
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 04 January 2012
Sec. Plant Physiology
Volume 2 - 2011 | https://doi.org/10.3389/fpls.2011.00107
This article is part of the Research Topic Arabidopsis 2010 and beyond – big science with a small weed View all 21 articles
The plant hormone gibberellin (GA) controls major aspects of plant growth such as germination, elongation growth, flower development, and flowering time. In recent years, a number of studies have revealed less apparent roles for GA in a surprisingly broad set of developmental as well as cell biological processes. The identification of GA receptor proteins on the one end of the signaling cascade, DELLA proteins as central repressors of the pathway and transcription regulators such as the phytochrome interacting factors and the GATA-type transcription factors GNC and CGA1/GNL on the current other end of the signaling cascade have extended our knowledge about how GA and DELLAs regulate a diverse set of plant responses.
The diterpenoid substances of the gibberellin (GA) class were first described and isolated in the 1920s and 1930s based on the property of a compound isolated from the rice pathogenic fungus Gibberella fujikuroi to induce strong elongation growth and other disease symptoms in rice. After their identification as causative agent of this so-called bakanae (foolish seedling) disease, it was later discovered that GAs are also synthesized by plants where they promote a number of important developmental processes besides elongation such as germination and flowering. In the following decades, GA biology gained particular attention because it was recognized that interfering with GA signaling by chemical or genetic means could be used to modulate plant growth and most importantly to control crop yield and quality (Peng et al., 1999; Rademacher, 2000; Hedden, 2003).
The mechanisms that underlie GA action in plant growth control have mainly been revealed through studies conducted in rice, Arabidopsis and other model species such as pea and tomato. There, the analysis of mutants with defects in GA biosynthesis and signaling as well as the availability of chemical GA biosynthesis inhibitors has allowed the identification of the molecular components that control GA response during germination (Lee et al., 2002; Cao et al., 2005; Penfield et al., 2006; Piskurewicz et al., 2008, 2009; Piskurewicz and Lopez-Molina, 2009), during hypocotyl elongation and hook formation (Achard et al., 2003, 2007b; Alabadi et al., 2004; Djakovic-Petrovic et al., 2007), in chlorophyll and anthocyanin accumulation (Jiang et al., 2007; Richter et al., 2010; Cheminant et al., 2011), in flower development and in flowering time control (Cheng et al., 2004; Tyler et al., 2004; Achard et al., 2007a) as well as in fertilization (Chhun et al., 2007). More recently, less apparent roles for GAs could be elucidated such as roles in cell proliferation (Achard et al., 2009), hypocotyl xylem expansion (Ragni et al., 2011), phosphate starvation response (Jiang et al., 2007), pathogen responses (Navarro et al., 2008), oxidative stress response (Achard et al., 2008), and the response to abiotic environmental cues (Achard et al., 2006).
In order to keep the complexity of the present minireview to an appropriate level, this review almost exclusively summarizes molecular results from rice and Arabidopsis thaliana, where the wealth of genetic resources has enabled gaining the most valuable insights into GA biology and signaling.
The discovery of the GA receptor based on the rice gibberellin insensitive dwarf1 (gid1) mutant in 2005 represented a major breakthrough in the understanding of the signaling pathway of this hormone (Ueguchi-Tanaka et al., 2005). While rice has only one GID1 gene, Arabidopsis has three functional GID1 orthologs, and the loss of all three Arabidopsis GID1 genes is required for a complete loss of GA response (Griffiths et al., 2006; Willige et al., 2007). Following hormone binding, the soluble GID1 proteins interact with the DELLA growth repressors such as SLENDER RICE1 (SLR1) in rice (Ikeda et al., 2001) and GIBBERELLIC ACID INSENSITIVE (GAI; Peng et al., 1997), REPRESSOR-OF-ga1-3 (RGA; Silverstone et al., 1998), and RGA-LIKE1 (RGL1), RGL2, and RGL3 in Arabidopsis (Lee et al., 2002; Wen and Chang, 2002; Cheng et al., 2004). In the absence of GA, these DELLA proteins repress germination, growth, and other GA-dependent processes. In the presence of GA, the GID1 interaction induces DELLA degradation via the rice SCFGID2 (SKP1-CULLIN-F-BOX complex with the F-box protein subunit GID2; Sasaki et al., 2003; Gomi et al., 2004) or the Arabidopsis SCFSLY1 or SCFSNE (SCF complexes with the F-box protein subunit SLEEPY1 or SNEEZY; Mcginnis et al., 2003; Dill et al., 2004; Fu et al., 2004; Dohmann et al., 2010; Ariizumi et al., 2011) E3 ubiquitin ligases and the 26S proteasome (Figure 1A).
