
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 28 September 2012
Sec. Plant Physiology
Volume 3 - 2012 | https://doi.org/10.3389/fpls.2012.00222
Flavonoids are widely distributed secondary metabolites with different metabolic functions in plants. The elucidation of the biosynthetic pathways, as well as their regulation by MYB, basic helix-loop-helix (bHLH), and WD40-type transcription factors, has allowed metabolic engineering of plants through the manipulation of the different final products with valuable applications. The present review describes the regulation of flavonoid biosynthesis, as well as the biological functions of flavonoids in plants, such as in defense against UV-B radiation and pathogen infection, nodulation, and pollen fertility. In addition, we discuss different strategies and achievements through the genetic engineering of flavonoid biosynthesis with implication in the industry and the combinatorial biosynthesis in microorganisms by the reconstruction of the pathway to obtain high amounts of specific compounds.
The pigments that color most flowers, fruits, and seeds are flavonoids. These secondary metabolites, widely distributed in plants, are classified in six major subgroups: chalcones, flavones, flavonols, flavandiols, anthocyanins, and proanthocyanidins or condensed tannins (Figure 1) and a seventh group is found in some species, the aurones (Winkel-Shirley, 2001, 2006). Legumes and a small number of nonlegume plants also synthesize specialized flavonoids such as the isoflavonoids (Yu and Mcgonigle, 2005; Miadoková, 2009; Du et al., 2010; Wang, 2011), while few species either produce 3-deoxyanthocyanins and phlobaphenes. Groups of unrelated species, including grape and peanut, synthesize stilbenes, compounds closely related to chalcones (Chong et al., 2009; Shen et al., 2009).
More than 6000 different flavonoids have been identified, and surely this number will increase (Ferrer et al., 2008). The different flavonoids have diverse biological functions, including protection against ultraviolet (UV) radiation and phytopathogens, signaling during nodulation, male fertility, auxin transport, as well as the coloration of flowers as a visual signal that attracts pollinators (Mol et al., 1998; Winkel-Shirley, 2002; Bradshaw and Schemske, 2003). Flavonoids are also responsible for the display of fall color in many plants, which may protect leaf cells from photooxidative damage, enhancing the efficiency of nutrient retrieval during senescence (Feild et al., 2001). Flavonols are probably the most important flavonoids participating in stress responses; they are the most ancient and widespread flavonoids, having a wide range of potent physiological activities (Stafford, 1991; Pollastri and Tattini, 2011).
Much effort has been made in elucidating the biosynthetic pathways of flavonoids from a genetic perspective. Mutants affecting flavonoid synthesis were isolated in a range of plant species. Maize (Zea mays), snapdragon (Antirrhinum majus), and petunia (Petunia hybrida) were established as the first major experimental models in this system, leading to the isolation of many structural and regulatory flavonoid genes (for a review, see Holton et al., 1993; Mol et al., 1998). More recently, Arabidopsis (Arabidopsis thaliana) has facilitated the analysis of the regulation and subcellular localization of the flavonoid pathway. An interesting aspect of using Arabidopsis for studying flavonoid biosynthesis is that single copy genes encode all enzymes of the central flavonoid metabolism, with the exception of flavonol synthase (FLS), which is encoded by six genes, but only two (FLS1 and FLS3) have demonstrated activity (Owens et al., 2008; Preuss et al., 2009). Genetic loci for both structural and regulatory genes have been identified largely based on mutations that abolish or reduce seed coat pigmentation; thus, the loci were named transparent testa or tt mutants (Koornneef, 1990; Borevitz et al., 2000). Consequently, most of the structural genes, as well as a number of regulatory genes, have been correlated with specific mutant loci in Arabidopsis. This species does not seem to use flavonoids in the same ways as some other species (for example, in defense or for male fertility); however, these mutants are helpful to define roles for these compounds in essential processes such as UV protection (Li et al., 1993; Landry et al., 1995) and the regulation of auxin transport (Murphy et al., 2000; Brown et al., 2001; Kuhn et al., 2011; Lewis et al., 2011).
Flavonoids are synthesized through the phenylpropanoid pathway, transforming phenylalanine into 4-coumaroyl-CoA, which finally enters the flavonoid biosynthesis pathway (Figure 2). The first enzyme specific for the flavonoid pathway, chalcone synthase, produces chalcone scaffolds from which all flavonoids derive. Although the central pathway for flavonoid biosynthesis is conserved in plants, depending on the species, a group of enzymes, such as isomerases, reductases, hydroxylases, and several Fe2+/2-oxoglutarate-dependent dioxygenases modify the basic flavonoid skeleton, leading to the different flavonoid subclasses (Martens et al., 2010). Last, tranferases modify the flavonoid backbone with sugars, methyl groups and/or acyl moieties, modulating the physiological activity of the resulting flavonoid by altering their solubility, reactivity and interaction with cellular targets (Bowles et al., 2005; Ferrer et al., 2008).
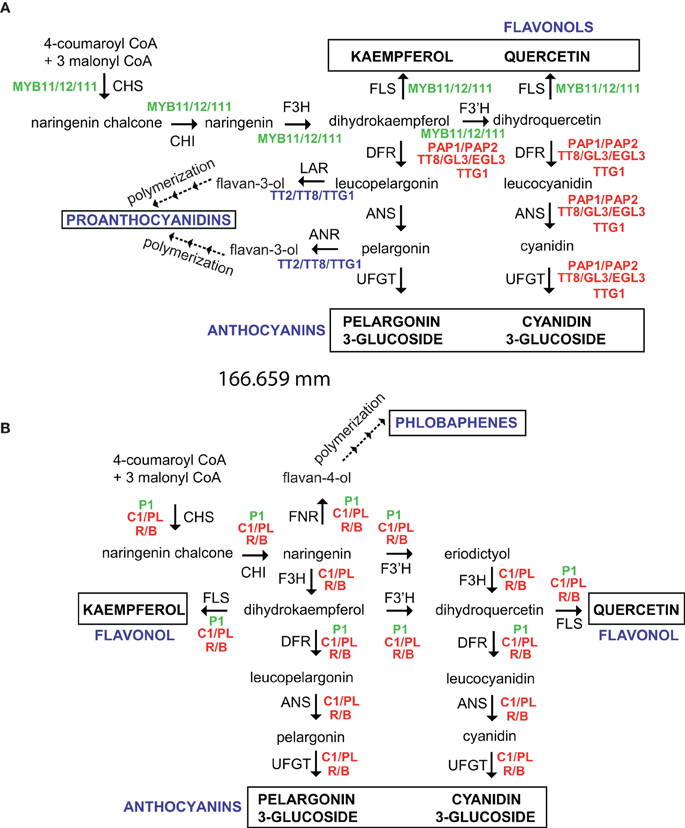
Figure 2. Regulation of the flavonoid pathway in Arabidopsis thaliana (A) and maize (B). Enzymes and intermediates are indicated in black and different regulators are indicated in color. End products are identified in capital letters. Dotted arrows indicate multiple steps. CHS, Chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid-3′-hydroxylase; DFR, dihydroflavonol 4-reductase; FNR, flavanone 4-reductase; ANS, anthocyanidin synthase; UFGT, UDP-glucose flavonoid 3-O glucosyltransferase; FLS, flavonol synthase; LAR, leucoanthocyanidin reductase; ANR, anthocyanidin reductase.
Evidence is emerging showing that consecutive enzymes of the phenylpropanoid and flavonoid biosynthesis are organized into macromolecular complexes that can be associated with endomembranes (Kutchan, 2005). Metabolic channeling in plant secondary metabolism enables plants to effectively synthesize specific natural products and thus avoid metabolic interference. The existence of cytochrome P450 monooxygenases (P450s)-related metabolons has been demonstrated: direct and indirect experimental data describe P450 enzymes in the phenylpropanoid, flavonoid, cyanogenic glucoside, and other biosynthetic pathways (Winkel, 2004; Ralston and Yu, 2006). Additional evidence for the channeling of intermediates between specific isoforms of phenylalanine ammonia lyase and cinnamate-4-hydroxylase has been provided using transgenic tobacco plants expressing epitope-tagged versions of two phenylalanine ammonia lyase isoforms (PAL1 and PAL2) and of cinnamate-4-hydroxylase (Achnine et al., 2004). Moreover, the existence of a multienzyme complex has been proposed for the anthocyanin pathway in rice by yeast-two hybrid experiments (Shih et al., 2008).
Most of the flavonoid synthesizing enzymes are recovered in soluble cell fractions; immunolocalization experiments suggest that they are loosely bound to the endoplasmic reticulum (ER), possibly in a multi-enzyme complex, whereas the pigments themselves accumulate in the vacuole (i.e., anthocyanins and proanthocyanidins) or the cell wall (i.e., phlobaphenes Winkel-Shirley, 2001). Flavonol synthase1 has recently been localized in Arabidopsis nuclei (Kuhn et al., 2011), as well as chalcone synthase and chalcone isomerase (Saslowsky et al., 2005). Interestingly, Antirrhinum majus aureusidin synthase, the enzyme that catalyzes aurone biosynthesis from chalcones, was localized in the vacuole, while the chalcone 4′-O-glucosyltransferase is localized in the cytoplasm, indicating that chalcones 4-O-glucosides are transported to the vacuole and therein converted to aurone 6-O-glucosides (Ono et al., 2006). Moreover, a flavonoid-3′-hydroxylase has been recently localized in the tonoplast in the hilum region of the soybean immature seed coat (Toda et al., 2012).
