- Laboratory Molekulare Ertragsphysiologie, Institut für Zierpflanzenbau und Gehölzwissenschaften, Leibniz Universität Hannover, Hannover, Germany
Three types of phospholipases, phospholipase D, secreted phospholipase A2, and patatin-related phospholipase A (pPLA) have functions in auxin signal transduction. Potential linkage to auxin receptors ABP1 or TIR1, their rapid activation or post-translational activation mechanisms, and downstream functions regulated by these phospholipases is reviewed and discussed. Only for pPLA all aspects are known at least to some detail. Evidence is gathered that all these signal reactions are located in the cytosol and seem to merge on regulation of PIN-catalyzed auxin efflux transport proteins. As a consequence, auxin concentration in the nucleus is also affected and this regulates the E3 activity of this auxin receptor. We showed that ABP1, PIN2, and pPLA, all outside the nucleus, have an impact on regulation of auxin-induced genes within 30 min. We propose that regulation of PIN protein activities and of auxin efflux transport are the means to coordinate ABP1 and TIR1 activity and that no physical contact between components of the ABP1-triggered cytosolic pathways and TIR1-triggered nuclear pathways of signaling is necessary to perform this.
Introduction
Phospholipases in Plants
Phospholipases hydrolyze phospholipids. They are categorized by the bonds they hydrolyze, which result in the products diacylglycerol and phosphorylated headgroup by phospholipase C (PLC), and phosphatidic acid (PA) and headgroup by phospholipase D (PLD; Meijer and Munnik, 2003; Li et al., 2009). A phospholipase A (PLA) is an acyl hydrolase which hydrolyzes phospholipids either at the hydroxyl at sn-1 position (phospholipase A1, PLA1) or at the sn-2 position (phospholipase A2; PLA2). Structurally quite different enzymes show PLA activity. PLA enzymes in plants are the small secreted phospholipases A2 (sPLA2; 14 kDa), the soluble or secreted patatin-related phospholipases A (pPLA; 45 kDa), and the lipase-like PLA1 (Scherer et al., 2010).
What is Signal Transduction?
Signals activate networks of biochemical reactions typically located in the cytosol (Figure 1). This is initiated by receptors which specifically recognize the signal molecules or absorb photons of a discrete energy (Figure 1). The type of enzymes participating in such networks is similar if not homologous in eukaryotic organisms, which means that generalizations can be drawn. Receptor activation first triggers the generation of second messengers by the activation of suitable enzymes within seconds or minutes. Whereas in higher animals G-protein-coupled receptors are most abundant, in plants there is only one G-alpha subunit and the true G-protein-coupled receptor has not been identified (Jones and Assmann, 2004). The most abundant type of receptor in plants is the receptor kinase. Among the enzymes generating second messengers in plants there are also phospholipases which generate phospholipid breakdown products as second messengers which, in plants, are still primarily potential second messengers because the exact identification of their downstream effectors often is still incomplete. Many other second messengers in plants have been established besides those generated by phospholipases. The established ones are cytosolic calcium, reactive oxygen species (ROS), nitric oxide (NO), cGMP, and cADPR, all of which are second messengers for plants. Furthermore, signal transduction mechanisms and networks include a number of characteristic enzymes, most prominently protein kinases and protein phosphatases. Another important principle in plant signal transduction is regulated proteolysis of critical protein by the proteasome. These critical proteins, which are often negatively acting transduction factors, are ubiquitinated by E3 ligases, and ubiquitinated proteins are hydrolyzed in the proteasome. Several plant receptors are coupled to or integrated in the ubiquitination and proteolysis machinery (Dreher and Callis, 2007; Stone and Callis, 2007).
All such mechanisms eventually aim at changing the activity of transcription factors by a multitude of different mechanisms (Schütze et al., 2008). These activity changes in transcription factors then induce long-lasting changes in gene expression and protein activities, providing new functions, and eliminate or decrease previous functions in a given tissue. This can usually be described as a new physiological response or a new status, which then becomes apparent as a visible change in the plant’s morphology or development.
It is important to realize what the time course of a biological response to a signal is. This was recently compared and evaluated for auxin signal transduction and the two main auxin receptors, ABP1 and TIR1 (Scherer, 2011). Elongation growth is the most rapid visible auxin response, starting with a lag phase of 10 min. Auxin-activated genes, at the earliest, may contribute after 8–10 min by synthesis of new protein to this most rapid “macroscopic” physiological response so that TIR1 can not be the receptor for such rapid responses. Within this first time span, however, a considerable number of rapid auxin responses have already been described, including activation of PLA and PLD activity. These reactions, starting with hormone binding to the receptor and prior to transcription comprise, by commonly accepted definition, the biochemical signal transduction (Figure 1). TIR1 provides the shortest possible signal transduction since it binds auxin as a receptor and employs its E3 activity to down-regulate Aux/IAA transcription factors by inducing hydrolysis in the proteasome (Mockaitis and Estelle, 2008). Even though an influence on the transcription of early auxin-induced genes as early as 2–5 min can be found (Ballas et al., 1993; Abel et al., 1994) these early auxin-induced genes are unlikely to generate direct physiological changes because they are transcription factors or they regulate hormone concentration. Moreover, often the time span for transcriptional increase of proteins is longer (Calderon-Villalobos et al., 2006). However, ABP1 and pPLA also influences early auxin-regulated genes at least as early as after 30 min (Effendi et al., 2011), so the question arises how the actions of the two receptors may meet or cooperate.
Review
Phospholipases in Auxin Signal Transduction
Several phospholipases have been implicated in auxin signaling: PLD, sPLA2, and pPLA. Data are still incomplete regarding mechanistic models and how and whether they can be activated by a receptor-driven post-translational mechanism. Activation can also be achieved by transcriptional/translational increase of activity which, however, is difficult to imagine happening any faster than within 10 min. Thus, timing is always an important aspect when analyzing signal transduction.
Phospholipase D and Auxin Functions
Phospholipases D are a gene family of nine genes in Arabidopsis, all of which seem to be cytosolic enzymes (Li et al., 2009). Auxin activation within 1 min of PLD activity led to PA accumulation (Lanteri et al., 2008). The relevant PLD isoform(s) have not yet been identified nor the relevant auxin receptor. This PLD activation was associated with equally rapid NO biosynthesis (Lanteri et al., 2008). The rapidity of activation kinetics eliminates TIR1 as the relevant receptor and suggests that ABP1 could be the receptor without proving this directly. For comparison, in abiotic stress responses, rapid activation kinetics of PLD within 2 min are also known (van der Luit et al., 2000; den Hartog et al., 2003; de Jong et al., 2004). Salicylic acid as another biotic signal leads to rapid PLD activation within 15 min but further details remain unknown (Krinke et al., 2009). Previously, PLD inactivation by GαGDP and an ABA receptor postulated to be a G-protein-coupled receptor was described (Zhao and Wang, 2004; Mishra et al., 2006). Thus, it seems possible that PLD could be receptor-activated in a post-translational mode even though the receptors for salicylic acid and auxin for the PLD pathway remain unknown at this time.
