
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Plant Sci. , 26 October 2012
Sec. Plant Metabolism and Chemodiversity
Volume 3 - 2012 | https://doi.org/10.3389/fpls.2012.00239
Lys-derived alkaloids, including piperidine, quinolizidine, indolizidine, and lycopodium alkaloids, are widely distributed throughout the plant kingdom. Several of these alkaloids have beneficial properties for humans and have been used in medicine. However, the molecular mechanisms underlying the biosynthesis of these alkaloids are not well understood. In the present article, we discuss recent advances in our understanding of Lys-derived alkaloids, especially the biochemistry, molecular biology, and biotechnology of quinolizidine alkaloid (QA) biosynthesis. We have also highlighted Lys decarboxylase (LDC), the enzyme that catalyzes the first committed step of QA biosynthesis and answers a longstanding question about the molecular entity of LDC activity in plants. Further prospects using current advanced technologies, such as next-generation sequencing, in medicinal plants have also been discussed.
Plant secondary metabolites play multiple roles in the interaction between plants and their environment (Dixon, 2001). Almost 100,000 secondary metabolites have been discovered from plant species (Verpoorte, 1998; Hostettmann and Terreaux, 2000; Afendi et al., 2012). Alkaloids are organic nitrogenous bases, usually in a heterocyclic ring, with characteristic toxicity and marked pharmacological effects in humans and animals (De Luca and St Pierre, 2000). Alkaloids are derived from the products of primary metabolism, with amino acids, such as Phe, Tyr, Trp, Orn, and Lys, serving as their main precursors (Facchini, 2001). Approximately 20% of plant species accumulate alkaloids and over 12,000 different alkaloids have been described, suggesting higher structural and biosynthetic diversity compared to other secondary metabolites (Croteau et al., 2000; De Luca and St Pierre, 2000).
Alkaloids derived from Lys are widely distributed throughout the plant kingdom and are subdivided into piperidine, quinolizidine, indolizidine, and lycopodium alkaloids. Piperidine alkaloids are found in plants in the order Piperales (e.g., piperine from Piper nigrum), Myrtales (e.g., (–)-pelletierine from Punica granatum), and Asterales (e.g., (–)-lobeline from Lobelia inflata). Quinolizidine (e.g., (–)-lupinine and (–)-sparteine from Lupinus luteus and Cytisus scoparius, respectively) and indolizidine (e.g., (–)-swainsonine from Swainsona canescens) alkaloids are found mainly in the order Fabales (Murakoshi et al., 1975, 1986; Colegate et al., 1979; Su and Horvat, 1981; Neuhofer et al., 1993; Felpin and Lebreton, 2004). Lycopodium alkaloids (e.g., (–)-huperzine A and lycopodine from Huperzia serrata) are found mainly in the genus Lycopodium (Figure 1; Liu et al., 1986; Ma and Gang, 2004). Several alkaloids have beneficial properties for humans and are used in medicine. For example, (–)-lobeline is used in the treatment of central nervous system disorders and drug abuse and (–)-huperzine A is used in the treatment of Alzheimer’s disease (Decker et al., 1992; Bai et al., 2000; Dwoskin et al., 2000; Dwoskin and Crooks, 2002).
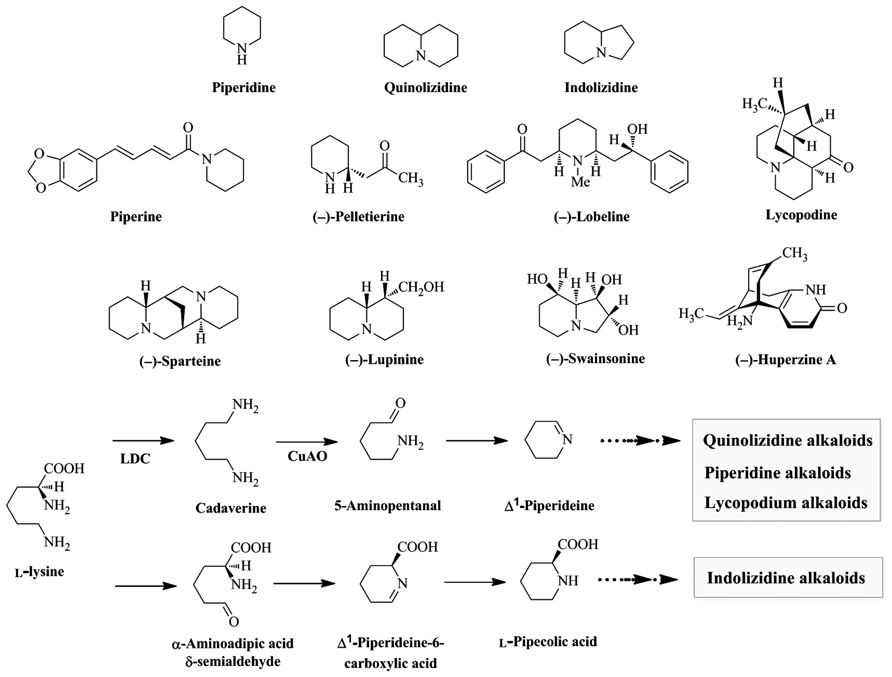
FIGURE 1. Biosynthesis of universal intermediates, skeleton structures, and some Lys-derived alkaloids found in plants. Piperine, (–)-pelletierine, and (–)-lobeline are piperidine alkaloids. (–)-Lupinine and (–)-sparteine are quinolizidine alkaloids. (–)-Swainsonine is an indolizine alkaloids. Lycopodine and (–)-huperzine A are lycopodium alkaloids. LDC, Lys decarboxylase; CuAO, copper amine oxidase.
