
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 08 March 2012
Sec. Plant Physiology
Volume 3 - 2012 | https://doi.org/10.3389/fpls.2012.00041
This article is part of the Research TopicLipid signaling in plantsView all 22 articles
Members of the family of JASMONATE ZIM-DOMAIN (JAZ) proteins are key regulators of the jasmonate (JA) hormonal response. The 12-member family is characterized by three conserved domains, an N-terminal domain, a TIFY-containing ZINC-FINGER EXPRESSED IN INFLORESCENCE MERISTEM domain, and a C-terminal Jas domain. JAZ proteins regulate JA-responsive gene transcription by inhibiting DNA-binding transcription factors in the absence of JA. JAZ proteins interact in a hormone-dependent manner with CORONATINE INSENSITIVE 1 (COI1), the recognition component of the E3 ubiquitin ligase, SCFCOI1, resulting in the ubiquitination and subsequent degradation of JAZs via the 26S proteasome pathway. Since their discovery in 2007, JAZ proteins have been implicated in protein–protein interactions with multiple transcription factors. These studies have shed light on the mechanism by which JAZs repress transcription, are targeted for degradation, modulate the JA signaling response, and participate in crosstalk with other hormone signaling pathways. In this review, we will take a close look at the recent discoveries made possible by the characterization JAZ protein–protein interactions.
Ellis Marsalis, primum mobile of the diverse and influential jazz family, knows that the interactions and connections made by family members provide insight into the operation and purpose of an extended social network. For plant scientists, identifying and characterizing the proteins that interact with members of the family of 12 JASMONATE ZIM-DOMAIN (JAZ) proteins in Arabidopsis has proved to be an extremely rewarding approach to understand the mechanism and networking of jasmonate (JA) hormone signaling. In this review, we will briefly summarize the advances that have been made through the identification and characterization of JAZ protein interactions.
In contrast to mammals, which are able to cope with a changing environment by changing their behavior, plants rely heavily on their ability to adjust their developmental and metabolic programs to adapt to their surroundings. Understanding how plant hormones such as auxin, gibberellic acid (GA), ethylene, and JA regulate these processes is of central importance to increasing our understanding of plant biology. JA is a plant-specific oxylipin signaling molecule derived from α-linolenic acid, 18:3, or the homologous 16:3 fatty acid, found in chloroplast membranes. It is chemically related to the prostaglandins, some of the most studied and important oxylipins produced by mammals (Browse, 2009a). JA signaling is initiated in response to environmental and developmental cues (Mandaokar et al., 2006; Dombrecht et al., 2007). The hormone has been implicated in the regulation of a range of biological processes including fertility (McConn and Browse, 1996; Stintzi and Browse, 2000), root growth (Staswick et al., 1992), fruit ripening (Perez et al., 1997), trichome development (Qi et al., 2011), and senescence (Xiao et al., 2004), as well as the responses to both biotic stressors, such as feeding by insects (Howe et al., 1996; McConn et al., 1997) and infection by necrotrophic pathogens (Feys et al., 1994), and abiotic stressors including the responses to ozone, UV radiation (Conconi et al., 1996), salt, drought (Zhu, 2002), and mechanical wounding (Farmer et al., 1992; Reymond et al., 2000). Historically, much of our knowledge regarding the JA signaling response has resulted from the study of mutants deficient in crucial steps of JA synthesis and perception (Browse, 2009b). Recent discoveries, however, have relied heavily on the study of protein–protein interactions to elucidate the mechanism by which JA regulates gene expression. Arabidopsis mutants deficient in JA synthesis or perception pathways are abnormally susceptible to fungal pathogens (Vijayan et al., 1998) and herbivorous insects (Howe et al., 1996; McConn et al., 1997), exhibit a reduced response to JA-mediated root growth inhibition (Staswick et al., 1992) and are male sterile (McConn and Browse, 1996; Stintzi and Browse, 2000). Understanding the mechanistic details of the JA signaling pathway can provide a means to study and manipulate these vital processes. It also has important applications with respect to engineering the production of industrially useful chemicals in plants, since JA induces the production of an array of secondary metabolites. The induction of pathways for secondary product synthesis by JA is accompanied by the direction of resources away from photosynthesis and other growth processes to strike a crucial balance between growth and defense (Browse, 2009a; Pauwels et al., 2009).
There has been increasing interest in understanding the mechanism by which the JA nuclear signaling pathway is initiated and regulated, as well as in its effects at the level of transcription. Several recent studies have connected JA-mediated transcriptional changes with the regulation of other hormone signals such as auxin (Tiryaki and Staswick, 2002), ethylene (Pre et al., 2008; Adams and Turner, 2011; Zhu et al., 2011), GA (Navarro et al., 2008; Hou et al., 2011), and brassinosteroids (BR; Ren et al., 2009). Several recent reviews (Gazzarrini and McCourt, 2003; Kazan and Manners, 2008; Koornneef and Pieterse, 2008; Grant and Jones, 2009; Peng, 2009; Hoffmann et al., 2011; Santner and Estelle, 2011) have undertaken the daunting task of discussing the complex crosstalk that takes place within the hormone signaling network in plants; however, these topics will not be discussed at length here. Instead we aim to focus on the JAZ family of repressor proteins and the recent discoveries that have been made possible by the investigation of protein–protein interactions in which they are participants (summarized in Table 1). These studies indicate that JAZ proteins, which function as transcriptional repressors of the JA signaling response, are not merely regulators of the JA signaling pathway, but, through interaction with other proteins, also serve as signaling hubs in the wider hormone regulatory network, affecting multiple pathways and aspects of plant growth and metabolic programming (Pauwels and Goossens, 2011; Kazan and Manners, 2012).
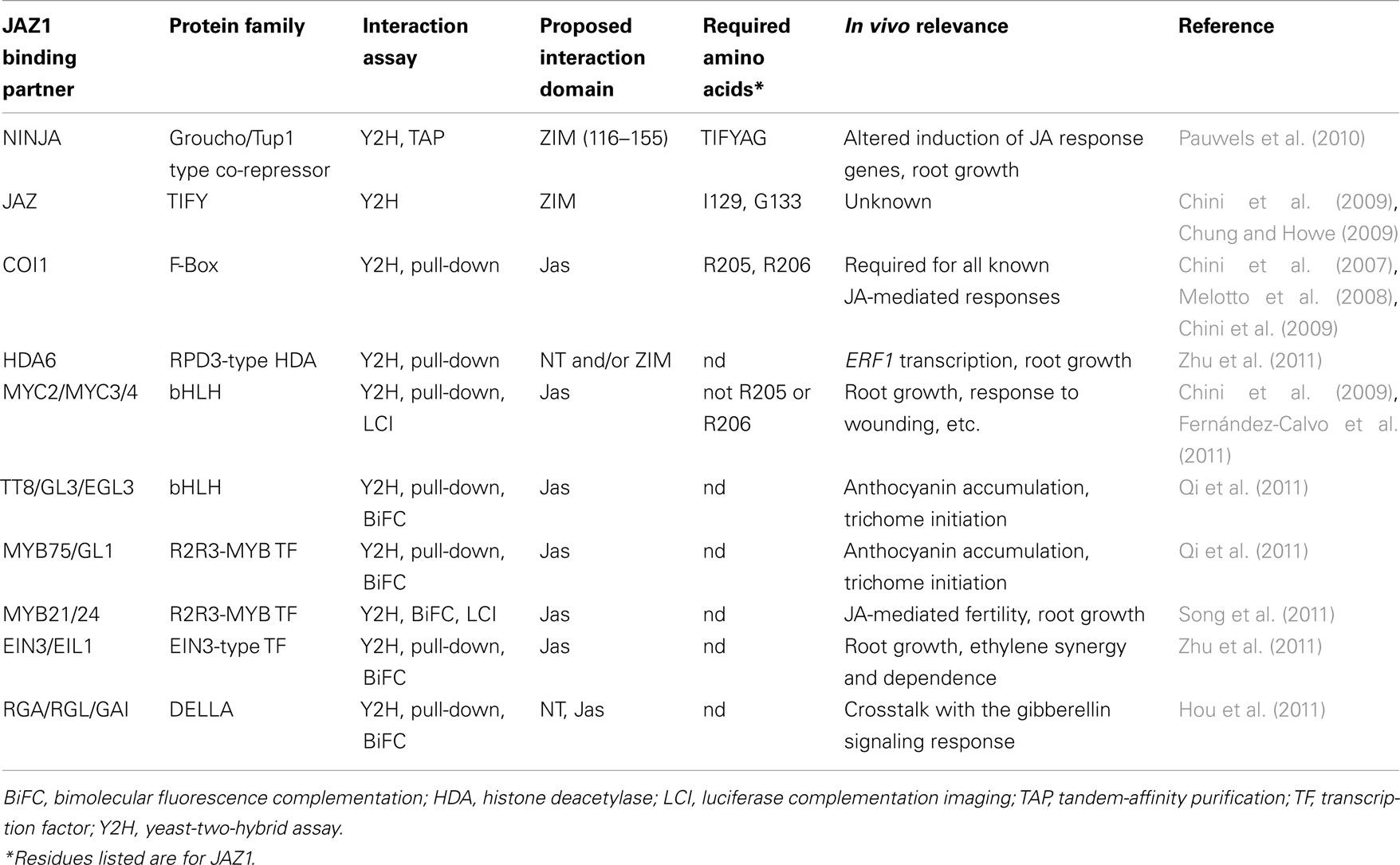
Table 1. Summary of our current knowledge of the protein–protein interactions in which one or more members of the JAZ family are participants.
