- Biomedical Research Centre, Sheffield Hallam University, Sheffield, UK
The general mechanisms of intracellular protein targeting are well established, and depend on a targeting sequence in the protein, which is recognized by a targeting factor. Once a membrane protein is delivered to the correct organelle its targeting sequence can be recognized by receptors and a translocase, leading to membrane insertion. However, the relative contribution of each step for generating fidelity and efficiency of the overall process has not been systematically addressed. Here, we use tail-anchored (TA) membrane proteins in cell-free competitive targeting assays to chloroplasts to show that targeting can occur efficiently and with high fidelity in the absence of all cytosolic components, suggesting that chloroplast envelope protein targeting is primarily dependent on events at the outer envelope. Efficiency of targeting was increased by the addition of complete cytosol, and by Hsp70 or Hsp90, depending on the protein, but none of these cytosolic components influenced the fidelity of targeting. Our results suggest that the main role of targeting factors in chloroplast localization is to increase targeting efficiency by maintaining recognition competency at the outer envelope.
Introduction
Protein localization to organelles is a vital aspect of cellular organisation, and we now have an excellent understanding of many the molecular mechanisms underlying each pathway. The central concept is the signal hypothesis (Blobel and Sabatini, 1971), which involves sequence information in the nascent protein promoting engagement with membrane receptors, leading to the protein entering or crossing the membrane. This hypothesis is supported by a vast number of studies involving diverse proteins and compartments (Wickner and Schekman, 2005), but most of our understanding is based on viewing the process as a series of essential steps, without quantifying the importance of each stage or the potential for bypassing some of the events. For example, N-terminal ER signal sequences can be selectively recognized by both the targeting factor, SRP, and the Sec61 translocon (Belin et al., 1996), yet the importance of each step for determining ER-specific localization has not been dissected.
Targeting processes can be split into four key stages: (1) Ribosome coordination, in which targeting sequences can be recognized more efficiently if targeting factors are present close to the ribosomal exit tunnel, exemplified by SRP (reviewed by Grudnik et al., 2009). This process is further promoted if targeting sequences are sensed in the exit tunnel, leading to enhanced affinity for targeting factors close to the exit site, which occurs for SRP (Berndt et al., 2009). (2) Targeting factors interact with the targeting sequence in the cytosol, which can chaperone hydrophobic targeting sequences, and also guide the nascent protein to a specific organelle via interaction with a cognate receptor. (3) Receptors at organelle surfaces bind the targeting factor bound to a nascent protein, and promote the transition of the protein to a translocase, typically due to an association between the receptor and the translocation. The receptor primarily binds the targeting factor, which may be the sole interaction as observed for SRP (reviewed by Grudnik et al., 2009), but may form additional interactions directly with the protein, as observed for Tom70 at mitochondria (MT) (Brix et al., 2000; Li et al., 2009). Association between the receptor and the translocase can be either stable or dynamic; for example Toc34 and Toc159 are permanent components of the chloroplast Toc complex, whereas Toc64 is more transiently associated (Soll, 2002; Qbadou et al., 2006). (4) Membrane translocases promote insertion into the membrane or transport across the membrane, although direct membrane insertion is possible in some cases, such as the insertion of cytochrome b5 into ER membranes (Brambillasca et al., 2005).
Each of these targeting steps has the potential to contribute to the efficiency and specificity of the process, and the prevailing view is that each step contributes to both of these outcomes (e.g., Borgese and Fasana, 2010). However, there are few studies where these two issues are dissected. The original signal hypothesis only included one recognition event for the signal sequence, and this was depicted as occurring at the membrane (Blobel and Sabatini, 1971). This logic suggests that any additional recognition events would make a lesser contribution, and could possibly be dispensed with. Hence, if translocases or associated receptors at the membrane are capable of discriminating between signal sequences the primary role of targeting factors may be to chaperone targeting sequences. The highly specific and coordinated interactions of SRP with its cognate receptor at the ER membrane (Grudnik et al., 2009) indicate that it plays a major role in targeting specificity, yet yeast are able to survive without SRP (Rothe and Lehle, 1998), and the targeting of ER proteins can occur instead by a posttranslational pathway involved Hsp70 (Ngosuwan et al., 2003), a molecular chaperone that is involved in protein targeting to all organelles (Zimmermann, 1998; Artigues et al., 2002; Soll, 2002; Abell et al., 2007; Kriechbaumer et al., 2012). A major reason why the specificity of protein targeting has been neglected is that protein targeting experiments commonly use cell free assays with single organelles, and thus focus on the efficiency of targeting.
One way of assessing the role of targeting factors in generating specificity is to test the effect of mutations or knockouts of targeting factors on mistargeting, but there are few examples. Perhaps the clearest case is the mistargeting of overexpressed ER tail-anchored (TA) proteins to mitochondria in yeast when Get3 is deleted. However, the secretory system is widely disrupted, and the majority of the non-targeted TA protein forms aggregates in the cytosol (Schuldiner et al., 2008), indicating that mistargeting only occurs under considerable cellular stress. Conversely, mitochondrial TA proteins are still targeted accurately to mitochondria in semi-permeabilized cells that lack cytosol (Setoguchi et al., 2006), showing that at least some targeting pathways are independent of targeting factors. How these mitochondrial TA proteins become targeted correctly is unclear because there is some uncertainty about which membrane components facilitate their targeting and insertion. Some studies provide evidence for the involvement of receptors Tom20 (Schleiff et al., 1997a,b; Motz et al., 2002), Tom70 (Otera et al., 2007) and SAM (Stojanovski et al., 2007), but another indicates that the process is dependent on other unidentified membrane proteins (Kemper et al., 2008).
When targeting pathways are overwhelmed by overexpression of proteins the usual result is aggregation of protein rather than mistargeting, one example being the plant nuclear envelope WIT1 protein, and this aggregation can be prevented by coexpression of Hsc70 (Brkljacic et al., 2009). A more subtle analysis of protein targeting is possible for those targeted to multiple organelles, such as the TA protein Fis1, which can target to both mitochondria and peroxisomes (Delille and Schrader, 2008). hFis targeting to peroxisomes is dependent upon the targeting factor Pex19, and in its absence will only target to mitochondria. However, overexpression of Pex19 has no effect on the balance of mitochondrial and peroxisomal targeting, indicating that it does not compete directly with mitochondrial targeting. A modified cytochrome b5, also a TA protein, was found to localize predominantly to mitochondria and partially to ER membranes in vivo, but when presented with only ER membranes in vitro could be targeted efficiently, showing that the balance of targeting can be shifted in non-native situations (Borgese et al., 2001). These examples suggest an influence of targeting factors on fidelity, but the complexities of in vivo experiments makes further mechanistic analysis difficult.