Over the past decade, intensive efforts have been made to identify the genes encoding the enzymes involved in the biosynthesis of isoquinoline, indole, tropane, and purine alkaloids (Croteau et al., 2000; Facchini et al., 2004). However, the molecular mechanisms underlying the biosynthesis of Lys-derived alkaloids are poorly understood. In this article, we discuss recent advances in our understanding of the Lys-derived alkaloids, especially the genes involved in the quinolizidine alkaloid (QA) biosynthetic pathway. We highlight the novel finding of Lys decarboxylase (LDC) from plants and discuss how it evolved from primary metabolism to alkaloid biosynthesis. Finally, we consider how to apply current advanced technologies to the study of unidentified alkaloid biosynthetic pathways.
L-Lys is the homologue of L-Orn, a precursor of pyrrolidine, tropane, and pyrrolizidine alkaloids. The extra methylene group in Lys participates in forming 6-membered piperidine rings, while Orn participates in forming 5-membered pyrrolizidine rings (Dewick, 2001; Poupon et al., 2011). Biosynthetic pathways of several potential Lys-derived alkaloids have been proposed based on tracer experiments with labeled precursors (Leistner and Spenser, 1973 and references therein). Feeding studies demonstrated that the first step in piperidine, quinolizidine, and lycopodium alkaloid biosynthesis is the decarboxylation of L-Lys into cadaverine by LDC (EC 4.1.1.18). Oxidative deamination of cadaverine by copper amine oxidase (CuAO, EC 1.4.3.22) yields 5-aminopentanal, which is spontaneously cyclized to Δ1-piperideine Schiff base (Leistner and Spenser, 1973; Gupta et al., 1979; Golebiewski and Spenser, 1988; Saito and Murakoshi, 1995; Ma and Gang, 2004). In contrast, indolizidine alkaloids are formed from L-Lys via L-pipecolic acid which is formed via the aldehyde and Schiff base, with retention of the nitrogen of the α-amino acid group (Figure 1; Harris et al., 1988a,b; Wickwire et al., 1990; Dewick, 2001). Δ1-Piperideine and L-pipecolic acid are universal intermediates that can be modified by condensation, hydrolysis, or methylation to produce diverse Lys-derived alkaloids (Figure 1; Evans, 2009).
The genes and enzymes involved in the biosynthesis of Lys-derived alkaloids are not well documented. The best-studied pathway at the genetic level is the one leading to the formation of QAs, which will be discussed in the following section. One other study addresses partially purified piperoyl-CoA: piperidine N-piperoyltransferase (EC 2.3.1.145) from P. nigrum which catalyzes the synthesis of piperine in the presence of piperoyl-CoA and piperidine (Geisler and Gross, 1990).
Quinolizidine alkaloids are a group of alkaloids possessing a quinolizidine ring or a piperidine ring (Saito and Murakoshi, 1995). QAs occur mainly in the family Leguminosae, especially in the genera Lupinus, Baptisia, Thermopsis, Genista, Cytisus, and Sophora (Ohmiya et al., 1995). More than 500 Lupinus species have been described and 200–300 species are distributed in North and South America (Wink et al., 1995). QAs are important as potential sources of medicine. They have a broad range of pharmacological properties, including cytotoxic, oxytocic, antipyretic, antibacterial, antiviral, and hypoglycemic activities, as determined by in vivo pharmacological screening (Saito and Murakoshi, 1995). Some QA-containing plants, for example Sophora flavescens, have been used as sources of crude drugs in Chinese–Japanese medicine (KAMPO; Tang and Eisenbrand, 1992).
Quinolizidine alkaloids are synthesized through the cyclization of the cadaverine unit (Golebiewski and Spenser, 1988), which is produced through the action of LDC (Figure 2; Saito and Murakoshi, 1995). Various QA skeletons are further modified by tailoring reactions (e.g., dehydrogenation, oxygenation, or esterification) to yield several hundred structurally related alkaloids (Ohmiya et al., 1995). Lupinus accumulates two types of QA esters, the derivatives of lupinine and lupanine. QA esters are assumed to be end products of biosynthesis and storage forms (Ohmiya et al., 1995). Two acyltransferases (ATs) involved in QA ester biosynthesis have been identified and characterized: (–)-13α-hydroxymultiflorine/(+)-13α-hydroxylupanine O-tigloyltransferase (HMT/HLT; EC 2.3.1.93), which catalyzes the transfer of the tigloyl group from tigloyl-CoA to (–)-13α-hydroxymultiflorine or (+)-13α-hydroxylupanine, and p-coumaroyl-CoA/feruloyl-CoA: (+)-epilupinine/(–)-lupinine O-coumaroyl/feruloyltransferase (ECT/EFT-LCT/LFT), which catalyzes the transfer of p-coumaroyl-CoA or feruloyl-CoA to the hydroxyl moiety of (+)-epilupinine or (–)-lupinine (Figure 2; Saito et al., 1992, 1993; Suzuki et al., 1994). However, only HMT/HLT has been cloned and characterized at the molecular level (Okada et al., 2005). HMT/HLT belongs to the BAHD super family, a plant acyl-CoA-dependent AT gene family that transfers acyl groups from their CoA esters to various plant metabolites (D’Auria, 2006).