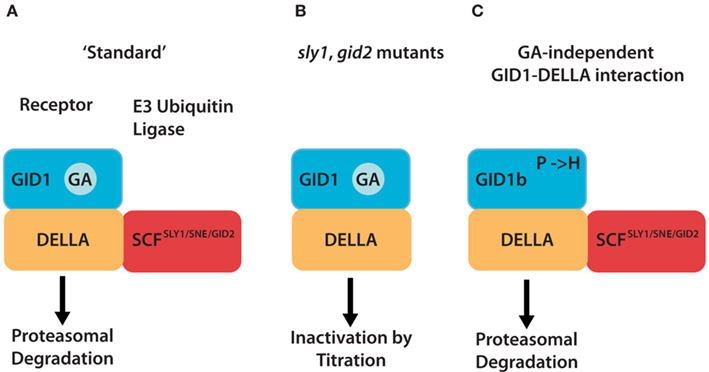
Figure 1. Different mechanism serve to inactivate DELLA repressors of the GA signaling pathway. (A) In the “standard” situation, GA-bound GID1 proteins interact with DELLA repressors and induce their ubiquitylation and degradation via E3 ubiquitin ligases such as Arabidopsis SCFSLY1/SNZ or rice SCFGID2. (B) DELLA ubiquitylation and degradation are defective in E3 ubiquitin ligase mutants such as sly1 or gid2. There, the GA-promoted GID1–DELLA interaction is sufficient to inactivate DELLAs and relieve DELLA-imposed growth restraints. (C) GID1 variants with a substitution of a conserved proline (P) residue can interact with DELLAs in a GA-independent manner and promote GA signaling independent from the hormone. Arabidopsis GID1b is a naturally occurring GID1 protein that has a histidine instead of the proline (P → H). GID1 mutant analyses additionally revealed that P → A or P → S substitutions render GID1 GA-independent.
In monocot and dicot species with only one DELLA protein, such as rice or tomato, the activity of GA signaling or the progression of GA response can be judged based on the abundance of the DELLA protein and GA responses can be completely uncoupled from GA signaling in DELLA gene mutants (Itoh et al., 2002; Bassel et al., 2004). In species with multiple DELLA proteins, such as Arabidopsis, the latter statement is complicated by the fact that homeostasis mechanisms are in place that regulate the overall abundance of the functionally redundant DELLA proteins via negative feedback mechanisms that at least in part function via GA biosynthesis (Peng et al., 1997). E.g., Arabidopsis mutants and transgenic lines that accumulate the DELLA protein GAI have reduced levels of the DELLA protein RGA when compared with wild type plants (Willige et al., 2007). At the same time it is known that the expression of GA biosynthesis genes is upregulated in such backgrounds and that reduced RGA levels are the consequence of increased GA-dependent protein turnover.
DELLA proteins are typically but not exclusively inactivated by protein degradation. Besides proteasomal degradation, it was shown that the GA-induced interaction with the GID1 receptors is sufficient to inhibit DELLA activity (Ariizumi et al., 2008; Ueguchi-Tanaka et al., 2008). This inactivating mechanism allows to explain the comparatively mild phenotypes of rice gid2 and Arabidopsis sly1 mutants, E3 ubiquitin ligase subunit mutants that accumulate very high levels of DELLA proteins. In these mutants, these inhibitory GA-induced GID1–DELLA protein interactions can still take place while DELLA ubiquitylation and proteasomal degradation are blocked (Figure 1B).
In addition, the analysis of a rice gid1 mutant suppressor mutation revealed that GID1 GA receptor variants exist that can interact with DELLAs in a GA-independent (or GA-hypersensitive) manner (Yamamoto et al., 2010). Biochemical and physiological data suggest that the substitution of a specific proline residue in GID1, e.g., by a serine or alanine allows interactions with the DELLA SLR1 in the absence of GA and that this mutant GID1 variant can therefore suppress GA-deficiency phenotypes (Figure 1C). Interestingly, one of the three GID1 proteins from Arabidopsis, GID1b, can also interact with DELLAs in a GA-independent manner (Griffiths et al., 2006; Nakajima et al., 2006). Intriguingly, the critical proline residue identified in rice GID1 corresponds to a histidine in Arabidopsis GID1b (but to proline in GID1a and GID1c). Even more so, the GID1b–DELLA protein interaction becomes entirely GA-dependent when the respective histidine of GID1b is replaced by a proline (Yamamoto et al., 2010). It can thus be postulated that GA-independent GA receptor function may be an integral part of GA signaling, in at least some plant species, where it may serve to fine tune GA-signal transduction by promoting GA-independent DELLA inactivation mechanisms.
The biochemical role of DELLA proteins as biological repressors of GA responses has been enigmatic for an entire decade (Peng et al., 1997). In recent years, however, several studies have elucidated the identity of DELLA-interaction partners and have allowed biochemical modes of action for DELLA function to be proposed. These studies revealed that the hypothesis about their predicted role as transcriptional regulators was correct, while at the same time, the hypothesis about them functioning as DNA-binding proteins most likely is incorrect.