Here, we are systematically testing the role of cytosolic targeting factors for both efficiency and specificity, using chloroplasts as a model system in competition with mitochondria and ER membranes. Previous studies with chloroplasts have either focused on the ability of various nascent protein complexes to promote import in single organelle assays (e.g., May and Soll, 2000), or have studied competitive targeting to multiple organelles in the presence of complete cytosolic lysate (e.g., Rudhe et al., 2002), but these approaches have not been combined. Our experimental approach is to test the role of cytosolic lysate and defined targeting factors in competitive cell free targeting assays, which have previously been shown to provide reliable targeting information compared to microscopy of GFP-tagged proteins in plant cells (Kriechbaumer et al., 2009). We specifically tested the effect of removing all targeting factors on the efficiency of targeting to the native organelle and potential mistargeting to other organelles. The selected proteins are TA and other membrane proteins associated with the chloroplast envelope, to focus the analysis on the first stages of chloroplast targeting, and to exploit our ability to prepare cytosol-free nascent proteins. We find that targeting is highly specific and efficient in the presence of a range of different cytosolic factors, and even in the absence of all cytosolic factors.
Materials and Methods
Cloning and Protein Expression
Clones for Hsp70 (At5g02500), Hsp90 (At5g56030), and Hsp40 (At3g44110) were obtained as cDNAs in plasmids from The Arabidopsis Information Resource (TAIR). The coding regions were cloned into a pET16b expression vector containing a 6-His-tag (Novagen, Madison, WI) and proteins were heterologously expressed in T7 Express Iq E.coli cells (NEB). Bacterial cells were harvested by centrifugation at 8000 g for 8 min, resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazol, pH 8.0), and treated with lysozyme (1 mg/ml) for 30 min on ice. The resulting lysate was sonicated three times at 200 W for 10 s and was centrifuged at 10,000 × g for 30 min at 4°C. His-tagged proteins were purified under native conditions using Ni-NTA agarose (Qiagen) according to the manufacturer's manual (The QIAexpressionist™). Dialysis was performed against 50 mM Tris-HCl pH 8.0 and the His-tag was cleaved, other than for pulldown experiments where indicated.
Transcription and Translation
Genes to be tested were fused to the pSPUTK SP6-promoter via overlapping extension-PCR (Urban et al., 1997). Transcriptions were performed with 15 μg PCR-fusion product and SP6-RNA-Polymerase (New England Biolabs) according to the manufacturer's instructions. Protein translations were performed in wheat germ extract (WGE; Promega) according to the manufacturer's manual using Easy Tag Express 35 S (Perkin Elmer).
RNC Preparation
RNCs were generated by translating transcripts lacking their stop codon for 7 min at 25°C. Reactions of 200 μl were supplemented with 2.5 mM cycloheximide and 500 mM KOAc. The sample was layered over 400 μl ice cold HSCC [500 mM sucrose, 500 mM KOAc, 5 mM Mg (OAc)2, 50 mM Hepes-KOH pH 7.9, 2.5 mM cycloheximide, 1 mM DTT]. After a centrifugation at 213,000 × g for 20 min the pellet was resuspended in 50 μl HSCC with reduced sucrose (100 mM), layered onto 150 μl HSCC, and centrifuged at 213,000 × g for 20 min. The pellet was finally resuspended in 40 μl LSC [100 mM sucrose, 100 mM KOAc, 5 mM Mg (OAc)2, 50 mM Hepes-KOH pH 7.9, 1 mM DTT].
For determination of protein-chaperone complexes 20 μl of RNC-released protein was incubated in a total volume of 200 μl import buffer (50 mM HEPES, pH 8.0, 330 mM sorbitol, 8.4 mM methionine, 13 mM ATP, 13 mM MgCl2) for 10 min at RT with and without 20 μl of WGE. The mixture was then centrifuged through 50 kDa molecular mass cutoff columns (Millipore) according to the manufacturer's instructions. The flow-through was collected and import buffer was added up to 200 μl to the retentate reservoir. Both flow-through and retentate were analysed by SDS-PAGE and Cyclon Phosphor Screen (Packard).
Organelle Preparation
For chloroplasts purification 3 g of pea leaves grown for approximately 14 days in soil were cut into small pieces (~2 mm ×2 mm) and placed into two 50-ml Falcon tubes with 30 ml ice-cold grinding buffer (2 mM EDTA, pH 8.0, 1 mM MgCl2, 1 mM MnCl2, 50 mM HEPES, pH 7.5, 330 mM sorbitol, 0.1% (w/vol) sodium ascorbate, 0.25% BSA). After homogenization for 30 s with a Polytron homogenizer (IKA T 18 basic ULTRA-TURRAX, intensity level 2), the homogenate was filtered through four layers of cheesecloth and centrifuged at 3000 × g for 2 min. The resulting pellets were resuspended with a paintbrush in 0.5 ml grinding buffer, layered onto a Percoll step gradient (3 ml of 80% Percoll in PBF [30% (w/vol) polyethylene glycol (PEG), 10% (w/vol) BSA, 10% (w/vol) Ficoll] overlaid with 9.5 ml of 40% Percoll in PBF) and centrifuged in a swinging bucket at 9000 × g and 4°C for 8 min. The lower band containing intact chloroplasts was transferred to a fresh tube, the volume adjusted to 25 ml with grinding buffer and centrifuged at 3000 × g for 4 min. The resulting pellet was resuspended in 0.5 ml 1 × HSM (50 mM HEPES, pH 8.0, 330 mM sorbitol, 8.4 mM methionine), and total protein concentration was determined by Bradford assay.
To isolate mitochondria, 3 g of 5-day-old maize coleoptiles were cut into small pieces and homogenized for 30 s in a Polytron homogenizer (IKA T 18 basic ULTRA-TURRAX, intensity level 2) in 30 ml of mitochondrial grinding buffer [50 mM TES, pH 7.5, 300 mM sucrose, 2 mM EDTA, 1 mM MgCl2, 1% (w/vol) PVP-40, 0.4% (w/vol) BSA, 4 mM cysteine]. The homogenate was filtered through four layers of prewetted muslin and centrifuged at 2960 × g for 3 min; the pellets were discarded. The supernatant was centrifuged at 17,700 × g for 20 min, the pellet was resuspended in 0.5 ml of wash buffer [20 mM TES, pH 7.5, 300 mM sucrose, 0.1% (w/vol) BSA] and loaded onto a continuous Percoll/PVP-40 gradient made up with 20 ml of heavy solution [10 ml of solution II consisting of 600 mM sucrose, 20 mM KH2/PO4, pH 7.5, 0.4% (w/vol) BSA, 5.5 ml Percoll and 4.5 ml 40% (w/vol) PVP-40] and 20 ml of light solution (10 ml of solution II, 5.5 ml Percoll and 4.5 ml H2O). The preparation was centrifuged at 39,200 × g for 40 min. Intact mitochondria formed a straw-coloured band near the bottom of the gradient, which was transferred to a fresh tube. Approximately 30 ml of wash medium was added to the mitochondrial fraction and centrifuged at 12,100 × g for 20 min. The pellet was gently resuspended in 0.5 ml 1 × HSM (50 mM HEPES, pH 8.0, 330 mM sorbitol, 8.4 mM methionine), and total protein concentration was determined by Bradford assay.
