
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
HYPOTHESIS AND THEORY article
Front. Plant Sci. , 04 January 2013
Sec. Plant Physiology
Volume 3 - 2012 | https://doi.org/10.3389/fpls.2012.00300
This article is part of the Research Topic Retrograde Signaling in Plants View all 26 articles
Plant organelles produce retrograde signals to alter nuclear gene expression in order to coordinate their biogenesis, maintain homeostasis, or optimize their performance under adverse conditions. Many signals of different chemical nature have been described in the past decades, including chlorophyll intermediates, reactive oxygen species (ROS), and adenosine derivatives. While the effects of retrograde signaling on gene expression are well understood, the initiation and transport of the signals and their mode of action have either not been resolved, or are a matter of speculation. Moreover, retrograde signaling should be considered as part of a broader cellular network, instead of as separate pathways, required to adjust to changing physiologically relevant conditions. Here we summarize current plastid retrograde signaling models in plants, with a focus on new signaling pathways, SAL1-PAP, methylerythritol cyclodiphosphate (MEcPP), and β-cyclocitral (β-CC), and outline missing links or future areas of research that we believe need to be addressed to have a better understanding of plant intracellular signaling networks.
Chloroplasts originated from free-living cyanobacteria that were engulfed by the eukaryotic cell ancestor (Delwiche, 1999). This endosymbiotic event resulted in the transfer of thousands of genes from the cyanobacterial plastid ancestor into the nuclear genome of the host (Goksoyr, 1967; Martin et al., 2002). As a result of this gene transfer, the stoichiometry of nuclear and chloroplastic encoded polypeptides functioning in the plastids requires the coordination of both genomes. One such regulatory mechanism, called anterograde signaling, entails the coordination of plastid gene expression (PGE) by nuclear encoded proteins (Woodson and Chory, 2008). The reverse mechanism, or retrograde signaling, requires the transfer of signals from the plastids to the nucleus to regulate the expression of genes encoding both plastid-localized (Strand et al., 2003) and other proteins involved in many cellular processes (op den Camp et al., 2003; Koussevitzky et al., 2007; Pesaresi et al., 2009).
The biogenesis and functioning of plant organelles are coordinated with nuclear gene expression. Early stages of chloroplast development require the establishment of “biogenic” signals to coordinate the production of photosynthetic complexes and membranes (Oelmuller et al., 1986; Pogson et al., 2008). On the other hand, “operational” signals are important for the normal functioning of chloroplasts in mature plants (Pogson et al., 2008). Mature chloroplasts can act as environmental sensors as adverse environmental conditions such as high light (HL) and drought can cause energy imbalances leading to oxidative stress that will impair organellar and cellular function (Wilson et al., 2006). As a result, nuclear and plastidic gene regulation is required to reach homeostasis.
The first evidence that chloroplasts can regulate nuclear gene expression was obtained in the albostrians barley mutants deficient in plastid ribosomes (Bradbeer et al., 1979) and in Brassica plants treated with spectinomycin, an inhibitor of organelle protein synthesis (Zubko and Day, 1998). In both cases, bleached leaves were produced with decreased amount of nuclear encoded chloroplast proteins. These observations lead to the proposal that perturbation in plastidic processes give rise to plastid products, or signals that can control cytosolic protein translation.
Since then, different types of retrograde signaling pathways, depending on the trigger sources and signals, have been reported. One signaling pathway is associated with tetrapyrrole biosynthesis intermediates, like Mg-ProtoporphyrinIX (Mg-ProtoIX) (Strand et al., 2003) and haem (Woodson et al., 2011). A second type is initiated by changes in redox potential at the electron transport chain (Fey et al., 2005; Pfannschmidt et al., 2009). The production of reactive oxygen species (ROS), such as hydrogen peroxide (H2O2) and singlet oxygen (1O−2) by excess oxidative power is a third mechanism that can trigger specific changes in nuclear gene expression (Apel and Hirt, 2004; Galvez-Valdivieso and Mullineaux, 2010; Suzuki et al., 2012). Finally, there is a type of retrograde signaling associated with PGE (Bradbeer et al., 1979; Nott et al., 2006).
The “classical,” or linear, model of retrograde signaling describes that specific signals produced in the organelles by different developmental and environmental cues are able to move into the nucleus where they elicit specific gene regulation. Although there is a good understanding of some of the triggers, the nature, and the final outcomes related to gene expression for some of these proposed retrograde signals, some of the signals are still debated or their mechanism of actions poorly understood. This Perspectives article presents a synopsis of the current knowledge of metabolite plant retrograde signals with a focus on the recent reports of novel signals. We also attempt to identify missing gaps in current models and provide suggestions for future directions of research. Readers are referenced to pertinent reviews for further details regarding other signaling pathways (Apel and Hirt, 2004; Pogson et al., 2008; Woodson and Chory, 2008; Galvez-Valdivieso and Mullineaux, 2010; Pfannschmidt, 2010; Barajas-López et al., 2012).
Classical retrograde signals in plants generally involved artificially stressing the plant cells by treating the plants with the herbicide norflurazon (NF), which is an inhibitor of carotenoid biosynthesis that can perturb chloroplast development (Foudree et al., 2010). A mutant screen for altered expression of the nuclear genes encoding plastidic proteins during chloroplast development gene led to the discovery of the GENOMES UNCOUPLED (GUN) mutants (Susek et al., 1993). gun mutants are defective in the chloroplast-to-nucleus signal transduction that represses the expression of photosynthesis-associated nuclear genes (PhANG) genes such as Light Harvesting Complex b (LHCB) during perturbations of chloroplast development by NF.