Phytochrome interacting factors (PIFs) are a subfamily of basic helix-loop-helix (bHLH) transcription factors that are characterized by the AP domain, a domain for the binding of the light-activated Pfr conformer of phytochromes (Leivar and Quail, 2010). DELLAs interact with and regulate the activity of PIF3 and PIF4 (De Lucas et al., 2008; Feng et al., 2008), and they may also regulate the activity of other PIFs (Gallego-Bartolome et al., 2010). Through these interactions, DELLAs seemingly prevent PIFs from binding to their cognate promoter binding sites, and they thereby interfere with the transcriptional activity of PIFs and ultimately their biological function, e.g., in promoting hypocotyl elongation (Figure 2A; De Lucas et al., 2008; Feng et al., 2008). Importantly, PIFs also integrate light signaling through their interaction with activated phytochrome, and this is followed by their proteasomal degradation. The effect of DELLA-dependent repression on PIF function is therefore most obvious in – but not necessarily restricted to – the dark where the effect of GA and DELLAs on PIF activity can be studied in the absence of the destabilizing effect of light on PIFs. The proposed DELLA–PIF interaction in the dark can very nicely explain the apparent photomorphogenic phenotype of dark-grown pif mutant seedlings (Leivar et al., 2008, 2009; Shin et al., 2009) or of seedlings with increased DELLA abundance and consequently increased DELLA-mediated PIF inactivation (De Lucas et al., 2008; Feng et al., 2008). Since it is known that GA levels decline when dark-grown seedlings are transferred to the light (Achard et al., 2007b), the resulting DELLA stabilization should lead to a repression of PIF function that is then followed by the light-induced PIF degradation. In the light, PIF proteins – albeit present at low levels – are expected to retain functionality in the light-grown seedling and plant. PIFs, namely PIF1, have also been proposed to directly promote the expression of the DELLA genes GAI and RGA, and to indirectly repress GA biosynthesis gene expression, and to thereby contribute to the homeostasis of the GA signaling pathway (Oh et al., 2007).
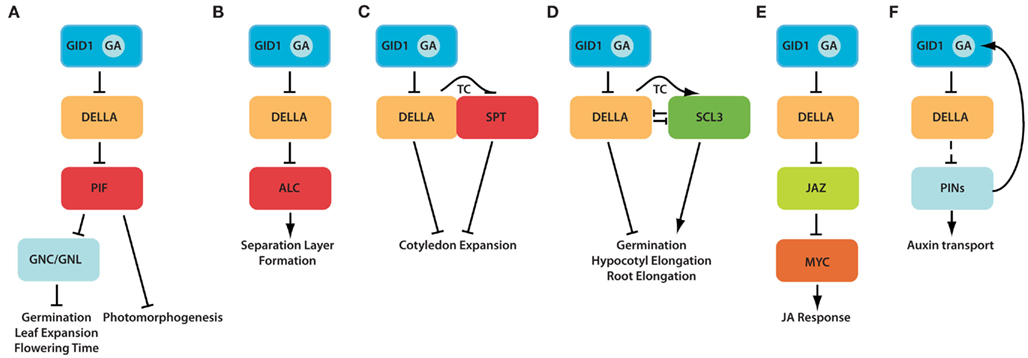
Figure 2. Different molecular modes of action of DELLA repressors. (A) GID1 induces DELLA inactivation by promoting DELLA degradation in response to hormone binding. DELLAs repress the DNA-binding activity of PIFs, which are also negatively regulated by light. GNC and CGA1/GNL are two important targets downstream from GA signaling, DELLAs, and PIFs. GNC and CGA1/GNL repress major but not all aspects of GA signaling. (B) DELLAs also repress the PIF-related protein ALC. Separation layer formation is dependent on the relief of DELLA repression on ALC and this is mediated by the induction of GA biosynthesis gene expression by the ALC upstream regulator IND. (C) SPT is closely related to ALC and PIFs but appears to be regulated by DELLAs in a differential manner from PIFs. The current hypothesis predicts that SPT and DELLAs interact and together repress cotyledon formation, thus SPT and DELLAs act in an analogous manner. DELLAs may also negatively control SPT transcription (transcriptional control, TC). (D) DELLAs also interact with the GRAS-family protein SCL3. DELLAs and SCL3 regulate each other in an antagonistic manner and have opposing effects on the biological processes that they control. (E) JAZ proteins have emerged as central repressors of JA signaling. Their activity is on the one side negatively controlled by DELLA interaction and on the other side by jasmonates (JA). JAZ proteins are repressors of MYC transcription factors. (F) DELLAs indirectly regulate the abundance of PIN proteins and thereby influence auxin transport and auxin transport-dependent development. Auxin (transport) positively feeds back on GA biosynthesis.