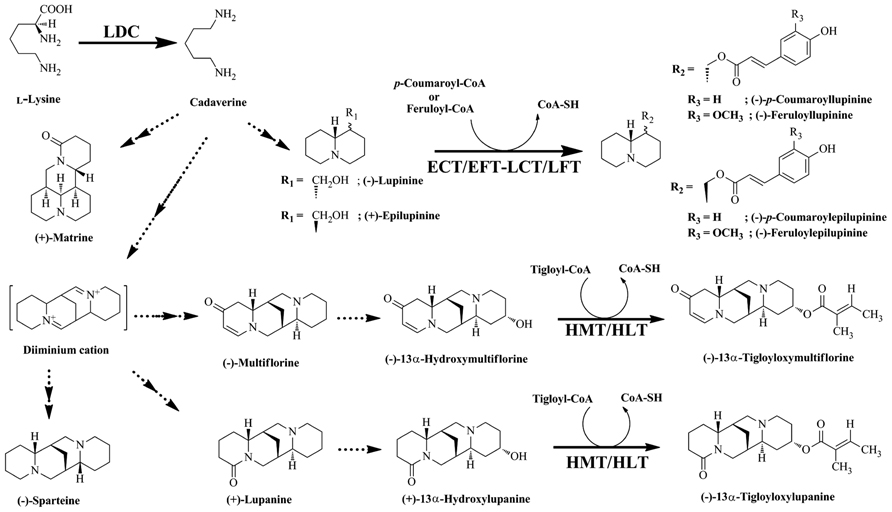
FIGURE 2. Biosynthetic pathway of quinolizidine alkaloids. Quinolizidine alkaloids are synthesized by the decarboxylation of Lys, yielding cadaverine, which is further modified by various reactions as shown. The metabolites derived from the pathway are shown. Enzymes involved in the synthesis of these alkaloids are indicated: LDC, Lys decarboxylase; HMT/HLT, tigloyl-CoA: (–)-13α-hydroxymultiflorine/(+)-13α-hydroxylupanine O-tigloyltransferase; ECT/EFT-LCT/LFT, p-coumaroyl-CoA/feruloyl-CoA: (+)-epilupinine/(–)-lupinine O-coumaroyl/feruloyltransferase.
An investigation of an alkaloid-rich “bitter” cultivar and an alkaloid-poor “sweet” cultivar of L. angustifolius using random amplified polymorphic DNA (RAPD) showed that total DNA polymorphisms did not differ substantially between the two cultivars (Hirai et al., 2000). A cDNA-amplified fragment length polymorphism (cDNA-AFLP) was then used to isolate the genes specifically expressed in the bitter plant; however, no bitter-specific gene was isolated (Hirai et al., 2000). The PCR-select subtraction technique was then used to profile differentially expressed genes in the two cultivars. Among 71 bitter-specific fragments, three genes exhibited homologies to Orn decarboxylase (ODC, EC 4.1.1.17), CuAO, and AT, representing potential candidates for structural genes encoding QA biosynthetic enzymes (Bunsupa et al., 2011). Further study showed that the AT might be involved either in the formation of QA esters or N-acylated polyamine conjugates (Bunsupa et al., 2011) and ODC was identified as Lys/Orn decarboxylase (L/ODC; see below) (Bunsupa et al., 2012). In addition, partial sequences encoding candidate transcription factors that might regulate the expression of genes in QA biosynthesis were obtained; these included DNA binding protein EREBP-3, leucine zipper protein, and coronatine-insensitive 1, which provide clues about the regulation of the QA biosynthetic pathway (Bunsupa et al., 2011). Genomic PCR of L. angustifolius AT and LDC indicated the presence of at least one copy of both genes in the genomes of both the bitter and sweet cultivars. However, transcripts of both genes were expressed only in the bitter cultivar. This finding is interesting in terms of the regulation of the entire QA biosynthetic pathway (Bunsupa et al., 2011, 2012).
Lys decarboxylase and ODC are key enzymes involved in the formation of cadaverine and putrescine by the decarboxylation of Lys and Orn, respectively. Cadaverine, putrescine, and other aliphatic polyamines, such as spermidine and spermine, are involved in a number of growth and developmental processes (Bagni and Tassoni, 2001). Unlike plant ODCs, plant LDCs have not been well studied at the molecular level. There are several studies on crude protein extracts or partially purified protein extracts from Glycine max (Kim et al., 1998; Ohe et al., 2009) and L. polyphyllus (Hartmann et al., 1980). There was speculation that a member of group IV of the PLP-dependent amino acid decarboxylases might be responsible for the production of cadaverine from Lys (Sandmeier et al., 1994) and that ODC may also have LDC activity in plants that make cadaverine-derived alkaloids (Sinclair et al., 2004).
Recently, we isolated a cDNA of L/ODC from L. angustifolius (La-L/ODC) using differential transcript screening in QA-producing and non-producing cultivars (Bunsupa et al., 2011, 2012). We also obtained L/ODCs cDNAs from S. flavescens and Echinosophora koreensis, which also produce QAs. Recombinant L/ODCs from QA-producing plants almost equally catalyzed the decarboxylation of L-Lys and L-Orn unlike the known ODCs that only accept Orn (Boeker and Fischer, 1983; Bunsupa et al., 2012). In vivo experiments of La-L/ODC expression in Arabidopsis thaliana and Nicotiana tabacum provided evidence that this enzyme is involved in the first step of QA biosynthesis (Bunsupa et al., 2012). This is the first plant LDC identified at the molecular level by cDNA cloning and the second QA biosynthesis enzyme for which the cDNA has been identified.
