- 1 Plant Biology Division, The Samuel Roberts Noble Foundation, Ardmore, OK, USA
- 2 Department of Biological Sciences, Center for Plant Lipid Research, University of North Texas, Denton, TX, USA
- 3 Department of Biological Sciences, East Tennessee State University, Johnson City, TN, USA
- 4 Plant Signal Research Group, Institute of Technology, University of Tartu, Tartu, Estonia
N-acylethanolamines (NAEs) are bioactive lipids derived from the hydrolysis of the membrane phospholipid N-acylphosphatidylethanolamine (NAPE). In animal systems this reaction is part of the “endocannabinoid” signaling pathway, which regulates a variety of physiological processes. The signaling function of NAE is terminated by fatty acid amide hydrolase (FAAH), which hydrolyzes NAE to ethanolamine and free fatty acid. Our previous work in Arabidopsis thaliana showed that overexpression of AtFAAH (At5g64440) lowered endogenous levels of NAEs in seeds, consistent with its role in NAE signal termination. Reduced NAE levels were accompanied by an accelerated growth phenotype, increased sensitivity to abscisic acid (ABA), enhanced susceptibility to bacterial pathogens, and early flowering. Here we investigated the nature of the early flowering phenotype of AtFAAH overexpression. AtFAAH overexpressors flowered several days earlier than wild type and AtFAAH knockouts under both non-inductive short day (SD) and inductive long day (LD) conditions. Microarray analysis revealed that the FLOWERING LOCUS T (FT) gene, which plays a major role in regulating flowering time, and one target MADS box transcription factor, SEPATALLA3 (SEP3), were elevated in AtFAAH overexpressors. Furthermore, AtFAAH overexpressors, with the early flowering phenotype had lower endogenous NAE levels in leaves compared to wild type prior to flowering. Exogenous application of NAE 12:0, which was reduced by up to 30% in AtFAAH overexpressors, delayed the onset of flowering in wild type plants. We conclude that the early flowering phenotype of AtFAAH overexpressors is, in part, explained by elevated FT gene expression resulting from the enhanced NAE hydrolase activity of AtFAAH, suggesting that NAE metabolism may participate in floral signaling pathways.
Introduction
N-acylethanolamines (NAEs) are bioactive lipids that function in the regulation of various physiological processes in animal systems. This includes the endocannabinoid signaling pathway that regulates several neurobehavioral and neurophysiological activities. Fatty acid derivatives that are amide-linked to an ethanolamine moiety make up the basic molecular structure of NAE, which differ in their acyl chain length and number of double bonds (Kim et al., 2010). NAE bioactivity has been shown to be terminated by the hydrolytic activity of fatty acid amide hydrolase (FAAH) where NAE is hydrolyzed into ethanolamine and its corresponding free fatty acid (McKinney and Cravatt, 2005; Fowler, 2006). In mammals, the importance of FAAH in regulating in vivo NAE levels was demonstrated using genetic approaches. For example, it was shown that FAAH knockout mice possess elevated levels of anandamide (NAE 20:4) compared with wild type mice. The higher levels of anandamide in FAAH knockout mice were accompanied by a variety of physiological changes, including hypomotility, increased analgesia, and significant catalepsy (Cravatt et al., 2001; Clement et al., 2003; Lichtman et al., 2004; Kilaru et al., 2010).
Like animals, recent research in plant systems showed that NAEs impact numerous physiological processes (Kim et al., 2010). In Arabidopsis thaliana, a homolog of the mammalian FAAH was identified and characterized (AtFAAH, locus At5g64440), and also was shown to possess NAE hydrolytic activity in vitro (Shrestha et al., 2003, 2006). The Arabidopsis FAAH encodes a protein of 607 amino acids with 37% identity to rat FAAH within the amidase signature (AS) domain (Shrestha et al., 2003). The AS family, consisting of more than 80 amidases, contain a highly conserved region of approximately 130 amino acids rich in serine, glycine, and alanine (Chebrou et al., 1996; Patricelli et al., 1999; Labahn et al., 2002; Gopalakrishna et al., 2004; Neu et al., 2007). Homologs of FAAH have been identified in multiple plant species and modeling of the AS region revealed a highly conserved active site (Shrestha et al., 2006). These studies suggest that similar to mammalian systems, the molecular machinery for terminating NAE signaling by FAAH also operates in plants (Blancaflor and Chapman, 2006).
Several lines of evidence support a role for FAAH in the catabolism of NAEs in plants. For instance, exogenous NAE at low micromolar concentrations produced a dose-dependent reduction in seedling growth (Blancaflor et al., 2003). Along with the arrested growth, seedlings displayed altered root cell, and cytoskeletal organization when treated with NAE (Blancaflor et al., 2003; Motes et al., 2005). When AtFAAH was over expressed in Arabidopsis seedlings, the growth inhibitory effects of exogenous NAE treatment were much reduced compared to wild type, whereas SALK T-DNA insertion AtFAAH knockout lines displayed an increased sensitivity to NAE-induced growth inhibition (Wang et al., 2006; Teaster et al., 2007; Cotter et al., 2011). The AtFAAH overexpressors generally displayed enhanced seedling growth and a tendency to flower earlier than wild type. On the other hand, AtFAAH knockouts did not show any other strong phenotype besides their greatly increased sensitivity to exogenous NAE (Wang et al., 2006). In addition to enhanced growth, AtFAAH overexpressors showed an increased sensitivity to the plant hormone, abscisic acid (ABA), indicating cross talk between NAE and ABA signaling pathways (Teaster et al., 2007; Cotter et al., 2011). As a trade-off for enhanced growth, AtFAAH overexpressors exhibited compromised resistance to a number of plant pathogens, including non-host pathogens that do not normally cause disease in Arabidopsis (Kang et al., 2008).
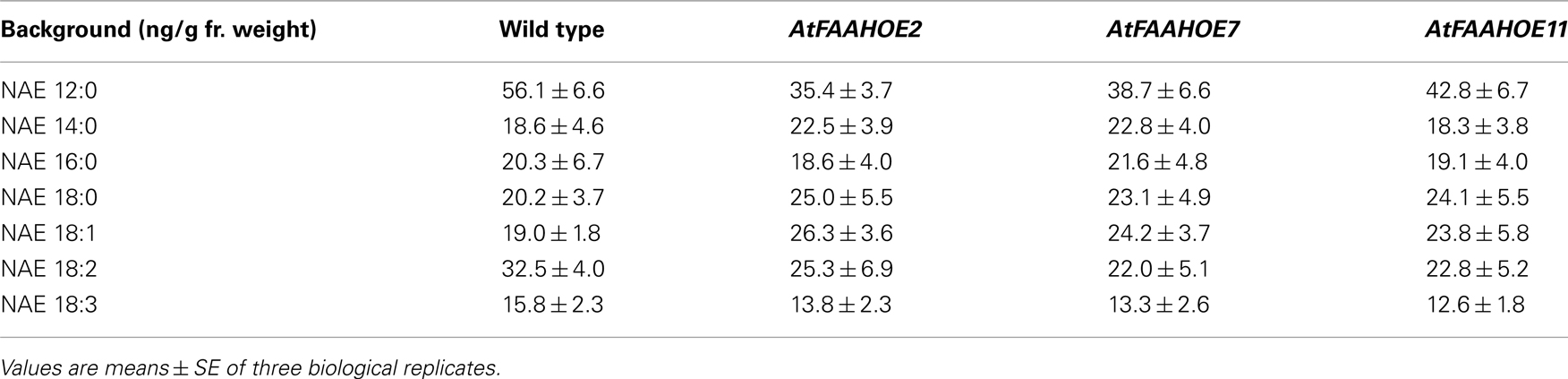
In this paper, we investigated the early flowering phenotype of AtFAAH overexpressors. We found that AtFAAH overexpressors flowered earlier than wild type and AtFAAH knockouts under both inductive long day (LD) and non-inductive short day (SD) conditions. Global gene expression profiling of 14-day-old Arabidopsis seedlings and verification by quantitative RT-PCR showed that FLOWERING LOCUS T (FT), which regulates flowering by activating other floral genes (Kardailsky et al., 1999; Corbesier et al., 2007; Turck et al., 2008; Seo et al., 2011), was elevated in AtFAAH overexpressors. We also found that the AtFAAH overexpressors grown under SD conditions for 14 days contained on average, about 9% less total NAE than wild type, and the levels of two specific types, NAE 12:0 and NAE 18:2, were reduced by up to 30% in AtFAAH overexpressors. Treatment of wild type Arabidopsis plants with exogenous NAE 12:0, delayed flowering. Taken together, our data suggest that the early flowering phenotype of AtFAAH overexpressors is attributed to altered NAE signaling.
