- Department of Biology and Biotechnology, Worcester Polytechnic Institute, Worcester, MA, USA
Kinesins are an ancient superfamily of microtubule dependent motors. They participate in an extensive and diverse list of essential cellular functions, including mitosis, cytokinesis, cell polarization, cell elongation, flagellar development, and intracellular transport. Based on phylogenetic relationships, the kinesin superfamily has been subdivided into 14 families, which are represented in most eukaryotic phyla. The functions of these families are sometimes conserved between species, but important variations in function across species have been observed. Plants possess most kinesin families including a few plant specific families. With the availability of an ever increasing number of genome sequences from plants, it is important to document the complete complement of kinesins present in a given organism. This will help develop a molecular framework to explore the function of each family using genetics, biochemistry, and cell biology. The moss Physcomitrella patens has emerged as a powerful model organism to study gene function in plants, which makes it a key candidate to explore complex gene families, such as the kinesin superfamily. Here we report a detailed phylogenetic characterization of the 71 kinesins of the kinesin superfamily in Physcomitrella. We found a remarkable conservation of families and subfamily classes with Arabidopsis, which is important for future comparative analysis of function. Some of the families, such as kinesins 14s are composed of fewer members in moss, while other families, such as the kinesin 12s are greatly expanded. To improve the comparison between species, and to simplify communication between research groups, we propose a classification of subfamilies based on our phylogenetic analysis.
Introduction
Kinesins are a superfamily of microtubule (MT) dependent motors that are present in all eukaryotes (Richardson et al., 2006). The critical importance for cell function of this superfamily is highlighted by its existence and diversification in the last common ancestor of plants, animals, and fungi. The members of the various families of kinesins perform a multitude of functions, but they are all related by their conserved motor domain (Miki et al., 2005).
The kinesin motor domain, or head, comprises approximately 360 amino acids, and contains the ATPase and MT binding activities. The motor domain can be located either at the C-terminus, N-terminus, or in the middle of the molecule. In addition to the motor domain, most kinesins have a neck region that contains family specific features, a coiled coil region that is important for dimerization, and a tail region that is thought to bind to specific cargo. The directionality of kinesin varies between families, and is sometimes correlated with the position of the motor. The directionality of all the kinesin is not known, but in general, members of the kinesin 1 and 2 families travel to the plus end of MTs, while members of the kinesin 14 family travel toward the minus ends.
Because of the large size of the kinesin superfamily, it has been important to unify the nomenclature across phyla to allow comparative analyses of function. A standardized nomenclature was proposed by a special interest subgroup of the American Society of Cell Biology (ASCB), which has been broadly adopted (Lawrence et al., 2004). This nomenclature separates all major kinesins into 14 families. Kinesins that do not belong to any of these families are considered orphans, but most kinesins identified can easily be assigned to a specific family. Together with the development of high throughput and next generation genomic sequencing, important efforts have taken place to use phylogenetic analysis and classification in diverse species ranging from unicellular to multicellular organisms to explore the large set of functions fulfilled by kinesins (Miki et al., 2005; Richardson et al., 2006).
In plants, kinesins have been implicated in a variety of cellular processes, including intracellular transport, spindle assembly, chromosome motility, phragmoplast assembly, MAP kinase regulation, and MT stability (Vale, 2003; Lee and Liu, 2004; Cai and Cresti, 2010; Zhu and Dixit, 2011a). Plants contain almost all the kinesin families, including kinesins important for flagellar development that are only present in plants with motile sperm, such as ferns and mosses (this study). Occasionally, the function of some members of a family do not appear to be conserved with its animal and fungal counterparts, and plants also contain specific kinesin families (Richardson et al., 2006).
The kinesin content has been determined in various plants. Reddy and co-workers identified 61, 52, 41, and 45 kinesins in Arabidopsis, poplar, and two cultivars of rice, respectively (Richardson et al., 2006), while the red algae Cyanidioschyzon merolae contains only five kinesins and the green algae Chlamydomonas reinhardtii 23 (Reddy and Day, 2001; Richardson et al., 2006). However, the full set of kinesins in basal land plants has yet to be investigated.
The moss Physcomitrella patens is a simple plant model organism that allows precise genetic manipulations and provides easy access to cells for high resolution microscopy (Cove, 2005). This makes it an ideal model system to study the participation of the MT cytoskeleton in many different processes. Surprisingly, only two kinesins, KINID1a and KINID1b, hereafter called Pp-KinesinOrph-IIa and Pp-KinesinOrph-IIb, have been studied in Physcomitrella and have been shown to be essential for the generation of interdigitated antiparallel MT in the phragmoplast (Hiwatashi et al., 2008). This highlights the need to have a complete inventory of the multitude of kinesins present in this organism to help perform future functional analysis. With an available genome sequence (Rensing et al., 2008) it is now possible to document all the kinesins present in this organism. In the present work, we perform a phylogenetic analysis of 71 kinesins from Physcomitrella, identified from their conserved motor domain.
Materials and Methods
Kinesin motor domain sequences were identified by BLAST against the cosmoss.org version 1.6; the 6th annotation of the Physcomitrella first genome assembly (Rensing et al., 2008), and the protein sequences were identified from predicted gene models. A total of 71 sequences were identified (Table 1), the head domain was extracted from the sequences by alignment comparison with a template based on the kinesin 1 head domain (Uniprot: P33176).
For phylogenetic comparison, the sequences were imported into Vector NTI Advance 11.5.1 (Invitrogen), and an alignment was generated using its AlignX program. The basic algorithm from AlignX is ClustalW; we maintained the default parameters as follows: gap opening penalty: 10, gap extension penalty: 0.05, gap separation penalty range: 8, percent identity for alignment delay: 40. The alignment was further improved by identifying the members of each family of kinesins using the fast neighbor distance-based algorithm from AlignX, and aligning the groups separately. The assignment to specific families was very consistent for the majority of the sequences identified. This was done to remove possible minor errors (Figure S1 in Supplementary Material) in the gene models, which were in general present at a low frequency. For the final alignment, the protein sequences of all the motor domains from all the families were used and the sequence for the globular tail domain of Physcomitrella’s myosin XIa (Uniprot: D6R266) was used as an outgroup.
Once a satisfactory alignment was completed, the alignment file was imported to Geneious (Biomatters Ltd.), where a tree was constructed using the plugin PhyML that applies the Maximum Likelihood method (Guindon et al., 2010). We maintained the default parameters as follows: substitution model: LG, proportion of invariable sites: 0-fixed, number of substitution rate categories: 1, no optimization, and a 1000 bootstrap resampling value. To help identify the various family groups, a representative member from human and all S. pombe and Saccharomyces cerevisiae kinesins were included in the alignment. In addition, the complete collection of the Arabidopsis kinesins was included for comparison.
Our preliminary trees constructed with the neighbor joining algorithm available in AlineX from Vector NTI Advance resulted in similar topologies for most classes. Furthermore, in the majority of the families, the human representative sequence is present, providing good support to our alignment and tree building strategy. In the tree that we present here, only nodes showing more than 50% bootstrap support are indicated, and the bootstrap support is shown.
We have used a nomenclature based, when available, on the kinesin family name designated by a number (Lawrence et al., 2004), followed by a class number (indicated by roman numerals; Table 1). To identify individual members of the classes we used letters in the case of Physcomitrella and numbers in the case of Arabidopsis in order to avoid possible future confusion when the classes are monophyletic and no clear orthologs are present between species.
Results and Discussion
In the following sections we report our findings on the number and class of kinesins for each subfamily and when possible discuss their predicted function based on comparison to other similar kinesins. A clear kinesin 1 member is not present in Physcomitrella, but interestingly kinesin 1 members are present in Arabidopsis and other seed plants (Richardson et al., 2006; Zhu and Dixit, 2011a). Because of their similarity with kinesin 1s, we decided to start our report with the Armadillo Repeat containing Kinesins (ARK), and we decided to not include them in the orphan section since they are well conserved across plants and can be clearly identified as a separate group. We were also not able to unequivocally assign moss proteins to families 3, 6, 10, and 11; a more detailed discussion about this is presented in the last section concerning orphan kinesins.
ARK Kinesins
This kinesin family is characterized and classified by the armadillo repeat motifs found within the protein’s C-terminal domain. Armadillo repeats are comprised of a repeating sequence of forty-two amino acids (Coates, 2003). This sequence contains three alpha helices; upon repetition these helices form a right handed super helix (Coates, 2003). Typically, these repeats are associated with cell signaling and the cytoskeleton. In Arabidopsis, it has been speculated that the armadillo regions bind to target proteins to aid in their MT based transport (Coates, 2003). Additionally, loss of function analysis of armadillo kinesins in Arabidopsis root hairs suggests that these proteins may play a significant role in actin and MT organization during polarized cell growth (Yang et al., 2007; Sakai et al., 2008).
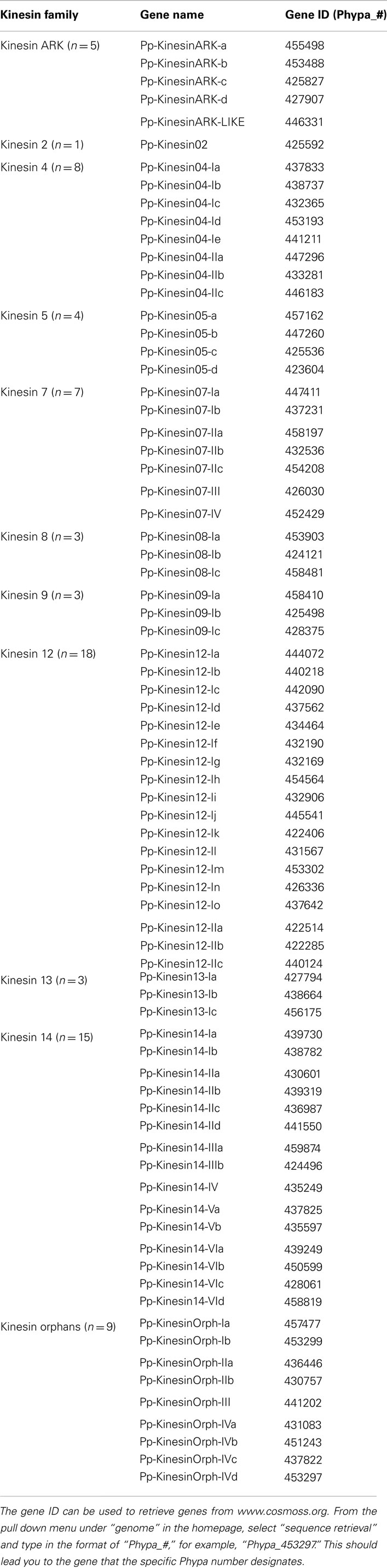
Phylogenetic analysis based on the motor domain indicates that there are five sequences in Physcomitrella related to the Arabidopsis ARK (Figure 1A). Four of these sequences are closely related to each other, forming a monophyletic group and their gene models show the presence of armadillo repeats (Figure 1B); we classified these as class I. The gene model for the fifth sequence is lacking the armadillo repeats that would confirm its identity as an armadillo repeat containing kinesin (Figure 1B); we tentatively classified this single kinesin as ARK-Like since the tree topology fails to confirm this kinesins as an ortholog of the lone At-kinesin 01. But it is intriguing that a very short gene model is also a landmark of this Arabidopsis kinesin 1 (Richardson et al., 2006).