Interestingly, lipids activate PLD (Li et al., 2009). The isoform PLDδ is activated by oleic acid (Wang and Wang, 2001), and μM Ca2+ and PIP2 activates the isoforms PLDβ1, PLDγ1, PLDγ2, and PLDδ (Li et al., 2009). Conceivably, the enzymes generating oleic acid and PIP2 and calcium increases could be activated/ inactivated by signals, e.g., stress signals (van der Luit et al., 2000; den Hartog et al., 2003; de Jong et al., 2004). PIP2 biosynthesis is the end product of phosphorylation by phosphatidylinositide kinases, but how these could be linked to signals or to auxin signaling in particular is still unknown.
So, there is no experimental evidence that activation of a pPLA (or sPLA2) by auxin could provide oleic acid to PLD but it is conceivably a possibility. This would constitute a second pathway to activate PLD’s in addition to calcium activation of PLD. At this point, the available evidence is too sketchy to identify a receptor for a clear activation pathway for PLD by auxin, but rapidity of activation indicates a post-translational activation mechanism (Figure 2).
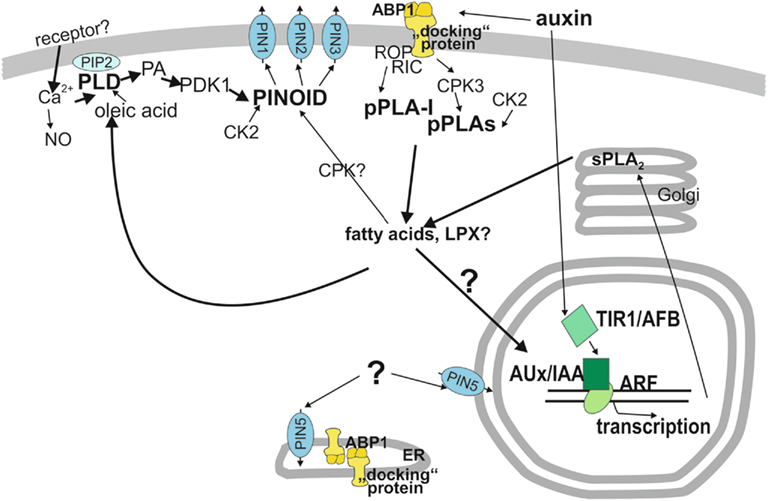
Figure 2. Model of functions of phospholipases in auxin signal transduction. Auxin perception occurs at two receptors, TIR1 (bottom half) and ABP1 (top half). In the upper half, ABP1 is the relevant receptor and reactions are assembled which occur faster than in 10 min, like the activation of phospholipase A and D. Other cytosolic components are also assembled in this cytosolic signal network even though some of them hypothetically where a linkage is only suggested. One important output of the cytosolic network is regulation of PIN protein activity, most likely by shifting the balance between endocytosis and exocytosis and /or shifting the subcellular localization of PIN proteins. The protein kinase PINOID is the main regulatory protein for PIN1, PIN2, and PIN3. Regulation of PIN5 by protein kinase or other cytosolic factors is unknown. How the pPLA hydrolysis products, fatty acids and lysolipids transmit their activity to other components is hypothetical. However, if pPLA activities are lacking, auxin functions are decreased or interrupted. Auxin perception occurs at the receptor TIR1 in the nucleus which leads to degradation of Aux/IAA proteins that act as repressors of ARF transcription factors. This initiates transcription of early auxin-induced genes. Because most early auxin-induced gene products do not carry out direct physiological reactions but rather regulate transcription and hormone concentrations gene products with other physiological functions need at least 10 min to become effective for physiological changes (Scherer, 2011). ABP1, AUXIN-BINDING-PROTEIN1; ARF, AUXIN RESPONSE FACTOR; Aux/IAA, auxin/indoleacetic acid proteins; CDPK, calcium-dependent protein kinase; CK2, casein kinase2; CPK3, calcium-dependent kinase3; “docking” protein, hypothetical transmembrane protein and co-receptor to ABP1; LPX, lysolipid; PA, phosphatidic acid; PDK1, phospholipid-dependent kinase1; PIN, pin-formed, auxin efflux transporter; PINOID, protein kinase; pPLA, patatin-related phospholipase A; pPLA-I, patatin-related phospholipase A-I; PLD, phospholipase D; sPLA2, secreted phospholipase A2; TIR1/AFB, TRANSPORT-INHIBITOR-RESISTANT1 and homologous AUXIN UP-REGULATED F-BOX PROTEIN.
Li and Xue (2007) investigated PLDζ2 mutants and found the enzyme is required for auxin responses. Here, a rapid time course of the enzymatic reaction in response to auxin was not described. Vesicle traffic is disturbed in knockout plants while vesicle flow and PIN cycling are slowed. Auxin sensitivity is decreased in knockout plants, root gravitropism is decreased, and hypocotyl elongation at 29°C is decreased as well. In overexpressing plants, the reverse phenotype is observed. The total absence of one PLD isoform might change critical lipid pools affecting membrane-associated functions so that a constant lack of input of this enzymatic activity, perhaps in only a few compartments, can possibly evoke such phenotypes. This means that certain lipids do indeed act as second messengers. A link to a receptor is not even necessary for PLDζ2 to influence general membrane properties. Regulatory input may proceed by transcription of PLDζ2, triggered by endogenous or exogenous cues. The absence of a critical activity which disturbs lipid pools and membrane functions such as vesicle flow, could be sufficient (however, see below).
A central theme in signal transduction is calcium. Therefore, and because there is fragmentary evidence for this, we have included a “Ca2+/PLD module” in conjunction with the evidence for calcium in auxin signal transduction in our scheme (Figure 2), not excluding other calcium functions in auxin signaling (Zhang et al., 2011). Activation by auxin of calcium influx was shown (Shishova et al., 2007; Monshausen et al., 2011). A calcium channel was hypothesized to be an interactor for ABP1 (Shishova and Lindberg, 2010) so that such a channel could even have the role of the hypothetical transmembrane protein (Klämbt, 1990). However, channel regulation can also be achieved by different mechanisms, and we favor a receptor kinase as a likely candidate for the hypothetical transmembrane protein (Figure 2).
A potential upstream integrator for PLD function in auxin signaling could be calcium and a downstream integrator could be PINOID, the kinase which phosphorylates PIN proteins and activates auxin transport (Dhonukshe et al., 2010; Huang et al., 2010). This is depicted as a “Ca2+/PLD module.” PLD can be assumed to be activated by calcium, which is an activator for isoforms PLDβ1, PLDγ1, PLDγ2, and PLDδ at μM concentrations. The effect of calcium on PLDζ2 has not been determined (Li et al., 2009). PA, the hydrolysis product of PLD, was shown to activate the 3-phosphoinositide-dependent protein kinase PDK1, which binds to a hydrophobic PDK1-interacting C-terminal domain in PINOID and phosphorylates PINOID (Zegzouti et al., 2006): This trans-phosphorylation increases autophosphorylation of PINOID, leading to the activation of PINOID. Activation of PINOID leads to phosphorylation of PIN proteins which, in turn, increases the basal polarity of PIN proteins and increases the efficiency of auxin transport (Friml et al., 2004; Michniewicz et al., 2007). Phosphorylation of PIN1 and PIN2 by PINOID have been shown and phosphorylation sites identified (Dhonukshe et al., 2010; Huang et al., 2010). This potential pathway to influence several auxin transport-dependent functions by PLDζ2 was already suggested by Li and Xue (2007); only the bridge to calcium is built here.