Materials and Methods
Plant Material and Measurement of Flowering Time
Three independent lines overexpressing AtFAAH (OE2, OE7, and OE11) and two AtFAAH T-DNA knockouts (SALK_118043 and SALK_095108) used in this study were described previously (Wang et al., 2006). For evaluating flowering, seeds were surface-sterilized with 95% ethanol, 30% bleach containing 0.1% Tween 20 and deionized water and planted on agar plates and stratified overnight at 4°C. Seeds were then moved to 22°C growth chambers set to SD (8/16 light/dark) or LD (16/8 light/dark) photoperiod, and grown for an additional 10 days. Individual seedlings were then transplanted to pots containing Metro mix and returned to their respective growth chamber. Plants were monitored daily after transfer to the growth chamber and the number of plants that flowered was recorded. The number of rosette leaves was recorded on particular days as indicated.
Gene Chip Microarray Experiments and Data Analysis
For microarrays, total RNA was extracted from rosette leaves of 14-day-old AtFAAH overexpressors and the AtFAAH T-DNA insertional mutant SALK_095108, and processed as described previously (Teaster et al., 2007). Seeds of all three genotypes were planted on MS media and seedlings grown for 14 days in 14/10 h light/dark cycle prior to RNA isolation. The microarray experiments were conducted with RNA isolated from three biological replicates using ATH1 Genome Arrays (Affymetrix).
For data analysis, the CEL file for each sample was exported from the Expression Console (Affymetrix) and normalized using robust multi-array average (RMA) as described by Irizarry et al. (2003). The presence/absence call for each probe set was obtained from dCHIP (Li and Wong, 2001). Differentially expressed genes in the AtFAAH overexpressor and SALK_095108 knockout in comparison with wild type control were selected using associative analysis (Dozmorov and Centola, 2003). Type I family wise error rate was reduced using a Bonferroni-corrected P-value of 2.19202E-06 representing a threshold of 0.05/N, where N represents number of probe sets present on the chip, which is 22810 for the Arabidopsis chip (Abdi, 2007). Microarray data was submitted to the European Molecular Biology Laboratory’s European Bioinformatics Institute (EMBL–EBI) ArrayExpress database under accession E-MEXP-3486.
Real Time Quantitative RT-PCR
Total RNA was isolated using an RNeasy Mini Kit (Qiagen) and reverse transcribed to first-strand cDNA with the Qiagen cDNA Synthesis Kit (Qiagen). First-strand cDNA was used as a template for quantitative PCR using gene-specific primers. Arabidopsis 18s rRNA, which served as a control for constitutive gene expression in plants, was amplified with primers (F) 5′-TCCTAGTAAGCGCGAGTCATCA-3′ (R) 5′-CGAACACTTCACCGGATCAT-3′ (Dean Rider et al., 2003). Gene-specific primers used were FT (At1g65480) (F) 5′-GGAGACGTTCTTGATCCGTTTAATAGATCAAT-3′ (R) 5′-ATAAACACGACACGATGAATTCCTGCAGT-3′; SEP3 (At1g24260) (F) 5′-GTGCCTTCAAGAGAGGCCTTAGCAGTT-3′ (R) 5′-TCTGAAGATCGTTGAGCTGGTCAAGCAT-3′. Relative expression levels using the formula for threshold cycle were calculated by the (2−ΔΔCt) method (Livak and Schmittgen, 2001). Means of four biological replicates with three technical replicates and SE were reported.
NAE Quantification
N-acylethanolamines were extracted from 100 to 250 mg of plant tissue and ground by bead beater in warm 2-propanol. The extract was combined with deuterated NAE standards (D4-NAE 16:0, Cayman Chemical Co.; 1 ppm each) and total lipids were extracted into chloroform. The organic phase was collected for further purification by solid phase extraction (SPE). Silica SPE cartridges (100 mg, 1.5 mL; Grace Davison Discovery Sciences) were conditioned with 2 mL methanol followed by 4 mL chloroform. Samples were applied to the column, washed with 2 mL chloroform, and NAEs were eluted with 2 mL of 1:1 (v/v) ethyl acetate:acetone. The eluate was collected, evaporated under nitrogen, and derivatized with 50 μL BSTFA (Fisher Scientific, Houston, TX, USA) for 30 min at 55°C. After derivatization, the samples were again evaporated under nitrogen and reconstituted in 50 μL hexane. NAEs were identified via selective ion monitoring and quantified against the internal deuterated standards (deuterated NAE 16:0) as TMS-ether derivatives by gas chromatography/mass spectrometry (Agilent model 6890 GC coupled with a 5973 mass selective detector) as described previously (Venables et al., 2005). NAE concentration was calculated based on fresh weight.
Evaluating Exogenous NAE 12:0 Effects on Flowering
Arabidopsis wild type seeds were germinated in soil and were maintained in a growth chamber at 22°C with 16/8 h light/dark cycle (60 μE m−2 s−1). Seven day-old seedlings (n = 15) were watered with 50 mL of 35 μM NAE 12:0 or 0.05% DMSO solution alone (solvent control) every 3 days until the plants were ready for drying. Time of flowering was recorded and inflorescence height was measured every 4 days. The number of rosette leaves was counted every 4 days from the time of germination until 24 days. The data presented are an average of three experiments with significance tested by Student’s t-test.
Results
Overexpression of AtFAAH Induces Early Flowering under Non-Inductive Short Days and Inductive Long Days
We evaluated the flowering time of three independent AtFAAH overexpressing lines described in Wang et al. (2006). We found that when grown under non-inductive SD conditions, AtFAAH overexpressors flowered earlier than wild type (Col-0) and AtFAAH knockouts (Figure 1A). Quantification of the timing of flowering revealed that more than 50% of all AtFAAH overexpressing lines flowered 28 days after planting with one line (AtFAAHOE11) showing more than 70% flowering. In contrast, less than 20% of wild type and AtFAAH knockouts flowered at day 28 (Figure 1B). These results were typical of three independent experiments with individual AtFAAH overexpressors flowering on average up to 10 days earlier than wild type and AtFAAH knockouts. Even under inductive LD conditions AtFAAH overexpressor flowered earlier than wild type or AtFAAH knockouts. At 16 days, more than 60% of all three AtFAAH overexpressors had already flowered whereas wild type and AtFAAH knockouts did not. At day 18, only 10% of wild type and AtFAAH knockouts flowered while 70–100% of AtFAAH overexpressors flowered (Figure 1B).