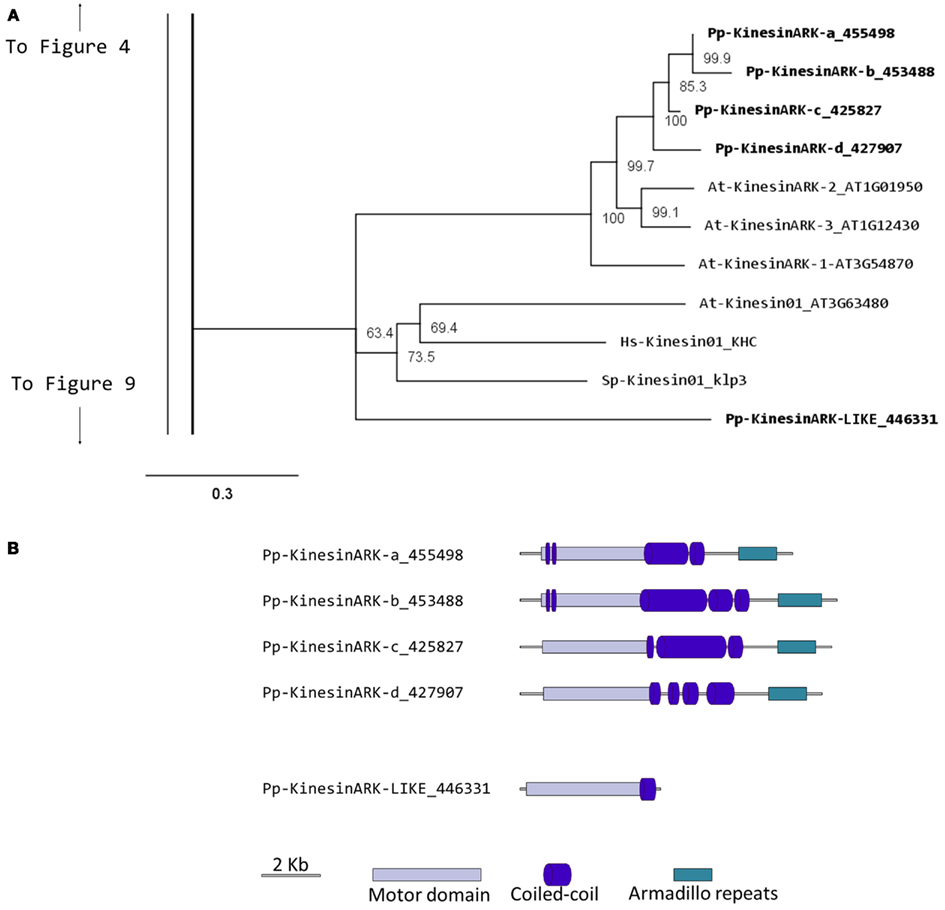
Figure 1. (A) Sub-region of the phylogenetic tree based on their motor domain showing kinesin 1s and armadillo repeat containing kinesins (ARKs). The amino acid sequences of the motor domain were aligned using ClustalW and the phylogenetic tree was constructed using the maximum likelihood method (PhyML) and a 1000 bootstrap resampling value. Numbers on the nodes show the statistical support of values above 50%. The scale shows the estimated branch length corresponding to the number of substitutions per site. The Physcomitrella numbers correspond to the Phypa number uniquely associated with each gene model (version 1.6) at cosmoss.org. (B) Gene models of kinesin 1s and ARKs. Schematic diagrams showing the structure and domain architecture of kinesin 1s and armadillo repeat containing kinesins (ARKs). Domains are indicated at the bottom of the diagrams. Armadillo repeats are comprised of a repeating sequence of forty-two amino acids that can form helices which upon repetition form a right handed super helix.
It will be interesting to investigate if the participation of ARK in cell polarization that has been documented in Arabidopsis (Yang et al., 2007; Sakai et al., 2008), is also conserved in mosses, which provide an excellent model system to study cell polarization and tip growth. Furthermore, comparative analysis of loss of function phenotypes may help understand how this family of molecules functions in the cell.
Kinesin 2
Kinesin 2s have previously been shown to be involved in neuronal organelle transport (Yamazaki et al., 1995; Setou et al., 2000), meiosis in spermatogenesis (Wang et al., 2010), and intraflagellar transport (Sloboda and Howard, 2007). One of the common characteristics of kinesin 2s is their ability to create both homo and heterodimers (Rashid et al., 1995). However, in the case of P. patens, there is only one kinesin 2 present (Figure 2), and therefore it will only homodimerize, unless it can associate with a different kinesin. The protein itself is relatively short, containing two short coiled coils, and one large coiled coil (Figure 3A). In Physcomitrella this protein is likely to participate in the de novo formation of flagella during spermatogenesis. Consistently, kinesin 2s are absent in Arabidopsis and other seed plants which do not have flagella.
Figure 2. Sub-region of the phylogenetic tree based on their motor domain showing kinesin 2s and kinesin 4s. The amino acid sequences of the motor domain were aligned using ClustalW and the phylogenetic tree was constructed using the maximum likelihood method (PhyML) and a 1000 bootstrap resampling value. Numbers on the nodes show the statistical support of values above 50%. The scale shows the estimated branch length corresponding to the number of substitutions per site. The Physcomitrella numbers correspond to the Phypa number uniquely associated with each gene model (version 1.6) at cosmoss.org.
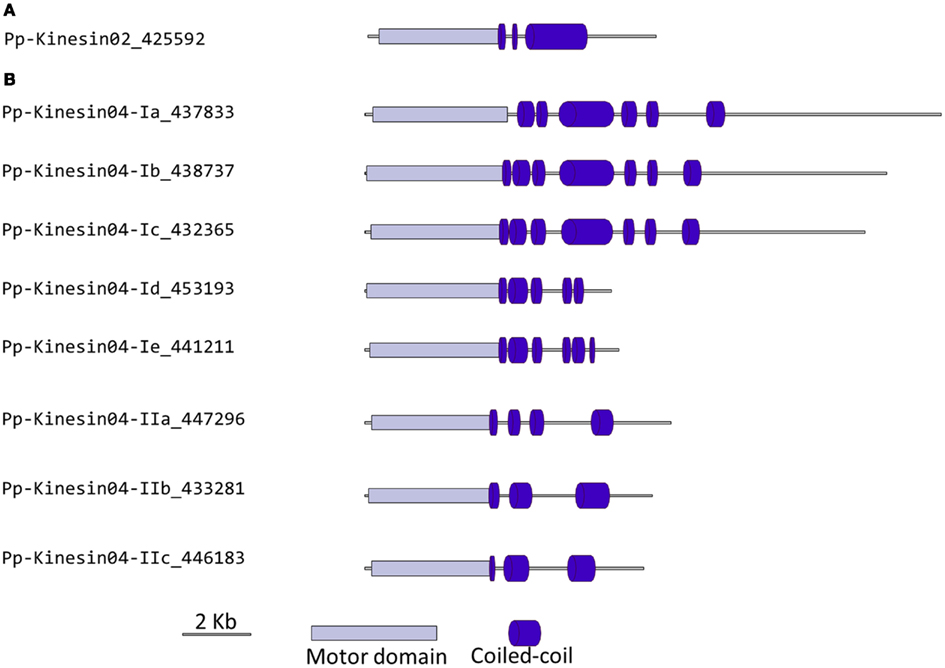
Figure 3. Gene models of kinesin 2s and 4s. Schematic diagrams showing the structure and domain architecture of (A) kinesin 2s and (B) kinesin 4s. Domains are indicated at the bottom of the diagrams.
Kinesin 4
This family is comprised of members that can bind to chromosomes in animals and are absent in budding and fission yeasts (Miki et al., 2005; Richardson et al., 2006). In animals, they are present in the nucleus as well as in the cytoplasm and they have been implicated in organelle and chromosomal transport (Miki et al., 2005). In plants, a member of this family was initially identified as a protein important for orienting cellulose microfibrils (FRA1). A mutation of the protein results in a fragile cell wall phenotype in Arabidopsis (Zhong et al., 2002); a similar mutant was also isolated in rice (Zhang et al., 2010). In addition, the rice kinesin was found to be nuclearly and cytoplasmically localized, and surprisingly it functions as a DNA binding protein important for gibberellin biosynthesis and cell elongation (Li et al., 2011). Single molecule analysis revealed that this molecule has unusually high processivity, suggesting a function in long-distance transport (Zhu and Dixit, 2011b).
Our phylogenetic analysis of kinesin 4s in Physcomitrella shows two well-defined classes (Figure 2), with class I clustering with the Arabidopsis kinesin 4s, including FRA1. Based on the available gene models, the five members of class I can be further subdivided into two classes, with Pp-kin04-Id and Pp-kin04-Ie having smaller C-terminal domains (Figure 3B). Class II is formed by three members, without counterparts in the Arabidopsis genome (Figure 2). This suggests the possibility that this class might carry out a function that is not present in seed plants. It would be interesting to determine whether the class I kinesin 4s have conserved a function in organizing the cell wall components in Physcomitrella, and whether both classes evolved similar or different functions.
It is interesting to note that there is an expanded collection of the kinesin 4s in Physcomitrella compared to Arabidopsis, the significance of this expansion remains to be elucidated.
Kinesin 5
Kinesin 5s are tetrameric kinesins important for spindle organization and mitosis (Ferenz et al., 2010). In yeast, null mutants display division phenotypes such as delayed anaphase, larger cells, and abnormal spindle morphology (Hagan and Yanagida, 1992; Straight et al., 1998). This family spans multiple kingdoms as it is found in mammals, fungi, and plants (Miki et al., 2005; Richardson et al., 2006; Bannigan et al., 2008). Using a conditional loss of function approach, a similar but expanded function has been documented in plants, where one member of this family in Arabidopsis (AtKRP125c) was found to be important for spindle and cortical MT organization (Bannigan et al., 2007).
In Physcomitrella, there are four kinesin 5 members, which cluster as a monophyletic group (Figure 4A). Based on their gene models, they have a very similar structure (Figure 4B). We anticipate that these kinesins will perform similar functions to their Arabidopsis, animal, and fungal counterparts. Nevertheless, it is interesting that a mutation in only one of the four genes of Arabidopsis results in an altered growth phenotype (Bannigan et al., 2007), suggesting a degree of specialization in some of the kinesin 5s in Arabidopsis. Future functional analyses of the four moss isoforms will help clarify whether a similar type of specialization is present or absent in Physcomitrella.
