- Biotechnology Research Lab, Miami Dade College, Miami, FL, USA
The number of plant mitochondrial genomes sequenced exceeds two dozen. However, for a detailed comparative study of different phylogenetic branches more plant mitochondrial genomes should be sequenced. This article presents sequencing data and comparative analysis of mitochondrial DNA (mtDNA) of the legume Vicia faba. The size of the V. faba circular mitochondrial master chromosome of cultivar Broad Windsor was estimated as 588,000 bp with a genome complexity of 387,745 bp and 52 conservative mitochondrial genes; 32 of them encoding proteins, 3 rRNA, and 17 tRNA genes. Six tRNA genes were highly homologous to chloroplast genome sequences. In addition to the 52 conservative genes, 114 unique open reading frames (ORFs) were found, 36 without significant homology to any known proteins and 29 with homology to the Medicago truncatula nuclear genome and to other plant mitochondrial ORFs, 49 ORFs were not homologous to M. truncatula but possessed sequences with significant homology to other plant mitochondrial or nuclear ORFs. In general, the unique ORFs revealed very low homology to known closely related legumes, but several sequence homologies were found between V. faba, Beta vulgaris, Nicotiana tabacum, Vitis vinifera, and even the monocots Oryza sativa and Zea mays. Most likely these ORFs arose independently during angiosperm evolution (Kubo and Mikami, 2007; Kubo and Newton, 2008). Computational analysis revealed in total about 45% of V. faba mtDNA sequence being homologous to the Medicago truncatula nuclear genome (more than to any sequenced plant mitochondrial genome), and 35% of this homology ranging from a few dozen to 12,806 bp are located on chromosome 1. Apparently, mitochondrial rrn5, rrn18, rps10, ATP synthase subunit alpha, cox2, and tRNA sequences are part of transcribed nuclear mosaic ORFs.
Introduction
The number of plant mitochondrial genomes being sequenced exceeds two dozen. Sequencing data analysis was well presented in different reviews (Kubo and Mikami, 2007; Kubo and Newton, 2008; Kitazaki and Kubo, 2010; Maréchal and Brisson, 2010; Woloszynska, 2010). The published data indicate that flowering plants contain the largest mitochondrial genomes reported so far, consisting of heterogeneous populations of mainly linear and sometimes circular DNA molecules. Interestingly, size and organization of these molecules differ not only among closely related plant species but even among lines of the same species (Bendich, 1993; Oldenburg and Bendich, 1996; Kubo and Mikami, 2007; Kubo and Newton, 2008). Sequences of plant mitochondrial genomes in most cases are organized as large circular molecules called “master chromosome,” usually containing several large (>500 bp) direct and a few inverted repeats. It has been suggested that mitochondrial DNA (mtDNA) is replicated in a recombination-dependent manner (Oldenburg and Bendich, 1996; Backert and Börner, 2000; Kubo and Newton, 2008). Intra- and inter-molecular recombination between large repeats can cause different isomeric forms or subgenomic versions of a master chromosome (Kubo and Mikami, 2007; Kubo and Newton, 2008; Kitazaki and Kubo, 2010; Maréchal and Brisson, 2010; Woloszynska, 2010; Chang et al., 2013). However, mitochondrial heteroplasmy is mainly determined by homologous recombinations between short repeats (<500 bp) (Vitart et al., 1992; Hartmann et al., 1994; Kanazawa et al., 1994; Bellaoui et al., 1998; Janska et al., 1998; Arrieta-Montiel et al., 2001; Albert et al., 2003; Woloszynska and Trojanowski, 2009; Alverson et al., 2011).
Differences in master chromosome size are due to unique sequences present in one and absent in another species’ mtDNA, or to repeat sequences or large duplications (mostly >1000 bp) representing up to 35% of the total genome size. For example, mitochondrial genome sizes of Zea mays lines range from 535,825 to 739,046 bp, while genome complexities range from 506,760 to 537,180 bp (Allen et al., 2007). Even unique sequences of mitochondrial genomes may differ intraspecifically by up to 7%.
Recombination between short repeats can lead to gain or loss of sequences. Sequence gain might come from chloroplast or nuclear genomes of the same plant, sometimes from mitochondrial plasmid DNA (Kubo et al., 2000; Kubo and Newton, 2008; McDermott et al., 2008; Kitazaki and Kubo, 2010) or even from viruses (Marienfeld et al., 1997; Goremykin et al., 2009), fungi, bacteria, or other plants. Sequence loss from mitochondrial genomes can be compensated by transfer to the nuclear genome (Kubo and Newton, 2008; Kitazaki and Kubo, 2010; Alverson et al., 2011).
Plant mitochondrial heteroplasmy covers a significant part of the mitochondrial genome. Less than 20% of the genome is represented by known protein, rRNA and tRNA encoding genes. The coding parts of these sequences are highly conserved. Lists of tRNA were found different for various plants, but every tRNA sequence is conserved and might have a mitochondrial or chloroplast origin (Marienfeld et al., 1997; Kubo and Newton, 2008; Kitazaki and Kubo, 2010; Alverson et al., 2011). In addition to known genes, every plant mitochondrial genome has an additional 10% or more of putative open reading frames (ORF). Some of these frames are conserved across several plant species, while others can be unique. Recombination events between short repeats in there majority do not alter the known coding sequences and ORFs. However, cases of altered ORFs or coding sequences were found (Marienfeld et al., 1997; Kubo and Newton, 2008; Kitazaki and Kubo, 2010; Alverson et al., 2011), some of these cause cytoplasmic male sterility (CMS). The mechanisms of CMS differ and are specific for each case (Allen et al., 2007; Kubo and Newton, 2008; Kitazaki and Kubo, 2010).
The study of plant mitochondrial genomes revealed important information regarding the evolution of these genomes (Kitazaki and Kubo, 2010) and of entire eukaryotic systems as well. Every plant mtDNA has some sequences in common with that of all other plants, as well as species- or group-specific sequences. Closely related plants usually share significant portions of mitochondrial sequences, but in some cases their mitochondrial sequences differ remarkably. More sequencing data are needed to supply sufficient information for a detailed comparative study of different phylogenetic groups of plants. Here sequencing data and comparative analysis are provided for the mitochondrial genome of the legume V. faba.
Materials and Methods
Mitochondrial DNA Isolation, Library Construction, Genome Sequencing, and Assembly
Mitochondria were isolated from 6 to 7 days, dark grown, etiolated seedlings of V. faba cultivar Broad Windsor (Territorial Seed Company, Cottage Grove, OR, USA) using DNAse I protocol. Purified mitochondria were lyzed and mtDNA was isolated as reported (Synenki et al., 1978). Three libraries were constructed:
1. mtDNA was digested by BamHI and cloned in BamHI digested pUC19 plasmid vector.