Two models have been proposed for the mechanism of anthocyanin transport from the ER to the vacuole storage sites: the ligandin transport and the vesicular transport (Grotewold and Davis, 2008; Zhao and Dixon, 2010). The ligandin transport model is based on genetic evidence showing that glutathione transferase (GST)-like proteins are required for vacuolar sequestration of pigments in maize, petunia and Arabidopsis (AtTT19) (Marrs et al., 1995; Alfenito et al., 1998). The vacuolar sequestration of anthocyanins in maize requires a multidrug resistance associated protein-type (MRP) transporter on the tonoplast membrane, which expression is co-regulated with the structural anthocyanin genes (Goodman et al., 2004). MRP proteins are often referred as glutathione S-X (GS-X) pumps because they transport a variety of glutathione conjugates. However, because anthocyanin–glutathione conjugate(s) have not been found, it is proposed that these GSTs might deliver their flavonoid substrates directly to the transporter, acting as a carrier protein or ligandin (Koes et al., 2005). This hypothesis is supported by the fact that Arabidopsis' GST (TT19), localized both in the cytoplasm and the tonoplast, can bind to glycosylated anthocyanins and aglycones but does not conjugate these compounds with glutathione (Sun et al., 2012). The vesicle-mediated transport model proposed is based on observations that anthocyanins and other flavonoids accumulate in the cytoplasm in discrete vesicle-like structures (anthocyanoplasts), and then they might be imported into the vacuole by an autophagic mechanism (Pourcel et al., 2010). Nevertheless, grape vesicle-mediated transport of anthocyanins involves a GST and two multidrug and toxic compound extrusion-type transporters (anthoMATEs). Thus, these observations point out to the coexistence of both mechanisms of transports, in which the participation of GSTs and transporters would be specific to cell and/or flavonoid-type (Gomez et al., 2011).
The flavonoid biosynthesis genes are regulated by the interaction of different families of transcription factors. Genes involved in the anthocyanin pathway are differentially regulated in monocot and dicot species by R2R3 MYB transcription factors, basic helix-loop-helix (bHLH), and WD40 proteins (Grotewold, 2005; Petroni and Tonelli, 2011). Thus, combinations of the R2R3-MYB, bHLH, and WD40 transcription factors and their interactions (MYB-bHLH-WD40 complex) determine the activation, and spatial and temporal expression of structural genes of anthocyanin biosynthesis. The regulation of anthocyanin biosynthesis in reproductive and other organs by MYB-bHLH-WD40 complex has been reviewed (Petroni and Tonelli, 2011). There are interesting differences in anthocyanin regulation between monocot and dicot species like Arabidopsis and maize. In Arabidopsis, TT2, TT8, and TTG1 form a ternary complex and activate proanthocyanidin biosynthesis in developing seeds, while, TTG1, a WD40 transcription factor, different bHLH (TT8, GL3, and EGL3) and MYB transcription factors (PAP1 and PAP2) interact to activate anthocyanin synthesis in vegetative tissues (Figure 2A) (Baudry et al., 2004; Feller et al., 2011). In maize, MYB and bHLH proteins are encoded by two multigene families (PL/C1 and B/R, respectively), and each member has a tissue- and developmental-specific pattern, while a WD40 protein PAC1 is required by both B1 or R1 proteins for full activation of anthocyanin biosynthetic genes in seeds and roots (Figure 2B) (Carey et al., 2004). Functional Arabidopsis TTG1 is required for anthocyanin accumulation during roots and trichomes development (Galway et al., 1994), and maize PAC1 can complement Arabidopsis ttg1 mutants; however, maize pac1 mutants only show a reduction in anthocyanin pigmentation in specific tissues (Carey et al., 2004). Even more, the regulation of flavonol biosynthesis exhibit important differences between both species. In Arabidopsis, three R2R3-MYB proteins, MYB12, MYB11, and MYB111 (PFG1-3), which exhibit differential spatial expression patterns, regulate AtFLS1 expression in a tissue- and developmental-specific manner (Stracke et al., 2007); while, ZmFLS1/2 are regulated by both P1 (R2R3-MYB) and the anthocyanin C1/PL1 and R/B regulators (Figure 2) (Falcone Ferreyra et al., 2012). Nevertheless, flavonols are essential for pollen germination and conditional male fertility in maize (Mo et al., 1992; Taylor and Hepler, 1997), but maize plants lacking the P1 and R/B+C1/PL1 anthocyanin regulators are fertile (Coe and Neuffer, 1988; Dooner et al., 1991; Neuffer et al., 1997). In addition, a PFG1-3-independent flavonol accumulation occurs in pollen and siliques/seeds in Arabidopsis (Stracke et al., 2010), suggesting that, in both species, additional regulators, not yet identified, are also involved in the regulation of FLS expression, and consequently, in flavonol accumulation.
In addition, the evolution of MYB and bHLH plant families has been deeply analyzed from structural and functional perspectives (Feller et al., 2011). Interestingly, the identification of a C1-like (MBF1) regulator in the gymnosperm Picea mariana (black spruce) further supports the idea that the regulation of anthocyanin pathway by a C1-like class of R2R3 MYB protein precedes the evolutionary separation of angiosperms from gymnosperms (Xue et al., 2003). The identification of both bHLH and MYB proteins in mosses further supports the hypothesis that the bHLH–MYB complex has evolved early during land plant evolution (Pires and Dolan, 2010).
Many R2R3 MYB transcription factors were first identified from several model plants, such as maize, Antirrhinum, petunia, and Arabidopsis. These transcription factors are involved in the regulation of the flavonoid biosynthesis pathway. The increasing availability of plant genomes has allowed the identification and isolation of a large number of MYB genes involved in the regulation of flavonoid biosynthesis from diverse non-model plant species such as grapevine (Vitis vinifera), strawberry (Fragaria x ananassa), apple (Malus domestica), cauliflower (Brassica oleracea var botrytis), potato (Solanum tuberosum L.), bayberry (Myrica rubra), mangosteen (Garcinia mangostana L.), pear (Pyrus pyrifolia), and purple kale (Brassica oleracea var. acephala f. tricolor) (Hichri et al., 2011). Furthermore, many of these regulators have been functionally characterized by transient experiments and stable expression in heterologous vegetal hosts. Table 1 shows examples of MYB transcription factors that regulate flavonoid biosynthesis.
A variety of derivatives of the initial phenylpropanoid scaffold serve vital roles in plant structural integrity, UV photoprotection, reproduction, and internal regulation of plant cell physiology and signaling. Phenylpropanoids also act as key chemical modulators of plant communication with insects and microbes, either as attractants or repellants, as phytoalexins against pathogens and herbivores, and as attractants to pollinators via flower color. They also induce root nodulation when excreted by symbiotic nitrogen-fixing rhizobia (Mandal et al., 2010).
The biological functions of flavonoids are linked to their potential cytotoxicity and their capacity to interact with enzymes through protein complexation. Some flavonoids provide stress protection, for example, acting as scavengers of free radicals such as reactive oxygen species (ROS), as well as chelating metals that generate ROS via the Fenton reaction (Williams et al., 2004). Flavonoids are also involved in the resistance to aluminum toxicity in maize. Roots of maize plants that were exposed to aluminum exuded high levels of phenolic compounds such as catechin and quercetin; indicating that their ability of chelating metals can be an in vivo mechanism to ameliorate aluminum toxicity (Kidd et al., 2001).
Evidence links flavonoids with the control of the polar transport of auxins. This hormone probably has a role in the stress response by controlling stomatal opening and by allocating resources under poor growth conditions (Peer and Murphy, 2007; Kuhn et al., 2011; Lewis et al., 2011). Flavonoids, such as quercetin, kaempferol, apigenin, and other aglycone molecules synthesized in the first steps of the flavonoid biosynthesis pathway, inhibit polar auxin transport and enhance consequent localized auxin accumulation in planta (Peer and Murphy, 2007; Kuhn et al., 2011; Lewis et al., 2011).
Flavonoids are involved in the nodulation process. Flavonoid-deficient roots of transgenic plants produced by RNA interference of chalcone synthase were unable to initiate nodules (Wasson et al., 2006). The complementation of the nodulation and flavonoid deficiency in roots by exogenous naringenin and liquiritigenin, precursors converted to the usual end product in Medicago truncatula and soybean, indicated that the lack of flavonoids was the reason for nodulation deficiency. In addition, these flavonoid-deficient roots had also increased auxin transport and lacked local inhibition of auxin transport at the site of nodulation (Wasson et al., 2006). However, an isoflavone-hypersensitive Rhizobium strain that requires very low levels of isoflavones to induce the nod genes is able to normally nodulate isoflavone-deficient roots, indicating that nodule primordia can form even in the absence of auxin transport inhibition by isoflavones in soybean roots. In conclusion, isoflavone-mediated auxin transport inhibition is not always essential for soybean nodulation (Subramanian et al., 2006). By silencing different M. truncatula flavonoid-biosynthesis enzymes, (isoflavone synthase, chalcone reductase, flavone synthase, and chalcone synthase), it was demonstrated that flavones and flavonols may act as internal inducers of rhizobial nod genes and auxin transport regulators during nodulation by Sinorhibium meliloti, respectively, (Zhang et al., 2009). In contrast to the increased auxin transport in isoflavone-null roots of soybean, isoflavone-null roots of M. truncatula showed unaltered auxin transport indicating that legumes use different flavonoid compounds to regulate auxin transport during nodulation (Zhang et al., 2009).
The UV-absorbing characteristics of flavonoids have long been considered as evidence for the role of flavonoids in UV protection. Studies in a wide range of species, such as Ligustrum vulgare, Vitis vinifera, petunia, and Arabidopsis have provided new evidence that UV light induces the synthesis of flavonol compounds (Ryan et al., 2002; Berli et al., 2010; Stracke et al., 2010; Agati et al., 2011; Kusano et al., 2011). Because the presence of the OH group in the 3-position of the flavonoid skeleton is the main structural feature responsible in chelating metal ions such as iron, copper, zinc, aluminum, and hence, inhibiting the formation of free radicals as well as to reduce ROS once formed, it was suggested that flavonols might play yet uncharacterized roles in the UV stress response (Verdan et al., 2011). Furthermore, grass species such as Deschampsia antarctica, Deschampsia borealis, and Calamagrostis epigeios that grow in regions with elevated levels of solar UV-B radiation have high constitutive levels of flavonoids like the flavones orientin and luteolin, that protect plants against this stress condition (Van De Staaij et al., 2002). Similarly, maize plants growing at high altitudes accumulate C-glycosyl flavones in leaves, maysin and its biosynthetic precursor rhamnosylisoorientin, flavones commonly found in silks, as a mechanism that prevents damage caused by high UV-B exposure (Zhang et al., 2003; Casati and Walbot, 2005). FLS genes are regulated by UV-B radiation in both high-altitude landraces and low-altitudes inbreds of maize. Higher transcript levels are present in high-altitude plants where there are high levels of UV-B radiation than at low-altitudes. Consequently, considering the protective role of flavonols to UV-B radiation, we hypothesize that the high transcript levels of ZmFLS genes may also contribute to the adaptation to this stress condition with higher UV-B fluxes (Falcone Ferreyra et al., 2012).