Competitive Targeting Assay
Competitive targeting assays were performed in a total reaction volume of 100 μl according to (Kriechbaumer et al., 2009), including 58 μl import buffer (50 mM HEPES, pH 8.0, 330 mM sorbitol, 8.4 mM methionine, 13 mM ATP, 13 mM MgCl2). Membrane insertion reactions comprised 2 μl of isolated RNC with varying additions. WGE was added at 10% vol/vol, recombinantly expressed chaperones Hsp70 (with Hsp40) and Hsp90 were added at 2.0 μM, and apyrase at 0.1 U/μ l. Following the addition of all cytosolic targeting factors and treatments, 1 mM puromycin was added. 10 μl chloroplasts (equivalent to 60 mg leaf material; total protein 300 μg), 10 μl mitochondria (equivalent to 60 mg coleoptile material; total protein 180 μg), or 2 μl canine pancreatic microsomes (at 50 A280 U ml−1; total protein 240 μg), were then added in various combinations, and incubation was performed at 30°C for 20 min. Organelles were repurified by two sequential centrifugation steps depending on the combination of organelles (2 min at 3000 × g to pellet chloroplasts, 20 min centrifugation at 17,000 × g to pellet mitochondria, and 20 min at 200,000 × g to pellet ER membranes, in this order) and washed with 0.1 M Na2CO3. Both fractions were analysed by SDS-PAGE and Cyclon Phosphor Screen (Packard); where dividing lines are shown on gel figures the samples were loaded on separate gels, but processed in parallel. Quantification of signal was performed using manufacturer's software (Packard) and processed to generate a mean, standard error, and significance of difference to the control import by One-Way ANOVA. In the case of thermolysin treatments, the import sample was treated with 40 U/ml thermolysin for 5 min at 30°C prior to analysis. For thermolysin and Triton X-100 treatment the samples were treated with 0.2% Triton X-100 (Sigma) for 5 min at 30°C before thermolysin treatment.
Pull-down of protein-chaperone complexes was performed before the addition of membranes, by first treating with 1 unit/ml apyrase on ice for 5 min, and then incubating with 20 μl NiNTA agarose beads (Promega) at 4°C for 60 min. Beads were washed twice with CG buffer (100 mM KOAc, 20 mM Hepes/KOH (pH 7.5), 5 mM MgOAc2, and 0.1 % Triton X-100).
Accession Numbers
Locus identifiers and ABRC clone numbers are given in brackets: Toc33 (At1g02280, 190I17); At1g17 (At1g17780, U24679); At3g63 (At3g63160, U83351); Tic22 (At4g33350, 144A17); Toc64 (At3g17970, 180A6); At3g58 (At3g58840, U83915); Hsp70 (At5g02500, DKLAT5G02500); Hsp90 (At5g56030, DKLAT5G56030); Hsp40 (At3g44110, DKLAT3G44110); Sec61β (At2g45070, 106L22).
Results
Our experimental approach to determining the role of cytosolic components in protein targeting was to use a defined cell free system comprising radiolabeled nascent proteins and purified organelles. To enable control of the targeting environment, we translated proteins lacking a stop codon, which locks the protein on the ribosome, and then separated the ribosome-nascent chain (RNC) complexes from cytosolic components by centrifugation. Once the RNC was resuspended in a physiological buffer the protein was released by puromycin treatment, and incubated with organelles and selected cytosolic components. This approach is particularly effective for TA membrane proteins because their targeting information is primarily contained within the C-terminal transmembrane domain, which is contained within the ribosomal exit tunnel until puromycin release, and therefore unavailable to targeting factors in the translation lysate (Abell et al., 2007). After targeting, the organelles were separated by centrifugation, and the proportion of protein targeted to each organelle was measured. We have used chloroplasts in the presence of either ER or mitochondrial membranes, which offers a choice of destination, and a test of whether mistargeting can occur to either of the major alternative membranes present in the cell. We treated organelles with sodium carbonate solution to disrupt non-specific association of proteins with the membrane, which could otherwise be misinterpreted as membrane insertion or translocation.
To test whether proteins released from RNCs can be efficiently targeted to chloroplasts, representative proteins were compared with their freshly translated forms in complete cytosolic lysate (wheat germ extract). The total targeting efficiency ranged 6–18%, depending on the protein and the method of synthesis, with RNCs supporting higher levels of targeting for Toc33 and Tic22, but lower levels for Toc64 (Figure 1A). These efficiencies show that RNC purification yields viable proteins for chloroplast targeting assays. It was also possible that free ribosomes would have some impact on the targeting, so we tested the efficiency of Toc64 targeting with and without a prespin to remove ribosomes. The efficiency of targeting was found to be similar, thereby ruling out any significant contribution from ribosomes (Figure 1B). Finally, to verify that nascent proteins purified via RNCs were free of cytosolic factors, two representative proteins were released into buffer or wheat germ extract and fractionated using 50 kDa molecular weight cutoff spin columns (Figure 1C). Proteins were recovered solely in the low molecular weight fraction in the presence of buffer, but were mostly present in the higher molecular weight fraction after incubation with wheat germ extract, thus indicating that complexes only form in the presence of added cytosolic factors.
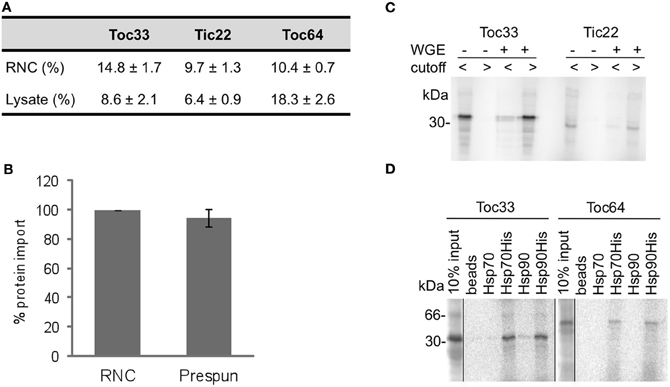
Figure 1. Protein import from RNCs to chloroplasts is efficient and ribosome independent. (A) Radio-labeled proteins (Toc33, Tic22, Toc64), either directly after translation in lysate or after RNC purification, were incubated with chloroplasts, and their import efficiency calculated as a percentage of the total protein input. Mean and standard error are shown (n = 3). (B) Toc64 was imported into chloroplasts with (RNC) or without (prespun) ribosomes present. Toc64 import was normalized to the RNC import, and mean and standard error are shown (n = 3). (C) Radio-labeled proteins (Toc33 or Tic22) with (+) and without (−) addition of WGE were fractionated by 50kDa molecular mass cutoff columns. Flow through (cutoff <), containing proteins smaller than 50 kDa, and retentate (cutoff >), with proteins and protein complexes of higher molecular weight than 50 kDa, were analysed by SDS-PAGE. (D) Radio-labeled proteins (Toc33 or Toc64) were released from RNCs in the presence of Hsp70 or Hsp90, either with or without a His-tag as indicated. After incubation, and apyrase treatment to stabilize interactions, complexes with His-tagged chaperone were pulled down using NiNTA agarose beads, and analysed by SDS-PAGE. 10% of protein input is shown.