At least two intermediates in the synthesis of photosynthetic pigments can act as plastidic signals to regulate nuclear gene expression. Treatment of wild type plants with NF not only inhibits the expression of the PhANG (Susek et al., 1993) but concomitantly induces 15-fold the levels of Mg-ProtoIX, the first committed precursor of chlorophyll. Genetic inhibition of Mg-ProtoIX production, such as in the gun2 and gun5 mutants (Mochizuki et al., 2001), which are defective in tetrapyrrole biosynthetic enzymes, results in misregulation of 70 out of 182 genes normally down-regulated in NF-treated wild type plants (Strand et al., 2003). Moreover, pharmacological approaches to accumulate Mg-ProtoIX, either by increasing its amount in the gun2 and gun5 mutants, or by feeding it to wild type plants, strongly support the hypothesis that Mg-ProtoIX is required for chloroplast-to-nucleus communication during early plant development (Strand et al., 2003; Kindgren et al., 2012).
Haem is a product of tetrapyrrole biosynthesis that acts as a positive retrograde signal from plastids in algae (von Gromoff et al., 2008). Evidence that haem could also be a potential signal in higher plants came from over expression of the Ferrochelatase 1 (FC) gene in the gain-of-function gun1-6D mutant. FC1 over expression leads to the accumulation of PhANGs in the presence of NF (Woodson et al., 2011). This “gun” phenotype can be rescued pharmacologically by decreasing the FC activity with Fe2+ chelator dipyridyl (DP). This response seems to be specific to the activity of FC1, as over expression of the other chloroplast-localized FC2 did not increase PhANG expression. Unexpectedly, and unlike Mg-ProtoIX, a reduction in total haem, rather than accumulation, occurs in the FC1 OX, and wild type plants after NF treatment (Woodson et al., 2011). This finding is in agreement with the ineffectiveness of haem feeding in seedlings to silence LHCB (Strand et al., 2003), but is in contrast with the effect of hemin (a more stable Fe substitute), which promoted global changes in gene expression in Chlamydomonas (von Gromoff et al., 2008; Voss et al., 2011). It is proposed that FC1 acts on specific chloroplastic haem pool, which can act as a positive retrograde signal exported from the chloroplasts (Woodson et al., 2011). Although haem can be exported from isolated chloroplasts (Thomas and Weinstein, 1990), the actual transport mechanism is unknown. The fact that there is no correlation between free haem levels and the gun phenotype indicates that the signaling haem may be bound to specific targets (Espinas et al., 2012). A possible scenario is that haem interacts with cytosolic or nuclear factors, such as in yeast (Zhang and Hach, 1999) or with haem-binding proteins to regulate gene expression. However, more work is required to identify downstream targets of haem and their mode of action.
Although initial evidence indicate that Mg-ProtoIX accumulates under oxidative stress in the cytosol and represses PhANG expression (Strand et al., 2003; Ankele et al., 2007; Pontier et al., 2007), some findings suggest a lack of correlation between the metabolite levels and gene expression (Gadjieva et al., 2005; Mochizuki et al., 2008; Moulin et al., 2008; Kakizaki et al., 2009; Zhang et al., 2012). Interestingly, another recent finding shows that oxidative stress induced by NF can induce transient accumulation of tetrapyrroles with concomitant repression of Lhcb in adult plants (Zhang et al., 2011). The contradictory results could be explained by differences in experimental conditions or technical issues related to the quantification of tetrapyrroles.
Mg-ProtoIX accumulation specifically inhibits the expression of genes harboring the CUF1 (G-Box) cis-element (Strand et al., 2003). Two alternative models have been proposed whereby an increase of Mg-ProtoIX promotes either the release of a transcriptional activator or the binding of a repressor (Gray, 2003; Strand et al., 2003). While the potential for Mg-ProtoIX to move into the cytosol has been actively debated, recently more details of a possible mechanism of action mechanism have been described. Mg-ProtoIX was found to bind to the cytosolic heat shock 90-type protein (HSP90) and inhibit the ATPase activity of HSP90 in vitro (Kindgren et al., 2012). Feeding and genetic experiments confirmed that the gun phenotype triggered by oxidative stress is partially suppressed when HSP90 is silenced and that it requires the action of Long Hypocotyl 5 (HY5) (Kindgren et al., 2012). Interestingly, HY5 binds to the promoter of photosynthetic genes (Lee et al., 2007). This type of signaling mechanism is analogous to that in yeast, where haem can interact with the complex HAP1-HSPs (Hon et al., 2001; Lee et al., 2002), which regulates proteins required for aerobic growth and oxidative damage control (Zhang and Hach, 1999). However, no direct interaction between HSP90 and HY5 has been reported yet in plants.
Despite a paucity of discovery of retrograde signals in the last decade, several signaling pathways have been recently proposed in the past 12 months. These novel signals include products of secondary metabolism (Estavillo et al., 2011; Xiao et al., 2012), oxidation products of carotenoids (Ramel et al., 2012), and dual localized proteins (Sun et al., 2011; Isemer et al., 2012) (Figure 1). These findings indicate that many metabolic pathways can act as potential sources of chloroplastic signals during different developmental stages of the plant or upon different stress responses. More importantly, unlike the previously discovered signals during artificial conditions, these new signals were identified during physiologically relevant stress responses like drought or HL. These new discoveries further support the concept that chloroplasts can indeed act as environmental sensors.