In summary, while a function of Ca2+ and PLD in auxin signaling is apparent, a future model will require identification of receptor(s) and auxin-relevant functions for particular isoforms, e.g., by investigating mutants. The only PLD isoform investigated for auxin functions is PLDζ2 but there the relevant receptor is not known (Lee et al., 2010).
Functions of sPLA2 in Auxin Signal Transduction
The data on functions of sPLA2 in auxin physiology are mostly derived from investigations on knockout and overexpressor mutants. Experimental observations are made after gravitropic stimulation after hours or after at least several days of seedling development so that functional linkage to a receptor is unclear. It has been shown that sPLA2α knockouts showed reduced elongation of stems and petioles and both knockouts and overexpressors were delayed in gravitropic bending of hypocotyls (Lee et al., 2003). In a more recent investigation, the phenotype of sPLA2α mutants suggests that they are involved in PIN protein cycling in roots and, thus, auxin transport regulation (Lee et al., 2010). It seems quite possible that the metabolites generated by sPLA2α, free fatty acids (FFAs) and/or lysophospholipids, exert effects on membrane vesicle transport-related functions which are the basis for auxin transport.
Time courses of activation of sPLA2α activity by auxin are not available. The transcriptional increase of RNA after 60 min by auxin has been reported (Lee et al., 2003). In fact, it is difficult to imagine how the secreted sPLA2 enzymes can be activated by a receptor on the extracytosolic side of membranes in Golgi dictyosomes (Lee et al., 2010). Even though ABP1 is also located on the extracytosolic side of ER and plasma membrane, there is no signal transmission paradigm known from the literature to suggest post-translational activation of an extracytosolic enzyme. Unfortunately, no enzyme inhibitors are available which can distinguish between sPLA2 and pPLA and could be used to test functions depending on either one. Summing up, while defects in auxin physiology in sPLA2 mutants were described a receptor cannot be pointed out and transcription/translation seems to be the mechanism of sPLA2 activation.
Sphingolipids and Auxin Functions
Other work on sphingolipid biosynthesis shows that a decrease to very long chain fatty acid levels – derived from sphingolipids – can affect several auxin functions, in particular, auxin-dependent inhibition of lateral root emergence and an abnormal patterning of the embryo apex, which leads to defective cotyledon organogenesis (Roudier et al., 2010; Markham et al., 2011). Defective organogenesis is thought to be associated with the mistargeting of the auxin efflux carrier PIN1 in specific cells, resulting in local alteration of polar auxin distribution. This is remedied by the addition of very long chain fatty acids. If very long chain fatty acids or (sphingo)lipids containing them are necessary for correct auxin transport, then the absence of the relevant enzyme in biosynthesis or hydrolysis is enough to disturb auxin functions. A link to a receptor is not necessary and not known here. Furthermore, regulation of expression of such enzymes by (other) signals could have an impact on phenotypes. Because of the many open questions no gene/enzyme of sphingolipid metabolism is added to Figure 2.
Patatin-Related Phospholipases a in Auxin Signal Transduction
The third group of phospholipases in auxin signal transduction are the pPLA. In contrast to PLD or sPLA2, receptor-triggered rapid activation and a post-translational activation mechanism can be suggested for pPLA’s, and defects in auxin-related functions in mutants of single isoforms were found. Receptor coupling, post-translational activation, and linkage to downstream functions are the hallmark of signal transduction enzymes.
The first report on auxin activating a non-identified PLA activity was probably on pPLA activation (Scherer and André, 1989). Later, by using selective inhibitors in physiological and transcriptional experiments (Scherer and Arnold, 1997; Paul et al., 1998; Holk et al., 2002; Scherer et al., 2007) and characterization of knockout mutants (Rietz et al., 2010), we showed that the auxin-activated PLA activity was a pPLA activity of one or more isoforms.
The rapid activation kinetics of pPLA activation within 3 min suggests that ABP1 is the relevant receptor (Scherer, 2011). The low discrimination between 2,3-D and 2,4-D of pPLA activation (Scherer and André, 1989; Paul et al., 1998) is reflected in the binding properties with low discrimination of ABP1 to these auxins (Ray, 1977).
In order to better understand the potential link between pPLA and the two receptors ABP1 and TIR1, we chose regulation of early auxin genes and the artificial auxin-activated gene DR5::GUS as a response to distinguish between the two receptors (Scherer et al., 2007). Blocking pPLA activity with two different enzyme inhibitors blocked both auxin-included DR5::GUS expression and early auxin-induced genes. TIR1-directed hydrolytic degradation of IAA17-luciferase hybrid protein, however, was not affected by the inhibitors. This allowed us to conclude that TIR1 was not a receptor for pPLA activation and ABP1 was the more likely receptor relevant for pPLA activation.
Coordination of Two Auxin Receptors, ABP1 and TIR1, by PIN Proteins Catalyzing Auxin Efflux Transport
Testing Rapid Auxin-Induced Gene Regulation in ABP1 and Other Mutants Indicates ABP1 as Receptor also for Gene Regulation
The need to decide which receptor could initiate rapid responses has led to investigations on ABP1. Whereas the homozygous abp1 knockout plants are embryo-lethal (Chen et al., 2001), heterozygous plants are viable and have a phenotype clearly deficient in physiological auxin responses. An even clearer indicator was, that auxin-induced gene regulation is disabled as soon as after 30 min of auxin application in abp1/+ (Effendi et al., 2011). Our data show that ABP1 regulates early auxin-induced genes just as TIR1 does. Furthermore, we have found that the pin2/eir1 mutant showed a high percentage of misregulation of early auxin-regulated genes including PIN genes (Effendi and Scherer, 2011) inasmuch as they are regulated by auxin (Vieten et al., 2005). Using quite different techniques, two other groups have shown in molecular detail that PIN1 and PIN2 protein activity is regulated by ABP1 but not by TIR1 within minutes by the slowing of endocytosis (Robert et al., 2010; Xu et al., 2010).
The main questions remain: if ABP1 and PIN2 (and also TIR1 as a well-known regulator of auxin-induced genes) have a rapidly established influence on auxin gene regulation, then how can pPLA be integrated in this? We tested this, quite similarly as before, by testing early auxin-induced gene regulation in pPLA knockout plants of the genes pPLAIIγ, pPLAIIδ, and pPLAIIε (Figure 3; methods as in Effendi et al., 2011). Of the seven genes tested, two to five genes were not up-regulated within 30 min. The affected number of misregulated genes is less than in abp1/+ or pin2/eir1 mutants, but is considerable nonetheless. Along with the data on auxin-related phenotypes of these three knockout mutants (Rietz et al., 2010), this provides a strong argument that pPLA is involved in auxin signal transduction. These pPLA-II genes themselves were not regulated by auxin (not shown).