2. mtDNA was digested by ApoI and cloned in EcoRI digested pUC19 plasmid vector.
3. A third library was generated by Genomex Biotechnology Company (Genomex appears to be a trading name of Amplicon Express, http://www.amplicon-express.com); mtDNA was mechanically nicked and 25–45 kb DNA fragments were cloned into the fosmid vector pEpiFOS-5. This library contains ∼1500 clones with an average insert size of 35 kb.
About 400 BamHI fragments were sequenced from both strands with sequence overlap of at least 100 bp for each primer. “Difficult” sequences with compression or large number of homonucleotide stretches were sequenced a few more times using different primers for both strands until the sequence became clear. For sequencing an Applied Biosystems four-capillary sequencing machine 3130 Genetic Analyzer with 55 cm column and corresponding sequencing kits was used. Among 400 sequenced fragments were 119 unique BamHI fragments.
From the ApoI library we isolated 1050 clones; 158 of these were selected containing at least one BamHI recognition site (∼30% of them contained inserts with two or three BamHI sites). These clones were thoroughly sequenced. Most of them overlapped partially or fully with already sequenced BamHI fragments and revealed the arrangement of these fragments. ApoI library sequencing led to additional 11 BamHI fragments.
Finally, 234 long fosmid clones ranging from 25 to 45 kb with average around 35 kb were sequenced. For direct sequencing of these fragments at a good quality, the PCR program was: 95°C for 5 min, followed by 50 cycles of 95°C for 30 s, 55°C for 10 s, 60°C for 4 min, and hold at 4°C.
For most of the sequencing cycles, primers known from BamHI and ApoI library sequencing were used, but at times new primers corresponding to new BamHI fragments were applied. By sequencing these fosmid DNA fragments 33 new unique BamHI fragments mainly representing recombinant versions of known fragments were found.
In total∼9 × 106 bp of V. faba mtDNA were sequenced corresponding a 15-fold coverage of the master chromosome. Of 163 unique BamHI fragments 144 could be included into the master chromosome sequence. Nineteen other fragments were recombinant versions between some of the 144 BamHI fragments belonging to the master chromosome. Computer alignment of all BamHI fragments and 234 fosmid DNA inserts allowed the construction of a 588,000 kb circular master chromosome that contained all unique sequences of V. faba mtDNA and was submitted to the GenBank database with accession number KC189947. Computation analysis was conducted using NCBI tools.
Results and Discussion
Vicia faba Mitochondrial DNA Sequencing Data Compared to Sequences Earlier Reported
Comparative analysis of our data and results previously published by Wahleithner and Wolstenholme (1988b), MacFarlane et al. (1990a,b), and Wahleithner et al. (1990), as expected, showed very high similarity. Few single nucleotide substitutions were found mainly in non-coding regions. Protein sequences of cob (392aa), atp9 (both 88 and 74aa), cox3 (265aa), and nad1 (325aa) were 100% identical. The atp6 protein (291aa) sequences had just one amino acid difference – isoleucine versus leucine.
A difference was found between rps14 coding sequences (100 amino acid length). In our version, it was glycine in position 85 instead of serine reported by Wahleithner and Wolstenholme (1988b). Multiple alignments between ribosomal protein S14 sequence and protein database showed that glycine is a standard amino acid on this position. The V. faba sequence is identical to rps14 of Pisum sativum (Hoffmann et al., 1999). The minor differences between known rps14 of different plant mitochondria never concerned glycine in this position.
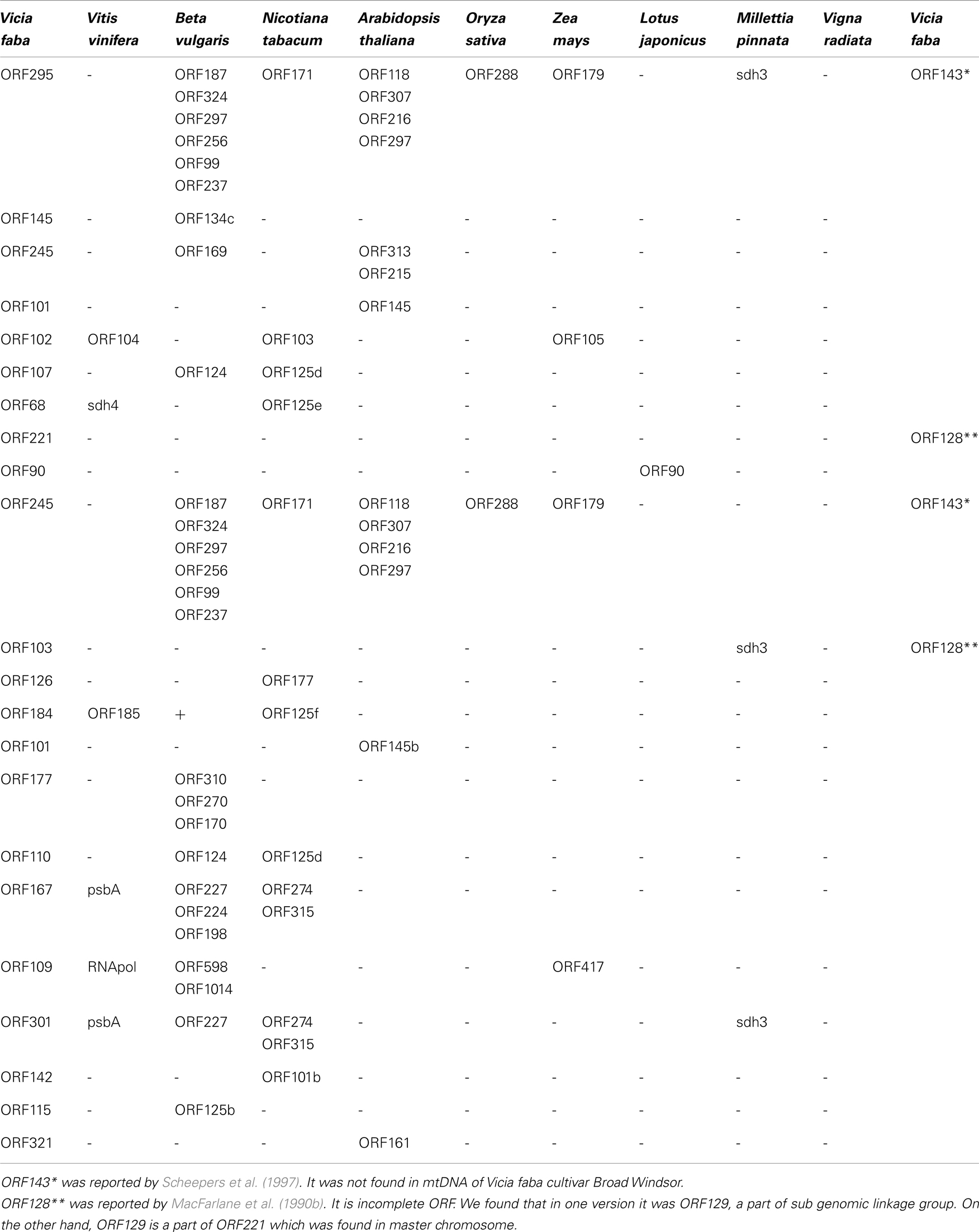
Significant differences of mtDNA sequences between cultivar Broad Windsor and another cultivar of V. faba were found as reported (Scheepers et al., 1997) around ORF143 near exon c of the nad5 gene. Actually, the Broad Windsor mitochondrial genome lacks a full size ORF143. Instead, it has ORF295 and ORF245. Amino acid sequences for nad5 protein exons c, d, and e related are identical.