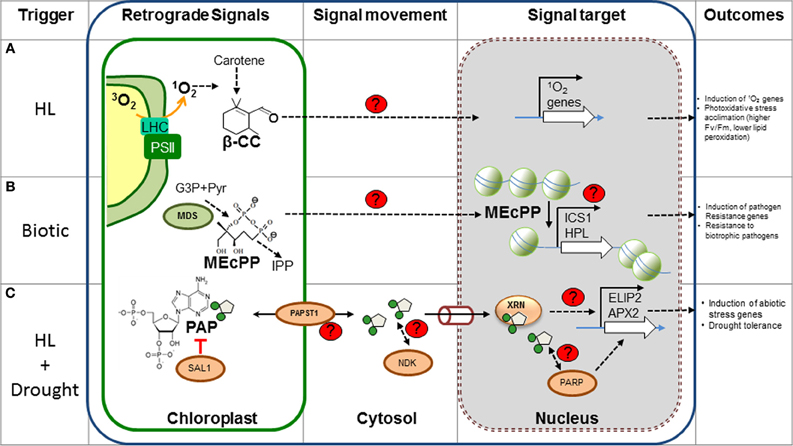
Figure 1. New metabolic retrograde signaling pathways. The different components of recently discovered plant retrograde signaling pathways are shown. (A) β-CC is most likely produced by oxidation of carotenes by 1O2 in the chloroplast and could diffuse through the membrane into the cytosol. Feeding β-CC results in the up-regulation of genes involved in stress responses, particularly those triggered by 1O2. It is speculated that the electrophilic carbonyl group could react with electron donors, such as sulfhydryl groups; however, the actual targets and mode of action are unknown. (B) MEcPP, an intermediary of isoprenoid precursors, is produced by MDS in the plastidic MEP pathway. MEcPP over accumulating mutants present high levels of SA and are resistant to biotrophic pathogens. MEcPP regulates the expression of the HPL and ICS1 gene, with concomitant production of SA. Although the transport and action mechanism are unknown, MEcPP could promote chromatin reorganization and induction of transcription of target genes. (C) PAP levels are catabolically regulated by SAL1 in the chloroplast and PAP transport is probably mediated by PAPST1. Cytosolic PAP could diffuse to the nucleus via the pores (cylinder) where it inhibits nuclear XRNs and affects gene regulation of stress inducible genes (i.e., APX2 and ELIP2). This mechanism is thought to play a role during drought, as PAP levels increase 30-fold. Other potential PAP targets (Nucleotide diphosphate kinase, NDK, and poly (ADP-ribose) polymerase, PARP, proteins) may mediate other aspects of signaling. The control mechanism of gene regulation by the XRNs proteins is a matter of investigation. Red lines, inhibition; black arrows, induction or activation; proteins involved in the signaling are indicated as ovals. Unknown components, processes or targets are indicated with red “?” or with dashed arrows. β-CC, β-cyclocitral; MEP, Methylerythritol phosphate pathway; MEcPP, methylerythritol cyclodiphosphate; MDS, MEcPP synthase; IPP, isopentenyl diphosphate; G3P, glyceraldehyde 3-phosphate; SA, salicylic acid; HL, high light. Figure adapted from Xiao et al. (2012) and Estavillo et al. (2011).
The dinucleotide 3′-phosphoadenosine 5′-phosphate (PAP) is a novel metabolite discovered to play a role as a plastid signal during drought and HL stress in Arabidopsis (Estavillo et al., 2011). PAP accumulates in plants lacking the phosphatase SAL1/FRY1, which degrades PAP into adenosine monophosphate (AMP) and phosphate (Chen et al., 2011; Estavillo et al., 2011). PAP is also increased by up to 30 fold in wild type plants during drought (Estavillo et al., 2011) and exhibits a smaller increase in response to HL. The sal1 mutants present up-regulation of 35% of the HL stress inducible genes, including Ascorbate Peroxidase 2 (APX2) and Early Light Induced Protein 2 (ELIP2), altered metabolome and 50% increased survival under water limiting conditions (Rossel et al., 2006; Wilson et al., 2009). PAP is produced as a byproduct of sulfonation reactions catalysed by cytosolic sulfo transferases (SOTs), whereby sulphate is transferred from 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to several metabolic substrates (Klein and Papenbrock, 2004). PAP levels are regulated by the dual localized SAL1 protein in both chloroplasts and mitochondria. Evidence that PAP can move between the chloroplast and nucleus was obtained by complementing PAP levels and APX2 expression in sal1 mutants with a SAL1 transgene targeted to the nucleus or chloroplast (Estavillo et al., 2011). Furthermore, a PAPS/PAP chloroplastic antiporter has now been reported that will facilitate exchange of PAP between the chloroplast and cytosol (Gigolashvili, 2012).
PAP most likely regulates gene expression by altering RNA metabolism mediated by 5–3′ exoribonucleases (XRNs). There are three XRNs in Arabidopsis; two encode the nuclear-localized XRN2 and XRN3 and are homologues of yeast Xrn2p/Rat1p. XRN2 and XRN3 act on uncapped RNAs like the excised hairpin loops in precursor miRNA transcripts (Kastenmayer and Green, 2000). The third gene, XRN4, encodes for a cytosolic XRN (homologous to yeast Xrn2p) that targets the resulting 3′ cleavage products of miRNA targets (Kastenmayer and Green, 2000; Souret et al., 2004). SAL1 and nuclear XRN mutants share similar morphological phenotypes (Gy et al., 2007), have improved drought tolerance and more than 50% of altered genes are co-regulated (Estavillo et al., 2011). Additionally, PAP can inhibit yeast XRN activity (Dichtl et al., 1997; van Dijk et al., 2011) and transcriptional regulation by HL of ELIP2 and APX2 genes is similar in Arabidopsis sal1 mutants (Estavillo et al., 2011). The mechanism by which XRNs modulate specific transcripts, such as APX2 during stress (i.e., impact on RNA post-transcriptional regulation, “degradome,” etc.) is unknown and merits further research.
A genetic screening for the constitutive expression of the stress inducible nuclear gene encoding the plastid-localized hydroxyperoxidase lyase (HPL) identified the isoprenoid precursor methylerythritol cyclodiphosphate (MEcPP) as a potential signal from the chloroplast (Xiao et al., 2012). MEcPP is converted to hydroxymethylbutenyl diphosphate (HMBPP) by 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase (HDS) during the production of universal isoprenoid precursors by the methylerithrytol phosphate (MEP) pathway in plastids (Rodríguez-Concepción, 2006). The mutant ceh1 (constitutively expressing HPL 1) accumulates MEcPP and HPL transcript, with up-regulation of salicylic acid (SA) production, and that of the ICS1 (encoding a SA-biosynthetic enzyme) transcript. This results in ceh1 mutants being more resistant to infection by Pseudomonas syringae, a biotrophic pathogen. Targeted silencing of all the enzymes in the MEP pathway indicates that only the accumulation of MEcPP is responsible for SA accumulation and induction of HPL. Moreover, feeding experiments demonstrates that MEcPP can directly regulate HPL. Thus, it seems that unlike the mechanism mediated by chlorophyll precursors where the flux through the pathway is important for retrograde signaling (Woodson et al., 2011), the induction of HPL is specifically triggered by MEcPP, and not by other intermediaries of the MEP pathway. Finally, the accumulation of MEcPP is also induced by wounding and HL stress with concomitant up-regulation of the HPL gene.
