
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 09 April 2012
Sec. Plant Development and EvoDevo
Volume 3 - 2012 | https://doi.org/10.3389/fpls.2012.00062
This article is part of the Research TopicSelaginellaView all 7 articles
Ammonium and urea are important nitrogen sources for autotrophic organisms. Plant genomes encode several families of specific transporters for these molecules, plus other uptake mechanisms such as aquaporins and ABC transporters. Selaginella and Physcomitrella are representatives of lycophytes and bryophytes, respectively, and the recent completion of their genome sequences provided us with an opportunity for comparative genome studies, with special emphasis on the adaptive processes that accompanied the conquest of dry land and the evolution of a vascular system. Our phylogenetic analysis revealed that the number of genes encoding urea transporters underwent a progressive reduction during evolution, eventually down to a single copy in vascular plants. Conversely, no clear evolutionary pattern was found for ammonium transporters, and their number and distribution in families varies between species. In particular Selaginella, similar to rice, favors the AMT2/MEP family of ammonium transporters over the plant-specific AMT1 type. In comparison, Physcomitrella presents several members belonging to both families.
Of all mineral elements required by plants, nitrogen is quantitatively the most important and thus often growth-limiting for many plants. Nitrogen is found in many organic compounds such as amino acids, nucleic acid, and consequently in proteins, as well as in nucleic acids. In most soils, nitrogen is heterogeneously distributed and found in various forms such as ammonium, nitrate, urea, amino acids, peptides, and even in water-insoluble forms (Jackson et al., 1990). Among the different nitrogen sources, ammonium is often preferred by plants, since its assimilation requires less energy. Moreover, ammonium is the predominant nitrogen form in anoxic soils, such as paddy rice fields, and it is abundantly used as fertilizer.
Due to the impermeability or poor permeability of the lipid bilayer of the plasma membrane to most nutrients including nitrogen compounds, uni and multicellular organisms developed a suite of nitrogen transporters. In 1979, electrophysiological studies in the unicellular algae Chara australis showed a positive inward current across the plasma membrane induced by ammonium addition (Walker et al., 1979). Fifteen years later, the first ammonium transporter, AtAMT1;1 from Arabidopsis, was cloned by yeast complementation (Ninnemann et al., 1994), in parallel with the yeast MEP orthologs (Marini et al., 1994). Since then, many para and orthologs were isolated either by yeast complementation or via homology cloning (Lauter et al., 1996; Gazzarrini et al., 1999; Simon-Rosin et al., 2003; Sonoda et al., 2003). Homologs have been found in bacteria, fungi, algae, plants, and animals (Marini et al., 1997, 2000; Gonzalez-Ballester et al., 2004) and the transport of the charged form of ammonium  was confirmed for the plant ammonium transporter AMT1;1 by electrophysiological characterization in Xenopus oocytes (Ludewig et al., 2002; Wood et al., 2006; Loqué et al., 2009).
was confirmed for the plant ammonium transporter AMT1;1 by electrophysiological characterization in Xenopus oocytes (Ludewig et al., 2002; Wood et al., 2006; Loqué et al., 2009).
Ammonium transporters fall into three subfamilies, Rhesus, Mep/AMTB (also called AMT2), and AMT1. Plant ammonium transporters belong to the two subclasses: AMT1 and AMT2 (Sohlenkamp et al., 2002). The AMT2 members are sequence-wise more closely related to fungal and bacterial ammonium transporters (Mep and AMTB) forming the Mep/AMTB/AMT2 subfamily (Ludewig et al., 2001). Bacterial AMTs, from Escherichia coli and from the archaebacterium Archaeoglobus fulgidus, have been crystallized, revealing a trimeric structure (Khademi et al., 2004; Andrade et al., 2005; Conroy et al., 2007).
In both Eubacteria and Archaea, AMT activity is regulated by the interaction with GlnK proteins (Coutts et al., 2002). Importantly, at least the bacterial AMTs, similar to the plant nitrate transporter CHL1 and the yeast glucose and amino acid transporters RGT2, SNF1, and GAP1 have a dual function as sensors and transporters, and thus can be defined as transceptors (Hyde et al., 2007; Ho et al., 2009; Thevelein and Voordeckers, 2009). Therefore, at least the bacterial AMTs link energy and nitrogen status of the cell with ammonium uptake to control ammonium-dependent transcription (Tremblay and Hallenbeck, 2009). Plants also contain GlnK homologs, however these are confined to the chloroplast (Hsieh et al., 1998). In plants, plasma membrane AMT activity is regulated by an allosteric auto-inhibition driven by the conserved cytosolic C-terminal tail, which connects the adjacent members in the trimeric complex (Loqué et al., 2007, 2009; Neuhauser et al., 2007; Lanquar et al., 2009; Lanquar and Frommer, 2010). An increase in the concentration of extracellular ammonium leads to the phosphorylation of a specific threonine residue in the cytosolic C-terminus of AtAMT1;1, resulting in closure of the pores of the transporter complex (Loqué et al., 2007; Lanquar et al., 2009). Since the C-terminus, and this regulatory threonine in particular, is highly conserved in plant AMT1s, it is conceivable that this feedback mechanism represents a common feature of plant ammonium transporters. We recently demonstrated that the Archaeglobus AMT1 also uses a trans-activation allosteric mechanism involving the conserved C-terminus, indicating that this unique regulatory system was developed early in evolution and has been maintained. This was surprising, because point mutations in the pore region were found that release the transporter from the strict dependence on trans-activation (Loqué et al., 2009). Such point mutations must have occurred during evolution, but apparently have not been fixed, suggesting a strong pressure to retain this regulatory mechanism, potentially to protect against accumulation of toxic levels of ammonium.