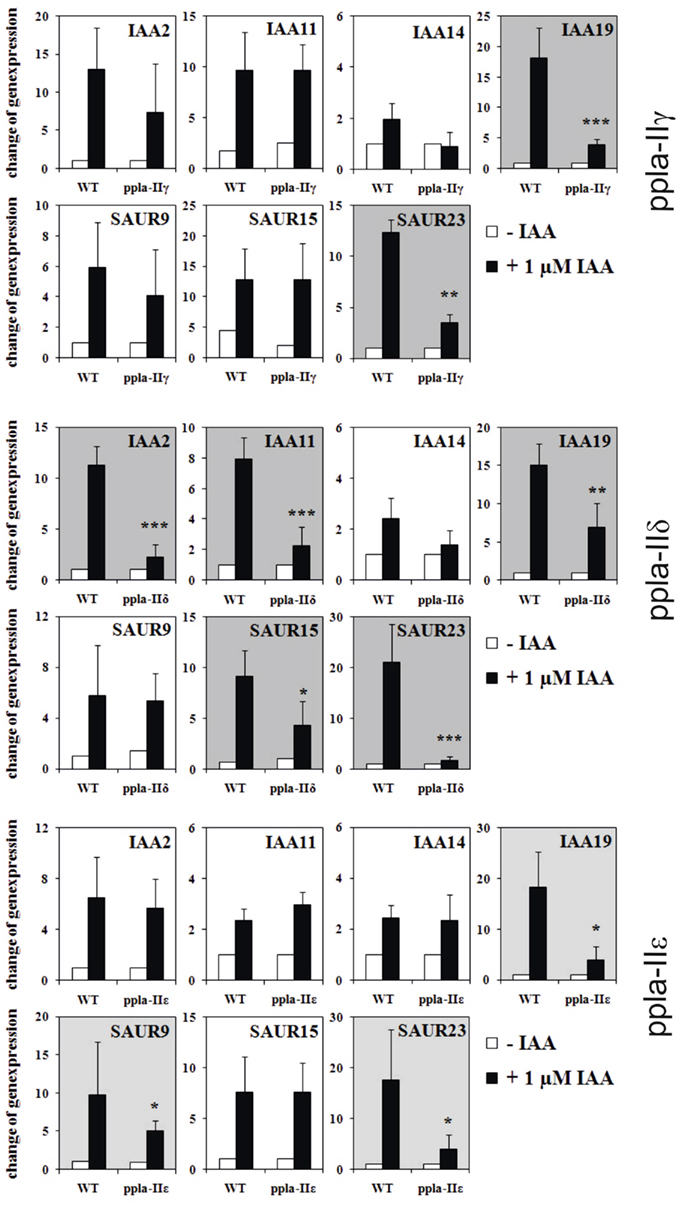
Figure 3. Regulation of early auxin-induced genes in light-grown pPLA group II knockouts and corresponding wild-type seedlings. Description of knockout plants was reported (Rietz et al., 2010). Mutant seedlings (ppla-IIγ, ppla-IIδ, ppla-IIε) and wild-type seedlings were grown in MS/2 liquid medium for 7 days under long day conditions. The seedlings were equilibrated for 4 h in fresh medium and treated either with 1 μM IAA or with MS/2 liquid medium for 30 min. The results are from four to six biological treatments with three technical repeats for each measurement. All other methods are as published (Effendi et al., 2011). Asterisks above bars indicate significant differences between the mutants and the corresponding wild-type treatments at *p < 0.05, **p < 0.01, and ***p < 0.001 level following the t-test.
A hypothetical picture emerges where calcium-dependent processes can be linked to PLD to one “module” and a second “module” where pPLA is an upstream component, associated with ABP1 as a receptor. TIR1 is viewed as a third and “nuclear module” regulated by auxin concentration. Thus our model emphasizes the regulation of PIN efflux-transport protein-activity by ABP1, which in turn will regulate cytosolic and/or nuclear auxin concentration, to which TIR1 reacts accordingly. It would be no surprise if other auxin transporters, e.g., the AUX1/LAX influx transporters or PGB auxin efflux transporters, will have to be welded into this concept.
Auxin efflux transport and hormone concentration regulation coupled to two receptors is the central idea of our new concept for auxin signal transduction.
In a Two Receptor Concept Regulation of Auxin Concentration by PIN Proteins is the Principle that Couples and Coordinates ABP1 and TIR1
If our model of coupling ABP1 to gene regulation executed by TIR1 is correct, this means that PIN activities are probably first regulated at the protein level, and then some of them later at the transcriptional level. Phosphorylation by the AGC3 kinases, among which PINOID seems to be the most important, leads to endocytosis inhibition, so more PIN protein remains at the plasma membrane (Dhonukshe et al., 2010; Huang et al., 2010) leading to rapid stimulation of auxin efflux (Paciorek et al., 2005). This mechanism also establishes a higher polarity of PIN localization at the basal or apical end or laterally (Friml et al., 2004; Michniewicz et al., 2007; Ding et al., 2011; Rakusová et al., 2011) and establishes polarity on an organ basis (Friml et al., 2003; Michniewicz et al., 2007). Our model refers only to the single cell situation where fewer PIN proteins – after auxin application – in the plasma membrane lead to lower efflux activity and the consequently higher cytosolic auxin concentration, thus exerting control in the nucleus of expression of TIR1-dependent genes. The multicellular effect of streamlined polarity of PIN proteins in tissues leading, for instance, to lateral root formation, needs coordination of many more reactions. Ectopic expression of PIN genes in root hairs showed that PIN1, PIN2, PIN3, and PIN8 (Ganguly et al., 2010) are the most actively regulated proteins in their root hair-growth test. As mentioned, PIN8 is probably not phosphorylated by AGC3 kinase so signal input into PIN8 regulation remains unclear. PIN2 and PIN3 can change their cellular polarity, PIN3 within 3 min (Friml et al., 2002; Harrison and Masson, 2008) which is the basis of tissue responses relying on polarity, like phototropism and gravitropism. Direct regulation of transport activity of PIN proteins as a possibility has not been reported.
PIN5 is different from all other PIN proteins. It is localized in the perinuclear ER (Mravec et al., 2009) and its expression is down-regulated by auxin (Mravec et al., 2009; Effendi and Scherer, 2011). PIN8 is expressed both in the ER and the plasma membrane (Mravec et al., 2009; Ganguly et al., 2010). Mravec et al. (2009) postulated that PIN5 concentrates auxin in the nucleus by an unknown mechanism; the first experimental evidence for this hypothesis was provided by Ganguly et al. (2010) in their root-hair over-expression assay. PIN5 and PIN8 do not posses the long cytosolic loop which carries the phosphorylation sites for the AGC kinases (Huang et al., 2010). The function of PIN5 may be to increase auxin concentration in the nucleus (Mravec et al., 2009), but the mode of regulatory input is currently unclear, and the question how an auxin gradient is kept up against the nuclear pores is – literally – open.
After several hours, auxin-induced changes in expression of most PIN proteins complicate the interpretation (Vieten et al., 2005). In contrast, the effect on transport was detectable as rapidly as after 4 min (Paciorek et al., 2005, Figure 2O), which is in line both with our predictions in our model in Figure 2 and the previously suggested model (Effendi et al., 2011). Therefore, only rapid methods or measurements will show the initial response pattern, later it may be obscured by additional responses such as transcription (Vieten et al., 2005; Braun et al., 2008). Our conclusion is that ABP1 and these pPLA-II genes are in the same signal transduction pathway because of the rapid effects on auxin-induced genes in abp/+ and ppla-II mutants.
