
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 09 December 2011
Sec. Plant Physiology
Volume 2 - 2011 | https://doi.org/10.3389/fpls.2011.00094
This article is part of the Research Topic Arabidopsis 2010 and beyond – big science with a small weed View all 21 articles
 Theresa Maria Singer
Theresa Maria Singer Cordula Moll and Rita Groß-Hardt*
Cordula Moll and Rita Groß-Hardt*Gene and genome duplications provide a playground for various selective pressures and contribute significantly to genome complexity. It is assumed that the genomes of all major eukaryotic lineages possess duplicated regions that result from gene and genome duplication. There is evidence that the model plant Arabidopsis has been subjected to at least three whole-genome duplication events over the last 150–200 million years. As a result, many cellular processes are governed by redundantly acting gene families. Plants pass through two distinct life phases with a haploid gametophytic alternating with a diploid sporophytic generation. This ontogenetic difference in gene copy number has important implications for the outcome of deleterious mutations, which are masked by the second gene copy in diploid systems but expressed in a dominant fashion in haploid organisms. As a consequence, maintaining the activity of duplicated genes might be particularly advantageous during the haploid gametophytic generation. Here, we describe the distinctive features associated with the alteration of generations and discuss how activity profiles of duplicated genes might get modulated in a life phase dependent fashion.
The genome represents the construction plan for all living organisms and has been subject to many different mutations over evolutionary time. In the last couple of decades, reverse genetic approaches have revealed that the majority of single loss-of-function mutants do not lead to apparent developmental defects (Thomas, 1993; Gu et al., 2003; Briggs et al., 2006). The toleration of genomic changes appears to be largely due to considerable genetic redundancy, which is predominantly but not exclusively, the result of gene and genome duplications (Ohno, 1970; Gu et al., 2003; Wagner, 2008). In 1970, Ohno proposed that whole-genome duplications occurred in the early history of all vertebrates. It now seems probable that all major lineages of eukaryotic genomes possess duplicated regions that may have resulted from genome duplications (Ohno, 1970; Lynch et al., 2001). According to their extent, the duplications have been categorized into small-scale duplications, which include local, tandem, and segmental duplications, and large-scale duplications of the entire genome, which result in a transient or stable increase in ploidy levels (Levasseur and Pontarotti, 2011) and a concomitant change in the expression of ploidy-sensitive genes (Guo et al., 1996; Galitski et al., 1999). Large-scale duplications occurred more often in plants than in animals (Li, 1997) and are especially widespread in angiosperms (Stebbins, 1950; Grant, 1981; Soltis and Soltis, 1999; Leitch and Leitch, 2008). It is estimated that 50–80% of angiosperms, including crop plants such as alfalfa, potato, wheat, and coffee, are polyploids (Wendel, 2000), and this polyploidy dates back to ancient duplication events (Grant, 1963; Masterson, 1994; Otto and Whitton, 2000). The analysis of the Arabidopsis bacterial artificial chromosomes (BACs) indicates that almost 60% of the genome consists of duplicated regions (Blanc et al., 2000), suggesting that Arabidopsis is an ancient polyploid. Synteny-based analysis of the Arabidopsis genome has provided evidence for at least three whole-genome duplications (Vision et al., 2000; Simillion et al., 2002; Bowers et al., 2003), and most genes in Arabidopsis appear to have duplicated approximately 65 million years ago (Lynch and Force, 2000). One obvious advantage of gene and genome duplications is the supply of raw genetic material, which provides a playground for various selective pressures. Classic evolutionary theory predicts that the typical fate of any gene duplicate is loss because duplicated genes should have redundant function immediately after formation, and full redundancy is considered to be genetically unstable (Thomas, 1993; Briggs et al., 2006; Hardtke, 2006). The duplicate can accumulate deleterious mutations and gradually develop into a pseudogene, a process termed non-functionalization (Haldane, 1933; Ohno, 1970; Tautz, 1992; Wagner, 1998; Lynch and Force, 2000), and expression analysis of polyploid plants has revealed that some duplicated loci become silenced after polyploidization (Pichersky et al., 1990; Ford and Gottlieb, 2002). Over evolutionary time, pseudogenes are either deleted or become so diverged from the ancestral gene that they are no longer identifiable (Walsh, 1995; Lynch and Conery, 2000; Lynch et al., 2001). A less likely scenario is for both copies to be stably maintained in the genome, which is possible if both copies differ in some aspects of their functions (Nowak et al., 1997). In a process termed subfunctionalization, one gene copy adopts partial aspects of the original function (Jensen, 1976; Orgel, 1977). This can be realized, for example, through evolution of temporally and spatially diverse expression patterns (Force et al., 1999). Duplicated genes can also evolve a different function, whereby one copy acquires advantageous mutations which become subject to selection, leading to the establishment of a novel function. This neofunctionalization represents the only mechanism by which the members can permanently escape mutational decay (Lynch and Force, 2000).