Strong evidence supports the hypothesis that MEcPP is an operational signal, and like PAP, demonstrates that mature chloroplasts can respond to external perturbations under physiologically relevant conditions. The gene response to MEcPP is specific and it does not involve regulation of PhANGs (Xiao et al., 2012). Comparative global gene expression analysis between ceh1 relative to wild type is needed to determine the extent of the signaling cascade by MEcPP and potential overlaps with other retrograde networks. Interestingly, oxidative stress in bacterial cultures induced MEcPP production (Ostrovsky et al., 1998) and MEcPP has been involved with the disruption of the interaction between chlamydial histone-like proteins and DNA. This would suggest that MEcPP could affect chromatin remodeling and gene expression (Grieshaber et al., 2006). However, direct evidence for MEcPP movement and its mechanism of action in plants is still lacking and deserves further investigation.
Carotenoids are considered to be one of the first lines of defense against 1O2 produced by the triplet excited chlorophyll during HL stress. Direct measurement of 1O2 and its regulated genes, indicates that production of 1O2 is one of the early responses to HL stress in Arabidopsis (González-Pérez et al., 2011; Ramel et al., 2012). Excess 1O2 under this condition leads to the production of oxidation products of carotenes. One of them, the volatile β-cyclocitral (β-CC), accumulates more than 1.5-fold after 1 h of stress treatment, which is accompanied by the induction of 1O2 stress-responsive genes (Ramel et al., 2012). More significantly, increasing doses of this volatile compound induces the up-regulation of 1O2, but not of H2O2, responsive genes. Microarray data also shows a striking specificity and similarity between gene expression profiles of β-CC plants and the flu mutant, which accumulates protochlorophyllide and presents a constitutive production of genes involved in oxidative stress by 1O2 (op den Camp et al., 2003). Finally, incubation of plants with β-CC prior to HL and cold stress prevented accumulation of lipid peroxidation and deterioration of PSII photochemical efficiency in a dose dependent manner, suggesting that β-CC increases photoxidative damage tolerance mechanisms. β-CC is a volatile short-chain compound, making diffusion a possible way of distribution from the place of origin to other parts of the cell. A likely action mechanism for β-CC is by reacting with sulfhydryl groups of proteins but more work is required to identify the target proteins and mode of action. Finally, it is possible that β-CC is a signal intermediate of the cascade triggered by 1O2 that relays the information to the cytosol.
The signals described above could be considered as metabolite signals, which in some instances are capable of moving out from the organelle to regulate nuclear gene expression. However, two examples of chloroplast-localized transcription factors (TF) that translocate to the nucleus and alter gene expression under specific conditions have been recently described. Arguably, metabolite signals that could be modulated by specific proteins better fit in the “signal” category (Leister, 2012), but identification of the existence of the mobile TF involved in retrograde signaling confirms the complexity of the mechanisms required to regulate nuclear gene expression.
The chloroplast envelope-bound PTM (PHD type transcription factor with transmembrane domains) is involved in retrograde signal pathways that regulate PhANGs expression under different types of stress conditions (Sun et al., 2011). The plant homeodomain (PHD) specifically binds methylated histones (H3K4me3) to promote transcription of downstream genes (De La Paz Sanchez and Gutierrez, 2009). Under normal conditions, PTM resides in the outer membrane of the chloroplast. However, a shorter version of PTM protein lacking the transmembrane domain can be found in the nucleus after stress treatment with NF, lincomycin and HL. Stress conditions induce proteolytic cleavage of the full length protein and its translocation to the nucleus. Interestingly, the ptm mutant has reduced expression of ABI4 and displays a gun phenotype (i.e., de-repression of LHCB in the presence of NF) similar to that of gun1, and abi4 during HL treatment. The involvement of PTM in the same pathway as GUN1 and ABI4 was confirmed by analyses of the corresponding double mutants. Interestingly, the levels of H3K4m3 methylation in the ABI4 promoter increased with the same stress treatments activating the PTM-dependent transcription of the ABI4 gene. An unexpected observation was the lack of constitutive repression of LHCB in the absence of NF in plant expressing soluble PTM. Although this may indicate that additional GUN1 mediated signals may be required, this work demonstrates that PTM mediates several retrograde signaling pathways.
Another mobile protein that could potentially be involved in retrograde signaling is Whirly1 protein (Isemer et al., 2012). Arabidopsis Whirly proteins (AtWhy1 and 3) are required for plastid genome stability (Marechal et al., 2009) and the barley homologue interacts with intron-containing plastidic RNA (Melonek et al., 2010). The barley Whirly 1 (HvWhy1) protein is dual localized to both chloroplast and mitochondria (Grabowski et al., 2008) with the same molecular weight for both forms. Unlike PTM, the AtWhirly1 protein of the same molecular weight was found in both chloroplasts and nucleus of the same cell. It was recently shown that recombinant AtWhirly1 can move from chloroplasts to the nucleus by transformation of tobacco plastids with a HA-AtWhirly1 fusion protein (Isemer et al., 2012). This clever approach demonstrated that HA-AtWhirly1 protein can translocate from the plastid to the nucleus via an unknown mechanism. Although pathogenesis related genes are up-regulated in the transgenic lines (Isemer et al., 2012), suggesting the possible involvement of Whirly1 in retrograde signaling during pathogen attack, the movement mechanism of Whirly [i.e., via stromules, diffusion, transport (Krause et al., 2012)], and gene targets have to be further investigated.
It is relatively easy to envision a linear type of mechanism whereby the available pool of signal interacts with target proteins to regulate gene expression (Estavillo et al., 2011; Kindgren et al., 2011; Xiao et al., 2012). The other option is that one signal can target many proteins or regulate the expression of many genes involved in different pathways. Rather than a linear succession of events, this would represent an intricate network which could provide more subtle levels of regulation under different and specific conditions (Leister, 2012). We outline some of the approaches utilized to discover new retrograde signaling pathways and components (Table 1).