The Master Chromosome Structure
Computer alignment of all V. faba mitochondrial BamHI fragments and 234 mtDNA fragments (25–45 kb) cloned in fosmids allowed to construct a 588,000 kb circular master chromosome with 45.04% GC content (Figure 1). V. faba shared about 40% similarity with mtDNA of the legumes Lotus japonicus, Millettia pinnata (Kazakoff et al., 2012), Glycine max (Chang et al., 2013), and Vigna radiata (Alverson et al., 2011). For all other known plant mitochondrial genomes homology was 25% and lower. Eleven large (>500 bp) repeats were found in the master chromosome: eight direct and three inverted ones. The largest repeat comprises 66,893/66,897 bp, the smallest 1,675 bp. Large repeats were highly similar (99%) or identical. Ten repeats have two, and one (the smallest) has three copies. The total size of large repeats covers 200,255 bp or 34% of the whole master chromosome size. The contribution of short (<500 bp) repeats has not been calculated but should not change significantly the complexity of the V. faba mitochondrial genome of 387,745 bp.
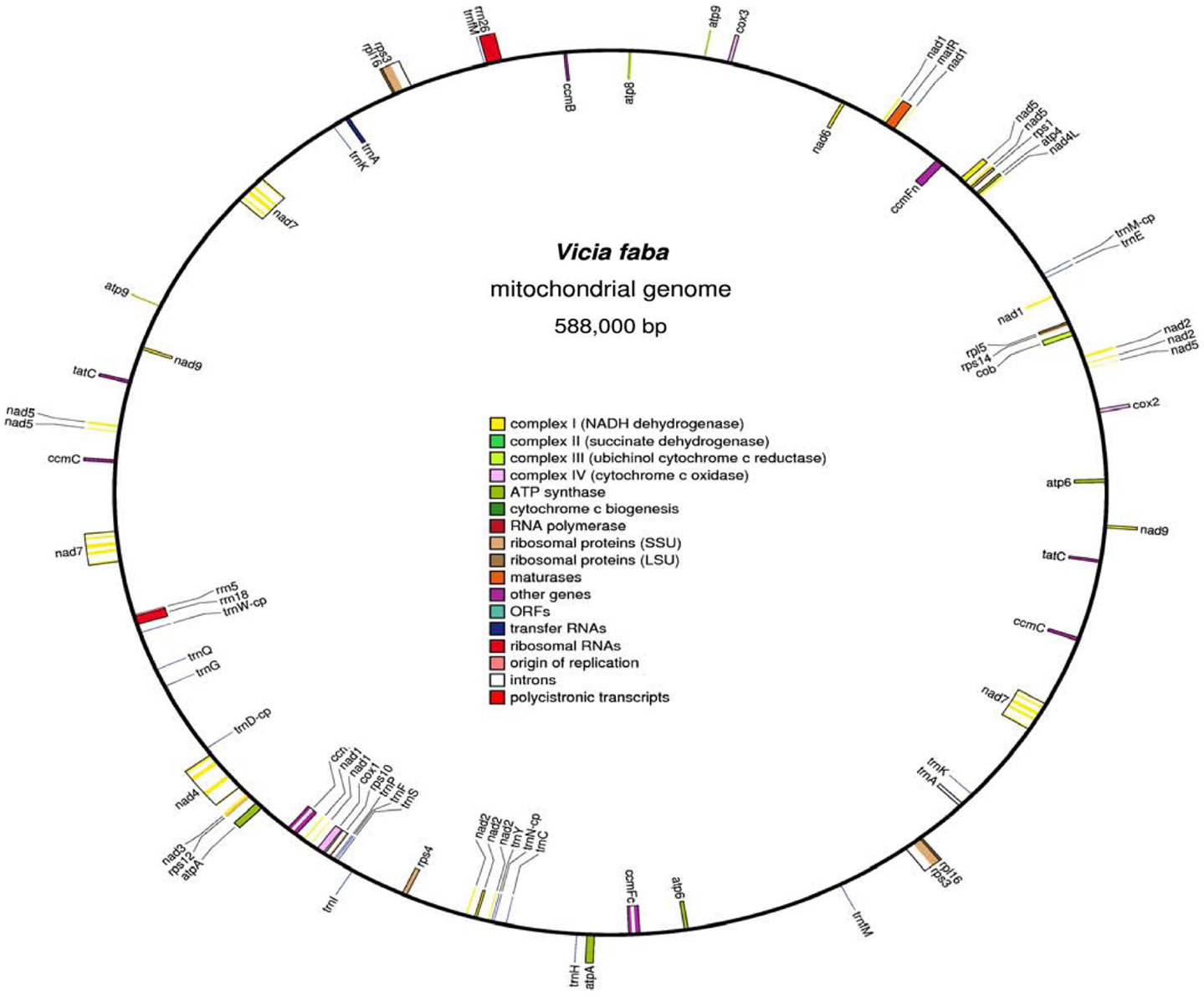
Figure 1. The circular map of Vicia faba mitochondrial genome. Clockwise and counter-clockwise strands are drawn on the inside and outside of the outer circle, respectively. GC content is shown in the inner circle. The map was drawn by using OGDRAW (Bock et al., 2007).
Three types of protein or putative polypeptide related sequences were found in Vicia faba mtDNA:
1. Conservative sequences, common to all other plants.
2. Aberrant putative ORFs unique to the specific plant or to a small group of plants.
3. Short fragments of known polypeptides which are not organized into ORF.
Thirty five mitochondrial genes encoding conserved proteins, 3 ribosomal RNAs and 17 tRNA genes were found. The V. faba master chromosome contained all nine nad genes of Complex I, the cob gene of Complex III, three cox genes of Complex IV, five atp genes of Complex V, four ccm genes of cytochrome c biogenesis, and 7 out of 16 possible ribosomal protein genes. For sdh3 and sdh4 genes of Complex II and for rps7 only fragments encoding the corresponding polypeptide sequences were found. For sdh3 and sdh4 these fragments appeared as parts of unknown ORFs.