Urea is another nitrogen source used by plants that is commonly available in nature. Urea is mainly derived from decomposition of nitrogen-containing molecules and from urine secretion by animals. Additionally, urea is widely used in agriculture as a fertilizer due to its stability, low toxicity and cost–effective production.
Urea transporters are present in most organisms and belong to different classes, some functioning as passive channels, others as secondary active transporters. Interestingly, urea transporters commonly found in animals and bacteria (UT, UreI, Yut) are absent in plant and fungal genomes (Wang et al., 2008). Plant and fungal genomes encode a different family of urea transporters, called DUR3. In 1993 the first plant urea transporter (Dur3) was cloned from Arabidopsis by homology to the yeast transporter DUR3 (Elberry et al., 1993). DUR3 proteins are secondary active transporters, structurally belonging to the sodium solute symporter (SSS) superfamily (Wang et al., 2008). Electrophysiological studies showed that a proton, rather than sodium, is co-transported with urea by AtDUR3 (Liu et al., 2003). In planta characterization showed that DUR3 genes from Arabidopsis and rice are induced by nitrogen deficiency (Liu et al., 2003; Wang et al., 2011). DUR3s have an important role in high affinity urea transport under nitrogen limiting growth conditions (Kojima et al., 2007; Wang et al., 2011). DUR3 is not the only mechanism for urea uptake, since several aquaporins in the TIP and NOD26 families have been shown to facilitate urea import through the plasma membrane (for review, see Kojima et al., 2006). Additionally, ABC transporters are known to transport urea in cyanobacteria (Valladares et al., 2002). Typically, plant genomes contain > 100 ABC transporter homologs; whether any of these transport urea remains an open question.
The genome sequence of the lycophyte Selaginella moellendorffii (Banks et al., 2011) provides a valuable tool for comparative genomic studies in plants. The lineage of this species arose ∼400 millions years ago early after vascular plants had evolved, and shares many primitive traits with bryophytes. This species adds to the increasing list of plants for which genome sequences have become available, including the moss Physcomitrella patens (Rensing et al., 2008).
Here, we compared the ammonium and urea transporter homologs of two dicots (Arabidopsis and poplar), one monocot (rice) a lycophyte (Selaginella), a bryophyte (Physcomitrella), and a unicellular alga (Chlamydomonas reinhardtii) with the aim of gaining more insight into the evolution of these important transporters. For Arabidopsis, poplar, rice and Chlamydomonas, AMT and DUR3 members have already been annotated and characterized elsewhere, although in the case of poplar annotation is still not complete (Table 1; Loqué and von Wirén, 2004; Cuturier et al., 2006; Fernandez and Galvan, 2007; Wang et al., 2008; Li et al., 2009). Our results indicate a progressive reduction of the number of genes encoding urea transporters during higher plant evolution, possibly suggesting a shift in nutrient form preference of angiosperms for other nitrogen sources. Analysis of number and distribution of ammonium transporters between the AMT1 and AMT2 families revealed that most species, with the exception of Physcomitrella, present a bias for one or the other subfamily (Arabidopsis: AMT1, Selaginella and rice: AMT2).