Sexually reproducing organisms pass through two distinct life phases. Meiosis results in the formation of haploid gametes, which fuse to produce a diploid life form. In plants, the haploid phase comprises a few to many cells, called gametophytes. The gametophyte is the dominant generation in mosses. By contrast, in flowering plants, the gametophyte is reduced to only a few cells, which might reflect an evolutionary trend (D’Amato, 1977). Thus, flowering plants are not only subject to phylogenetic ploidy changes but also encounter ploidy changes on an ontogenetic level. Diploid organisms have one copy more of each chromosome than haploid organisms and theoretical considerations have suggested various implications associated with the respective genomic settings (Mable and Otto, 1998). One obvious advantage of a diploid organism over a haploid is that recessive deleterious mutations are masked. As a side effect, deleterious mutations can accumulate in the gene pool of diploid organisms, which can negatively influence the quality of the gene pool. However, mutations also increase genomic variability and can lead to the evolution of novel gene functions. By contrast, haploid genomes have by definition a lower redundancy level. As a consequence, deleterious mutations are dominantly expressed and hence not tolerated. As in haploid gametophytes, early embryo development can rely on a single gene copy in those cases, where one parental allele is silenced, a process referred to as imprinting. In the respective developmental stages mutations of essential gene functions are similarly dominant as in haploid organisms (Grossniklaus et al., 1998; Luo et al., 1999; Ohad et al., 1999), unless the intact allele is reactivated upon loss of the other. Recently, substantial new collections of imprinted genes have been identified (Autran et al., 2011; Hsieh et al., 2011; Luo et al., 2011; McKeown et al., 2011; Wolff et al., 2011), and genome wide expression profiling has suggested that, in the early two to four celled-embryo, more than 30% of all gene products are exclusively maternally contributed (Autran et al., 2011).
The haploid and diploid life phases of flowering plants might have conflicting interests with respect to the retention or removal of extra gene copies resulting from gene and genome duplications. Due to gene dosis constraints, a given segmental gene duplication might result in a net loss of fitness (Chernoff et al., 1992). The extra gene copy might, however, also result in a net gain of fitness in haploid tissue, where deleterious mutations can be masked (Figure 1). As an adaption to potentially conflicting demands for extra gene copies, it is tempting to speculate that plants might have evolved compensatory mechanisms to preferentially maintain the activity of duplicated genes in haploid tissue. In this respect, it is interesting that the three Arabidopsis pre-mRNA splice factors AtBRR2, AtPRP8, and GFA1 are not only encoded by ubiquitously expressed loci but also by a gene copy that is preferentially active in the female gametophyte (Figure 2). In theory, the activity status of a given gene can be regulated on different levels. The establishment of transcriptional differences implies the presence of life phase specific cis-regulatory elements, which is supported by the substantial number of genes enriched in gametophytic tissue (Honys and Twell, 2003, 2004; Sprunck et al., 2005; Yu et al., 2005; Johnston et