- 1Plant Physiology, Department of Horticulture, College of Agriculture, Food and the Environment, University of Kentucky, Lexington, KY, USA
- 2Department of Biosystems and Agricultural Engineering, University of Kentucky, Lexington, KY, USA
- 3Enterprise Institute for Renewable Fuels, Donald Danforth Plant Science Center, St. Louis, MO, USA
Second generation feedstocks for bioethanol will likely include a sizable proportion of perennial C4 grasses, principally in the Panicoideae clade. The Panicoideae contain agronomically important annual grasses including Zea mays L. (maize), Sorghum bicolor (L.) Moench (sorghum), and Saccharum officinarum L. (sugar cane) as well as promising second generation perennial feedstocks including Miscanthus × giganteus and Panicum virgatum L. (switchgrass). The underlying complexity of these polyploid grass genomes is a major limitation for their direct manipulation and thus driving a need for rapidly cycling comparative model. Setaria viridis (green millet) is a rapid cycling C4 panicoid grass with a relatively small and sequenced diploid genome and abundant seed production. Stable, transient, and protoplast transformation technologies have also been developed for Setaria viridis making it a potentially excellent model for other C4 bioenergy grasses. Here, the lignocellulosic feedstock composition, cellulose biosynthesis inhibitor response and saccharification dynamics of Setaria viridis are compared with the annual sorghum and maize and the perennial switchgrass bioenergy crops as a baseline study into the applicability for translational research. A genome-wide systematic investigation of the cellulose synthase-A genes was performed identifying eight candidate sequences. Two developmental stages; (a) metabolically active young tissue and (b) metabolically plateaued (mature) material are examined to compare biomass performance metrics.
Introduction
As the depth of knowledge ever increasingly grows in the model plant Arabidopsis and the utility of this information proves translatable, there remains a need for additional model plants, to decode and translate traits that are absent in Arabidopsis (Rensink and Buell, 2004). Monocotyledonous taxa (grasses) display both marked phenotypic and metabolic variability. Moreover, they produce the majority of the world food crops and a growing need for biomass for biofuel production is being driven toward perennial grasses. Thus, a notable need exists for a range of model species to ask a great many questions underscoring agronomically important traits. Existing examples of model grasses include Brachypodium distachyon and rice (Oryza sativa), which have served as tractable model plants for gene discovery and translational biology (Ouyang et al., 2007; Vogel, 2008). However, there are also important physiological differences among the grasses. A principle difference exists between mechanisms of photosynthesis. Rice and Brachypodium utilize C3 (carbon fixation) photosynthesis whereas the existing and most promising bioenergy feedstocks including maize (Zea mays), sorghum (Sorghum bicolor), switchgrass (Panicum virgatum L.), or Miscanthus (Miscanthus × giganteus) employ C4 (carbon fixation) photosynthesis (Schmitt and Edwards, 1981; Oula, 2008; Wang et al., 2009). While Setaria has been proposed as models for C4 photosynthesis (Doust et al., 2009; Brutnell et al., 2010; Li and Brutnell, 2011), based on it’s close phylogenetic relationship and genetic synteny, it also exists as an excellent model for the lignocellulosic biomass crops. For perspective, maize has a large genome comparable in size to the human genome, while Setaria is one-fifth the size and contains all the genes necessary for C4 photosynthesis and primary metabolic processes. Primary cell walls in dicots and monocots are inherently different. For instance in dicots, cellulose fibers are encased in a matrix of xyloglucan, pectin, and structural proteins. On the contrary, a typical monocot primary cell wall still has cellulose microfibrils as the main structural biopolymer, but has a hemicellulose structure comprised of glucuronoarabinoxylans along with hydroxycinnamates and a very low contribution of pectin (see Vogel, 2008). Differences between the C3 and C4 plants, in our case focused solely monocotyledonous (monocots) examples (some dicotyledonous C4 plants exist), involve structural (physiologically) and metabolic advances in leaf anatomy and atmospheric carbon during photosynthesis. Despite the large evolutionary adaptation in plant physiology between C3 and C4 metabolism, only minor differences are noted in the cell walls, such as suberized bundle sheath cell walls to prevent CO2 leakage (Langdale, 2011). Little evidence exists that C3 versus C4 metabolism is accompanied by any marked shifts in the composition of cell wall polysaccharides comprising lignocellulosic biomass. However, the primary metabolic response(s) to environmental constraints, such as drought (Doust et al., 2009), are notable different in plants with C3 versus C4 metabolism and therefore the feedback regulation of carbon through primary metabolism is feasibly influenced. Such shifts could in turn influence cell wall processes. Indeed, in a study by Hibberd and Quick (2002), the spatially discreet evolution of C4 metabolism around xylem vessels in a C3 plant was associated with both a pH shift and provision of carbon skeletons into lignin synthesis for rapid incorporation in lignified xylem vessels. Regardless, the development of physiological, compositional, and genetic resources in Setaria as a model organism for lignocellulosic feedstocks should complement resources in Arabidopsis and Brachypodium. Briefly, Setaria, is the weedy counterpart to the grain crop Setaria italica (millet), more commonly known as the green foxtail. Many crop grasses have vast and repetitive genome structure (Vicient et al., 2001; Bennetzen, 2007). By contrast, Setaria viridis has a relatively manageable genome and yet displays synteny to the Panicoideae grasses, which include the major biofuel grasses (Li and Brutnell, 2011). Setaria (as referred to from this point onward) displays a life cycle of around 8 weeks, produces abundant seed each generation, is self-compatible and is a diploid providing for genetic tractability (Doust et al., 2009).
As an initial step in the development of Setaria as a model for bioenergy grasses, the cell wall carbohydrate composition of Setaria was compared with other crop species in the Panicoideae at two developmental stages; (a) metabolically active young tissue and (b) metabolically plateaued (mature) material (see Figure 1 for example). Furthermore, employing current genomic data we phylogenetically examine the cellulose synthase-A (CESA) gene family. Metabolically active tissue was examined because such a sampling time is representative of an early season perennial forage production system [Panicoideae clade species such as switchgrass, eastern gamagrass (Tripsacum dactyloides) and Miscanthus × giganteus] and due to being a temporally favorable timeframe to examine phenotypes in a research setting. Secondly, an important comparison is that of mature tissue where genetic manipulations for persistent biomass traits can be compared and potentially translated.

FIGURE 1. Representative sample points exemplified during the developmental program of Setaria and sorghum. Plants were sampled at two stages: immature tissue denoted as stage 1 (S1) and mature tissue denoted as stage 2 (S2), exemplified here for Setaria (A) and sorghum (B).
Results and Discussion
Setaria as a Model for Cell Wall Synthesis in the Panicoideae: The Identification of two Developmental Stages that Present Similar Cell Wall Composition
Cellulose, Neutral Sugars, and Lignin
It is recognized that the relative abundance of cellulose, lignin, and hemicellulose varies depending on tissue type, age, and environmental/biological condition of the plant tissue (Meinert and Delmer, 1977; Rogers and Campbell, 2004). Cellulose, a natural highly abundant macromolecule made up of glucose 1, 4-β-linked is a principle substrate for bioethanol conversion (Hidayat et al., 2012). Further, hemicellulose is also a convertible carbohydrate substrate to alcohol fuels (Saha, 2003). By contrast, the presence of lignin inhibits enzymatic processes (Li et al., 2010; Kim et al., 2011) and it’s down-regulation through genetic approaches has been shown to improve the saccharification efficiency (Chen and Dixon, 2007; Saathoff et al., 2011; Wang et al., 2012). Therefore, we surveyed cellulose, lignin, and neutral sugar composition in Setaria and compared these with other annual and perennial members of the Panicoideae. Cellulose and lignin content of acid-insoluble residue (AIR biomass) was quantified. As defined in the methods and materials, we sought two developmental stages for comparative studies. Analysis of cellulose content in immature biomass revealed no significant change (P > 0.05) in relative content among the three annual panicoid species (Setaria, maize, and sorghum). By contrast, lower cellulose content was measured in switchgrass samples (Figures 2A,B; P > 0.05). There are several plausible explanations for these data. Firstly, switchgrass is a perennial warm season grass whereas the other Panicoideae clade species are annual grasses. The modest variation in cellulose accumulation may reflect the ecological and genetic distances between annual and perennial crops. Alternatively, the capacity to synchronize temporal staging (sampling time) may have been congruent for the annual species, but the perennial switchgrass may display minor developmental variation. In prior analysis of numerous upland and lowland switchgrass cultivars, modest variability in cellulose content was observed (Stork et al., 2009), and therefore species level variation could also exist. Comparison of cellulose content in the mature (S2) compared with immature (S1) biomass revealed an expected and consistent proportional decrease during development (Figures 2A,B; P > 0.05). This proportional decrease in cellulose content may be a target for future transcriptional re-wiring efforts that aim to increase cellulose without significant cost to the fitness of the plant. It was also found that the degree to which the cellulose content was reduced in mature tissue relative to the metabolically active tissue was greatest in the two highly domesticated species, maize and sorghum.