Amino acid sequence alignment revealed an 110aa rps7-related mitochondrial polypeptide for Pisum sativum and Lotus japonicus. In V. faba it was divided by a stop codon instead of serine in position 73 into two polypeptides of 72 and 37aa which were not organized into ORF. The genes nad9, atp6, atp9, ccmC, rpl16, rps3, and tatC were duplicated. Gene nad5 exons D and E were also duplicated. The gene nad7 with all five exons was triplicated. Additionally, exon 5 of nad7 was copied to other locations.
Comparative analysis between the master chromosome and minicircles (Wahleithner and Wolstenholme, 1987) did not reveal any significant sequence homology longer than a few dozen nucleotides. The origin and direction of replication for V. faba mitochondrial plasmids determined by Wahleithner and Wolstenholme (1988a) were aligned with the master chromosome sequence. Few dozens of 10–13 bp sequences, homologous to the core motif AGGAA, with few nucleotides flanking this motif were found on the master chromosome.
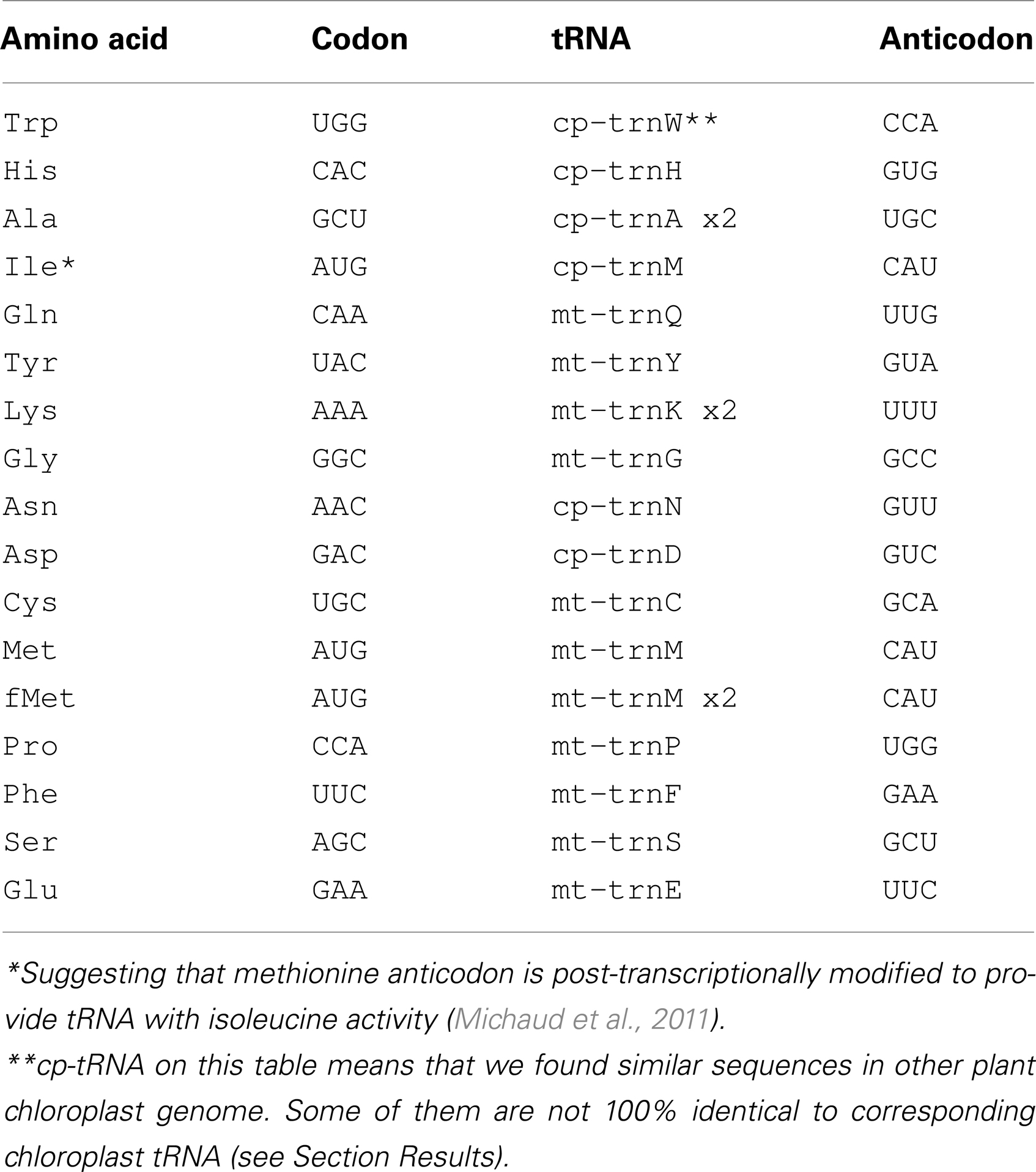
Six of the tRNA genes (Table 1) were highly homologous to chloroplast genome sequences.
Retrotransposon-Related Sequences of V. faba Mitochondrial DNA
All three classes of nuclear retrotransposon-related sequences were found within the V. faba mitochondrial genome; the Ty1/copia subclass, a non-LTR retroelement reverse transcriptase and an ORF with notable homology to the RNase H and reverse transcriptase domains of the Ty3/gypsy superfamily. These sequences were represented by relatively short ORFs (encoding less than 200aa) with high similarity to large nuclear ORFs (>1000aa), similar as described previously (Knoop et al., 1996).
Mitovirus Related Sequences
The open reading frame ORF128, in position 4678–5064 of V. faba mtDNA revealed high similarity to the RNA-dependent RNA polymerase region of virus pfam05919 belonging to Mitoviruses of the family Narnaviridae. Mitoviruses are simple viruses that invade fungal mitochondria without forming true capsids (Cole et al., 2000). Their genomes consist of one gene encoding RNA-dependent RNA polymerase. ORFs representing part of mitoviral RNA polymerase were found in mitochondrial genomes of Arabidopsis thaliana (Marienfeld et al., 1997; Hong et al., 1998), Brassica napus (Tuomivirta and Hantula, 2005), and Vitis vinifera (Goremykin et al., 2009). A small part of mitovirus related sequences was reported previously for V. faba mtDNA (Marienfeld et al., 1997).