The two functionally homologous GATA family transcription factors GLUCOSE NITROGEN CARBON (GNC) and CYTOKININ-INDUCED GATA FACTOR1/GNC-LIKE (CGA1/GNL) were identified as putative GA pathway regulators based on their transcriptional regulation by GA. The expression of GNC and GNL is repressed by PIFs and their transcriptional regulation by GA and PIFs was found to require the GID1 GA receptors and DELLA degradation (Figure 2A; Richter et al., 2010). GNC and CGA1/GNL repress a range of well-established GA responses including germination, elongation growth, and flowering, and they promote greening. GNC or CGA1/GNL overexpression results in plant growth as well as gene expression phenotypes that largely phenocopy GA-deficiency as well as PIF gene deficiency. Most importantly, the loss of GNC and CGA1/GNL function partially suppresses the phenotype of the GA-deficient ga1 mutant. Based on these criteria, it can be postulated that GNC and GNL are major GA pathway regulators downstream of the DELLAs and the PIFs. Since the expression of these two GATA factors is also controlled – possibly via the PIFs – by light, glucose, nitrogen, and cytokinin, it may be argued that GNC and CGA1/GNL are central integrators of growth controlling signals (Bi et al., 2005; Naito et al., 2007).
The PIF-related bHLH protein ALCATRAZ (ALC) was initially discovered based on its role in the formation of the separation layer of the valve margin in Arabidopsis siliques (Rajani and Sundaresan, 2001). ALC contains a DELLA-binding domain but lacks the AP-binding site for phytochrome interaction (Leivar and Quail, 2010). The transcription factor INDEHISCENCE (IND) is an upstream regulator of ALC and it induces the expression of a critical GA biosynthesis gene in the separation layer where ALC functionality is essential (Arnaud et al., 2010). In analogy to the interaction of DELLAs with PIF3 and PIF4, genetic, and protein–protein interaction studies clearly suggest that DELLAs also interact with ALC and that GA relieves the DELLA-imposed repression on ALC by promoting DELLA proteolysis and thereby allowing proper valve margin development (Figure 2B).
SPATULA (SPT) is the closest homolog of ALC in Arabidopsis and, similarly to PIFs and ALC, SPT is able to bind the DELLAs RGA and GAI (Gallego-Bartolome et al., 2010). SPT was identified based on its role in septum, style, and stigma development in the flower (Alvarez and Smyth, 1999). SPT was subsequently also shown to control germination in response to cold temperature in Arabidopsis seeds and to repress cotyledon expansion (Penfield et al., 2005; Josse et al., 2011). In the context of DELLA function and GA signaling, the role of SPT as a repressor of cotyledon expansion is important (Josse et al., 2011). In contrast to the antagonistic DELLA–PIF and DELLA–ALC interactions, DELLAs and SPT function in an analogous manner as repressors of cotyledon elongation (Figure 2C; Josse et al., 2011). Loss-of-function mutants of SPT as well as of DELLAs have enlarged cotyledons and this phenotype is enhanced in mutants lacking SPT as well as DELLAs. Interestingly, DELLA abundance is negatively correlated with SPT abundance, suggesting that a homeostasis mechanism is in place that controls the overall abundance of the DELLA and SPT repressors. Since GA treatment cause an increase in SPT transcript abundance also when SPT gene expression is under control of an overexpression promoter, it was postulated that an SPT transcript stabilizing mechanism is responsible for the accumulation of SPT in the presence of GA (Josse et al., 2011). At present, it remains to be seen whether such a regulation requires the interaction between DELLAs and SPT and which other molecular players contribute to this interaction.
The five Arabidopsis DELLA proteins belong to the superfamily of GRAS (GAI, RGA, SCARECROW) transcription regulators that in Arabidopsis additionally includes SCARECROW (SCR) and 27 SCARECROW-like (SCL) proteins (Pysh et al., 1999; Lee et al., 2008). The DELLA domain and the adjacent VHYNP domain allow to distinguish the DELLAs from the remaining SCR/SCL proteins (Pysh et al., 1999; Willige et al., 2007). A number of studies have tried to understand the evolutionary and functional relationship between SCR/SCL proteins and DELLAs. SCL3 attracted the attention of GA biologists because its transcription was repeatedly found to be strongly repressed by GA and to be induced by DELLAs (Willige et al., 2007; Zentella et al., 2007). Genetic analyses of SCL3-deficient mutants have allowed positioning SCL3 into the GA signaling pathway. SCL3 and DELLA (RGA) interact and SCL3 positively regulates GA reponses (Heo et al., 2011; Zhang et al., 2011). The primary effect of the interaction between SCL3 and DELLA is antagonistic, however, this appears to be more complicated because DELLA induces the transcription of its SCL3 antagonist and, inversely, because SCL3 possibly stabilizes its DELLA antagonist by modulating GA biosynthesis (Figure 2D).