Upstream Activators and Downstream Consequences of pPLA Activation by Auxin
Protein Kinases and Other Proteins Potential Upstream Activators of PPLA
A potential molecular mechanism to activate group II pPLA enzymes by phosphorylation at the termini has been shown (Rietz et al., 2010). So far, three group II enzymes have been investigated and all were phosphorylated by CPKs. Two, pPLA-IIδ and pPLA-IIε, were activated by phosphorylation by CPK3. Auxin-induced calcium influx could activate CPK3 (Monshausen et al., 2011). When we inspected the C-termini of all 10 pPLA isoforms, several potential phosphorylation sites were suggested which, however, appear to be different for groups II and III enzymes (Figure 4). Among amino acids of conserved serines in the C-terminus of group II enzymes, lysines and arginines are abundant (relative position 30 and 40 in Figure 4), which could fit to the substrate preferences of basophilic CPKs (de la Fuente van Bentem et al., 2008). The serine at relative position 39 was indeed phosphorylated by CPK3, which activated pPLA-IIγ and pPLA-IIε enzymatic activity (Rietz et al., 2010). The conserved serines at position 29 (Figure 4) in group II enzymes are followed by two acidic glutamates, which means that this site can be phosphorylated by the acidophilic casein kinase2 (de la Fuente van Bentem et al., 2008). In group III enzymes, the surrounding of conserved serines/threonines is quite different. The preference for basic amino acids is much less pronounced (relative position 17 and 43) and, conspicuously, proline is associated with the serine at position 43 (Figure 4). Further upstream, a conserved serine is present at relative position 6 in Figure 4 in all 10 sequences which, however, does not show any known features of substrate peptides of kinases. Thus, multiple pathways of activation by phosphorylation could be operating on pPLA enzymes.
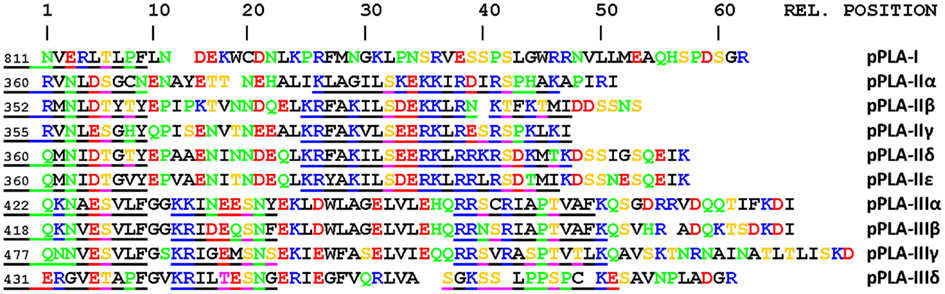
Figure 4. Potential phosphorylation sites in the C-termini of pPLA enzymes. Sequences are aligned to highlight maximize homology. The number at the beginning of sequences indicates the position of the first amino acid shown within this sequence. The scale on top is used to refer to positions of potential phosphorylation sites. Some N-termini are at the right are deleted. The color code is black for aliphatic or lipophilic amino acids, green for hydrophilic amino acids, red for acidic amino acids, and blue for basic amino acids. Serines and threonines are in yellow. Sites with obvious homologous serines/threonines are underlined. Phosphorylation sites for basophilic CDKs are K/R-x2-pS-h-D/E, for SnF1-related kinase K/R-x2-K/R-x2-pS, for the acidophilic CK2 it is pS-D/E-D/E-D/E (de la Fuente van Bentem et al., 2008). Phosphorylation at relative position 40 was shown with CPK3 in pPLA-IIγ, pPLA-IIε, and pPLA-IIδ and phosphorylation activated pPLA-IIγ and pPLA-IIε (Rietz et al., 2010). Other phosphorylation sites are hypothetical.
Therefore, we integrated both CPK3 and CK2 into the model upstream of pPLA (Figure 2). CK2 is not known to be activated by calcium or second messengers (Cozza et al., 2010; Montenarh, 2010). The ck2 dexamethasone-induced antisense plants have a pleiotropic auxin phenotype (Marquès-Bueno et al., 2011). However, one should keep in mind that other targets for CK2 could exist, e.g., PDK1 phosphorylating PINOID, explaining a pleiotropic auxin phenotype. Therefore, CK2 is also placed upstream of PDK1 into a different context in Figure 2.
The single group I enzyme, pPLA-I, is the evolutionarily oldest in the group and does not possess the suggested phosphorylation sites as highlighted for the other sequences (Figure 4). In the N-terminal non-enzymatic domain of this isoform, we found a region homologous to binding sites of small G proteins and also homologous to parts of importin α (unpublished). Interestingly, the knockouts of this gene have agravitropic and aphototropic hypocotyls and many early auxin-induced genes were not up-regulated by 10 μM auxin after 30 min in ppla-I knockouts and the overall phenotype of abp1/+ and ppla-I knockouts are quite similar (Y. Effendi, K. Raddatz, S. Rietz, C. Labusch, M. Wimalasekera, M. Zeidler, and G.F.E. Scherer, in preparation). Small G proteins have their place in auxin signal transduction (Tao et al., 2002, 2005; Wu et al., 2011) and, more recently, have been associated with ABP1 as the relevant receptor (Xu et al., 2010). We hypothesize that ABP1 could be functionally linked to pPLA-I via small G protein(s) (Figures 2 and 4).
pPLA Hydrolysis Products, Free Fatty Acids and Lysolipids Participating in Auxin Signaling
The hydrolysis products of phospholipases are FFAs and lysophospholipids. Either they themselves or their derivatives should have a second messenger function. They are postulated to carry out functions downstream from pPLA in auxin signal transduction. Metabolites generated by sPLA2 enzymes must be viewed as equivalent provided they are identical and present in identical compartments.
Accumulation of lysolipids as a response to auxin treatment could only be observed at very high auxin concentration (Scherer and André, 1989; Scherer, 1995; Paul et al., 1998), so that lysolipids seem not to be potential second messengers for physiological concentrations of auxin as a signal. Reacylation of lysolipid is quite active (Schwartze and Roos, 2008), which may explain why LPC accumulation is found only at high auxin concentrations. That PLA hydrolysis is followed by rapid conversion, e.g., to acyl-CoA of FFA or reacylation of lysolipids (Larsson et al., 2007; Schwartze and Roos, 2008), may be a necessary consequence because both FFA and lysolipids could be membrane-perturbing substances at higher concentrations. Reacylation also prevents an overshooting response to the signal. If the pPLA isoforms preferentially hydrolyze lipids having a specific fatty acid composition (possibly determined also by the headgroup), it is not an average set of FFA which is liberated, but a selected mixture of FFAs. This could be one possibility to provide unique functions to each pPLA isoform.