Unlike the case of photosynthetic derived signals, it is less clear how the different triggers are sensed in most retrograde signaling pathways. For example, how is the protease mediated cleavage of PTM regulated by HL (Sun et al., 2011), or the accumulation and transport of PAP induced by drought and HL (Estavillo et al., 2011)? These are difficult questions to address as they may involve the interaction of many different signaling networks.
The movement of the signal between organelles is a key feature of a metabolite, or protein capable of conveying information into the nucleus. Passive diffusion and active transport are the two most likely scenarios depending on the type of signal. However, monitoring this movement can be technically challenging. Analytical methods for metabolic profiling, such as cell fractionation in conjunction with non-aqueous fractionation, GC/MS- and LC/MS-based, and HPLC, or confocal microscopy for specific signals can be attempted. For instance, the observation that Mg-ProtoIX can accumulate in both chloroplast and cytosol suggests that this metabolite can be exported from site of origin, which may result in the repression of photosynthetic genes under stress (Ankele et al., 2007; Zhang et al., 2011). However, such approaches may be too harsh for labile signals or may not be possible due to cross-contamination of organelles (Estavillo et al., 2011; Woodson et al., 2011).
An alternative to direct measurements of the signal is the use of genetic approaches. Genetically encoded biosensors specific for putative signals, such as those developed for sugars and hormones (Frommer et al., 2009), could be used to measure the dynamics of the signal movement in different compartments. In a similar approach, the levels of the signal can be manipulated by targeting “sensors,” or “modulator” proteins to specific compartments and monitoring the changes in phenotypes. For example, targeting of a PAP degrading enzyme to the nucleus complements the molecular and morphological phenotype of sal1 mutants, supporting the hypothesis that PAP moves in between organelles (Estavillo et al., 2011). Clearly, multiple lines of evidence are required to understand signal movement.
Identification of transporters that regulate the flux of the signal from the plastid into the cytosol is critical for understanding a signal transduction pathway. Although some of the proposed signals, like β-CC, could freely diffuse from the site of origin (Ramel et al., 2012), movement of others, like the highly charged PAP, or tetrapyrrole intermediates must necessitate specific transporters. There is good evidence that haem can leave intact chloroplasts (Thomas and Weinstein, 1990) and that Mg-ProtoIX can be found in the cytoplasm under stress conditions (Ankele et al., 2007). Although a couple of transport mechanisms have been proposed for Mg-ProtoIX (Moller et al., 2001; Larkin et al., 2003; Ankele et al., 2007), no chloroplast transporter has been described for haem. Very recently, co-expression analyses of genes related to glucosinolate metabolism lead to identification of PAPS transporter 1 (PAPST1), a transporter belonging to the mitochondrial carrier family that localizes to both the thylakoid, and plastid envelope (Gigolashvili, 2012). PAPST1 can transport ADP/ATP or PAPS /ATP, or PAP in an antiport manner in vitro. It still remains unclear whether this transporter plays any role in the PAP signaling pathway, especially during abiotic stress. Modulation of the transporter activity could be a major point of flux control, as also considered elsewhere (Leister, 2012).
Affinity chromatography against putative signals coupled to proteomic analyses is a logical approach to dissect the components of the retrograde signaling pathways. This strategy rendered a large number of proteins associated to oxidative stress that bound to Mg-ProtoIX covalently linked gel matrix (Kindgren et al., 2011). This led to the proposal that a regulatory complex composed of HSP90 and Mg-ProtoIX could mediate gene expression (Kindgren et al., 2012). However, results can be misleading by non-specific interaction and proper controls and other additional lines of evidence are required (i.e., mutant analyses of potential targets, etc.).
Some of the retrograde mechanisms have been identified using inducible reporter genes, a common strategy for dissecting signaling pathways (Susek et al., 1993; Rossel et al., 2006; Estavillo et al., 2011). Another strategy to dissect new signaling components in order to understand mechanism of action is by performing reverse genetic screens of known signaling mutants (Šimková et al., 2012). However, the associated phenotypic screening can be misleading due to the potential pleiotropic effects of the signals. Additionally, study of co-regulated genes in a pathway can lead to the discovery of newer components (Gigolashvili, 2012).
Although the general view of retrograde metabolic signals is that of the “classical” model, where one signal acts through a rather linear pathway, it is likely that a “one signal, many targets” scenario is more common than anticipated, especially when considering metabolites that could bind to proteins. For example, 60% of genes misregulated in the SAL1 mutant alx8 mutant do not seem to be affected by XRNs (Estavillo et al., 2011). This implicates that additional PAP targets exist in plants that may be important in controlling gene expression during stress. In fact, in plants and other organisms PAP can bind to other proteins, such as SOTs (Klaassen and Boles, 1997), nucleoside diphosphate kinase (Schneider et al., 1998) and poly(ADP-ribose) polymerase 1 (Toledano et al., 2012).
There are several recent examples of some classical retrograde signals converging with other networks such as light signaling (Ruckle and Larkin, 2009), plant immune signaling (Nomura et al., 2012), transition from cell proliferation to cell expansion (Andriankaja et al., 2012), cold acclimation (Crosatti et al., 2012), and ABA signaling (Koussevitzky et al., 2007) to name a few. It would be interesting to investigate whether the newer metabolic signals interact with other pathways (such as stomata regulation) that could be modulated by ABA, which levels increase upon HL, or drought stress.