Under normal growth conditions JA hormone levels are low and JA-mediated responses are kept in a repressed state by JAZ proteins. JAZs function as inhibitors of transcription factors that, when free of JAZ inhibition, act to activate transcription from JA-response genes. In response to stresses, such as those that result from insect feeding or necrotrophic pathogen infection, an increase in levels of JA–Ile allows the hormone to facilitate interaction between the JAZ repressors and the F-box protein CORONATINE INSENSITIVE 1 (COI1), the recognition component of the E3 ubiquitin ligase SCFCOI1. This interaction targets JAZs for ubiquitination and degradation via the 26S proteasome pathway, releasing DNA-binding transcription factors and allowing for the induction of JA responsive genes (Figure 1; Chini et al., 2007; Thines et al., 2007). Though protein–protein interactions have been required to elucidate the JA sensing mechanism, the COI1 protein was identified through the study of mutants. One of the first JA signaling mutants, coi1-1 (Feys et al., 1994), was identified in a screen for Arabidopsis seedlings resistant to root growth inhibition by coronatine, a bacterial toxin bearing structural similarity to JA–Ile, the isoleucine-conjugated derivative of JA later identified as the active form of the hormone. coi1-1 mutants were male sterile, resistant to root growth inhibition by JA, and deficient in all JA-related responses (Feys et al., 1994; Xie et al., 1998).
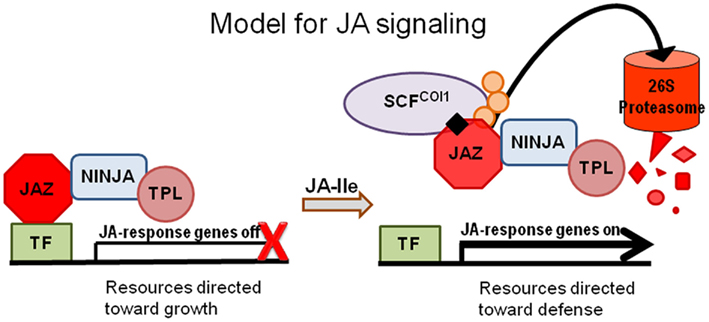
Figure 1. JAZ proteins bind transcription factors (TFs) and recruit co-repressors, including NINJA and TOPLESS (TPL), to repress gene transcription in the absence of the active form of jasmonate (JA), JA–Ile (black diamond). In the presence of JA–Ile, the hormone facilitates interaction between JAZ proteins and the F-box protein COI1, the recognition component of the E3 ubiquitin ligase SCFCOI1. As a result, JAZ proteins are ubiquitinated (orange circles) and subsequently degraded in the 26S proteasome releasing TFs from inhibition and activating JA-responsive gene transcription.
CORONATINE INSENSITIVE 1 is an F-box protein that is closely related to TIR1, the recognition component of SCFTIR1, an E3 ligase that targets AUX/IAA proteins for degradation via the 26S proteasome pathway in response to the growth hormone auxin (Gray et al., 2001; Dharmasiri et al., 2005; Browse, 2009a). This homology, together with protein–protein interaction data showing that COI1 associates with SKP2/ASK1 and CULLIN, components of the SCF complex (Devoto et al., 2002), suggested that JA signaling also depended on the action of an E3 ubiquitin ligase (Gray et al., 2001; Dharmasiri et al., 2005; Browse, 2009a). Further work identifying MYC2 as a transcription factor required for full JA responses provided researchers with enough information to propose a relief-of-repression model for JA signaling (Figure 1) in which transcription factors, such as MYC2, are inhibited by repressor proteins (Berger et al., 1996; Lorenzo et al., 2004). According to the model, in response to JA these repressor proteins would be targeted for degradation through the SCFCOI1/26S proteasome pathway, liberating DNA-binding transcription factors and allowing for transcription of JA-responsive genes (Chico et al., 2008; Browse, 2009a).
Mutant screens were less helpful in the search for targets of the SCFCOI1 complex. The identity of the repressors of JA-mediated responses remained unclear until 2007, when three research groups independently published work identifying the JAZ family of proteins as the much sought after targets of COI1. JAZ proteins were identified by transcriptional profiling of a JA synthesis mutant, opr3, by analysis of the dominant JA-resistant mutant jai3-1, and by characterization of a splice variant of JAZ10, JAZ10.3. This work showed that degradation of JAZ proteins is essential for induction of JA-mediated responses, an action that is dependent on the F-box protein COI1 and the 26S proteasome (Mandaokar et al., 2006; Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). These findings provided the basis for the current model for JA signaling in which JA–Ile serves as a signal that activates transcription of JA responsive genes by enhancing interaction between JAZ repressor proteins and COI1, the recognition component of the SCFCOI1 E3 ligase (Devoto et al., 2002; Staswick et al., 2002; Melotto et al., 2008). This action leads to the ubiquitination of JAZs and their subsequent degradation in the 26S proteasome. This action releases transcription factors, such as MYC2, from inhibition and provides a mechanism for the activation of JA-mediated transcriptional responses (Figure 1; Devoto et al., 2002; Lorenzo et al., 2004; Chini et al., 2007; Thines et al., 2007).
Protein–protein interactions play important roles in nearly all cellular processes. They make possible the formation of large complexes of transcriptional regulators, aid in signal transduction, and facilitate cellular protein targeting. JAZ proteins are members of the larger TIFY group, which also includes ZINC-FINGER EXPRESSED IN INFLORESCENCE MERISTEM (ZIM), ZIM-LIKE 1 (ZML1), ZIM-LIKE 2 (ZML2), PEAPOD proteins, PPD1 and PPD2, as well as the protein TIFY8 (Vanholme et al., 2007; Bai et al., 2011). The ZIM and ZML proteins are putative transcription factors containing a GATA-Zinc finger, but JAZ repressors lack an identifiable DNA-binding domain; thus their function as transcriptional regulators must rely on their ability to be recruited through protein–protein interactions (Vanholme et al., 2007).