The initial test was to determine whether proteins could target to organelles in the absence of any cytosolic components, and then various combinations of components were added, including complete wheat germ extract, which is widely used to support native cell free targeting of proteins. Since molecular chaperones have been implicated in chloroplast targeting processes (Fellerer et al., 2011; Schwenkert et al., 2011) we also tested the addition of various combinations of Hsp40, Hsp70, and Hsp90. Hsp40 stimulates the activity of Hsp70, so these were used in combination in all cases. To test whether molecular chaperones could bind proteins in the import assay, His-tagged Hsp70 (with Hsp40) or Hsp90 were incubated with Toc33 and Toc64 after their release from RNCs, and precipitated using NiNTA agarose beads. A significant fraction of each protein was pelleted with each chaperone, but not with chaperones lacking their His tag or beads alone (Figure 1D), confirming that Hsp70 and Hsp90 are functional for protein binding, although the binding of Hsp90 by Toc64 may be restricted to the TPR domain of Toc64 (Qbadou et al., 2006). As an additional control for genuine chaperone function in import assays, the ATP dependence of chaperone action was tested by apyrase treatment.
Several chloroplast TA proteins were selected, and the first one to be tested was the TA protein Toc33, which is a component of the chloroplast translocase, and has been studied extensively along with closely related Toc34. However, there is no consensus on the targeting mechanism, since some studies provide evidence for spontaneous membrane insertion (reviewed by Hofmann and Theg, 2005), whilst others detect requirements for pre-existing Toc33 and AKR2A targeting factor (Dhanoa et al., 2010). The targeting information is provided by the tail anchor, but there is also an important contribution from the N-terminal domain, a feature that is atypical for TA proteins (Dhanoa et al., 2010).
When Toc33 is incubated with chloroplasts and ER, targeting is specific for chloroplasts in the absence of any lysate or chaperones (Figures 2A and B), with accumulation of Toc33 ~10 fold greater in chloroplasts than ER membranes. The low levels of apparent insertion at ER membranes are likely to represent non-specific association of Toc33 that is not efficiently dispersed by sodium carbonate. The level of Toc33 targeting to chloroplasts is stimulated slightly by the addition of lysate, although this is not sensitive to apyrase treatment, suggesting that the stimulation is not due to the action of NTP-dependent components. Targeting is inhibited weakly by Hsp70/40, and is strongly promoted by Hsp90 (~2 fold), both effects being sensitive to apyrase. Taken together, these data suggest that Toc33 is able to target efficiently and selectively to chloroplasts in the absence of any cytosolic factors, but efficiency of targeting to the chloroplast membrane is enhanced by Hsp90 in the presence of ATP. The small stimulatory effect of complete lysate suggests that stimulatory components such as Hsp90 are counterbalanced by inhibitory components such as Hsp70/40. A similar pattern of data was obtained when Toc33 was incubated with chloroplasts and mitochondria (Figures 2C and D), providing further support for the conclusion that cytosolic components do not have a significant impact on targeting selectivity. Competency of mitochondria and ER preparations was tested by using TA proteins that are known to target to these organelles: At3g58 for mitochondria (Kriechbaumer et al., 2009); and Arabidopsis Sec61β for ER (Figure 2E). Both proteins targeted to the correct organelle and not to chloroplasts.
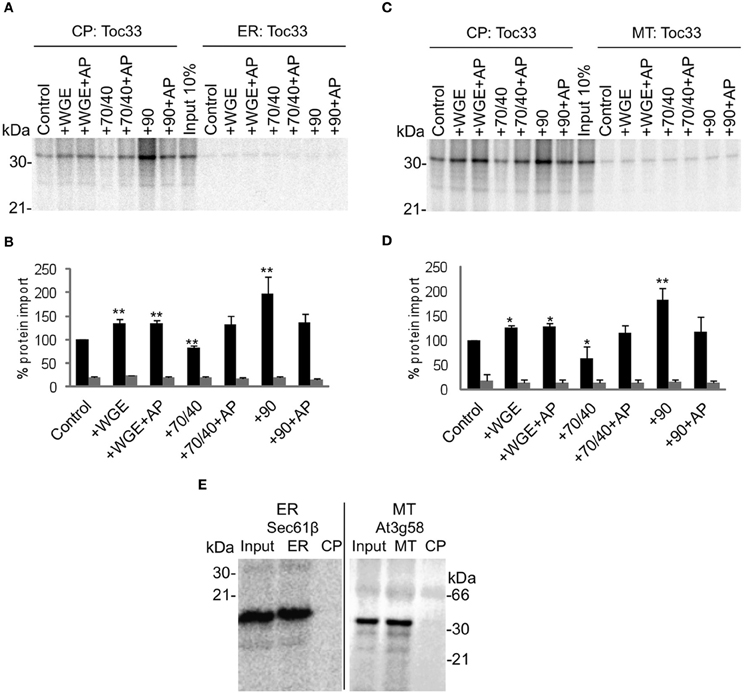
Figure 2. Toc33 targets to chloroplasts efficiently and selectively under a wide range of cytosolic conditions. (A) Radio-labeled Toc33 protein derived from RNCs was incubated in a competitive targeting assay with chloroplasts (CP) and ER, with the addition of cytosolic components as indicated; control is buffer only, WGE is wheat germ extract, chaperones (Hsp) are indicated by their numbers (70/40 or 90), and AP represents apyrase treatment. Membranes were refractionated and washed with carbonate prior to analysis by SDS-PAGE. 10% of protein input is shown, and the protein molecular weight markers are indicated in kDa. (B) Quantification of protein import in part A. Mean and standard error are shown, normalized to the control import into chloroplasts (n = 3), and significant difference from the control import is indicated by one star (p = 0.05) or two stars (p = 0.01). (C) Competitive targeting assay for Toc33 import into chloroplasts (CP) and mitochondria (MT), conducted as described in part A. (D) Quantification of protein import in part C. Mean and standard error are shown, normalized to the control import into chloroplasts (n = 3), and significant difference from the control import is indicated by one star (p = 0.05) or two stars (p = 0.01). (E) Competitive targeting assay for Arabidopsis Sec61β import into ER and chloroplasts (CP), and for At3g58 import into mitochondria (MT) and chloroplasts (CP), conducted as described in part A.