Several new metabolite retrograde signals have been recently proposed (Estavillo et al., 2011; Ramel et al., 2012; Xiao et al., 2012). New knowledge about the mechanism of other signaling pathways has also been gained (Fischer et al., 2012; Kindgren et al., 2012; Maruta et al., 2012; Šimková et al., 2012). Yet, none of the pathways are complete and for some transport or movement has yet to be demonstrated. Consequently, signal sensing and modulation is an important area of research. Additional efforts in analytical techniques for signal quantification and movement are critical for assessing the true “signal” nature of a metabolite. The fact that two new signaling pathways involve post-transcriptional regulation (Estavillo et al., 2011) and histone modifications (Sun et al., 2011) open an unexplored area for research: are there other cases of chloroplast-to-nucleus regulation where these types of gene regulation occur? The combination of deep sequencing technologies with the traditional reporter gene screen and multiple mutant approaches (including revertants) can give a more detailed picture of specific and overlapping networks for gene regulation. Finally, the development of new experimental conditions (i.e., controlled drought, finer manipulation of light quality and intensity) and technologies (i.e., biosensors, phenomics, revertant screening, and proteomics) will be instrumental in the discovery of new, true signaling components.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was funded by the Australian Research Council Centre of Excellence in Plant Energy of Biology (CE0561495), an ARC Linkage Grant (LP110200257), an Australian Commonwealth International Postgraduate Research Scholarship for Kai Xun Chan, and an the Australian National University PhD Merit Scholarship for Su Yin Phua.
Andriankaja, M., Dhondt, S., De bodt, S., Vanhaeren, H., Coppens, F., De milde, L., et al. (2012). Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev. Cell 22, 64–78.
Ankele, E., Kindgren, P., Pesquet, E., and Strand, A. (2007). In vivo visualization of Mg-protoporphyrinIX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell 19, 1964–1979.
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399.
Barajas-López, J. D. D., Blanco, N. E., and Strand, Å. (2012). Plastid-to-nucleus communication, signals controlling the running of the plant cell. Biochim. Biophys. Acta. doi: 10.1016/j.bbamcr.2012.06.020. [Epub ahead of print].
Bradbeer, J. W., Atkinson, Y. E., Borner, T., and Hagemann, R. (1979). Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesized Rna. Nature 279, 816–817.
Brautigam, K., Dietzel, L., Kleine, T., Stroher, E., Wormuth, D., Dietz, K. J., et al. (2009). Dynamic plastid redox signals integrate gene expression and metabolism to induce distinct metabolic states in photosynthetic acclimation in Arabidopsis. Plant Cell 21, 2715–2732.
Chan, K. X., Crisp, P. A., Estavillo, G. M., and Pogson, B. J. (2010). Chloroplast-to-nucleus communication: current knowledge, experimental strategies and relationship to drought stress signaling. Plant Signal. Behav. 5, 1–8.
Chen, H., Zhang, B., Hicks, L. M., and Xiong, L. (2011). A nucleotide metabolite controls stress-responsive gene expression and plant development. PLoS ONE 6:e26661. doi: 10.1371/journal.pone.0026661
Crosatti, C., Rizza, F., Badeck, F. W., Mazzucotelli, E., and Cattivelli, L. (2012). Harden the chloroplast to protect the plant. Physiol. Plant. 147, 55–63.
De La Paz Sanchez, M., and Gutierrez, C. (2009). Arabidopsis ORC1 is a PHD-containing H3K4me3 effector that regulates transcription. Proc. Natl. Acad. Sci. U.S.A. 106, 2065–2070.
Delwiche, C. F. (1999). Tracing the thread of plastid diversity through the tapestry of life. Am. Nat. 154, S164–S177.
Dichtl, B., Stevens, A., and Tollervey, D. (1997). Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 16, 7184–7195.
Espinas, N. A., Kobayashi, K., Takahashi, S., Mochizuki, N., and Masuda, T. (2012). Evaluation of unbound free heme in plant cells by differential acetone extraction. Plant Cell Physiol. 53, 1344–1354.
Estavillo, G. M., Crisp, P. A., Pornsiriwong, W., Wirtz, M., Collinge, D., Carrie, C., et al. (2011). Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23, 3992–4012.
Fey, V., Wagner, R., Brautigam, K., Wirtz, M., Hell, R., Dietzmann, A., et al. (2005). Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J. Biol. Chem. 280, 5318–5328.
Fischer, B. B., Ledford, H. K., Wakao, S., Huang, S. G., Casero, D., Pellegrini, M., et al. (2012). SINGLET OXYGEN RESISTANT 1 links reactive electrophile signaling to singlet oxygen acclimation in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 109, E1302–E1311.
Foudree, A., Aluru, M., and Rodermel, S. (2010). PDS activity acts as a rheostat of retrograde signaling during early chloroplast biogenesis. Plant Signal. Behav. 5, 1629–1632.
Frommer, W. B., Davidson, M. W., and Campbell, R. E. (2009). Genetically encoded biosensors based on engineered fluorescent proteins. Chem. Soc. Rev. 38, 2833–2841.
Gadjieva, R., Axelsson, E., Olsson, U., and Hansson, M. (2005). Analysis of gun phenotype in barley magnesium chelatase and Mg-protoporphyrin IX monomethyl ester cyclase mutants. Plant Physiol. Biochem. 43, 901–908.
Galvez-Valdivieso, G., and Mullineaux, P. M. (2010). The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol. Plant. 138, 430–439.
Gigolashvili, T. (2012). Much more than a thylakoid ADP/ATP carrier– enlightening a role of TAAC in plastidic phosphoadenosine 50-phosphosulfate (PAPS) supply to the cytosol. Plant Cell 21, 1813.
González-Pérez, S., Gutiérrez, J., García-García, F., Osuna, D., Dopazo, J., Lorenzo, Ó., et al. (2011). Early transcriptional defense responses in Arabidopsis cell suspension culture under high-light conditions. Plant Physiol. 156, 1439–1456.
Grabowski, E., Miao, Y., Mulisch, M., and Krupinska, K. (2008). Single-stranded DNA-binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol. 147, 1800–1804.
Gray, J. C. (2003). Chloroplast-to-nucleus signalling: a role for Mg-protoporphyrin. Trends Genet. 19, 526–529.
Grieshaber, N. A., Sager, J. B., Dooley, C. A., Hayes, S. F., and Hackstadt, T. (2006). Regulation of the Chlamydia trachomatis histone H1-Like protein Hc2 is IspE dependent and IhtA independent. J. Bacteriol. 188, 5289–5292.