FIGURE 2. Cellulose and lignin determination of acid-insoluble residue (AIR). Cellulose content was determined in aboveground (A) and belowground (B) material for Setaria, maize, sorghum, and switchgrass at both stage S1 and S2. The acid-insoluble lignin portion of aboveground (C) and belowground (D) and acid-soluble lignin (E,F) was quantified. n = 3 biological replicates. Error bars represent the standard error from the mean. One-way ANOVA with a post hoc Tukey test was performed across the four species per stage. Convention adopted is small letters for stage 1 (S1) and capital letters for stage 2 (S2). Same letters indicate non-significance (P > 0.05) different letters indicate significant differences (P < 0.05).
The majority of lignin was measured as insoluble lignin. Lignin content increased significantly between the first and second sampling in both aerial (Figure 2C) and root tissue (Figure 2D). Increased lignification during development was consistent with diversification of the primary cell wall into lignified secondary cell walls in maturing stems, leaves, and roots (Redfearn and Nelson, 2003; Parrish and Fike, 2005; Stork et al., 2009). Soluble lignin although representing a minor proportion of cell wall biomass was measured and found to be less predictable (Figures 2E,F). For example, in aerial tissue, soluble content did not significantly change in Setaria or maize (P > 0.05) and significantly decreased in sorghum and switchgrass between S1 and S2 (P < 0.05). Collectively, data highlight a contiguous developmental program during the deposition patterns for cellulose and lignin. These data broadly support the utility of Setaria to provide model organism characteristics to above-mentioned crop species, taking note of potential differences when comparing annual and perennial grasses.
Biomass was examined using X-ray diffraction (XRD) to infer a relative crystallinity index (RCI; Segal et al., 1959). The RCI is an empirical measure of the relative proportion of the crystalline versus non-crystalline (amorphous) biomass components, and is not a measure of the crystallinity of cellulose (Andersson et al., 2003). All samples fell in a comparable range of 59–69%, consistent with prior studies (Harris and DeBolt, 2008). Interestingly, there was a general trend of increased RCI from S1 to S2 (Table 1). Lignification during secondary cell wall synthesis would increase the amorphous signal at 2-theta (Θ) 18° and subsequently influence the RCI determination. Of further note, a large peak consistent with calcium oxalate was evident in all samples at 2-theta (Θ) 26.6° and was more pronounced in S2 (data not shown).
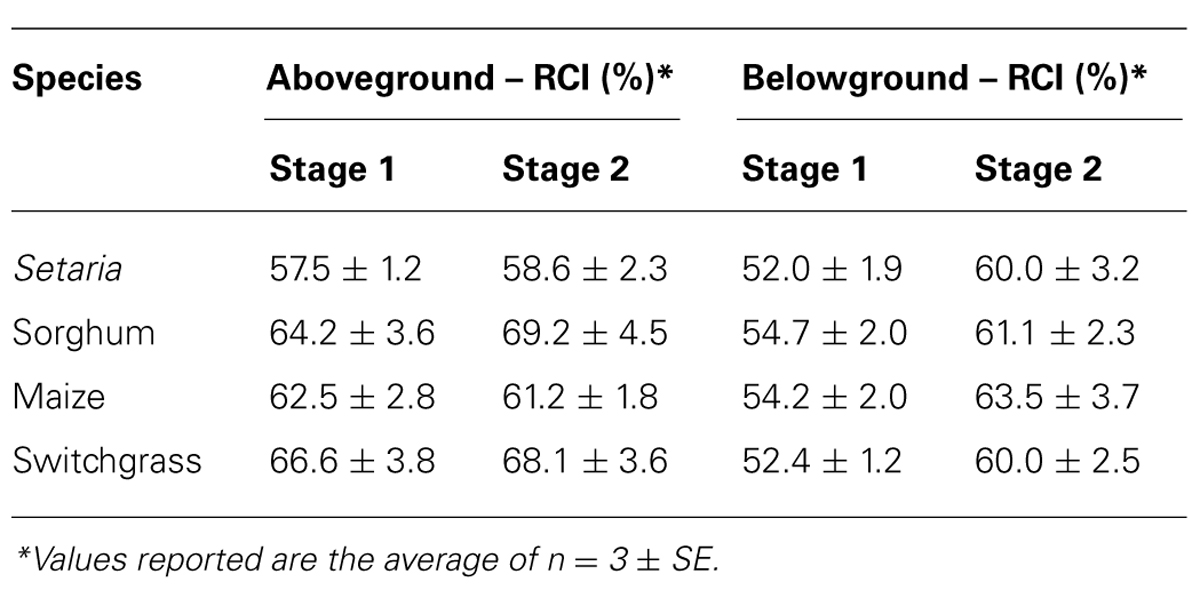
TABLE 1. Relative crystallinity index (RCI) determined for specified spatial and temporal regions of each species.
To further understand how the cell wall was changing over time, we measured neutral sugars in S1 (Figures 3A,B ) and S2 (Figures 3C,D). Neutral sugars collectively comprise the building blocks for hemicellulose biosynthesis in the plant cell walls (Bauer et al., 1973) such as xyloglucans, xylans, mannans and glucomannans, and β-(1 → 3,1 → 4)-glucans (Scheller and Ulvskov, 2010). These polysaccharides make up a third class of cell wall biomass component, in addition to cellulose and lignin and comprise arabinose, mannose, rhamnose, galactose, fucose, xylose, and soluble glucose. In S1 aboveground biomass, rhamnose was more abundant in the switchgrass biomass relative to annual species (Figure 3A). By contrast, arabinose, galactose, fucose, glucose, and xylose did not display notable shifts in relative abundance (% AIR or mg g-1). For stage S2, some variability in xylose, glucose, and rhamnose content was observed among genotypes, but this was also most pronounced in switchgrass (Figure 3). Mannose and galactose were not abundantly represented in our analysis. These can be explained by (glucurono)arabinoxylans being the predominant component of hemicelluloses in the grasses, while xyloglucans dominate in the cell walls of dicotyledons (including gymnosperms; Scheller and Ulvskov, 2010). The proportional shifts between species herein were minor and no pronounced shifts in total neutral sugars between S1 and S2 were observed. These data support that the increased RCI in S2 (Table 1) may be consistent with increased order or abundance of crystalline cellulose within the cell wall rather than a proportional shift in hemicellulose or lignin. The use of hemicellulosic neutral sugars as a feedstock for enzymatic conversion to biofuel requires different degradation enzymes than cellulose (Saha, 2003). The xylose and arabinose sugar backbones represented the most abundant non-glucose sugars available for bioconversion (Figure 3). Alternatively, recent innovations show that integrated thermochemical deconstruction of cellulosic and hemicellulosic fractions of lignocellulosic biomass can be simultaneously converted to levulinic acid and furfural, respectively (Alonso et al., 2013), and would not require separate bioprocesses. Ultimately, thermochemical deconstruction of lignocellulosic biomass to biofuel may negate modest compositional shifts, but technical hurdles remain in this maturing technology.
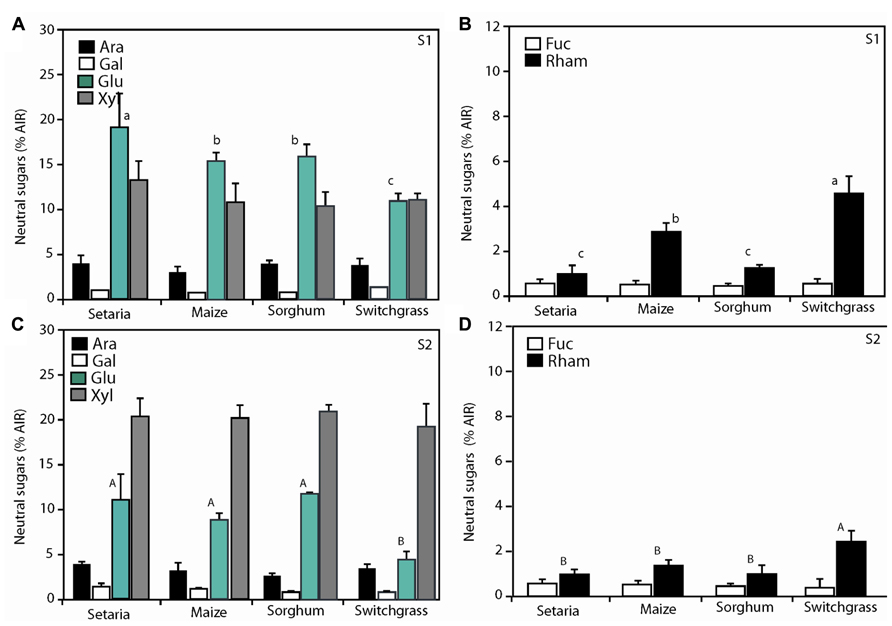
FIGURE 3. Neutral sugar monosaccharide analysis for aboveground material. The relative abundance of each sugar was compared to a true standard and determined Setaria, maize, sorghum, and switchgrass at S1 (A,B) and S2 (C,D). n = 3 biological replicates. Error bars represent the standard error from the mean. One-way ANOVA with a post hoc Tukey test was performed across the four species per stage. Convention adopted is small letters for stage 1 (S1) and capital letters for stage 2 (S2). Same letters indicate non-significance (P > 0.05) different letters indicate significant differences (P < 0.05).