Members of the JAZ family exhibit high sequence variability, but possess three conserved domains which comprise the distinguishing characteristics of the group. The N-terminal (NT) region contains a weakly conserved NT domain. Recent studies have implicated this domain in a small set of protein–protein interactions, but the domain remains largely uncharacterized (Hou et al., 2011). The ZIM domain, consisting of 30 amino acids within the central portion of the JAZ peptide sequence, contains a highly conserved TIFY motif (TIF[F/Y]XG; Vanholme et al., 2007). The TIFY motif has been shown to be required for the repressor activity of several JAZ proteins, as well as the formation homo- and heterodimers within the JAZ family (Chini et al., 2009; Chung and Howe, 2009; Pauwels et al., 2010). No function has yet been ascribed to the larger, C-terminal portions of the ZIM domain (boxed in Figure 2). The C-terminal Jas domain is strongly conserved across the JAZ family with 12 of 29 amino acid residues identical or with conservative substitutions across all 12 Arabidopsis JAZ proteins (Chini et al., 2007; Thines et al., 2007). The domain is involved in a wide range of protein–protein interactions (Chini et al., 2009; Hou et al., 2011; Song et al., 2011; Zhu et al., 2011), contains within it a degron responsible for the degradation of JAZs in the presence of JA–Ile (Chini et al., 2007; Thines et al., 2007; Melotto et al., 2008), and may play a role in nuclear localization (Figure 2; Thines et al., 2007; Grunewald et al., 2009).
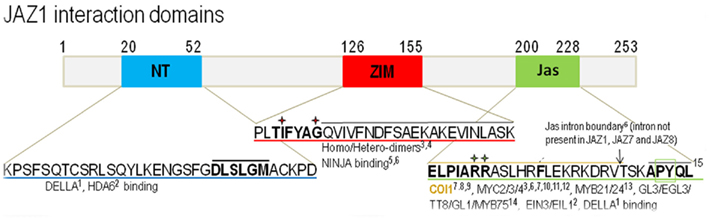
Figure 2. Schematic of JAZ1 interaction domains. Colored boxes indicate conserved domains with the sequence, below. Proteins proposed to bind within each domain are listed below. The yellow bar indicates residues sufficient for COI1 binding. Red stars mark residues required for forming homo- and/or heterodimers (ZIM domain). Green stars mark residues required for COI1 binding (Jas domain). The black bars over bold residues mark a conserved EAR-like motif (NT domain) and a proposed nuclear localization signal (Jas domain). The black bar over residues 134–155 marks a conserved region with no ascribed function. References: 1 Hou et al. (2011), 2 Zhu et al. (2011), 3 Chini et al. (2009), 4 Chung and Howe (2009), 5 Pauwels et al. (2009), 6Chung et al. (2010), 7 Chini et al. (2007), 8 Melotto et al. (2008), 9 Sheard et al. (2011), 10 Niu et al. (2011), 11 Cheng et al. (2011), 12 Fernández-Calvo et al. (2011), 13 Song et al. (2011), 14 Qi et al. (2011), 15 Grunewald et al. (2009).
JASMONATE ZIM-DOMAIN proteins lacking all or part of the C-terminal Jas domain exhibit decreased COI1 binding and are referred to as JAZΔJas proteins (Chung et al., 2009). Researchers responsible for the initial identification of the JAZ family of proteins capitalized on this feature by characterizing plants expressing stabilized versions of JAZ1, JAZ3, and JAZ10 that exhibited reduced JA sensitivity relative to wild-type (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). Further, experiments with plants expressing both JAZ1–GUS and JAZ3–GFP indicated that these proteins are destabilized in the presence of JA, an action that is dependent on COI1, the 26S proteasome, and the C-terminal Jas domain (Chini et al., 2007; Thines et al., 2007). Yeast-two-hybrid and in vitro pull-down assays confirmed that JAZs interact with COI1 through their C-terminal Jas domain and that the interaction is dependent on JA–Ile (or coronatine; Thines et al., 2007; Melotto et al., 2008). Mutational analysis identified two basic amino acids within the Jas degron in JAZ1 and JAZ9, R205, R206, and R223, K224, respectively, that are required for interaction with COI1 (Figure 2). Ectopic expression of either JAZ1ΔJas or the alanine-substituted version of JAZ1(A205A206) resulted in the production of male sterile plants, a defect characteristic of JA-insensitive mutants (Melotto et al., 2008). This protein–protein interaction data, along with the complementary genetic work, strongly supported the hypothesis that JAZ proteins are destabilized in response to JA–Ile through a mechanism dependent on COI1–JAZ interactions.
Expression of dominant JAZΔJas repressor proteins also seems to be a strategy employed by plants, possibly for the modulation and/or attenuation of the energetically costly JA response. Using sequence comparisons of JAZ genes, Chung et al. (2011) identified a conserved intron present in 9 of 12 Arabidopsis JAZ proteins. The intron resides within the Jas domain, splitting it into NT and C-terminal portions (Figure 2). RT-PCR analysis of JA-treated seedlings revealed the presence alternatively spliced forms of JAZ transcripts that encode proteins in which the Jas intron is retained. These splice variants contain ∼20 amino acids comprising the NT portion of the Jas domain, but reach a premature stop codon as a result of retention of the intron. Consequently, these alternatively spliced proteins lack the conserved X5PY sequence at the C-terminal end of their Jas domains and are referred to by Chung et al. (2011) as ΔPY JAZs.
Yeast-two-hybrid and pull-down assays revealed that both JAZ2ΔPY and JAZ3ΔPY have diminished COI1 binding capabilities in the presence of JA–Ile relative to the corresponding full-length variants, JAZ2.1 and JAZ3.1. To test whether the conserved PY (boxed in Figure 2) residues at the C-terminal end of the Jas domain are required for full interaction with COI1 alanine substitutions were made in both JAZ2 and JAZ10. According to these experiments, a JAZ2 P227A Y228A mutant and a JAZ10 P191A Y192A mutant both interacted fully with COI1 in the presence of JA–Ile or coronatine. This result suggests that other (non-conserved) residues in the X5PY peptide may be required for COI1–JAZ interaction (Chung et al., 2011). Alternative splicing likely affects the ability of JAZ proteins to interact with COI1, and could provide a mechanistic explanation for the modulation of the JA signaling response.
The crystal structure of COI1 complexed with JA–Ile and the Jas peptide from JAZ1 provided further insights into the molecular mechanism of JA hormone action (Sheard et al., 2011). In the crystal structure, JA–Ile (or coronatine) interacts in a vertical orientation with 11 residues of COI1 in the leucine-rich-repeat/loop structure that constitutes the ligand binding pocket in this class of F-box proteins, while three additional residues form a cavity that accommodates binding of the active (3R, 7S) isomer of JA–Ile, but not the inactive (3R, 7R) isomer. Much of the JA–Ile ligand is buried by these COI1 residues surrounding the binding site, but the keto group of JA and the carboxyl group of Ile are exposed and available for interaction with the Jas domain of JAZ. Despite the large number of identified interactions between JA–Ile and COI1, results from radioligand binding assays revealed that JA–Ile binds only very weakly to COI1. Inclusion of JAZ1 or JAZ6 in these assays increased binding specificity 50-fold, indicating that COI1 and JAZ are mutually dependent co-receptors for the JA hormone. The portion of JAZ1 that binds COI1 and JA–Ile (residues 200–220) has a bipartite structure. The NT portion (ELPIA) forms a loop which interacts both with the carbonyl of JA–Ile and with residues of COI1, effectively closing the binding site of the JA–Ile co-receptor. The C-terminal portion of the Jas peptide forms an α-helix that interacts with residues lining the central tunnel of the leucine-rich repeat ring of COI1. The additional binding here is essential for co-receptor function (Sheard et al., 2011).
Additionally, an inositol 1,2,4,5,6-pentakisphosphate (IP5) molecule appears to have a key role in assembly and function of the hormone receptor (Mosblech et al., 2011; Sheard et al., 2011). The IP5 molecule was identified in the crystal structure located close to the JA–Ile) binding pocket of COI1, where it coordinates three arginine residues of COI1 and R206 of the Jas peptide (Figure 2; Sheard et al., 2011). Removal of IP5 by dialysis inactivated the receptor complex and the inactive form could be reactivated by the addition of IP5. These observations fully explain the persistence of JAZΔJas proteins and may also provide an explanation for the dominant, JA-insensitive phenotypes that their expression produces in transgenic plants. They also demonstrate the value of crystallographic approaches to advancing our understanding of protein–protein interactions and the mechanistic details of JA hormone sensing.