Identification of La-L/ODC answers a longstanding question about the molecular entity of LDC activity in plants. We found LDCs in plant species that produce QAs. A point mutation from His-344 to Phe-344 (La-L/ODC numbering) is key for La-L/ODC to exhibit substrate promiscuity for Lys and Orn. This amino acid substitution has presumably occurred via adaptive evolution of LDC within the QA-producing plants to expand substrate accessibility to Lys (Bunsupa et al., 2012). These results reveal how plants have been opportunistic in adapting primary metabolic processes to permit the diversification of secondary metabolism, in this case, substrate promiscuity. Accordingly, the mutation in this position may be found not only in QA-producing Leguminosae but also in plants that produce cadaverine-derived alkaloids. This finding is a breakthrough in the understanding of both QA biosynthesis and LDC occurrence and function in plants.
A major safety issue of lupine-based foods is the presence of QAs, which are toxic to human and mammals. Genetic mapping of L. angustifolius and L. albus has been completed, which enables the mapping of the genes controlling key domestication traits, including alkaloid production, water permeability, flowering time, pod shattering, pigment production in seeds, and anthracnose resistance (Boersma et al., 2005; Nelson et al., 2006; Phan et al., 2006). This information will be useful not only to the Lupinus research community but also to other legume research communities studying molecular breeding and evolution in leguminous plants. The identified La-L/ODC provides a good candidate enzyme as an entry-point enzyme of QA biosynthesis for metabolic engineering to manipulate the production of QAs.
The subcellular compartmentalization of alkaloid biosynthetic enzymes is as complex as the cell type-specific localization of gene transcripts, enzymes, and metabolites (Ziegler and Facchini, 2008). The majority of QAs are synthesized in shoot tissues. Of the QAs that accumulate in seeds, half are synthesized in situ and half are translocated primarily by phloem (Lee et al., 2007). The concentration of QAs in seeds decreases during germination and development and increases as soon as new leaves are formed (Wink and Witte, 1985). The biosynthesis of QAs occurs mainly in the green parts of plants where the enzymes for Lys, cadaverine, and lupanine synthesis are present (Wink and Hartmann, 1979, 1982; Wink and Mende, 1987). La-L/ODC is localized in chloroplasts, while HMT/HLT localized in mitochondria. ECT is assumed to be present in another organelle, but not in mitochondria (Suzuki et al., 1996; Bunsupa et al., 2012).
The concentration of QAs in cell suspension cultures of Lupinus and related plants is approximately 100–1000 times lower than that of concentration in leaves of differentiated plants (Hartmann et al., 1980; Hernandez et al., 2011). In cell suspension cultures, the storage tissue is probably repressed; thus, the QAs cannot be stored in large quantities and are degraded rapidly either inside the cells or in the cell culture medium (Wink, 1987). In callus, the concentration of QAs is correlated to the amount of chlorophyll in the cells (Saito et al., 1989a,b).
Lupinus spp. are the major grain legumes in Australia and have been used as feed supplements. In the past decade, they have gained attention as ingredients for human food because of their high percentage of protein (34–43% of dry matter) and acceptable content of essential amino acids (Sujak et al., 2006). To genetically engineer plants successfully, a routine transformation system is needed. Thus, it is necessary to develop a transformation system for Lupinus lines to confer desired traits, such as herbicide resistance, resistance to certain pathogens, increased pod and seed set, and improved seed quality (Pigeaire et al., 1997). Many transformation systems for Lupinus sp. have been developed; however, only a few have been successful. For example, shoot regeneration using Agrobacterium-mediated gene transfer to preorganized meristematic tissue combined with axillary regeneration has been successful in L. angustifolius (Pigeaire et al., 1997), L. luteus (Li et al., 2000), and L. mutabilis (Babaoglu et al., 2000). However, transformation efficiency is poor and transformation in some Lupinus species and even in different cultivars of the same species is difficult. In L. mutabilis, the transformed roots synthesize isoflavones, but not QAs (Babaoglu et al., 2004). Genetic engineering of L. angustifolius by over expressed sun flower seed albumin gene has succeeded to enhance methionine levels and increase nutritive value of seeds of transgenic Lupinus plants (Molvig et al., 1997).
In the past few years, progress has been made in the elucidation of alkaloid biosynthetic pathways and their regulation. However, only two genes, HMT/HLT and L/ODC, have been isolated and characterized so far. Rapid progress in the development of integrative approaches, such as genomics, transcriptomics, proteomics, and metabolomics, has provided essential information for the understanding of many complex biological processes at different levels (Fukushima et al., 2009; Yonekura-Sakakibara and Saito, 2009). Comparative and integrative analyses of such “omics” data will facilitate the determination of gene function and pinpoint rate limiting steps and factors in the biosynthetic pathway of secondary metabolites. Transcriptomic and metabolomic approaches, particularly transcriptome coexpression network analysis, have been used to identify genes involved in specific pathways not only in Arabidopsis but also in crops and medicinal plants. Such strategies have proven to be useful in fundamental plant biology and applied biotechnology (Ziegler et al., 2006; Hirai et al., 2007; Saito et al., 2008: Seki et al., 2008; Yonekura-Sakakibara et al., 2008, 2012; Liscombe et al., 2009; Okazaki et al., 2009; Saito and Matsuda, 2010; Okazaki and Saito, 2012).