Cellulose Biosynthesis Inhibitor Response Studies
A chemical inhibitor can be a useful research tool, particularly when combined with genomic tools and model system resources (Robert et al., 2009; Töth and van der Hoorn, 2010). Cellulose biosynthesis inhibitors (CBI) have been effectively used with the dicotyledonous model system Arabidopsis thaliana to identify mutations in CESA (Scheible et al., 2001; Desprez et al., 2002), some of which have led to beneficial deconstruction characteristics of cellulose (Harris et al., 2012). CBIs have also been used to learn important information about the delivery mechanisms of cellulose biosynthetic machinery (Crowell et al., 2009; Gutierrez et al., 2009), which will ultimately be important to regulate cellulose synthesis in plants.
In plants, directional cell growth is facilitated by a rigid, yet extensible cell wall, which acts to collectively constrain internal turgor pressure. Cellulose forms the central load-bearing component of this wall and therefore inhibiting cellulose biosynthesis causes radially swollen cells in seedlings providing a robust visual phenotype drug activity. A panel of CBIs was tested for potency herein, due to their possible use in future studies on biomass traits. These CBIs fell into three different classes of inhibition mechanism in Arabidopsis (summarized in Brabham and DeBolt, 2013), clearance of CESA (isoxaben (N-[3-(1-ethyl-1-methylpropyl)-1,2-oxazol-5-yl]-2,6-dimethoxybenzamide)), stopping CESA (2,6-dichlorobenzonitrile – DCB) and co-targeting cortical microtubules and CESA [morlin (7-ethoxy-4-methyl, coumarin); Figure 4]. The LD50 dose rates for isoxaben, morlin, and DCB in Arabidopsis are 4 nM, 500 nM, and 5 μM (Heim et al., 1989; DeBolt et al., 2007a,b). Results showed that isoxaben failed to induce radial LD50 root length suppression until 50–100 nM (Figure 4), which represented a 15- to 20-fold reduction in activity. Morlin also displayed a 10-fold reduction in activity, with an LD50 of approximately 50 μM. Based on these data, DCB alone remained a highly potent CBI inhibitor for the panicoid with dose rate similar to those observed in Arabidopsis. The lower degree of CBI potency observed for isoxaben and morlin, but not for DCB, requires future exploration. Indeed, 14C-isoxaben studies from Heim et al. (1993) suggest that isoxaben uptake is not blocked at the cuticle in monocotyledons. Further, the action of DCB does not support a non-specific biochemical drug tolerance such as more abundant cytochrome P450 enzymes (Morant et al., 2003). Hence, future chemical genetic studies employ mutagenesis populations to map based clone resistance loci to CBIs are needed. Such studies will be aided by a genetic model such as Setaria.
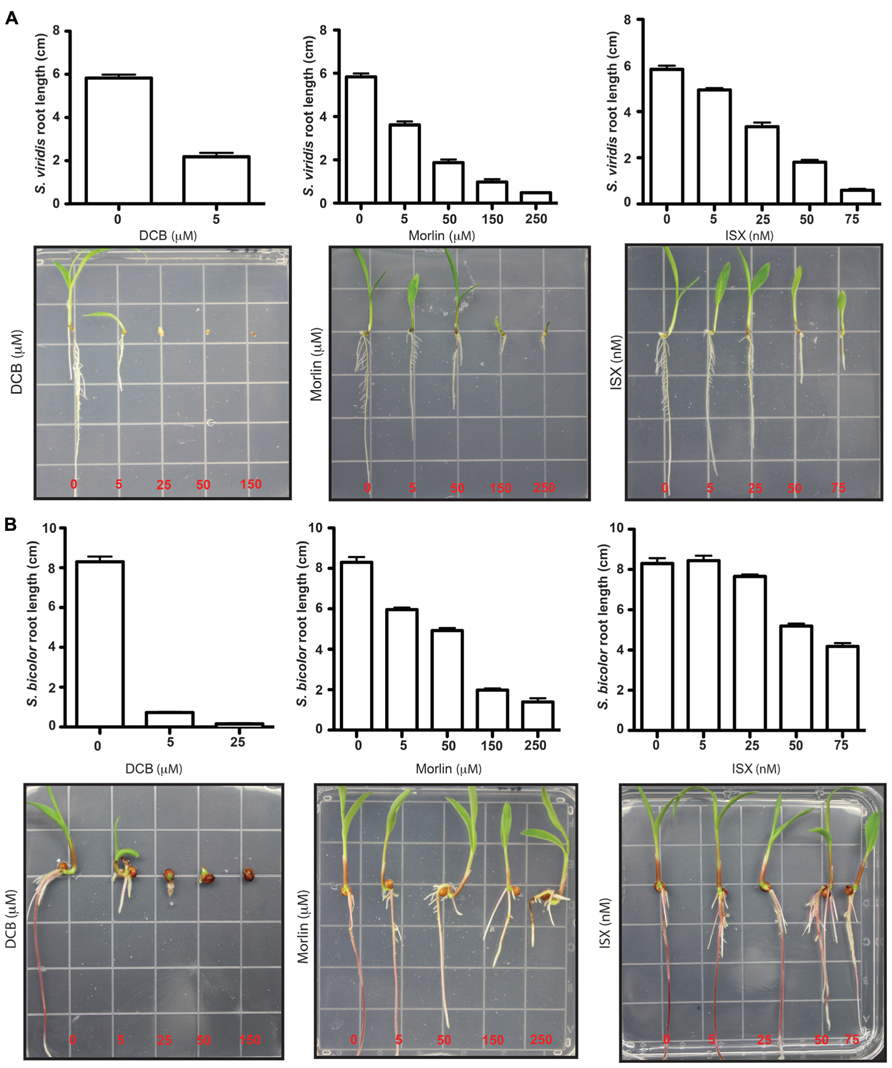
FIGURE 4. Comparing response to cellulose biosynthesis inhibitors (CBIs) in Setaria and sorghum. Response to 2,6-dichlorobenzonitrile (DCB), morlin (7-ethoxy-4-methyl chromen-2-one), and isoxaben (N-[3-(1-ethyl-1-methylpropyl)-1,2-oxazol-5-yl]-2, 6-dimethoxybenzamide) from switchgrass (A) and sorghum (B) was examined during early seedling growth using in plate assays on sterile Murashige and Skoog (MS)-agar media. Average root length was measured (n ≥ 10) and error bars represent standard error from the mean.
Phylogenetic Analysis of CESA Genes
In order to ask whether similarities in cellulose biosynthesis were reflected by similar organization of the CESA gene family, we performed a simple phylogenetic analysis of annotated genes. To aid in providing confidence in the bioinformatic analysis, two separate phylogenetic trees were established using distinct protein prediction compared with genomic sequence. Homology-based associations were made between CESA identified in the genomic data derived from A. thaliana (AT), Setaria italica (SI), Z. mays (ZM), Sorghum bicolor (SB), and P. vulgare (PV). Firstly, the MUSCLE multiple alignment tool (Edgar, 2004) was utilized followed by the PhyML program (Guindon et al., 2010) to create the phylogenetic tree based on the translational predictions (protein sequences) of the putative CESAs. GARLI was utilized in parallel to produce a phylogenetic tree based on genomic sequence information. When grouping CESA genes or gene products within the broad CESA phylogenetic tree, it was necessary to visualize groups (sub-clades) of putative CESAs within the tree, and therefore they were referred to based on the closest homology to the established Arabidopsis CESA nomenclature of Richmond (2000). The phylogenetic tree inferred by protein sequences (Figure 5) clustered in five classes with the Arabidopsis CESA1, 3, 7, 6-like, and 4. It was perhaps noteworthy that the Arabidopsis CESA6-like clade, which also comprised proteins similar to Arabidopsis CESA2, 5, and 9 (Persson et al., 2007) was the most highly represented cluster. By contrast, the Panicoideae homologs with Arabidopsis secondary cell wall CESA4 were least common, which only Z. mays and Setaria containing putative members of the CESA4-associated clade. With the exception of the Arabidopsis CESA4 clade-comprising of only two putative CESA members, the remaining clades contain at least one putative CESA member from each of the five species investigated. Using genomic data to assemble and predict phylogenetic relationships (Figure 5), the putative CESAs divide readily into four distinct clades, clustering around Arabidopsis CESA1 (primary cell wall; Arioli et al., 1998), CESA3 (primary cell wall; Scheible et al., 2001), CESA4/7/8 (vascular secondary cell wall; Taylor et al., 2003), and CESA6-like (CESA2, 5, and 9, included in primary and seed coat secondary cell wall; Stork et al., 2010; Mendu et al., 2011; Sullivan et al., 2011). It was found that at least one putative CESA member from each of the five species was represented within each sub-clade. However, in the heterotrimeric model for cellulose synthase complex (CSC) formation in the primary and secondary cell wall (Delmer and Amor, 1995; Tanaka et al., 2003), the lack of at least three distinct CESAs within the CESA4, 7, and 8 sub-clade suggests either divergence among heterotrimeric associations or an example of large incompletions within the genome sequencing for all of the Panicoideae. In comparing the two phylogenetic trees, there were no marked differences in the CESAs that comprised a particular sub-clade. For example, in both phylogenetic trees the Arabidopsis CESA6-like CESA2, 5, 6, and 9 cluster closely with each other and were broadly represented. The discrepancy in the number of clades between the two trees can be attributed to the fact that within the protein sequence phylogenetic tree, the Arabidopsis CESA4 clade and CESA7 clade are separated while they are combined into one clade within the genetic sequence phylogenetic tree. Ultimately, targeted reverse genetics are urgently needed in the model grasses to elucidate questions related to the tissue specificity, functionality, redundancy, and stoichiometry of the CESA genes. Further, next generation sequencing to produce in depth transcriptional patterns for spatially and temporally diverse tissues could reveal whether differentiation during caryopsis development utilizes CESA6-like sequences in a non-redundant manner as observed in Arabidopsis (Mendu et al., 2011; Sullivan et al., 2011).