Flavonoids protect plants against pathogen and herbivores. According to the phytochemical co-evolution theory, the secondary metabolites are likely the most important mediators of plant-insect interactions. Thus, both plants and insect herbivores have evolved leading to the plant defense (i.e., plant secondary metabolites) and herbivore offense (i.e., detoxification ability) (Cornell and Hawkins, 2003; Kliebenstein, 2004; Bidart-Bouzat and Imeh-Nathaniel, 2008). Human-induced changes in abiotic environmental factors such as atmospheric CO2 and ozone (O3) levels, UV light, changes in precipitation patterns or temperature may directly affect the concentration of secondary chemicals in plants, which in turn may influence levels of herbivory or pathogen attack. For example, UV-B radiation modifies the production of secondary metabolites with photoprotective qualities (Mazza et al., 2000; Bassman, 2004), such as anthocyanins, isoflavonoids, and flavonol glycosides. The induction of UV-absorbing chemicals is shared with plant responses to other stresses, such as herbivore or pathogen attack, and this induction may act either positively or negatively on the levels of phytochemical production. Genes associated with the response of Nicotiana longiflora plants to insect herbivory were also induced by UV-B radiation (Izaguirre et al., 2003). Conversely, there is evidence that the induction of the flavonoid biosynthesis pathway by UV light can be inhibited by pathogen-induced defense responses in parsley (Petroselinum crispum) (Logemann and Hahlbrock, 2002). Furthermore, genes regulating the phenylpropanoid pathway leading to the synthesis of phenolic compounds like flavonoids are regulated by both UV light levels and herbivory (reviewed in Stratmann, 2003). Thus, Arabidopsis plants respond to the combination of biotic (bacterial elicitor flg22) and abiotic stresses (UV-B radiation) through the synthesis of defense-related compounds such as phytoalexins and lignin as structural barriers to restrict pathogen spread, and modify the expression of genes involved in the production of protective metabolites such as flavonols. This crosstalk involves antagonist regulation of two MYB transcription factors, the positive and negative regulators, MYB12 and MYB4, respectively (Schenke et al., 2011).
Genes expressed in incompatible and compatible plant-microbe interactions were identified by using a large-scale transcript profiling analysis of soybean and M. truncatula. There was a sharp and rapid up-regulation of genes encoding enzymes involved in the phenylpropanoid pathway, in particular for the synthesis of isoflavones and isoflavanones (Samac and Graham, 2007). The responses of soybean to avirulent and virulent strains of the bacterial pathogen P. syringae pv. glycinea, differing in the presence or absence of avrB, were investigated using a cDNA array (Zou et al., 2005). Decreased levels of transcripts specific to the anthocyanin branch of the flavonoid pathway were observed. The largest group of up-regulated genes included genes involved in the flavone and isoflavone biosynthesis. Thus, it is suggested that the opposite regulation of these branches enhances production of isoflavones that act as antioxidants and antimicrobial compounds vs. those responsible for color (Samac and Graham, 2007).
The response of susceptible soybean “Essex” and a partially resistant recombinant inbred line (RIL23) to the fungal pathogen Fusarium solani f. sp. glycines, the causal agent of Sudden Death Syndrome, was studied (Iqbal et al., 2005). Up-regulation of several genes encoding enzymes in the phenylpropanoid pathway was observed in RIL23, suggesting that the products of this pathway participate in the resistance to Sudden Death Syndrome. The transcription of phenylpropanoid metabolism genes was up-regulated (along with others) after challenging susceptible soybean with the pathogen Phytophthora sojae (Moy et al., 2004). Isoflavones are also important in R-gene-mediated resistance to P. sojae; RNAi down-regulated isoflavone synthase genes in soybean roots resulted in a 95% reduction in isoflavone accumulation and an enhanced susceptibility to the pathogen (Subramanian et al., 2005).
Sustained up-regulation of genes involved in the phenylpropanoid metabolism has been associated with R-gene-mediated resistance responses in M. truncatula responding to foliar pathogens. Expression profiling of the response of two M. truncatula genotypes (one susceptible and one resistant) to the fungal pathogen Colletotrichum trifolii showed that both genotypes respond to the infection by up regulation of genes encoding phenylpropanoid pathway enzymes (Torregrosa et al., 2004). In a similar way, up-regulation of genes involved in the phenylpropanoid pathway, particularly those leading to isoflavone and isoflavonoid compounds was reported in the response of M. truncatula to abiotrophic pathogen, Erysiphe pisi, the causal agent of powdery mildew (Foster-Hartnett et al., 2007).
Corn earworm, Helicoverpa zea, is a major pest of maize (Ortega et al., 1980). Thus, interest in achieving maize with resistance to corn earworm has increased. One type of natural resistance is associated with the presence in silks of a C-glycosyl flavone: maysin, as well as related compounds: apimaysin and methoxymaysin (Waiss et al., 1979; Elliger et al., 1980; Snook et al., 1994). These compounds are insecticidal to H. zea larvae and are thought to interfere with the amino acid metabolism in the insect gut through their subsequent conversion to more toxic quinones. Quinones reduce the availability of free amino acids and proteins by binding to –SH and –NH2 groups (Byrne et al., 1997). Using flavone synthesis as a model quantitative trait locus (QTL) system, it was shown that in a population segregating for functional and nonfunctional p1 alleles, the p1 locus is the gene underlying the major QTL for maysin concentration and activity against the earworm (Byrne et al., 1996, 1997). Transgenic maize over-expressing the p1 gene had increased silk maysin level (Johnson et al., 2007). The transgenic plants were more resistant to earworm larvae, increasing insect mortality levels and decreasing mean weights of surviving larvae. Larval weight was significantly negatively correlated with maysin level in transgenic silks.
The unique structure and combination of different flavonoids in each species produce yellow pollen with a range of visible and UV reflection spectra that can be detected by the targeted insects and larger animals, facilitating successful pollination. The flavonoids impart a distinctive yellow color to pollen and can be 2–4% of the dry weight (Zerback et al., 1989; Van Der Meer et al., 1992). The existence of “white pollen” has been reported in species as diverse as bristle cone pine and morning glory. The correlation between pollen fertility and flavonoids was first established in wind pollinated maize, with its numerous and well-characterized anthocyanin mutants (Mo et al., 1992). Flavonoid-deficient mutants lacking chalcone synthase were generated in maize and petunia to elucidate the roles of flavonoids in pollen (Pollak et al., 1993). These mutants were not only deficient in flavonoids but were also male sterile due to a failure to produce a functional pollen tube. This deficiency could be reversed by adding the flavonol kaempferol at pollination (Mo et al., 1992). These mutant plants are conditionally male fertile, as flavonoid-deficient pollen does not function in self-crosses but it is partially functional on wild-type stigmas containing flavonols (Mo et al., 1992).
The silencing of chalcone synthase gene results in parthenocarpy in tomato, but it was not identified if the cause of this phenomenon was the lack of flavonan-3-ols and/or flavonols (Schijlen et al., 2007). The silencing of a FLS in tobacco causes production of less-seeded fruits, and silenced lines had lower flavonol and anthocyanidins levels, while the flavan-3-ol content is increased. In addition, the pollen of these silenced lines was unable to produce functional pollen tubes. This capacity can be reversed with quercetin (in vivo and in vitro); implying that flavonols (in particular quercetin) have essential roles in pollen germination and consequently in plant fertility (Mahajan et al., 2011). Moreover, the results described indicate that the silencing of FLS could represent a new strategy to obtain plants with seedless/less-seeded fruits but fertility can be chemically restored for seed production.
Arabidopsis mutant plants in the chalcone synthase gene, which are completely devoid of flavonols in flowers and stamens, are fertile and have no pollen tube growth aberrations, indicating that flavonols are not universally essential for pollen fertility (Burbulis et al., 1996; Ylstra et al., 1996). The Arabidopsis polyketide synthases (LAP5 and LAP6) are required for pollen development and sporopollenin biosynthesis (Kim et al., 2010). Single and double mutants in LAP5/6 have reduced to undetectable levels of different flavonoids in developing anthers and lack exine deposition causing male sterility. Both anther-specific enzymes produce the triketide and the tetraketide α-pyrones required for sporopollenin synthesis by condensation of hydroxyl fatty acyl-CoAs with malonyl-CoA. Although it was suggested that both enzymes could be involved in the synthesis of alkyl pyrones and phenolic constituents of sporopollenin in exine (Dobritsa et al., 2010), flavonoids are not produced by the action of these enzymes (Kim et al., 2010). Therefore, pollen grain cell walls defective in exine in the lap mutants may be deficient in the deposition of extracellular pollen coat tryphine that contains flavonoids. Consequently, the reduced flavonoid levels can be an indirect result of reduced deposition of flavonoid-containing tryphine.
The engineering of the flavonoid pathway for the purposeful accumulation of compounds has been extensively used in industry (Tanaka et al., 2008). More than 20 years have passed since the color of a plant was modified for the first time through genetic engineering: the brick red petunia accumulating pelargonidin that expressed the maize dihydroflavonol reductase gene (Meyer et al., 1987; Nakamura et al., 2006). A huge number of genetic engineering attempts have been described to produce novel flower colors in several plant species, such as petunia, gerbera, rose, carnation, lisianthus, and torenia, by modifying the flavonoid biosynthesis pathway, either by transcriptional down-regulation, by inactivating the key enzymes of the anthocyanin pathway, or by heterologous expression of key enzymes (Tanaka et al., 2009, 2010; Nishihara and Nakatsuka, 2011). The choice of the gene source is important for an efficient change of the pathway. However, although some successful examples of color modification have been described in rose, carnation and Nierembergia, undesirable results were also reported probably because tissue-specific promoters were not used, adding to the difficulty in obtaining stable phenotypes (Tanaka et al., 2009, 2010). Passiflora species have economical importance due to the use as ornamentals, having an enormous floral diversity among species, including variation in size, morphology, fusion of floral organs, and a wide range of pigmentation patterns of the corona filaments provided by different types of anthocyanin molecules. EST databases were used to assemble sequences of P. edulis and P. suberosa corresponding to 15 different genes of the anthocyanin biosynthesis pathway as well as regulatory factors, providing useful resources for research and manipulation of secondary metabolism using transgenic approaches (Aizza and Dornelas, 2011). The results of such engineering are predictable at least to some extent. However, it is not easy to predict the amount of the compounds accumulated. In addition, the engineering of flavonoid biosynthetic pathways may affect other metabolic pathways in plants and may result in detrimental effects (Tanaka et al., 2008).