Open Reading Frames of V. faba Mitochondrial DNA
The total number of genes encoding conservative proteins, rRNAs, and tRNAs was 52. In addition, we found 114 unnamed ORF, in there majority more than 100aa long. NCBI BLAST analysis revealed three groups of ORFs:
1. 36 ORFs with no significant homology to any known proteins
2. 29 ORFs with significant homology to Medicago truncatula nuclear genome. Some of them were also homologous to other plant mitochondrial ORFs
3. 49 ORFs with significant homology to other plant mitochondrial or nuclear ORFs.
Some ORFs from groups 2 and 3 might represent novel alternative splicing variants formed by exonization of non-coding DNA sequences (Chen et al., 2012). Other ORFs consisted in part of known conservative protein encoded by non-mosaic genes together with parts of unknown sequences as part of ORFs from V. faba, or from other plant mitochondria.
V. faba ORFs with Significant Homology to Other Plant Mitochondrial ORFs
NCBI BLAST analysis of unique mtORFs of V. faba revealed very low similarity to those of known closely related legumes. Three different V. faba ORFs showed homology to parts of Millettia pinnata sdh3, one to the ORF90 of Lotus japonicus and none to Vigna radiata. Nevertheless, a several sequence homologies were found between V. faba, Beta vulgaris, Nicotiana tabacum, Vitis vinifera, and even the monocots Oryza sativa, Zea mays (Table 2).
V. faba ORF143 was reported by Scheepers et al. (1997). Both Ad/N and Ad/447 line mitochondria possessed ORF143 but it was not found in Broad Windsor. Instead we found two longer ORF295 and ORF245. In the case of ORF295, the first 28 amino acids are highly homologous to the first 28 amino acids of nad3. The central part of ORF245 has a high homology, with the central part of ORF295 but both N and C ends of these ORFs are different. Note that both Beta vulgaris and Oryza sativa mitochondria also possess multiple ORFs, partially homologous to V. faba ORF295, ORF245, and ORF143 (Table 2). We also found that some of our unique ORFs were homologous to ORFs in mitochondrial genomes of Daucus carota, Citrullus lanatus, Lupinus luteus, Brassica napus, Boea hygrometrica, Phoenix dactylifera, Glycine max, Phaseolus vulgaris.
Chloroplast-Specific Insertions in Vicia faba Mitochondrial DNA
The chloroplast genome of V. faba has not yet been sequenced. Therefore, chloroplast sequences of Medicago truncatula, Arabidopsis thaliana, and Glycine max were used to find chloroplast-derived insertions in V. faba mtDNA. Analysis of M. truncatula chloroplast-specific (cp) sequences revealed 10 fragments (four of them duplicated) in V. faba mtDNA, ranging from 77 to 1389 nt with similarity of 74–97%. The sequences homologous to cpDNA comprise 1.1% of V. faba mtDNA. Six of the 10 cpDNA fragments contained tRNA genes (tRNAAla, tRNATrp, tRNAAsn, tRNAAsp, tRNAHis, and tRNAMet). The chloroplast-encoded tRNAAla gene contained one intron. Both exons of tRNAAla, as well as the tRNATrp gene sequence, were 100% homologous to V. faba mtDNA. For four other cp-tRNA genes the identity was <100%. Almost all of these sequences were also found within the M. truncatula nuclear DNA. In addition to the tRNA genes, cp, and mtDNA share homologous sequences encoding fragments of 16S and 23S ribosomal RNA as well as fragments of proteins rpl12 and ycf68.
Sequence Homology between Vicia faba Mitochondrial DNA and Medicago truncatula Nuclear Genome
Homology analysis between V. faba mtDNA and the nuclear genome of A. thaliana using NCBI BLAST search revealed about 20% of homologous mtDNA. For the related legume Glycine max, the homology was slightly higher (∼27%).
A high level of co-linearity was found earlier between the nuclear linkage groups of the legumes V. faba and M. truncatula, despite the large differences in genome size (Ellwood et al., 2008; Young et al., 2011; Alghamdi et al., 2012). Chromosome mapping demonstrated an evidence of shared macrosynteny between V. faba and M. truncatula nuclear genomes (Ellwood et al., 2008). The nuclear genome of M. truncatula has recently been sequenced (Young et al., 2011). So, it was logical to look at possible similarities between V. faba and M. truncatula on the nucleotide and amino acid sequence level. NCBI BLAST search revealed about 45% of V. faba mtDNA sequence being homologous to M. truncatula nuclear sequences. This is more homology than found with any sequenced plant mitochondrial genome. Thirty five percent of homologous sequences range from a few dozen to 12,806 bp and are located on chromosome 1.
In this publication we present some data interesting in aspect of sequence relationships between mitochondrial and nuclear genomes. When we analyzed homology between these two genomes in the area of mitochondrial 5S (rrn5), 18S (rrn18), and rps10 genes we found significant (99%) DNA sequence homology overlapping 5S, 18S, ORF134, ORF198, trnW-cp, and about 8000 bp of following uninterrupted sequence homology (positions, complement 320126–333187 bp). We found in this area large ORF1152 annotated as putative ribosomal protein S10 in Medicago truncatula chromosome 1 (Figure 2) (sequence encoding this ORF overlapped genes of rrn5 and rrn18 ribosomal RNA with 99% homology as well as ORF135). It was a transcribed mosaic gene with 18 exons. ORF1152 amino acid sequence was fused with V. faba mtDNA gene of rps10 highly homologous to mitochondrial genomes of many plants (position, complement (383419–384753). Thus, we found a transcribed nuclear genome sequence organized into ORF1152, which contained sequences of rrn5 and rrn18, and fused with amino acid sequence covering mitochondrial rps10 gene located 63000 bp apart. And it was not just a single case. We found two more transcribed ORFs: ORF1116 and ORF856, in different positions of Medicago truncatula chromosome 1 (Figure 3). First halves of these ORF are similar and alternatively spliced. Second halves are different. Both of them are fusions between ATP synthase subunit alpha and cox2 genes but for ORF856 homology to cox2 gene was much more significant (Figure 3). In addition to ATP synthase subunit alpha and cox2 genes two unnamed protein products ORF134 and ORF198 were found in V. faba mtDNA. ORF134 and ORF198 were homologous to second half of ORF1116 but not to ORF856. At the same time, ORF1152 and ORF1116 nucleotide sequences shared around (99%) of 1700 bp complementary nucleotide sequence homology.