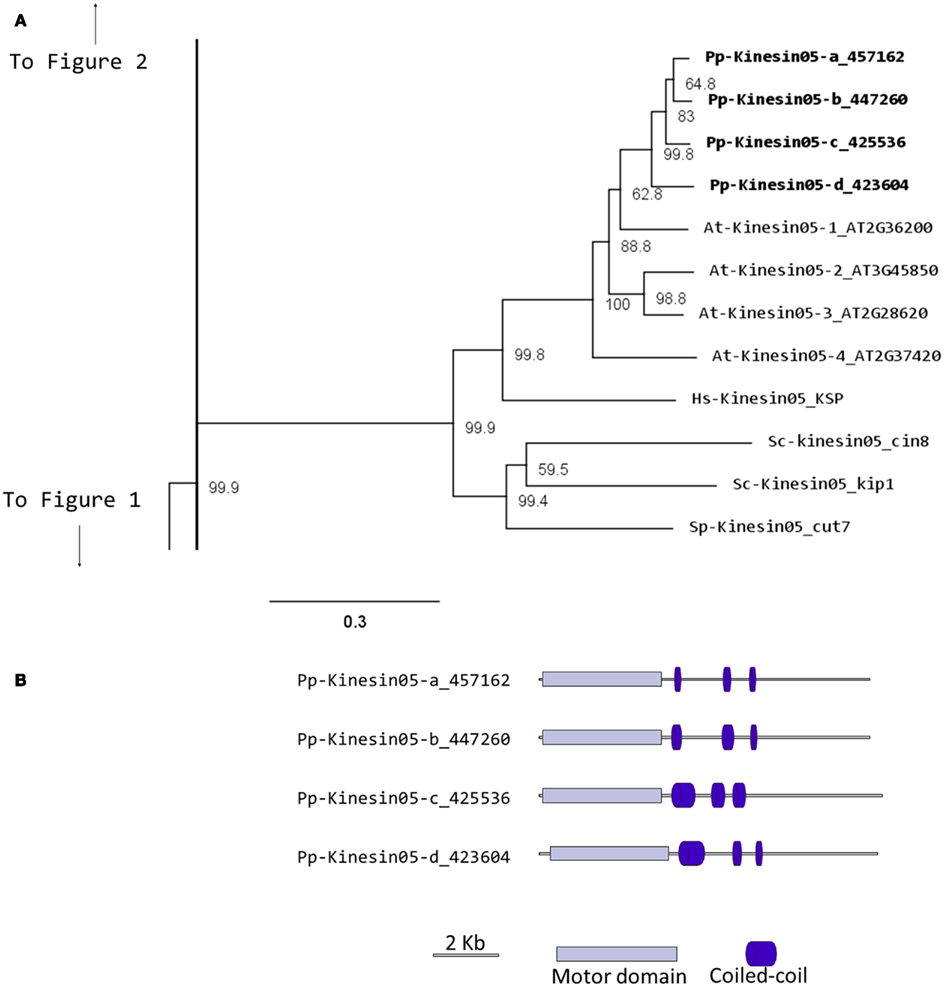
Figure 4. (A) Sub-region of the phylogenetic tree based on their motor domain showing kinesin 5s. The amino acid sequences of the motor domain were aligned using ClustalW and the phylogenetic tree was constructed using the maximum likelihood method (PhyML) and a 1000 bootstrap resampling value. Numbers on the nodes show the statistical support of values above 50%. The scale shows the estimated branch length corresponding to the number of substitutions per site. The Physcomitrella numbers correspond to the Phypa number uniquely associated with each gene model (version 1.6) at cosmoss.org. (B) Gene models of kinesin 5s. Schematic diagrams showing the structure and domain architecture of kinesin 5s. Domains are indicated at the bottom of the diagrams.
Kinesin 7
Members of the kinesin 7 family have been implicated in the transport of chromosomes and nuclear migration in animal and yeast (Miki et al., 2005). This family is greatly expanded in Arabidopsis with 14 members (Richardson et al., 2006; Zhu and Dixit, 2011a). Functional analysis of some of its members has shown a participation in cell division. For example, loss of function of AtNACK1 and AtNACK2 results in inhibition of cytokinesis (Tanaka et al., 2004; Takahashi et al., 2010); a similar phenotype was found in rice, when the expression of the single OsNACK gene is reduced in a leaky mutant (Sazuka et al., 2005). Other members of this family have a mitochondrial signaling sequence, but their function has yet to be investigated (Itoh et al., 2001).
Our phylogenetic analysis shows a smaller size for this family in Physcomitrella with seven members compared with Arabidopsis (Figure 5); nevertheless the classes found seem to be conserved between species. We identified four classes, with class I containing the MKRP-related kinesins, that could be associated with organelles (Itoh et al., 2001). Physcomitrella has only two representatives for this class, compared with five for Arabidopsis. Class II has three representatives in Physcomitrella, compared with seven in Arabidopsis. The moss class II kinesin 7s seem to represent an independent monophyletic group with no specific clustering to the Arabidopsis subgroups. Unfortunately this makes it difficult to clearly define a functional ortholog to the well-characterized NACK kinesins (Tanaka et al., 2004; Takahashi et al., 2010; Sasabe et al., 2011), and further functional analysis will be needed to determine whether the moss class II kinesin 7s are also involved in cytokinesis.
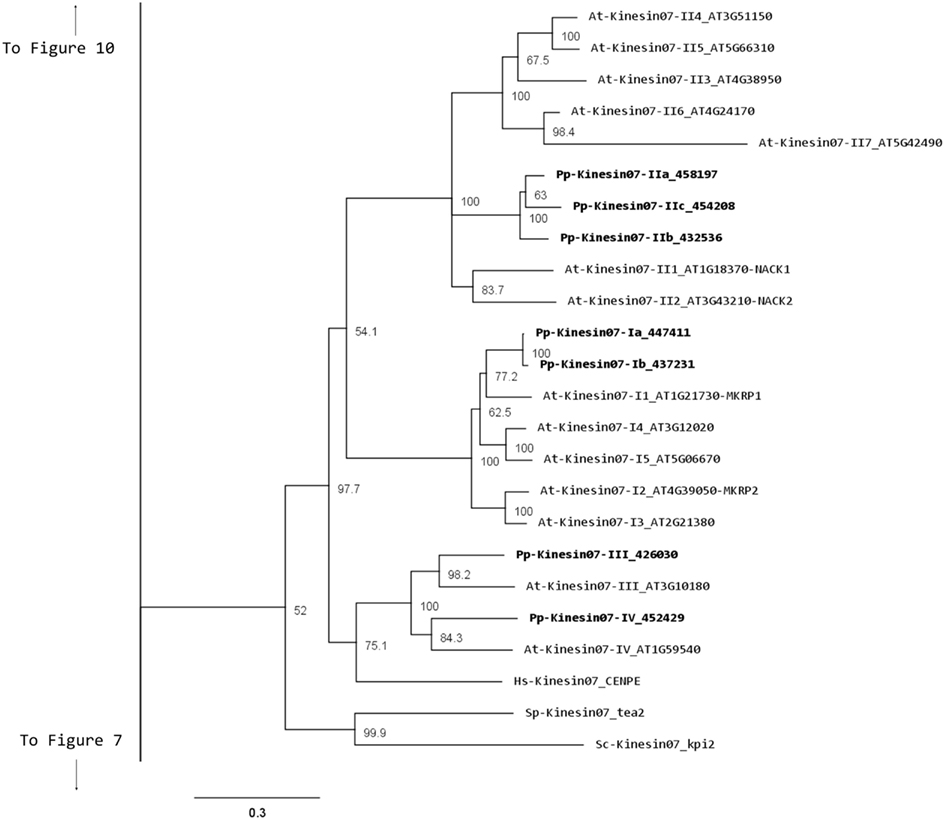
Figure 5. Sub-region of the phylogenetic tree based on their motor domain showing kinesin 7s. The amino acid sequences of the motor domain were aligned using ClustalW and the phylogenetic tree was constructed using the maximum likelihood method (PhyML) and a 1000 bootstrap resampling value. Numbers on the nodes show the statistical support of values above 50%. The scale shows the estimated branch length corresponding to the number of substitutions per site. The Physcomitrella numbers correspond to the Phypa number uniquely associated with each gene model (version 1.6) at cosmoss.org.
Classes III and IV are closely related at the motor domain level (Figure 5), but their C-terminal domains are very different, with class III containing a much longer coiled coil rich domain (Figure 6). Interestingly, orthologs exist for both classes in Arabidopsis, an indication that the common ancestor of mosses and vascular plants contained these two classes. These classes are more closely related to CENPE and may share some of its function on kinetochore capture (Weaver et al., 2003).
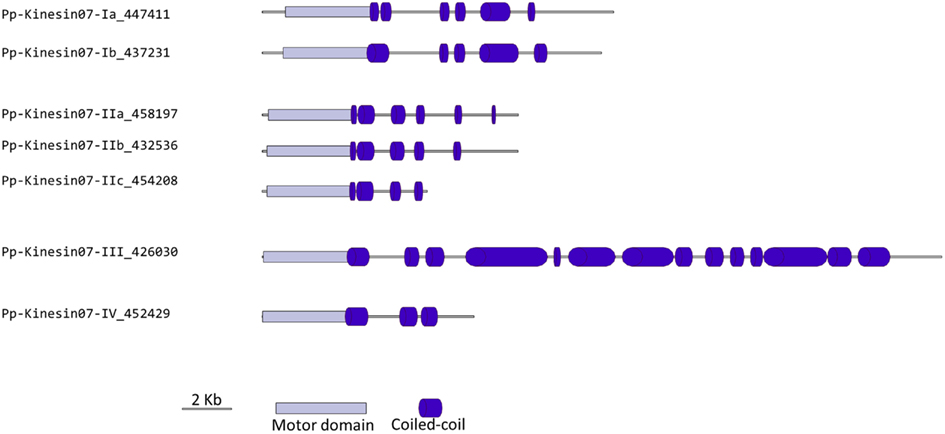
Figure 6. Gene models of kinesin 7s. Schematic diagrams showing the structure and domain architecture of kinesin 7s. Domains are indicated at the bottom of the diagrams.
Kinesin 8
Although nothing is known about kinesin 8s in plant systems, significant research has been conducted on these kinesins in animal and fungi. Some of the group’s functions include mitochondrial transport in Drosophila, mitotic chromosome segregation in yeast, and MT destabilization in humans (Miki et al., 2005; Peters et al., 2010). Our phylogenetic analysis indicates that there are two kinesin 8 classes for both moss and Arabidopsis (Figure 7). Of these two, class I contains a single Arabidopsis kinesin and two moss kinesins; class II contains a single moss and Arabidopsis kinesin. The gene model for the moss class II kinesin shows an extended N-terminal domain (Figure 8A). Because of their similarity to other kinesin 8 members from animals and fungi, we anticipate these kinesins will have a conserved function. Nevertheless, the existence of two ortholog genes in plants suggests that diversification of function was already present, to some degree, in the last common ancestor of bryophytes and vascular plants.