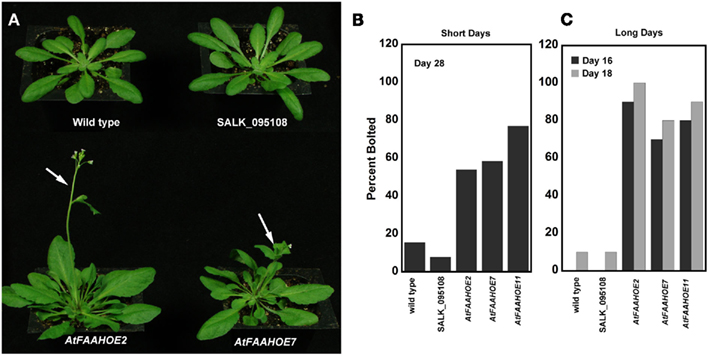
Figure 1. Overexpression of AtFAAH induces early flowering of Arabidopsis under short and long days. (A) Representative images of 30-day-old wild type, SALK_095108 AtFAAH knockout and two AtFAAH overexpressors (OE2 and OE7) grown during short day conditions, 8/16 h (light/dark). Note that the two AtFAAH overexpressors have already flowered (arrows). The percent of wild type, AtFAAH knockout and AtFAAH overexpressors that had flowered by day 28 under non-inductive SDs (B) and by day 16 and day 18 under inductive LDs (C). The graph is representative of three experiments with n > 10.
We measured the number of rosette leaves and rosette diameter of SD-grown plants, on day 21 and day 28 after planting, to determine if a more rapid growth was responsible for the early flowering phenotype of the AtFAAH overexpressors. The total number of rosette leaves and rosette diameter did not differ significantly for wild type and AtFAAH overexpressors (Figures 2A,B). After 28 days of SD growth, >50% of AtFAAH overexpressors had flowered and had 9–10 rosette leaves. Under LD-growth conditions, all plants, regardless of genotype, had flowered, and had seven to nine rosette leaves. Accelerated growth under LDs is expected because of greater photosynthesis, but the similar number of leaves on AtFAAH overexpressors and LD-grown plants indicates that the plants flowered at nearly the same plastochron age (i.e., morphological age rather than chronological age) (Erickson and Michelini, 1957).
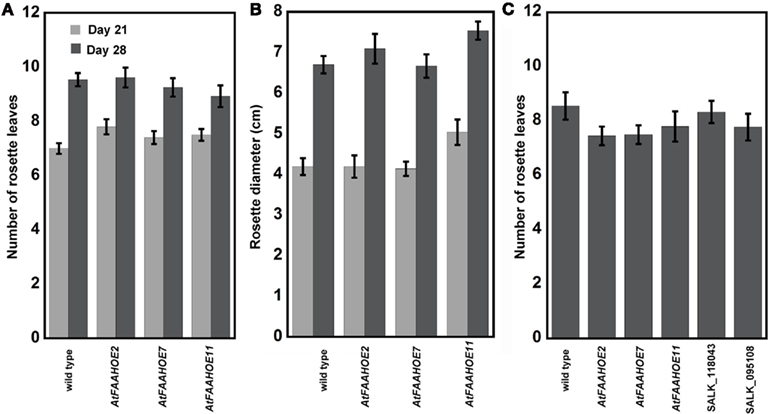
Figure 2. The number of rosette leaves and rosette diameter is not affected by altered AtFAAH expression. Number of rosette leaves (A) and rosette diameter (B) of wild type and three AtFAAH overexpressors under SDs after 21 days, when no plants were flowering, and after 28 days, when some wild type and most AtFAAH overexpressors were flowering. (C) Number of rosette leaves under longs days 28 days after planting in wild type, two AtFAAH knockouts (SALK_118043 and SALK_095108) and three AtFAAH overexpressors (OE2, 7, and 11); all plants were flowering.
Transcriptional Profiling Links Elevated Flowering Locus (FT) Expression to Early Flowering of AtFAAH Overexpressors
To better understand the molecular basis for the early flowering phenotype of AtFAAH overexpressors, microarray analysis was conducted to compare transcript profiles of wild type with that of the AtFAAH overexpressors and AtFAAH knockouts (SALK_095108). Consistent with the fact that AtFAAH knockouts did not have any obvious phenotype (Wang et al., 2006; Figures 1 and 2), a less dramatic difference in the overall gene expression profiles were observed between wild type and AtFAAH knockouts as indicated by a heat map generated from ratios of transcript levels from AtFAAH knockouts (SALK_095108) and wild type (Figure 3A). In contrast, there were more distinct differences in the overall transcript profiles when wild type was compared to AtFAAH overexpressors (AtFAAHOE11; Figure 3A). With a Bonferroni-corrected P-value threshold of 2.19202E-06, we found 157 genes to be differentially regulated by twofold or more in AtFAAH overexpressors compared with wild type. Out of 157 genes, 50 were upregulated and 107 were downregulated in the AtFAAH overexpressor line (Table S1 in Supplementary Material). Only 28 genes were differentially expressed by twofold or more between AtFAAH knockouts and wild type, of which 7 were upregulated and 21 were downregulated (Table S1 in Supplementary Material). As expected, the AtFAAH gene itself was downregulated in AtFAAH knockouts and upregulated in AtFAAH overexpressors, which helped to validate our microarray results. Seven genes were downregulated and one gene was upregulated by twofold or more in both AtFAAH overexpressors and knockouts (Figure 3B). The seven genes downregulated by twofold or more in both AtFAAH overexpressors and knockouts are presented in Table S2 in Supplementary Material. Among the seven genes downregulated in both AtFAAH overexpressors and knockouts, only circadian clock associated (CCA1) has been clearly implicated in flowering. CCA1 is a transcription factor involved in plant circadian rhythms and overexpressing CCA1 causes delayed flowering in Arabidopsis (Wang and Tobin, 1998). The downregulation of CCA1 in AtFAAH overexpressors could disrupt circadian rhythms important for flowering; however, its downregulation in AtFAAH knockouts where no flowering phenotype was apparent is not clear.
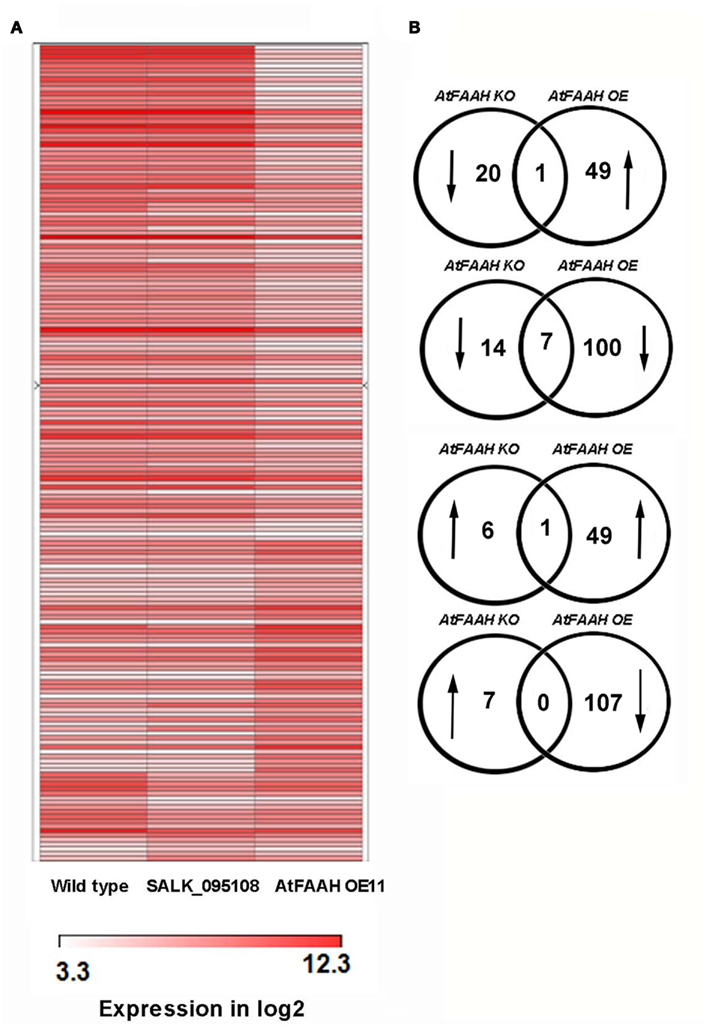
Figure 3. AtFAAH overexpression triggers a more dramatic change in gene expression than AtFAAH knockouts. (A) Heat map of gene expression in wild type, AtFAAH knockout (SALK_095108) and AtFAAH overexpressor (OE11). (B) Venn diagrams showing the number of genes differentially regulated in AtFAAH knockouts and AtFAAH overexpressors relative to wild type.