Tandem affinity purification (TAP) is an alternative to the less stringent and less physiologically relevant yeast-two-hybrid screen. It is a method for protein purification which utilizes mass spectrometry in order to identify new members of a protein–protein interaction complex. The method eliminates some of the problems associated with the generation of false positive results seen with the yeast-two-hybrid method, because proteins are expressed under physiological conditions in planta and are purified using a two-step method involving binding to two subsequent affinity columns (Puig et al., 2001).
Using this method, Pauwels et al. (2010) identified the NOVEL INTERACTOR OF JAZ (NINJA) protein. TAP-tagged JAZ1 additionally identified the known signaling regulators, COI1, MYC3, and JAZ12. A subsequent TAP experiment designed to elucidate the function of NINJA within the signaling pathway showed that NINJA co-purified with the Groucho/Tup1 co-repressor TOPLESS (TPL) as well as the TPL-related proteins TPR2 and TPR3. TPL and TPR proteins had already been implicated in the regulation of auxin signaling, in which AUX/IAA proteins recruit TPL and/or TPR proteins to auxin-responsive promoters through interaction with DNA-binding AUXIN RESPONSE FACTOR (ARF) proteins (Osmont and Hardtke, 2008; Szemenyei et al., 2008). This work provided a plausible mechanistic explanation for the repression activity of JAZ proteins in which they recruit the co-repressors NINJA and TPL/TPR to JA-responsive promoters through interaction with MYC transcription factors.
Pauwels et al. (2010) used yeast-two-hybrid assays to demonstrate that NINJA interacts with most JAZs. Truncation of JAZ1 revealed a portion of the protein located within the ZIM domain (consisting of amino acids 116–155) sufficient for JAZ1–NINJA interaction (Figure 3). Deletion of the highly conserved TIFY motif within this region (TIFYAG in JAZ1) eliminated JAZ1–NINJA interaction (Pauwels et al., 2010). JAZ7 and JAZ8, the only two JAZs incapable of NINJA interaction (Pauwels et al., 2010), possess a TIFYXG sequence. However, both JAZ7 and JAZ8 have an Asn residue in the fifth position of the TIFYXG motif (Figure 3). This residue, absent from the TIFY motif in all other JAZ proteins, may hinder interaction with NINJA. No experimental work, however, has been done testing the significance of this residue.
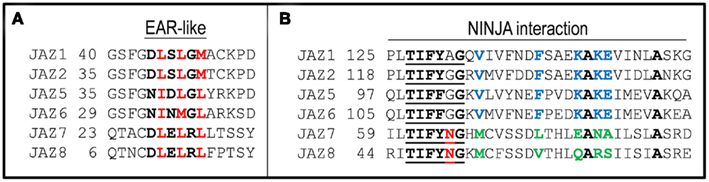
Figure 3. (A) Alignment of the semi-conserved EAR-like motif present in the NT domain of select JAZ proteins. Specific residues comprising the EAR-like motif are shown in bold. Red residues indicate the key (leucine or similar). (B) Alignment of the C-terminal portion of JAZs sufficient for NINJA. The TIFYXG motif required interaction (Pauwels et al., 2009) is underlined. This contains an N residue (red) in JAZ7 and JAZ8, the only two JAZs incapable of NINJA interaction. Residues conserved across all six JAZs shown are bold, residues conserved in JAZ1, JAZ2, JAZ5, and JAZ6, but not JAZ7 and JAZ8 are colored blue or green, respectively.
Within the context of auxin signaling, it has been suggested that TPL may function as a co-repressor involved in the recruitment of general repressive machinery, such as histone deacetylases, that modify chromatin in order to keep it in a closed state, negatively affecting gene transcription. This hypothesis is supported by studies involving a temperature sensitive, dominant-negative TPL mutant, tpl-1. tpl-1 mutants have a point mutation resulting in an N176H change in the protein. tpl-1 interacts genetically with both the histone modifying mutant histone deacetylase 19 (hda19) and the histone acetyltransferase mutant, hag1. Like tpl-1 mutants, hda19 mutants are temperature sensitive and their seedlings display distinct developmental defects when grown at restrictive temperature, including the formation of monocots, tubes and pins. Dominant-negative tpl-1 mutants interact genetically with the transcriptional co-activator, GCN5, a histone acetyl transferase. The GCN5 mutant hag1 partially rescues the tpl-1 fused cotyledon phenotype in Arabidopsis seedlings (Long et al., 2006; Szemenyei et al., 2008). These phenotypic effects are believed to be consequences of changes in the extent of histone acetylation.
To elucidate the physiologic role of NINJA, plants expressing altered levels of the NINJA protein were tested for their ability to respond to JA in root growth assays. 35S:NINJA over-expressing plants showed slightly reduced sensitivity to JA, while ninja-RNA interference (RNAi) plants were slightly hypersensitive to JA according to the assay. Additionally, the tpl-1 mutant was hypersensitive to JA according to root growth assays, further supporting the hypothesis that the TPL protein is a co-repressor involved in the negative regulation of JA signaling (Pauwels et al., 2010). Although phenotypic analyses of JA-mediated root growth inhibition suggest that NINJA plays a physiological role the JA response, the relatively weak phenotypes observed also raise the question of whether JAZs employ other mechanisms for repressing transcription.
A possible NINJA-independent mechanism for repression by JAZs could be the direct recruitment of the transcriptional co-repressor, TPL. Analysis of the JAZ peptide sequence revealed that several JAZ proteins contain an EAR (ethylene-responsive element binding factor-associated amphiphilic repression)-like motif (often, but not exclusively within their NT region; Figure 3). JAZ EAR-like elements closely resemble the EAR motif of AUX/IAA proteins used to directly recruit TPL and TPL-related proteins (Tiwari et al., 2004; Szemenyei et al., 2008). Additionally, protein–protein interaction studies performed by the Arabidopsis Interactome Mapping Consortium (2011) have identified both JAZ5 and JAZ8 as TPL-interacting proteins. Further work will be required to determine whether JAZ–TPL interactions have any biological significance with respect to the regulation of JA signaling.
An alternate NINJA-independent mechanism for JAZ protein repression of the JA-mediated transcriptional response has been proposed by Zhu et al. (2011). They showed that JAZ1 interacts with the chromatin modifying enzyme, HISTONE DEACETYLASE 6 (HDA6; Zhu et al., 2011). Both yeast-two-hybrid assays and in vitro pull-down experiments showed that JAZ1 interacted directly with the HDA6. To test whether this interaction is specific to JAZ1 or a general feature of the JAZ family, JAZ3 and JAZ9 were tested via yeast-two-hybrid assays in which they were also found to be capable of HDA6 interaction. Additional yeast-two-hybrid assays using truncated JAZ1 identified a C-terminally truncated JAZ1 peptide (amino acids 1–190) capable of HDA6 interaction and an N-terminally truncated peptide (amino acids 160–250) that is not. These results indicate that interaction with HDA6 is likely mediated by residues within the NT or ZIM domain of JAZ1, but additional experiments will be required to identify precise regions of JAZ proteins that are important for interaction with HDA6 (Zhu et al., 2011).
The physiological relevance of HDA6 to the JA signaling response was shown by analysis of hda6 mutants. The hda6 loss-of-function mutant, axe1-4, exhibits increased expression from the JA-inducible ERF1 locus. Conversely, HDA6 over-expressing plants exhibited decreased ERF1 expression, suggesting that HDA6 is involved in the negative regulation of ERF1 transcription. These findings are strikingly similar to those found when characterizing HDA19 overexpressors and HDA19 RNAi plants. Altering levels of HDA19 correlated with transcriptional changes in the ERF1 locus, with 35S:HDA19 transgenics exhibiting decreased ERF1 expression and hda19-RNAi mutants exhibiting increased transcription from the ERF1 locus (Zhou et al., 2005). Taken together, these results indicate that histone de-acetylation likely plays a role in the transcriptional regulation of JA responsive genes, specifically as a co-repressor involved in the regulation of the ERF1 locus.