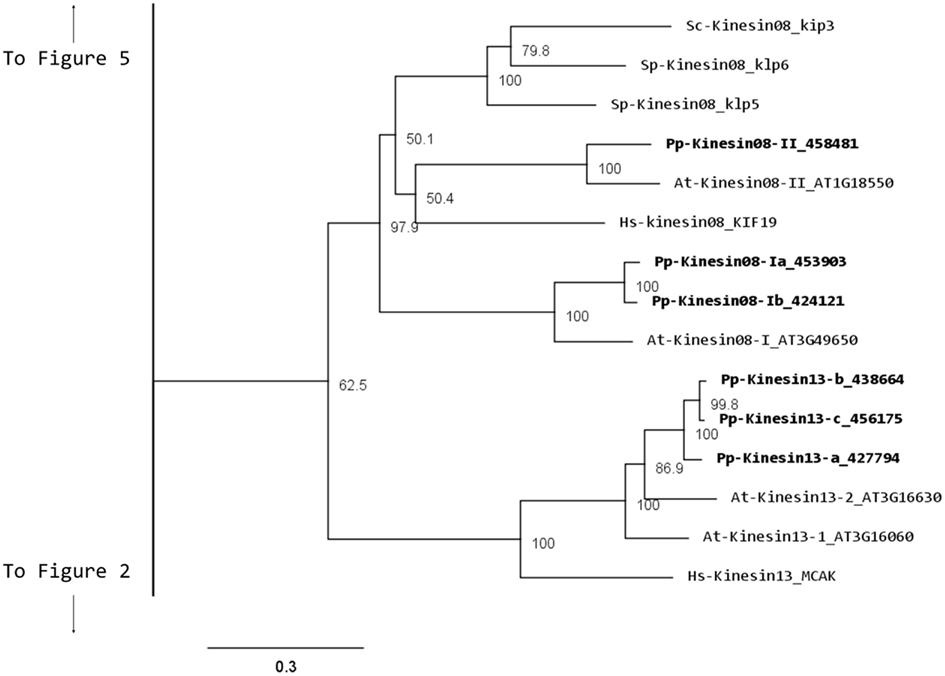
Figure 7. Sub-region of the phylogenetic tree based on their motor domain showing kinesin 8s and kinesin 13s. The amino acid sequences of the motor domain were aligned using ClustalW and the phylogenetic tree was constructed using the maximum likelihood method (PhyML) and a 1000 bootstrap resampling value. Numbers on the nodes show the statistical support of values above 50%. The scale shows the estimated branch length corresponding to the number of substitutions per site. The Physcomitrella numbers correspond to the Phypa number uniquely associated with each gene model (version 1.6) at cosmoss.org.
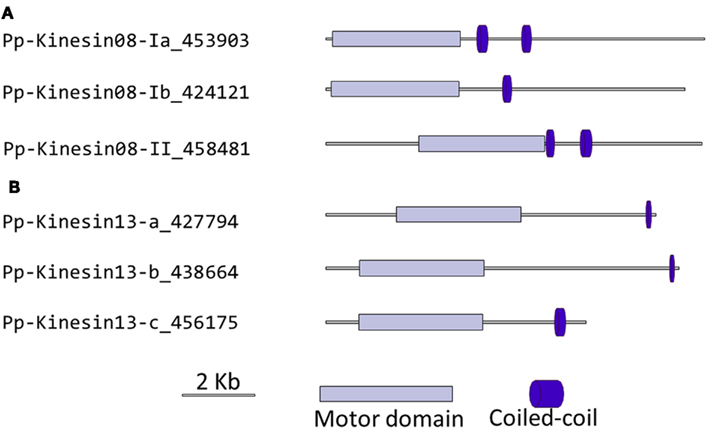
Figure 8. Gene models of (A) kinesin 8s and (B) kinesin 13s. Schematic diagrams showing the structure and domain architecture of kinesin 8s and 13s. Domains are indicated at the bottom of the diagrams.
Kinesin 9
Kinesin 9s have been implicated in flagella regulation, structure, and construction (Bernstein et al., 1994; Yokoyama et al., 2004; Demonchy et al., 2009), and consistent with this function, they are absent in flowering plants, yeast, and invertebrates (Richardson et al., 2006). Our phylogenetic analysis identified three kinesin 9s in Physcomitrella (Figure 9A). Two of the three gene models found in the cosmoss.org database (Pp-Kinesin09-b and c) seem to be inaccurate because of some abnormal insertions and gaps are present when compared to other kinesin 9 sequences. The genomic sequences corresponding to the questionable regions were examined in more detailed and it was found that some exons are not present in the latest proteome version in cosmoss.org (version 1.6) and that some introns were incorrectly spliced (Figure 9B and Figure S1 in Supplementary Material). We were not able to determine if additional problems exist in the gene models of the regions after the motor domain, which due to reduced conservation are harder to identify and their detailed description is beyond the scope of this manuscript. Similarly to kinesin 2 (Sloboda and Howard, 2007), we anticipate kinesin 9s will participate in the de novo assembly of flagella during spermatogenesis in moss.
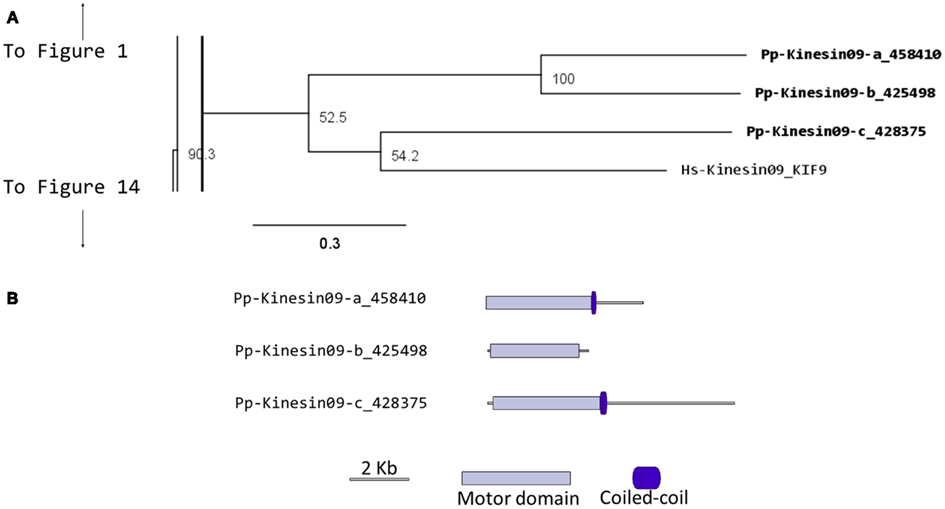
Figure 9. (A) Sub-region of the phylogenetic tree based on their motor domain showing kinesin 9s. The amino acid sequences of the motor domain were aligned using ClustalW and the phylogenetic tree was constructed using the maximum likelihood method (PhyML) and a 1000 bootstrap resampling value. Numbers on the nodes show the statistical support of values above 50%. The scale shows the estimated branch length corresponding to the number of substitutions per site. The Physcomitrella numbers correspond to the Phypa number uniquely associated with each gene model (version 1.6) at cosmoss.org. (B) Gene models of kinesin 9s. Schematic diagrams showing the structure and domain architecture of kinesin 9s. Domains are indicated at the bottom of the diagrams.
Kinesin 10
The kinesin 10 family members are commonly referred as “Kid” in human (Tokai et al., 1996) and “KIF 22” in mouse (Yang et al., 1997). They have been suggested to be involved in spindle formation and chromosome movement (Miki et al., 2005). It is notable that members of the kinesin 10 family, which are present in Arabidopsis (two members) are absent in Physcomitrella (Figure 14). Although PAKRP2 has been sometimes grouped in the kinesin 10 family (Richardson et al., 2006; Zhu and Dixit, 2011a), it is more appropriate to be classified as an orphan kinesin based on our analysis (Figure 14). A detailed discussion about this classification can be found at the section for orphan kinesins below. The presence of orthologs of the Arabidopsis kinesin 10s in other basal plant species may provide clues about essential developmental processes present in a common ancestor but lost in mosses.
Kinesin 12
In animals, kinesin 12s have been implicated in bipolar spindle assembly (Rogers et al., 2000; Tanenbaum et al., 2009) and neuron development and axon growth (Liu et al., 1996; Buster et al., 2003). In plants, kinesin 12s have been found to be involved in phragmoplast organization and orientation (Lee and Liu, 2000; Pan et al., 2004; Muller et al., 2006). In general, kinesin 12s have an N-terminus head with a long C-terminus tail abundant in coiled coils (Miki et al., 2005).
Our phylogenetic analysis shows two classes of kinesins 12s; class I kinesin 12s, which are related to the phragmoplast orienting kinesins or POKs (Muller et al., 2006), and class II kinesin 12s, which are related to the phragmoplast-associated kinesin 1s or PAKRP1s (Figure 10). We found a surprisingly large number of class I kinesin 12s in Physcomitrella: a total of 18 genes, compared with only three in Arabidopsis. Some of the gene models corresponding to regions after the motor domain seem to be incomplete, but the majority of the class I sequences show long C-terminal domains with abundant coiled coils (Figure 11). The significance of this large number of kinesins is not understood and presents a challenging problem due to the likelihood of functional redundancy between its members. Nevertheless, due to their similarity to Arabidopsis POKs, these proteins are probably important for phragmoplast orientation. In contrast to the large number of class I kinesins 12s, there are only three class II kinesin 12s in Physcomitrella, forming a monophyletic group (Figure 10). The gene models for these kinesins show very similar C-terminal structures with abundant coiled coil structures, but not of the large magnitude of the ones present in class I (Figure 11). Again, we anticipate that these kinesins will play a similar role in phragmoplast organization as that of their Arabidopsis counterparts.