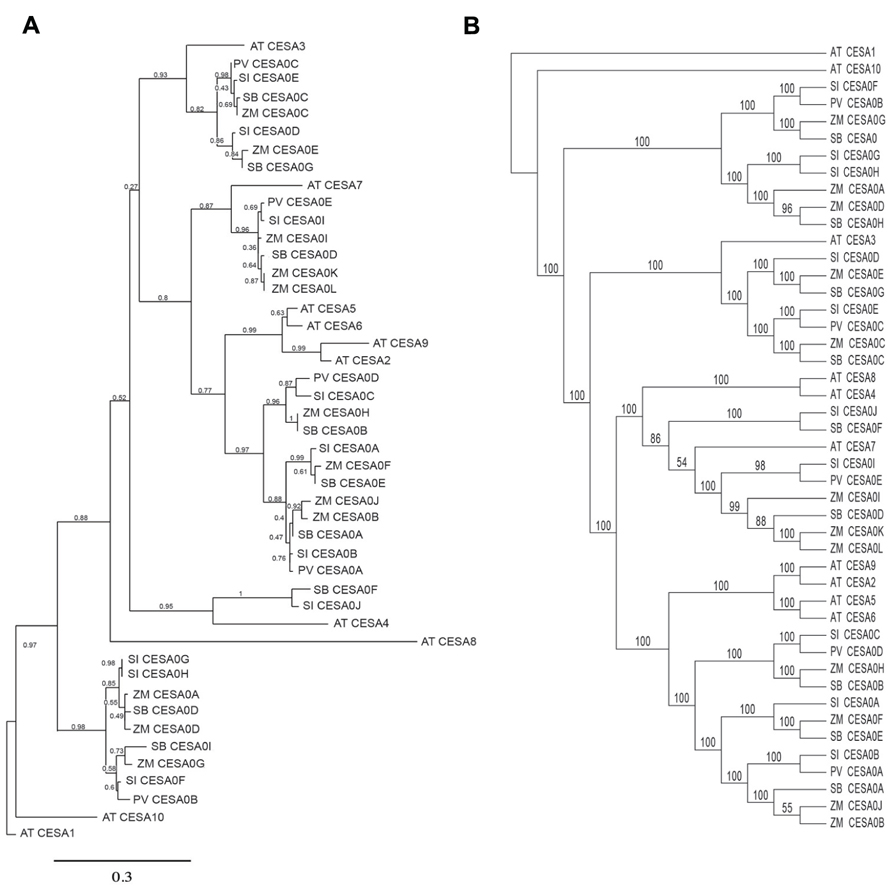
FIGURE 5. Phylogenetic trees of CESA proteins (A) and CESA genes (B). Phylogenetic relationships were determined for A. thaliana (AT), Setaria italica (SI), Z. mays (ZM), Sorghum bicolor (SB), and P. vulgare (PV).
Degradability
Saccharification efficiency was measured for the four species as a criterion for utilizing lignocellulose biomass to produce alcohol-based biofuels. Several different tests were performed to determine the dynamics and cross species consistency of digestibility. Firstly, we examined the pseudo-apparent kinetics of digestibility in semi-purified cellulose (Harris et al., 2009) from the mature stem (S2) of each panicoid (Figure 6). Results showed a consistent rate of conversion of cellulose to fermentable sugars, with sorghum displaying a notable increase in digestibility.
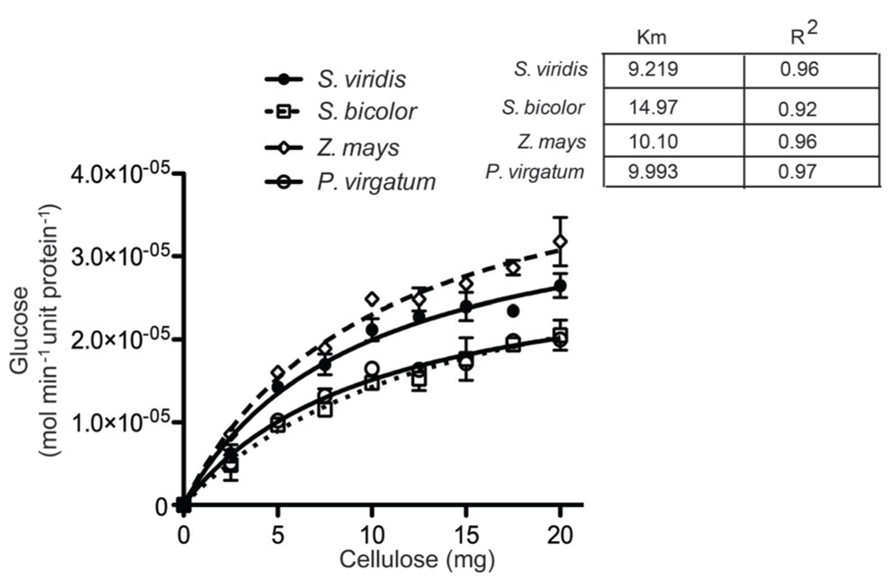
FIGURE 6. Pseudo-apparent kinetics and cellulosome biodegradation of total biomasses. Michaelis–Menten enzymatic kinetics were determined for semi-purified cellulose for the aboveground stage 2 (S2) of Setaria viridis (SV); Z. mays (ZM); Sorghum bicolor (SB), and P. vulgare (PV).
We subsequently used a cellulosome digestion via Clostridium thermocellum and measured the output of carbon from raw unprocessed biomass. During the saccharification reaction two main products are generated, glucose and cellobiose. Additionally, C. thermocellum is capable of metabolizing glucose and cellobiose, and acetate, lactate and ethanol are products of this metabolism. It is possible to estimate the total amount of carbon released from the biomass as the result of the simultaneous saccharification and fermentation process by the inclusion of all the products, thus accounting for the total carbon signature of the converted biomass. Time course fermentation (from 0 to 240 h) released an increased amount of carbon as time progressed (Figure 7A). For both stages, we observed a similar increase in the temporal series; although, as expected for stage 2, a lesser amount of total carbon was released (Figure 7A). Three noteworthy trends were observed; (1) Setaria displayed the second highest rate in stage 1 and the highest total carbon conversion for the root material. (2) The highest rates of total carbon release was, for both stages, observed in switchgrass, and (3) the values determined for maize and sorghum were intermediate. The most interesting aspect, when single fermentation products were taken into account was related to the production of acetate whereby the highest release was associated with the fermentation of the root biomass independently of the species considered although, it was most obvious at the 240 h time point than the other time assayed (Figure 7B). This was the only case where the root biomass conversion products were higher than the conversion products from the shoot biomass.