Another chloroplast TA protein, At1g17 (Kriechbaumer et al., 2009), was tested in the same assay, and found to display similar properties to Toc33 (Figure 3). Its targeting to chloroplasts was stimulated by lysate and Hsp90, and inhibited by Hsp70/40. Compared to Toc33, the stimulation by Hsp90 was weaker, and the inhibition by Hsp70/40 was stronger. Again, these findings were similar in the presence of either ER or mitochondria. A third chloroplast TA protein, At3g63 (Kriechbaumer et al., 2009), behaved like Toc33 and At1g17, but with milder effects of both Hsp70/40 and Hsp90 (Figure 4), suggesting that it may interact with other targeting factors present in the cytosol, or may simply not interact appreciably with any targeting factors.
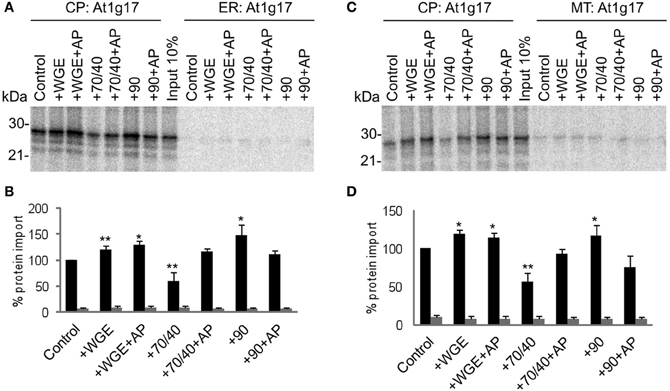
Figure 3. At1g17 targets to chloroplasts efficiently and selectively under a wide range of cytosolic conditions. (A) Radio-labeled At1g17 protein derived from RNCs was incubated in a competitive targeting assay with chloroplasts (CP) and ER, with the addition of cytosolic components as indicated; control is buffer only, WGE is wheat germ extract, chaperones (Hsp) are indicated by their numbers (70/40 or 90), and AP represents apyrase treatment. Membranes were refractionated and washed with carbonate for analysis by SDS-PAGE. 10% of protein input is shown, and the protein molecular weight markers are indicated in kDa. (B) Quantification of protein import in part A. Mean and standard error are shown, normalized to the control import into chloroplasts (n = 3), and significant difference from the control import is indicated by one star (p = 0.05) or two stars (p = 0.01). (C) Competitive targeting assay for At1g17 import into chloroplasts (CP) and mitochondria (MT), conducted as described in part A. (D) Quantification of protein import in part C. Mean and standard error are shown, normalized to the control import into chloroplasts (n = 3), and significant difference from the control import is indicated by one star (p = 0.05) or two stars (p = 0.01).
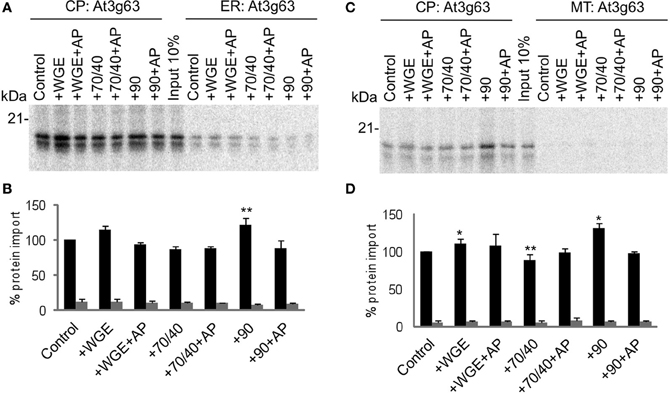
Figure 4. At3g63 targets to chloroplasts efficiently and selectively under a wide range of cytosolic conditions. (A) Radio-labeled At3g63 protein derived from RNCs was incubated in a competitive targeting assay with chloroplasts (CP) and ER, with the addition of cytosolic components as indicated; control is buffer only, WGE is wheat germ extract, chaperones (Hsp) are indicated by their numbers (70/40 or 90), and AP represents apyrase treatment. Membranes were refractionated and washed with carbonate before analysis by SDS-PAGE. 10% of protein input is shown, and the protein molecular weight markers are indicated in kDa. (B) Quantification of protein import in part A. Mean and standard error are shown, normalized to the control import into chloroplasts (n = 3), and significant difference from the control import is indicated by one star (p = 0.05) or two stars (p = 0.01). (C) Competitive targeting assay for At3g63 import into chloroplasts (CP) and mitochondria (MT), conducted as described in part A. (D) Quantification of protein import in part C. Mean and standard error are shown, normalized to the control import into chloroplasts (n = 3), and significant difference from the control import is indicated by one star (p = 0.05) or two stars (p = 0.01).
Two other types of membrane proteins were tested in the competitive targeting assay: the intermembrane space protein Tic22, which possesses an N-terminal targeting sequence (Figure 5); and the multi-spanning outer envelope receptor Toc64, which possesses an internal targeting sequence (Figure 6). These also had no requirement for lysate to achieve efficient and selective targeting. However, Toc64 is affected differently by chaperone types, its targeting being promoted ~50% by Hsp70/40, and inhibited ~30% by Hsp90, with both effects being sensitive to apyrase. Although Hsp90 is able to bind the TPR clamp domain of Toc64 (Qbadou et al., 2006), this type of binding is independent of ATP, and therefore the targeting inhibition by Hsp90 is not likely to occur via this interaction. Moreover, a similar inhibition by Hsp90 is observed for Tic22 targeting (Figure 5), suggesting that Hsp90 inhibits the targeting process via its chaperone activity. Tic22 targeting is also promoted by Hsp70/40, but only weakly. We conclude that nascent proteins have a preference for either Hsp70/40 or Hsp90, and that the presence of the non-preferred chaperone inhibits targeting.