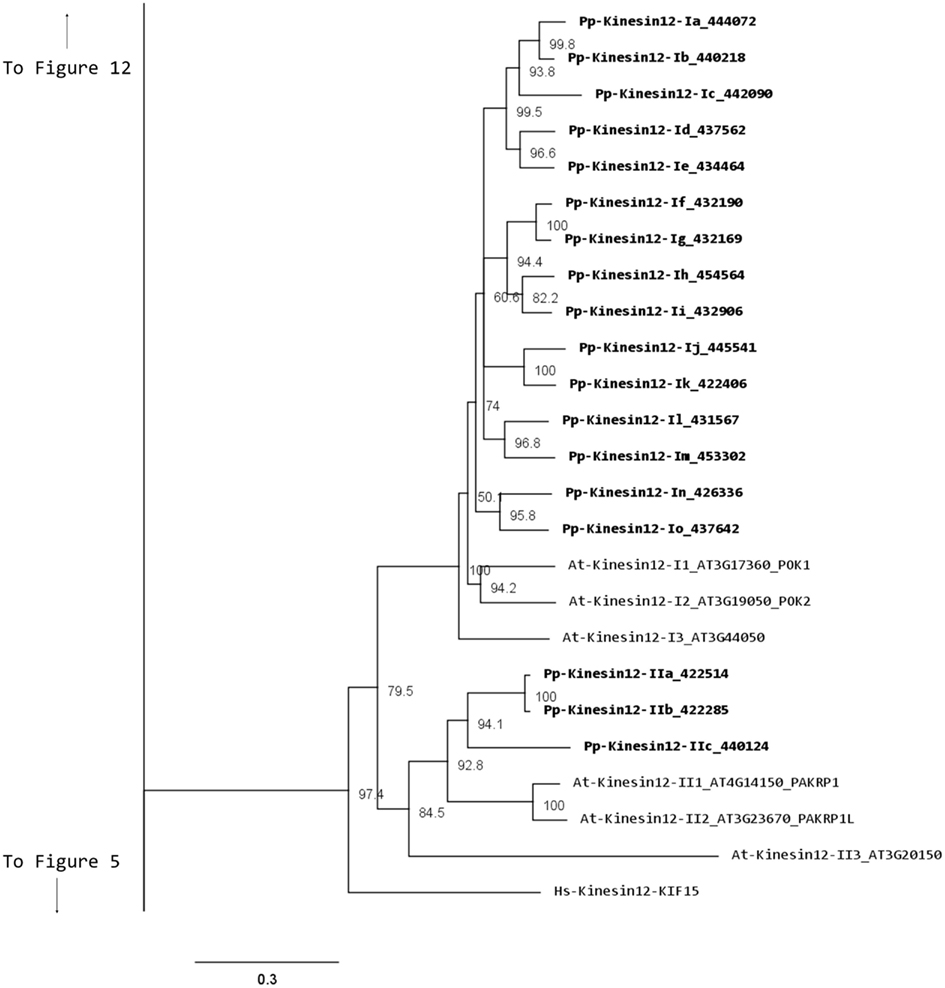
Figure 10. Sub-region of the phylogenetic tree based on their motor domain showing kinesin 12s. The amino acid sequences of the motor domain were aligned using ClustalW and the phylogenetic tree was constructed using the maximum likelihood method (PhyML) and a 1000 bootstrap resampling value. Numbers on the nodes show the statistical support of values above 50%. The scale shows the estimated branch length corresponding to the number of substitutions per site. The Physcomitrella numbers correspond to the Phypa number uniquely associated with each gene model (version 1.6) at cosmoss.org.

Figure 11. Gene models of kinesin 12s. Schematic diagrams showing the structure and domain architecture of kinesin 12s. Domains are indicated at the bottom of the diagrams.
Kinesin 13
Similar to kinesin 8s, the animal kinesin 13s have been reported to destabilize MT and to function in intracellular transport (Miki et al., 2005). Although both plant and animal kinesin 13s share similar motor domain sequences, the kinesin 13s in plants do not have a lysine-rich neck domain present in animal kinesin 13s (Lee and Liu, 2004). The absence of this structural motif suggests that the plant kinesins may have a different function than their animal counterparts (Lee and Liu, 2004). Consistently, the Arabidopsis kinesin-13-2, which has been shown to be associated with Golgi stacks, provides further evidence that plant kinesin 13s may differ from animal kinesin 13s in functionality (Lu et al., 2005).
Our phylogenetic analysis shows that in Arabidopsis there are two kinesin 13s while in Physcomitrella, there are three kinesin 13 members, which cluster as a monophyletic group (Figure 7). Based on their gene models, the moss kinesin 9s have a very similar structure (Figure 8B). Interestingly, for the three moss kinesin 9s, the sole coiled coil region is located at the extreme C-terminus of the molecule away from the motor domain; while in most of the kinesins, the coiled coil regions are generally closer to the motor domain (Figure 8B). It would be interesting to determine whether this structural characteristic is important for the function of these kinesins. The Arabidopsis kinesin-13-2 is closely related to those found in moss (Figure 7), which suggests possible functional conservation between the Arabidopsis kinesin-13-2 and all the moss kinesin 13s.
Kinesin 14
Members of this family have been associated with functions in re-arrangement of the MT arrays at various stages of the cell cycle as well as in organelle transport (Miki et al., 2005; Richardson et al., 2006; Zhu and Dixit, 2011a). Kinesin 14s were initially divided in two groups, kinesin 14A and kinesin 14B, according to their structure and function (Miki et al., 2005). However, this family is vastly expanded in plants with 21 and 15 members in Arabidopsis and Physcomitrella, respectively, compared to 4 in humans (Richardson et al., 2006). In addition, the fact that plant kinesins 14 display some specific structural motifs prompted us to propose a new classification for the plant kinesin 14s, divided in six different classes (Figure 12).
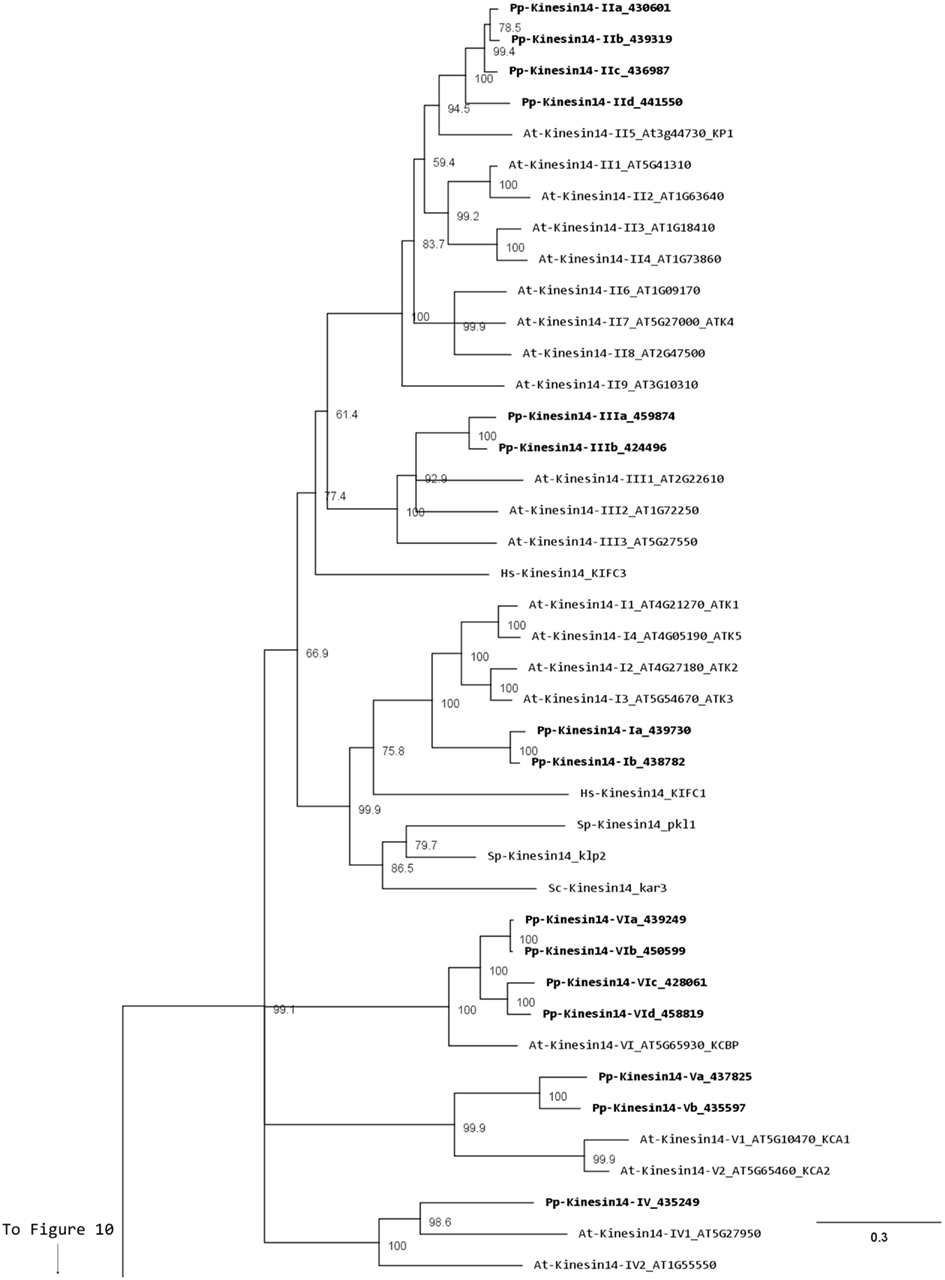
Figure 12. Sub-region of the phylogenetic tree based on their motor domain showing kinesin 14s. The amino acid sequences of the motor domain were aligned using ClustalW and the phylogenetic tree was constructed using the maximum likelihood method (PhyML) and a 1000 bootstrap resampling value. Numbers on the nodes show the statistical support of values above 50%. The scale shows the estimated branch length corresponding to the number of substitutions per site. The Physcomitrella numbers correspond to the Phypa number uniquely associated with each gene model (version 1.6) at cosmoss.org.
Class I kinesin 14s are related to KIFC1 which is associated with the nuclear membrane in mammalian cells and is important for acrosome biogenesis and possibly for vesicle transport (Yang and Sperry, 2003; Yang et al., 2006; Nath et al., 2007), and to Kar3p which is essential for nuclear fusion during mating in S. cerevisiae, by mediating MT sliding (Meluh and Rose, 1990). Among the four homologs present in Arabidopsis (Figure 12), ATK1 and ATK5 have been well studied. They share similar functions during mitosis by controlling the MT organization at the cortex, the preprophase band, the spindle, and the phragmoplast, and ATK1 also play a major role in male meiosis (Liu et al., 1996; Chen et al., 2002; Marcus et al., 2003; Ambrose et al., 2005; Ambrose and Cyr, 2007). Interestingly, ATK5 possesses a second, ATP independent, MT binding site on its N-terminal region, that could be important for bundling, a property already reported for the kinesin Ncd in Drosophila (Furuta and Toyoshima, 2008). Class I in Physcomitrella is composed of two members (Figure 12), which contain a C-terminal motor domain (Figure 13), and therefore, similar to ATK5 (Ambrose et al., 2005), are likely to be minus-end directed motors.