MYC2 was identified in two independent mutant screens (Berger et al., 1996; Boter et al., 2004; Lorenzo et al., 2004) and was for several years the only DNA-binding transcription factor known to interact with members of the JAZ family of proteins (Chini et al., 2007). MYC2 is a protein belonging to the 133-member basic helix-loop-helix (bHLH) family characterized by its bHLH domain responsible for DNA binding and for the formation of homo- and/or heterodimers with other bHLH proteins. As a MYC-related protein, MYC2 has a partially conserved leucine-zipper (ZIP) motif adjacent to its bHLH domain, which may stabilize protein dimers. Previous work has demonstrated that MYC2 differentially regulates the JA response through competitive interaction with the ethylene-responsive transcription factor ERF1. MYC2 induces a subset of early JA-response genes, but inhibits a set of defense genes such as PDF1.2, PR4, and PR1 (Lorenzo et al., 2004). In contrast to coi1 mutants, which completely lack JA responses, myc2 mutants remain fertile, and exhibit only partial insensitivity to JA-mediated growth and defense (Fernández-Calvo et al., 2011).
Chini et al. (2007) provided evidence that MYC2 interacts with JAZ3 directly. This direct interaction supports the mechanistic model for the role of MYC2 as a transcription factor in JA signaling (shown in Figure 1). In this model, JAZ repressor proteins, in the absence of JA hormone, bind to MYC2 and inhibit its activity. In the presence of hormone, JAZ proteins are degraded and MYC2 is then able to induce a set of JA-responsive genes (Berger et al., 1996; Lorenzo et al., 2004; Chini et al., 2007). Despite the considerable evidence for the involvement of MYC2 in the up-regulation of JA response genes, the relatively weak phenotype of myc2 mutants compared to coi1 mutants suggests that MYC2 must not be the only relevant transcription factor involved in the regulation of the JA-induced transcriptional response.
Investigating the interaction partners of the JAZ family of proteins has recently proven to be useful in identifying additional transcription factors that are responsible for the early JA response. Three groups of researchers independently identified close homologs of MYC2, MYC3 and MYC4, via yeast-two-hybrid screens using JAZ proteins as bait (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011). MYC3 and MYC4 are phylogenetically closely related to MYC2; and together with MYC2 and bHLH28/MYC5 they constitute the IIIe subgroup of bHLH proteins (Fernández-Calvo et al., 2011). Further, interaction assays in yeast showed that, like MYC2, MYC3 and MYC4 both interact with the majority of JAZ proteins. In MYC2 and MYC3, a region of ∼60 aa was shown to be sufficient for interaction with eleven JAZ proteins and was thus designated as the JAZ-interaction domain (JID; Fernández-Calvo et al., 2011).
myc3 and myc4 T-DNA insertion mutants as well as myc3-RNAi and myc4-RNAi plants were indistinguishable from wild-type when tested for JA-mediated root growth inhibition, a result that can be explained by redundancy between members this family of MYC transcription factors (Lorenzo et al., 2004; Cheng et al., 2011; Niu et al., 2011). To gain insights into the binding determinants for MYC2 and related proteins, Fernández-Calvo et al. (2011) identified a consensus sequence for MYC2, MYC3, and MYC4 DNA binding using an 11-mer microarray. According to these experiments, all three transcription factors most tightly bind a canonical G-box consisting of the sequence CACGTG, providing further support for the hypothesis that MYC transcription factors have at least partially redundant functions (Fernández-Calvo et al., 2011).
Two approaches were taken to overcome the proposed redundancy; phenotypic evaluation of double and triple mutants and analysis of transgenic plants over-expressing MYC3 or MYC4. Both MYC3 and MYC4 over-expressing plants are hypersensitive to JA-mediated root growth inhibition, accumulate excess anthocyanin and exhibit altered expression of a set of JA-responsive genes including JAZ-family members and the wound-responsive genes, LOX3, VSP2, and TAT3 (Cheng et al., 2011; Niu et al., 2011). Mutational analysis showed that myc3 and myc4 intensified the severity of phenotypes observed in myc2, with myc2 myc3 myc4 triple mutants exhibiting JA-related phenotypes that closely mimic coi1 mutants, in most respects. However, the myc2 myc3 myc4 triple mutants were fully fertile and did not exhibit full resistance to JA-mediated inhibition of root growth. These observations indicate that other transcription factors, in addition to MYC2, MYC3, and MYC4 must be required for full induction of JA responses (Fernández-Calvo et al., 2011). Interestingly, a dominant mutation at the Arabidopsis MYC3 locus, atr2D, results in the induction of stress-response genes (Smolen et al., 2002). The mutation encodes a D94N change at a conserved site in the JID. The dominant phenotype may well result from activity of the altered MYC-D94N protein that is no longer bound and inhibited by JAZ proteins.
Characterization of JAZ-interacting proteins identified in yeast-two-hybrid screens have lead to the recent identification of several additional transcription factors targeted by JAZ repressors. MYB21 and MYB24 are two R2R3 MYB transcription factors identified as JAZ-interacting proteins via yeast-two-hybrid screen using JAZ8 as bait (Song et al., 2011). Further yeast-two-hybrid assays testing all members of the JAZ family of proteins showed that interaction with MYB21 and MYB24 was not a unique feature of JAZ8. Strong interaction was also observed between the two MYB proteins and JAZ1, JAZ11 and, to a lesser degree, JAZ10 via yeast-two-hybrid assay. Further, protein–protein interaction assays were employed successfully to confirm these interactions in vivo, including firefly luciferase complementation imaging (LCI), and bimolecular fluorescence complementation (BiFC; Song et al., 2011).
Results from yeast-two-hybrid experiments utilizing both truncated MYB21 and MYB24, as well as truncated JAZ8 and JAZ11 proteins indicate that interaction between JAZ and MYB proteins is likely dependent on the Jas domain (of JAZs) and the NT, R2R3 portion of MYB21 and MYB24 (Song et al., 2011). MYB21 and MYB24 had already been implicated in JA-mediated male fertility via transcriptional profiling and mutant analysis of the early JA response. In these studies, myb21 mutants exhibited reduced fertility as a result of effects in essential reproductive processes such as stamen filament elongation, pollen viability, and anther dehiscence. Although myb24 alone did not produce a phenotype, myb24 exacerbated the myb21 mutant phenotype, suggesting that the two proteins have at least partially overlapping roles (Mandaokar et al., 2003, 2006). Over-expression of MYB21 in a coi1 mutant background was able to partially restore stamen filament length, as well as restore anther dehiscence and provided low levels of fertility. Interestingly, over-expression of MYB21 did not restore other JA-related defects in coi1 mutants, such as insensitivity to JA-mediated root growth inhibition, anthocyanin accumulation or susceptibility to Bradysia impatiens. These results suggest that MYB21 may perform a role that is directed toward fertility and may be a less-important regulator of other JA-mediated growth and defense responses (Song et al., 2011).
Another yeast-two-hybrid screen using JAZ1 as bait identified GLABARA3 (GL3) and MYB75, transcription factors belonging to the bHLH and MYB families, respectively. Further yeast-two-hybrid showed that both GL3 and MYB75 interact with most JAZ family members. Further, the MYB transcription factor GLABRA1 (GL1), and the bHLH-family proteins, ENHANCER OF GLABARA3 (EGL3), and TRANSPARENT TESTA8 (TT8) also interacted with multiple JAZ proteins according to yeast-two-hybrid assays (Qi et al., 2011). Yeast-two-hybrid assays using truncated versions of both JAZ8 and JAZ11 indicated that the interaction was dependent on the C-terminal, JAS-containing region of both proteins.