The three major anthocyanins pelargonidin, cyanidin, and delphinidin, contribute to orange to red, red to magenta, and magenta to purple colors, respectively (Figure 3). Methylation of cyanidin and delphinidin leads to three additional classes of anthocyanins: peonidin, petudin, and malvidin. The final color of a flower is determined by different factors that contribute to the spectrum and intensity of color: (1) the type and level of anthocyanins (determined by the anthocyanin biosynthetic pathway); (2) the complexing of anthocyanins with metal ions (producing a variety of bluish or purple hues); (3) the presence of co-pigments (flavones or flavonols); (4) the modification of basic anthocyanins (hydroxylation, methylation, acylation or conjugation); and, (5) the variation in vacuolar pH (Yu and Mcgonigle, 2005). In order to achieve a specific color by accumulating a corresponding compound, it is necessary to up-regulate the pathway leading to the compound and down-regulate the competing pathways. Sometimes, the existence of feedback inhibition in the flavonoid pathway, or certain down-regulation of a biosynthetic enzyme might destabilize a suggested metabolic biosynthetic complex (Winkel, 2004; Tanaka and Ohmiya, 2008). Consequently, engineering a pathway may have unexpected effects on a different pathway, especially if there is cross-talk between them. Different strategies have been applied to the modification of the flavonoid pathway, such as antisense, sense suppression (co-suppression), and RNAi for the down-regulation. An increase in the amount of flavonoids can be achieved by the over-expression of one of the biosynthesis or regulatory genes. Some transgenes in plants are prone to be silenced by methylation, producing variations at the transcriptional level, phenotypic differences and consequently affecting the levels of accumulation of a determined compound (Elomaa et al., 1995). Some examples of genetically modified plants in the flavonoid pathway are described below.
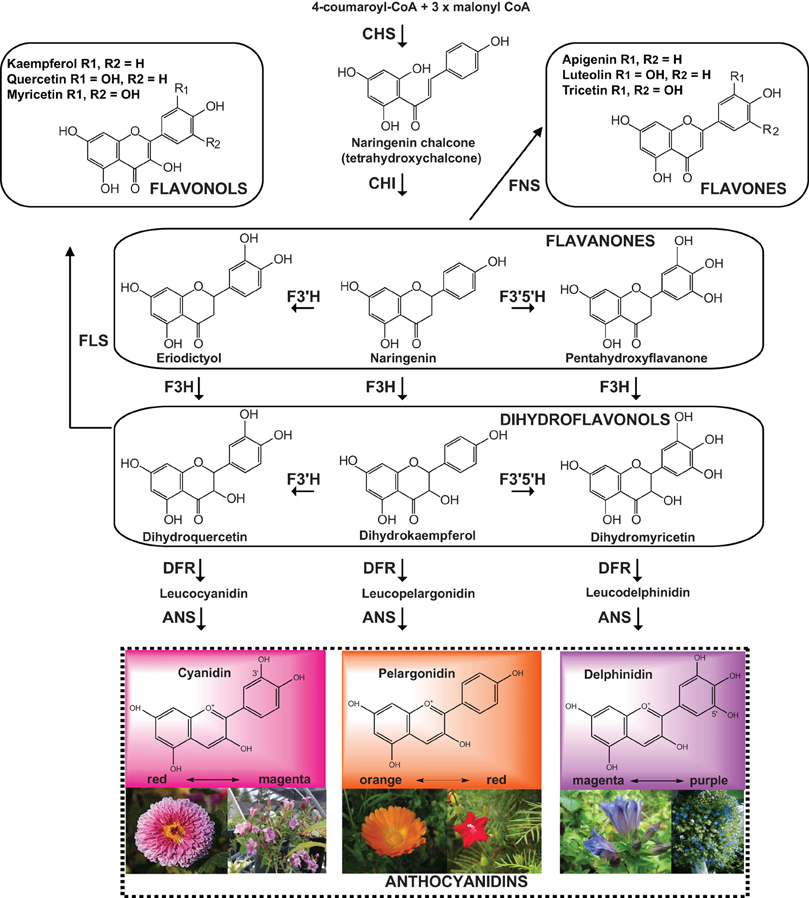
Figure 3. Biosynthesis of anthocyanidins: cyanidin, pelargonidin and delphinidin. The simplified scheme comprises the anthocyanidin branch and other flavonoid end products (flavonols and flavones). The enzymes catalyzing each step are indicated in bold. CHS, Chalcone synthase; CHI, chalcone isomerase; FNS, flavone synthase; F3H, flavonoid-3-hydroxylase; F3′H, flavonoid-3′-hydroxylase; F3′5′H, flavonoid 3′,5′-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase. The pictures of flowers correspond to (left to right): Chrysanthemum morifolium, pink gentian (Gentiana scabra), Calendula officianalis, Ipomoea quamochit, blue gentian (Gentiana triflora) and Evolvulus pilosus.
The substrate specificity of an enzyme can greatly contribute to determining the B-ring hydroxylation pattern. This variation influences the color of anthocyanins and, therefore, the flower/fruit color. For example, dihydroflavonol reductase from petunia efficiently utilizes dihydromyricetin but does not utilize dihydrokaempferol, and thus, this species does not accumulate pelargonidin, resulting in varieties lacking orange or bright-red flowers (Figure 3). In the case of maize and gerbera, dihydroflavonol reductase can utilize dihydrokaempferol as a substrate; thus, the generation of transgenic petunia plants expressing maize or gerbera dihydroflavonol reductase allowed the accumulation of pelargonidin, bearing brick red and orange flowers, respectively (Meyer et al., 1987). Rosa hybrida lacks violet to blue flower varieties due to the absence of delphinidin-based anthocyanins, usually the major constituents of purple and blue flowers, because roses do not possess flavonoid 3′, 5′-hydroxylase, a key enzyme for delphinidin biosynthesis. Expression of F3′5′H gene from violet (Viola x wittrockiana) resulted in the accumulation of a high percentage of delphinidin and a novel bluish flower color in rose petals. In addition, the endogenous dihydroflavonol reductase gene was down-regulated, and the dihydroflavonol reductase gene from Iris hollandica and the violet flavonoid 3′,5′-hydroxylase gene were over-expressed in a rose cultivar, producing a dominant accumulation of delphinidin in the petals. The heritability of delphinidin accumulation was confirmed in F1 and F2 progeny by hybridization breeding of transgenic roses with seed parent non-transgenic roses (Chandler and Tanaka, 2007; Katsumoto et al., 2007).
The modification of the expression of genes that encode transcription factors regulating the flavonoid pathway is also commonly used. For example, the co-expression of the Delila (Del) and Rosea1 (Ros1) cDNAs, each under the control of the fruit-specific E8 promoter, led to high levels of anthocyanin throughout the fruit tissues, which were consequently purple colored (Butelli et al., 2008). This result demonstrates that the anthocyanin biosynthetic pathway can be fully switched on in fruits if activated appropriately. Some additional examples of engineering of the flavonoid biosynthesis pathway and the phenotypes obtained are described in Table 2.
Secondary metabolism is intimately linked with other aspects of plant differentiation, in which transcription factors play a key coordinating role. Recent findings illustrate the complexity of regulatory networks that control flavonoid biosynthesis in Arabidopsis and other species. They also underline the close relationship between secondary metabolism and epidermal and seed differentiation in Arabidopsis, and the central role played by conserved WD40 proteins in regulating these processes (Broun, 2005). Evidence of this connection is provided by the phenotype of Arabidopsis plants that over-express the RED gene, which encodes a bHLH transcription factor that regulates anthocyanin biosynthesis in maize. RED over-expression is sufficient to correct all defects in the ttg1 (transparent testa glabra1, WD-like protein) mutant, restoring trichomes, root-hair initiation, seed-coat development, and flavonoid production (Broun, 2005).
Plant secondary metabolites are thought to be the result of sophisticated evolution. The relatively large number of secondary metabolites that are biologically active are of interest in drug discovery; and the biological activity and potency of secondary metabolites are derived largely from their complex structures. Various phenylpropanoids, including flavonoids and stilbenes, possess extraordinary antioxidant activity and estrogenic, antiviral, antibacterial, and anticancer activities. The putative health-protecting functions of flavonoids have stimulated significant research toward the elucidation of their biosynthetic networks, as well as the development of production platforms using genetically tractable hosts.
One way to produce flavonoids to be used in the pharmaceutical industry is through combinatorial biosynthesis. This technique consists of an approach in which genes from different organisms are assembled in an artificial gene cluster construct for the production of a bioactive compound. For example, Escherichia coli and Saccharomyces cerevisiae systems carrying artificial biosynthesis pathways for production of plant-specific medicinal polyketides, such as flavonoids, stilbenoids, isoflavonoids, and curcuminoids have been designed (for a review, see Katsuyama et al., 2007c; Horinouchi, 2009; Marienhagen and Bott, 2012). Starting with amino acids tyrosine and phenylalanine as substrates, these different heterologous systems yielded naringenin, resveratrol, genistein, and curcumin. Also, supplementation of E. coli cells with various carboxylic acids as precursors resulted in the production of previously unknown compounds; and the addition of modification enzymes to the artificial pathways led to the production of known and unknown flavonols and flavones (Miyahisa et al., 2006). These microbial systems are promising for the construction of flavonoid libraries employing enzymes of various origins as members of the artificial pathway, and for efficient use of the potential microorganism hosts. One of the advantages in assembling a biosynthesis pathway for a certain product is that replacing a single enzyme chalcone synthase or stilbene synthase, belonging to different species, gives a different product, the structure of which depends on the catalytic properties of the enzyme, the number of extensions, extender substrates employed, and mode of cyclization of the extended intermediate (Horinouchi, 2009).