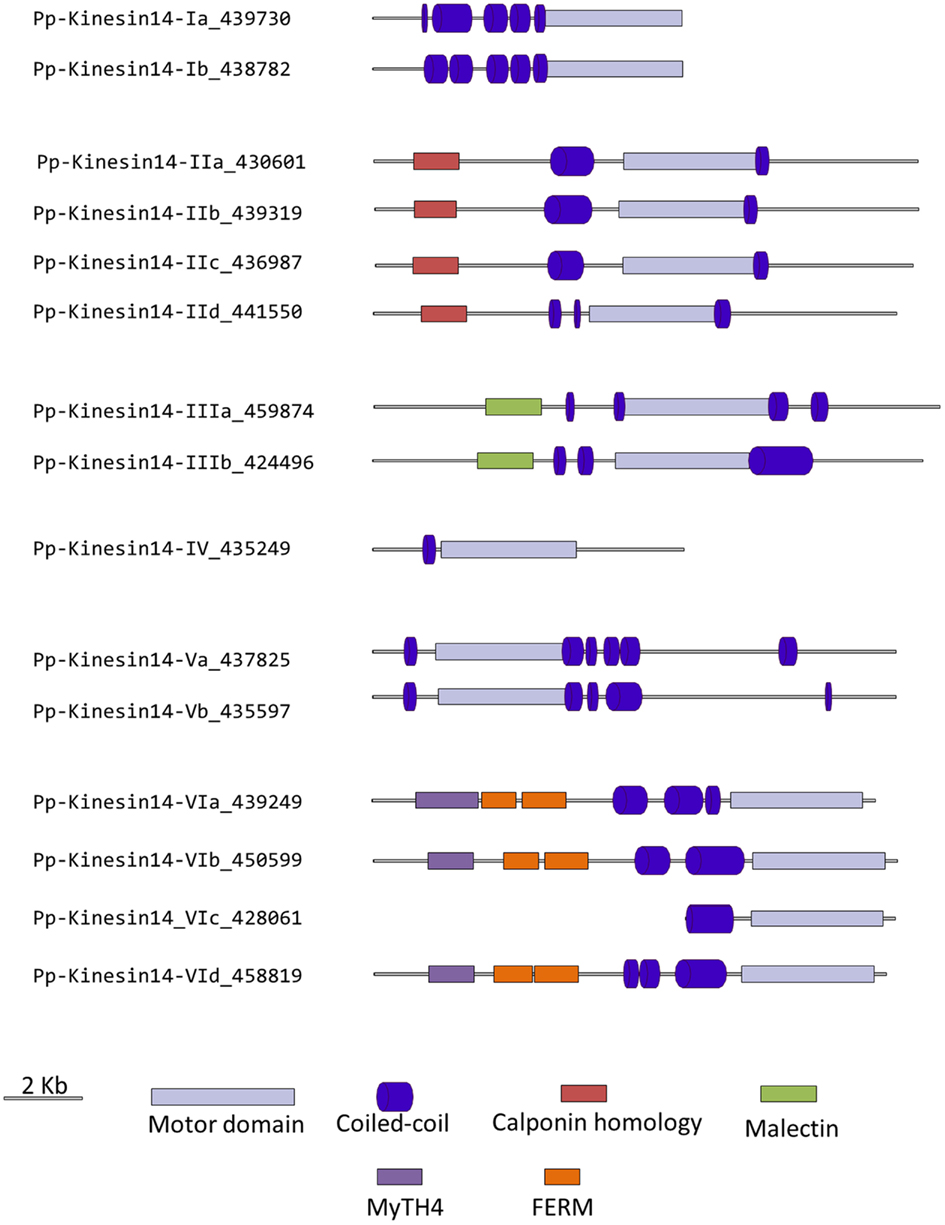
Figure 13. Gene models of kinesin 14s. Schematic diagrams showing the structure and domain architecture of kinesin14s. Domains are indicated at the bottom of the diagrams. Calponin homology domains mediate interaction with F-actin; Malectin domains allow binding to carbohydrate such as di-glucose; and Myosin Tail Homology domain 4 (MyTH4) and FERM domains (motif named after proteins that contains it: 4.1 protein, Ezrin, Radixin, Moesin) also called talin-like region, are known to bind microtubules.
Members of classes II and III are related to KIFC3, which is involved in Golgi positioning and integration in mouse (Xu et al., 2002). Class II is the largest class with eight members in Arabidopsis and four in Physcomitrella (Figure 12). In Arabidopsis, AtKP1 has been shown to organize the cortical MT array (Ni et al., 2005; Yang et al., 2011), and to regulate mitochondrial functions (Yang et al., 2011). ATK4 and homologs in cotton and rice, possess a Calponin Homology (CH) domain that mediates interaction with F-actin (Tamura et al., 1999; Preuss et al., 2004; Frey et al., 2009; Xu et al., 2009), and could be important for regulating the motor activity and coordinating the activities of MT and actin microfilaments during premitotic nuclear migration (Umezu et al., 2011). Interestingly, the four members of this class in Physcomitrella also contain the CH domain (Figure 13), strongly suggesting that the binding to actin microfilaments is conserved in moss.
Class III has three representatives in Arabidopsis and two in Physcomitrella (Figure 12). However, the function of these kinesins still remains unknown. Intriguingly, these kinesins possess a malectin domain (Figure 13), which allows binding to carbohydrate such as di-glucose (Schallus et al., 2008). This domain has been identified in malectins, which are conserved proteins of the endoplasmic reticulum in animals and involved in protein N-glycosylation (Schallus et al., 2008), as well as in plasma membrane-located leucin-rich repeat receptor kinases such as FERONIA in Arabidopsis, where it is thought to regulate cell growth in response to cell wall changes (Zou et al., 2011). The functional significance of the malectin domains for kinesins is unclear and further investigation will be needed to decipher the role of class III kinesin 14s in plants.
Class IV kinesin 14s are plant specific. The homolog in tobacco, TBK5, is thought to function in relocating and gathering newly formed MTs and/or MTs nucleating units (Goto and Asada, 2007). Physcomitrella possesses only one member compared to two in Arabidopsis (Figure 12), which makes it a great system to gain more insight into the function(s) fulfilled by these kinesins in plants.
Class V kinesin 14s KAC1 and KAC2 have been recently identified in Arabidopsis in a genetic screen for chloroplast movement in response to light intensity changes (Suetsugu et al., 2010). Interestingly, they show no MT binding activity or detectable ATPase activity. Instead, they are thought to interact with actin microfilaments and mediate chloroplast movement in an actin-dependent manner. However, the precise mechanism by which they regulate chloroplast movement still needs to be determined. Physcomitrella also contains two members of this class (Figure 12) and whether or not they interact with MTs and/or actin filaments to move chloroplasts is not known. It will be interesting to investigate whether these two members of the class V kinesin 14 have similar functions as their Arabidopsis homologs. We want to note that gene model corresponding to the N-terminal sequence for kinesin 14-Vb is incorrect in Phytozome due to an incorrect prediction of a splicing site. This does not affect the motor domain sequence that was used for our phylogenetic tree. We provide in the supplementary material what we believe is the correct protein sequence for this molecule (Figure S1 in Supplementary Material).
Surprisingly, class VI encompasses only one member in Arabidopsis, KCBP, compared to four in Physcomitrella (Figure 12). KCBP, which contains a calmodulin-binding domain, participates in cortical MT organization (Oppenheimer et al., 1997), and is involved in the different stages of mitosis by regulating bundling and sliding of MT (Bowser and Reddy, 1997; Vos et al., 2000). The four moss homologs contain a myosin tail homology domain 4 (MyTH4) and two FERM domains (motif named after proteins that contains it: 4.1 protein, Ezrin, Radixin, Moesin) also called talin-like region (Figure 13), which are known to bind MTs (Narasimhulu et al., 1997). Therefore, moss class VI kinesin 14s are likely to function in cross-linking or bundling of MTs.
Orphan kinesins
We have grouped the remainder moss kinesins into four classes based on the similarity of their motor domain. Class I is composed of two related kinesins with no homologs in Arabidopsis or animals (Figure 14). The gene models for the region outside of the motor domain may not be well predicted due to limited transcript sequence information, so it is difficult to deduce any specific function from the available gene model sequence (Figure 15). A similar situation is present for the single member of class III (Figures 14 and 15). Due to the small number of members in these classes it should be relatively simple to evaluate their function using the various loss of function techniques available in Physcomitrella. However, it is also relevant to mention that at this point it is hard to rule out the possibility that these genes might be pseudogenes.
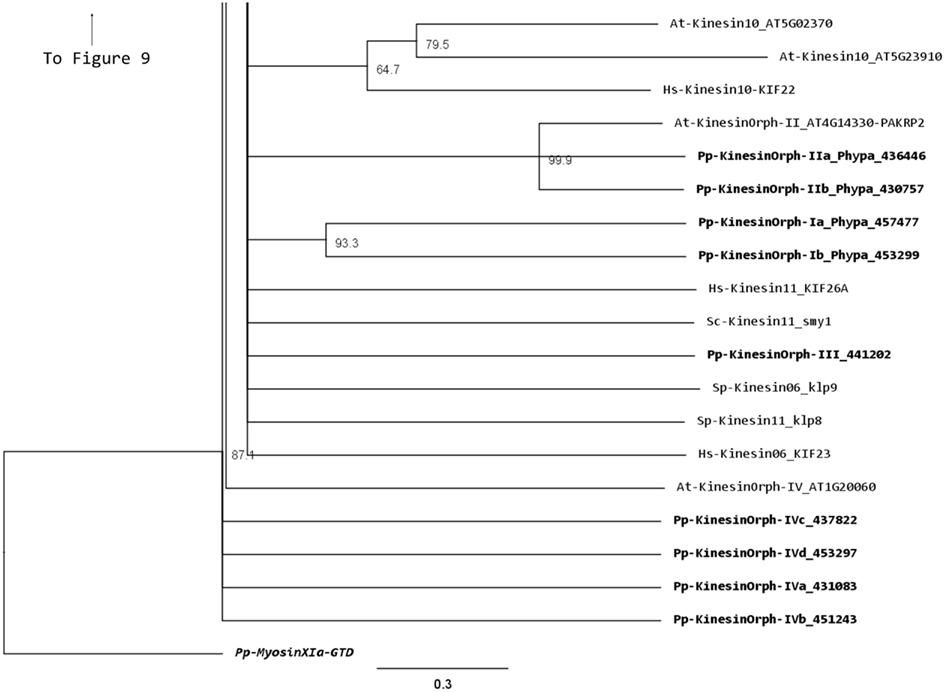
Figure 14. Sub-region of the phylogenetic tree based on their motor domain showing kinesin 6s, 10s, 11s, and orphan kinesins. The amino acid sequences of the motor domain were aligned using ClustalW and the phylogenetic tree was constructed using the maximum likelihood method (PhyML) and a 1000 bootstrap resampling value. Numbers on the nodes show the statistical support of values above 50%. The scale shows the estimated branch length corresponding to the number of substitutions per site. The Physcomitrella numbers correspond to the Phypa number uniquely associated with each gene model (version 1.6) at cosmoss.org.
