- 1 Biology Department, University of Massachusetts, Amherst, MA, USA
- 2 Plant Biology Graduate Program, University of Massachusetts, Amherst, MA, USA
Secondary cell wall synthesis occurs in specialized cell types following completion of cell enlargement. By virtue of mechanical strength provided by a wall thickened with cellulose, hemicelluloses, and lignin, these cells can function as water-conducting vessels and provide structural support. Several transcription factor families regulate genes encoding wall synthesis enzymes. Certain NAC and MYB proteins directly bind to the SNBE and AC elements upstream of structural genes and other transcription factors. The most detailed model of this regulatory network is established predominantly for a eudicot, Arabidopsis thaliana. In grasses, both the patterning and the composition of secondary cell walls are distinct from that of eudicots. These differences suggest transcriptional regulation is similarly distinct. Putative rice and maize orthologs of several eudicot cell wall regulators genetically complement mutants of A. thaliana or result in wall defects when constitutively overexpressed; nevertheless, aside from a maize, ZmMYB31, and a switchgrass protein, PvMYB4, function has not been tested in a grass. Similar to the seminal work conducted in A. thaliana, gene expression profiling in maize, rice, and other grasses implicates additional genes as regulators. Characterization of these genes will continue to elucidate the relationship between the transcription regulatory networks of eudicots and grasses.
Introduction
Plant cell walls are diverse in their polymer composition, which vary among plant species as well as cell types within a species. The primary cell wall, consisting mostly of cellulose, hemicelluloses, pectin, and proteins demarcates each plant cell. Secondary cell walls, composed mostly of cellulose, hemicelluloses, and lignin, are deposited between the plasma membrane and primary wall in specialized cell types following cessation of cell enlargement. Even though all plant cell walls contain a cell wall matrix made up of cellulose microfibrils, hemicelluloses, and other cell wall polymers, there are significant differences in polymer types and their relative abundance between dictos and monocots (Vogel, 2008). For instance, relatively high amounts of xyloglucan, pectins, and structural proteins are typically present in the primary walls of eudicots, non-commelinoid monocots, and gymnosperms. The commelinoid monocots like grasses contain glucuronoarabinoxylans, small quantities of pectin and structural proteins and high levels of hydroxycinnamates (Vogel, 2008). Apart from wall composition, eudicots such as Arabidopsis thaliana and monocots such as rice (Oryza sativa L.) exhibit distinct morphological characteristics in addition to their namesake two and single cotyledons. Pinnate or palmate venation is characteristic of eudicots while monocots posses parallel venation. Other morphological and anatomical distinctions exist in their vasculature, tissues highly enriched in secondary cell walls. Stem radial thickening in eudicots is derived from a specialized cell layer called the cambium that differentiates into the phloem and xylem. On the other hand, monocots lack a specialized cambium layer and therefore do not undergo secondary growth (Figure 1). The vascular bundles of grasses are often well defined by a single layer of bundle sheath cells surrounding the xylem and phloem and the organization of the bundles is distinct from eudicots. In A. thaliana, the vascular bundles are arranged as a ring along the periphery of the stem in a pattern known as eustelic, whereas monocot bundles posses an atactostele arrangement characterized by several circles around the periphery of a stem, as in rice and Brachypodium distachyon (Figure 1), or scattered throughout the stem as in maize (Zea mays L.; Kiesselbach, 1949).