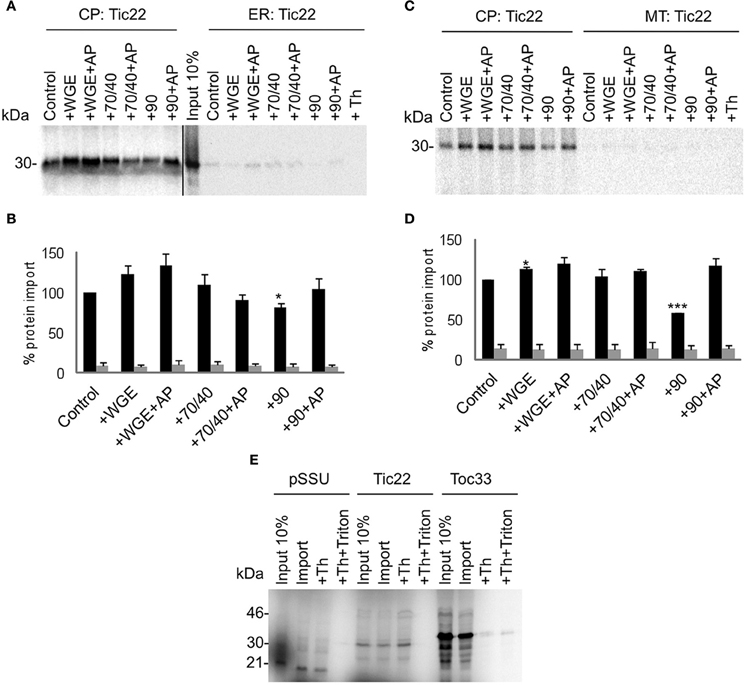
Figure 5. Tic22 targets to chloroplasts efficiently and selectively under a wide range of cytosolic conditions. (A) Radio-labeled Tic22 protein derived from RNCs was incubated in a competitive targeting assay with chloroplasts (CP) and ER, with the addition of cytosolic components as indicated; control is buffer only, WGE is wheat germ extract, chaperones (Hsp) are indicated by their numbers (70/40 or 90), and AP represents apyrase treatment. Membranes were refractionated, treated with thermolysin where indicated (Th), and washed with carbonate for analysis by SDS-PAGE. 10% of protein input is shown, and the protein molecular weight markers are indicated in kDa. (B) Quantification of protein import in part A. Mean and standard error are shown, normalized to the control import into chloroplasts (n = 3), and significant difference from the control import is indicated by one star (p = 0.05) or two stars (p = 0.01). (C) Competitive targeting assay for Tic22 import into chloroplasts (CP) and mitochondria (MT), conducted as described in part A. (D) Quantification of protein import in part C. Mean and standard error are shown, normalized to the control import into chloroplasts (n = 3), and significant difference from the control import is indicated by one star (p = 0.05) or three stars (p = 0.001). (E) Targeting assays for the small subunit of Rubisco (pSSU), Tic22, and Toc33 into chloroplasts (CP) were conducted as described in part A. Samples were treated after import with Thermolysin (Th) or thermolysin combined with Triton X-100 (Th + Triton) as indicated.
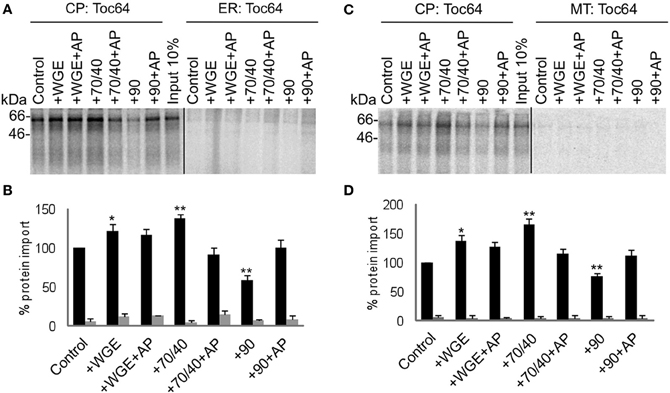
Figure 6. Toc64 targets to chloroplasts efficiently and selectively under a wide range of cytosolic conditions. (A) Radio-labeled Toc64 protein derived from RNCs was incubated in a competitive targeting assay with chloroplasts (CP) and ER, with the addition of cytosolic components as indicated; control is buffer only, WGE is wheat germ extract, chaperones (Hsp) are indicated by their numbers (70/40 or 90), and AP represents apyrase treatment. Membranes were refractionated and washed with carbonate prior to analysis by SDS-PAGE. 10% of protein input is shown, and the protein molecular weight markers are indicated in kDa. (B) Quantification of protein import in part A. Mean and standard error are shown, normalized to the control import into chloroplasts (n = 3), and significant difference from the control import is indicated by one star (p = 0.05) or two stars (p = 0.01). (C) Competitive targeting assay for Toc64 import into chloroplasts (CP) and mitochondria (MT), conducted as described in part A. (D) Quantification of protein import in part C. Mean and standard error are shown, normalized to the control import into chloroplasts (n = 3), and significant difference from the control import is indicated by one star (p = 0.05) or two stars (p = 0.01).
The localization of Tic22 to the intermembrane space allowed the nature of the apparent non-native targeting to be tested using protease protection analysis. Although the N-terminal targeting sequence of Tic22 has the potential to be cleaved in the intermembrane space, this is known to occur with low efficiency (Kouranov et al., 1999; Vojta et al., 2007), and we also observed minimal processing in our assays. Therefore, imported mature form is expected to be observed upon protease treatment. Application of thermolysin to ER or mitochondrial fractions after their separation caused complete degradation of protein (Figures 5A and C), confirming that the residual levels associated with non-target organelles do not represent translocated protein. Thermolysin treatment of the chloroplast fraction did not result in protein degradation, confirming that Tic22 is fully imported into the chloroplast (Figure 5E). Similarly, the chloroplast stromal protein pSSU is cleaved to its mature form and protected from protease, whilst Toc33 is largely exposed to protease. In all cases, the addition of Triton X-100 with protease results in the loss of all protein, which confirms the effectiveness of the protease and the role of membranes in providing protection.
Our data show that when given a choice including their native organelle, proteins are targeted accurately in the absence of any cytosolic factors. To test the limits of this specificity, we tested whether proteins could be targeted to a single non-native organelle (Figure 7). The chloroplast protein Toc33 does not target significantly to mitochondria (Figure 7A, pellet fraction), either without cytosol or in the presence of chaperones (import <1% of input). Likewise, the mitochondria TA protein At3g58 does not target significantly to chloroplasts (Figure 7B). Hence, proteins do not rely on cytosolic components to avoid mistargeting, indicating that key selection events occur at the organelle.
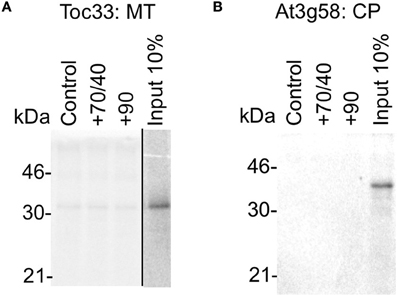
Figure 7. Protein mistargeting to single organelles does not occur in the absence of cytosolic components. (A) Radio-labeled Toc33 protein derived from RNCs was incubated in a targeting assay with mitochondria (MT), with the addition of cytosolic components as indicated; control is buffer only, and chaperones (Hsp) are indicated by their numbers (70/40 or 90). Membranes were refractionated and washed with carbonate for analysis by SDS-PAGE. 10% of protein input is shown, and the protein molecular weight markers are indicated in kDa. (B) Radio-labeled At3g58 protein derived from RNCs was incubated in a targeting assay with chloroplasts (CP), and otherwise processed as described for part A.
Discussion
We have shown that a selection of chloroplast membrane proteins can target specifically and with high efficiency in the absence of cytosolic factors, although efficiency can typically be boosted by ~20% with cytosolic lysate, and by ~50% with chaperones alone. We therefore conclude that cytosolic factors do not generally play a major role in determining the selectivity of targeting, which must be determined directly at the organelle.