al., 2007; Steffen et al., 2007). Promoters conferring expression during female gametogenesis have been identified (Yu et al., 2005), however the responsible transcription factor binding sites have not yet been defined. Haerizadeh et al. (2006) have shown that a gamete-restrictive silencing factor, GRSF, which is expressed in accessory cells, stably represses gamete specific gene expression in Arabidopsis. Additionally, a 77-bp comprising cis-regulatory element responsible for egg apparatus specific expression of the enhancer detector line ET253 was identified (Yang et al., 2005), and the specific expression of members of the type I MADS domain family in the central cell and antipodal cells of the female gametophyte suggests the presence of a chalaza-determining cis-regulatory element (Bemer et al., 2010). Additionally, the analysis of a battery of MYB98 downstream targets has identified two distinct cis-regulatory elements associated with MYB98 induced synergid expression (Punwani et al., 2007; Punwani and Drews, 2008). Corresponding work on MYB- and MADS-box family transcription factors in the male gametophyte have identified target sites necessary for pollen specific expression (Verelst et al., 2007a,b). Additionally, the dissection of the vegetative cell specific LAT52 promoter has delineated three activator domains, each sufficient to confer pollen specific expression when combined with the minimal CaMV 35S promoter (Bate and Twell, 1998) and promoters conferring sperm cell specific expression have been identified (Mori et al., 2006; von Besser et al., 2006). That genes of the haploid life phase can jointly be targeted, is also suggested by the recently identified ouroboros mutant of Ectocarpus siliculosus, which exhibits a sporophyte to gametophyte conversion (Coelho et al., 2011). As a consequence, the characteristic alterations between haploid and diploid generation is replaced by a reiteration of the gametophytic phase. Although the respective single recessive locus has not yet been identified, this example shows that the developmental programs of a given life phase can converge in a single homeotic master regulator.
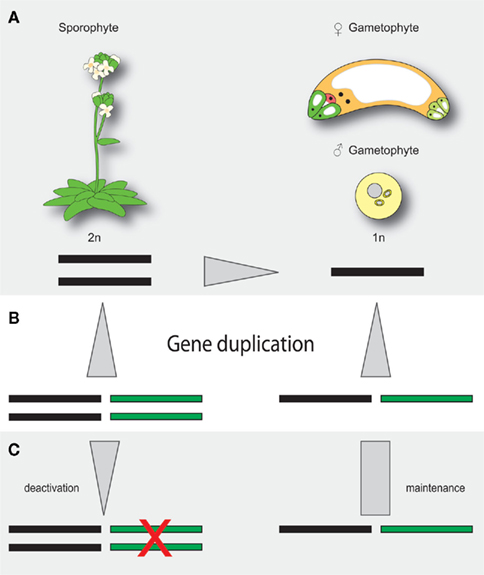
Figure 1. Hypothetical model of life phase dependent redundancy adjustment. (A) The diploid sporophyte and the haploid gametophytes exhibit, by definition, different redundancies for a given locus. Deleterious mutations are consequently masked in the sporophyte but dominantly expressed in the gametophyte. (B) Gene duplication increases redundancy levels in the haploid and diploid phase. (C) If the activity of a given gene is preferentially maintained in the gametophyte, different redundancy levels between the sporophyte and the gametophytic life phase might be buffered. Black bar, ancient locus; green bar, duplicated locus, gray arrows indicate redundancy differences.
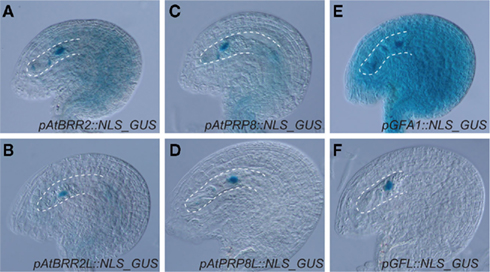
Figure 2. Expression patterns of three pre-mRNA splice factor encoding gene pairs in the ovule. Recent work in the authors’ laboratory indicate that AtBRR2, AtPRP8, and GFA1 are encoded by genes (At1g20960, At1g80070 and At1g06220, respectively) expressed in sporophytic and gametophytic tissue (A,C,E), but also by a gene copy, AtBRR2L (At2g42270), AtPRP8L (At4g38780), and GFL (At5g25230) that is preferentially active in the female gametophyte (B,D,F). The micropylar half of the female gametophyte is in focus and encircled by a dottet line.