Gy, I., Gasciolli, V., Lauressergues, D., Morel, J.-B., Gombert, J., Proux, F., et al. (2007). Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19, 3451–3461.
Hon, T., Lee, H. C., Hach, A., Johnson, J. L., Craig, E. A., Erdjument-Bromage, H., et al. (2001). The Hsp70-Ydj1 molecular chaperone represses the activity of the heme activator protein Hap1 in the absence of heme. Mol. Cell. Biol. 21, 7923–7932.
Isemer, R., Mulisch, M., Schäfer, A., Kirchner, S., Koop, H.-U., and Krupinska, K. (2012). Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett. 586, 85–88.
Kakizaki, T., Matsumura, H., Nakayama, K., Che, F.-S., Terauchi, R., and Inaba, T. (2009). Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 151, 1339–1353.
Kastenmayer, J. P., and Green, P. J. (2000). Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 97, 13985–13990.
Kindgren, P., Eriksson, M.-J., Benedict, C., Mohapatra, A., Gough, S. P., Hansson, M., et al. (2011). A novel proteomic approach reveals a role for Mg-protoporphyrin IX in response to oxidative stress. Physiol. Plant. 141, 310–320.
Kindgren, P., Norén, L., Barajas López, J. D. D., Shaikhali, J., and Strand, Å. (2012). Interplay between Heat Shock Protein 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Mol. Plant 5, 901–913.
Klaassen, C., and Boles, J. (1997). Sulfation and sulfotransferases 5: the importance of 3'-phosphoadenosine 5'-phosphosulfate (PAPS) in the regulation of sulfation. FASEB J. 11, 404–418.
Klein, M., and Papenbrock, J. (2004). The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. J. Exp. Bot. 55, 1809–1820.
Koussevitzky, S., Nott, A., Mockler, T. C., Hong, F., Sachetto-Martins, G., Surpin, M., et al. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719.
Krause, K., Oetke, S., and Krupinska, K. (2012). Dual targeting and retrograde translocation: regulators of plant nuclear gene expression can be sequestered by plastids. Int. J. Mol. Sci. 13, 11085–11101.
Larkin, R. M., Alonso, J. M., Ecker, J. R., and Chory, J. (2003). GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906.
Lee, H. C., Hon, T., and Zhang, L. (2002). The molecular chaperone Hsp90 mediates heme activation of the yeast transcriptional activator Hap1. J. Biol. Chem. 277, 7430–7437.
Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., et al. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749.
Leister, D. (2012). Retrograde signalling in plants: from simple to complex scenarios. Front. Plant Sci. 3:135. doi: 10.3389/fpls.2012.00135
Marechal, A., Parent, J.-S., Veronneau-Lafortune, F., Joyeux, A., Lang, B. F., and Brisson, N. (2009). Whirly proteins maintain plastid genome stability in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 14693–14698.
Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., et al. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. 99, 12246–12251.
Maruta, T., Noshi, M., Tanouchi, A., Tamoi, M., Yabuta, Y., Yoshimura, K., et al. (2012). H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. J. Biol. Chem. 287, 11717–11729.
Melonek, J., Mulisch, M., Schmitz-Linneweber, C., Grabowski, E., Hensel, G., and Krupinska, K. (2010). Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids. Planta 232, 471–481.
Mochizuki, N., Brusslan, J. A., Larkin, R., Nagatani, A., and Chory, J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. U.S.A. 98, 2053–2058.
Mochizuki, N., Tanaka, R., Tanaka, A., Masuda, T., and Nagatani, A. (2008). The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105, 15184–15189.
Moller, S. G., Kunkel, T., and Chua, N. H. (2001). A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev. 15, 90–103.
Moulin, M., McCormac, A. C., Terry, M. J., and Smith, A. G. (2008). Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl. Acad. Sci. U.S.A. 105, 15178–15183.
Nomura, H., Komori, T., Uemura, S., Kanda, Y., Shimotani, K., Nakai, K., et al. (2012). Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 3:926. doi: 10.1038/ncomms1926
Nott, A., Jung, H.-S., Koussevitzky, S., and Chory, J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57, 739–759.
Oelmuller, R., Levitan, I., Bergfeld, R., Rajasekhar, V. K., and Mohr, H. (1986). Expression of nuclear genes as affected by treatments acting on the plastids. Planta 168, 482–492.
op den Camp, R. G. L., Przybyla, D., Ochsenbein, C., Laloi, C., Kim, C., Danon, A., et al. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15, 2320–2332.
Ostrovsky, D., Diomina, G., Lysak, E., Matveeva, E., Ogrel, O., and Trutko, S. (1998). Effect of oxidative stress on the biosynthesis of 2-C-methyl-d-erythritol-2, 4-cyclopyrophosphate and isoprenoids by several bacterial strains. Arch. Microbiol. 171, 69–72.
Pesaresi, P., Hertle, A., Pribil, M., Kleine, T., Wagner, R., Strissel, H., et al. (2009). Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21, 2402–2423.
Pfannschmidt, T. (2010). Plastidial retrograde signalling a true plastid factor or just metabolite signatures? Trends Plant Sci. 15, 427–435.
Pfannschmidt, T., Bräutigam, K., Wagner, R., Dietzel, L., Schröter, Y., Steiner, S., et al. (2009). Potential regulation of gene expression in photosynthetic cells by redox and energy state: approaches towards better understanding. Ann. Bot. 103, 599–607.
Pogson, B. J., Woo, N. S., Förster, B., and Small, I. D. (2008). Plastid signalling to the nucleus and beyond. Trends Plant Sci. 13, 602–609.
Pontier, D., Albrieux, C., Joyard, J., Lagrange, T., and Block, M. A. (2007). Knock-out of the magnesium protoporphyrin IX methyltransferase gene in Arabidopsis. J. Biol. Chem. 282, 2297–2304.
Ramel, F., Birtic, S., Ginies, C., Soubigou-Taconnat, L., Triantaphylidès, C., and Havaux, M. (2012). Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5535–5540.