Gene expression studies and pathobiological assays had suggested that GA controls JA-responsive gene expression (Cao et al., 2006; Hou et al., 2008) and JA-mediated plant immune responses (Navarro et al., 2008). In an analysis of the underlying molecular causes it was found that GA attenuates the JA-induced expression of a number of JA-responsive genes (Hou et al., 2010). The expression of these genes is dependent on the transcription activators MYC2, MYC3, and MYC4 and their negative regulators, the dimerizing jasmonate ZIM domain proteins (JAZ; Chini et al., 2007; Fernandez-Calvo et al., 2011). Via NINJA proteins, JAZ proteins are linked to the TOPLESS(-RELATED) corepressors (Pauwels et al., 2010) and the repressive activity of JAZs is relieved in the presence of JA by their proteasomal degradation via the E3 ligase SCFCOI1 (Thines et al., 2007). It was recently shown that the DELLA proteins RGA, GAI, RGL1, and RGL2 directly bind to and thereby inhibit the (representative) JAZ proteins JAZ1, JAZ3, and JAZ9 (Hou et al., 2010). Hierarchically, JA-mediated JAZ degradation has thus to be seen as the major control element of the pathway, while DELLAs modulate JA responses by repressing JAZ activity (Figure 2E).
Most of the DELLA-dependent regulatory events that have been identified so far involve a cascade of – typically repression – events that ultimately lead to gene expression changes that – somehow – are responsible for GA-mediated growth control. Few of these signaling events allow hypothesizing about how GA controls growth at the cell biological level. At least two reports link GA signaling to auxin transport and these findings help at least in part to explain how the GA signal is influenced by or is influencing transport of the plant hormone auxin and ultimately plant growth. Auxin transport controls organ initiation and development as well as tropic responses: auxin is synthesized in the shoot apex and transported toward the root tip via the activity of PIN-FORMED (PIN) auxin efflux carriers. When the shoot and consequently the shoot-derived auxin source is removed, roots cease to grow and fail to respond properly to GA. This reduced GA-responsiveness can be suppressed when the auxin indole-3-acetic acid (IAA) is applied to the site of shoot removal. Genetic and cell biological analyses showed that the cessation of root growth correlates with the accumulation of DELLAs (RGA) in the root and is suppressed in DELLA gene loss-of-function mutants (Fu and Harberd, 2003). Thus, auxin and auxin transport may control DELLA abundance and control (root) growth. At the same time it was recently found that GA controls auxin transport and PIN protein abundance (Willige et al., 2011). GA promotes the degradation of the auxin efflux carriers PIN1 and PIN2, at least in root tips and inflorescence stems, possibly by increased targeting of these transporter proteins for degradation in the vacuole (Vieten et al., 2007; Willige et al., 2011). At the physiological level, the downregulation of PIN proteins as observed in GA and GA signaling-deficient mutants correlates with a reduction in auxin transport in inflorescence stems, defective embryo development and reduced root gravitropism. Since GA-dependent changes in PIN protein abundance do not correlate with GA-dependent changes in PIN gene transcription, and since PIN protein abundance at the same time is increased when PIN protein degradation is blocked, it has to be argued that GA controls PIN abundance at the post-translational level (Figure 2F; Willige et al., 2011).
This review provides an overview of the current knowledge on the mechanisms governing GA signaling. Certainly, it has become clear that especially in recent years major advances have been made in understanding the molecular mode of action of the DELLA proteins as key repressors of the pathway. A number of obvious questions remain to be answered, some of which have already been phrased in the above paragraphs. At least two additional major issues that have puzzled the GA signaling field for some time will require clarification in the near future: the O-Linked N-acetylglucosaminyltransferase (OGT) SPINDLY (SPY) was identified repeatedly as a major repressor of GA signaling (Jacobsen and Olszewski, 1993; Wilson and Somerville, 1995; Jacobsen et al., 1996; Shimada et al., 2006). SPYs repression function in Arabidopsis GA signaling was recapitulated in other plant species including tomato and rice (Greb et al., 2002; Shimada et al., 2006). Most importantly, spy mutant alleles fully suppress GA-deficiency as well as defects in GA signaling and this phenotype can best be explained by a direct control of DELLA reperessor activity by SPY-mediated O-Linked N-acetylglucosamine-modification. A proof for this or an alternative mode of action urgently needs to be brought about.
Several reports have identified DELLA proteins as phosphoproteins. Although it had initially been claimed that this modification is essential for the proteasomal degradation of DELLAs (Sasaki et al., 2003; Gomi et al., 2004), this view was subsequently corrected (Itoh et al., 2005). On the other side, in vitro data using pharmacological inhibitors suggest that phosphoregulation may be involved in DELLA protein turnover (Wang et al., 2009). Thus, it remains to be seen whether and how DELLA phosphorylation regulates DELLA activity. The mutation of predicted phosphorylation sites in Arabidopsis RGL2 impairs GA-induced protein degradation but whether or not this effect is ultimately due to changes in RGL2 phosphorylation rather than RGL2 functionality could not be fully clarified (Hussain et al., 2005, 2007). The recently identified casein kinase, CKI, is a candidate kinase that may phosphorylate DELLAs and regulate their activity in vivo to control flowering time in rice (Dai and Xue, 2011). Further experiments will bring light into the identity of the in vivo targets of this kinase and may identify further kinases that interfere with DELLA activity.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was funded by the DFG grant SCHW751/7-1 as part of the Arabidopsis Functional Genomics Network (AFGN).