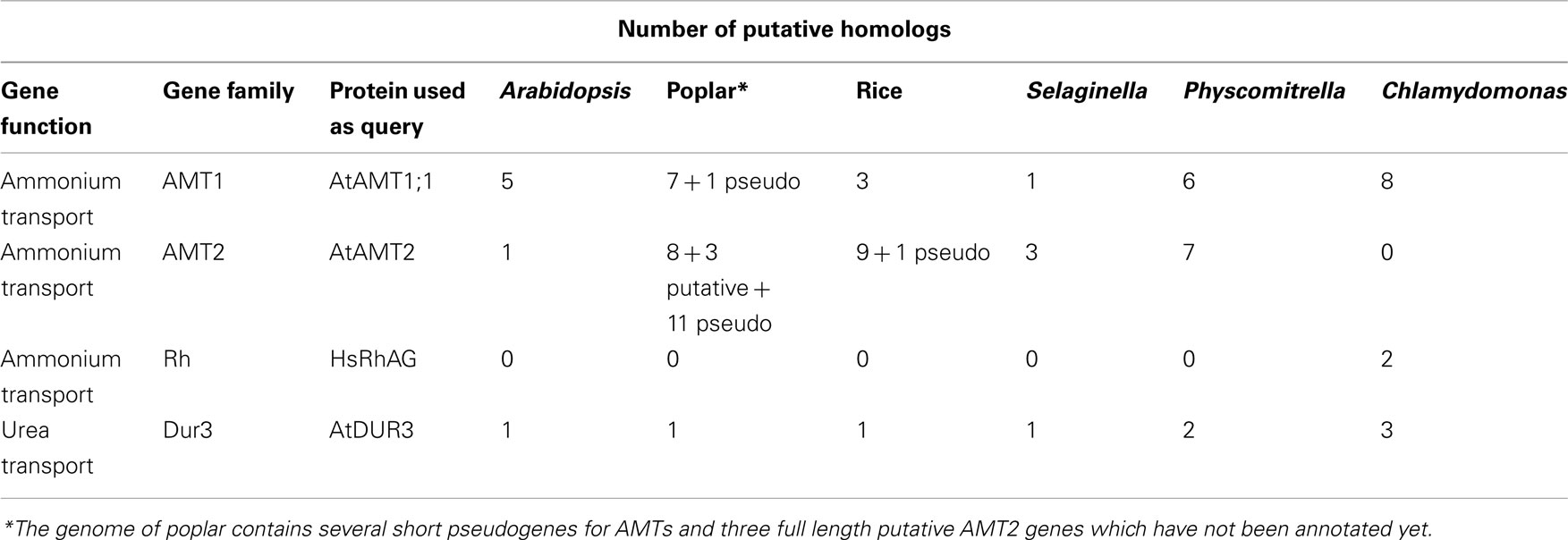
Table 1. Comparison of number of homologs for ammonium and urea transporters in various plant and algal species.
Protein sequences of the six members of Arabidopsis AMTs and AtDUR3 were used to retrieve homologs of the ammonium and urea transporter families in Selaginella, Physcomitrella, rice, poplar, and Chlamydomonas (Table 1). For poplar, only the paralogs already annotated as AMT (Cuturier et al., 2006) were further considered in the phylogenic analysis. Sequences were blasted against a translated-nucleotide sequence database in the Ensembl Genome browser (www.ensemblgenomes.org). Searches for the urea transporter classes UT-A, -B, -C retrieved no hits in the plant databases. Within a species, coding and flanking sequences of the paralogs were carefully analyzed to distinguish between gene duplications and alleles. Protein sequences were aligned by ClustalW and poorly conserved N- and C-terminal domains (corresponding to amino acids 1–39 and 475–501 for AtAMT1;1 and 1–27 and 660–704 for AtDUR3) were trimmed (Figures A1 and A2 in Appendix). Phylogenetic trees were created on the basis of these alignments by the Mega5.05 program (Tamura et al., 2011) using a Neighbor-Joining, Poisson-corrected method, with pairwise deletion option and 1000 bootstraps. Additional trees were generated by Maximum-Likelihood method, Poisson-corrected, with 1000 bootstraps (Figures A3 and A4 in Appendix).
Analysis of the S. moellendorffii genome indicates the presence of four ammonium transporters, with little variation between the two haplotypes sequenced (Tables 1 and 2). One AMT (SmAMT1) belongs to the AMT1 family and are closely related to the Physcomitrella AMTs (Figure 1; Figure A3 in Appendix). The three other Selaginella AMTs fall into the AMT2 subfamily.
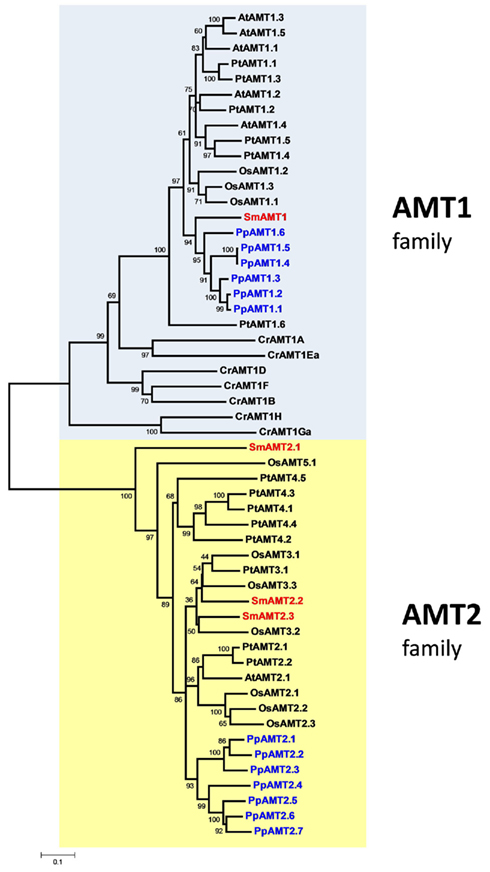
Figure 1. Unrooted phylogenetic tree of the ammonium transporters in Arabidopsis thaliana (At), Populus trichocarpa (Pt), Oryza sativa (Os), Selaginella moellendorffii (Sm), Physcomitrella patens (Pp), and Chlamydomonas reinhardtii (Cr). Red indicates Selaginella AMTs, blue Physcomitrella. The tree was constructed by aligning the protein sequences by ClustalW and generated with the Neighborhood-Joining method.