Transcriptional profiling did not show an overwhelming representation of flowering-related genes that were different between wild type and AtFAAH overexpressors. However, it was noteworthy that FLOWERING LOCUS T (FT), which is a pivotal gene in the transition from vegetative growth to flowering (Corbesier et al., 2007; Giakountis and Coupland, 2008), was elevated in AtFAAH overexpressors. Furthermore, the expression of SEPTELLA3 (SEP3) gene, which encodes a MADs box transcription factor that is regulated by FT in the photoperiodic flowering pathway (Teper-Bamnolker and Samach, 2005), also was elevated in AtFAAH overexpressors (Table 1; Table S1 in Supplementary Material). To verify results from microarray experiments, we conducted quantitative, real time RT-PCR of FT and SEP3 transcripts using three independent AtFAAH overexpressing lines grown for 14 days in LD and SD conditions. Consistent with the microarray results, both LD- and SD-grown AtFAAH overexpressing seedlings showed elevated transcript levels of FT and SEP3. Under SD conditions, FT and SEP3 in three AtFAAH overexpressors were threefold higher relative to wild type (Figure 4A). A similar trend was observed under LD conditions except for one AtFAAH overexpressor (line OE7), which was only twofold elevated relative to wild type (Figure 4B).
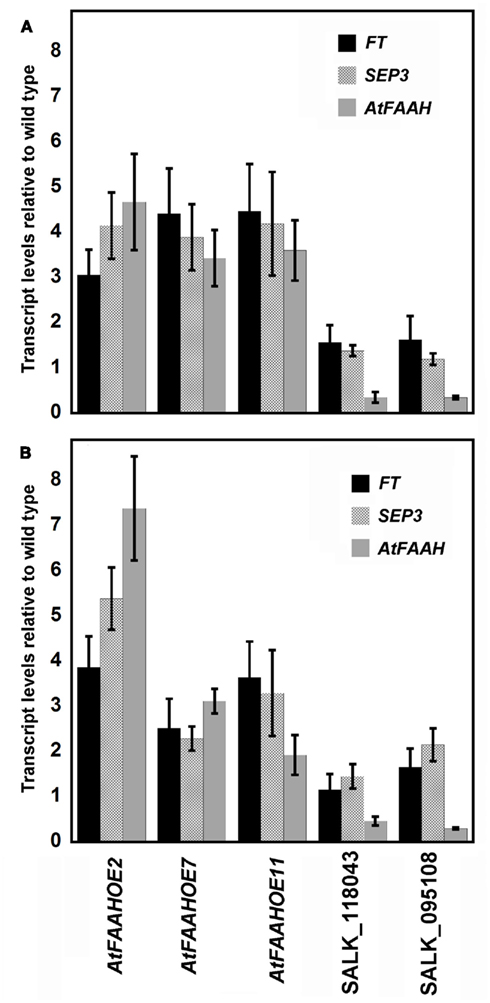
Figure 4. Relative expression profiles of FT, SEP3, and AtFAAH in three AtFAAH overexpressors and two AtFAAH knockouts (SALK_118043 and SALK_095108) determined by quantitative RT-PCR. Fourteen day-old plants grown under short days (A) and long days (B). Values plotted were normalized to 18S rRNA and are the mean of four biological replicates ± SE.
FT and SEP3 were the only upregulated genes in AtFAAH overexpressors that had a known association with flowering (Table 1). To determine whether other genes differentially regulated by AtFAAH overexpression are related to flowering, we compared AtFAAH overexpression transcript data (Table S1 in Supplementary Material) with data from global expression analysis of the shoot apical meristem upon photoperiodic induction (i.e., transfer to LDs; Schmid et al., 2003). This study uncovered a large number of floral repressors that were downregulated upon floral photoperiodic induction. Interestingly, four genes namely At3g16460, At3g27200, At5g43270, and At3g20370, which were downregulated in AtFAAH overexpressors (Table S1 in Supplementary Material), were downregulated upon floral induction by photoperiod and therefore suggested to be potential flowering repressors (Schmid et al., 2003). One gene, namely At3g28500, was upregulated in AtFAAH overexpressors (Table 1) and by photoperiodic flower induction (Schmid et al., 2003). Interestingly in silico analysis using the Genevestigator database1 , 2 (Zimmermann et al., 2004) and the Arabidopsis eFP Browser3 (Winter et al., 2007), indicated that At3g28500, which encodes a 60S acidic ribosomal protein, was preferentially expressed in all stages of flower development as well as the inflorescence shoot apex (Figure A1 in Appendix). Hence there may be additional clues yet to mine related to the molecular regulation of floral transition in the microarray data from AtFAAH overexpressors.
Endogenous NAE Levels are Lower in AtFAAH Overexpressors
To determine if increased NAE hydrolytic activity could explain the early flowering phenotype in AtFAAH overexpressors, we quantified endogenous NAE levels of 14-day-old wild type and three AtFAAH overexpressor lines grown under SDs. Consistent with the overexpression of FAAH, total endogenous NAE content in the three AtFAAH overexpressors was less than that of wild type (Figure 5A). We then examined the levels of the different NAE species to determine which NAE type contributed most to the decline in total NAE levels in AtFAAH overexpressors. We found that the levels of NAE 14:0, NAE 16:0, NAE 18:0, NAE 18:1, and NAE 18:3 were similar between wild type and the three AtFAAH overexpressors (Table 2). On the other hand, endogenous NAE 12:0 and NAE 18:2 were significantly less (up to a 30% reduction) in AtFAAH overexpressors compared to wild type (Table 2; Figure 5B), suggesting that these two NAE types may play a role as lipid mediators in the regulation of flowering time.
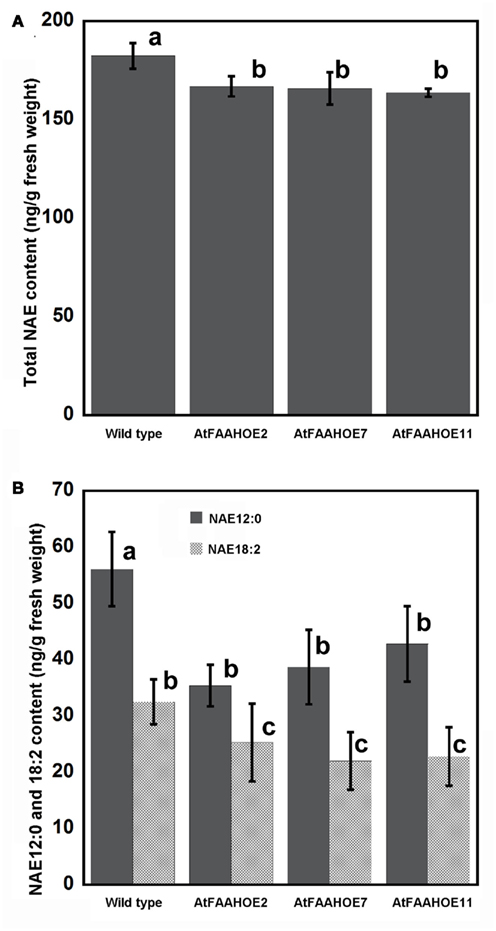
Figure 5. Comparison of endogenous NAE profiles in 14-day-old wild type and AtFAAH overexpressors. NAEs were quantified by isotope-dilution mass spectrometry and summed for total NAE content (A) and total NAE 12:0 and NAE 18:2 (B). Values represent the mean ± SE of three individual extractions from 14-day-old seedlings grown under short days. Mean ± SE with different letters are significantly different (p < 0.005; Tukey’s test).