Downstream Effectors of FFA and Lysolipids Could Be Lipid-Activated or Calcium-Activated Protein Kinases
In earlier work, lipid regulation of protein kinase and protein phosphatase was investigated. Knockout mutants or other mutants were generally not available before about 2000, so molecular identification of the kinases and phosphatases was not reported in this early work (reviewed in Scherer, 1996; Scherer, 2010). A calcium-dependent CPK activity was reported to be activated by fatty acids, but only in the absence of calcium (Klucis and Polya, 1987; Lucantoni and Polya, 1987). In as much as AGC kinases have homology to the animal lipid-activated protein kinase C one might even speculate that AGC kinases cold be activated by lipids binding to their hydrophobic pocket (Frödin et al., 2002; Bögre et al., 2003). We showed that lysolipids phosphorylate specific proteins in isolated membrane fractions (Martiny-Baron and Scherer, 1989), but lipid specificity was limited (Scherer et al., 1993), and the relevant kinases could not be identified. In summary, lipid-activated protein kinase(s) are an eagerly awaited link in plant signal transduction.
Conclusion and Outlook
The postulate of a regulatory triad, ABP1, TIR1, and PIN proteins (or auxin transport in general, perhaps also catalyzed by other auxin transporters, respectively), is the novel aspect of our model. This provides pPLA’s and PLD a place in auxin signal transduction and several more seemingly disparate topics in auxin signal transduction can be combined into one model. Regulation of auxin transport is initiated and conducted by cytosol-based enzymatic post-translational regulatory reactions. Transcriptional regulation – initiated by TIR1 – of early auxin-induced genes is dependent on auxin concentration changes. This makes auxin-induced gene regulation a third “module” in signal transduction besides the “Ca2+/PLD model” and the “pPLA module.” Whether, alternatively, there exists a regulatory pathway from ABP1 more directly to the nucleus remains open. We envisage regulation of plasma membrane-located PIN proteins as important in regulating cytosolic auxin concentration (Scherer, 2011). PIN proteins are regulated by endocytosis/exocytosis balances which, in turn, are regulated by PINOID-catalyzed protein phosphorylation (Kleine-Vehn and Friml, 2008; Huang et al., 2010).
The potential targets of lysolipids and fatty acids are summarized in Figure 5. The lack of a precise function of fatty acids and lysolipids in auxin signaling is a tremendous challenge in auxin lipid signaling to be resolved in future research. CPK activation by fatty acids is one possibility, as it could lead to phosphorylation of critical proteins. The second possibility is that desaturated fatty acids activate PLDδ (Wang and Wang, 2001) and generate PA to activate PDK1 and, eventually, PINOID (Zegzouti et al., 2006). The third hypothesis would be that lipidic messengers reach the nucleus and influence transcription within 10 min (Figure 4), which is a remote possibility, though its simplicity makes it rather attractive.
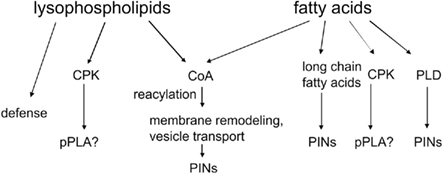
Figure 5. Potential downstream effectors of pPLA or sPLA2 metabolites. Explanations are in the text.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Support from the Deutsches Zentrum für Luft- und Raumfahrt (contract number 50WB0627 and 50BW0933) and from the Deutsche Forschungsgemeinschaft (Sche207/24-1) is gratefully acknowledged. K. Hunter helped with language corrections.
References
Abel, S., Oeller, P. W., and Theologis, A. (1994). Early auxin-induced genes encode short-lived nuclear proteins. Biochemistry 33, 326–330.
Ballas, N., Wong, L.-M., and Theologis, A. (1993). Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum). J. Mol. Biol. 233, 580–596.
Bögre, L., Okrész, L., Henriques, R., and Anthony, R. G. (2003). Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci. 8, 424–431.
Braun, N., Wyrzykowska, J., Muller, P., David, K., Couch, D., Perrot-Rechenmann, C., and Fleming, A. J. (2008). Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic development in Arabidopsis and tobacco. Plant Cell 20, 2746–2762.
Calderon-Villalobos, L. I., Kuhnle, C., Li, H., Rosso, M., Weisshaar, B., and Schwechheimer, C. (2006). LucTrap vectors are tools to generate luciferase fusions for the quantification of transcript and protein abundance in vivo. Plant Physiol. 141, 3–14.
Chen, J. G., Ullah, H., Young, J. C., Sussman, M. R., and Jones, A. M. (2001). ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 15, 902–911.
Cozza, G., Bortolato, A., and Moro, S. (2010). How druggable is protein kinase CK2? Med. Res. Rev. 30, 419–462.
de Jong, C. F., Laxalt, A. M., Bargmann, B. O., de Wit, P. J., Joosten, M. H., and Munnik, T. (2004). Phosphatidic acid accumulation is an early response in the Cf-4/Avr4 interaction. Plant J. 39, 1–12.
de la Fuente van Bentem, S., Anrather, D., Dohnal, I., Roitinger, E., Csaszar, E., Joore, J., Buijnink, J., Carreri, A., Forzani, C., Lorkovic, Z. J., Barta, A., Lecourieux, D., Verhounig, A., Jonak, C., and Hirt, H. (2008). Site-specific phosphorylation profiling of Arabidopsis proteins by mass spectrometry and peptide chip analysis. J. Proteome Res. 7, 2458–2470.
den Hartog, M., Verhoef, N., and Munnik, T. (2003). Nod factor and elicitors activate different phospholipid signaling pathways in suspension-cultured alfalfa cells. Plant Physiol. 132, 311–317.
Dhonukshe, P., Huang, F., Galvan-Ampudia, C. S., Mähönen, A. P., Kleine-Vehn, J., Xu, J., Quint, A., Prasad, K., Friml, J., Scheres, B., and Offringa, R. (2010). Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137, 3245–3255.
Ding, Z., Galván-Ampudia, C. S., Demarsy, E., Langowski, L., Kleine-Vehn, J., Fan, Y., Morita, M. T., Tasaka, M., Fankhauser, C., Offringa, R., and Friml, J. (2011). Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat. Cell Biol. 13, 4474–4452.
Dreher, K., and Callis, J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. 99, 787–822.
Effendi, Y., Rietz, S., Fischer, U., and Scherer, G. F. (2011). The heterozygous abp1/ABP1 insertional mutant has defects in functions requiring polar auxin transport and in regulation of early auxin-regulated genes. Plant J. 65, 282–294.
Effendi, Y., and Scherer, G. F. (2011). AUXIN BINDING-PROTEIN1 (ABP1), a receptor to regulate auxin transport and early auxin genes in an interlocking system with PIN proteins and the receptor TIR1. Plant Signal. Behav. 6, 1101–1103.
Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jürgens, G.. (2003). Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426, 147–153.
Friml, J., Wisniewska, J., Benkova, E., Mendgen, K., and Palme, K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809.
Friml, J., Yang, X., Michniewicz, M., Weijers, D., Quint, A., Tietz, O., Benjamins, R., Ouwerkerk, P. B., Ljung, K., Sandberg, G., Hooykaas, P. J., Palme, K., and Offringa, R. (2004). A PINOID-dependent binary switch in apical–basal PIN polar targeting directs auxin efflux. Science 306, 862–865.
Frödin, M., Antal, T. L., Dümmler, B. A., Jensen, C. J., Deak, M., Gammeltoft, S., and Biondi, R. M. (2002). A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J. 21, 5396–5407.