Advances in sequencing technology, next-generation sequencing, and computer technology for processing of large data sets are opening a new era in plant science. It is estimated that the cost of DNA sequencing drops at a faster rate than the cost of computer data processing (Ehrhardt and Frommer, 2012). Therefore, the development of sequencing techniques with high-throughput will likely be applied to alkaloid-producing plants and provide the basis for many sequence-based approaches that are now limited to model plants such as Arabidopsis and rice (Ziegler and Facchini, 2008). Today, many researchers are making progress in plant genome and transcriptome sequencing. For instance, the 1KP project aims to generate large-scale gene sequence information for 1000 different plants, including a wide variety of species from algae to land and aquatic plants, to understand the vast biodiversity that has barely been touched by genomics to date1. In addition, the medicinal plant consortium project focuses on providing transcriptomic and metabolic resources for 14 key medicinal plants, for instance Atropa belladonna and Camptotheca acuminata, to the worldwide research community for the advancement of drug production and development2. Recently, we started the deep transcriptome sequencing of Japanese medicinal plants and both cultivars of L. angustifolius to obtain more information about the enzymes and transcription factors involved in the production of plant secondary metabolites. Such publicly accessible databases will provide novel information for the research community and ultimately accelerate the progress of the study of plant science.
The predicted function or in vitro characterization of alkaloid biosynthetic enzymes and other components must be confirmed in vivo (Ziegler and Facchini, 2008). However, there is no reliable and highly efficient transformation protocol for Lupinus and related plants. Virus induced gene silencing (VIGS), a technology that exploits an RNA-mediated antiviral defense mechanism, is a fast method for the generation of transient transformed plants (Lu et al., 2003). VIGS has been used widely in plants for analysis of gene function and has been adapted for high-throughput functional genomics (Burch-Smith et al., 2004). Thus, VIGS might be a powerful tool for assessing and characterizing the function of candidate genes in the QA biosynthetic pathway. Further investigations on inter- and intracellular transport of pathway intermediates and products and on cellular and subcellular localization of enzymes are also important because they are crucial factors in controlling the accumulation of target metabolites in plants (Facchini et al., 2004).
Knowledge of the biosynthetic pathway of Lys-derived alkaloids is limited. The best-studied pathway at the genetic level is QA biosynthesis; however, only two genes have been identified. Application of current technologies, such as next-generation sequencing, high-end proteomics, and metabolomics approaches, is needed to study secondary metabolic pathways for a better understanding of metabolic networks. Such information will aid in the development of successful strategies in genetic engineering of secondary metabolites in plants.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported in part by the Grants-in-Aid for Scientific Research (KAKENHI) from The Ministry of Education, Culture, Sports, Science, and Technology (MEXT).
Afendi, F. M., Okada, T., Yamazaki, M., Hirai-Morita, A., Nakamura, Y., Nakamura, K., et al. (2012). KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 53, e1.
Babaoglu, M., Davey, M. R., Power, J. B., Sporer, F., and Wink, M. (2004). Transformed roots of Lupinus mutabilis: induction, culture and isoflavone biosynthesis. Plant Cell Tissue Organ Cult. 78, 29–36.
Babaoglu, M., McCabe, M. S., Power, J. B., and Davey, M. R. (2000). Agrobacterium-mediated transformation of Lupinus mutabilis L. using shoot apical explants. Acta Physiol. Plant. 22, 111–119.
Bagni, N., and Tassoni, A. (2001). Biosynthesis, oxidation and conjugation of aliphatic polyamines in higher plants. Amino Acids 20, 301–317.
Bai, D. L., Tang, X. C., and He, X. C. (2000). Huperzine A, a potential therapeutic agent for treatment of Alzheimer’s disease. Curr. Med. Chem. 7, 355–374.
Boeker, E. A., and Fischer, E. H. (1983). “[28] Lysine decarboxylase (Escherichia coli B),” in Methods in Enzymology, eds H. Tabor, and C. W. Tabor (London: Academic Press), 180–184.
Boersma, J. G., Pallotta, M., Li, C., Buirchell, B. J., Sivasithamparam, K., and Yang, H. (2005). Construction of a genetic linkage map using MFLP and identification of molecular markers linked to domestication genes in narrow-leafed Lupin (Lupinus angustifolius L.). Cell. Mol. Biol. Lett. 10, 331–344.
Bunsupa, S., Katayama, K., Ikeura, E., Oikawa, A., Toyooka, K., Saito, K., et al. (2012). Lysine decarboxylase catalyzes the first step of quinolizidine alkaloid biosynthesis and coevolved with alkaloid production in leguminosae. Plant Cell 24, 1202–1216.
Bunsupa, S., Okada, T., Saito, K., and Yamazaki, M. (2011). An acyltransferase-like gene obtained by differential gene expression profiles of quinolizidine alkaloid-producing and non-producing cultivars of Lupinus angustifolius. Plant Biotechnol. 28, 89–94.
Burch-Smith, T. M., Anderson, J. C., Martin, G. B., and Dinesh-Kumar, S. P. (2004). Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 39, 734–746.