Achard, P., Baghour, M., Chapple, A., Hedden, P., Van Der Straeten, D., Genschik, P., Moritz, T., and Harberd, N. P. (2007a). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. U.S.A. 104, 6484–6489.
Achard, P., Liao, L., Jiang, C., Desnos, T., Bartlett, J., Fu, X., and Harberd, N. P. (2007b). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143, 1163–1172.
Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., Van Der Straeten, D., Peng, J., and Harberd, N. P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94.
Achard, P., Gusti, A., Cheminant, S., Alioua, M., Dhondt, S., Coppens, F., Beemster, G. T., and Genschik, P. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19, 1188–1193.
Achard, P., Renou, J. P., Berthome, R., Harberd, N. P., and Genschik, P. (2008). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18, 656–660.
Achard, P., Vriezen, W. H., Van Der Straeten, D., and Harberd, N. P. (2003). Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15, 2816–2825.
Alabadi, D., Gil, J., Blazquez, M. A., and Garcia-Martinez, J. L. (2004). Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 134, 1050–1057.
Alvarez, J., and Smyth, D. R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386.
Ariizumi, T., Lawrence, P. K., and Steber, C. M. (2011). The role of two F-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol. 155, 765–775.
Ariizumi, T., Murase, K., Sun, T.-P., and Steber, C. M. (2008). Proteolysis-independent down-regulation of DELLA repression by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF 1. Plant Cell 20, 2447–2459.
Arnaud, N., Girin, T., Sorefan, K., Fuentes, S., Wood, T. A., Lawrenson, T., Sablowski, R., and Ostergaard, L. (2010). Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 24, 2127–2132.
Bassel, G. W., Zielinska, E., Mullen, R. T., and Bewley, J. D. (2004). Down-regulation of DELLA genes is not essential for germination of tomato, soybean, and Arabidopsis seeds. Plant Physiol. 136, 2782–2789.
Bi, Y. M., Zhang, Y., Signorelli, T., Zhao, R., Zhu, T., and Rothstein, S. (2005). Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 44, 680–692.
Cao, D., Cheng, H., Wu, W., Soo, H. M., and Peng, J. (2006). Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 142, 509–525.
Cao, D., Hussain, A., Cheng, H., and Peng, J. (2005). Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223, 105–113.
Cheminant, S., Wild, M., Bouvier, F., Pelletier, S., Renou, J. P., Erhardt, M., Hayes, S., Terry, M. J., Genschik, P., and Achard, P. (2011). DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23, 1849–1860.
Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D. E., Cao, D., Luo, D., Harberd, N. P., and Peng, J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055–1064.
Chhun, T., Aya, K., Asano, K., Yamamoto, E., Morinaka, Y., Watanabe, M., Kitano, H., Ashikari, M., Matsuoka, M., and Ueguchi-Tanaka, M. (2007). Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell 19, 3876–3888.
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., Garcia-Casado, G., Lopez-Vidriero, I., Lozano, F. M., Ponce, M. R., Micol, J. L., and Solano, R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671.
Dai, C., and Xue, H. W. (2011). Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 29, 1916–1927.
De Lucas, M., Daviere, J. M., Rodriguez-Falcon, M., Pontin, M., Iglesias-Pedraz, J. M., Lorrain, S., Fankhauser, C., Blazquez, M. A., Titarenko, E., and Prat, S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484.
Dill, A., Thomas, S. G., Hu, J., Steber, C. M., and Sun, T. P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16, 1392–1405.
Djakovic-Petrovic, T., De Wit, M., Voesenek, L. A., and Pierik, R. (2007). DELLA protein function in growth responses to canopy signals. Plant J. 51, 117–126.
Dohmann, E. M., Nill, C., and Schwechheimer, C. (2010). DELLA proteins restrain germination and elongation growth in Arabidopsis thaliana COP9 signalosome mutants. Eur. J. Cell Biol. 89, 163–168.
Feng, S., Martinez, C., Gusmaroli, G., Wang, Y., Zhou, J., Wang, F., Chen, L., Yu, L., Iglesias-Pedraz, J. M., Kircher, S., Schafer, E., Fu, X., Fan, L. M., and Deng, X. W. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479.
Fernandez-Calvo, P., Chini, A., Fernandez-Barbero, G., Chico, J. M., Gimenez-Ibanez, S., Geerinck, J., Eeckhout, D., Schweizer, F., Godoy, M., Franco-Zorrilla, J. M., Pauwels, L., Witters, E., Puga, M. I., Paz-Ares, J., Goossens, A., Reymond, P., De Jaeger, G., and Solano, R. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715.
Fu, X., and Harberd, N. P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421, 740–743.
Fu, X., Richards, D. E., Fleck, B., Xie, D., Burton, N., and Harberd, N. P. (2004). The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16, 1406–1418.
Gallego-Bartolome, J., Minguet, E. G., Marin, J. A., Prat, S., Blazquez, M. A., and Alabadi, D. (2010). Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol. Biol. Evol. 27, 1247–1256.
Gomi, K., Sasaki, A., Itoh, H., Ueguchi-Tanaka, M., Ashikari, M., Kitano, H., and Matsuoka, M. (2004). GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 37, 626–634.