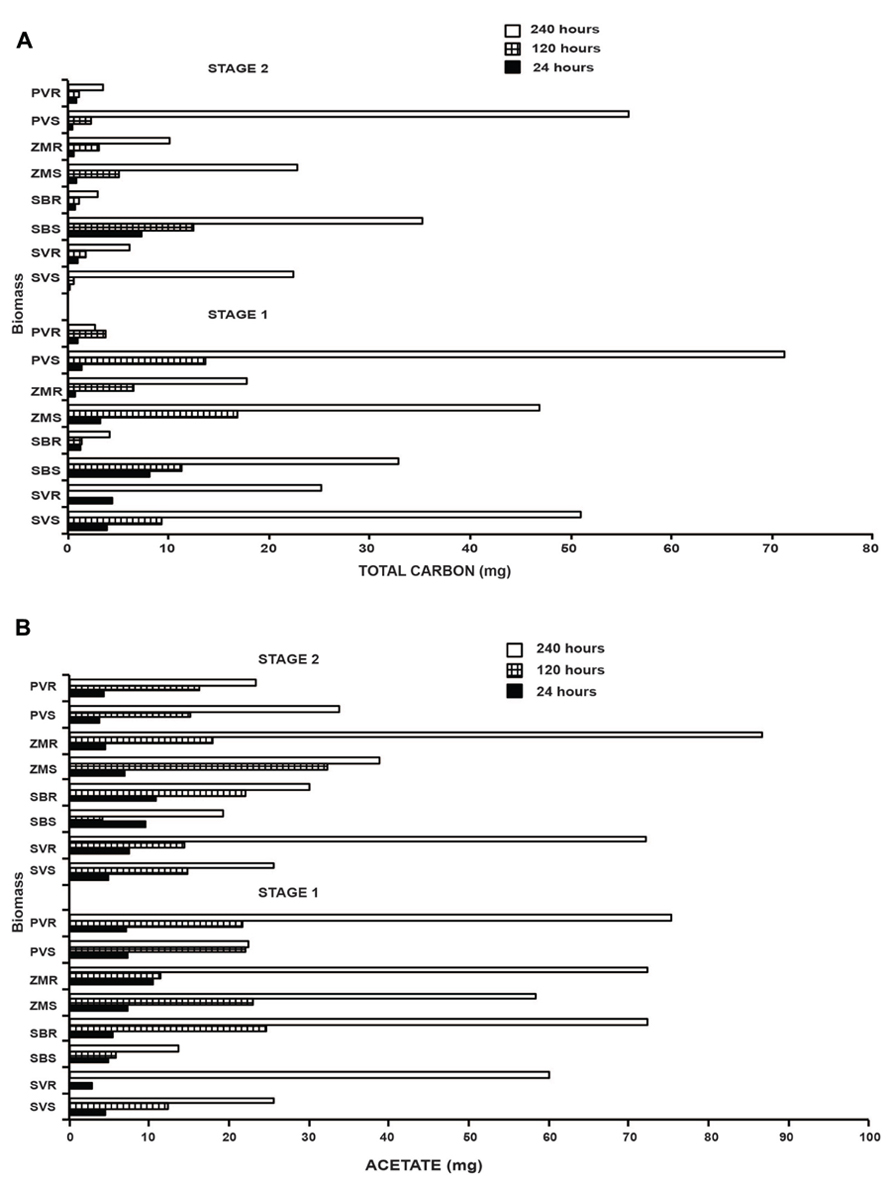
FIGURE 7. Total carbon (A) and lactate (B) production obtained through Clostridium thermocellum system during biomass deconstruction. In (A), biological degradation of the four grasses was completed through Clostridium thermocellum system. The saccharification efficiency was determined for stage 1 and 2 (S1, S2) and for aboveground and belowground material for Setaria viridis (SV); Z. mays (ZM); Sorghum bicolor (SB), and P. vulgare (PV). In (B), The graph displays the acetate production for stage 1 and 2 and for three time points (24, 120, and 240 h) representing the initial stage the middle and the final stage of the process. The convention employed in the graph was: name of species plus S (S denotes shoot) for the aboveground and R (R denotes root) for the belowground material.
Conclusion
Several studies highlight Setaria as a model organism for the panicoid (see Li and Brutnell, 2011), complementing the existing resources for C3 grasses via rice and Brachypodium. In this study, we examined the baseline composition, CBI response, and CESA gene family in Setaria. Data indicated that commonly used CBIs for dicot studies, such as isoxaben as less effective for Setaria, whereas DCB displays similar LD50 responses. Eight members of the CESA gene family were identified for functional genomic characterization. The use of two growth stages demonstrated the potential translational role of Setaria as a model species for lignocellulosic panicoid crops. To summarize, aboveground Setaria biomass was similar to the comparative (non-model) species.
Materials and Methods
Plant Growth Condition and Material Collection
Four grasses species were considered and include maize-hybrid 684GENSS, sorghum var. Della, Panicum virgatum switchgrass var. Alamo and Setaria. Seeds were sown on soil less media (MetroMix 360, SunGro Industries Bellevue, WA, USA). Plants were grown and maintained in a temperature-controlled glasshouse at 24°C, on a 16 h:8 h light:dark cycle with additional supplemental light if natural light was absent. MetroMix 360 media was maintained at field water conditions and fertilized with 3 g of osmocote (The Scotts Company, Marysville, OH, USA) integrated into the media when planting. A physiological staging was found to be most appropriate, as opposed to temporal staging due to inherent variation in reproductive cycle. Stage 1 was defined by four fully developed leaves (not including the cotyledon) and evidence of culm elongation and stage 2 (mature) determined by the onset of apical inflorescence protrusion. Plant specimen were divided into aboveground and belowground material. Each sub-sample was air dried for two days at 26°C then oven dried at 45°C for a further 7 days. Approximately 10 samples were pooled for each replicate. Four biological replicates of randomly harvested material were sampled per species and per developmental stage. Each biological replicate was made of pooled samples per species and per stage. After oven drying, shoot and root material was ground using to 1 mm fragments (Arthur H. Thomas Co Scientific, Philadelphia, PA, USA) and used for further analytical determinations detailed below.
Cell Wall Carbohydrate Compositional Studies
Cellulose content for all samples was measured colorimetrically (Updegraff, 1969). Neutral and acidic sugar composition was determined by high performance liquid chromatography (HPLC; Mendu et al., 2011). In brief, samples were homogenized using a grinder (Arthur H. Thomas Co Scientific, Philadelphia, PA, USA) equipped with a 1-mm sieve. Twenty-five milligrams plant material were destarched as by the specification in National Renewable Energy Laboratory (NREL), LAP-004 (1996) guidelines by incubating the samples in 1 ml 70% ethanol overnight at 65°C, washed twice with 1 ml 70% ethanol for 1 h and once with 1 ml acetone for 5 min. The samples were dried overnight at 30°C with shakings. Cellulose content was determined by weighing out 5 mg of dry biomass extract in quadruplicate and boiled in acetic-nitric reagent (acetic acid:nitric acid:water 8:1:2) for 30 min to remove lignin and hemicellulosic carbohydrates (Updegraff, 1969). Remaining material was then washed twice with 8 ml MQ-water and 4 ml acetone and dried under vacuum. The cellulose samples were then hydrolyzed in 67% sulfuric acid for 1 h. The glucose content of the samples was determined by the anthrone method (Updegraff, 1969). Ten microliters of the sulfuric acid hydrolyzed samples were mixed with 490 μl water and 1 ml 0.3% anthrone in concentrated sulfuric acid on ice. The samples were boiled for 5 min then placed immediately back on ice. The absorbance of the samples was measured using a Bio-Mate Thermo Scientific spectrophotometer (Thermo Fischer, Waltham, MA, USA) set at OD 620 nm and the cellulose content was calculated by multiplying the measured glucose concentration of each sample by the total volume of the assay and then by the hydration correction factor of 0.9 to correct for the water molecule added during hydrolysis of the cellulose polymer.
For monosaccharide analysis, 4 mg of ethanol–acetone washed plant biomass was measured into glass tubes, a total of 36.5 μl of 72% (w/v) H2SO4 was added including the internal standard Ribose. The samples were incubated on ice for 2 h, mixing every 30 min with the vortex. Samples were diluted to 4% (w/v) H2SO4 by adding 963.5 μl of water and autoclaved at 121°C for 1 h. Neutral sugar standards (Fuc, Ara, Rha, Gal, Glc, Man, and Xyl) and acid sugar standards (GalUA) were also processed in the same way at the same time. Neutral and acidic cell wall sugars were identified and quantified by pulsed electrochemical detection using a Dionex ED50 (Dionex, Thermoscientific). Sugar separation was achieved using a BioLC GS50 HPLC device and a CarboPAC-PAl pellicular anion-exchange column with column guard (Dionex). Column and detector temperature was maintained at 30°C using a Dionex LC25 chromatography oven. Samples were introduced to the column from a Dionex AS50 autosampler and neutral sugars were eluted isocratically in 22 mM NaOH over a 30-min period, after which the column was washed in 1 M NaOAc and 200 mM NaOH for 5 min. The column was recharged by passing 200 mM NaOH through it for 10 min and then reequilibrating the column in 22 mM NaOH for 10 min prior to injection of the next sample. This protocol resulted in optimal (near-baseline) resolution of Gal, Glc, Man, and Xyl (Downie and Bewley, 1996). Acidic sugar separation and quantification used the same HPLC procedure but followed the eluent profile of Dean et al. (2007). For total biomass monosaccharides, the neutral sugars were normalized to the amount of plant material (aboveground and belowground) used for the sample preparation.