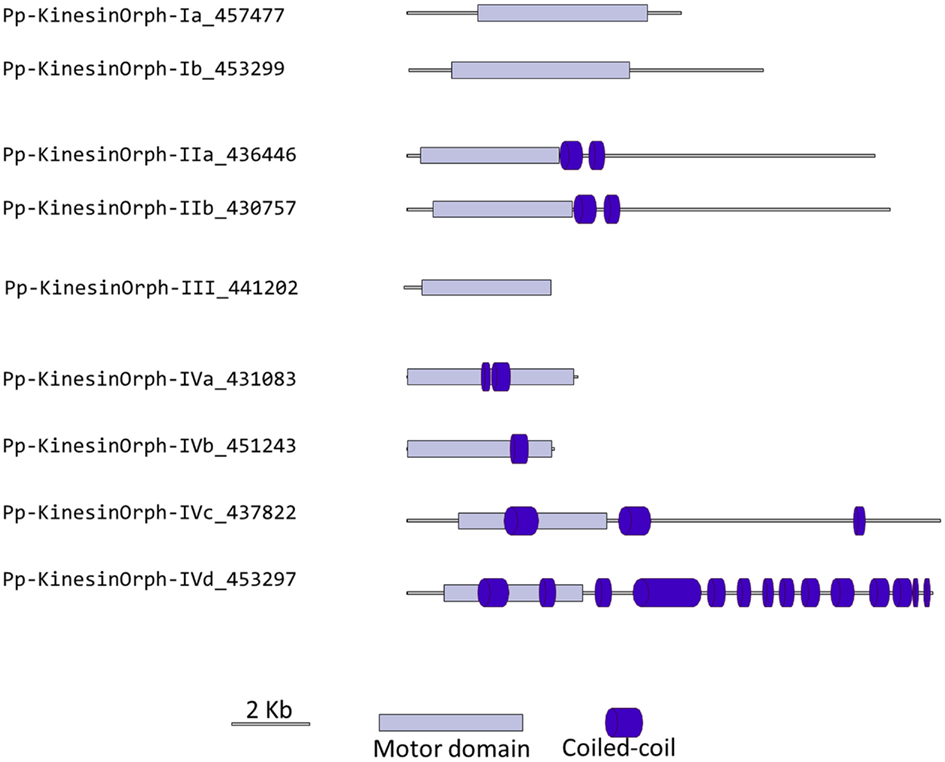
Figure 15. Gene models of orphan kinesins. Schematic diagrams showing the structure and domain architecture of orphan kinesins. Domains are indicated at the bottom of the diagrams.
The class II orphan kinesins are composed of a pair of kinesins (KINID1a and KINID1b) that have been shown to be important for interdigitation of phragmoplast MTs and cell plate expansion in moss (Hiwatashi et al., 2008). These kinesins are orthologs of PAKRP2 from Arabidopsis, which is predicted to function in the transport of Golgi-derived vesicles in the phragmoplast (Lee et al., 2001); nevertheless a conserved function between the moss and Arabidopsis orthologs has so far not been established (Hiwatashi et al., 2008). In previous analysis, where PAKRP2 was classified as a kinesin 10, the clade that PAKRP2 belonged to was parallel with other kinesin families as well as the second clade of Kinesin 10. In addition, that particular clade was not resolved in three of the four methods used to build the tree and had a low score in the methods that resolved the clade (Richardson et al., 2006). Based on our analysis, we suggest an orphan classification for PAKRP2 because it groups with the moss orthologs with a high bootstrap score, but not with bona fide kinesin 10s. Furthermore, the class II orphan kinesins have much longer C-terminal domains when compared with kinesin 10s. It is therefore more appropriate for these kinesins to be classified as orphan kinesins.
There is only one class III kinesin found in Physcomitrella. This kinesin is highly divergent since its relationship cannot be resolved between kinesin 6s and kinesin 11s from human and yeasts in our phylogenetic tree (Figure 14). Whether this kinesin will have conserved function to kinesin 6 or 11 is not clear form our tree, but kinesin 6s and 11s are known to be highly divergent (Miki et al., 2005). In addition, the lacking of gene model information on the C-terminal after the motor domain casts shadow on the possibility that it might also be a pseudogene (Figure 15).
The final group of kinesins is class IV; these kinesins have the most divergent motor domains and tend to cluster with kinesin 11 members from animals and yeasts (Figure 14). In yeast, kinesin 11 or Smy1p does not bind to MTs and it seems to regulate myosin V function (Lillie and Brown, 1998; Beningo et al., 2000). The four class IV members in Physcomitrella have coiled coils in their head domains (Figure 15), suggesting a non-functional motor similar to yeast. Therefore these four kinesins are classified as class IV of the orphan kinesins. Besides the relatively low homology in the motor domains, there is little additional similarity in the rest of the molecules from this class (Figure 15). Analogous to class I and III kinesins, it will be important to determine their phylogenetic distribution and conservation in other species, but a detailed loss of function and biochemical analyzes will be required to determine their function.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members from the Vidali Lab for support and discussion, in particular to Erin Agar for careful reading of the manuscript and her comments. Support was provided by WPI-startup fund and a National Science Foundation grant to Luis Vidali (IOS-1002837).
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Plant_Cell_Biology/10.3389/fpls.2012.00230/abstract
Figure S1. Updated sequences for Pp-Kinesin09-b, Pp-Kinesin09-c, and Pp-Kinesin14-Vb. The bold sequences indicate exons absent from the Phytozome database. These sequences were updated from the genomic sequence information available on JGI (www.jgi.doe.gov) by aligning representative kinesins from multiple species.
References
Ambrose, J. C., and Cyr, R. (2007). The kinesin ATK5 functions in early spindle assembly in Arabidopsis. Plant Cell 19, 226–236.
Ambrose, J. C., Li, W., Marcus, A., Ma, H., and Cyr, R. (2005). A minus-end-directed kinesin with plus-end tracking protein activity is involved in spindle morphogenesis. Mol. Biol. Cell 16, 1584–1592.
Bannigan, A., Lizotte-Waniewski, M., Riley, M., and Baskin, T. I. (2008). Emerging molecular mechanisms that power and regulate the anastral mitotic spindle of flowering plants. Cell Motil. Cytoskeleton 65, 1–11.
Bannigan, A., Scheible, W. R., Lukowitz, W., Fagerstrom, C., Wadsworth, P., Somerville, C., et al. (2007). A conserved role for kinesin-5 in plant mitosis. J. Cell. Sci. 120, 2819–2827.
Beningo, K. A., Lillie, S. H., and Brown, S. S. (2000). The yeast kinesin-related protein Smy1p exerts its effects on the class V myosin Myo2p via a physical interaction. Mol. Biol. Cell 11, 691–702.
Bernstein, M., Beech, P. L., Katz, S. G., and Rosenbaum, J. L. (1994). A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J. Cell Biol. 125, 1313–1326.
Bowser, J., and Reddy, A. S. (1997). Localization of a kinesin-like calmodulin-binding protein in dividing cells of Arabidopsis and tobacco. Plant J. 12, 1429–1437.
Buster, D. W., Baird, D. H., Yu, W. Q., Solowska, J. M., Chauviere, M., Mazurek, A., et al. (2003). Expression of the mitotic kinesin Kif15 in postmitotic neurons: implications for neuronal migration and development. J. Neurocytol. 32, 79–96.
Cai, G., and Cresti, M. (2010). Microtubule motors and pollen tube growth – still an open question. Protoplasma 247, 131–143.
Chen, C., Marcus, A., Li, W., Hu, Y., Calzada, J. P., Grossniklaus, U., et al. (2002). The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development 129, 2401–2409.
Coates, J. C. (2003). Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol. 13, 463–471.
Demonchy, R., Blisnick, T., Deprez, C., Toutirais, G., Loussert, C., Marande, W., et al. (2009). Kinesin 9 family members perform separate functions in the trypanosome flagellum. J. Cell Biol. 187, 615–622.
Ferenz, N. P., Gable, A., and Wadsworth, P. (2010). Mitotic functions of kinesin-5. Semin. Cell Dev. Biol. 21, 255–259.
Frey, N., Klotz, J., and Nick, P. (2009). Dynamic bridges – a calponin-domain kinesin from rice links actin filaments and microtubules in both cycling and non-cycling cells. Plant Cell Physiol. 50, 1493–1506.
Furuta, K., and Toyoshima, Y. Y. (2008). Minus-end-directed motor Ncd exhibits processive movement that is enhanced by microtubule bundling in vitro. Curr. Biol. 18, 152–157.
Goto, Y., and Asada, T. (2007). Excessive expression of the plant kinesin TBK5 converts cortical and perinuclear microtubules into a radial array emanating from a single focus. Plant Cell Physiol. 48, 753–761.
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321.
Hagan, I., and Yanagida, M. (1992). Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature 356, 74–76.
Hiwatashi, Y., Obara, M., Sato, Y., Fujita, T., Murata, T., and Hasebe, M. (2008). Kinesins are indispensable for interdigitation of phragmoplast microtubules in the moss Physcomitrella patens. Plant Cell 20, 3094–3106.
Itoh, R., Fujiwara, M., and Yoshida, S. (2001). Kinesin-related proteins with a mitochondrial targeting signal. Plant Physiol. 127, 724–726.
Lawrence, C. J., Dawe, R. K., Christie, K. R., Cleveland, D. W., Dawson, S. C., Endow, S. A., et al. (2004). A standardized kinesin nomenclature. J. Cell Biol. 167, 19–22.
Lee, Y. R., Giang, H. M., and Liu, B. (2001). A novel plant kinesin-related protein specifically associates with the phragmoplast organelles. Plant Cell 13, 2427–2439.
Lee, Y. R., and Liu, B. (2000). Identification of a phragmoplast-associated kinesin-related protein in higher plants. Curr. Biol. 10, 797–800.
Lee, Y. R., and Liu, B. (2004). Cytoskeletal motors in Arabidopsis. Sixty-one kinesins and seventeen myosins. Plant Physiol. 136, 3877–3883.
Li, J., Jiang, J., Qian, Q., Xu, Y., Zhang, C., Xiao, J., et al. (2011). Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell 23, 628–640.
Lillie, S. H., and Brown, S. S. (1998). Smy1p, a kinesin-related protein that does not require microtubules. J. Cell Biol. 140, 873–883.
Liu, B., Cyr, R. J., and Palevitz, B. A. (1996). A kinesin-like protein, KatAp, in the cells of Arabidopsis and other plants. Plant Cell 8, 119–132.
Lu, L., Lee, Y. R., Pan, R., Maloof, J. N., and Liu, B. (2005). An internal motor kinesin is associated with the Golgi apparatus and plays a role in trichome morphogenesis in Arabidopsis. Mol. Biol. Cell 16, 811–823.