The genome of the moss P. patens revealed a higher and more diversified number of ammonium transporters, with six members of the AMT1 subfamily and seven members of the AMT2 subfamily (Table 1). AMT paralogs often differ with respect to affinities for ammonium and regulation. The existence of multiple paralogs may either help to increase transport capacity, ensure flexibility for regulation, and help cover a wider range of ammonium levels. In both AMT1 and AMT2 clades, the members of the Physcomitrella AMTs group closely together, suggesting a recent origin of the paralogs within the group (Figure 1; Figure A3 in Appendix). Three of the AMT1 members (named PpAMT1;1, PpAMT1;2, and PpAMT1;3), highly similar, correspond to recent gene duplications as they occur in tandem at the same genetic locus. The persistence of several paralogs in Physcomitrella is not limited to the AMT family, and in fact it seems to be a common feature derived from genome duplication events (Lang et al., 2005; Rensing et al., 2007).
The unicellular alga C. reinhardtii has eight AMTs, all belonging to the AMT1 family, and sharing only 31–46% identity with AMT1s of Arabidopsis or Selaginella. Two of them (CrAMT-E and -G) also show alternative splicing (Fernandez and Galvan, 2007). Regarding phylogeny, they fall into three subclades, separated from other plant genes. This divergence is emphasized also by the lack of the conserved regulatory threonine in the C-terminus (Loqué et al., 2007). Other residues, usually conserved in AMT1s but missing or misplaced in some Chlamydomonas AMTs are the N-terminal cysteines, which have been shown to be important to promote interactions among monomers in the trimeric complex (Graff et al., 2011). It will be interesting to study how AMTs in this organism assemble and are regulated. Apart from the AMT genes, the genome of Chlamydomonas also contains homologs of the animal Rhesus ammonia transporters (Rh), that are distantly related to the AMT family (Soupene et al., 2004). Rh genes are however absent in Selaginella, Physcomitrella, and in any plant species sequenced so far.
An interesting observation is that the proportion of AMT1 and AMT2 genes in the various species is different (Table 1). The Arabidopsis genome possesses five AMT1 members and one AMT2, but in rice the proportion is inverted (three AMT1s and nine AMT2s). Similarly, the Selaginella genome is more enriched in AMT2s (three members) and a single AMT1. Similar to poplar, Physcomitrella is more balanced, with six AMT1 and seven AMT2 types; finally, Chlamydomonas is so far the species containing more AMT1 members (eight) and totally lacking the AMT2 type. It is interesting to observe that the C-terminus of the AMT1 family is highly conserved also in Selaginella and Physcomitrella (but not Chlamydomonas) as is the equivalent of threonine-460 in the C-terminus AtAMT1;1, which is involved in ammonium-dependent allosteric regulation (Loqué et al., 2007; Lanquar et al., 2009), suggesting that the regulatory mechanism via phosphorylation of AMT1 members is maintained in all multicellular plants.
Analysis of the Selaginella genome suggests the presence of a unique DUR3 homolog (named SmDUR3), similar to the one found in Arabidopsis, poplar, or rice genomes (Table 1). Whereas vascular plants have a single urea transporter, the moss Physcomitrella possesses two DUR3 homologs (PpDUR3A and B), with 85% identity between each other. Going further back in the evolutionary tree, Chlamydomonas has three different copies, named DUR3A, B and C, more closely related to each other (about 80% identity) than with the DUR3 members of multicellular plants (Figure 2; Figure A4 in Appendix). Other algae, as well as fungi, are known to have additional copies of DUR3 genes (Kakinuma et al., 2008; Morel et al., 2008).
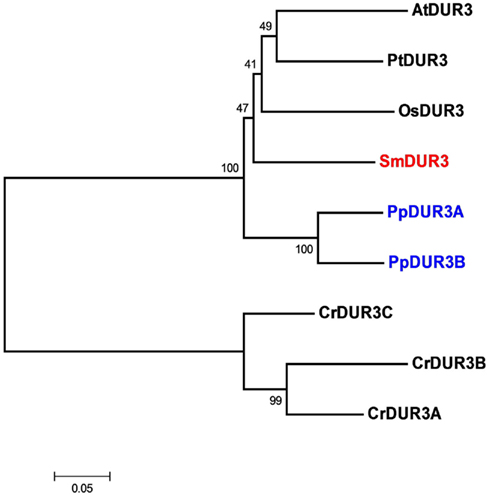
Figure 2. Unrooted phylogenetic tree of the urea transporters in Arabidopsis thaliana (At), Populus trichocarpa (Pt), Oryza sativa (Os), Selaginella moellendorffii (Sm), Physcomitrella patens (Pp), and Chlamydomonas reinhardtii (Cr). Red indicates Selaginella DUR3, blue Physcomitrella. The tree was constructed by aligning the protein sequences by ClustalW and generated with the Neighborhood-Joining method.