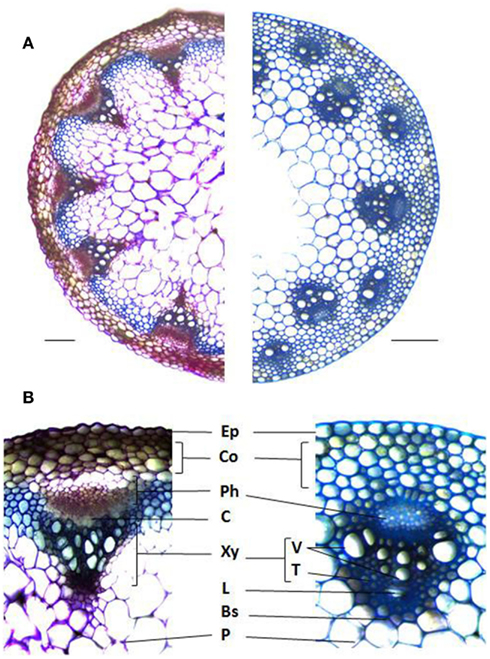
Figure 1. Stem cross sections illustrating the different cell types and arrangements between dicots and monocots. (A) Arabidopsis thaliana (left) and Brachypodium distachyon (right) stained with Toluidine blue. (B) Vascular bundle anatomy of A. thaliana (left) and B. distachyon (right). Ep, Epidermis; Co, Cortex; Ph, Phloem; C, Cambium; Xy, Xylem; V, Vessels; T, Tracheads; L, Lacuna; Bs, Bundle Sheath; P, Pith. Bars = 0.1 mm.
With distinctions and similarities abound, it is decidedly unclear how the transcriptional regulation of secondary cell wall biosynthesis in eudicots and grasses relate. Plant cell wall biosynthesis is regulated at different molecular and cellular levels. Current evidence supports a complex regulatory network consisting of a handful of proteins from only a small portion of over 65 different transcription factor families (Figure 2A). Phylogenetic analysis has identified close homologs of A. thaliana regulators from both vascular and non-vascular plants and some of those regulators were capable of complementing A. thaliana cell wall mutants (Zhong et al., 2011). These findings suggest the evolutionary conservation of the transcriptional regulators in secondary cell wall biosynthesis. On the other hand, due to the pronounced differences between eudicot and monocot secondary wall composition and anatomy, there are likely unique aspects of the regulatory network yet to be resolved.
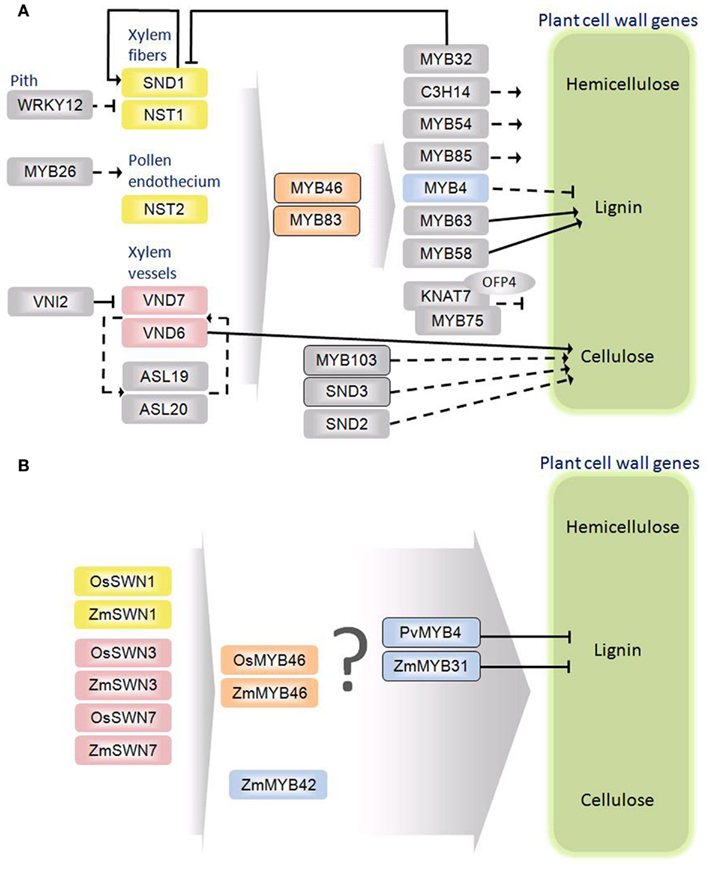
Figure 2. Schematic diagrams of the secondary cell wall regulatory networks in Arabidopsis thaliana (A) and monocots (B). Rectangles represent transcription factors. The oval indicates an interacting protein. Solid arrows and bordered rectangles signify evidence for direct interactions. Dashed arrows indicate no evidence for direct interaction. Orthology between A. thaliana and grasses is denoted by color.