Lignin Quantification
Acid Hydrolysis
Acid-soluble lignin, acid-insoluble lignin, and ash were measured using the laboratory analytical protocols NREL, LAP-004 (1996). Briefly, approximately 300 mg sample-1 (±0.01 g) of dry plant tissue were placed into 30 ml test tubes with three replicates followed by an addition of 3 ml of sulfuric acid (72% v/v) to each test tube and stirred thoroughly with a glass rod. Samples were incubated in a water bath at 30°C for 2 h and stirred every 15 min. After hydrolysis, samples were diluted with 84 ml of deionized water (4% concentration) into a 250-ml Erlenmeyer flask that was autoclaved at 121°C at 15 psi for 1 h.
Acid-Soluble Lignin Measurement
Acid-soluble lignin was measured using NREL, LAP-004 (1996). Briefly, the autoclaved solution was allowed to cool to room temperature before withdrawing 1 ml of the hydrolyzate without disturbing any solid particles in the solution. A 4% solution of sulfuric acid was used as a reference blank. To accommodate the absorbance range (0.2–0.7) of the spectrophotometer at 240 nm, both the hydrolyzate and reference 4% sulfuric acid solution were diluted 1:30 with deionized water. The absorbance of the hydrolyzate at 240 nm using a 1-cm light path cuvette was used to calculate the acid-soluble lignin.
Acid-Insoluble Lignin Measurement
Porcelain filter crucibles containing a glass microfiber filter (Whatman 934-AH) were placed in a furnace at 575°C for 4 h. The crucibles were removed from the furnace and placed into a desiccator to cool for a minimum of 1 h before being weighed. The autoclaved samples were vacuum filtered through the crucibles. The crucibles and their contents were dried in an oven at 105°C overnight before transferring into a desiccator to cool. The weight of the crucibles were recorded and placed in a furnace at 575°C for 4 h before reducing the temperature to 105°C overnight. The crucibles were removed and placed into a desiccator to cool and weighed.
Enzyme Kinetic of Semi-Purified Cellulose
Crude cell walls were prepared from total biomass of each species (stage 2) as by published protocol (Reiter et al., 1993) and it was scaled up in order to meet the requirements for enzyme kinetics. Cellulose which was purified according to the method by Updegraff (1969) was homogenized using an Arthur H. Thomas Co Scientific grinder (Philadelphia, PA, USA) equipped with a 0.5-mm sieve. A cocktail of cellulase and cellobiose (Sigma, USA), equivalent to 2 FPU (filter paper unit), were used to digest an increased amount of cellulose substrates (2, 5, 7.5, 10, 12.5, 15, 17.5, and 20 mg) in a 50-mM citrate buffer (pH 4.8) at 50°C for 2 h. Prior to glucose determination, the enzymes were heat-inactivated by boiling the samples for 5 min at 100°C. Glucose quantification was done by using a Yellow Springs Instruments (YSI)-glucose analyzer. The Inability to exactly calculate the number of catalytic ends in the complex mixture of cell wall biomass allowed only for the calculation of relative estimation, expressed as relative kinetics (relative Vmax and Km ). The glucose values were converted in mol min-1 unit protein-1 and used to determine the apparent kinetics values using the program GRAPHPAD PRISM-4 (Graphpad, La Jolla, CA, USA).
Cellulose Biosynthesis Inhibitor Studies
The effect of three CBI, isoxaben (N-[3-(1-ethyl-1-methylpropyl)-1,2-oxazol-5-yl]-2,6-dimethoxybenzamide; Dow Chemical Company, Midland, MI, USA), DCB (Sigma Aldrich, St Louis, MO, USA), and morlin (7-ethoxy-4-methyl, coumarin Sigma Aldrich, St Louis, MO, USA) were employed to evaluate the plant responses to increased concentration on an in plate assay. Due to the large-sized maize seeds and the poor germination rate of switchgrass seeds, the assay was carried only for Setaria viridis and Sorghum bicolor. Seeds were surface sterilized in 1% bleach and 0.1% sodium dodecyl sulfate (SDS), for 15 min, twice with a water wash in between. An additional 70% ethanol wash was completed for 5 min; the seeds were then rinsed 10 times with sterile water. For Sorghum Bicolor, an average of 15 seeds were plated onto Hoagland’s based media containing either isoxaben (concentration range: 5, 25, 50, and 75 nM), DCB (concentration range: 5, 25, 50, and 150 μM) and morlin (concentration range: 5, 50, 150, and 250 μM) as well as a mock control plate (dimethyl sulfoxide, DMSO). A similar protocol was employed for Setaria viridis, but the number of seeds increased to between 30 and 40. Plates were incubated in a climate-controlled chamber (Conviron, Winnipeg, Canada) at 22°C, with a 16 h:8 h light:dark regimen. Each drug concentration was replicated (n = 3) and the effects of the drugs were evaluated on the root length and determined 8 days post-germination.
Anaerobic Fermentation Analysis
Inoculum Preparation and Chemicals
Clostridium thermocellum ATCC 27405 was provided by Michael Flythe (USDA, Lexington, KY, USA) and was maintained in our laboratory, as described by Johnson et al. (1981). Chemically defined thermophile medium was prepared as described by Dhamagadda et al. (2010). A bacterial culture was grown for 24 h in Balch tubes (pH 6.7) at 63°C containing cellulose strips (Whatman Grade 1 filter paper) suspended in 8.5 ml basal medium. This initial culture was used to inoculate 54 ml of cellulose fiber in 100-ml sealed serum vials (Bellco Biotechnology, Vineland, NJ, USA) under CO2 atmosphere. After 18 h of growth, the secondary culture was diluted with fresh basal media (substrate free) to prepare the standard inoculum stock (final optical density of 0.162 OD 600; ~0.078 g dry cells l-1).
Biomass Microbial Cultures and Sampling
One milliliter of prepared inoculum culture was inserted by syringe into a test tube containing 8.5 ml of thermophile medium and 0.5 g of plant material to achieve a 5% w/w bacterial suspension. Test tubes containing plant material and media were tightly sealed with CO2 headspace and autoclaved at 120°C for 20 min prior to inoculation. Cultures were incubated at 63°C in a water bath. The control consisted of substrate (0.5 g Avicel), media, and bacteria. One milliliter supernatant (triplicate samples without replacement) was collected after 0, 24, 72, 120, 168, and 240 h of fermentation. Each sample collected was centrifuged (5000 × g, 20 min, 4°C) and aliquots of cell free supernatant were stored at -20°C until analysis by HPLC. The samples were HPLC analyzed for the presence of the five main products known to be produced by the bacterium. These were matched to the retention time of true standards. Products were: cellobiose, glucose, acetate, ethanol, and lactate.
X-ray Diffraction Analysis
Finely ground biomass samples were then contained in a custom-built sample holder of pressed boric acid. In brief, plant material was placed into a mold, containing a sleeve and hand pressed with a solid metal plug forming a disk shape. The sleeve and plug were removed and a boric acid (Fischer, Madison, WI, USA) base was then formed by pouring the boric acid over the bottom and sides of the sample and applying 40,000 psi of pressure to the 40 mm × 40 mm mold using a Carver Autopellet Press (Wabash, IN, USA). Samples were pressed to create an even horizontal surface. A Bruker-AXS Discover D8 Diffractometer (Bruker-AXS USA, Madison, WI, USA) was used for wide angle XRD with Cu Ka radiation generated at 30 mA and 40 kV. The experiments were carried out using Bragg-Brentano geometries (symmetrical reflection). Diffractograms were collected between 5° and 35° (for samples with little baseline drift), with 0.02° resolution and 2-s exposure time interval for each step. Sample rotation to redirect the X-ray beam diffraction site was achieved per replicate. The data analysis was carried out using the calculation for RCI of: RCI = I002 - Iam /I002 × 100, where I002 is the maximum peak height above baseline at approximately 22.5° and Iam is the minimum peak height above the baseline at ~18°. For assessment of experimental accuracy, the pressed samples were examined using reflective geometries at 22.5° 2-theta with the sample scanned rotationally (360°) and in an arc (90°) to obtain an intensity/spatial orientation plot of a sample for which the RCI had already been established. The range of reflective intensities was then used to estimate the accuracy of the RCI determination using a 95% cutoff across the plot range. Diffractograms were collected in Diffrac-Plus-XRD Commander software (Bruker-AXS, Karlsruhe, Germany) and minimally processed (baseline identification, noise correction, 3D display, and cropping of RCI signature region) using the EVA and TexEval (Bruker-AXS Karlsruhe, Germany) software.