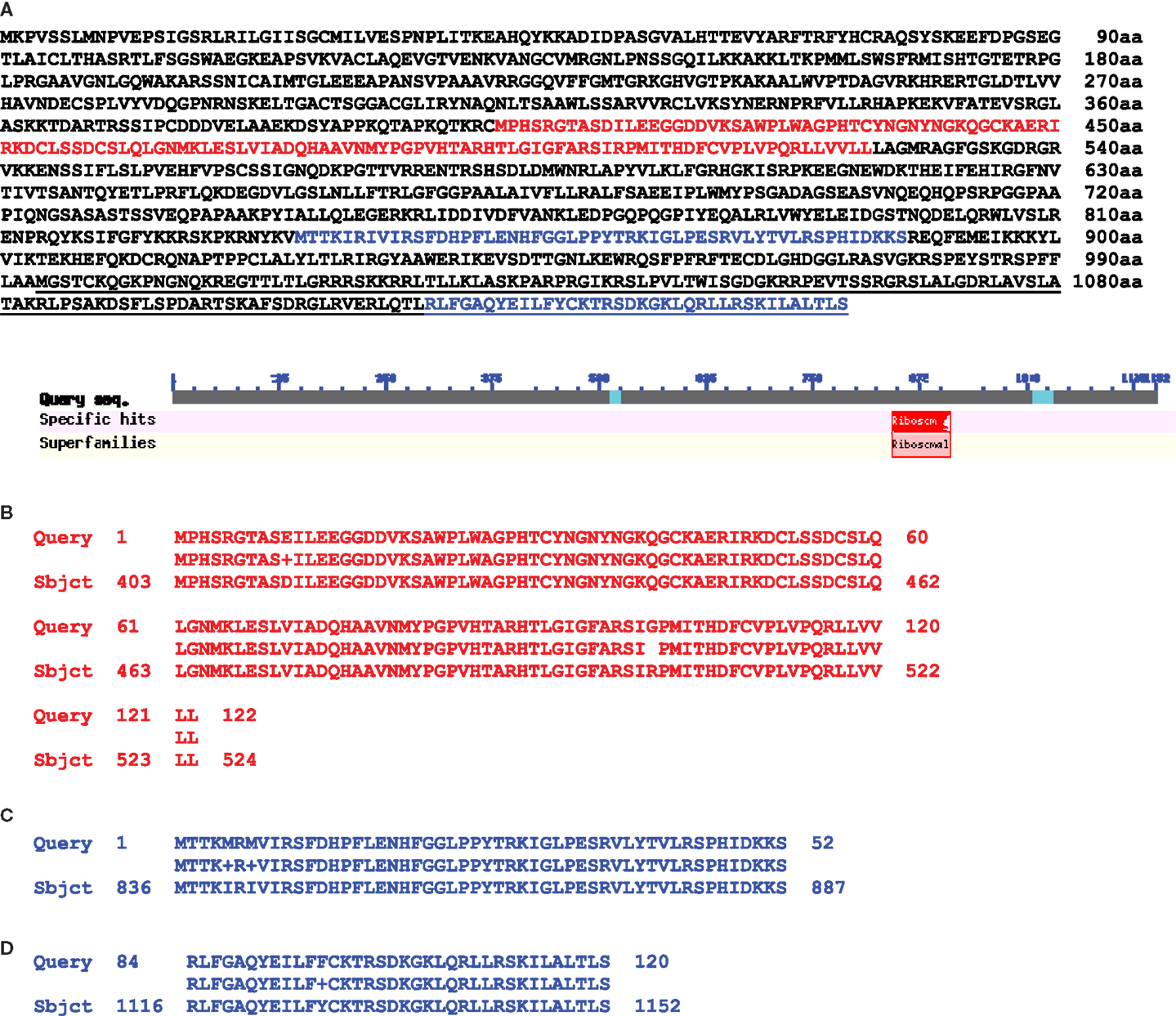
Figure 2. Protein alignment between M. truncatula nuclear putative ORF1152 and Vicia faba mitochondrial ORF135 and gene rps10. (A) Amino acid sequence of M. truncatula chromosome 1 putative ORF1152 (NCBI BLAST). (B) Red colored sequence which is a part of mitochondrial ORF135. (C,D) Blue colored parts of the sequence which is a Vicia faba CDS of rps10 gene. (C) An exon 1 sequence and (D) is an exon 2 sequence. Underlined is a sequence of nuclear alternative gene encoding putative rps10 (159aa).

Figure 3. Protein alignment between M. truncatula nuclear putative ORF1116 and ORF856 and Vicia faba mitochondrial genes atpA and cox2. (A) Amino acid sequence of M. truncatula chromosome 1 putative ORF1116 and (B) ORF856 (NCBI BLAST). Red colored is a homologous sequence between ORF1116 and ORF856. Black colored are different amino acids within area of homology between ORF1116 and ORF856 (red colored). Underlined is a perfect (99%) homology between Vicia faba mitochondrion gene of ATP synthase F1 subunit 1 and ORF1116. (C) A blue colored homology between mitochondrial ORF134 and ORF1116. (D) A green colored homology between mitochondrial ORF198 and ORF1116. (E) Homology between Vicia faba mitochondrion cox2 gene (259aa) and ORF1116. (F) Homology between Vicia faba mitochondrion cox2 gene (259aa) and ORF856.
tRNA Related Sequences in Medicago truncatula Nuclear Genome
Here we present data only for tRNAs found common for chloroplast and mitochondrial genomes. For mitochondrial tRNAAla, we found homology with M. truncatula nuclear genome only for exon 2 of tRNAAla. All other full size tRNA sequences common both for chloroplast and mitochondrial genomes were found in M. truncatula nuclear genome. Four copies of V. faba tRNATrp sequence found in chromosome 1, and 1 copy in chromosome 3, 4, and 8 each. In chromosome 1, tRNATrp sequence was found as a part of genes encoding ORF76 (2 copies in opposite orientation), ORF321 and ORF329. All three genes were transcribed and had mosaic structure. ORF76 had 2 exons, ORF321 and ORF329 had 6 exons each. For ORF76 and ORF321 positions of exon 1, following intron and exon 2 were the same. In all three ORFs sequence complementary to tRNATrp gene covered exon 1, starting from nucleotide 8 until the end, and part of the following intron. For ORF76 and ORF321, tRNATrp sequence was 100% homologous to mitochondrial and chloroplast sequences. For ORF329, it was a one point mutation (Figure 4). Amino acid sequence alignment showed that there is a difference between amino acid sequences of ORF76 and ORF321 compared to ORF329 which could be a result of alternative splicing (Figure 4B).

Figure 4. Nucleotide (A) and protein (B) alignment between M. truncatula nuclear putative ORF76, ORF321, and ORF329 and Vicia faba mitochondrial gene tRNATrp. (A) A nucleotide sequence of exon 1 (red color) in putative ORF76, ORF321, and ORF329 (NCBI BLAST). Highlighted with yellow is tRNATrp in complementary orientation. Black color is intron sequence. Green color shows point mutation. (B) An alignment between exon 1 (first 18 amino acids) and part of exon 2 sequences. Query is a sequence of ORF76 or ORF321, Sbjct is a sequence of ORF329.
tRNATrp gene copies, as well as other four tRNA genes common for chloroplast and mitochondrial genomes, were located in chromosomes 1 and 4. For tRNATrp it was also found in chromosomes 3 and 8, for tRNAAsp – in chromosome 5, for tRNAHis – in chromosome 7, for tRNAMet – in chromosomes 3, 5, and 7, for tRNAAsn – in chromosome 7.