Apart from the obvious regulation through diversification of cis-regulatory elements, activity profiles can be modulated at further levels. This includes the epigenetic regulation of genetic loci and the regulation of mRNA and protein levels by tissue specific mRNA and protein-degradation machineries. In this respect, work by Bate et al. (1996) is very interesting. The group has studied the role of the LAT52 5′UTR in transient expression assays, which strongly enhances LAT52 translation in the pollen. By contrast, no such effect has been observed in sporophytic tissue, suggesting that the respective regulatory network is differentially active in male gametophytic versus diploid tissue. Similarly, Ylstra and McCormick (1999) have reported on two mRNAs which exhibit tissue specific stability differences in tobacco: GUT15 mRNA is unstable in BY2 cells but very stable in pollen and GRP2 mRNA was found to be readily degraded in pollen but is stable in BY2 cells, suggesting life phase dependent differences in the regulation of mRNA levels.
To this end it is not clear whether gene duplicates are more likely kept active in the haploid gametophyte than in sporophytic tissue, however research of the last decades has shown that there is substantial genetic redundancy covering distinct aspects of male and female gametophyte development. In the female gametophyte, important cytoskeletal functions are governed by homologous gene pairs: NACK1/HINKEL and STUD/TETRASPORE/AtNACK2 code for kinesin-like proteins and NACK1/HINKEL plays a critical role in embryogenesis (Strompen et al., 2002), whereas STUD/TETRASPORE/AtNACK2 is required for meiosis (Hulskamp et al., 1997; Spielman et al., 1997). Notably, plants defective in both, NACK1/HINKEL and STUD/TETRASPORE/AtNACK2, exhibit female gametophytic defects with abnormal nuclei size and positioning (Tanaka et al., 2004), suggesting that these processes are regulated in a redundant manner. Similarly, the c-tubulin TUBG1 together with TUBG2 regulates nuclei size and positioning in the female gametophyte (Pastuglia et al., 2006).
Also, fusion of central cell nuclei, which is a prerequisite for the formation of triploid endosperm, is regulated in a redundant manner: MIRO1 is a GTPase, which has been shown to play a role during pollen tube growth and embryogenesis. The Arabidopsis genome contains three MIRO GTPases, and mutations in MIRO1 affect pollen tube growth and early embryo development (Yamaoka and Leaver, 2008). By contrast, homozygous miro2 mutants don’t exhibit developmental abnormalities, however the double mutant MIRO1/miro1, miro2/miro2 strongly enhances the pollen tube growth defect and nuclei fusion in the central cell of the female gametophyte is disturbed (Sormo et al., 2011). Similarly, the BiP protein, a molecular chaperone Hsp70 in the endoplasmic reticulum, is required for polar nuclei fusion (Maruyama et al., 2010): Arabidopsis contains three BiP proteins, with BiP1 and BiP2 being 99% identical. Whereas bip single mutants do not exhibit any defects, central cell nuclei fail to fuse in bip1 bip2 double mutants. Functional redundancy does also occur in pollen as evidenced, for example, by the analysis of members of the pollen specific MIKC* class of MADS domain transcription factors (Verelst et al., 2007b) and characterization of apyrases AtAPY1 and AtAPY2, which together regulate pollen germination (Steinebrunner et al., 2003). The example of RPT5a, which constitutes one of six AAA-ATPases of the regulatory particle (RPT) demonstrates that the function of the second allele can critically depend on accession specific backgrounds (Gallois et al., 2009): RPT5 is encoded by two genes as a result of a gene duplication event. Both genes, RPT5a and RPT5b are redundantly required for the development of the female gametophyte and double mutants in the Wassilewskija (WS) accession exhibit a development arrest at the one- to two-nucleate embryo sac. By contrast, male gametogenesis is already perturbed in the rpt5a single mutants in the Wassilewskija background. Intriguingly, a rpt5a single mutation in Columbia does not affect male gametogenesis due to a complementary function of the Columbia RPT5b gene. Another hint for substantial redundancy in gametophytic tissue can be inferred from transcriptome studies: Yu et al. (2005) have screened for genes enriched in the female gametophyte and selected six genes for functional analysis through T-DNA insertion lines. Notably, they did not observe a defect in any of the lines, which is explained with possible redundant functions of homologous genes.