Marcus, A. I., Li, W., Ma, H., and Cyr, R. J. (2003). A kinesin mutant with an atypical bipolar spindle undergoes normal mitosis. Mol. Biol. Cell 14, 1717–1726.
Meluh, P. B., and Rose, M. D. (1990). KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell 60, 1029–1041.
Miki, H., Okada, Y., and Hirokawa, N. (2005). Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 15, 467–476.
Muller, S., Han, S., and Smith, L. G. (2006). Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Curr. Biol. 16, 888–894.
Narasimhulu, S. B., Kao, Y. L., and Reddy, A. S. N. (1997). Interaction of Arabidopsis kinesin-like calmodulin binding protein with tubulin subunits: modulation by Ca2+-calmodulin. Plant J. 12, 1139–1149.
Nath, S., Bananis, E., Sarkar, S., Stockert, R. J., Sperry, A. O., Murray, J. W., et al. (2007). Kif5B and Kifc1 interact and are required for motility and fission of early endocytic vesicles in mouse liver. Mol. Biol. Cell 18, 1839–1849.
Ni, C. Z., Wang, H. Q., Xu, T., Qu, Z., and Liu, G. Q. (2005). AtKP1, a kinesin-like protein, mainly localizes to mitochondria in Arabidopsis thaliana. Cell Res. 15, 725–733.
Oppenheimer, D. G., Pollock, M. A., Vacik, J., Szymanski, D. B., Ericson, B., Feldmann, K., et al. (1997). Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 94, 6261–6266.
Pan, R. Q., Lee, Y. R., and Liu, B. (2004). Localization of two homologous Arabidopsis kinesin-related proteins in the phragmoplast. Planta 220, 156–164.
Peters, C., Brejc, K., Belmont, L., Bodey, A. J., Lee, Y., Yu, M., et al. (2010). Insight into the molecular mechanism of the multitasking kinesin-8 motor. EMBO J. 29, 3437–3447.
Preuss, M. L., Kovar, D. R., Lee, Y. R., Staiger, C. J., Delmer, D. P., and Liu, B. (2004). A plant-specific kinesin binds to actin microfilaments and interacts with cortical microtubules in cotton fibers. Plant Physiol. 136, 3945–3955.
Rashid, D. J., Wedaman, K. P., and Scholey, J. M. (1995). Heterodimerization of the two motor subunits of the heterotrimeric kinesin, KRP85/95. J. Mol. Biol. 252, 157–162.
Reddy, A. S., and Day, I. S. (2001). Kinesins in the Arabidopsis genome: a comparative analysis among eukaryotes. BMC Genomics 2, 2. doi:10.1186/1471-2164-2-2
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69.
Richardson, D. N., Simmons, M. P., and Reddy, A. S. (2006). Comprehensive comparative analysis of kinesins in photosynthetic eukaryotes. BMC Genomics 7, 18. doi:10.1186/1471-2164-7-18
Rogers, G. C., Chui, K. K., Lee, E. W., Wedaman, K. P., Sharp, D. J., Holland, G., et al. (2000). A kinesin-related protein, KRP(180), positions prometaphase spindle poles during early sea urchin embryonic cell division. J. Cell Biol. 150, 499–512.
Sakai, T., Honing, H., Nishioka, M., Uehara, Y., Takahashi, M., Fujisawa, N., et al. (2008). Armadillo repeat-containing kinesins and a NIMA-related kinase are required for epidermal-cell morphogenesis in Arabidopsis. Plant J. 53, 157–171.
Sasabe, M., Boudolf, V., De Veylder, L., Inze, D., Genschik, P., and Machida, Y. (2011). Phosphorylation of a mitotic kinesin-like protein and a MAPKKK by cyclin-dependent kinases (CDKs) is involved in the transition to cytokinesis in plants. Proc. Natl. Acad. Sci. U.S.A. 108, 17844–17849.
Sazuka, T., Aichi, I., Kawai, T., Matsuo, N., Kitano, H., and Matsuoka, M. (2005). The rice mutant dwarf bamboo shoot 1: a leaky mutant of the NACK-type kinesin-like gene can initiate organ primordia but not organ development. Plant Cell Physiol. 46, 1934–1943.
Schallus, T., Jaeckh, C., Feher, K., Palma, A. S., Liu, Y., Simpson, J. C., et al. (2008). Malectin: a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol. Biol. Cell 19, 3404–3414.
Setou, M., Nakagawa, T., Seog, D. H., and Hirokawa, N. (2000). Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science 288, 1796–1802.
Sloboda, R. D., and Howard, L. (2007). Localization of EB1, IFT polypeptides, and kinesin-2 in Chlamydomonas flagellar axonemes via immunogold scanning electron microscopy. Cell Motil. Cytoskeleton 64, 446–460.
Straight, A. F., Sedat, J. W., and Murray, A. W. (1998). Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 143, 687–694.
Suetsugu, N., Yamada, N., Kagawa, T., Yonekura, H., Uyeda, T. Q., Kadota, A., et al. (2010). Two kinesin-like proteins mediate actin-based chloroplast movement in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 8860–8865.
Takahashi, Y., Soyano, T., Kosetsu, K., Sasabe, M., and Machida, Y. (2010). HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol. 51, 1766–1776.
Tamura, K., Nakatani, K., Mitsui, H., Ohashi, Y., and Takahashi, H. (1999). Characterization of katD, a kinesin-like protein gene specifically expressed in floral tissues of Arabidopsis thaliana. Gene 230, 23–32.
Tanaka, H., Ishikawa, M., Kitamura, S., Takahashi, Y., Soyano, T., Machida, C., et al. (2004). The AtNACK1/HINKEL and STUD/TETRASPORE/AtNACK2 genes, which encode functionally redundant kinesins, are essential for cytokinesis in Arabidopsis. Genes Cells 9, 1199–1211.
Tanenbaum, M. E., Macurek, L., Janssen, A., Geers, E. F., Alvarez-Fernandez, M., and Medema, R. H. (2009). Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr. Biol. 19, 1703–1711.
Tokai, N., Fujimoto-Nishiyama, A., Toyoshima, Y., Yonemura, S., Tsukita, S., Inoue, J., et al. (1996). Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle. EMBO J. 15, 457–467.
Umezu, N., Umeki, N., Mitsui, T., Kondo, K., and Maruta, S. (2011). Characterization of a novel rice kinesin O12 with a calponin homology domain. J. Biochem. 149, 91–101.
Vos, J. W., Safadi, F., Reddy, A. S. N., and Hepler, P. K. (2000). The kinesin-like calmodulin binding protein is differentially involved in cell division. Plant Cell 12, 979–990.
Wang, W., Dang, R., Zhu, J. Q., and Yang, W. X. (2010). Identification and dynamic transcription of KIF3A homologue gene in spermiogenesis of Octopus tankahkeei. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 157, 237–245.
Weaver, B. A., Bonday, Z. Q., Putkey, F. R., Kops, G. J., Silk, A. D., and Cleveland, D. W. (2003). Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162, 551–563.
Xu, T., Qu, Z., Yang, X., Qin, X., Xiong, J., Wang, Y., et al. (2009). A cotton kinesin GhKCH2 interacts with both microtubules and microfilaments. Biochem. J. 421, 171–180.
Xu, Y., Takeda, S., Nakata, T., Noda, Y., Tanaka, Y., and Hirokawa, N. (2002). Role of KIFC3 motor protein in Golgi positioning and integration. J. Cell Biol. 158, 293–303.
Yamazaki, H., Nakata, T., Okada, Y., and Hirokawa, N. (1995). Kif3a/B – a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J. Cell Biol. 130, 1387–1399.
Yang, G., Gao, P., Zhang, H., Huang, S., and Zheng, Z. L. (2007). A mutation in MRH2 kinesin enhances the root hair tip growth defect caused by constitutively activated ROP2 small GTPase in Arabidopsis. PLoS ONE 2, e1074. doi:10.1371/journal.pone.0001074
Yang, W. X., Jefferson, H., and Sperry, A. O. (2006). The molecular motor KIFC1 associates with a complex containing nucleoporin NUP62 that is regulated during development and by the small GTPase RAN. Biol. Reprod. 74, 684–690.
Yang, W. X., and Sperry, A. O. (2003). C-terminal kinesin motor KIFC1 participates in acrosome biogenesis and vesicle transport. Biol. Reprod. 69, 1719–1729.
Yang, X. Y., Chen, Z. W., Xu, T., Qu, Z., Pan, X. D., Qin, X. H., et al. (2011). Arabidopsis kinesin KP1 specifically interacts with VDAC3, a mitochondrial protein, and regulates respiration during seed germination at low temperature. Plant Cell 23, 1093–1106.
Yang, Z., Hanlon, D. W., Marszalek, J. R., and Goldstein, L. S. (1997). Identification, partial characterization, and genetic mapping of kinesin-like protein genes in mouse. Genomics 45, 123–131.
Yokoyama, R., O’toole, E., Ghosh, S., and Mitchell, D. R. (2004). Regulation of flagellar dynein activity by a central pair kinesin. Proc. Natl. Acad. Sci. U.S.A. 101, 17398–17403.
Zhang, M., Zhang, B., Qian, Q., Yu, Y., Li, R., Zhang, J., et al. (2010). Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J. 63, 312–328.
Zhong, R., Burk, D. H., Morrison, W. H. III, and Ye, Z. H. (2002). A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14, 3101–3117.
Zhu, C., and Dixit, R. (2011a). Functions of the Arabidopsis kinesin superfamily of microtubule-based motor proteins. Protoplasma 249, 887–899.
Zhu, C., and Dixit, R. (2011b). Single molecule analysis of the Arabidopsis FRA1 kinesin shows that it is a functional motor protein with unusually high processivity. Mol. Plant 4, 879–885.
Keywords: phylogenetic analysis, kinesin, microtubule, moss, phragmoplast, gene knockout, intracellular motility, intracellular transport
Citation: Shen Z, Collatos AR, Bibeau JP, Furt F and Vidali L (2012) Phylogenetic analysis of the kinesin superfamily from Physcomitrella. Front. Plant Sci. 3:230. doi: 10.3389/fpls.2012.00230
Received: 20 June 2012; Accepted: 25 September 2012;
Published online: 16 October 2012.
Edited by:
Bo Liu, University of California-Davis, USAReviewed by:
Ram Dixit, Washington University in St. Louis, USASherryl Bisgrove, Simon Fraser University, Canada
Giampiero Cai, Università degli Studi di Siena, Italy
Copyright: © 2012 Shen, Collatos, Bibeau, Furt and Vidali. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Luis Vidali, Department of Biology and Biotechnology, Worcester Polytechnic Institute, 100 Institute Road, Worcester, MA 01609, USA. e-mail:bHZpZGFsaUB3cGkuZWR1