Ganguly, A., Lee, S. H., Cho, M., Lee, O. R., Yoo, H., and Cho, H. T. (2010). Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiol. 153, 1046–1061.
Harrison, B. R., and Masson, P. H. (2008). ARL2, ARG1 and PIN3 define a gravity signal transduction pathway in root statocytes. Plant J. 53, 380–392.
Holk, A., Rietz, S., Zahn, M., Paul, R. U., Quader, H., and Scherer, G. F. E. (2002). Molecular identification of cytosolic, patatin-related phospholipases A from Arabidopsis with potential functions in plant signal transduction. Plant Physiol. 130, 90–101.
Huang, F., Zago, M. K., Abas, L., van Marion, A., Galván-Ampudia, C. S., and Offringa, R. (2010). Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22, 1129–1142.
Jones, A. M., and Assmann, S. M. (2004). Plants: the latest model system for G-protein research. EMBO Rep. 5, 572–578.
Klämbt, D. (1990). A view about the function of auxin-binding proteins at plasma membranes. Plant Mol. Biol. 14, 1045–1050.
Kleine-Vehn, J., and Friml, J. (2008). Polar targeting and endocytic recycling in auxin-dependent plant development. Annu. Rev. Cell Dev. Biol. 24, 447–473.
Klucis, E., and Polya, G. M. (1987). Calcium-independent activation of two plant leaf calcium-regulated protein kinases by fatty acids. Biochem. Biophys. Res. Commun. 147, 1041–1047.
Krinke, O., Flemr, M., Vergnolle, C., Collin, S., Renou, J. P., Taconnat, L., Yu, A., Burketová, L., Valentová, O., Zachowski, A., and Ruelland, E. (2009). Phospholipase D activation is an early component of the salicylic acid signaling pathway in Arabidopsis cell suspensions. Plant Physiol. 150, 424–436.
Lanteri, L. L., Laxalt, A. M., and Lamattina, L. (2008). Nitric oxide triggers phosphatidic acid accumulation via phospholipase D during auxin-induced adventitious root formation in cucumber. Plant Physiol. 147, 188–198.
Larsson, K. E., Kjellberg, J. M., Tjellström, H., and Sandelius, A. S. (2007). LysoPC acyltransferase/PC transacylase activities in plant plasma membrane and plasma membrane-associated endoplasmic reticulum. BMC Plant Biol. 7, 64. doi:10.1186/1471-2229-7-64
Lee, H. Y., Bahn, S. C., Kang, Y.-M., Lee, K. H., Kim, H. J., Noh, E. K., Palta, J. P., Shin, J. S., and Ryu, S. B. (2003). Secretory low molecular weight phospholipase A plays important roles in cell elongation and shoot gravitropism in Arabidopsis. Plant Cell 15, 1990–2002.
Lee, O. R., Kim, S. J., Kim, H. J., Hong, J. K., Ryu, S. B., Lee, S. H., Ganguly, A., and Cho, H. T. (2010). Phospholipase A2 is required for PIN-FORMED protein trafficking to the plasma membrane in the Arabidopsis root. Plant Cell 22, 1812–1825.
Li, G., and Xue, H. W. (2007). Arabidopsis PLDzeta2 regulates vesicle trafficking and is required for auxin response. Plant Cell 19, 281–295.
Li, M., Hong, Y., and Wang, X. (2009). Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochim. Biophys. Acta 1791, 927–935.
Lucantoni, A., and Polya, G. M. (1987). Activation of wheat embryo Ca2+-regulated protein kinase by unsaturated fatty acids in the presence and absence of calcium. FEBS Lett. 221, 33–36.
Markham, J. E., Molino, D., Gissot, L., Bellec, Y., Hématy, K., Marion, J., Belcram, K., Palauqui, J. C., Satiat-Jeunemaître, B., and Faure, J. D. (2011). Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell 23, 2362–2378.
Marquès-Bueno, M. M., Moreno-Romero, J., Abas, L., De Michele, R., and Martínez, M. C. (2011). A dominant negative mutant of protein kinase CK2 exhibits altered auxin responses in Arabidopsis. Plant J. 67, 169–180.
Martiny-Baron, G., and Scherer, G. F. E. (1989). Phospholipid-stimulated protein kinase in plants. J. Biol. Chem. 264, 18052–18059.
Meijer, H. J., and Munnik, T. (2003). Phospholipid-based signaling in plants. Annu. Rev. Plant Biol. 54, 265–306.
Michniewicz, M., Zago, M. K., Abas, L., Weijers, D., Schweighofer, A., Meskiene, I., Heisler, M. G., Ohno, C., Zhang, J., Huang, F., Schwab, R., Weigel, D., Meyerowitz, E. M., Luschnig, C., Offringa, R., and Friml, J. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130, 1044–1056.
Mishra, G., Zhang, W., Deng, F., Zhao, J., and Wang, X. (2006). A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312, 264–266.
Mockaitis, K., and Estelle, M. (2008). Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24, 55–80.
Monshausen, G. B., Miller, N. D., Murphy, A. S., and Gilroy, S. (2011). Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high resolution imaging with automated computer-vision-based analysis. Plant J. 65, 309–318.
Mravec, J., Skupa, P., Bailly, A., Hoyerová, K., Krecek, P., Bielach, A., Petrásek, J., Zhang, J., Gaykova, V., Stierhof, Y. D., Dobrev, P. I., Schwarzerová, K., Rolcík, J., Seifertová, D., Luschnig, C., Benková, E., Zazímalová, E., Geisler, M., and Friml, J. (2009). Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459, 1136–1140.
Paciorek, T., Zazimalova, E., Rudthardt, N., Petrasek, J., Stierhof, Y.-D., Klein-Vehn, J., Morris, D. A., Emans, N., Jürgens, G., Geldner, N., and Friml, J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435, 1251–1256.
Paul, R., Holk, A., and Scherer, G. F. E. (1998). Fatty acids and lysophospholipids as potential second messengers in auxin action. Rapid activation of phospholipase A2 activity by auxin in suspension-cultured parsley and soybean cells. Plant J. 16, 601–611.
Rakusová, H., Gallego-Bartolomé, J., Vanstraelen, M., Robert, H. S., Alabadí, D., Blázquez, M. A., Benková, E., and Friml, J. (2011). Polarization of PIN3-dependent auxin transport for hypocotyl gravitropic response in Arabidopsis thaliana. Plant J. 67, 817–826.
Ray, P. M. (1977). Specificity of auxin-binding sites on maize coleoptile membranes as possible receptor sites for auxin action. Plant Physiol. 60, 585–591.
Rietz, S., Dermendjiev, G., Oppermann, E., Tafesse, F. G., Effendi, Y., Holk, A., Parker, J. E., Teige, M., and Scherer, G. F. E. (2010). Roles of Arabidopsis patatin-related phospholipases A in root development related to auxin response and phosphate deficiency. Mol. Plant 3, 524–538.
Robert, S., Kleine-Vehn, J., Barbez, E., Sauer, M., Paciorek, T., Baster, P., Vanneste, S., Zhang, J., Simon, S., Covanová, M., Hayashi, K., Dhonukshe, P., Yang, Z., Bednarek, S. Y., Jones, A. M., Luschnig, C., Aniento, F., Zažímalová, E., and Friml, J. (2010). ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143, 111–121.