Exogenous NAE 12:0 Delays the Onset of Flowering in Wild Type Arabidopsis Plants
Given that NAE 12:0 is about 30% reduced in AtFAAH overexpressors, we asked if exogenous application of NAE 12:0 delays flowering time in wild type Arabidopsis plants. Seven days after planting directly onto soil, 50 mL of 35 μM NAE 12:0 was applied directly to the base of the plants every 3 days until senescence. We found that at 24 days, rosette leaf number in NAE 12:0-treated wild type plants were slightly less than solvent controls (Figure 6A). NAE-treated plants showed a delay in flowering by at least 6 days and this occurred whether the treatment began at day 7 or on day 20 (few days prior to flowering under these conditions). We also quantified inflorescence stem height when NAE 12:0 was applied beginning at 7 days or beginning at 20 days. We found that plants treated with exogenous NAE 12:0 regardless of the age of the plant, had shorter inflorescence stems compared to plants treated with the solvent control solution (Figures 6B,C). Hence, it appeared that application of NAE to plants, either early or late, had two general effects – one, a reduction in growth and two, a delay in flowering. The similar impact on both flowering and inflorescence elongation between early (7 day) and late (20 day) treatments suggests that these impacts perhaps result from a signal transduction-mediated transition event and not simply a delay in development.
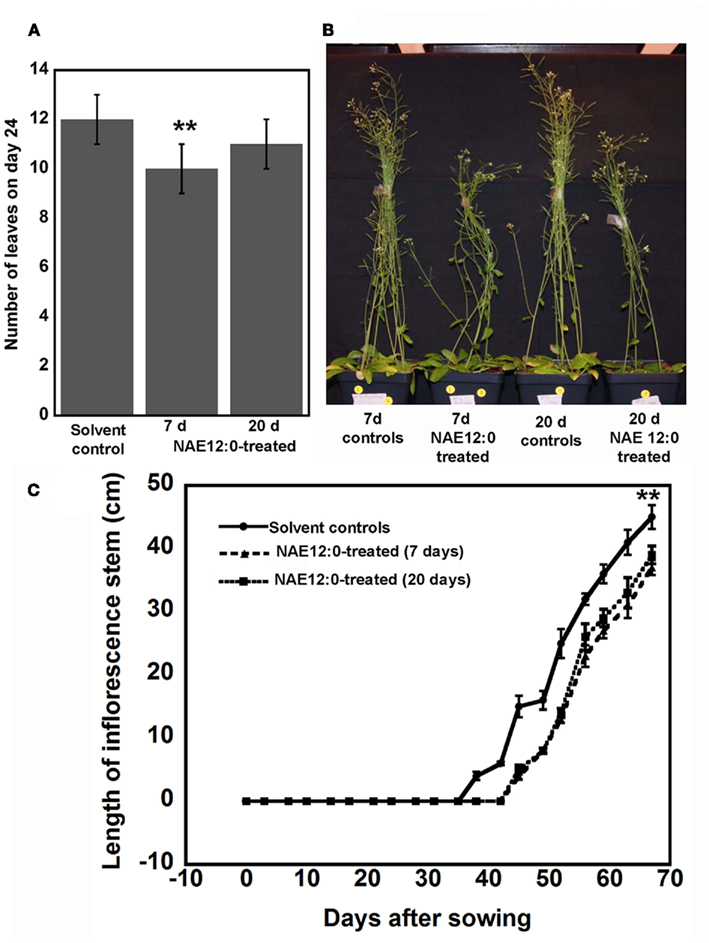
Figure 6. Exogenous NAE 12:0 application delays flowering time in wild type plants. (A) Number of leaves of in 24-day-old plants treated with solvent control solution of 35 μM NAE 12:0 at 7 or 20 days after planting. (B) Representative images of wild type Arabidopsis plants exposed continuously to NAE 12:0 beginning at 7 or 20 days after planting. Note the shorter inflorescence stems of NAE 12:0-treated plants. (C) Quantification of the length of the inflorescence stem after the initiation of flowering. Note that flowering in NAE 12:0-treated plants was delayed by about 6 days regardless of whether the plant was treated at 7 or 20 days after planting. Asterisks indicate statistically significant difference according to Student’s t-test (p < 0.01).
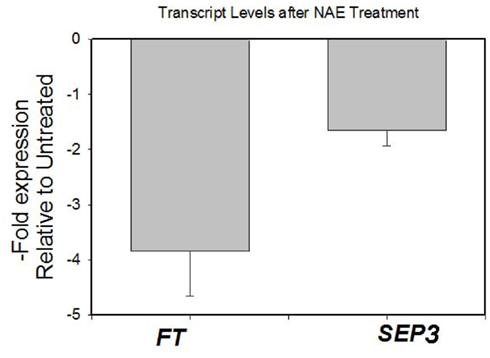
We also treated 7-day-old seedlings with 50 μM NAE 12:0 and examined the expression of FT and SEP3 by quantitative, real time RT-PCR. Consistent with the delayed flowering time of NAE 12:0-treated plants and elevated expression of FT in AtFAAH overexpressors, we found that exogenous NAE 12:0 reduced FT expression by about threefold. SEP3 expression on the other hand was reduced by only 1.3-fold (Figure A2 in Appendix).
Discussion
Although early flowering in AtFAAH overexpressors was noted previously (Wang et al., 2006), it was unclear how the development of this phenotype is triggered. In this paper, we showed that early flowering of AtFAAH overexpressors was associated with increased expression of the key flowering integrator gene FT, under both inductive LD and non-inductive SDs. These results are not entirely surprising, given that FT is well established to be a potent promoter of the transition to flowering by coordinating the expression of a complex network of genes within the shoot apex (for review see Giakountis and Coupland, 2008). Some of the genes in the FT pathway act as repressors of FT transcription during non-inductive SDs (Gómez-Mena et al., 2001; Piñeiro et al., 2003; Takada and Goto, 2003) or are transcriptionally activated by FT during inductive LDs (Ruiz-Garcia et al., 1997; Teper-Bamnolker and Samach, 2005). However, despite the elevated expression of FT, many of the genes that have been associated with FT function did not change dramatically in AtFAAH overexpressors (Table S1 in Supplementary Material). Only the floral organ identity gene, SEP3, was elevated in AtFAAH overexpressors by twofold or more, which is consistent with a previous report showing that FT regulates SEP3 accumulation (Teper-Bamnolker and Samach, 2005). These observations suggest that FT might have other downstream target genes that promote flowering or unknown repressors that regulate FT transcription in the AtFAAH overexpression background. One possible target of FT is the 60S acidic ribosomal protein encoded by the At3g28500 gene, which was upregulated twofold in our microarrays (Table 1) and also reported to be induced in LDs in an FT-dependent manner (Schmid et al., 2003). Moreover, the At3g28500 gene was upregulated fourfold when HEME ACTIVATOR PROTEIN 3b (HAP3b), which encodes a CCAAT-binding transcription factor, is overexpressed and downregulated more than twofold in hap3b mutants. HAP3b overexpressors display early flowering whereas hap3b mutants are late flowering under LDs (Cai et al., 2007) suggesting that At3g28500 is an important component of flowering under LDs. Although the significance of overexpressing a 60S acidic ribosomal protein gene for promoting flowering is unknown, it is tempting to speculate that the translation of select mRNA in the cell particularly those involved in flowering might be affected, and thus partly explain early flowering in AtFAAH overexpressors.