Transcriptional Regulation in A. thaliana
Transcriptional regulation is one of the most important processes controlling plant cell wall biosynthesis, mediated by the interaction and interplay of cis-regulatory DNA elements and the trans-acting transcription factor proteins. Recent evidence suggests the involvement of an AP2 family protein SHINE/WAX INDUCER 1 (SHN) as a global level regulator of cell wall biosynthesis (Ambavaram et al., 2011). Constitutive overexpression of A. thaliana SHN in rice results in the activation of cellulose and other cell wall-associated genes and the repression of lignin pathway genes. This protein is also capable of activating key NAC and MYB regulators and electrophoretic mobility shift assays (EMSA) demonstrate the direct binding of SHN protein to promoters of rice cell wall-associated transcription factors (Ambavaram et al., 2011). A WRKY family transcription factor, WRKY12, acts as a global repressor of secondary wall biosynthesis (Wang et al., 2010). Transcripts of this gene are abundant in the cortex and pith cells of A. thaliana that lack secondary walls. Loss-of-function wrky12 mutants exhibit increased expression of transcription factors associated with secondary wall biosynthesis as well as ectopic depositions of lignin, cellulose, and xylan and consequently, and overall increase in plant biomass (Wang et al., 2010). In the presence of WRKY12 protein, pith stem cells are maintained parenchymatous and the deposition of secondary cell walls is repressed. MYB32 similarly acts as a repressor of secondary wall biosynthesis, but in cells where this pathway has been activated; thus, it may provide negative feedback (Preston et al., 2004). Transgenic overexpression of MYB32 resulted in the repression of SECONDARY WALL-ASSOCIATED NAC-DOMAIN PROTEIN 1 (SND1), a higher order activator of secondary wall biosynthesis (Wang et al., 2011). Interestingly, the SND1 protein directly binds the MYB32 promoter to activate gene expression (Wang et al., 2011). A group of NAC-domain transcription factors, NAC SECONDARY WALL THICKENING FACTOR 1 (NST1), NST2, SND1 (also known as NST3), VASCULAR-RELATED NAC-DOMAIN 6 (VND6), and VND7, collectively known as the secondary wall NACs (SWNs) are implicated as positively acting master regulators in a variety of tissues (Demura and Fukuda, 2007; Zhong and Ye, 2007). Among the NACs, SND1 functions as a key switch governing the regulation of all secondary wall polymers (Zhong et al., 2006; Mitsuda et al., 2007). Overexpression of SND1 leads to ectopic deposition and activation of cellulose, hemicellulose, and lignin biosynthesis genes. Conversely, dominant repression of SND1 results in the absence of secondary wall development in vascular and interfascicular fibers (Zhong et al., 2006; Mitsuda et al., 2007). It also directly activates itself and is repressed by MYB transcription factors under the direct influence of MYB46 (Wang et al., 2011). The SND1 protein directly binds the cis-regulatory regions of MYB46, MYB83, and C3H14 genes to activate their expression (Zhong et al., 2008; Ko et al., 2009; McCarthy et al., 2009). It also acts as a direct regulator of F5H, a gene encoding a key enzyme involved in lignin biosynthesis (Zhao et al., 2010). Another direct target of SND1 is the KNOX type Homeodomain transcription factor, KNAT7, which interacts with OVATE FAMILY PROTEIN 4 to repress secondary wall biosynthesis (Li et al., 2011). Loss-of-function mutants of knat7 display an increase in cell wall gene expression, wall thickening in interfascicular fiber cells, and an increase in lignin content (Li et al., 2011). Interestingly, a weak activator of cell wall gene expression, MYB75, was found to physically interact with KNAT7 protein to repress cell wall biosynthesis (Bhargava et al., 2010). These results imply that wall regulators can play multiple roles by interacting with different trans-acting factors in different cell types to provide more flexibility and complexity to the regulatory network.