The efficiency boost provided by cytosolic lysate was mostly independent of apyrase treatment, which could be due to the involvement of non-NTP dependent targeting factors such as AKR2A (Bae et al., 2008; Dhanoa et al., 2010). Alternatively, the two ATP-dependent chaperones Hsp70 and Hsp90 may counter each other, so that when apyrase is added the loss of activity of one is balanced by the loss of inhibition by the other; this notion is supported by the opposite effects that Hsp70 and Hsp90 had on each protein. Targeting activity in the lysate may also be masked by the general molecular crowding of the wide variety of other proteins present, which may interact non-specifically with the nascent protein or cause aggregation that inhibits targeting. Although lysate does not fully replicate the cytosolic environment, it contains all of the potential factors required for chloroplasts, and its minimal effect on targeting efficiency supports the overall conclusion that there is no absolute requirement for targeting factors.
The different responses of each protein to Hsp70 and Hsp90 are consistent with numerous studies showing that targeting factors tend to promote the targeting of a subset of proteins (e.g., Fellerer et al., 2011) (and reviewed by Jarvis, 2008; Schwenkert et al., 2011). For example, some proteins bind Hsp90, whilst others bind Hsp70 alone, or the guidance complex of Hsp70 and 14-3-3 protein. Most proteins appear to rely on Toc34 at a late stage of recognition, but Hsp90-bound proteins are first recognized by Toc64 (Qbadou et al., 2006), and many Hsp70-bound proteins can be recognized by OEP61 (Von Loeffelholz et al., 2011). However, there is no clear correlation between the features or the destination of the protein and its chaperone dependence (Kriechbaumer et al., 2012). If targeting sequences are modified to reduce Hsp70 or 14-3-3 protein binding no impact can be seen on cell free targeting (Rial et al., 2000; Nakrieko et al., 2004), and Toc64 knockout does not result in an observable phenotype (Aronsson et al., 2007). These observations may reflect either redundancy between targeting pathways, or the importance of direct recognition at the organelle. Another targeting factor, AKR2A, is involved in the targeting of chloroplast TA proteins and other outer envelope proteins such as OEP7 (Bae et al., 2008; Dhanoa et al., 2010), although its role in the targeting of other types of chloroplast protein has not been tested. Future studies could use purified AKR2A in this assay system to systematically test its role in targeting. Despite this complexity and redundancy in chloroplast targeting pathways, our findings suggest that the control of localization is exerted at a later stage.
Although our study finds cytosolic factors to play a minor role in protein targeting to chloroplasts, the nascent protein is being released into an uncrowded environment and does not have the opportunity to aggregate with other cytosolic proteins. In a typical cellular environment crowded by other proteins, targeting factors may be required to protect the targeting sequence, reduce premature folding, and prevent aggregation (Ellis and Minton, 2006), and would therefore assume a greater importance for efficiency. The role of molecular crowding in the requirement for targeting factors could potentially be tested by simulating cytosolic conditions using reagents such as PEG and Ficoll (Ellis and Minton, 2006).
Ultimately the nascent protein must be delivered to a translocase or membrane lipids in a conformation that is suitable for insertion or translocation at the membrane. This final recognition step may be the key event for generating specificity of targeting, a notion that is supported by the ability of Toc75 to directly recognize chloroplast targeting sequences (Hinnah et al., 2002). Similarly, ER targeting sequences can be recognized directly at the ER membrane (Belin et al., 1996), suggesting that our conclusions may apply more generally to other cellular organelles. The only conceptually different targeting pathway is cotranslational SRP-mediated targeting to ER membranes, which occurs by a sequence of tightly regulated steps from the ribosome to the membrane (Grudnik et al., 2009). However, SRP knock-outs in yeast are not lethal, and proteins are able to use an Hsp70-mediated posttranslational pathway (Rothe and Lehle, 1998). Therefore, it appears that protein targeting generally requires cytosolic components to promote delivery to an organelle, but that acceptance at the organelle depends primarily on membrane components.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank ABRC for supplying plasmids. This work was supported by a BBSRC Research Grant awarded to Ben M. Abell [BB/E01559X/1].
References
Abell, B. M., Rabu, C., Leznicki, P., Young, J. C., and High, S. (2007). Post-translational integration of tail-anchored proteins is facilitated by defined molecular chaperones. J. Cell Sci. 120, 1743–1751.
Aronsson, H., Boij, P., Patel, R., Wardle, A., Topel, M., and Jarvis, P. (2007). Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J. 52, 53–68.
Artigues, A., Iriarte, A., and Martinez-Carrion, M. (2002). Binding to chaperones allows import of a purified mitochondrial precursor into mitochondria. J. Biol. Chem. 277, 25047–25055.
Bae, W., Lee, Y. J., Kim, D. H., Lee, J., Kim, S., Sohn, E. J., and Hwang, I. (2008). AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat. Cell. Biol. 10, 220–227.
Belin, D., Bost, S., Vassalli, J. D., and Strub, K. (1996). A two-step recognition of signal sequences determines the translocation efficiency of proteins. EMBO J. 15, 468–478.
Berndt, U., Oellerer, S., Zhang, Y., Johnson, A. E., and Rospert, S. (2009). A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc. Natl. Acad. Sci. U.S.A. 106, 1398–1403.
Blobel, G., and Sabatini, D. (1971). Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl. Acad. Sci. U.S.A. 68, 390–394.
Borgese, N., and Fasana, E. (2010). Targeting pathways of C-tail-anchored proteins. Biochim. Biophys. Acta 1808, 937–946.
Borgese, N., Gazzoni, I., Barberi, M., Colombo, S., and Pedrazzini, E. (2001). Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol. Biol. Cell 12, 2482–2496.
Brambillasca, S., Yabal, M., Soffientini, P., Stefanovic, S., Makarow, M., Hegde, R. S., and Borgese, N. (2005). Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. EMBO J. 24, 2533–2542.
Brix, J., Ziegler, G. A., Dietmeier, K., Schneider-Mergener, J., Schulz, G. E., and Pfanner, N. (2000). The mitochondrial import receptor Tom70, identification of a 25 kDa core domain with a specific binding site for preproteins. J. Mol. Biol. 303, 479–488.
Brkljacic, J., Zhao, Q., and Meier, I. (2009). WPP-domain proteins mimic the activity of the HSC70-1 chaperone in preventing mistargeting of RanGAP1-anchoring protein WIT1. Plant Physiol. 151, 142–154.
Delille, H. K., and Schrader, M. (2008). Targeting of hFis1 to peroxisomes is mediated by Pex19p. J. Biol. Chem. 283, 31107–31115.
Dhanoa, P. K., Richardson, L. G., Smith, M. D., Gidda, S. K., Henderson, M. P., Andrews, D. W., and Mullen, R. T. (2010). Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope. PLoS ONE 5:e10098. doi: 10.1371/journal.pone.0010098
Ellis, R. J., and Minton, A. P. (2006). Protein aggregation in crowded environments. Biol. Chem. 387, 485–497.