Phylogenetic Analysis
The sequences of the A. thaliana cellulose synthase genes were obtained from the TAIR database1 and were utilized as the initial reference in order to obtain the putative CESA genes of the grasses. The cDNA sequences of the ten cellulose synthase genes of A. thaliana were employed to conduct the BLASTX (Altschul et al., 1997) search within the Setaria italica, Sorghum bicolor, Panicum virgatum, and Z. mays genomes. BLASTX searches were conducted using the phytozome database provided by JGI2. Due to the very few number of putative CESA genes for Panicum virgatum that resulted from the BLASTX search, an additional BLASTP search was conducted using the 12 CESA genes of the Z. mays to determine if there were any additional putative genes. After the identification of the putative CESA genes, the protein sequences of each of the putative genes were screened to determine if they contained the CxxC motif (Richmond, 2000) considered to be unique to the CESA genes and distinguishing them from the cellulose synthase like genes. A total of 36 putative cellulose synthase genes were identified. The putative CESA genes that contained the CxxC motif of the four monocot species as well as the known CESA genes of A. thaliana were then uploaded onto the PhyML program (Guindon et al., 2010). The putative CESA gene protein sequences that contained the CxxC motif of the four monocot species above as well as the CESA protein sequences of A. thaliana were analyzed using PhyML (v.3.0; Guindon et al., 2010). The advanced setting within PhyML conducts a sequence alignment using MUSCLE (v.3.7; Edgar, 2004), then automatically utilizes the alignment to create the phylogenetic tree. A total of 500 bootstrap replicates were conducted under the WAG (Whelan And Goldman) protein substitution model (Whelan and Goldman, 2001).
In addition to the amino acid-based tree, a tree was constructed using the DNA sequences of the 36 putative CESA genes as well as the 10 Arabidopsis CESA genes. The sequences were obtained from the Phytozome database and the TAIR database, respectively. The sequences were submitted to the Guidance web server (Penn et al., 2010) and aligned with MAFFT (Katoh et al., 2005). Guidance is a program that both aligns sequences and assesses confidence in each position in the alignment, in order to objectively identify and remove regions of ambiguous alignment prior to phylogenetic analysis. Any column in the alignment that had a confidence score of less than 95% was removed by Guidance. GARLI (v. 2.0; Zwickl, 2006) was used to analyze the resulting alignment under the GTR + I + G model (Rodriguez et al., 1990), in which a best tree search (five replicates) and a 200-replicate bootstrap analysis were conducted, both using the default settings. The resulting maximum likelihood tree was then visualized via FIGTREE3.
Statistical Analysis
Statistical analysis consisting of one-way analysis of variance (ANOVA) with a post hoc Tukey test was performed by using PRISM4 (Graphpad, La Jolla, CA, USA). The outcomes are indicated in letters on the corresponding graphs. Same letter signify non-significance (P > 0.05) whereas different letters indicate significance (P < 0.05).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to acknowledge Eric G. Chapman for his insight in the phylogenetic analysis, Aubrey Shea and Jozsef Stork for technical support. This work was supported by grants from the Department of Energy (DOE-FOA 10-0000368 to Seth DeBolt and Tom Brutnell) and the United States Department of Agriculture BDRI program (to Seth DeBolt and Sue Nokes).
Footnotes
References
Alonso, M. D., Wettstein, S. G., Mellmer, M. A., Gurbuz, E. I., and Dumesic, J. A. (2013). Integrated conversion of hemicellulose and cellulose from lignocellulosic biomass. Energy Environ. Sci. 6, 76–80. doi: 10.1039/c2ee23617f
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Andersson, S., Serimaa, R., Paakkari, T., Saranpää, P., and Pesonen, E. (2003). Crystallinity of wood and the size of cellulose crystallites in Norway spruce (Picea abies). J. Wood Sci. 49, 531–537. doi: 10.1007/s10086-003-0518-x
Arioli, T., Peng, L., Betzner, A. S., Burn, J., Wittke, W., Herth, W., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. doi: 10.1126/science.279.5351.717
Bauer, W. D., Talmadge, K. W., Keegstra, K., and Albersheim, P. (1973). The structure of plant cell walls: II. The hemicellulose of the walls of suspension-cultured sycamore cells. Plant Physiol. 51, 174–187. doi: 10.1104/pp.51.1.174
Bennetzen, J. L. (2007). Patterns in grass genome evolution. Curr. Opin. Plant Biol. 10, 176–181. doi: 10.1016/j.pbi.2007.01.010
Brabham, C., and DeBolt, S. (2013). Chemical genetics to examine cellulose biosynthesis. Front. Plant Sci. 3:309. doi: 10.3389/fpls.2012.00309
Brutnell, T., Wang, L., Swartwood, K., Goldschmidt, A., Jackson, D., Zhu, X.-G., et al. (2010). Setaria viridis: a model for C4 photosynthesis. Plant Cell 22, 2537–2544. doi: 10.1105/tpc.110.075309
Chen, F., and Dixon, R. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761. doi: 10.1038/nbt1316
Crowell, E. F., Bischoff, V., Desprez, T., Rolland, A., Stierhof, Y.-D., Schumacher, K., et al. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21, 1141–1154. doi: 10.1105/tpc.108.065334
Dean, G., Zheng, H., Tewari, J., Huang, J., Young, D., Hwang, Y., et al. (2007). The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19, 4007–4021. doi: 10.1105/tpc.107.050609
DeBolt, S., Gutierrez, R., Ehrhardt, D. W., Melo, C. V., Ross, L., Cutler, S. R., et al. (2007a). Morlin, an inhibitor of cortical microtubule dynamics and cellulose synthase movement. Proc. Natl. Acad. Sci. U.S.A. 104, 5854–5859. doi: 10.1073/pnas.0700789104
DeBolt, S., Gutierrez, R., Ehrhardt, D. W., and Somerville, C. (2007b). Nonmotile cellulose synthase subunits repeatedly accumulate within localized regions at the plasma membrane in Arabidopsis hypocotyl cells following 2,6-dichlorobenzonitrile treatment. Plant Physiol. 145, 334–338. doi: 10.1104/pp.107.104703
Desprez, T., Vernhettes, S., Fagard, M., Refrégier, G., Desnos, T., Aletti, E., et al. (2002). Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 128, 482–490. doi: 10.1104/pp.010822
Dhamagadda, V. S., Nokes, S. E., Strobel, H. J., and Flythe, M. D. (2010). Investigation of the metabolic inhibition observed in solid substrate cultivation of Clostridium thermocellum on cellulose. Bioresour. Technol. 101, 6039–6044. doi: 10.1016/j.biortech.2010.02.097
Doust, A. N., Kellogg, E. A., Devos, K. M., and Bennetzen, J. L. (2009). Foxtail millet: a sequence-driven grass model system. Plant Physiol. 149, 137–141. doi: 10.1104/pp.108.129627
Downie, B., and Bewley, J. D. (1996). Dormancy in white spruce (Picea glauca [Moench.] Voss.) seeds is imposed by tissues surrounding the embryo. Seed Sci. Res. 6, 9–15. doi: 10.1017/S0960258500002944
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Gutierrez, R., Lindeboom, J. J., Paredez, A. R., Emons, A. M. C., and Ehrhardt, D. W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11, 797–806. doi: 10.1038/ncb1886
Harris, D., and DeBolt, S. (2008). Relative Crystallinity of plant biomass: studies on assembly, adaptation and acclimation. PLoS ONE 3:e2897. doi: 10.1371/journal.pone.0002897
Harris, D., Stork, J., and Debolt, S. (2009). Genetic modification in cellulose-synthase reduces crystallinity and improves biochemical conversion to fermentable sugar. Glob. Change Biol. Bioenergy 1, 51–61. doi: 10.1111/j.1757-1707.2009.01000.x
Harris, D. M., Corbin, K., Wang, T., Gutierrez, R., Bertolo, A. L., Petti, C., et al. (2012). Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 109, 4098–4103. doi: 10.1073/pnas.1200352109
Heim, D. R., Bjelk, L. A., James, J., Schneegurt, M. A., and Larrinua, I. M. (1993). Mechanism of isoxaben tolerance in Agrostis palustris var. Penncross. J. Exp. Bot. 44, 1185–1189. doi: 10.1093/jxb/44.7.1185
Heim, D. R., Roberts, J. L., Pike, P. D., and Larrinua, I. M. (1989). Mutation of a locus of Arabidopsis thaliana confers resistance to the herbicide isoxaben. Plant Physiol. 90, 146–150. doi: 10.1104/pp.90.1.146
Hibberd, J. M., and Quick, P. W. (2002). Characteristics of C4 photosynthesis in stems and petioles of C3 flowering plants. Nature 415, 451–454. doi: 10.1038/415451a
Hidayat, B., Felby, C., Johansen, K., and Thygesen, L. G. (2012). Cellulose is not just cellulose: a review of dislocations as reactive sites in the enzymatic hydrolysis of cellulose microfibrils. Cellulose 19, 1481–1493. doi: 10.1007/s10570-012-9740-2
Johnson, E. A., Madia, A., and Demain, A. L. (1981). Chemically defined minimal medium for growth of the anaerobic cellulolytic thermophile Clostridium thermocellum. Appl. Environ. Microbiol. 41, 1060–1062.