The SND1 homologs NST1 and NST2 play a crucial role in the A. thaliana anther endothecium (Mitsuda et al., 2005). In this tissue, these proteins are activated by MYB26 which is essential for anther dehiscence and proper pollen release (Yang et al., 2007). Other SWNs, in particular VND6 and VND7, regulate metaxylem and protoxylem development and are repressed by the VND-INTERACTING2 NAC protein (Kubo et al., 2005; Yamaguchi et al., 2010). The VND proteins VND6 and VND7 driven by SND1 promoter were capable of complementing the snd1/nst1 mutant phenotype implying their conserved functionality (Zhong et al., 2007a). They are positively regulated by ASYMMETRIC LEAVES2-LIKE19 (ASL19) and ASL20 (Soyano et al., 2008). Transgenic overexpression of ASL19 and ASL20 induces trans-differentiation of non-vascular tissues into treachery elements and an increased cell wall thickening in mutant lines. They are also able to partially recover the dominant negative effect of the VND6 and VND7 repressor lines (Soyano et al., 2008). The SWNs, SND1, NST1, NST2, VND6, and VND7 activate a cascade of downstream transcription factors such as SND2, SND3, MYB103, MYB85, MYB54, MYB46, MYB69, MYB63, MYB83, and KNAT7 (Zhong et al., 2008; Ko et al., 2009; McCarthy et al., 2009). Some of the downstream regulators, SND2, SND3, and MYB103 exclusively activate cellulose biosynthesis where as the others such as MYB63, and MYB58 regulate lignin biosynthesis (Zhong et al., 2008). Even though direct protein–DNA interactions have been shown for some of the cellulose and lignin specific regulators, further characterization of many downstream regulators is needed.
Discovery of the trans-acting transcription factors of cell wall biosynthesis facilitated the opportunity to identify common cis-elements shared among the master regulators. The tracheary-element-regulating cis-element (TERE) is one such 11-bp motif, CTT/(C)NAAA/(C)GCNA(T), involved in tissue specific cell wall biosynthesis and programmed cell death. First identified in the Zinnia cysteine protease 4 promoter and is present in numerous cell death and xylem differentiation genes such as Cysteine protease 1 (XCP1), XCP2, Serine protease 1, and several other genes associated with wall function that include xylanases and acetyltransferases (Pyo et al., 2007). More recent studies demonstrated the physical interaction between VND6 protein and the TERE (Ohashi-Ito et al., 2010). Other SWNs bind a 19-bp imperfect palindromic sequence (T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T) referred to as the secondary wall NAC binding element (SNBE; Zhong et al., 2010). A synthetic promoter harboring six copies of the SNBE fused to a GUS reporter revealed specific expression in xylem and interfascicular fibers, phenocopying native MYB46 promoter behavior (Zhong et al., 2007b). Other direct targets of SND1, including MYB83, MYB103, SND3, and KNAT7 also posses the SNBE element (Zhong et al., 2010). A similar cis-element, TACNTTNNNNATGA, was identified recently in the SND1 promoter and is the target of binding that serves as a target of positive feedback from SND1 itself (Wang et al., 2011).