The alternation of generations realized in flowering plants combines two generation on a single organism. Here we have discussed the possibility that the associated differences in gene copy number might result in different demands of both generations to maintain a given duplicated gene in its active state. Notably, the respective data sets to address this issue have already been generated through comprehensive transcriptome profiling approaches of diploid and haploid tissues. Future work will hopefully show, whether the number of active gene family members covering a given gene function is higher in gametophytic than in sporophytic tissue.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was funded by the DFG grant GR1833/3-1 within the framework of the Arabidopsis Functional Genomics Network (AFGN). We thank David Haig for helpful discussions and Felicity de Courcy and members of the Groß-Hardt laboratory for critical reading of the manuscript.
Autran, D., Baroux, C., Raissig, M. T., Lenormand, T., Wittig, M., Grob, S., Steimer, A., Barann, M., Klostermeier, U. C., Leblanc, O., Vielle-Calzada, J. P., Rosenstiel, P., Grimanelli, D., and Grossniklaus, U. (2011). Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145, 707–719.
Bate, N., Spurr, C., Foster, G. D., and Twell, D. (1996). Maturation-specific translational enhancement mediated by the 5′-UTR of a late pollen transcript. Plant J. 10, 613–623.
Bate, N., and Twell, D. (1998). Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol. Biol. 37, 859–869.
Bemer, M., Heijmans, K., Airoldi, C., Davies, B., and Angenent, G. C. (2010). An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol. 154, 287–300.
Blanc, G., Barakat, A., Guyot, R., Cooke, R., and Delseny, M. (2000). Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12, 1093–1101.
Bowers, J. E., Chapman, B. A., Rong, J., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438.
Briggs, G. C., Osmont, K. S., Shindo, C., Sibout, R., and Hardtke, C. S. (2006). Unequal genetic redundancies in Arabidopsis – a neglected phenomenon? Trends Plant Sci. 11, 492–498.
Chernoff, Y. O., Inge-Vechtomov, S. G., Derkach, I. L., Ptyushkina, M. V., Tarunina, O. V., Dagkesamanskaya, A. R., and Ter-Avanesyan, M. D. (1992). Dosage-dependent translational suppression in yeast Saccharomyces cerevisiae. Yeast 8, 489–499.
Coelho, S. M., Godfroy, O., Arun, A., Le Corguille, G., Peters, A. F., and Cock, J. M. (2011). OUROBOROS is a master regulator of the gametophyte to sporophyte life cycle transition in the brown alga Ectocarpus. Proc. Natl. Acad. Sci. U.S.A. 108, 11518–11523.
Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545.
Ford, V. S., and Gottlieb, L. D. (2002). Single mutations silence PGiC2 genes in two very recent allotetraploid species of Clarkia. Evolution 56, 699–707.
Galitski, T., Saldanha, A. J., Styles, C. A., Lander, E. S., and Fink, G. R. (1999). Ploidy regulation of gene expression. Science 285, 251–254.
Gallois, J. L., Guyon-Debast, A., Lecureuil, A., Vezon, D., Carpentier, V., Bonhomme, S., and Guerche, P. (2009). The Arabidopsis proteasome RPT5 subunits are essential for gametophyte development and show accession-dependent redundancy. Plant Cell 21, 442–459.
Grossniklaus, U., Vielle-Calzada, J. P., Hoeppner, M. A., and Gagliano, W. B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280, 446–450.
Gu, Z., Steinmetz, L. M., Gu, X., Scharfe, C., Davis, R. W., and Li, W. H. (2003). Role of duplicate genes in genetic robustness against null mutations. Nature 421, 63–66.
Guo, M., Davis, D., and Birchler, J. A. (1996). Dosage effects on gene expression in a maize ploidy series. Genetics 142, 1349–1355.
Haerizadeh, F., Singh, M. B., and Bhalla, P. L. (2006). Transcriptional repression distinguishes somatic from germ cell lineages in a plant. Science 313, 496–499.
Hardtke, C. S. (2006). Root development – branching into novel spheres. Curr. Opin. Plant Biol. 9, 66–71.
Honys, D., and Twell, D. (2003). Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132, 640–652.
Honys, D., and Twell, D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5, R85.