The success in the fermentative production of plant specific phenylpropanoids by E. coli carrying an artificially assembled pathway was the first example showing that a complete plant biosynthesis pathway can be established in an heterologous microorganism for production of flavanones from the amino acid precursors, phenylalanine, and tyrosine (Hwang et al., 2003; Miyahisa et al., 2005). This was also possibly accomplished in yeast, where p-coumaric acid was produced by expressing phenylalanine ammonia lyase and cinnamate-4-hydroxylase. Moreover, by expressing a 4-coumarate-CoA ligase and a chalcone synthase, a racemic mixture of flavanones was produced in yeast (Chemler and Koffas, 2008). The addition of chalcone isomerase and chalcone reductase along with the flavanone pathway then led to the production of both 5-deoxyflavanones and 5-hydroxyflavanones (Chemler and Koffas, 2008). Additional expression of a flavone synthase with the flavanone pathway produced several flavones in both yeast and E. coli, while expression of a flavanone 3-hydroxylase along with either a flavonol synthase or a dihydroflavonol 4-reductase with a leucocyanidin reductase yielded flavonols or catechins, respectively (Miyahisa et al., 2006; Katsuyama et al., 2007a). Moreover, 7-O-methyl apigenin, genkwanin, having antibacterial activity against Vibrio cholera and Enterococcus faecalis and anti-inflammatory activity, was synthesized from naringenin using E. coli expressing flavone synthase I and flavone 7-O-methyltransferase genes from poplar (Min et al., 2009).
Metabolic engineering has also been employed to greatly improve the efficiency of E. coli to produce various flavonoids by focusing on increasing intracellular pools of biosynthetic pathway cofactors. For instance, anthocyanin synthesis relies on UDP-glucose as the sugar donor. Over-expression of UDP-glucose biosynthesis genes with an anthocyanidin synthase-3-O-glucosyltransferase (ANS-3GT) fusion in E. coli yielded high anthocyanin concentrations (Yan et al., 2007). Further yield improvements were obtained by deleting competitive pathways for UDP-glucose (He et al., 2008). Nevertheless, the production in large scale of flavonoid glycosides was successfully optimized by bioconversion assays in E. coli expressing plant UDP-sugar: glycosyltransferases, allowing the relatively low specificity of enzymes toward the substrate acceptors to obtain a large range of glycoside products (He et al., 2008).
Other examples of combinatorial biosynthesis are the production of 5-deoxyflavanones, a natural raspberry ketone, and anthocyanin in E. coli (Beekwilder et al., 2007; Yan et al., 2007, 2008). The genetic design used was an artificial phenylpropanoid pathway assembling enzyme from various organisms in E. coli, and adding further modification enzymes. In addition, the yields were greatly increased when the two subunits of acetyl-CoA carboxylase from Corynebacterium glutamicum were also expressed, presumably because enhanced expression of the acetyl-CoA carboxylase greatly increases the intracellular pool of malonyl-CoA, a precursor for the synthesis (Miyahisa et al., 2005). Genistein, an isoflavone, was also synthesized by combinatorial biosynthesis; its interest resides because of its phytoestrogen activity and chemopreventive actions against cancer, osteoporosis, and cardiovascular disease (Dixon and Steele, 1999; Dixon and Ferreira, 2002). Microbial production of isoflavonoids had been difficult because isoflavone synthase, a key enzyme catalyzing the production of isoflavonoids from naringenin, is a membrane-bound cytochrome P450 enzyme that requires a specific electron transfer system. Thus, yeast or fungi were used to overcome the difficulty in expressing functionally active microsomal cytochrome P450 enzymes, which are usually difficult to express in active form in bacterial cells. Therefore, genistein was produced by the use of multiple microorganisms: first, naringenin was produced from tyrosine in the E. coli system, and then genistein was converted from naringenin using the yeast system (Katsuyama et al., 2007c). Moreover, a high-level production of both natural and non-natural isoflavones was later achieved in yeast fed with natural and synthetic flavanones (Chemler et al., 2010). In addition, Streptomyces venezuelae can be used as a heterologous host for the production of plant polyketides such as flavones and flavonols from naringenin by the introduction of flavone synthase I from Petroselium crispum, flavanone 3-hydroxylase from Citrus sinensis and a flavonol synthase gene from Citrus unshius, respectively (Ryeol et al., 2010).
A different example of combinatorial biosynthesis is to produce previously unknown flavonoids by precursor-directed biosynthesis. A precursor is supplied to a mutant that is blocked in the early stage of the biosynthesis of a natural product. This technique was successfully applied to the production of novel secondary metabolites by supplementation of analogs of natural precursors, which, in turn, gave novel compounds. For production of previously unknown flavanones by the system, unnatural carboxylic acids, such as fluorocinnamic acids, furyl, thienyl, pyridyl, and naphthyl acrylic acids, gave the corresponding unnatural flavanones and stilbenes (Katsuyama et al., 2007a,b).
Many enzymes of bacterial origin, as well as of plant origin, have a potential to catalyze flavonoid modification, for example, to prenylated, hydroxylated and glycosylated compounds (Winkel-Shirley, 2001; Vogel and Heilmann, 2008; Caputi et al., 2012). These modification enzymes can be readily incorporated as members in the artificial biosynthesis pathways, which should lead to the construction of larger libraries of natural and novel flavonoids, stilbenoid, and curcuminoid compounds. It was possible to modify the flavanones produced in the E. coli system by further introducing the flavanone 3-hydroxylase/flavonol synthase or flavone synthase genes into the biosynthetic pathway. Introduction of flavanone 3-hydroxylase and flavonol synthase genes from Citrus species in E. coli led to the production of flavonols: kaempferol from tyrosine and galangin from phenylalanine (Miyahisa et al., 2006). Similarly, introduction of a flavone synthase I gene from P. crispum in E. coli led to the production of flavones: apigenin from tyrosine and chrysin from phenylalanine. Finally, some of the novel flavanones synthesized were further modified by flavone synthase resulting in the production of novel flavones (Miyahisa et al., 2006), suggesting that the flavone synthase enzyme can modify a broad spectrum of flavanone substrates.
Most of the major enzymes and genes involved in the flavonoid pathways have been characterized. However, different aspects of flavonoid biology still remain unknown. For example, the expression patterns and the activities of some of the transcription factors that regulate this branched pathway have not yet been identified. In addition, there is little evidence about the existence of protein–protein interactions that form metabolic channels that increase the efficiency of this pathway; and there is little information about the transport of flavonoids into the vacuoles. The answers to these questions will be of great importance in order to achieve efficient engineering of the flavonoid pathway in plants. In addition, the use of A. thaliana plants as a genetic tool clearly has helped in studying different aspects of plant secondary metabolism. Data obtained using this species will allow extrapolation to other plants of commercial and agronomic interest.
The great biodiversity of plants that arose during evolution has generated a concomitant variety of flavonoid structures known to date and many to be discovered. Further analysis of different plant species will allow the discovery of novel structures and possibly new metabolic pathways. Future studies will also contribute to the improvement of floricultural, food, pharmaceutical, and chemical industries. Moreover, evidence of beneficial functions of flavonoids in human health and the use of natural compounds for the prevention and treatment of different pathologies is continuously increasing in the world; and interest will continue to grow among researchers in the coming years.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by FONCyT grants PICT-2006-00957 and PICT-2010-00105 to Paula Casati. Paula Casati, María Lorena Falcone Ferreyra and Sebastián Pablo Rius are members of the Research Career of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) of Argentina.
Achnine, L., Blancaflor, E. B., Rasmussen, S., and Dixon, R. A. (2004). Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydrozylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 16, 3098–3109.
Agati, G., Biricolti, S., Guidi, L., Ferrini, F., Fini, A., and Tattini, M. (2011). The biosynthesis of flavonoids is enhanced similarly by UV radiation and root zone salinity in L. vulgare leaves. J. Plant Physiol. 168, 204–212.
Aizza, L. C., and Dornelas, M. C. (2011). A genomic approach to study anthocyanin synthesis and flower pigmentation in Passion flowers. J. Nucleic Acids. doi: 10.4061/2011/371517. [Epub ahead of print].
Alfenito, M. R., Souer, E., Goodman, C. D., Buell, R., Mol, J., Koes, R., and Walbot, V. (1998). Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10, 1135–1150.
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380.
Bassman, J. H. (2004). Ecosystem consequences of enhanced solar ultraviolet radiation: secondary plant metabolites as mediators of multiple trophic interactions in terrestrial plant communities. Photochem. Photobiol. 79, 382–398.
Beekwilder, J., Van Der Meer, I. M., Sibbesen, O., Broekgaarden, M., Qvist, I., Mikkelsen, J. D., and Hall, R. D. (2007). Microbial production of natural raspberry ketone. Biotechnol. J. 2, 1270–1279.
Berli, F. J., Moreno, D., Piccoli, P., Hespanhol-Viana, L., Silva, M. F., Bressan-Smith, R., Cavagnaro, J. B., and Bottini, R. (2010). Abscisic acid is involved in the response of grape (Vitis vinifera L.) cv. Malbec leaf tissues to ultraviolet-B radiation by enhancing ultraviolet-absorbing compounds, antioxidant enzymes and membrane sterols. Plant Cell Environ. 33, 1–10.
Bidart-Bouzat, M. G., and Imeh-Nathaniel, A. (2008). Global change effects on plant chemical defenses against insect herbivores. J. Integr. Plant Biol. 50, 1339–1354.
Boase, M., Lewis, D., Davies, K., Marshall, G., Patel, D., Schwinn, K., and Deroles, S. (2010). Isolation and antisense suppression of flavonoid 3′, 5′-hydroxylase modifies flower pigments and colour in cyclamen. BMC Plant Biol. 10, 107.