Greb, T., Schmitz, G., and Theres, K. (2002). Isolation and characterization of the Spindly homologue from tomato. J. Exp. Bot. 53, 1829–1830.
Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z. L., Powers, S. J., Gong, F., Phillips, A. L., Hedden, P., Sun, T. P., and Thomas, S. G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18, 3399–3414.
Heo, J. O., Chang, K. S., Kim, I. A., Lee, M. H., Lee, S. A., Song, S. K., Lee, M. M., and Lim, J. (2011). Funneling of gibberellin signaling by the GRAS transcription regulator SCARECROW-LIKE3 in the Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 108, 2166–2171.
Hou, X., Hu, W. W., Shen, L., Lee, L. Y., Tao, Z., Han, J. H., and Yu, H. (2008). Global identification of DELLA target genes during Arabidopsis flower development. Plant Physiol. 147, 1126–1142.
Hou, X., Lee, L. Y., Xia, K., Yan, Y., and Yu, H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19, 884–894.
Hussain, A., Cao, D., Cheng, H., Wen, Z., and Peng, J. (2005). Identification of the conserved serine/threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J. 44, 88–99.
Hussain, A., Cao, D., and Peng, J. (2007). Identification of conserved tyrosine residues important for gibberellin sensitivity of Arabidopsis RGL2 protein. Planta 226, 475–483.
Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., Matsuoka, M., and Yamaguchi, J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13, 999–1010.
Itoh, H., Sasaki, A., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Hasegawa, Y., Minami, E., Ashikari, M., and Matsuoka, M. (2005). Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol. 46, 1392–1399.
Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70.
Jacobsen, S. E., Binkowski, K. A., and Olszewski, N. E. (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 93, 9292–9296.
Jacobsen, S. E., and Olszewski, N. E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5, 887–896.
Jiang, C., Gao, X., Liao, L., Harberd, N. P., and Fu, X. (2007). Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 145, 1460–1470.
Josse, E. M., Gan, Y., Bou-Torrent, J., Stewart, K. L., Gilday, A. D., Jeffree, C. E., Vaistij, F. E., Martinez-Garcia, J. F., Nagy, F., Graham, I. A., and Halliday, K. J. (2011). A DELLA in disguise: SPATULA restrains the growth of the developing Arabidopsis seedling. Plant Cell 23, 1337–1351.
Lee, M. H., Kim, B., Song, S. K., Heo, J. O., Yu, N. I., Lee, S. A., Kim, M., Kim, D. G., Sohn, S. O., Lim, C. E., Chang, K. S., Lee, M. M., and Lim, J. (2008). Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol. Biol. 67, 659–670.
Lee, S., Cheng, H., King, K. E., Wang, W., He, Y., Hussain, A., Lo, J., Harberd, N. P., and Peng, J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16, 646–658.
Leivar, P., Monte, E., Oka, Y., Liu, T., Carle, C., Castillon, A., Huq, E., and Quail, P. H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823.
Leivar, P., and Quail, P. H. (2010). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28.
Leivar, P., Tepperman, J. M., Monte, E., Calderon, R. H., Liu, T. L., and Quail, P. H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21, 3535–3553.
Mcginnis, K. M., Thomas, S. G., Soule, J. D., Strader, L. C., Zale, J. M., Sun, T. P., and Steber, C. M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15, 1120–1130.
Naito, T., Kiba, T., Koizumi, N., Yamashino, T., and Mizuno, T. (2007). Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 71, 1557–1560.
Nakajima, M., Shimada, A., Takashi, Y., Kim, Y. C., Park, S. H., Ueguchi-Tanaka, M., Suzuki, H., Katoh, E., Iuchi, S., Kobayashi, M., Maeda, T., Matsuoka, M., and Yamaguchi, I. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46, 880–889.
Navarro, L., Bari, R., Achard, P., Lison, P., Nemri, A., Harberd, N. P., and Jones, J. D. (2008). DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18, 650–655.
Oh, E., Yamaguchi, S., Hu, J., Yusuke, J., Jung, B., Paik, I., Lee, H. S., Sun, T. P., Kamiya, Y., and Choi, G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19, 1192–1208.
Pauwels, L., Barbero, G. F., Geerinck, J., Tilleman, S., Grunewald, W., Perez, A. C., Chico, J. M., Bossche, R. V., Sewell, J., Gil, E., Garcia-Casado, G., Witters, E., Inze, D., Long, J. A., De Jaeger, G., Solano, R., and Goossens, A. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791.
Penfield, S., Gilday, A. D., Halliday, K. J., and Graham, I. A. (2006). DELLA-mediated cotyledon expansion breaks coat-imposed seed dormancy. Curr. Biol. 16, 2366–2370.
Penfield, S., Josse, E. M., Kannangara, R., Gilday, A. D., Halliday, K. J., and Graham, I. A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15, 1998–2006.