A series of MYB transcription factor family proteins are also implicated in the cell wall regulatory network, a majority of which act downstream of the SWNs. One such protein, MYB46, acts subsequent the SWNs (Figure 2A). Lignin biosynthesis is specifically regulated by MYB63 and MYB58 interacting with the AC/Pal-box promoter sequences. This motif was first identified in the promoter of parsley PHENYLALANINE AMMONIA-LYASE 1 and subsequently identified as three AC rich elements AC-I (ACCTACC), AC-II (ACCAACC), AC-III (ACCTAAC) involved in lignin gene regulation (Lois et al., 1989; Hatton et al., 1995; Raes et al., 2003). Binding of MYB proteins to the AC elements trans-activates the respective promoters thus, activating the genes in a xylem specific manner repressing the expression of the same genes in phloem or the cortical cells (Hatton et al., 1995). The consensus sequence of the AC element was recently expanded to include four more forms interchanging a T with the C at the last position; thus, ACC(T/A)A(A/C)(C/T) (Zhong and Ye, 2012). This 7-bp sequence, the secondary wall MYB responsive element (SMRE), is bound by both MYB46 and MYB83 proteins and is sufficient for the activation of a suite of transcription factor and cell wall biosynthetic genes (Zhong and Ye, 2012). The MYB46-responsive cis-element (M46RE) is an 8-bp sequence (A/G)(G/T)T(A/T)GGT(A/G) found in the C3H14 promoter, which is a direct target of MYB46 (Kim et al., 2012). Trans-activation assays coupled with EMSA reveled M46RE is required and sufficient for the activation of C3H14. The 8-bp core sequence was present in nearly 43% of the genes in the A. thaliana genome but was enriched in the downstream genes activated by MYB46 along with secondary cell wall related structural genes (Kim et al., 2012). Apart from the key SWNs and MYBs, a handfull of downstream MYBs, NACs, and transcription factors from other families are involved in this complex cell wall regulatory network. A closely related homolog of MYB32, MYB4, functions as a repressor of CINNAMATE-4-HYDROXYLASE (C4H; Jin et al., 2000). Loss-of-function mutants of MYB4 exhibited elevated levels of sinapoyl malate, a component in the lignin pathway, and an increase in C4H expression. Collectively, these findings suggest a complex and hierarchical transcription regulatory network for eudicot cell wall biosynthesis (Figure 2A). While this review primarily discusses discoveries in A. thaliana, it should be noted that a number of regulators have been characterized in other eudicot species such as Populus trichocarpa, Eucalyptus gunnii, Nicotiana tabacum, Antirrhinum majus, Pinus taeda, Vitis vinifera, and Medicago truncatula.
Grass Cell Wall Regulators
From the time of eudicot and monocot divergence 140–150 million year ago, the transcription factor families have disproportionately expanded and the regulatory networks have likely diverged; thus, the existing eudicot network model for transcription regulation is not wholly generalizable to monocots (Chaw et al., 2004; Shiu et al., 2005). Conversely, recent functional characterization of grass transcription factors implies great commonality in how a similar network could regulate grass cell wall biosynthesis. While many cell wall genes have been characterized in grasses, almost nothing is known about the regulation of walls in monocots. The A. thaliana model, which is by far the best developed, is admittedly nascent. This stands in stark contrast to the model for grasses that consists of only a few genes (Figure 2B). Maize CAFFEIC ACID-O-METHYL TRANSFERASE (COMT) is a key lignin pathway gene with an AC-III element recognized by R2R3-MYB transcription factors (Vignols et al., 1995; Fornalé et al., 2010). Group four R2R3-MYB transcription factors are described as repressors and based on sequence homology to the know A. thaliana MYB repressors, five maize transcription factors, ZmMYB31, ZmMYB42, ZmMYB2, ZmMYB8, and ZmMYB39 were identified as candidates for direct repression of ZmCOMT (Fornalé et al., 2006). When overexpressed in A. thaliana, ZmMYB31 and ZmMYB42 resulted in down regulation of lignin associated genes and subsequently reduced lignin content (Fornalé et al., 2010). Overexpression of ZmMYB42 caused a reduction in leaf size, an adaxial curvature indicative of less tertiary vein formation, reduction of the syringyl lignin monomers, and dwarfism in A. thaliana (Sonbol et al., 2009). The absence of results in a monocot is likely due to the relative