Bogs, J., Jaffe, F. W., Takos, A. M., Walker, A. R., and Robinson, S. P. (2007). The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 143, 1347–1361.
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394.
Bovy, A. G., De Vos, R., Kemper, M., Schijen, E., Pertejo, M. A., Muir, S. R., Collins, G., Robinson, S., Verhoeyen, M., and Hughes, S. (2002). High flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes Lc and C1. Plant Cell 14, 2509–2526.
Bowles, D., Isayenkova, J., Lim, E. K., and Poppenberger, B. (2005). Glycosyltransferases: managers of small molecules. Curr. Opin. Plant Biol. 8, 254–263.
Bradley, J. M., Davies, K. M., Deroles, S. C., Bloor, S. J., and Lewis, D. H. (1998). The maize Lc regulatory gene up-regulates the flavonoid biosynthetic pathway of Petunia. Plant J. 13, 381–392.
Bradshaw, H. D., and Schemske, D. W. (2003). Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 426, 176–178.
Broun, P. (2005). Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8, 272–279.
Brown, D. E., Rashotte, A. M., Murphy, A. S., Normanly, J., Tague, B. W., Peer, W. A., Taiz, L., and Muday, G. K. (2001). Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126, 524–535.
Bruce, W., Folkerts, O., Garnaat, C., Crasta, O., Roth, B., and Bowen, B. (2000). Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors CRC and P. Plant Cell 12, 65–80.
Burbulis, I. E., Iacobucci, M., and Shirley, B. W. (1996). A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. Plant Cell 8, 1013–1025.
Butelli, E., Titta, L., Giorgio, M., Mock, H.-P., Matros, A., Peterek, S., Schijlen, E. G., Hall, R. D., Bovy, A. G., Luo, J., and Martin, C. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308.
Byrne, P. F., Darrah, L. L., Snook, M. E., Wiseman, B. R., Widstrom, N. W., Moellenbeck, D. J., and Barry, B. D. (1996). Maize silk-browning, maysin content, and antibiosis to the corn earworm, Helicoverpa zea (Boddie). Maydica 41, 13–18.
Byrne, P. F., Mcmullen, M. D., Wiseman, B. R., Snook, M. E., Musket, T. A., Theuri, J. M., Widstrom, N. W., and Coe, E. H. (1997). Identification of maize chromosome regions associated with antibiosis to corn earworm (Lepidoptera: Noctuidae) larvae. J. Econ. Entomol. 90, 1039–1045.
Caputi, L., Malnoy, M., Goremykin, V., Nikiforova, S., and Martens, S. (2012). A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 69, 1030–1042.
Carey, C. C., Strahle, J. T., Selinger, D. A., and Chandler, V. L. (2004). Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell 16, 450–464.
Casati, P., and Walbot, V. (2005). Differential accumulation of maysin and rhamnosylisoorientin in leaves of high-altitude landraces of maize after UV-B exposure. Plant Cell Environ. 28, 788–799.
Coe, E. H., and Neuffer, M. G. (1988). “The genetics of corn,” in Corn and Corn Improvement, eds G. F. Sprague and J. W. Dudley (Madison, WI: American Society of Agronomy), 181–258.
Cornell, H. V., and Hawkins, B. A. (2003). Herbivore responses to plant secondary compounds: a test of phytochemical coevolution theory. Am. Nat. 161, 507–522.
Czemmel, S., Stracke, R., Weisshaar, B., Cordon, N., Harris, N. N., Walker, A. R., Robinson, S. P., and Bogs, J. (2009). The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 151, 1513–1530.
Chandler, S., and Tanaka, Y. (2007). Genetic modification in floriculture. Crit. Rev. Plant Sci. 26, 169–197.
Chemler, J. A., and Koffas, M. A. (2008). Metabolic engineering for plant natural product biosynthesis in microbes. Curr. Opin. Plant Biol. 19, 597–605.
Chemler, J. A., Lim, C. G., Daiss, J. L., and Koffas, M. A. (2010). A versatile microbial system for biosynthesis of novel polyphenols with altered estrogen receptor binding activity. Chem. Biol. 17, 392–401.
Chiu, L. W., Zhou, X., Burke, S., Wu, X., Prior, R. L., and Li, L. (2010). The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol. 154, 1470–1480.
Chong, J., Poutaraud, A., and Hugueney, P. (2009). Metabolism and roles of stilbenes in plants. Plant Sci. 117, 143–155.
Deluc, L., Bogs, J., Walker, A. R., Ferrier, T., Decendit, A., Merillon, J. M., Robinson, S. P., and Barrieu, F. (2008). The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 147, 2041–2053.
Dixon, R. A., and Steele, C. L. (1999). Flavonoids and isoflavonoids – a gold mine for metabolic engineering. Trends Plant Sci. 4, 394–400.
Dobritsa, A. A., Lei, Z., Nishikawa, S., Urbanczyk-Wochniak, E., Huhman, D. V., Preuss, D., and Sumner, L. W. (2010). LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol. 153, 937–955.
Dooner, H. K., Robbins, T. P., and Jorgensen, R. A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25, 173–199.
Du, H., Huang, Y., and Tang, Y. (2010). Genetic and metabolic engineering of isoflavonoid biosynthesis. Appl. Microbiol. Biotechnol. 86, 1293–1312.
Du, H., Zhang, L., Liu, L., Tang, X.-F., Yang, W.-J., Wu, Y.-M., Huang, Y.-B., and Tang, Y.-X. (2009). Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry (Mosc.) 74, 1–11.
Elomaa, P., Helariutta, Y., Kotilainen, M., Teeri, T., Griesbach, R., and Seppänen, P. (1995). Transgene inactivation in Petunia hybrida is influenced by the properties of the foreign gene. Mol. Gen. Genet. 248, 649–656.
Elliger, C. A., Chan, B. G., Waiss, A. C., Lundin, R. E., and Haddon, W. F. (1980). C-glycosylflavones from Zea mays that inhibit insect development. Phytochemistry 19, 293–297.
Falcone Ferreyra, M. L., Casas, M. I., Questa, J., Herrera, L., Deblasio, S., Wang, J., Jackson, D., Grotewold, E., and Casati, P. (2012). Evolution and expression of tandem duplicated maize flavonol synthase genes. Front. Plant Sci. 3:101. doi: 10.3389/fpls.2012.00101
Ferreyra, M. L. F., Rius, S. P., Emiliani, J., Pourcel, L., Feller, A., Morohashi, K., Casati, P., and Grotewold, E. (2010). Cloning and characterization of a UV-B-inducible maize flavonol synthase. Plant J. 62, 77–91.
Feild, T. S., Lee, D. W., and Holbrook, N. M. (2001). Why leaves turn red in autumn. The role of anthocyanins in senescing leaves of red-osier dogwood. Plant Physiol. 127, 566–574.
Feller, A., Machemer, K., Braun, E. L., and Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116.
Ferrer, J., Austin, M., Stewart, C. J., and Noel, J. (2008). Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 46, 356–370.
Foster-Hartnett, D., Danesh, D., Peñuela, S., Sharopova, N., Endre, G., Vandenbosch, K. A., Young, N. D., and Samac, D. A. (2007). Molecular and cytological responses of Medicago truncatula to Erysiphe pisi. Mol. Plant Pathol. 8, 307–319.
Galway, M. E., Masucci, J. D., Lloyd, A. M., Walbot, V., Davis, R. W., and Schiefelbein, J. W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166, 740–754.
Gomez, C., Conejero, G., Torregrosa, L., Cheynier, V., Terrier, N., and Ageorges, A. (2011). In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant J. 67, 960–970.
Goodman, C. D., Casati, P., and Walbot, V. (2004). A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell 16, 1812–1826.
Grotewold, E. (2005). Plant metabolic diversity: a regulatory perspective. Trends Plant Sci. 10, 57–62.
Grotewold, E., Chamberlin, M., Snook, M., Siame, B., Butler, L., Swenson, J., Maddock, S., Clair, G. S., and Bowen, B. (1998). Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10, 721–740.
Grotewold, E., and Davis, K. (2008). Trafficking and sequestration of anthocyanins. Nat. Prod. Comm. 3, 1251–1258.
He, X.-Z., Li, W.-S., Blount, J. W., and Dixon, R. A. (2008). Regioselective synthesis of plant (iso)flavone glycosides in Escherichia coli. Appl. Microbiol. Biotechnol. 80, 253–260.
Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S., and Lauvergeat, V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62, 2465–2483.
Holton, T. A., Brugliera, F., and Tanaka, Y. (1993). Cloning and expression of flavonol synthase from Petunia hybrida. Plant J. 4, 1003–1010.
Horinouchi, S. (2009). Combinatorial biosynthesis of plant medicinal polyketides by microorganisms. Curr. Opin. Chem. Biol. 13, 197–204.
Hwang, E. I., Kaneko, M., Ohnishi, Y., and Horinouchi, S. (2003). Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl. Environ. Microbiol. 69, 2699–2706.
Iqbal, M., Yaegashi, S., Ahsan, R., Shopinski, K., and Lightfoot, D. (2005). Root response to Fusarium solani f. sp. glycines: temporal accumulation of transcripts in partially resistant and susceptible soybean. Theor. Appl. Genet. 110, 1429–1438.
Izaguirre, M. M., Scopel, A. L., Baldwin, I. T., and Ballare, C. L. (2003). Convergent responses to stress. Solar ultraviolet-B radiation and Manduca sexta herbivory elicit overlapping transcriptional responses in field-grown plants of Nicotiana longiflora. Plant Physiol. 132, 1755–1767.
Johnson, E. T., Berhow, M. A., and Dowd, P. F. (2007). Expression of a maize myb transcription factor driven by a putative silk-specific promoter significantly enhances resistance to Helicoverpa zea in transgenic maize. J. Agric. Food Chem. 55, 2998–3003.
Jung, C. S., Griffiths, H. M., De Jong, D. M., Cheng, S., Bodis, M., Kim, T. S., and De Jong, W. S. (2009). The potato developer (D) locus encodes an R2R3 MYB transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin. Theor. Appl. Genet. 120, 45–57.