Truncation analysis of the MYC transcription factors TT8 and EGL3 indicates that they both interact with JAZ1 and JAZ8 through a C-terminal portion that includes the bHLH domain (Qi et al., 2011). Interactions of the MYB transcription factors, MYB75 and GL1, with JAZ1 or JAZ8 was independent of the R2R3 domain and instead require the C-terminal portion of these proteins (Qi et al., 2011). These interactions suggest that these bHLH and MYB proteins may be direct targets of JAZ repressor proteins (Table 1), functioning downstream of COI1 in the JA-induced signaling cascade which regulates anthocyanin accumulation and trichome initiation. This hypothesis is supported by additional data showing that over-expression of GL3 in coi1-2 mutant plants unmistakably rescues trichome initiation, and weakly rescues anthocyanin accumulation (Qi et al., 2011). This study provided a plausible mechanistic explanation for previous reports implicating WD-repeat/bHLH/MYB protein complexes in the regulation of subset of JA responsive genes defense genes (Zhang et al., 2003; Baudry et al., 2004; Zimmermann et al., 2004).
Ethylene, another important plant hormone, works together with JA to regulate defense against necrotrophic pathogens and control some developmental processes (Wang et al., 2002). Binding of ethylene to its receptor, encoded by the ETHYLENE RESPONSE1 gene family, allows for the downstream inactivation of CONSTITUTIVE ETHYLENE RESPONSE1 (CTR1), a raf-like kinase. This action subsequently facilitates the activation of a diverse set of ethylene-responsive transcription factors, which regulate expression of ethylene-responsive genes (Guo and Ecker, 2004). Though researchers have been able to successfully document many of the effects of the crosstalk that takes place between ethylene and JA, until recently there has not been a mechanistic explanation for this crosstalk.
ETHYLENE INSENSITIVE 3 (EIN3), and its nearest homolog, EIN3-LIKE 1 (EIL1), are two transcription factors largely responsible for primary gene induction downstream of ethylene sensing (Guo and Ecker, 2004). Recent work by Zhu et al. (2011) exploring protein–protein interactions has shed light on the means by which the JA-ethylene synergy and dependence takes place. This work showed that JAZ1, JAZ3, and JAZ9 are all capable of interacting with the ethylene signaling regulators EIN3 and EIL1 in yeast-two-hybrid assays. Interaction was confirmed in the case of JAZ1–EIN3 and JAZ1–EIL1 with in vitro pull-down experiments as well as by BiFC. Further characterization showed that a C-terminal, Jas-containing fragment of JAZ1 is sufficient to pull down EIN3 (Zhu et al., 2011).
The transcription factors EIN3 and EIL1 activate a subset of pathogen-defense and developmental regulators. These transcriptional regulators are stabilized in the presence of ethylene and this stabilization allows them to induce expression of ethylene-responsive genes. However, binding of JAZ proteins to EIN3 and EIL1 partially inhibits that activity, thus providing a second level of control though JA crosstalk. In the presence of JA, JAZ degradation allows full activation of the transcription factors, thus providing for synergy between JA and ethylene in activating defenses against necrotrophic pathogens. This recent work exploring protein–protein interactions has shed light on the means by which the JA-ethylene synergy and mutual dependence occurs in planta (Zhu et al., 2011).
Gibberellins are a class of diterpene hormones required for many growth processes including germination, stem and hypocotyl elongation, and flower development. DELLA proteins function as important regulators of GA signaling, and are responsible for inhibiting expression of GA-induced genes in the absence of hormone. Upon hormone sensing by the GA receptor, GID1, it binds DELLAs, targeting them for ubiquitination by the SCFSLY1 complex, an E3 ligase, and subsequent degradation via the 26S proteasome pathway. As a result, activation of the GA-mediated transcriptional response occurs (Gao et al., 2011).
It has previously been shown that DELLA proteins, whose primary role is in the regulation of GA signaling, also affect expression of JA-responsive genes (Cheng et al., 2009; Peng, 2009). The mechanism for this crosstalk, however, was not elucidated until recently, when a yeast-two-hybrid screen using a portion of RGA, a DELLA protein, as bait identified JAZ1 as an RGA-interacting protein. JAZ3 and JAZ9 also interact with RGA according to yeast-two-hybrid assays. Interaction assays using NT, ZIM, and Jas domain deletions showed that interaction between JAZ and DELLA proteins does not depend upon the ZIM domain, but is diminished upon deletion of both the NT and Jas domain. To determine the role of DELLAs in the regulation of JA signaling researchers tested whether JAZ, RGA, and MYC2 are capable of forming a complex. In these experiments it was shown that, GST-JAZ1co-purified, with MYC2, an action that was attenuated in a dose-dependent manner by the addition of RGA. In a reciprocal experiment, JAZ1 co-purified with RGA, an interaction that was attenuated by the addition of MYC2. These pull-down experiments indicate that RGA and MYC2 compete for binding to JAZ1, rather than form a complex. The work provides plausible mechanistic explanation for DELLA-mediated relief of JAZ repression in which DELLAs sequester JAZ proteins and diminish JAZ–MYC2 interaction (Hou et al., 2011).
Jasmonate-mediated root growth inhibition was tested using seedlings harboring loss-of-function DELLA mutants. In these experiments, which were all done in a ga1-3 (a GA synthesis mutant) background, loss of DELLA function resulted in a loss of JA responsiveness. Transient expression of 2x35S:JAZ1-HA, 2x35S:MYC2, and 2x35S:RGA in Arabidopsis mesophyll protoplasts resulted in increased expression of GUS from the pTAT1:GUS reporter relative to co-expression of JAZ1 and MYC2 constructs alone, a phenomenon that is attenuated by the addition of GA (Hou et al., 2011). Given the growth promoting role of GA versus the defense promoting role of JA, it may be in the interest of efficiency that these hormones act in such a way that each counteracts the transcriptional responses of the other. Identification and characterization of the interactions in which JAZs take part, has begun to shed light on the mechanism that plants use to modulate these types of regulatory responses.
The research we have reviewed indicates that JAZ proteins interact and influence many transcription factors, but there are two broad questions that will require a more-detailed understanding of these interactions. The bHLH transcription factors, MYC2, MYC3, and MYC4, bind to JAZ proteins via a well-defined, NT JID (residues 93–160 in MYC2; Fernández-Calvo et al., 2011). Even though TT8 and EGL3 are related bHLH transcription factors with a relatively well-conserved JID (Fernández-Calvo et al., 2011), it is the C-terminal portion (which includes the bHLH domain) that was found to mediate interaction of these proteins with JAZ1 and JAZ8 (Qi et al., 2011). For the MYB transcription factors, MYB75 (PAP1) and GL1 binding to JAZ occurs through the C-terminal portion of the protein (Qi et al., 2011), but for MYB21 and MYB24, interaction was through an NT portion that is composed primarily of the R2 and R3 domains (Song et al., 2011). It is not yet clear whether these different findings reflect a genuine diversity in JAZ-interaction sites, or if further tests of the specific residues and motifs necessary and sufficient for JAZ binding will provide a smaller set of sequence determinants.
All these bHLH and MYB proteins as well as EIN3 and EIL1 have been shown to bind to the C-terminus of JAZ and in some cases specifically to the Jas domain (Chini et al., 2007, 2009; Song et al., 2011; Zhu et al., 2011). [For the DELLA proteins, binding at both Jas and the N-terminus has been proposed (Hou et al., 2011).] Because Jas is the only C-terminal sequence that is conserved among the 12 JAZ proteins, it is likely that this domain contains the binding site(s) for these transcription factors. The Jas domain also contains the JA–Ile/COI1 binding site as defined by protein-interaction studies and the crystal structure (Sheard et al., 2011). It is possible that transcription factors compete for the Jas domain with formation of the JAZ/JA–Ile/COI1 complex. However, we do know that the JAZ1 binding sites for COI1 (in the presence of coronatine) and for MYC2 are not congruent because the mutations R205A and/or R206A in the JAZ1 Jas domain abolish COI interaction but have no discernible effect on interaction with MYC2 (Melotto et al., 2008).