It was reported earlier that tRNAs in addition to their traditionally known role in translation might be involved in the regulation of transcript profiles (Irmer et al., 2010; Rogers et al., 2012). Computation analysis of tRNA sequences found in V. faba mitochondrion revealed that in the M. truncatula nuclear genome these tRNA sequences can be found as a part of different ORFs. Some of them were a part of exon or complementary to the part of exon; others were on the exon-intron junction point or a part of introns. All these sequences were part of transcripts, which suggests some active role. This role may be different in each specific case, but what attracted our attention is the fact that many of these tRNA sequence copies (not all) in M. truncatula nuclear genome were a part of some kind of transcribed ORF.
Possible Sequence Relationship between Plant Mitochondrial and Nuclear Genomes
Summarizing data related to mitochondrial rrn5, rrn18, rps10, ATP synthase subunit alpha, cox2, and tRNA sequences as a part of nuclear transcribed ORFs led to the following conclusions:
1. V. faba mtDNA sequences can be organized into Medicago truncatula nuclear ORFs comprising various mitochondrial gene fragments. We present in this article genes rps10, atpA and cox2, ORF135, ORF134, and ORF198. But we found more such examples.
2. These ORF sequences are transcribed and spliced in the nuclear genome.
3. These ORFs may occur in nuclear genome in several versions representing variants of the same gene as result of alternative splicing or of recombination between ancestral ORFs.
4. Nuclear ORF genes comprise not only fragments of sequences encoding V. faba mitochondrial proteins but also rrn5, rrn18, or tRNA genes being analyzed in this work.
5. It is not clear whether rRNA or tRNA related sequences are translated (not previously reported) but their transcripts suggest a role in gene regulation.
6. We suggest that at least part of these ORFs could via gene duplication, recombination, and alternative splicing contribute to evolutionary innovation of genomes (Chen et al., 2012).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Author wishes to express his gratefulness and gratitude to Dr. I. Schubert, Dr. J.-H. Weil, Dr. L. Maréchal-Drouard, Dr. Marc Boutry, and Dr. H. Belmont for their support, help, and critical reading and comments on an earlier version of the manuscript. This work was supported by grants from US Department of Education (STEM Ladder to Student Success Project, Award # P031C110011 and STEM-TRAC PR Award # P031C110190).
References
Albert, B., Lelandais, C., Pla, M., Leuret, C., Vitart, V., Mathieu, C., et al. (2003). Amplification of Nicotiana sylvestris mitochondrial subgenomes is under nuclear control and is associated with phenotypic changes. Genetica 117, 17–25.
Alghamdi, S. S., Migdadi, H. M., Ammar, M. H., Paull, J. G., and Siddique, K. H. M. (2012). Faba bean genomics: current status and future prospects. Euphytica 186, 609–624.
Allen, J. O., Fauron, C. M., Minx, P., Roark, L., Oddiraju, S., Lin, G. N., et al. (2007). Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics 177, 1173–1192.
Alverson, A. J., Zhuo, S., Rice, D. W., Sloan, D. B., and Palmer, J. D. (2011). The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS ONE 6:e16404. doi:10.1371/journal.pone.0016404
Arrieta-Montiel, M., Lyznik, A., Woloszynska, M., Janska, H., Tohme, J., and MacKenzie, S. A. (2001). Tracing evolutionary and developmental implications of mitochondrial stoichiometric shifting in the common bean. Genetics 158, 851–864.
Backert, S., and Börner, T. (2000). Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr. Genet. 37, 304–314.
Bellaoui, M., Martin-Canadell, A., Pelletier, G., and Budar, F. (1998). Low-copy-number molecules are produced by recombination, actively maintained and can be amplified in the mitochondrial genome of Brassicaceae: relationship to reversion of the male sterile phenotype in some cybrids. Mol. Gen. Genet. 257, 177–185.
Bendich, A. J. (1993). Reaching for the ring: the study of mitochondrial genome structure. Curr. Genet. 24, 279–290.
Bock, R., Lohse, M., and Drechsel, O. (2007). Organellar Genome DRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 52, 267–274.
Chang, S., Wang, Y., Lu, J., Gai, J., Li, J., Chu, P., et al. (2013). The mitochondrial genome of soybean reveals complex genome structures and gene evolution at intercellular and phylogenetic levels. PLoS ONE 8:e56502. doi:10.1371/journal.pone.0056502
Chen, L., Tovar-Coronam, J. M., and Urrutia, A. O. (2012). Alternative splicing: a potential source of functional innovation in the eukaryotic genome. Int. J. Evol. Biol. 2012, 596274.
Cole, T. E., Hong, Y., Brasier, C. M., and Buck, K. W. (2000). Detection of an RNA-dependent RNA polymerase in mitochondria from a mitovirus-infected isolate of the Dutch Elm disease fungus, Ophiostoma novo-ulmi. Virology 15, 239–243.
Ellwood, S. R., Phan, H. T., Jordan, M., Hane, J., Torres, A. M., Avila, C. M., et al. (2008). Construction of a comparative genetic map in faba bean (Vicia faba L.); conservation of genome structure with Lens culinaris. BMC Genomics 9:380. doi:10.1186/1471-2164-9-380
Goremykin, V. V., Salamini, F., Velasco, R., and Viola, R. (2009). Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol. Biol. Evol. 26, 99–110.
Hartmann, C. H., Recipon, H., Jubier, M. F., Valon, C., Delcher-Besin, E., Henry, Y., et al. (1994). Mitochondrial DNA variability detectable in a single wheat regenerant involves a rare recombinant event across a short repeat. Curr. Genet. 25, 456–464.
Hoffmann, M., Dombrowski, S., Guha, C., and Binder, S. (1999). Cotranscription of the rpl5-rps14-cob gene cluster in pea mitochondria. Mol. Gen. Genet. 261, 537–545.
Hong, Y., Cole, T. E., Brasier, C. M., and Buck, K. W. (1998). Evolutionary relationships among putative RNA-dependent RNA polymerases encoded by a mitochondrial virus-like RNA in the Dutch elm disease fungus, Ophiostoma novo-ulmi, by other viruses and virus-like RNAs and by the Arabidopsis mitochondrial genome. Virology 20, 158–169.