Katsumoto, Y., Fukuchi-Mizutani, M., Fukui, Y., Brugliera, F., Holton, T. A., Karan, M., Nakamura, N., Yonekura-Sakakibara, K., Togami, J., Pigeaire, A., Tao, G.-Q., Nehra, N. S., Lu, C.-Y., Dyson, B. K., Tsuda, S., Ashikari, T., Kusumi, T., Mason, J. G., and Tanaka, Y. (2007). Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 48, 1589–1600.
Katsuyama, Y., Funa, N., Miyahisa, I., and Horinouchi, S. (2007a). Synthesis of unnatural flavonoids and stilbenes by exploiting the plant biosynthetic pathway in Escherichia coli. Chem. Biol. 14, 613–621.
Katsuyama, Y., Matsuzawa, M., Funa, N., and Horinouchi, S. (2007b). In vitro synthesis of curcuminoids by type III polyketide synthase from Oryza sativa. J. Biol. Chem. 282, 37702–37709.
Katsuyama, Y., Miyahisa, I., Funa, N., and Horinouchi, S. (2007c). One-pot synthesis of genistein from tyrosine by coincubation of genetically engineered Escherichia coli and Saccharomyces cerevisiae cells. Appl. Microbiol. Biotechnol. 73, 1143–1149.
Kidd, P. S., Llugany, M., Poschenrieder, C., Gunse, B., and Barcelo, J. (2001). The role of root exudates in aluminium resistance and silicon-induced amelioration of aluminium toxicity in three varieties of maize (Zea mays L.). J. Exp. Bot. 52, 1339–1352.
Kim, S. S., Grienenberger, E., Lallemand, B., Colpitts, C. C., Kim, S. Y., Souza Cde, A., Geoffroy, P., Heintz, D., Krahn, D., Kaiser, M., Kombrink, E., Heitz, T., Suh, D. Y., Legrand, M., and Douglas, C. J. (2010). LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl alpha-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22, 4045–4066.
Kliebenstein, D. J. (2004). Secondary metabolites and plant/environment interactions: a view through Arabidopsis thaliana tinged glasses. Plant Cell Environ. 27, 675–684.
Koca, U., Berhow, M. A., Febres, V. J., Champ, K. I., Carrillo-Mendoza, O., and Moore, G. A. (2009). Decreasing unpalatable flavonoid components in Citrus: the effect of transformation construct. Physiol. Plant 137, 101–114.
Koes, R., Verweij, W., and Quattrocchio, F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242.
Koornneef, M. (1990). Mutations affecting the testa colour in Arabidopsis. Arab. Inf. Serv. 27, 1–4.
Kuhn, B. M., Geisler, M., Bigler, L., and Ringli, C. (2011). Flavonols accumulate asymmetrically and affect auxin transport in Arabidopsis. Plant Physiol. 156, 585–595.
Kusano, M., Tohge, T., Fukushima, A., Kobayashi, M., Hayashi, N., Otsuki, H., Kondou, Y., Goto, H., Kawashima, M., Matsuda, F., Niida, R., Matsui, M., Saito, K., and Fernie, A. R. (2011). Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV-B light. Plant J. 67, 354–369.
Kutchan, T. M. (2005). A role for intra- and intercellular translocation in natural product biosynthesis. Curr. Opin. Plant Biol. 8, 292–300.
Landry, L. G., Chapple, C. C. S., and Last, R. L. (1995). Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 109, 1159–1166.
Lewis, D. R., Ramirez, M. V., Miller, N. D., Vallabhaneni, P., Ray, W. K., Helm, R. F., Winkel, B. S., and Muday, G. K. (2011). Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol. 156, 144–164.
Li, J., Ou-Lee, T. M., Raba, R., Amundson, R. G., and Last, R. L. (1993). Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5, 171–179.
Li, X., Qin, J. C., Wang, Q. Y., Wu, X., Lang, C. Y., Pan, H. Y., Gruber, M. Y., and Gao, M. J. (2011). Metabolic engineering of isoflavone genistein in Brassica napus with soybean isoflavone synthase. Plant Cell Rep. 30, 1435–1442.
Logemann, E., and Hahlbrock, K. (2002). Crosstalk among stress responses in plants: pathogen defense overrides UV protection through an inversely regulated ACE/ACE type of light-responsive gene promoter unit. Proc. Natl. Acad. Sci. U.S.A. 99, 2428–2432.
Lloyd, A. M., Walbot, V., and Davis, R. W. (1992). Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258, 1773–1775.
Mahajan, M., Ahuja, P. S., and Yadav, S. K. (2011). Post-transcriptional silencing of flavonol synthase mRNA in tobacco leads to fruits with arrested seed set. PLoS ONE 6:e28315. doi: 10.1371/journal.pone.0028315
Mandal, S. M., Chakraborty, D., and Dey, S. (2010). Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal. Behav. 5, 359–368.
Marienhagen, J., and Bott, M. (2012). Metabolic engineering of microorganisms for the synthesis of plant natural products. J. Biotechnol. [Epub ahead of print].
Marrs, K. A., Alfenito, M. R., Lloyd, A. M., and Walbot, V. (1995). A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375, 397–400.
Martens, S., Preuss, A., and Matern, U. (2010). Multifunctional flavonoid dioxygenases: flavonols and anthocyanin biosynthesis in Arabidopsis thaliana L. Phytochemistry 71, 1040–1049.
Mathews, H., Clendennen, S. K., Caldwell, C. G., Liu, X. L., Connors, K., Matheis, N., Schuster, D. K., Menasco, D. J., Wagoner, W., and Lightner, J. (2003). Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15, 1689–1703.
Mazza, C. A., Boccalandro, H. E., Giordano, C. V., Battista, D., Scopel, A. L., and Ballare, C. L. (2000). Functional significance and induction by solar radiation of ultraviolet-absorbing sunscreens in field-grown soybean crops. Plant Physiol. 122, 117–126.
Meyer, P., Heidemann, I., Forkmann, G., and Saedler, H. (1987). A new petunia flower colour generated by transformation of a mutant with a maize gene. Nature 330, 677–678.
Miadoková, E. (2009). Isoflavonoids - an overview of their biological activities and potential health benefits. Interdiscip. Toxicol. 2, 211–218.
Min, J. Y., Kim, B. G., and Ahn, J.-H. (2009). Biological synthesis of 7-O-Methyl apigenin from naringenin using Escherichia coli expressing two genes. J. Microbiol. Biotechnol. 19, 491–494.
Miyahisa, I., Funa, N., Ohnishi, Y., Martens, S., Moriguchi, T., and Horinouchi, S. (2006). Combinatorial biosynthesis of flavones and flavonols in Escherichia coli. Appl. Microbiol. Biotechnol. 71, 53–58.
Miyahisa, I., Kaneko, M., Funa, N., Kawasaki, H., Kojima, H., Ohnishi, Y., and Horinouchi, S. (2005). Efficient production of (2 S)-flavanones by Escherichia coli containing an artificial biosynthetic gene cluster. Appl. Microbiol. Biotechnol. 68, 498–504.
Mo, Y., Nagel, C., and Taylor, L. P. (1992). Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc. Natl. Acad. Sci. U.S.A. 89, 7213–7217.
Mol, J., Grotewold, E., and Koes, R. (1998). How genes paint flowers and seeds. Trends Plant Sci. 3, 212–217.
Moy, P., Qutob, D., Chapman, B. P., Atkinson, I., and Gijzen, M. (2004). Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol. Plant Microbe Interact. 17, 1051–1062.
Murphy, A., Peer, W. A., and Taiz, L. (2000). Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 211, 315–324.
Nakamura, N., Fukuchi-Mizutani, M., Suzuki, K., Miyazaki, K., and Tanaka, Y. (2006). RNAi suppression of the anthocyanidin synthase gene in Torenia hybrida yields white flowers with higher frequency and better stability than antisense and sense suppression. Plant Biotechnol. 23, 13–17.
Neuffer, M. G., Coe, E. H., and Wessler, S. R. (1997). Mutants of Maize. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
Nishihara, M., and Nakatsuka, T. (2011). Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 33, 433–441.
Ono, E., Fukuchi-Mizutani, M., Nakamura, N., Fukui, Y., Yonekura-Sakakibara, K., Yamaguchi, M., Nakayama, T., Tanaka, T., Kusumi, T., and Tanaka, Y. (2006). Yellow flowers generated by expression of the aurone biosynthetic pathway. Proc. Natl. Acad. Sci. U.S.A. 103, 11075–11080.
Ortega, A., Vasal, S. K., Mihm, J., and Hershey, C. (1980). Breeding for Insect Resistance in Maize. New York, NY: J. Wiley and Sons.
Owens, D. K., Alerding, A. B., Crosby, K. C., Bandara, A. B., Westwood, J. H., and Winkel, B. S. J. (2008). Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol. 147, 1046–1061.
Pattanaik, S., Kong, Q., Zaitlin, D., Werkman, J. R., Xie, C. H., Patra, B., and Yuan, L. (2010). R2R3 MYB regulator from tobacco. Isolation and functional characterization of a floral tissue-specific. Planta 231, 1061–1076.
Peer, W. A., and Murphy, A. S. (2007). Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci. 12, 556–563.
Petroni, K., and Tonelli, C. (2011). Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181, 219–229.
Pires, N., and Dolan, L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874.
Pollak, P. E., Vogt, T., Mo, Y., and Taylor, L. P. (1993). Chalcone synthase and flavonol accumulation in stigmas and anthers of Petunia hybrida. Plant Physiol. 102, 925–932.
Pollastri, S., and Tattini, M. (2011). Flavonols: old compounds for old roles. Ann. Bot. 108, 1225–1233.
Pourcel, L., Irani, N. G., Lu, Y., Riedl, K., Schwartz, S., and Grotewold, E. (2010). The formation of anthocyanic vacuolar inclusions in Arabidopsis thaliana and implications for the sequestration of anthocyanin pigments. Mol. Plant 3, 78–90.