Accurately mapping binding determinants on the JAZ proteins and their transcription-factor partners may help to solve an enduring puzzle in the JA hormone mechanism. Over-expression of JAZΔJas proteins, including JAZ10.4, a naturally occurring splice variant of JAZ10 pre-mRNA (Chung et al., 2009, 2011), result in dominant, JA-insensitive phenotypes (Melotto et al., 2008; Chung and Howe, 2009). These JAZΔJas proteins do not interact with COI1 and are not degraded in plants treated with JA (Melotto et al., 2008) so it has been proposed that they continue to act as repressors by inhibiting the transcription factors that mediate expression of JA-responsive genes. However, if the transcription factors bind to JAZ (only) via the Jas domain, how can inhibition be achieved? A model proposing poisoning of the SCFCOI1 ubiquitin ligase by JAZΔJas protein, allowing persistence of full-length JAZ proteins in the presence of JA hormone (Chini et al., 2007), was based on a conclusion that JAZ proteins interacted with COI1 through the ZIM domain. Subsequent findings, and especially the COI1 crystal structure (Sheard et al., 2011), with COI1 binding to the Jas domain, make it doubtful that COI1–ZIM interactions are physiologically relevant, making this explanation unlikely. Since there is extensive dimerization (or oligomerization) among JAZ proteins (mediated by the TIFY motif of the ZIM domain; Chini et al., 2009; Chung and Howe, 2009), a second possibility is that JAZΔJas proteins protect full-length JAZ from SCFCOI1-mediated degradation through dimerization (Chini et al., 2009). However, it is not clear why JAZΔJas–JAZ complexes should be more resistant to degradation than JAZ–JAZ complexes.
More data are needed on the JAZ isoforms that persist in JA-treated, 355:JAZΔJas plants. In addition however, protein–protein interaction experiments including detailed mapping of the binding determinants in JAZ for MYC2 and other transcription factors (as well as for COI1) will help in formulating and testing hypotheses about the molecular mechanisms through which JAZΔJas proteins produce strong JA-insensitive phenotypes.
JASMONATE ZIM-DOMAIN proteins are capable of a multitude of biologically relevant protein–protein interactions required for regulation of JA responses as well as for interaction with other signaling pathways interconnected with JA signaling through the expansive regulatory network (Table 1). In recent years, our knowledge of JA signaling has greatly increased in no small part because of our increased understanding of the interactions involving the JAZ family of proteins. Understanding the nature of protein–protein interactions that take place within the core nuclear JA signaling module, such as those which occur between JAZs and MYC2, COI1 and NINJA, has provided researchers with the clues required to elucidate the mechanics of the JA sensing mechanism. Additionally, techniques such as TAP and the yeast-two-hybrid screen have aided in the identification of new transcription factors involved in the JA response.
In all likelihood more JAZ interactions will be discovered in the future. Based on the examples covered in this review we expect that these new interactions will identify additional components of the signaling network that responds to JA hormone. Protein–protein interactions provide a means by which plants can add complexity to signaling networks, and other aspects of cellular biochemistry. The individual roles for each of the JAZ proteins have not been fully elucidated, and mapping the interactions of individual JAZs could aid in this understanding. Additionally, the biological relevance of JAZ–JAZ homo- and heterodimers has not been determined. Understanding the interactions involved may be essential for understanding how JA signaling is modulated in order to respond to a diverse set of stressors and developmental cues. Furthermore, published protein interaction screens (Pauwels et al., 2010; Hou et al., 2011) and a recently published large-scale documentation of the Arabidopsis interactome (Arabidopsis Interactome Mapping Consortium, 2011) identify putative JAZ targets, opening the door for additional discoveries to be made elucidating the mechanism by which JAZs regulate a wide array of processes.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Research on jasmonate in the authors’ laboratory is funded by Grant DE-FG-99ER20323 from the Office of Basic Energy Sciences at the U.S. Department of Energy and by the Agricultural Research Center at Washington State University. Amanda Wager was supported by NIH Training Grant 5T32GM083864-02.
Adams, E., and Turner, J. (2011). COI1, a jasmonate receptor, is involved in ethylene-induced inhibition of Arabidopsis root growth in the light. J. Exp. Bot. 61, 4373–4386.
Arabidopsis Interactome Mapping Consortium. (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607.
Bai, Y., Meng, Y., Huang, D., Qi, Y., and Chen, M. (2011). Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98, 128–136.
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380.
Berger, S., Bell, E., and Mullet, J. E. (1996). Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111, 525–531.
Boter, M., Ruã Z-Rivero, O., Abdeen, A., and Prat, S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18, 1577–1591.
Browse, J. (2009a). Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205.
Browse, J. (2009b). The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry 70, 1539–1546.
Cheng, H., Song, S., Xiao, L., Soo, H. M., Cheng, Z., Xie, D., and Peng, J. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5, e1000440. doi:10.1371/journal.pgen.1000440
Cheng, Z., Sun, L., Qi, T., Zhang, B., Peng, W., Liu, Y., and Xie, D. (2011). The bHLH transcription factor MYC3 interacts with the JASMONATE ZIM-DOMAIN proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4, 279–288.
Chico, J. M., Chini, A., Fonseca, S., and Solano, R. (2008). JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 11, 486–494.
Chini, A., Fonseca, S., Chico, J. M., Fernández-Calvo, P., and Solano, R. (2009). The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59, 77–87.
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., Garcia-Casado, G., Lopez-Vidriero, I., Lozano, F. M., Ponce, M. R., Micol, J. L., and Solano, R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671.
Chung, H. S., Cooke, T. F., Depew, C. L., Patel, L. C., Ogawa, N., Kobayashi, Y., and Howe, G. A. (2011). Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63, 613–622.
Chung, H. S., and Howe, G. A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell Online 21, 131–145.
Chung, H. S., Niu, Y., Browse, J., and Howe, G. A. (2009). Top hits in contemporary JAZ: an update on jasmonate signaling. Phytochemistry 70, 1547–1559.
Conconi, A., Smerdon, M. J., Howe, G. A., and Ryan, C. A. (1996). The octadecanoid signalling pathway in plants mediates a response to ultraviolet radiation. Nature 383, 826–829.
Devoto, A., Nieto-Rostro, M., Xie, D. X., Ellis, C., Harmston, R., Patrick, E., Davis, J., Sherratt, L., Coleman, M., and Turner, J. G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin–ligase complex in Arabidopsis. Plant J. 32, 457–466.
Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445.
Dombrecht, B., Xue, G. P., Sprague, S. J., Kirkegaard, J. A., Ross, J. J., Reid, J. B., Fitt, G. P., Sewelam, N., Schenk, P. M., Manners, J. M., and Kazan, K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245.
Farmer, E. E., Johnson, R. R., and Ryan, C. A. (1992). Regulation of expression of proteinase inhibitor genes by methyl jasmonate and jasmonic acid. Plant Physiol. 98, 995–1002.
Fernández-Calvo, P., Chini, A., Fernández-Barbero, G., Chico, J.-M., Gimenez-Ibanez, S., Geerinck, J., Eeckhout, D., Schweizer, F., Godoy, M., Franco-Zorrilla, J. M., Pauwels, L., Witters, E., Puga, M. I., Paz-Ares, J., Goossens, A., Reymond, P., De Jaeger, G., and Solano, R. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell Online 23, 701–715.
Feys, B. J. F., Benedetti, C. E., Penfold, C. N., and Turner, J. G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759.
Gao, X.-H., Xiao, S.-L., Yao, Q.-F., Wang, Y.-J., and Fu, X.-D. (2011). An updated GA signaling “relief of repression” regulatory model. Molecular Plant 4, 601–606.
Gazzarrini, S., and McCourt, P. (2003). Cross-talk in plant hormone signalling: what Arabidopsis mutants are telling us. Ann. Bot. 91, 605–612.