It should be noted that the early flowering phenotype exhibited by AtFAAH overexpressors is not typical of other early flowering plants in Arabidopsis. For example, early flowering in FT overexpressors under SDs is typically accompanied by smaller rosette leaves and a reduced number of rosette leaves (Kardailsky et al., 1999; Kobayashi et al., 1999; Abe et al., 2005). A reduced number of rosette leaves is also a characteristic found in several flowering mutants that enhance the expression of FT (Piñeiro et al., 2003; Yoo et al., 2005; Cai et al., 2007; Seo et al., 2011), and reflects the shorter period of vegetative growth. The number of leaves of AtFAAH overexpressors and wild type at 21 and 28 days, under SD conditions, were the same, indicating that vegetative development was not accelerated. However, by 28 days most AtFAAH overexpressors had transitioned to flowering with 9–10 rosette leaves, while most wild type plants were vegetative and still producing leaves. AtFAAH overexpressors therefore behaved more like plants grown under LD conditions, which, irrespective of genotype, produced seven to nine leaves before flowering (Figure 2). These data indicate that the early flowering in AtFAAH overexpressors is independent of the rate of rosette leaf production and is not a product of enhanced growth, but results from uncoupling flowering from photoperiod, presumably via FT activation. In addition to FT, overexpression of other genes in the flowering pathway uncouple flowering from photoperiod including CONSTANS (CO), a transcription factor that activates FT (Takada and Goto, 2003), and the downstream meristem identity genes LEAFY (LFY; Schultz and Haughn, 1991) and APETALA1 (AP1; Mandel et al., 1992). If AtFAAH overexpression bypasses photoperiodic induction by elevating FT directly, elevated LFY, and AP1 would be expected. However, these genes did not change significantly in microarray studies of AtFAAH overexpressors.
Recent work in tomato (Shalit et al., 2009) and maize (Danilevskaya et al., 2010), define FT and its orthologs as general determinacy factors that function to limit vegetative growth – with smaller leaves and the transition to reproductive growth being only two manifestations of the broader impacts. However, the reduced leaf size that commonly accompanies FT overexpression was not observed in AtFAAH overexpressors. It is possible that elevated AtFAAH, which previously was shown to enhanced vegetative growth in Arabidopsis (Wang et al., 2006), can overcome the negative effects of FT overexpression on leaf development by reducing endogenous levels of NAE (Figure 5). Consistent with this notion is the finding that exogenous application of NAE 12:0 to wild type plants, which was more than 30% less in AtFAAH overexpressors than wild type, delayed flowering in wild type plants (Figure 6).
A global expression study of Arabidopsis shoot apices uncovered a large number of potential floral repressors that were downregulated upon photoperiodic induction compared to upregulated genes (Schmid et al., 2003). A similar trend was observed in AtFAAH overexpressors in that there were more genes downregulated than upregulated (Figure 3; Table 1). Interestingly, four of the potential floral repressors identified by Schmid et al. (2003) were also downregulated in AtFAAH overexpressors including genes encoding mannose-binding lectin superfamily protein (At3g16460), cupredoxin superfamily protein (At3g27200), squamosa-promoter binding protein-like (SPL; At5g43270), and TRAF-like family protein (At3g20370). Although members of the SPL gene family are known to regulate floral transition through microRNAs (miRNAs) and FT activation (Wang et al., 2009), the significance of the downregulation of the aforementioned four genes for the early flowering phenotype of AtFAAH overexpressors is not clear. Activation of FT expression also is well known to be regulated by the B-box zinc finger transcription factor CO, particularly under LD conditions (An et al., 2004; Ayre and Turgeon, 2004). Expression of CO was not significantly changed in AtFAAH overexpressors but a gene homologous to CO (At3g02380) was twofold downregulated (Table S1 in Supplementary Material). Because it is the timing of CO expression and CO protein stability that is crucial for photoperiodic flower induction (Imaizumi, 2010), we cannot rule out the possibility that microarray analysis of AtFAAH overexpressors was not able to detect significant changes in CO expression given that RNA from only one time point was collected. Alternatively, there are reports that induction of FT expression and flowering could be facilitated by CO-independent pathways. For example, the miRNA172 pathway was shown to induce FT expression and flowering in Arabidopsis despite the absence of functional CO (Jung et al., 2007). The depletion of endogenous NAEs by AtFAAH overexpression could bypass the requirement for CO to trigger FT expression through yet to be characterized CO-independent pathways.
N-lauroylethanolamine (NAE 12:0) elicits a number of growth inhibitory effects when applied to Arabidopsis seedlings (Blancaflor et al., 2003; Motes et al., 2005). This NAE type is comparatively low in concentration in desiccated seeds, but is among the most prevalent NAE type in seedling and leaf tissues (Wang et al., 2006; Figure 5). Seed germination and post-germinative growth is characterized by the targeted depletion of the predominant polyunsaturated NAEs such that the NAE composition and content of vegetative tissues differs substantially from that of seeds (Kim et al., 2010). While there is much to be learned about the relationship of individual NAE species with physiological function in plants, this manuscript provides new information about NAE 12:0, namely the delay in flowering time in Arabidopsis. Plants watered with NAE 12:0-containing solutions (35 μM) were delayed in flowering (Figure 6). This delay was observed whether the exogenous application of NAE 12:0 began at 7 or 20 days after sowing indicating that the timing (prior to flowering) but not the length of exposure to NAE 12:0 was required to delay their transition to flowering. Perhaps more significantly, the endogenous level of NAE 12:0 was the NAE type that was most altered in the AtFAAH overexpressing lines, suggesting that the specific metabolism of NAE 12:0 by AtFAAH is related to flowering transition time. Response to application of NAE 12:0 solutions to the soil implies that NAE 12:0 is likely taken up through the roots and translocated to the shoots to where it exerts its effect. It is tempting to speculate that FT may be a direct target of NAE 12:0, since FT has homology to known phosphatidylethanolamine binding proteins (PEBP). To our knowledge there have been no studies to examine the interaction of acylethanolamines with FT or FT-related proteins. In fact the annotation of FT as a member of the PEBP family comes from sequence homology rather than from functional binding data. Certainly future experiments to probe the role of NAE signaling in flowering time via FT should include assays of direct interaction between FT and NAE 12:0 and other ethanolamine-containing lipids. Whether the action of NAE 12:0 on FT is direct or indirect, the data presented here provide the first indication that NAE metabolism may play a role in the signal transduction events that lead to FT-mediated transition from vegetative to reproductive growth in Arabidopsis. It remains to be tested whether or not the NAE pathway represents a bypass of other floral transition mechanisms such as CO.
Author Contributions
N. D. Teaster performed flowering assays, quantitative RT-PCR and assisted in gathering plant material for NAE quantification. J. Keereetaweep conducted the NAE quantification assays. Y.-S. Wang and Y. Tang conducted the microarray studies and A. Kilaru and C. N.-Q. Tran performed the external NAE experiments on flowering time. N. D. Teaster wrote the paper and analyzed data with the assistance of B. G. Ayre, K. D. Chapman, and E. B. Blancaflor.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant from the United States Department of Energy, Office of Basic Energy Sciences (BES, grant number DE-FG02-05ER15647) to K. D. Chapman and E. B. Blancaflor and grants from the Estonia Science Foundation (ETF7869) and Ministry of Education and Research (SF0180071S07) to Y.-S. Wang. C. N.-Q. Tran was supported by a summer research fellowship from the Texas Academy of Math and Sciences, University of North Texas.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Plant_Physiology/10.3389/fpls.2012.00032/abstract
Table S1| Differentially expressed genes in AtFAAH overexpressor and AtFAAH knockouts compared to wild type. These data were deposited with the EMBL–EBI ArrayExpress database under accession E-MEXP-3486.