Irmer, H., Hennings, I., Bruchhaus, I., and Tannich, E. (2010). tRNA gene sequences are required for transcriptional silencing in Entamoeba histolytica. Eukaryotic Cell 9, 306–314.
Janska, H., Sarria, R., Woloszynska, M., Arrieta-Montiel, M., and MacKenzie, S. A. (1998). Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell 10, 1163–1168.
Kanazawa, A., Tsutsumi, N., and Hirai, A. (1994). Reversible changes in the composition of the population of mt DNAs during dedifferentiation and regeneration in tobacco. Genetics 138, 865–870.
Kazakoff, S. H., Imelfort, M., Edwards, D., Koehorst, J., Biswas, B., Batley, J., et al. (2012). Capturing the biofuel wellhead and powerhouse: the chloroplast and mitochondrial genomes of the leguminous feedstock tree Pongamia pinnata. PLoS ONE 7:e51687. doi:10.1371/journal.pone.0051687
Kitazaki, K., and Kubo, T. (2010). Cost of having the largest mitochondrial genome: evolutionary mechanism of plant mitochondrial genome. J. Bot. 2010, 1–13.
Knoop, V., Unseld, M., Marienfeld, J., Brandt, P., Sünkel, S., Ullrich, H., et al. (1996). Copia-, gypsy- and LINE-like retrotransposon fragments in the mitochondrial genome of Arabidopsis thaliana. Genetics 142, 579–585.
Kubo, T., and Mikami, T. (2007). Organization and variation of angiosperm mitochondrial genome. Physiol. Plant 129, 6–13.
Kubo, T., and Newton, K. (2008). Angiosperm mitochondrial genomes and mutations. Mitochondrion 8, 5–14.
Kubo, T., Nishizawa, S., Sugawara, A., Itchoda, N., Estiati, A., and Mikamia, T. (2000). The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNACys(GCA). Nucleic Acids Res. 28, 2571–2576.
MacFarlane, J. L., Wahleithner, J. A., and Wolstenholme, D. R. (1990a). A broad bean mitochondrial atp6 gene with an unusually simple, non-conserved 5’ region. Curr. Genet. 18, 87–91.
MacFarlane, J. L., Wahleithner, J. A., and Wolstenholme, D. R. (1990b). A gene for cytochrome c oxidase subunit III (COXIII) in broad bean mitochondrial DNA: structural features and sequence evolution. Curr. Genet. 17, 33–40.
Maréchal, A., and Brisson, N. (2010). Recombination and the maintenance of plant organelle genome stability. New Phytol. 186, 299–317.
Marienfeld, J. R., Unseld, M., Brandt, P., and Brennicke, A. (1997). Viral nucleic acid sequence transfer between fungi and plants. Trends Genet. 13, 260–261.
McDermott, P., Connolly, V., and Kavanagh, T. A. (2008). The mitochondrial genome of a cytoplasmic male sterile line of perennial ryegrass (Lolium perenne L.) contains an integrated linear plasmid-like element. Theor. Appl. Genet. 11, 459–470.
Michaud, M., Cognat, V., Duchéne, A. M., and Maréchal-Drouard, L. (2011). A global picture of tRNA genes in plant genomes. Plant J. 66, 80–93.
Oldenburg, D. J., and Bendich, A. J. (1996). Size and structure of replicating mitochondrial DNA in cultured tobacco cells. Plant Cell 8, 447–461.
Rogers, T. E., Ataidem, S. F., Dare, K., Katz, A., Seveau, S., Roy, H., et al. (2012). A pseudo-tRNA modulates antibiotic resistance in Bacillus cereus. PLoS ONE 7:e41248. doi:10.1371/journal.pone.0041248
Scheepers, D., Hong, L., and Boutry, M. (1997). Variant mitochondrial transcripts of a broad bean line are associated with two point mutations located upstream of the nad5 exon c. Plant Sci. 129, 203–212.
Synenki, R. M., Levings, C. S., and Shah, D. M. (1978). Physicochemical characterization of mitochondrial DNA from soybean. Plant Physiol. 61, 460–464.
Tuomivirta, T. T., and Hantula, J. (2005). Three unrelated viruses occur in a single isolate of Gremmeniella abietina var. abietina type A. Virus Res. 110, 31–39.
Vitart, V., De Paepe, R., Mathieu, C., Chetrit, P., and Vedel, F. (1992). Amplification of substoichiometric recombinant mitochondrial in a nuclear, male sterile mutant regenerated from protoplast culture in Nicotiana sylvestris. Mol. Gen. Genet. 233, 193–200.
Wahleithner, J. A., MacFarlane, J. L., and Wolstenholme, D. R. (1990). A sequence encoding a maturase-related protein in a group II intron of a plant mitochondrial nad1 gene. Proc. Natl. Acad. Sci. U.S.A. 87, 548–552.
Wahleithner, J. A., and Wolstenholme, D. R. (1987). Mitochondrial plasmid DNAs of broad bean: nucleotide sequences, complex secondary structures, and transcription. Curr. Genet. 12, 55–67.
Wahleithner, J. A., and Wolstenholme, D. R. (1988a). Origin and direction of replication in mitochondrial plasmid DNAs of broad bean, Vicia faba. Curr. Genet. 14, 163–170.
Wahleithner, J. A., and Wolstenholme, D. R. (1988b). Ribosomal protein S14 genes in broad bean mitochondrial DNA. Nucleic Acids Res. 16, 6897–6913.
Woloszynska, M. (2010). Heteroplasmy and stoichiometric complexity of plant mitochondrial genomes – though this be madness, yet there’s method in it. J. Exp. Bot. 61, 657–657.
Woloszynska, M., and Trojanowski, T. (2009). Counting mtDNA molecules in Phaseolus vulgaris: sublimons are constantly produced by recombination via short repeats and undergo rigorous selection during substoichiometric shifting. Plant Mol. Biol. 70, 511–521.
Keywords: Vicia faba, mitochondrial genome, sequencing, Medicago truncatula, nuclear genome
Citation: Negruk V (2013) Mitochondrial genome sequence of the legume Vicia faba. Front. Plant Sci. 4:128. doi: 10.3389/fpls.2013.00128
Received: 14 March 2013; Accepted: 19 April 2013;
Published online: 07 May 2013.
Edited by:
Scott Jackson, University of Georgia, USAReviewed by:
Peter Langridge, Australian Centre for Plant Functional Genomics, AustraliaEiji Nambara, University of Toronto, Canada
Copyright: © 2013 Negruk. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Valentine Negruk, Biotechnology Research Lab, Miami Dade College, North Campus, 11380 NW, 27th Avenue, Miami, FL 33167, USA. e-mail:dm5lZ3JvdWtAbWRjLmVkdQ==