Fellerer, C., Schweiger, R., Schongruber, K., Soll, J., and Schwenkert, S. (2011). Cytosolic HSP90 cochaperones HOP and FKBP interact with freshly synthesized chloroplast preproteins of Arabidopsis. Mol. Plant 4, 1133–1145.
Grudnik, P., Bange, G., and Sinning, I. (2009). Protein targeting by the signal recognition particle. Biol. Chem. 390, 775–782.
Hinnah, S. C., Wagner, R., Sveshnikova, N., Harrer, R., and Soll, J. (2002). The chloroplast protein import channel Toc75, pore properties and interaction with transit peptides. Biophys. J. 83, 899–911.
Hofmann, N. R., and Theg, S. M. (2005). Chloroplast outer membrane protein targeting and insertion. Trends Plant Sci. 10, 450–457.
Jarvis, P. (2008). Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 179, 257–285.
Kemper, C., Habib, S. J., Engl, G., Heckmeyer, P., Dimmer, K. S., and Rapaport, D. (2008). Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J. Cell Sci. 121, 1990–1998.
Kouranov, A., Wang, H., and Schnell, D. J. (1999). Tic22 is targeted to the intermembrane space of chloroplasts by a novel pathway. J. Biol. Chem. 274, 25181–25186.
Kriechbaumer, V., Shaw, R., Mukherjee, J., Bowsher, C. G., Harrison, A. M., and Abell, B. M. (2009). Subcellular distribution of tail-anchored proteins in Arabidopsis. Traffic 10, 1753–1764.
Kriechbaumer, V., Von Loffelholz, O., and Abell, B. M. (2012). Chaperone receptors: guiding proteins to intracellular compartments. Protoplasma 249, 21–30.
Li, J., Qian, X., Hu, J., and Sha, B. (2009). Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J. Biol. Chem. 284, 23852–23859.
May, T., and Soll, J. (2000). 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell 12, 53–64.
Motz, C., Martin, H., Krimmer, T., and Rassow, J. (2002). Bcl-2 and porin follow different pathways of TOM-dependent insertion into the mitochondrial outer membrane. J. Mol. Biol. 323, 729–738.
Nakrieko, K. A., Mould, R. M., and Smith, A. G. (2004). Fidelity of targeting to chloroplasts is not affected by removal of the phosphorylation site from the transit peptide. Eur. J. Biochem. 271, 509–516.
Ngosuwan, J., Wang, N. M., Fung, K. L., and Chirico, W. J. (2003). Roles of cytosolic Hsp70 and Hsp40 molecular chaperones in post-translational translocation of presecretory proteins into the endoplasmic reticulum. J. Biol. Chem. 278, 7034–7042.
Otera, H., Taira, Y., Horie, C., Suzuki, Y., Suzuki, H., Setoguchi, K., Kato, H., Oka, T., and Mihara, K. (2007). A novel insertion pathway of mitochondrial outer membrane proteins with multiple transmembrane segments. J. Cell Biol. 179, 1355–1363.
Qbadou, S., Becker, T., Mirus, O., Tews, I., Soll, J., and Schleiff, E. (2006). The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64. EMBO J. 25, 1836–1847.
Rial, D. V., Arakaki, A. K., and Ceccarelli, E. A. (2000). Interaction of the targeting sequence of chloroplast precursors with Hsp70 molecular chaperones. Eur. J. Biochem. 267, 6239–6248.
Rothe, C., and Lehle, L. (1998). Sorting of invertase signal peptide mutants in yeast dependent and independent on the signal-recognition particle. Eur. J. Biochem. 252, 16–24.
Rudhe, C., Chew, O., Whelan, J., and Glaser, E. (2002). A novel in vitro system for simultaneous import of precursor proteins into mitochondria and chloroplasts. Plant J. 30, 213–220.
Schleiff, E., Shore, G. C., and Goping, I. S. (1997a). Human mitochondrial import receptor, Tom20p. Use of glutathione to reveal specific interactions between Tom20-glutathione S-transferase and mitochondrial precursor proteins. FEBS Lett. 404, 314–318.
Schleiff, E., Shore, G. C., and Goping, I. S. (1997b). Interactions of the human mitochondrial protein import receptor, hTom20, with precursor proteins in vitro reveal pleiotropic specificities and different receptor domain requirements. J. Biol. Chem. 272, 17784–17789.
Schuldiner, M., Metz, J., Schmid, V., Denic, V., Rakwalska, M., Schmitt, H. D., Schwappach, B., and Weissman, J. S. (2008). The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134, 634–645.
Schwenkert, S., Soll, J., and Bolter, B. (2011). Protein import into chloroplasts–how chaperones fea-ture into the game. Biochim. Biophys. Acta 1808, 901–911.
Setoguchi, K., Otera, H., and Mihara, K. (2006). Cytosolic factor- and TOM-independent import of C-tail-anchored mitochondrial outer membrane proteins. EMBO J. 25, 5635–5647.
Stojanovski, D., Guiard, B., Kozjak-Pavlovic, V., Pfanner, N., and Meisinger, C. (2007). Alternative function for the mitochondrial SAM complex in biogenesis of alpha-helical TOM proteins. J. Cell Biol. 179, 881–893.
Urban, A., Neukirchen, S., and Jaeger, K. E. (1997). A rapid and efficient method for site-directed mutagenesis using one-step overlap extension PCR. Nucleic Acids Res. 25, 2227–2228.
Vojta, L., Soll, J., and Bolter, B. (2007). Protein transport in chloroplasts—targeting to the intermembrane space. FEBS J. 274, 5043–5054.
Von Loeffelholz, O., Kriechbaumer, V., Ewan, R. A., Jonczyk, R., Lehmann, S., Young, J. C., and Abell, B. M. (2011). OEP61 is a chaperone receptor at the plastid outer envelope. Biochem. J. 438, 143–153.
Wickner, W., and Schekman, R. (2005). Protein translocation across biological membranes. Science 310, 1452–1456.
Keywords: protein localization, tail-anchored protein, targeting factors, molecular chaperones, competitive targeting
Citation: Kriechbaumer V and Abell BM (2012) Chloroplast envelope protein targeting fidelity is independent of cytosolic components in dual organelle assays. Front. Plant Sci. 3:148. doi: 10.3389/fpls.2012.00148
Received: 04 April 2012; Accepted: 14 June 2012;
Published online: 28 June 2012.
Edited by:
Robert Mullen, University of Guelph, CanadaReviewed by:
Inhwan Hwang, Pohang University of Science and Technology, South KoreaSerena Schwenkert, LMU Munich, Germany
Copyright: © 2012 Kriechbaumer and Abell. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Ben M. Abell, Biomedical Research Centre, Sheffield Hallam University, Howard Street, Sheffield S1 1WB, UK. e-mail: b.abell@shu.ac.uk