Table S2| Genes downregulated twofold or more in both AtFAAH overexpressors and AtFAAH knockouts.
Footnotes
References
Abdi, H. (2007). “Bonferroni and Sidak corrections for multiple comparisons,” in Encyclopedia of Measurement and Statistics, ed. N. J. Salkind (Thousand Oaks, CA: Sage), 103–107.
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056.
An, H., Roussot, C., Suárez-López, P., Corbesier, L., Vincent, C., Piñeiro, M., Hepworth, S., Mouradov, A., Justin, S., and Turnbull, C. (2004). Constans acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131, 3615–3626.
Ayre, B. G., and Turgeon, R. (2004). Graft transmission of a floral stimulant derived from constans. Plant Physiol. 135, 2271–2278.
Blancaflor, E. B., and Chapman, K. D. (2006). “Similarities between endocannabinoid signaling in animal systems and N-acylethanolamine metabolism in plants,” in Communication in Plants, eds F. Baluska, S. Mancuso, and D. Volkmann (Berlin: Springer-Verlag), 205–219.
Blancaflor, E. B., Hou, G., and Chapman, K. D. (2003). Elevated levels of N-lauroylethanolamine, an endogenous constituent of desiccated seeds, disrupt normal root development in Arabidopsis thaliana seedlings. Planta 217, 206–217.
Cai, X., Ballif, J., Endo, S., Davis, E., Liang, M., Chen, D., Dewald, D., Kreps, J., Zhu, T., and Wu, Y. (2007). A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol. 145, 98.
Chebrou, H., Bigey, F., Arnaud, A., and Galzy, P. (1996). Study of the amidase signature group. Biochim. Biophys. Acta 1298, 285–293.
Clement, A. B., Hawkins, E. G., Lichtman, A. H., and Cravatt, B. F. (2003). Increased seizure susceptibility and proconvulsant activity of anandamide in mice lacking fatty acid amide hydrolase. J. Neurosci. 23, 3916–3923.
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., Giakountis, A., Farrona, S., Gissot, L., Turnbull, C., and Coupland, G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033.
Cotter, M. Q., Teaster, N. D., Blancaflor, E., and Chapman, K. D. (2011). N-acylethanolamine (NAE) inhibits growth in Arabidopsis thaliana seedlings via ABI3-dependent and-independent pathways. Plant Signal. Behav. 6, 671–679.
Cravatt, B. F., Demarest, K., Patricelli, M. P., Bracey, M. H., Giang, D. K., Martin, B. R., and Lichtman, A. H. (2001). Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc. Natl. Acad. Sci. U.S.A. 98, 9371–9376.
Danilevskaya, O. N., Meng, X., and Ananiev, E. V. (2010). Concerted modification of flowering time and inflorescence architecture by ectopic expression of TFL1-like genes in maize. Plant Physiol. 153, 238–251.
Dean Rider, S., Henderson, J. T., Jerome, R. E., Edenberg, H. J., Romero-Severson, J., and Ogas, J. (2003). Coordinate repression of regulators of embryonic identity by pickle during germination in Arabidopsis. Plant J. 35, 33–43.
Dozmorov, I., and Centola, M. (2003). An associative analysis of gene expression array data. Bioinformatics 19, 204–211.
Fowler, C. J. (2006). The cannabinoid system and its pharmacological manipulation – a review, with emphasis upon the uptake and hydrolysis of anandamide. Fundam. Clin. Pharmacol. 20, 549–562.
Giakountis, A., and Coupland, G. (2008). Phloem transport of flowering signals. Curr. Opin. Plant Biol. 11, 687–694.
Gómez-Mena, C., Piñeiro, M., Franco-Zorrilla, J. M., Salinas, J., Coupland, G., and Martínez-Zapater, J. M. (2001). Early bolting in short days: an Arabidopsis mutation that causes early flowering and partially suppresses the floral phenotype of leafy. Plant Cell 13, 1011–1024.
Gopalakrishna, K. N., Stewart, B. H., Kneen, M. M., Andricopulo, A. D., Kenyon, G. L., and Mcleish, M. J. (2004). Mandelamide hydrolase from Pseudomonas putida: characterization of a new member of the amidase signature family. Biochemistry 43, 7725–7735.
Imaizumi, T. (2010). Arabidopsis circadian clock and photoperiodism: time to think about location. Curr. Opin. Plant Biol. 13, 83–89.
Irizarry, R. A., Bolstad, B. M., Collin, F., Cope, L. M., Hobbs, B., and Speed, T. P. (2003). Summaries of affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15.
Jung, J. H., Seo, Y. H., Seo, P. J., Reyes, J. L., Yun, J., Chua, N. H., and Park, C. M. (2007). The gigantea-regulated microRNA172 mediates photoperiodic flowering independent of constans in Arabidopsis. Plant Cell 19, 2736–2748.
Kang, L., Wang, Y. S., Uppalapati, S. R., Wang, K., Tang, Y., Vadapalli, V., Venables, B. J., Chapman, K. D., Blancaflor, E. B., and Mysore, K. S. (2008). Overexpression of a fatty acid amide hydrolase compromises innate immunity in Arabidopsis. Plant J. 56, 336–349.
Kardailsky, I., Shukla, V. K., Ahn, J. H., Dagenais, N., Christensen, S. K., Nguyen, J. T., Chory, J., Harrison, M. J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965.
Kilaru, A., Tamura, P., Garg, P., Isaac, G., Baxter, D., Duncan, S. R., Venables, B. J., Welti, R., Koulen, P., and Chapman, K. D. (2010). Changes in N-acylethanolamine pathway related metabolites in a rat model of cerebral ischemia/reperfusion. Lipids 45, 863–875.
Kim, S. C., Chapman, K. D., and Blancaflor, E. B. (2010). Fatty acid amide lipid mediators in plants. Plant Sci. 178, 411–419.
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962.
Labahn, J., Neumann, S., Büldt, G., Kula, M. R., and Granzin, J. (2002). An alternative mechanism for amidase signature enzymes. J. Mol. Biol. 322, 1053–1064.
Li, C., and Wong, W. H. (2001). Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. U.S.A. 98, 31–36.
Lichtman, A. H., Shelton, C. C., Advani, T., and Cravatt, B. F. (2004). Mice lacking fatty acid amide hydrolase exhibit a cannabinoid receptor-mediated phenotypic hypoalgesia. Pain 109, 319–327.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-[delta][delta]CT method. Methods 25, 402–408.
Mandel, M. A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M. F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene apetala1. Nature 360, 273–277.
McKinney, M. K., and Cravatt, B. F. (2005). Structure and function of fatty acid amide hydrolase. Annu. Rev. Biochem. 74, 411–432.
Motes, C. M., Pechter, P., Yoo, C. M., Wang, Y. S., Chapman, K. D., and Blancaflor, E. B. (2005). Differential effects of two phospholipase D inhibitors, 1-butanol and N-acylethanolamine, on in vivo cytoskeletal organization and Arabidopsis seedling growth. Protoplasma 226, 109–123.
Neu, D., Lehmann, T., Elleuche, S., and Pollmann, S. (2007). Arabidopsis amidase 1, a member of the amidase signature family. FEBS J. 274, 3440–3451.