Roudier, F., Gissot, L., Beaudoin, F., Haslam, R., Michaelson, L., Marion, J., Molino, D., Lima, A., Bach, L., Morin, H., Tellier, F., Palauqui, J. C., Bellec, Y., Renne, C., Miquel, M., Dacosta, M., Vignard, J., Rochat, C., Markham, J. E., Moreau, P., Napier, J., and Faure, J. D. (2010). Very-long-chain fatty acids are involved in polar auxin transport and developmental patterning in Arabidopsis. Plant Cell 22, 364–375.
Scherer, G. F. E. (1995). Activation of phospholipase A by auxin and mastoparan in hypocotyl segments from zucchini and sunflower. J. Plant Physiol. 145, 483–490.
Scherer, G. F. E. (1996). Phospholipid signalling and lipid-derived second messengers in plants. Plant Growth Regul. 18, 125–133.
Scherer, G. F. E. (2010). “Phospholipase A in plant signal transduction,” in Lipid Signaling in Plants. Plant Cell Monographs, Vol. 16, ed. T. Munnik (Springer: Heidelberg), 3–22.
Scherer, G. F. E. (2011). AUXIN-BINDING-PROTEIN1, the second auxin receptor: what is the significance of a two-receptor concept in plant signal transduction? J. Exp. Bot. 65, 282–294.
Scherer, G. F. E., and André, B. (1989). A rapid response to a plant hormone: auxin stimulates phospholipase A2 in vivo and in vitro. Biochem. Biophys. Res. Commun. 163, 111–117.
Scherer, G. F. E., and Arnold, B. (1997). Auxin-induced growth is inhibited by phospholipase A2 inhibitors. Implications for auxin-induced signal transduction. Planta 202, 462–469.
Scherer, G. F. E., Führ, A., and Schütte, M. (1993). “Activation of membrane-associated protein kinase by lipids, its substrates, and its function in signal transduction,” Society for Experimental Biology Seminar Series 53: Post-translational Modifications in Plants, Vol. 53, eds N. H. Battey, H. G. Dickinson, and A. M. Hetherington (Cambridge, UK: Cambridge University Press), 109–121.
Scherer, G. F. E., Ryu, S. B., Wang, X., Matos, A. R., and Heitz, T. (2010). Patatin-related phospholipase A: nomenclature, subfamilies, and functions in plants. Trends Plant Sci. 15, 693–700.
Scherer, G. F. E., Zahn, M., Callis, J., and Jones, A. M. (2007). A role for phospholipase A in auxin-regulated gene expression. FEBS Lett. 581, 4205–4211.
Schütze, K., Harter, K., and Chaban, C. (2008). Post-translational regulation of plant bZIP factors. Trends Plant Sci. 13, 247–255.
Schwartze, W., and Roos, W. (2008). The signal molecule lysophosphatidylcholine in Eschscholzia californica is rapidly metabolized by reacylation. Planta 229, 183–191.
Shishova, M., and Lindberg, S. (2010). A new perspective on auxin perception. J. Plant Physiol. 167, 417–422.
Shishova, M., Yemelyanov, V., Rudashevskaya, E., and Lindberg, S. (2007). A shift in sensitivity to auxin within development of maize seedlings. J. Plant Physiol. 164, 1323–1330.
Stone, S. L., and Callis, J. (2007). Ubiquitin ligases mediate growth and development by promoting protein death. Curr. Opin. Plant Biol. 10, 624–632.
Tao, L. Z., Cheung, A. Y., Nibau, C., and Wu, H. M. (2005). RAC GTPases in tobacco and Arabidopsis mediate auxin-induced formation of proteolytically active nuclear protein bodies that contain AUX/IAA proteins. Plant Cell 17, 2369–2383.
Tao, L. Z., Cheung, A. Y., and Wu, H. M. (2002). Plant Rac-like GTPases are activated by auxin and mediate auxin-responsive gene expression. Plant Cell 14, 2745–2760.
van der Luit, A. H., Piatti, T., van Doorn, A., Musgrave, A., Felix, G., Boller, T., and Munnik, T. (2000). Elicitation of suspension-cultured tomato cells triggers the formation of phosphatidic acid and diacylglycerol pyrophosphate. Plant Physiol. 123, 1507–1516.
Vieten, A., Vanneste, S., Wisniewska, J., Benková, E., Benjamins, R., Beeckman, T., Luschnig, C., and Friml, J. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132, 4521–4531.
Wang, C., and Wang, X. (2001). A novel phospholipase D of Arabidopsis that is activated by oleic acid and associated with the plasma membrane. Plant Physiol. 127, 1102–1112.
Wu, H. M., Hazak, O., Cheung, A. Y., and Yalovsky, S. (2011). RAC/ROP GTPases and auxin signaling. Plant Cell 23, 1208–1218.
Xu, T., Wen, M., Nagawa, S., Fu, Y., Chen, J.-G., Wu, M.-J., Perrot-Rechenmann, C., Friml, J., Jones, A. M., and Yang, Z. (2010). Cell surface- and Rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143, 99–110.
Zegzouti, H., Anthony, R. G., Jahchan, N., Bögre, L., and Christensen, S. K. (2006). Phosphorylation and activation of PINOID by the phospholipid signaling kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 6404–6409.
Zhang, J., Vanneste, S., Brewer, P. B., Michniewicz, M., Grones, P., Kleine-Vehn, J., Löfke, C., Teichmann, T., Bielach, A., Cannoot, B., Hoyerová, K., Chen, X., Xue, H. W., Benková, E., Zažímalová, E., and Friml, J. (2011). Inositol trisphosphate-induced Ca2+ signaling modulates auxin transport and PIN polarity. Dev. Cell 20, 855–866.
Keywords: AUXIN-BINDING-PROTEIN1, auxin signal transduction, patatin-related phospholipase A, phospholipase D, PIN-FORMED PROTEIN, secreted phospholipase A2, TRANSPORT-INHIBITOR-RESISTANT1
Citation: Scherer GFE, Labusch C and Effendi Y (2012) Phospholipases and the network of auxin signal transduction with ABP1 and TIR1 as two receptors: a comprehensive and provocative model. Front. Plant Sci. 3:56. doi: 10.3389/fpls.2012.00056
Received: 10 November 2011; Paper pending published: 07 January 2012;
Accepted: 04 March 2012; Published online: 09 April 2012.
Edited by:
Xuemin Wang, University of Missouri-St Louis and Donald Danforth Plant Science Center, USAReviewed by:
Stephen B. Ryu, Korea Research Institute of Bioscience and Biotechnology, South KoreaKirk L. Pappan, Edenspace Systems Corporation, USA
Copyright: © 2012 Scherer, Labusch and Effendi. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Günther F. E. Scherer, Laboratory Molekulare Ertragsphysiologie, Institut für Zierpflanzenbau und Gehölzwissenschaften, Leibniz Universität Hannover, Herrenhäuser Str. 2, D30419 Hannover, Germany. e-mail:c2NoZXJlckB6aWVyLnVuaS1oYW5ub3Zlci5kZQ==