recalcitrance of crop species to genetic study; thus, A. thaliana serves as an imperfect heterologous system to study grass gene function. The maize gene, ZmMYB31 is the first wall specific regulator characterized in a grass (Fornalé et al., 2010). Chromatin immunoprecipitation demonstrated the direct interaction between ZmMYB31 and ZmCOMT promoter and an AC element similar to AC-II was identified as the binding motif. The switchgrass (Panicum virgatum L.) protein PvMYB4, an ortholog to A. thaliana MYB4, is another recently characterized repressor of lignin (Shen et al., 2012). Ectopic expression of PvMYB4 in switchgrass resulted in a reduction in total lignin and altered lignin monomer ratio (Shen et al., 2012). The AC element is also implicated as the binding site of PvMYB4, which results in repression of lignin pathway genes (Shen et al., 2012). In addition, rice and maize orthologs of A. thaliana SWNs and MYB46 were shown to activate secondary wall biosynthesis when overexpressed in A. thaliana (Zhong and Ye, 2012). Moreover, OsSWNs and ZmSWNs were able to complement and partially rescue the pendant stem phenotype of the A. thaliana snd1/nst1 double mutant. Similarly, OsMYB46 and ZmMYB46 under the control of AtMYB46 promoter were able to complement the loss of helical secondary wall thickening in vessels of A. thaliana myb46/myb83 double mutant. In addition, SNBEs were identified in OsMYB46 and ZmMYB46 promoters and were bound and activated by the rice and maize SWNs in A. thaliana transient protoplast assays (Zhong and Ye, 2012). While studies in a heterologous system have proven informative, further functional characterization in a grass species is necessary. Contrary to expectations of distinct aspects of monocot cell wall regulation, the existing model is populated exclusively by homologs of known A. thaliana genes.
Gene expression profiling was critical in identifying many of the candidates that formed the foundation of the existing A. thaliana cell wall regulatory network (Oh et al., 2003; Ehlting et al., 2005; Kubo et al., 2005; Zhao et al., 2005). Similar tools can be applied to grasses to identify candidates for functional characterization, especially those specific to monocots. Accordingly, a comparison of the transcriptome of maize elongating and non-elongating internodes revealed several transcription factors that are feasibly involved in cell wall regulation (Bosch et al., 2011). Likewise, the expression of three barley (Hordeum vulgare L.) NACs, HvNAC033, HvNAC034, and HvNAC039, were significantly greater in stem tissue where extensive secondary cell wall biosynthesis occurs. One of these proteins, HvNAC033, is the closest barley homolog to A. thaliana NST1, further supporting the possible role of this protein in grass cell wall regulation (Christiansen et al., 2011). Comparison of expression networks across species similarly reinforces the notion of shared features across eudicots and monocots and revealed potential distinctions. Genes with expression patterns similar to secondary wall CELLULOSE SYNTHASE A genes in rice included those most similar to A. thaliana MYB63, MYB103, NST1, SND2, and KNAT7 (Ruprecht et al., 2011). Conversely, several rice transcription factors were co-expressed with structural genes that do not share sequence homology to co-expressed or characterized A. thaliana genes. These include MYB and NAC as well as bZip and AP2 family genes (Ruprecht et al., 2011). As with ZmMYB42 and PvMYB4, candidates identified using sequence similarity and expression profiling require further characterization in the native systems in order to solidify both overlap and divergence between eudicot and grass cell wall regulatory networks.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Office of Science (BER) Department of Energy Grant DE-FG02-08ER64700DE. We wish to thank Dominick Matos for assistance with figure preparation.
References
Ambavaram, M. M. R., Krishnan, A., Trijatmiko, K. R., and Pereira, A. (2011). Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol. 155, 916–931.
Bhargava, A., Mansfield, S. D., Hall, H. C., Douglas, C. J., and Ellis, B. E. (2010). MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol. 154, 1428–1438.
Bosch, M., Mayer, C.-D., Cookson, A., and Donnison, I. S. (2011). Identification of genes involved in cell wall biogenesis in grasses by differential gene expression profiling of elongating and non-elongating maize internodes. J. Exp. Bot. 62, 3545–3561.