Patricelli, M. P., Lovato, M. A., and Cravatt, B. F. (1999). Chemical and mutagenic investigations of fatty acid amide hydrolase: evidence for a family of serine hydrolases with distinct catalytic properties. Biochemistry 38, 9804–9812.
Piñeiro, M., Gómez-Mena, C., Schaffer, R., Martínez-Zapater, J. M., and Coupland, G. (2003). Early bolting in short days is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT. Plant Cell 15, 1552–1562.
Ruiz-Garcia, L., Madueño, F., Wilkinson, M., Haughn, G., Salinas, J., and Martinez-Zapater, J. M. (1997). Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9, 1921–1934.
Schmid, M., Uhlenhaut, N. H., Godard, F., Demar, M., Bressan, R., Weigel, D., and Lohmann, J. U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130, 6001–6012.
Schultz, E. A., and Haughn, G. W. (1991). Leafy, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell 3, 771–781.
Seo, E., Yu, J., Ryu, K. H., Lee, M. M., and Lee, I. (2011). Werewolf, a regulator of root hair pattern formation, controls flowering time through the regulation of FT mRNA stability. Plant Physiol. 156, 1867–1877.
Shalit, A., Rozman, A., Goldshmidt, A., Alvarez, J. P., Bowman, J. L., Eshed, Y., and Lifschitz, E. (2009). The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. U.S.A. 106, 8392–8397.
Shrestha, R., Dixon, R. A., and Chapman, K. D. (2003). Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis thaliana. J. Biol. Chem. 278, 34990–34997.
Shrestha, R., Kim, S., Dyer, J. M., Dixon, R. A., and Chapman, K. D. (2006). Plant fatty acid (ethanol) amide hydrolases. Biochim. Biophys. Acta 1761, 324–334.
Takada, S., and Goto, K. (2003). Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15, 2856–2865.
Teaster, N. D., Motes, C. M., Tang, Y., Wiant, W. C., Cotter, M. Q., Wang, Y. S., Kilaru, A., Venables, B. J., Hasenstein, K. H., Gonzalez, G., Blancaflor, E. B., and Chapman, K. D. (2007). N-acylethanolamine metabolism interacts with abscisic acid signaling in Arabidopsis thaliana seedlings. Plant Cell 19, 2454–2469.
Teper-Bamnolker, P., and Samach, A. (2005). The flowering integrator FT regulates sepallata3 and fruitfull accumulation in Arabidopsis leaves. Plant Cell 17, 2661–2675.
Turck, F., Fornara, F., and Coupland, G. (2008). Regulation and identity of florigen: flowering locus T moves center stage. Annu. Rev. Plant Biol. 59, 573–594.
Venables, B. J., Waggoner, C. A., and Chapman, K. D. (2005). N-acylethanolamines in seeds of selected legumes. Phytochemistry 66, 1913–1918.
Wang, J. W., Czech, B., and Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749.
Wang, Y. S., Shrestha, R., Kilaru, A., Wiant, W., Venables, B. J., Chapman, K. D., and Blancaflor, E. B. (2006). Manipulation of Arabidopsis fatty acid amide hydrolase expression modifies plant growth and sensitivity to N-acylethanolamines. Proc. Natl. Acad. Sci. U.S.A. 103, 12197–12202.
Wang, Z.-Y., and Tobin, E. M. (1998). Constitutive expression of the circadian clock associated (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217.
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., and Provart, N. J. (2007). An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2, e718. doi:10.1371/journal.pone.0000718
Yoo, S. K., Chung, K. S., Kim, J., Lee, J. H., Hong, S. M., Yoo, S. J., Yoo, S. Y., Lee, J. S., and Ahn, J. H. (2005). Constans activates suppressor of overexpression of constans 1 through flowering locus T to promote flowering in Arabidopsis. Plant Physiol. 139, 770–778.
Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632.
Appendix
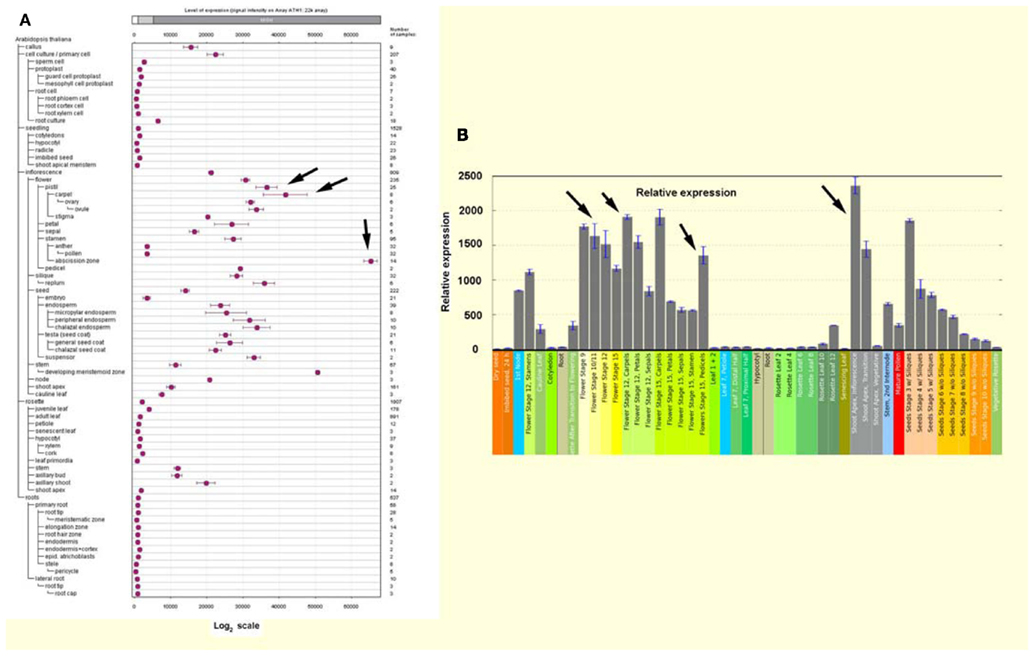
Figure A1. At3g28500 is strongly expressed in flowers and inflorescence meristem (arrows). In silico expression profiling of At3g28500 based on publicly available microarray data sets from Genevestigator [(A), Zimmermann et al., 2004] and Arabidopsis eFP browser [(B), Winter et al., 2007].
Keywords: Arabidopsis, fatty acid amide hydrolase, flowering, FLOWERING LOCUS T, lipid signaling, N-acylethanalomanine
Citation: Teaster ND, Keereetaweep J, Kilaru A, Wang Y-S, Tang Y, Tran CN-Q, Ayre BG, Chapman KD and Blancaflor EB (2012) Overexpression of fatty acid amide hydrolase induces early flowering in Arabidopsis thaliana. Front. Plant Sci. 3:32. doi: 10.3389/fpls.2012.00032
Received: 25 November 2011; Paper pending published: 20 December 2011;
Accepted: 01 February 2012; Published online: 20 February 2012.
Edited by:
Xuemin Wang, University of Missouri St Louis, USAReviewed by:
Sona Pandey, Donald Danforth Plant Science Center, USAJin Chen, Michigan State University, USA
Copyright: © 2012 Teaster, Keereetaweep, Kilaru, Wang, Tang, Tran, Ayre, Chapman and Blancaflor. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Elison B. Blancaflor, Plant Biology Division, The Samuel Roberts Noble Foundation, 2510 Sam Noble Parkway, Ardmore, OK, USA. e-mail:ZWJsYW5jYWZsb3JAbm9ibGUub3Jn
†Present address: Neal D. Teaster, Crop Production Systems Research Unit, Agricultural Research Service, United States Department of Agriculture, PO Box 350, Stoneville, MS 38776, USA.