Hsieh, T. F., Shin, J., Uzawa, R., Silva, P., Cohen, S., Bauer, M. J., Hashimoto, M., Kirkbride, R. C., Harada, J. J., Zilberman, D., and Fischer, R. L. (2011). Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. U.S.A. 108, 1755–1762.
Hulskamp, M., Parekh, N. S., Grini, P., Schneitz, K., Zimmermann, I., Lolle, S. J., and Pruitt, R. E. (1997). The STUD gene is required for male-specific cytokinesis after telophase II of meiosis in Arabidopsis thaliana. Dev. Biol. 187, 114–124.
Jensen, R. A. (1976). Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 30, 409–425.
Johnston, A. J., Meier, P., Gheyselinck, J., Wuest, S. E., Federer, M., Schlagenhauf, E., Becker, J. D., and Grossniklaus, U. (2007). Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol. 8, R204.
Leitch, A. R., and Leitch, I. J. (2008). Perspective – genomic plasticity and the diversity of polyploid plants. Science 320, 481–483.
Levasseur, A., and Pontarotti, P. (2011). The role of duplications in the evolution of genomes highlights the need for evolutionary-based approaches in comparative genomics. Biol. Direct 6, 11.
Luo, M., Bilodeau, P., Koltunow, A., Dennis, E. S., Peacock, W. J., and Chaudhury, A. M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 96, 296–301.
Luo, M., Taylor, J. M., Spriggs, A., Zhang, H., Wu, X., Russell, S., Singh, M., and Koltunow, A. (2011). A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 7, e1002125. doi:10.1371/journal.pgen.1002125
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155.
Lynch, M., and Force, A. (2000). The probability of duplicate gene preservation by subfunctionalization. Genetics 154, 459–473.
Lynch, M., O’Hely, M., Walsh, B., and Force, A. (2001). The probability of preservation of a newly arisen gene duplicate. Genetics 159, 1789–1804.
Mable, B. K., and Otto, S. P. (1998). The evolution of life cycles with haploid and diploid phases. Bioessays 20, 453–462.
Maruyama, D., Endo, T., and Nishikawa, S. (2010). BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 1684–1689.
Masterson, J. (1994). Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264, 421–424.
McKeown, P. C., Laouielle-Duprat, S., Prins, P., Wolff, P., Schmid, M. W., Donoghue, M. T., Fort, A., Duszynska, D., Comte, A., Lao, N. T., Wennblom, T. J., Smant, G., Kohler, C., Grossniklaus, U., and Spillane, C. (2011). Identification of imprinted genes subject to parent-of-origin specific expression in Arabidopsis thaliana seeds. BMC Plant Biol. 11, 113. doi:10.1186/1471-2229-11-113
Mori, T., Kuroiwa, H., Higashiyama, T., and Kuroiwa, T. (2006). Generative cell specific 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8, 64–71.
Nowak, M. A., Boerlijst, M. C., Cooke, J., and Smith, J. M. (1997). Evolution of genetic redundancy. Nature 388, 167–171.
Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J. J., Goldberg, R. B., and Fischer, R. L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11, 407–416.
Orgel, L. E. (1977). Gene-duplication and the origin of proteins with novel functions. J. Theor. Biol. 67, 773.
Otto, S. P., and Whitton, J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437.
Pastuglia, M., Azimzadeh, J., Goussot, M., Camilleri, C., Belcram, K., Evrard, J. L., Schmit, A. C., Guerche, P., and Bouchez, D. (2006). Gamma-tubulin is essential for microtubule organization and development in Arabidopsis. Plant Cell 18, 1412–1425.
Pichersky, E., Soltis, D., and Soltis, P. (1990). Defective chlorophyll a/b-binding protein genes in the genome of a homosporous fern. Proc. Natl. Acad. Sci. U.S.A. 87, 195–199.
Punwani, J. A., and Drews, G. N. (2008). Development and function of the synergid cell. Sex. Plant Reprod. 21, 7–15.
Punwani, J. A., Rabiger, D. S., and Drews, G. N. (2007). MYB98 positively regulates a battery of synergid-expressed genes encoding filiform apparatus localized proteins. Plant Cell 19, 2557–2568.
Simillion, C., Vandepoele, K., Van Montagu, M. C., Zabeau, M., and Van de Peer, Y. (2002). The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99, 13627–13632.