The progressive loss of genes for urea transporters could indicate that the evolution of the vascular system shifted the preference for other nitrogen sources, such as ammonium, nitrate, and amino acids, all present in the xylem sap of vascular plants (Smirnoff and Stewart, 1985; Atkins, 2000; Schjoerring et al., 2002).
The comparative analysis of ammonium transporters revealed that different lineages of plants vary in the number of AMTs belonging to either family, and in many cases they show a bias toward one of the two AMT subfamilies, with some organisms particularly enriched in the plant-specific AMT1 clade (Arabidopsis, Chlamydomonas), while others rely more on the AMT2 family, typical for bacteria and fungi. The physiological and evolutionary relevance of these shifts remains to be determined. The number of urea transporters decreased during evolution, maybe indicating a preference of vascular plants for other nitrogen sources or the evolution of yet uncharacterized urea transporters.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by a grant from the National Science Foundation (MCB-1021677) to Wolf B. Frommer.
Andrade, S. L., Dickmanns, A., Ficner, R., and Einsle, O. (2005). Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. U.S.A. 102, 14994–14999.
Atkins, C. (2000). Biochemical aspects of assimilate transfers along the phloem path: N-solutes in lupins. Aust. J. Plant Physiol. 27, 531–537.
Banks, J. A., Nishiyama, T., Hasebe, M., Bowman, J. L., Gribskov, M., Depamphilis, C., Albert, V. A., Aono, N., Aoyama, T., Ambrose, B. A., Ashton, N. W., Axtell, M. J., Barker, E., Barker, M. S., Bennetzen, J. L., Bonawitz, N. D., Chapple, C., Cheng, C., Correa, L. G., Dacre, M., Debarry, J., Dreyer, I., Elias, M., Engstrom, E. M., Estelle, M., Feng, L., Finet, C., Floyd, S. K., Frommer, W. B., Fujita, T., Gramzow, L., Gutensohn, M., Harholt, J., Hattori, M., Heyl, A., Hirai, T., Hiwatashi, Y., Ishikawa, M., Iwata, M., Karol, K. G., Koehler, B., Kolukisaoglu, U., Kubo, M., Kurata, T., Lalonde, S., Li, K., Li, Y., Litt, A., Lyons, E., Manning, G., Maruyama, T., Michael, T. P., Mikami, K., Miyazaki, S., Morinaga, S., Murata, T., Mueller-Roeber, B., Nelson, D. R., Obara, M., Oguri, Y., Olmstead, R. G., Onodera, N., Petersen, B. L., Pils, B., Prigge, M., Rensing, S. A., Riano-Pachon, D. M., Roberts, A. W., Sato, Y., Scheller, H. V., Schulz, B., Schulz, C., Shakirov, E. V., Shibagaki, N., Shinohara, N., Shippen, D. E., Sorensen, I., Sotooka, R., Sugimoto, N., Sugita, M., Sumikawa, N., Tanurdzic, M., Theissen, G., Ulvskov, P., Wakazuki, S., Weng, J. K., Willats, W. W., Wipf, D., Wolf, P. G., Yang, L., Zimmer, A. D., Zhu, Q., Mitros, T., Hellsten, U., Loque, D., Otillar, R., Salamov, A., Schmutz, J., Shapiro, H., Lindquist, E., Lucas, S., Rokhsar, D., and Grigoriev, I. V. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963.
Conroy, M. J., Durand, A., Lupo, D., Li, X. D., Bullough, P. A., Winkler, F. K., and Merrick, M. (2007). The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc. Natl. Acad. Sci. U.S.A. 104, 1213–1218.
Coutts, G., Thomas, G., Blakey, D., and Merrick, M. (2002). Membrane sequestration of the signal transduction protein GlnK by the ammonium transporter AmtB. EMBO J. 21, 536–545.
Cuturier, J., Montanini, B., Martin, F., Brun, A., Blaudez, D., and Chalot, M. (2006). The expanded family of ammonium transporters in the perennial poplar plant. New Phytol. 174, 137–150.
Elberry, H. M., Majumdar, M. L., Cunningham, T. S., Sumrada, R. A., and Cooper, T. G. (1993). Regulation of the urea active transporter gene (DUR3) in Saccharomyces cerevisiae. J. Bacteriol. 175, 4688–4698.
Fernandez, E., and Galvan, A. (2007). Inorganic nitrogen assimilation in Chlamydomonas. J. Exp. Bot. 58, 2279–2287.