Peng, J., Carol, P., Richards, D. E., King, K. E., Cowling, R. J., Murphy, G. P., and Harberd, N. P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205.
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., Beales, J., Fish, L. J., Worland, A. J., Pelica, F., Sudhakar, D., Christou, P., Snape, J. W., Gale, M. D., and Harberd, N. P. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261.
Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y., and Lopez-Molina, L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745.
Piskurewicz, U., and Lopez-Molina, L. (2009). The GA-signaling repressor RGL3 represses testa rupture in response to changes in GA and ABA levels. Plant Signal. Behav. 4, 63–65.
Piskurewicz, U., Tureckova, V., Lacombe, E., and Lopez-Molina, L. (2009). Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO J. 28, 2259–2271.
Pysh, L. D., Wysocka-Diller, J. W., Camilleri, C., Bouchez, D., and Benfey, P. N. (1999). The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18, 111–119.
Rademacher, W. (2000). GROWTH RETARDANTS: effects on gibberellin biosynthesis and other metabolic pathways. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 501–531.
Ragni, L., Nieminen, K., Pacheco-Villalobos, D., Sibout, R., Schwechheimer, C., and Hardtke, C. S. (2011). Mobile gibberellin directly stimulates Arabidopsis hypocotyl xylem expansion. Plant Cell 23, 1322–1336.
Rajani, S., and Sundaresan, V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11, 1914–1922.
Richter, R., Behringer, C., Müller, I. K., and Schwechheimer, C. (2010). The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 24, 2093–2104.
Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D. H., An, G., Kitano, H., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299, 1896–1898.
Shimada, A., Ueguchi-Tanaka, M., Sakamoto, T., Fujioka, S., Takatsuto, S., Yoshida, S., Sazuka, T., Ashikari, M., and Matsuoka, M. (2006). The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 48, 390–402.
Shin, J., Kim, K., Kang, H., Zulfugarov, I. S., Bae, G., Lee, C. H., Lee, D., and Choi, G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 106, 7660–7665.
Silverstone, A. L., Ciampaglio, C. N., and Sun, T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169.
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., Nomura, K., He, S. Y., Howe, G. A., and Browse, J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665.
Tyler, L., Thomas, S. G., Hu, J., Dill, A., Alonso, J. M., Ecker, J. R., and Sun, T. P. (2004). DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135, 1008–1019.
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T. Y., Hsing, Y. I., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698.
Ueguchi-Tanaka, M., Hirano, K., Hasegawa, Y., Kitano, H., and Matsuoka, M. (2008). Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20, 2437–2446.
Vieten, A., Sauer, M., Brewer, P. B., and Friml, J. (2007). Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 12, 160–168.
Wang, F., Zhu, D., Huang, X., Li, S., Gong, Y., Yao, Q., Fu, X., Fan, L. -M., and Deng, X. W. (2009). Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21, 2378–2390.
Wen, C. K., and Chang, C. (2002). Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14, 87–100.
Willige, B. C., Ghosh, S., Nill, C., Zourelidou, M., Dohmann, E. M., Maier, A., and Schwechheimer, C. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19, 1209–1220.
Willige, B. C., Isono, E., Richter, R., Zourelidou, M., and Schwechheimer, C. (2011). Gibberellin regulates PIN-FORMED abundance and is required for auxin transport-dependent growth and development in Arabidopsis thaliana. Plant Cell 23, 2184–2195.
Wilson, R. N., and Somerville, C. R. (1995). Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol. 108, 495–502.
Yamamoto, Y., Hirai, T., Yamamoto, E., Kawamura, M., Sato, T., Kitano, H., Matsuoka, M., and Ueguchi-Tanaka, M. (2010). A rice gid1 suppressor mutant reveals that gibberellin is not always required for interaction between its receptor, GID1, and DELLA proteins. Plant Cell 22, 3589–3602.
Zentella, R., Zhang, Z. L., Park, M., Thomas, S. G., Endo, A., Murase, K., Fleet, C. M., Jikumaru, Y., Nambara, E., Kamiya, Y., and Sun, T. P. (2007). Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19, 3037–3057.
Keywords: gibberellin, bHLH, DELLA protein, GATA transcription factor, jasmonate ZIM domain protein, phytochrome interacting factor, receptor, signal transduction
Citation: Schwechheimer C (2012) Gibberellin signaling in plants – the extended version. Front. Plant Sci. 2:107. doi: 10.3389/fpls.2011.00107
Received: 30 September 2011;
Paper pending published: 11 October 2011;
Accepted: 13 December 2011;
Published online: 04 January 2012.
Edited by:
Andreas P. M. Weber, University of Duesseldorf, GermanyReviewed by:
Brad Binder, University of Tennessee-Knoxville, USACopyright: © 2012 Schwechheimer. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Claus Schwechheimer, Plant Systems Biology, Center for Life and Food Sciences Weihenstephan, Technische Universität München, Emil-Ramann-Str. 4, 85354 Freising, Germany. e-mail:Y2xhdXMuc2Nod2VjaGhlaW1lckB3encudHVtLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.