Y., and Zhang, W. H. (2011). Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234, 331–345. doi: 10.1007/s00425-011-1403-2
Souer, E., van Houwelingen, A., Kloos, D., Mol, J., and Koes, R. (1996). The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. doi: 10.1016/S0092-8674(00)81093-4
Sperotto, R. A., Boff, T., Duarte, G. L., Santos, L. S., Grusak, M. A., and Fett, J. P. (2010). Identification of putative target genes to manipulate Fe and Zn concentrations in rice grains. J. Plant Physiol. 167, 1500–1506. doi: 10.1016/j.jplph.2010.05.003
Sperotto, R. A., Ricachenevsky, F. K., Duarte, G. L., Boff, T., Lopes, K. L., Sperb, E. R., et al. (2009). Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 230, 985–1002. doi: 10.1007/s00425-009-1000-9
Sperotto, R. A., Ricachenevsky, F. K., and Fett, J. P. (2007). Iron deficiency in rice shoots: identification of novel induced genes using RDA and possible relation to leaf senescence. Plant Cell Rep. 26, 1399–1411. doi: 10.1007/s00299-007-0330-y
Sperotto, R. A., Ricachenevsky, F. K., Waldow, V. A., and Fett, J. P. (2012a). Iron biofortification in rice: it’s a long way to the top. Plant Sci. 190, 24–39. doi: 10.1016/j.plantsci.2012.03.004
Sperotto, R. A., Vasconcelos, M. W., Grusak, M. A., and Fett, J. P. (2012b). Effects of different Fe supplies on mineral partitioning and remobilization during the reproductive development of rice (Oryza sativa L.). Rice 5:27. doi: 10.1186/1939-8433-5-27
Takasaki, H., Maruyama, K., Kidokoro, S., Ito, Y., Fujita, Y., Shinozaki, K., et al. (2010). The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genomics 284, 173–183. doi: 10.1007/s00438-010-0557-0
Thomas, H. (2013). Senescence, ageing and death of the whole plant. New Phytol. 197, 696–711. doi: 10.1111/nph.12047
Tran, L. S., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Uauy, C., Distelfeld, A., Fahima, T., Blechl, A., and Dubcovsky, J. (2006). A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314, 1298–1301. doi: 10.1126/science.1133649
Wang, H., Zhao, Q., Chen, F., Wang, M., and Dixon, R. A. (2011). NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J. 68, 1104–1114. doi: 10.1111/j.1365-313X.2011.04764.x
Waters, B. M., Chu, H. H., Didonato, R. J., Roberts, L. A., Eisley, R. B., Lahner, B., et al. (2006). Mutations in Arabidopsis yellow stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol. 141, 1446–1458. doi: 10.1104/pp.106.082586
Waters, B. M., and Grusak, M. A. (2008). Whole-plant mineral partitioning throughout the life cycle in Arabidopsis thaliana ecotypes Columbia, Landsberg erecta, Cape Verde Islands, and the mutant line ysl1ysl3. New Phytol. 177, 389–405.
Waters, B. M., Uauy, C., Dubcovsky, J., and Grusak, M. A. (2009). Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. J. Exp. Bot. 60, 4263–4274. doi: 10.1093/jxb/erp257
White, P. J., and Broadley, M. R. (2005). Biofortifying crops with essential mineral elements. Trends Plant Sci. 10, 586–593. doi: 10.1016/j.tplants.2005.10.001
Wingler, A., von Schaewen, A., Leegood, R. C., Lea, P. J., and Quick, W. P. (1998). Regulation of leaf senescence by cytokinin, sugars, and light. Effects on NADH-dependent hydroxypyruvate reductase. Plant Physiol. 116, 329–335. doi: 10.1104/pp.116.1.329
Wirth, J., Poletti, S., Aeschlimann, B., Yakandawala, N., Drosse, B., Osorio, S., et al. (2009). Rice endosperm iron biofortification by targeted and synergistic action of nicotianamine synthase and ferritin. Plant Biotechnol. J. 7, 631–644. doi: 10.1111/j.1467-7652.2009.00430.x
Xie, Q., Frugis, G., Colgan, D., and Chua, N. H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. doi: 10.1101/gad.852200
Yoshida, S. (2003). Molecular regulation of leaf senescence. Curr. Opin. Plant Biol. 6, 79–84. doi: 10.1016/S1369526602000092
Zavaleta-Mancera, H. A., Franklin, K. A., Ougham, H. J., Thomas, H., and Scott, I. M. (1999). Regreening of senescent Nicotiana leaves. I. Reappearance of NADPH-protochlorophyllide oxidoreductase and light-harvesting chlorophyll a/b-binding protein. J. Exp. Bot. 50, 1677–1682. doi: 10.1093/jxb/50.340.1677
Keywords: biofortification, iron, NAC transcription factor, nutrient remobilization, senescence, zinc
Citation: Ricachenevsky FK, Menguer PK and Sperotto RA (2013) kNACking on heaven’s door: how important are NAC transcription factors for leaf senescence and Fe/Zn remobilization to seeds? Front. Plant Sci. 4:226. doi: 10.3389/fpls.2013.00226
Received: 12 March 2013; Accepted: 10 June 2013;
Published online: 01 July 2013.
Edited by:
Michael A. Grusak, United States Department of Agriculture-Agricultural Research Service, Children’s Nutrition Research Center, USAReviewed by:
Trevor Garnett, Australian Centre for Plant Functional Genomics, AustraliaChristina B. Garcia, Baylor College of Medicine, USA
Copyright: © 2013 Ricachenevsky, Menguer and Sperotto. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Raul Antonio Sperotto, Centro de Ciências Biológicas e da Saúde, Programa de Pós-Graduação em Biotecnologia, Centro Universitário UNIVATES, Rua Avelino Tallini 171, Lajeado, Rio Grande do Sul 95.900-000, Brazil e-mail:cmFzcGVyb3R0b0B1bml2YXRlcy5icg==