Soltis, D. E., and Soltis, P. S. (1999). Polyploidy: origins of species and genome evolution. Trends Ecol. Evol. (Amst.) 14, 348–352.
Sormo, C. G., Brembu, T., Winge, P., and Bones, A. M. (2011). Arabidopsis thaliana MIRO1 and MIRO2 GTPases are unequally redundant in pollen tube growth and fusion of polar nuclei during female gametogenesis. PLoS ONE 6, e18530. doi:10.1371/journal.pone.0018530
Spielman, M., Preuss, D., Li, F. L., Browne, W. E., Scott, R. J., and Dickinson, H. G. (1997). TETRASPORE is required for male meiotic cytokinesis in Arabidopsis thaliana. Development 124, 2645–2657.
Sprunck, S., Baumann, U., Edwards, K., Langridge, P., and Dresselhaus, T. (2005). The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L). Plant J. 41, 660–672.
Steffen, J. G., Kang, I. H., Macfarlane, J., and Drews, G. N. (2007). Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51, 281–292.
Steinebrunner, I., Wu, J., Sun, Y., Corbett, A., and Roux, S. J. (2003). Disruption of apyrases inhibits pollen germination in Arabidopsis. Plant Physiol. 131, 1638–1647.
Strompen, G., El Kasmi, F., Richter, S., Lukowitz, W., Assaad, F. F., Jurgens, G., and Mayer, U. (2002). The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr. Biol. 12, 153–158.
Tanaka, H., Ishikawa, M., Kitamura, S., Takahashi, Y., Soyano, T., Machida, C., and Machida, Y. (2004). The AtNACK1/HINKEL and STUD/TETRASPORE/AtNACK2 genes, which encode functionally redundant kinesins, are essential for cytokinesis in Arabidopsis. Genes Cells 9, 1199–1211.
Verelst, W., Saedler, H., and Munster, T. (2007a). MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters. Plant Physiol. 143, 447–460.
Verelst, W., Twell, D., de Folter, S., Immink, R., Saedler, H., and Munster, T. (2007b). MADS-complexes regulate transcriptome dynamics during pollen maturation. Genome Biol. 8, R249.
Vision, T. J., Brown, D. G., and Tanksley, S. D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117.
von Besser, K., Frank, A. C., Johnson, M. A., and Preuss, D. (2006). Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133, 4761–4769.
Wolff, P., Weinhofer, I., Seguin, J., Roszak, P., Beisel, C., Donoghue, M. T., Spillane, C., Nordborg, M., Rehmsmeier, M., and Kohler, C. (2011). High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 7, e1002126. doi:10.1371/journal.pgen.1002126
Yamaoka, S., and Leaver, C. J. (2008). EMB2473/MIRO1, an Arabidopsis micro GTPase, is required for embryogenesis and influences mitochondrial morphology in pollen. Plant Cell 20, 589–601.
Yang, W., Jefferson, R. A., Huttner, E., Moore, J. M., Gagliano, W. B., and Grossniklaus, U. (2005). An egg apparatus-specific enhancer of Arabidopsis, identified by enhancer detection. Plant Physiol. 139, 1421–1432.
Ylstra, B., and McCormick, S. (1999). Analysis of mRNA stabilities during pollen development and in BY2 cells. Plant J. 20, 101–108.
Keywords: gene duplication, flowering plants, alternation of generations, haploid, diploid
Citation: Singer TM, Moll C and Groß-Hardt R (2011) When double is not twice as much. Front. Plant Sci. 2:94. doi: 10.3389/fpls.2011.00094
Received: 16 September 2011;
Paper pending published: 10 October 2011;
Accepted: 22 November 2011;
Published online: 09 December 2011.
Edited by:
Andreas P. M. Weber, University of Duesseldorf, GermanyCopyright: © 2011 Singer, Moll and Groß-Hardt. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Rita Groß-Hardt, Developmental Genetics, Center for Plant Molecular Biology, University of Tuebingen, Auf der Morgenstelle 1, 72076 Tuebingen, Germany. e-mail:cml0YS5ncm9zcy1oYXJkdEB6bWJwLnVuaS10dWViaW5nZW4uZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.