Grant, M. R., and Jones, J. D. G. (2009). Hormone (dis)harmony moulds plant health and disease. Science 324, 750–752.
Gray, W. M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276.
Grunewald, W., Vanholme, B., Pauwels, L., Plovie, E., Inze, D., Gheysen, G., and Goossens, A. (2009). Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 10, 923–928.
Guo, H., and Ecker, J. R. (2004). The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7, 40–49.
Hoffmann, M., Hentrich, M., and Pollmann, S. (2011). Auxin-oxylipin crosstalk: relationship of antagonists. J. Integr. Plant Biol. 53, 429–445.
Hou, X., Lee, L. Y. C., Xia, K., Yan, Y., and Yu, H. (2011). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19, 884–894.
Howe, G. A., Lightner, J., Browse, J., and Ryan, C. A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8, 2067–2077.
Kazan, K., and Manners, J. M. (2008). Jasmonate signaling: toward an integrated view. Plant Physiol. 146, 1459–1468.
Kazan, K., and Manners, J. M. (2012). JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17, 22–31.
Koornneef, A., and Pieterse, C. M. J. (2008). Cross talk in defense signaling. Plant Physiol. 146, 839–844.
Long, J. A., Ohno, C., Smith, Z. R., and Meyerowitz, E. M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523.
Lorenzo, O., Chico, J. M., Sanchez-Serrano, J. J., and Solano, R. (2004). Jasmonate-insensitive1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950.
Mandaokar, A., Kumar, V. D., Amway, M., and Browse, J. (2003). Microarray and differential display identify genes involved in jasmonate-dependent anther development. Plant Mol. Biol. 52, 775–786.
Mandaokar, A., Thines, B., Shin, B., Lange, B. M., Choi, G., Koo, Y. J., Yoo, Y. J., Choi, Y. D., and Browse, J. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46, 984–1008.
McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416.
McConn, M., Creelman, R. A., Bell, E., Mullet, J. E., and Browse, J. (1997). Jasmonate is essential for insect defense Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 94, 5473–5477.
Melotto, M., Mecey, C., Niu, Y., Chung, H. S., Katsir, L., Yao, J., Zeng, W., Thines, B., Staswick, P., Browse, J., Howe, G. A., and He, S. Y. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55, 979–988.
Mosblech, A., Thurow, C., Gatz, C., Feussner, I., and Heilmann, I. (2011). Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 65, 949–957.
Navarro, L., Bari, R., Achard, P., Lisón, P., Nemri, A., Harberd, N. P., and Jones, J. D. G. (2008). DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18, 650–655.
Niu, Y., Figueroa, P., and Browse, J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62, 2143–2154.
Osmont, K. S., and Hardtke, C. S. (2008). The topless plant developmental phenotype explained! Genome Biol. 9, 219.
Pauwels, L., Barbero, G. F., Geerinck, J., Tilleman, S., Grunewald, W., Perez, A. C., Chico, J. M., Bossche, R. V., Sewell, J., Gil, E., Garcia-Casado, G., Witters, E., Inze, D., Long, J. A., De Jaeger, G., Solano, R., and Goossens, A. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791.
Pauwels, L., Inzé, D., and Goossens, A. (2009). Jasmonate-inducible gene: what does it mean? Trends Plant Sci. 14, 87–91.
Pauwels, L., and Goossens, A. (2011). The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089–3100.
Peng, J. R. (2009). Gibberellin and jasmonate crosstalk during stamen development. J. Integr. Plant Biol. 51, 1064–1070.
Perez, A. G., Sanz, C., Olias, R., and Olias, J. M. (1997). Effect of methyl jasmonate on in vitro strawberry ripening. J. Agric. Food Chem. 45, 3733–3737.
Pre, M., Atallah, M., Champion, A., De Vos, M., Pieterse, C. M. J., and Memelink, J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357.
Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., and Séraphin, B. (2001). The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229.
Qi, T., Song, S., Ren, Q., Wu, D., Huang, H., Chen, Y., Fan, M., Peng, W., Ren, C., and Xie, D. (2011). The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell Online 23, 1795–1814.
Ren, C., Han, C., Peng, W., Huang, Y., Peng, Z., Xiong, X., Zhu, Q., Gao, B., and Xie, D. (2009). A leaky mutation in DWARF4 reveals an antagonistic role of brassinosteroid in the inhibition of root growth by jasmonate in Arabidopsis. Plant Physiol. 151, 1412–1420.
Reymond, P., Weber, H., Damond, M., and Farmer, E. E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell Online 12, 707–720.
Santner, A., and Estelle, M. (2011). The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 61, 1029–1040.
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., Kobayashi, Y., Hsu, F.-F., Sharon, M., Browse, J., He, S. Y., Rizo, J., Howe, G. A., and Zheng, N. (2011). Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 468, 400–405.
Smolen, G. A., Pawlowski, L., Wilensky, S. E., and Bender, J. (2002). Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis. Genetics 161, 1235–1246.
Song, S., Qi, T., Huang, H., Ren, Q., Wu, D., Chang, C., Peng, W., Liu, Y., Peng, J., and Xie, D. (2011). The JASMONATE-ZIM DOMAIN proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell Online 23, 1000–1013.
Staswick, P. E., Su, W., and Howell, S. H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. U.S.A. 89, 6837–6840.
Staswick, P. E., Tiryaki, I., and Rowe, M. L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415.
Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. U.S.A. 97, 10625–10630.
Szemenyei, H., Hannon, M., and Long, J. A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386.
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G. H., Nomura, K., He, S. Y., Howe, G. A., and Browse, J. (2007). JAZ repressor proteins are targets of the SCFCO11 complex during jasmonate signalling. Nature 448, 661–665.
Tiryaki, I., and Staswick, P. E. (2002). An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 130, 887–894.
Tiwari, S. B., Hagen, G., and Guilfoyle, T. J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16, 533–543.
Vanholme, B., Grunewald, W., Bateman, A., Kohchi, T., and Gheysen, G. (2007). The tify family previously known as ZIM. Trends Plant Sci. 12, 239–244.
Vijayan, P., Shockey, J., Levesque, C. A., Cook, R. J., and Browse, J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 7209–7214.
Wang, K. L. C., Li, H., and Ecker, J. R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell Online 14, S131–S151.
Xiao, S., Dai, L., Liu, F., Wang, Z., Peng, W., and Xie, D. (2004). COS1: an Arabidopsis coronatine insensitive1 suppressor essential for regulation of jasmonate-mediated plant defense and senescence. Plant Cell Online 16, 1132–1142.
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., and Turner, J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094.
Yan, Y. X., Stolz, S., Chetelat, A., Reymond, P., Pagni, M., Dubugnon, L., and Farmer, E. E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483.
Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869.
Zhou, C., Zhang, L., Duan, J., Miki, B., and Wu, K. (2005). HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell Online 17, 1196–1204.
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273.
Zhu, Z., An, F., Feng, Y., Li, P., Xue, L., A, M., Jiang, Z., Kim, J.-M., To, T. K., Li, W., Zhang, X., Yu, Q., Dong, Z., Chen, W.-Q., Seki, M., Zhou, J.-M., and Guo, H. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 12539–12544.
Keywords: Arabidopsis, hormone action, jasmonate, JAZ
Citation: Wager A and Browse J (2012) Social network: JAZ protein interactions expand our knowledge of jasmonate signaling. Front. Plant Sci. 3:41. doi: 10.3389/fpls.2012.00041
Received: 23 December 2011; Paper pending published: 14 January 2012;
Accepted: 18 February 2012; Published online: 08 March 2012.
Edited by:
Xuemin Wang, University of Missouri-St Louis and Donald Danforth Plant Science Center, USAReviewed by:
Masanori Arita, University of Tokyo, JapanCopyright: © 2012 Wager and Browse. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: John Browse, Institute of Biological Chemistry, Washington State University, Pullman, WA 99164, USA. e-mail:jab@wsu.edu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.