Chaw, S.-M., Chang, C.-C., Chen, H.-L., and Li, W.-H. (2004). Dating the monocot–dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol. 58, 424–441.
Christiansen, M., Holm, P., and Gregersen, P. (2011). Characterization of barley (Hordeum vulgare L.) NAC transcription factors suggests conserved functions compared to both monocots and dicots. BMC Res. Notes 4, 302. doi:10.1186/1756-0500-4-302
Demura, T., and Fukuda, H. (2007). Transcriptional regulation in wood formation. Trends Plant Sci. 12, 64–70.
Ehlting, J., Mattheus, N., Aeschliman, D. S., Li, E., Hamberger, B., Cullis, I. F., Zhuang, J., Kaneda, M., Mansfield, S. D., Samuels, L., Ritland, K., Ellis, B. E., Bohlmann, J., and Douglas, C. J. (2005). Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 42, 618–640.
Fornalé, S., Shi, X., Chai, C., Encina, A., Irar, S., Capellades, M., Fuguet, E., Torres, J.-L., Rovira, P., Puigdomènech, P., Rigau, J., Grotewold, E., Gray, J., and Caparrós-Ruiz, D. (2010). ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 64, 633–644.
Fornalé, S., Sonbol, F.-M., Maes, T., Capellades, M., Puigdomènech, P., Rigau, J., and Caparrós-Ruiz, D. (2006). Down-regulation of the maize and Arabidopsis thaliana caffeic acid O-methyl-transferase genes by two new maize R2R3-MYB transcription factors. Plant Mol. Biol. 62, 809–823.
Hatton, D., Sablowski, R., Yung, M.-H., Smith, C., Schuch, W., and Bevan, M. (1995). Two classes of cis sequences contribute to tissue-specific expression of a PAL2 promoter in transgenic tobacco. Plant J. 7, 859–876.
Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., Tonelli, C., Weisshaar, B., and Martin, C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161.
Kiesselbach, T. A. (1949). The Structure and Reporduction of Corn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
Kim, W.-C., Ko, J.-H., and Han, K.-H. (2012). Identification of a cis-acting regulatory motif recognized by MYB46, a master transcriptional regulator of secondary wall biosynthesis. Plant Mol. Biol. 78, 489–501.
Ko, J.-H., Kim, W.-C., and Han, K.-H. (2009). Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. 60, 649–665.
Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., Mimura, T., Fukuda, H., and Demura, T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860.
Li, E., Wang, S., Liu, Y., Chen, J.-G., and Douglas, C. J. (2011). Ovate family protein4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J. 67, 328–341.
Lois, R., Dietrich, A., and Schulz, W. (1989). A phenylalanine ammonia-lyase gene from parsley: structure, regulation and identification of elicitor and light responsive cis-acting elements. EMBO J. 8, 1641–1648.
McCarthy, R. L., Zhong, R., and Ye, Z.-H. (2009). MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50, 1950–1964.
Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19, 270–280.
Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006.
Oh, S., Park, S., and Han, K.-H. (2003). Transcriptional regulation of secondary growth in Arabidopsis thaliana. J. Exp. Bot. 54, 2709–2722.
Ohashi-Ito, K., Oda, Y., and Fukuda, H. (2010). Arabidopsis vascular-related NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461–3473.
Preston, J., Wheeler, J., Heazlewood, J., Li, S. F., and Parish, R. W. (2004). AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 40, 979–995.
Pyo, H., Demura, T., and Fukuda, H. (2007). TERE; a novel cis-element responsible for a coordinated expression of genes related to programmed cell death and secondary wall formation during differentiation of tracheary elements. Plant J. 51, 955–965.
Raes, J., Rohde, A., Christensen, J., Peer, Y., and Boerjan, W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133, 1051–1071.