Colegate, S. M., Dorling, P. R., and Huxtable, C. R. (1979). A spectroscopic investigation of swainsonine: an α-mannosidase inhibitor isolated from Swainsona canescens. Aust. J. Chem. 32, 2257–2264.
Croteau, R., Kutchan, T., and Lewis, N. (2000). “Natural products (secondary metabolites),” in Biochemistry and Molecular Biology of Plants, eds B. Buchanan, W. Gruissem, and R. Jones (Rockville, MD: American Society of Plant Physiologists), 1250–1318.
D’Auria, J. C. (2006). Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 9, 331–340.
Decker, M. W., Majchrzak, M. J., and Anderson, D. J. (1992). Effects of nicotine on spatial memory deficits in rats with septal lesions. Brain Res. 572, 281–285.
De Luca, V., and St Pierre, B. (2000). The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci. 5, 168–173.
Dewick, P. M. (2001). Medicinal Natural Products: A Biosynthetic Approach, West Sussex, England: John Wiley & Sons Ltd.
Dwoskin, L. P., and Crooks, P. A. (2002). A novel mechanism of action and potential use for lobeline as a treatment for psychostimulant abuse. Biochem. Pharmacol. 63, 89–98.
Dwoskin, L. P., Xu, R., Ayers, J. T., and Crooks, P. A. (2000). Recent developments in neuronal nicotinic acetylcholine receptor antagonists. Expert Opin. Ther. Pat. 10, 1561–1581.
Ehrhardt, D. W., and Frommer, W. B. (2012). New technologies for 21st century plant science. Plant Cell 24, 374–394.
Facchini, P. J. (2001). Alkaloid biosynthesis in plants: biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 29–66.
Facchini, P. J., Bird, D. A., and St-Pierre, B. (2004). Can Arabidopsis make complex alkaloids? Trends Plant Sci. 9, 116–122.
Felpin, F. X., and Lebreton, J. (2004). History, chemistry and biology of alkaloids from Lobelia inflata. Tetrahedron 60, 10127–10153.
Fukushima, A., Kusano, M., Redestig, H., Arita, M., and Saito, K. (2009). Integrated omics approaches in plant systems biology. Curr. Opin. Chem. Biol. 13, 532–538.
Geisler, J. G., and Gross, G. G. (1990). The biosynthesis of piperine in Piper nigrum. Phytochemistry 29, 489–492.
Golebiewski, W. M., and Spenser, I. D. (1988). Biosynthesis of the lupine alkaloids. II. Sparteine and lupanine. Can. J. Chem. 66, 1734–1748.
Gupta, R. N., Horsewood, P., Koo, S. H., and Spenser, I. D. (1979). The biosynthesis of the Lythraceae alkaloids. I. The lysine-derived fragment. Can. J. Chem. 57, 1606–1614.
Harris, C. M., Campbell, B. C., Molyneux, R. J., and Harris, T. M. (1988a). Biosynthesis of swainsonine in the diablo locoweed (Astragalus oxyphyrus). Tetrahedron Lett. 29, 4815–4818.
Harris, C. M., Schneider, M. J., Ungemach, F. S., Hill, J. E., and Harris, T. M. (1988b). Biosynthesis of the toxic indolizidine alkaloids slaframine and swainsonine in Rhizoctonia leguminicola: metabolism of 1-hydroxyindolizidines. J. Am. Chem. Soc. 110, 940–949.
Hartmann, T., Schoofs, G., and Wink, M. (1980). A chloroplast-localized lysine decarboxylase of Lupinus polyphyllus: the first enzyme in the biosynthetic pathway of quinolizidine alkaloids. FEBS Lett. 115, 35–38.
Hernandez, E. M., Rangel, M. L., Angel, J. A., Lopez, J. A., Sporer, F., Wink, M., et al. (2011). Quinolizidine alkaloid composition in different organs of Lupinus aschenbornii. Rev. Bras. Farmacogn. 21, 824–828.
Hirai, M. Y., Sugiyama, K., Sawada, Y., Tohge, T., Obayashi, T., Suzuki, A., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 6478–6483.
Hirai, M. Y., Suzuki, H., Yamazaki, M., and Saito, K. (2000). Biochemical and partial molecular characterization of bitter and sweet forms of Lupinus angustifolius, an experimental model for study of molecular regulation of quinolizidine alkaloid biosynthesis. Chem. Pharm. Bull. 48, 1458–1461.
Hostettmann, K., and Terreaux, C. (2000). Search for new lead compounds from higher plants. Chimia 54, 652–657.
Kim, H. S., Kim, B. H., and Cho, Y. D. (1998). Purification and characterization of monomeric lysine decarboxylase from soybean (Glycine max) axes. Arch. Biochem. Biophys. 354, 40–46.
Lee, M. J., Pate, J. S., Harris, D. J., and Atkins, C. A. (2007). Synthesis, transport and accumulation of quinolizidine alkaloids in Lupinus albus L. and L. angustifolius L. J. Exp. Bot. 58, 935–946.
Leistner, E., and Spenser, I. D. (1973). Biosynthesis of the piperidine nucleus. Incorporation of chirally labeled (1-3H) cadaverine. J. Am. Chem. Soc. 95, 4715–4725.
Li, H., Wylie, S. J., and Jones, M. G. K. (2000). Transgenic yellow lupin (Lupinus luteus). Plant Cell Rep. 19, 634–637.