Gazzarrini, S., Lejay, L., Gojon, A., Ninnemann, O., Frommer, W. B., and Von Wiren, N. (1999). Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11, 937–947.
Gonzalez-Ballester, D., Camargo, A., and Fernandez, E. (2004). Ammonium transporter genes in Chlamydomonas: the nitrate-specific regulatory gene Nit2 is involved in Amt1;1 expression. Plant Mol. Biol. 56, 863–878.
Graff, L., Obrdlik, P., Yuan, L., Loque, D., Frommer, W. B., and Von Wiren, N. (2011). N-terminal cysteines affect oligomer stability of the allosterically regulated ammonium transporter LeAMT1;1. J. Exp. Bot. 62, 1361–1373.
Ho, C. H., Lin, S. H., Hu, H. C., and Tsay, Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194.
Hsieh, M. H., Lam, H. M., Van De Loo, F. J., and Coruzzi, G. (1998). A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proc. Natl. Acad. Sci. U.S.A. 95, 13965–13970.
Hyde, R., Cwiklinski, E. L., Macaulay, K., Taylor, P. M., and Hundal, H. S. (2007). Distinct sensor pathways in the hierarchical control of SNAT2, a putative amino acid transceptor, by amino acid availability. J. Biol. Chem. 282, 19788–19798.
Jackson, R. B., Manwaring, J. H., and Caldwell, M. M. (1990). Rapid physiological adjustment of roots to localized soil enrichment. Nature 344, 58–60.
Kakinuma, M., Coury, D. A., Nakamoto, C., Sakaguchi, K., and Amano, H. (2008). Molecular analysis of physiological responses to changes in nitrogen in a marine macroalga, Porphyra yezoensis (Rhodophyta). Cell Biol. Toxicol. 24, 629–639.
Khademi, S., O’Connell, J. III, Remis, J., Robles-Colmenares, Y., Miercke, L. J., and Stroud, R. M. (2004). Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A. Science 305, 1587–1594.
Kojima, S., Bohner, A., Gassert, B., Yuan, L., and Von Wiren, N. (2007). AtDUR3 represents the major transporter for high-affinity urea transport across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J. 52, 30–40.
Kojima, S., Bohner, A., and Von Wiren, N. (2006). Molecular mechanisms of urea transport in plants. J. Membr. Biol. 212, 83–91.
Lang, D., Eisinger, J., Reski, R., and Rensing, S. A. (2005). Representation and high-quality annotation of the Physcomitrella patens transcriptome demonstrates a high proportion of proteins involved in metabolism in mosses. Plant Biol. 7, 238–250.
Lanquar, V., and Frommer, W. B. (2010). Adjusting ammonium uptake via phosphorylation. Plant Signal Behav. 5, 736–738.
Lanquar, V., Loque, D., Hormann, F., Yuan, L., Bohner, A., Engelsberger, W. R., Lalonde, S., Schulze, W. X., Von Wiren, N., and Frommer, W. B. (2009). Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell 21, 3610–3622.
Lauter, F. R., Ninnemann, O., Bucher, M., Riesmeier, J. W., and Frommer, W. B. (1996). Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proc. Natl. Acad. Sci. U.S.A. 93, 8139–8144.
Li, B.-Z. M. M., Li, S.-M., Li, H.-Y., Zhu, S.-W., Shi, W.-M., and Su, Y.-H. (2009). Molecular basis and regulation of ammonium transporter in rice. Rice Sci. 16, 314–322.
Liu, L. H., Ludewig, U., Frommer, W. B., and Von Wiren, N. (2003). AtDUR3 encodes a new type of high-affinity urea/H+ symporter in Arabidopsis. Plant Cell 15, 790–800.
Loqué, D., Lalonde, S., Looger, L. L., Von Wirén, N., and Frommer, W. B. (2007). A cytosolic trans-activation domain essential for ammonium uptake. Nature 446, 195–198.
Loqué, D., Mora, S. I., Andrade, S. L., Pantoja, O., and Frommer, W. B. (2009). Pore mutations in ammonium transporter AMT1 with increased electrogenic ammonium transport activity. J. Biol. Chem. 284, 24988–24995.
Loqué, D., and von Wirén, N. (2004). Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 55, 1293–1305.
Ludewig, U., Von Wiren, N., and Frommer, W. B. (2002). Uniport of NH4+ by the root hair plasma membrane ammonium transporter LeAMT1;1. J. Biol. Chem. 277, 13548–13555.
Ludewig, U., Von Wiren, N., Rentsch, D., and Frommer, W. B. (2001). Rhesus factors and ammonium: a function in efflux? Genome Biol. 2, 1–5.
Marini, A. M., Matassi, G., Raynal, V., Andre, B., Cartron, J. P., and Cherif-Zahar, B. (2000). The human Rhesus-associated RhAG protein and a kidney homologue promote ammonium transport in yeast. Nat. Genet. 26, 341–344.