Ruprecht, C., Mutwil, M., Saxe, F., Eder, M., Nikoloski, Z., and Persson, S. (2011). Large-scale co-expression approach to dissect secondary cell wall formation across plant species. Front. Plant Sci. 2:23. doi:10.3389/fpls.2011.00023
Shen, H., He, X., Poovaiah, C. R., Wuddineh, W. A., Ma, J., Mann, D. G. J., Wang, H., Jackson, L., Tang, Y., Neal Stewart, C., Chen, F., and Dixon, R. A. (2012). Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 193, 121–136.
Shiu, S.-H., Shih, M.-C., and Li, W.-H. (2005). Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol. 139, 18–26.
Sonbol, F.-M., Fornalé, S., Capellades, M., Encina, A., Touriño, S., Torres, J.-L., Rovira, P., Ruel, K., Puigdomènech, P., Rigau, J., and Caparrós-Ruiz, D. (2009). The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol. Biol. 70, 283–296.
Soyano, T., Thitamadee, S., Machida, Y., and Chua, N.-H. (2008). ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 20, 3359–3373.
Vignols, F., Rigau, J., Torres, M., Capellades, M., and Puigdomenech, P. (1995). The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7, 407–416.
Wang, H., Avci, U., Nakashima, J., Hahn, M. G., Chen, F., and Dixon, R. A. (2010). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. U.S.A. 107, 22338–22343.
Wang, H., Zhao, Q., Chen, F., Wang, M., and Dixon, R. A. (2011). NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J. 68, 1104–1114.
Yamaguchi, M., Ohtani, M., Mitsuda, N., Kubo, M., Ohme-Takagi, M., Fukuda, H., and Demura, T. (2010). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22, 1249–1263.
Yang, C., Xu, Z., Song, J., Conner, K., Vizcay Barrena, G., and Wilson, Z. A. (2007). Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19, 534–548.
Zhao, C., Craig, J. C., Petzold, H. E., Dickerman, A. W., and Beers, E. P. (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138, 803–818.
Zhao, Q., Wang, H., Yin, Y., Xu, Y., Chen, F., and Dixon, R. A. (2010). Syringyl lignin biosynthesis is directly regulated by a secondary cell wall master switch. Proc. Natl. Acad. Sci. U.S.A. 107, 14496–14501.
Zhong, R., Demura, T., and Ye, Z.-H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158–3170.
Zhong, R., Lee, C., McCarthy, R. L., Reeves, C. K., Jones, E. G., and Ye, Z.-H. (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 52, 1856–1871.
Zhong, R., Lee, C., and Ye, Z. H. (2010). Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant 3, 1087–1103.
Zhong, R., Lee, C., Zhou, J., McCarthy, R. L., and Ye, Z.-H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782.
Zhong, R., Richardson, E., and Ye, Z.-H. (2007a). Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225, 1603–1611.
Zhong, R., Richardson, E. A., and Ye, Z. H. (2007b). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19, 2776–2792.
Zhong, R., and Ye, Z.-H. (2007). Regulation of cell wall biosynthesis. Curr. Opin. Plant Biol. 10, 564–572.
Keywords: transcription factors, secondary cell wall
Citation: Handakumbura PP and Hazen SP (2012) Transcriptional regulation of grass secondary cell wall biosynthesis: playing catch-up with Arabidopsis thaliana. Front. Plant Sci. 3:74. doi: 10.3389/fpls.2012.00074
Received: 05 March 2012; Accepted: 31 March 2012;
Published online: 23 April 2012.
Edited by:
Jose Manuel Estevez, University of Buenos Aires and CONICET, ArgentinaReviewed by:
Uener Kolukisaoglu, University of Tuebingen, GermanyVasileios Fotopoulos, Cyprus University of Technology, Cyprus
Copyright: © 2012 Handakumbura and Hazen. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Samuel P. Hazen, Biology Department, University of Massachusetts, 221 Morrill Science Center, Amherst, MA 01003, USA. e-mail: hazen@bio.umass.edu