Liscombe, D. K., Ziegler, J., Schmidt, J., Ammer, C., and Facchini, P. J. (2009). Targeted metabolite and transcript profiling for elucidating enzyme function: isolation of novel N-methyltransferases from three benzylisoquinoline alkaloid-producing species. Plant J. 60, 729–743.
Liu, J. S., Yu, C. M., Zhou, Y. Z., Han, Y. Y., Wu, F. W., Qi, B. F., et al. (1986). Study on the chemistry of huperzine A and B. Acta. Chim. Sin. (Engl. Ed.) 44, 1035–1040.
Lu, R., Martin-Hernandez, A. M., Peart, J. R., Malcuit, I., and Baulcombe, D. C. (2003). Virus-induced gene silencing in plants. Methods 30, 296–303.
Molvig, L., Tabe, L. M., Eggum, B. O., Moore, A. E., Craig, S., Spencer, D., et al. (1997). Enhanced methionine levels and increased nutritive value of seeds of transgenic lupins (Lupinus angustifolius L.) expressing a sunflower seed albumin gene. Proc. Nat. Acad. Sci. U.S.A. 94, 8393–8398.
Murakoshi, I., Sugimoto, K., Haginiwa, J., Ohmiya, S., and Otomasu, H. (1975). (–)-(Trans-4′-hydroxycinnamoyl) lupinine. A new alkaloid in Lupinus seedlings. Phytochemistry 14, 2714–2715.
Murakoshi, I., Yamashita, Y., Ohmiya, S., and Otomasu, H. (1986). (–)-3β-13α-dihydroxylupanine from Cytisus scoparius. Phytochemistry 25, 521–524.
Nelson, M. N., Phan, H. T., Ellwood, S. R., Moolhuijzen, P. M., Hane, J., Williams, A., et al. (2006). The first gene-based map of Lupinus angustifolius L.-location of domestication genes and conserved synteny with Medicago truncatula. Theor. Appl. Genet. 113, 225–238.
Neuhofer, H., Witte, L., and Gorunovic, M. (1993). Alkaloids in the bark of Punica granatum L. (pomegranate) from Yugoslavia. Pharmazie 48, 389–391.
Ohe, M., Scoccianti, V., Bagni, N., Tassoni, A., and Matsuzaki, S. (2009). Putative occurrence of lysine decarboxylase isoforms in soybean (Glycine max) seedlings. Amino Acids 36, 65–70.
Ohmiya, S., Saito, K., and Murakoshi, I. (1995). “Lupine Alkaloids,” in The Alkaloids: Chemistry and Pharmacology, Vol. 47, ed. G. A. Cordell (London: Academic Press), 1–114.
Okada, T., Hirai, M. Y., Suzuki, H., Yamazaki, M., and Saito, K. (2005). Molecular characterization of a novel quinolizidine alkaloid O-tigloyltransferase: cDNA cloning, catalytic activity of recombinant protein and expression analysis in Lupinus plants. Plant Cell Physiol. 46, 233–244.
Okazaki, Y., Shimojima, M., Sawada, Y., Toyooka, K., Narisawa, T., Mochida, K., et al. (2009). A chloroplastic UDP-Glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 21, 892–909.
Okazaki, Y., and Saito, K. (2012). Recent advances of metabolomics in plant biotechnology. Plant Biotechnol. Rep. 6, 1–15.
Phan, H. T., Ellwood, S. R., Ford, R., Thomas, S., and Oliver, R. (2006). Differences in syntenic complexity between Medicago truncatula with Lens culinaris and Lupinus albus. Funct. Plant Biol. 33, 775–782.
Pigeaire, A., Abernethy, D., Smith, P. M., Simpson, K., Fletcher, N., Lu, C. Y., et al. (1997). Transformation of a grain legume (Lupinus angustifolius L.) via Agrobacterium tumefaciens-mediated gene transfer to shoot apices. Mol. Breed. 3, 341–349.
Poupon, E., Salame, R., and Yan, L. H. (2011). “Biomimetic synthesis of ornithine/arginine and lysine-derived alkaloids: selected examples,” in Biomimetic Organic Synthesis, ed. E. Poupon and B. Nay (Weinheim, Germany: Wiley-VCH), 1–60.
Saito, K., Hirai, M. Y., and Yonekura-Sakakibara, K. (2008). Decoding genes by coexpression networks and metabolomics – `majority report by precogs’. Trends Plant Sci. 13, 36–43.
Saito, K., Koike, Y., Suzuki, H., and Murakoshi, I. (1993). Biogenetic implication of lupin alkaloid biosynthesis in bitter and sweet forms of Lupinus luteus and L. albus. Phytochemistry 34, 1041–1044.
Saito, K., and Matsuda, F. (2010). Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 61, 463–489.
Saito, K., and Murakoshi, I. (1995). “Chemistry, biochemistry and chemotaxonomy of lupine alkaloids in the Leguminosae,” in Studies in Natural Products Chemistry, Vol. 15, Structure and Chemistry (Part C), ed. A. U. Rahman (Amsterdam: Elsevier), 519–550.
Saito, K., Suzuki, H., Takamatsu, S., and Murakoshi, I. (1992). Acyltransferases for lupin alkaloids in Lupinus hirsutus. Phytochemistry 32, 87–91.