Marini, A. M., Soussiboudekou, S., Vissers, S., and Andre, B. (1997). A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 4282–4293.
Marini, A. M., Vissers, S., Urrestarazu, A., and André, B. (1994). Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 13, 3456–3463.
Morel, M., Jacob, C., Fitz, M., Wipf, D., Chalot, M., and Brun, A. (2008). Characterization and regulation of PiDur3, a permease involved in the acquisition of urea by the ectomycorrhizal fungus Paxillus involutus. Fungal Genet. Biol. 45, 912–921.
Neuhauser, B., Dynowski, M., Mayer, M., and Ludewig, U. (2007). Regulation of NH4+ transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiol. 143, 1651–1659.
Ninnemann, O., Jauniaux, J. C., and Frommer, W. B. (1994). Identification of a high-affinity NH4+ transporter from plants. EMBO J. 13, 3464–3471.
Rensing, S. A., Ick, J., Fawcett, J. A., Lang, D., Zimmer, A., Van de Peer, Y., and Reski, R. (2007). An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evol. Biol. 2, 130. doi:10.1186/1471-2148-7-130
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., Nishiyama, T., Perroud, P. F., Lindquist, E. A., Kamisugi, Y., Tanahashi, T., Sakakibara, K., Fujita, T., Oishi, K., Shin, I. T., Kuroki, Y., Toyoda, A., Suzuki, Y., Hashimoto, S., Yamaguchi, K., Sugano, S., Kohara, Y., Fujiyama, A., Anterola, A., Aoki, S., Ashton, N., Barbazuk, W. B., Barker, E., Bennetzen, J. L., Blankenship, R., Cho, S. H., Dutcher, S. K., Estelle, M., Fawcett, J. A., Gundlach, H., Hanada, K., Heyl, A., Hicks, K. A., Hughes, J., Lohr, M., Mayer, K., Melkozernov, A., Murata, T., Nelson, D. R., Pils, B., Prigge, M., Reiss, B., Renner, T., Rombauts, S., Rushton, P. J., Sanderfoot, A., Schween, G., Shiu, S. H., Stueber, K., Theodoulou, F. L., Tu, H., Van De Peer, Y., Verrier, P. J., Waters, E., Wood, A., Yang, L., Cove, D., Cuming, A. C., Hasebe, M., Lucas, S., Mishler, B. D., Reski, R., Grigoriev, I. V., Quatrano, R. S., and Boore, J. L. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69.
Schjoerring, J. K., Husted, S., Mack, G., and Mattsson, M. (2002). The regulation of ammonium translocation in plants. J. Exp. Bot. 53, 883–890.
Simon-Rosin, U., Wood, C., and Udvardi, M. K. (2003). Molecular and cellular characterisation of LjAMT2; 1, an ammonium transporter from the model legume Lotus japonicus. Plant Mol. Biol. 51, 99–108.
Smirnoff, N., and Stewart, G. R. (1985). Nitrate assimilation and translocation by higher-plants – comparative physiology and ecological consequences. Physiol. Plant. 64, 133–140.
Sohlenkamp, C., Wood, C. C., Roeb, G. W., and Udvardi, M. K. (2002). Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol. 130, 1788–1796.
Sonoda, Y., Ikeda, A., Saiki, S., Von Wiren, N., Yamaya, T., and Yamaguchi, J. (2003). Distinct expression and function of three ammonium transporter genes (OsAMT1;1-1;3) in rice. Plant Cell Physiol. 44, 726–734.
Soupene, E., Inwood, W., and Kutzu, S. (2004). Lack of the Rhesus protein Rh1 impairs growth of the green alga Chlamydomonas reinhardtii at high CO2. Proc. Natl. Acad. Sci. U.S.A. 101, 7787–7782.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Thevelein, J. M., and Voordeckers, K. (2009). Functioning and evolutionary significance of nutrient transceptors. Mol. Biol. Evol. 26, 2407–2414.
Tremblay, P. L., and Hallenbeck, P. C. (2009). Of blood, brains and bacteria, the Amt/Rh transporter family: emerging role of Amt as a unique microbial sensor. Mol. Microbiol. 71, 12–22.
Valladares, A., Montesinos, M. L., Herrero, A., and Flores, E. (2002). An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol. Microbiol. 43, 703–715.
Walker, N. A., Beilby, M. J., and Smith, F. A. (1979). Amine uniport at the plasmalemma of charophyte cells.1. Current-voltage curves, saturation kinetics, and effects of unstirred layers. J. Membr. Biol. 49, 21–55.
Wang, W. H., Kohler, B., Cao, F. Q., Liu, G. W., Gong, Y. Y., Sheng, S., Song, Q. C., Cheng, X. Y., Garnett, T., Okamoto, M., Qin, R., Müller-Röber, B., Tester, M., and Liu, L. H. (2011). Rice DUR3 mediates high-affinity urea transport and plays an effective role in improvement of urea acquisition and utilization when expressed in Arabidopsis. New Phytol. 193, 432–444.