Katoh, K., Kuma, K., Toh, H., and Miyata, T. (2005). MAFFT version 5: improvement in 1060 accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Kim, Y., Mosier, N., Ladisch, M., Pallapolu, V., Lee, Y., Garlock, R., et al. (2011). Comparative study on enzymatic digestibility of switchgrass varieties and harvests processed by leading pretreatment technologies. Bioresour. Technol. 102, 11089–11096. doi: 10.1016/j.biortech.2011.06.054
Langdale, J. A. (2011). C4 cycles: past, present, and future research on C4 photosynthesis. Plant Cell 23, 3879–3892. doi: 10.1105/tpc.111.092098
Li, P., and Brutnell, T. (2011). Setaria viridis and Setaria italica, model genetic systems for panicoid grasses. J. Exp. Bot. 62, 3031–3037. doi: 10.1093/jxb/err096
Li, X., Ximenes, E., Kim, Y., Slininger, M., Meilan, R., Ladisch, M., et al. (2010). Lignin monomer composition affects Arabidopsis cell-wall degradability after liquid hot water pretreatment. Biotechnol. Biofuels 3:27. doi: 10.1186/1754-6834-3-27
Meinert, M., and Delmer, D. (1977). Changes in biochemical composition of the cell wall of the cotton fiber during development. Plant Physiol. 59, 1088–1097. doi: 10.1104/pp.59.6.1088
Mendu, V., Griffiths, J. S., Persson, S., Stork, J., Downie, A. B., Voiniciuc, C., et al. (2011). Subfunctionalization of cellulose synthases in seed coat epidermal cells mediates secondary radial wall synthesis and mucilage attachment. Plant Physiol. 157, 441–453. doi: 10.1104/pp.111.179069
Morant, M., Bak, S., Møller, B. L., and Werck-Reichhart, D. (2003). Plant cytochromes P450: tools for pharmacology, plant protection and phytoremediation. Curr. Opin. Biotechnol. 14, 151–162. doi: 10.1016/S0958-1669(03)00024-7
Oula, G. (2008). C4 photosynthesis and water stress. Ann. Bot. 103, 635–644. doi: 10.1093/aob/mcn093
Ouyang, S., Zhu, W., Hamilton, J., Lin, H., Campbell, M., Childs, K., et al. (2007). The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 35, D883–D887. doi: 10.1093/nar/gkl976
Parrish, D., and Fike, J. (2005). The biology and agronomy of switchgrass for biofuels. Crit. Rev. Plant Sci. 24, 423–459. doi: 10.1080/07352680500316433
Penn, O., Privman, E., Landan, G., Graur, D., and Pupko, T. (2010). An alignment confidence score capturing robustness to guide tree uncertainty. Mol. Biol. Evol. 27, 1759–1767. doi: 10.1093/molbev/msq066
Persson, S., Paredez, A., Carroll, A., Palsdottir, H., Doblin, M., Poindexter, P., et al. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 15566–15571. doi: 10.1073/pnas.0706592104
Redfearn, D. D., and Nelson, C. J. (2003). “Grasses for southern areas,” in An Introduction to Grassland Agriculture, ed. R. F. Barnes (Ames, IA: Blackwell Publishing Company), 149–170.
Reiter, W.-D., Chapple, C. C. S., and Somerville, C. R. (1993). Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science 261, 1032–1035. doi: 10.1126/science.261.5124.1032
Rensink, W., and Buell, C. (2004). Arabidopsis to rice. Applying knowledge from a weed to enhance our understanding of a crop species. Plant Physiol. 135, 622–629. doi: 10.1104/pp.104.040170
Richmond, T. (2000). Higher plant cellulose synthases. Genome Biol. 1, 1–6. doi: 10.1186/gb-2000-1-4-reviews3001
Robert, S., Raikhel, N. V., and Hicks, G. R. (2009). Powerful partners: Arabidopsis and chemical genomics. Arabidopsis Book 7:e0109. doi: 10.1199/tab.0109
Rodriguez, F., Oliver, J. L., Marin, A., and Medina, J. R. (1990). The general stochastic model of nucleotide substitution. J. Theor. Biol. 142, 485–501. doi: 10.1016/S0022-5193(05)80104-3
Rogers, L., and Campbell, M. (2004). The genetic control of lignin deposition during plant growth and development. New Phytol. 164, 17–30. doi: 10.1111/j.1469-8137.2004.01143.x
Saathoff, A., Sarath, G., Chow, E., Dien, B., and Tobias, C. (2011). Downregulation of cinnamyl-alcohol dehydrogenase in switchgrass by RNA silencing results in enhanced glucose release after cellulase treatment. PLoS ONE 1:e16416. doi: 10.1371/journal.pone.0016416
Saha, B. (2003). Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 30, 279–291. doi: 10.1007/s10295-003-0049-x
Scheible, W.-R., Eshed, R., Richmond, T., Delmer, D., and Somerville, C. (2001). Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc. Natl. Acad. Sci. U.S.A. 98, 10079–10084. doi: 10.1073/pnas.191361598
Scheller, H. V., and Ulvskov, P. (2010). Hemicelluloses. Annu. Rev. Plant Biol. 61, 263–289. doi: 10.1146/annurev-arplant-042809-112315
Schmitt, M. R., and Edwards, G. E. (1981). Photosynthetic capacity and nitrogen use efficiency of maize, wheat, and rice: a comparison between C3 and C4 photosynthesis. J. Exp. Bot. 32, 459–466. doi: 10.1093/jxb/32.3.459
Segal, L., Creely, J. J., Martin, A. E., and Conrad, C. M. (1959). An empirical method for estimating the degree of crystallinity of native cellulose using the X-ray diffractometer. Tex. Res. J. 29, 786–794. doi: 10.1177/004051755902901003
Stork, J., Harris, D., Griffiths, J., Williams, B., Beisson, F., Li-Beisson, Y., et al. (2010). CELLULOSE SYNTHASE9 serves a nonredundant role in secondary cell wall synthesis in Arabidopsis epidermal testa cells. Plant Physiol. 153, 580–589. doi: 10.1104/pp.110.154062
Stork, J., Montross, M., Smith, R., Schwer, L., Chen, W., Reynolds, M., et al. (2009). Regional examination shows potential for native feedstock options for cellulosic biofuel production. Glob. Change Biol. Bioenergy 1, 230–239. doi: 10.1111/j.1757-1707.2009.01015.x
Sullivan, S., Ralet, M.-C., Berger, A., Diatloff, E., Bischoff, V., Gonneau, M., et al. (2011). CESA5 is required for the synthesis of cellulose with a role in structuring the adherent mucilage of Arabidopsis seeds. Plant Physiol. 156, 1725–1739. doi: 10.1104/pp.111.179077
Tanaka, K., Murata, K., Yamazaki, M., Onosato, K., Miyao, A., and Hirochika, H. (2003). Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol. 133, 73–83. doi: 10.1104/pp.103.022442
Taylor, N. G., Howells, R. M., Huttly, A. K., Vickers, K., and Turner, S. R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 1450–1455. doi: 10.1073/pnas.0337628100
Töth, R. K., and van der Hoorn, R. (2010). Emerging principles in plant chemical genetics. Trends Plant Sci. 15, 81–88. doi: 10.1016/j.tplants.2009.11.005
Updegraff, D. M. (1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32, 420–424. doi: 10.1016/S0003-2697(69)80009-6
Vicient, C., Jaaskelainen, M., Kalendar, R., and Schulman, A. (2001). Active retrotransposons are a common feature of grass genomes. Plant Physiol. 125, 1283–1292. doi: 10.1104/pp.125.3.1283
Vogel, J. (2008). Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 11, 301–307. doi: 10.1016/j.pbi.2008.03.002
Wang, X., Gowik, U., Tang, H., Bowers, J., Westhoff, P., and Paterson, A. (2009). Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biol. 10, R68. doi: 10.1186/gb-2009-10-6-r68
Wang, Z., Li, R., Xu, J., Marita, J., Hatfield, R., Qu, R., et al. (2012). Sodium hydroxide pretreatment of genetically modified switchgrass for improved enzymatic release of sugars. Bioresour. Technol. 110, 364–370. doi: 10.1016/j.biortech.2012.01.097
Keywords: Setaria, cell wall, lignocellulose, Panicoideae, biofuel, cellulose synthase
Citation: Petti C, Shearer A, Tateno M, Ruwaya M, Nokes S, Brutnell T and DeBolt S (2013) Comparative feedstock analysis in Setaria viridis L. as a model for C4 bioenergy grasses and panicoid crop species. Front. Plant Sci. 4:181. doi: 10.3389/fpls.2013.00181
Received: 29 March 2013; Accepted: 20 May 2013;
Published online: 19 June 2013.
Edited by:
Samuel P. Hazen, University of Massachusetts, USAReviewed by:
Markus Pauly, University of California at Berkeley, USAChristian Voigt, University of Hamburg, Germany
Copyright: © 2013 Petti, Shearer, Tateno, Ruwaya, Nokes, Brutnell and DeBolt. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Seth DeBolt, Plant Physiology, Department of Horticulture, College of Agriculture, Food and the Environment, University of Kentucky, N318 Agricultural Science Center North, Lexington, KY 40546, USA e-mail:c2RlYm8yQHVreS5lZHU=

 Andrew Shearer1
Andrew Shearer1