Rodríguez-Concepción, M. (2006). Early steps in isoprenoid biosynthesis: multilevel regulation of the supply of common precursors in plant cells. Phytochem. Rev. 5, 1–15.
Rossel, J. B., Walter, P. B., Hendrickson, L., Chow, W. S., Poole, A., Mullineaux, P. M., et al. (2006). A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ. 29, 269–281.
Ruckle, M. E., and Larkin, R. M. (2009). Plastid signals that affect photomorphogenesis in Arabidopsis thaliana are dependent on GENOMES UNCOUPLED 1 and cryptochrome 1. New Phytol. 182, 367–379.
Schneider, B., Xu, Y. W., Janin, J., Véron, M., and Deville-Bonne, D. (1998). 3′-Phosphorylated nucleotides are tight binding inhibitors of nucleoside diphosphate kinase activity. J. Biol. Chem. 273, 28773–28778.
Schwarzländer, M., König, A.-C., Sweetlove, L. J., and Finkemeier, I. (2012). The impact of impaired mitochondrial function on retrograde signalling: a meta-analysis of transcriptomic responses. J. Exp. Bot. 63, 1735–1750.
Šimková, K., Moreau, F., Pawlak, P., Vriet, C., Baruah, A., Alexandre, C., et al. (2012). Integration of stress-related and reactive oxygen species-mediated signals by Topoisomerase VI in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 109, 16360–16365.
Souret, F. F., Kastenmayer, J. P., and Green, P. J. (2004). AtXRN4 Degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15, 173–183.
Strand, A., Asami, T., Alonso, J., Ecker, J. R., and Chory, J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421, 79–83.
Sun, X., Feng, P., Xu, X., Guo, H., Ma, J., Chi, W., et al. (2011). A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2:477. doi: 10.1038/ncomms1486
Susek, R. E., Ausubel, F. M., and Chory, J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799.
Suzuki, N., Koussevitzky, S., Mittler, R. O. N., and Miller, G. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270.
Thomas, J., and Weinstein, J. D. (1990). Measurement of heme efflux and heme content in isolated developing chloroplasts. Plant Physiol. 94, 1414–1423.
Toledano, E., Ogryzko, V., Danchin, A., Ladant, D., and Mechold, U. (2012). 3′-5′ phosphoadenosine phosphate is an inhibitor of PARP-1 and a potential mediator of the lithium-dependent inhibition of PARP-1 in vivo. Biochem. J. 443, 485–490.
van Dijk, E. L., Chen, C. L., D'Aubenton-Carafa, Y., Gourvennec, S., Kwapisz, M., Roche, V., et al. (2011). XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475, 114–117.
von Gromoff, E. D., Alawady, A., Meinecke, L., Grimm, B., and Beck, C. F. (2008). Heme, a plastid-derived regulator of nuclear gene expression in chlamydomonas. Plant Cell 20, 552–567.
Voss, B., Meinecke, L., Kurz, T., Al-Babili, S., Beck, C. F., and Hess, W. R. (2011). Hemin and magnesium-protoporphyrin IX induce global changes in gene expression in Chlamydomonas reinhardtii. Plant Physiol. 155, 892–905.
Wagner, D., Przybyla, D., op den Camp, R., Kim, C., Landgraf, F., Lee, K. P., et al. (2004). The genetic basis of singlet oxygen–induced stress responses of Arabidopsis thaliana. Science 306, 1183–1185.
Wilson, K. E., Ivanov, A. G., Öquist, G., Grodzinski, B., Sarhan, F., and Huner, N. P. A. (2006). Energy balance, organellar redox status, and acclimation to environmental stress. Can. J. Bot. 84, 1355–1370.
Wilson, P. B., Estavillo, G. M., Field, K. J., Pornsiriwong, W., Carroll, A. J., Howell, K. A., et al. (2009). The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J. 58, 299–317.
Woodson, J. D., and Chory, J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395.
Woodson, J. D., Perez-Ruiz, J. M., and Chory, J. (2011). Heme synthesis by plastid ferrochelatase i regulates nuclear gene expression in plants. Curr. Biol. 21, 897–903.
Xiao, Y., Savchenko, T., Baidoo, E. E., Chehab, W. E., Hayden, D. M., Tolstikov, V., et al. (2012). Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149, 1525–1535.
Zhang, L., and Hach, A. (1999). Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cell. Mol. Life Sci. 56, 415–426.
Zhang, N.-H., Zhao, Y., Liang, H.-G., Sun, J.-Y., and Du, L.-F. (2012). A new gun mutant of oilseed rape with a reduced porphyrin flux through Mg-chelatase. Acta Physiol. Plant. doi: 10.1007/s11738-012-1049-0
Zhang, Z.-W., Yuan, S., Feng, H., Xu, F., Cheng, J., Shang, J., et al. (2011). Transient accumulation of Mg-protoporphyrin IX regulates expression of PhANGs– New evidence for the signaling role of tetrapyrroles in mature Arabidopsis plants. J. Plant Physiol. 168, 714–721.
Keywords: retrograde signaling, metabolite, drought, high light, gene regulation, 3′-phosphoadenosine 5′-phosphate, methylerythritol cyclodiphosphate, β-cyclocitral
Citation: Estavillo GM, Chan KX, Phua SY and Pogson BJ (2013) Reconsidering the nature and mode of action of metabolite retrograde signals from the chloroplast. Front. Plant Sci. 3:300. doi: 10.3389/fpls.2012.00300
Received: 30 October 2012; Accepted: 12 December 2012;
Published online: 04 January 2013.
Edited by:
Dario Leister, Ludwig-Maximilians-University Munich, GermanyReviewed by:
Tatjana Kleine, Ludwig-Maximilians-Universität München, GermanyCopyright © 2013 Estavillo, Chan, Phua and Pogson. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Barry J. Pogson, ARC Centre of Excellence in Plant Energy of Biology, Research School of Biology, The Australian National University, Canberra, ACT 0200, Australia. e-mail:YmFycnkucG9nc29uQGFudS5lZHUuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.