Wang, W. H., Kohler, B., Cao, F. Q., and Liu, L. H. (2008). Molecular and physiological aspects of urea transport in higher plants. Plant Sci. 175, 467–477.
Wood, C. C., Poree, F., Dreyer, I., Koehler, G. J., and Udvardi, M. K. (2006). Mechanisms of ammonium transport, accumulation, and retention in ooyctes and yeast cells expressing Arabidopsis AtAMT1;1. FEBS Lett. 580, 3931–3936.
Figure A1. Alignment of protein sequences of ammonium transporters by ClustalW. In gray squares are the regions excluded by analyses.
Figure A2. Alignment of protein sequences of urea transporters by ClustalW. In gray squares are the regions excluded by analyses.
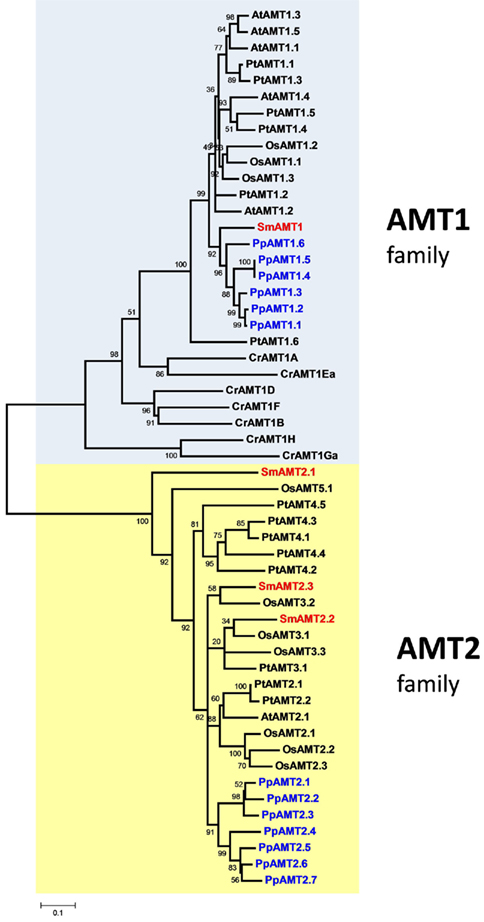
Figure A3. Unrooted phylogenetic tree of the ammonium transporters in Arabidopsis thaliana (At), Populus trichocarpa (Pt), Oryza sativa (Os), Selaginella moellendorffii (Sm), Physcomitrella patens (Pp), and Chlamydomonas reinhardtii (Cr). Red indicates Selaginella AMTs, blue Physcomitrella. The tree was constructed by aligning the protein sequences by ClustalW and generated with the Maximum-Likelihood method.
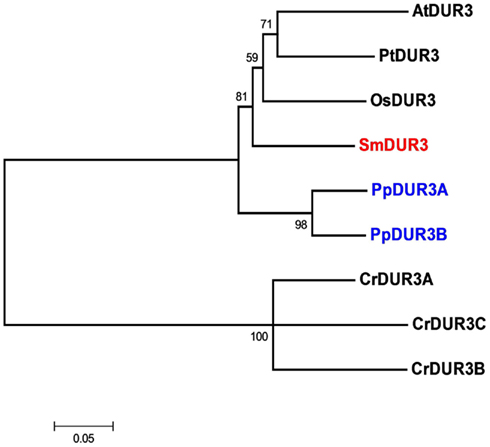
Figure A4. Unrooted phylogenetic tree of the urea transporters in Arabidopsis thaliana (At), Populus trichocarpa (Pt), Oryza sativa (Os), Selaginella moellendorffii (Sm), Physcomitrella patens (Pp), and Chlamydomonas reinhardtii (Cr). Red indicates Selaginella DUR3, blue Physcomitrella. The tree was constructed by aligning the protein sequences by ClustalW and generated with the Maximum-Likelihood method.
Keywords: Selaginella, Physcomitrella, ammonium, urea, transporter, uptake
Citation: De Michele R, Loqué D, Lalonde S and Frommer WB (2012) Ammonium and urea transporter inventory of the Selaginella and Physcomitrella genomes. Front. Plant Sci. 3:62. doi: 10.3389/fpls.2012.00062
Received: 19 December 2011; Accepted: 14 March 2012;
Published online: 09 April 2012.
Edited by:
Amy Litt, The New York Botanical Garden, USAReviewed by:
Biao Ding, The Ohio State University, USACopyright: © 2012 De Michele, Loqué, Lalonde and Frommer. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Wolf B. Frommer, Department of Plant Biology, Carnegie Institution for Science, 260 Panama Street, Stanford, CA 94305, USA. e-mail:d2Zyb21tZXJAY2FybmVnaWVzY2llbmNlLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.