Preuss, A., Stracke, R., Weisshaar, B., Hillebrecht, A., Matern, U., and Martens, S. (2009). Arabidopsis thaliana expresses a second functional flavonol synthase. FEBS Lett. 583, 1981–1986.
Ralston, L., and Yu, O. (2006). Metabolons involving plant cytochrome P450s. Phytochem. Rev. 5, 459–472.
Rimando, A. M., Pan, Z., Polashock, J. J., Dayan, F. E., Mizuno, C. S., Snook, M. E., Liu, C.-J., and Baerson, S. R. (2012). In planta production of the highly potent resveratrol analogue pterostilbene via stilbene synthase and O-methyltransferase co-expression. Plant Biotechnol. J. 10, 269–283.
Ryan, K. G., Swinny, E. E., Markham, K. R., and Winefield, C. (2002). Flavonoid gene expression and UV photoprotection in transgenic and mutant Petunia leaves. Phytochemistry 59, 23–32.
Ryeol, P. S., Paik, J. H., Ahn, M. S., Park, J. W., and Yoon, Y. J. (2010). Biosynthesis of plant-specific flavones and flavonols in streptomyces venezuelae. J. Microbiol. Biotechnol. 20, 1295–1299.
Samac, D. A., and Graham, M. A. (2007). Recent advances in legume-microbe interactions: recognition, defense response, and symbiosis from a genomic perspective. Plant Physiol. 144, 582–587.
Saslowsky, D. E., Warek, U., and Winkel, B. S. (2005). Nuclear localization of flavonoid enzymes in Arabidopsis. J. Biol. Chem. 280, 23735–23740.
Schenke, D., Böttcher, C., and Scheel, D. (2011). Crosstalk between abiotic ultraviolet-B stress and biotic (flg22) stress signalling in Arabidopsis prevents flavonol accumulation in favor of pathogen defence compound production. Plant Cell Environ. 34, 1849–1864.
Schijlen, E. G., De Vos, C. H., Martens, S., Jonker, H. H., Rosin, F. M., Molthoff, J. W., Tikunov, Y. M., Angenent, G. C., Van Tunen, A. J., and Bovy, A. G. (2007). RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol. 144, 1520–1530.
Shen, T., Wang, X. N., and Lou, H. X. (2009). Natural stilbenes: an overview. Nat. Prod. Rep. 26, 916–935.
Shih, C. H., Chu, H., Tang, L. K., Sakamoto, W., Maekawa, M., Chu, I. K., Wang, M., and Lo, C. (2008). Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 228, 1043–1054.
Snook, M. E., Widstrom, N. W., Wiseman, B. R., Gueldner, R. C., Wilson, R. L., Himmelsbach, D. S., Harwood, J. S., and Costello, C. E. (1994). “New flavonol-C-glycosides from corn (Zea mays L.) for the control of the corn earworm (Helicoverpa zea),” in Bioregulators for Crop Protection and Pest Control, ed P. A. Hedin, ACS Symposium Series No. 557 (Washington DC: American Chemical Society), 122–135.
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., and Weisshaar, B. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677.
Stracke, R., Jahns, O., Keck, M., Tohge, T., Niehaus, K., Fernie, A. R., and Weisshaar, B. (2010). Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol. 188, 985–1000.
Stratmann, J. (2003). Ultraviolet-B radiation co-opts defense signaling pathways. Trends Plant Sci. 8, 526–533.
Subramanian, S., Graham, M. Y., Yu, O., and Graham, T. L. (2005). RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae. Plant Physiol. 137, 1345–1353.
Subramanian, S., Stacey, G., and Yu, O. (2006). Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J. 48, 261–273.
Sun, Y., Li, H., and Huang, J.-R. (2012). Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant 5, 387–400.
Tanaka, Y., Brugliera, F., and Chandler, S. (2009). Recent progress of flower colour modification by biotechnology. Int. J. Mol. Sci. 10, 5350–5369.
Tanaka, Y., Brugliera, F., Kalc, G., Senior, M., Dyson, B., Nakamura, N., Katsumoto, Y., and Chandler, S. (2010). Flower color modification by engineering of the flavonoid biosynthetic pathway: practical perspectives. Biosci. Biotechnol. Biochem. 74, 1760–1769.
Tanaka, Y., and Ohmiya, A. (2008). Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Chem. Biol. 19, 190–197.
Tanaka, Y., Sasaki, N., and Ohmiya, A. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54, 733–749.
Taylor, L. P., and Hepler, P. K. (1997). Pollen germination and tube growth. Ann. Rev. Plant Physiol. Plant Mol. Biol. 48, 461–491.
Toda, K., Kuroiwa, H., Senthil, K., Shimada, N., Aoki, T., Ayabe, S. I., Shimada, S., Sakuta, M., Miyazaki, Y., and Takahashi, R. (2012). The soybean F3′H protein is localized to the tonoplast in the seed coat hilum. Planta 236, 79–89.
Torregrosa, C., Cluzet, S., Fournier, J., Huguet, T., Gamas, P., Prospéri, J.-M., Esquerré-Tugayé, M.-T., Dumas, B., and Jacquet, C. (2004). Cytological, genetic, and molecular analysis to characterize compatible and incompatible interactions between Medicago truncatula and Colletotrichum trifolii. Mol. Plant Microbe Interact. 17, 909–920.
Van De Staaij, J., De Bakker, N. V., Oosthoek, A., Broekman, R., Van Beem, A., Stroetenga, M., Aerts, R., and Rozema, J. (2002). Flavonoid concentrations in three grass species and a sedge grown in the field and under controlled environment conditions in response to enhanced UV-B radiation. J. Photochem. Photobiol. B 66, 21–29.
Van Der Meer, I. M., Stam, M. E., Van Tunen, A. J., Mol, J. N., and Stuitje, A. R. (1992). Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 4, 253–262.
Verdan, A. M., Wang, H. C., García, C. R., Henry, W. P., and Brumaghim, J. L. (2011). Iron binding of 3-hydroxychromone, 5-hydroxychromone, and sulfonated morin: implications for the antioxidant activity of flavonols with competing metal binding sites. J. Inorg. Biochem. 105, 1314–1322.
Vogel, S., and Heilmann, J. (2008). Synthesis, cytotoxicity, and antioxidative activity of minor prenylated chalcones from Humulus lupulus. J. Nat. Prod. 71, 1237–1241.
Waiss, A. C., Chan, B. G., Elliger, C. A., Wiseman, B. R., McMillian, W. W., Widstrom, N. W., Zuber, M. S., and Keaster, A. J. (1979). Maysin, a flavone glycoside from corn silks with antibiotic activity toward corn earworm. J. Econ. Entomol. 72, 256–258.
Wang, X. (2011). Structure, function, and engineering of enzymes in isoflavonoid biosynthesis. Funct. Integr. Genomics 11, 13–22.
Wasson, A. P., Pellerone, F. I., and Mathesius, U. (2006). Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by Rhizobia. Plant Cell 18, 1617–1629.
Williams, R., Spencer, J., and Rice-Evans, C. (2004). Flavonoids: antioxidants or signalling molecules? free radic. Biol. Med. 36, 838–849.
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493.
Winkel-Shirley, B. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5, 218–223.
Winkel-Shirley, B. (2006). “The biosynthesis of flavonoids,” in The Science of Flavonoids, ed E. Grotewold (New York, NY: Springer), 71–95.
Xue, B., Charest, P. J., Devantier, Y., and Rutledge, R. G. (2003). Characterization of a MYBR2R3 gene from black spruce (Picea mariana) that shares functional conservation with maize C1. Mol. Genet. Genomics 270, 78–86.
Yan, Y., Huang, L., and Koffas, M. A. (2007). Biosynthesis of 5-deoxyflavanones in microorganisms. Biotechnol. J. 2, 1250–1262.
Yan, Y., Li, Z., and Koffas, M. A. (2008). High-yield anthocyanin biosynthesis in engineered Escherichia coli. Biotechnol. Bioeng. 100, 126–140.
Ylstra, B., Muskens, M., and Tunen, A. J. (1996). Flavonols are not essential for fertilization in Arabidopsis thaliana. Plant Mol. Biol. 32, 1155–1158.
Yu, O., and Mcgonigle, B. (2005). Metabolic engineering of isoflavone biosynthesis. Adv. Agron. 86, 147–190.
Zerback, R., Dressler, K., and Hess, D. (1989). Flavonoid compounds from pollen and stigma of Petunia hybrida: inducers of the vir region of the Agrobacterium tumefaciens Ti plasmid. Plant Sci. 62, 83–91.
Zhang, J., Subramanian, S., Stacey, G., and Yu, O. (2009). Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J. 57, 171–183.
Zhang, P., Wang, Y., Zhang, J., Maddock, S., Snook, M., and Peterson, T. (2003). A maize QTL for silk maysin levels contains duplicated Myb-homologous genes which jointly regulate flavone biosynthesis. Plant Mol. Biol. 52, 1–15.
Zhao, J., and Dixon, R. A. (2010). The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 15, 72–80.
Zou, J., Rodriguez-Zas, S., Aldea, M., Li, M., Zhu, J., Gonzalez, D. O., Vodkin, L. O., Delucia, E., and Clough, S. J. (2005). Expression profiling soybean response to Pseudomonas syringae reveals new defense-related genes and rapid HR-specific downregulation of photosynthesis. Mol. Plant Microbe Interact. 18, 1161–1174.
Keywords: flavonoid, transcription factors, genetic engineering, defense, combinatorial biosynthesis
Citation: Falcone Ferreyra ML, Rius SP and Casati P (2012) Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3:222. doi: 10.3389/fpls.2012.00222
Received: 15 August 2012; Paper pending published: 28 August 2012;
Accepted: 11 September 2012; Published online: 28 September 2012.
Edited by:
Hanjo A. Hellmann, Washington State University, USAReviewed by:
Sanja Roje, Washington State University, USACopyright © 2012 Falcone Ferreyra, Rius and Casati. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Paula Casati, Centro de Estudios Fotosintéticos y Bioquímicos, Universidad Nacional de Rosario, Suipacha 531, 2000 Rosario, Argentina. e-mail:Y2FzYXRpQGNlZm9iaS1jb25pY2V0Lmdvdi5hcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.