Saito, K., Yamazaki, M., Takamatsu, S., Kawaguchi, A., and Murakoshi, I. (1989a). Greening induced production of (+)-lupanine in tissue culture of Thermopsis lupinoides. Phytochemistry 28, 2341–2344.
Saito, K., Yamazaki, M., Yamakawa, K., Fujisawa, S., Takamatsu, S., Kawaguchi, A., and Murakoshi, I. (1989b). Lupin alkaloids in tissue culture of Sophora flavescens var. angustifolia: greening induced production of matrine. Chem. Pharm. Bull. 37, 3001–3004.
Sandmeier, E., Hale, T. I., and Christen, P. (1994). Multiple evolutionary origin of pyridoxal-5′-phosphate-dependent amino acid decarboxylases. Eur. J. Biochem. 221, 997–1002.
Seki, H., Ohyama, K., Sawai, S., Mizutani, M., Ohnishi, T., Sudo, H., et al. (2008). Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc. Natl. Acad. Sci. U.S.A. 105, 14204–14209.
Sinclair, S. J., Johnson, R., and Hamill, J. D. (2004). Analysis of wound-induced gene expression in Nicotiana species with contrasting alkaloid profiles. Funct. Plant Biol. 31, 721–729.
Su, H. C. F., and Horvat, R. (1981). Isolation, identification, and insecticidal properties of Piper nigrum amides. J. Agric. Food Chem. 29, 115–118.
Sujak, A., Kotlarz, A., and Strobel, W. (2006). Compositional and nutritional evaluation of several lupin seeds. Food Chem. 98, 711–719.
Suzuki, H., Koike, Y., Murakoshi, I., and Saito, K. (1996). Subcellular localization of acyltransferases for quinolizidine alkaloid biosynthesis in Lupinus. Phytochemistry 42, 1557–1562.
Suzuki, H., Murakoshi, I., and Saito, K. (1994). A novel O-tigloyltransferase for alkaloid biosynthesis in plants. Purification, characterization, and distribution in Lupinus plants. J. Biol. Chem. 269, 15853–15860.
Tang, W., and Eisenbrand, G. (1992).Chinese drugs of plant origin: Chemistry, Pharmacology, and Use in Traditional and Modern Medicine. Berlin: Springer.
Verpoorte, R. (1998). Exploration of nature’s chemodiversity: the role of secondary metabolites as leads in drug development. Drug Discov. Today 3, 232–238.
Wickwire, B. M., Harris, C. M., Harris, T. M., and Broquist, H. P. (1990). Pipecolic acid biosynthesis in Rhizoctonia leguminicola. J. Biol. Chem. 265, 14742–14747.
Wink, M. (1987). Quinolizidine alkaloids: biochemistry, metabolism, and function in plants and cell suspension cultures. Planta Med. 53, 509–514.
Wink, M., and Hartmann, T. (1979). Cadaverine-pyruvate transamination: the principal step of enzymatic quinolizidine alkaloid biosynthesis in Lupinus polyphyllus cell suspension cultures. FEBS Lett. 101, 343–346.
Wink, M., and Hartmann, T. (1982). Localization of the enzymes of quinolizidine alkaloid biosynthesis in leaf chloroplasts of Lupinus polyphyllus. Plant Physiol. 70, 74–77.
Wink, M., and Mende, P. (1987). Uptake of lupanine by alkaloid-storing epidermal cells of Lupinus polyphyllus. Planta Med. 53, 465–469.
Wink, M., and Witte, L. (1985). Quinolizidine alkaloids as nitrogen source for lupin seedlings and cell cultures. Z. Naturforsch. 40c, 767–775.
Wink, M., Meißner, C., and Witte, L. (1995). Patterns of quinolizidine alkaloids in 56 species of the genus Lupinus. Phytochemistry 38, 139–153.
Yonekura-Sakakibara, K., Fukushima, A., Nakabayashi, R., Hanada, K., Matsuda, F., Sugawara, S., et al. (2012). Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 69, 154–167.
Yonekura-Sakakibara, K., and Saito, K. (2009). Functional genomics for plant natural product biosynthesis. Nat. Prod. Rep. 26, 1466–1487.
Yonekura-Sakakibara, K., Tohge, T., Matsuda, F., Nakabayashi, R., Takayama, H., Niida, R., et al. (2008). Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20, 2160–2176.
Ziegler, J., and Facchini, P. J. (2008). Alkaloid biosynthesis: metabolism and trafficking. Annu. Rev. Plant Biol. 59, 735–769.
Keywords: Lupinus, lysine decarboxylase, lysine-derived alkaloids, o-tigloyltransferase, quinolizidine alkaloids
Citation: Bunsupa S, Yamazaki M and Saito K (2012) Quinolizidine alkaloid biosynthesis: recent advances and future prospects. Front. Plant Sci. 3:239. doi: 10.3389/fpls.2012.00239
Received: 07 August 2012; Paper pending published: 29 August 2012;
Accepted: 09 October 2012; Published online: 26 October 2012.
Edited by:
Gustavo Bonaventure, Max Planck Institute for Chemical Ecology, GermanyReviewed by:
Masami Y. Hirai, RIKEN Plant Science Center, JapanCopyright: © 2012 Bunsupa, Yamazaki and Saito. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Kazuki Saito, Graduate School of Pharmaceutical Sciences, Chiba University, Inohana 1-8-1, Chuo-ku, Chiba 260-8675, Japan. e-mail:a3NhaXRvQGZhY3VsdHkuY2hpYmEtdS5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.