- 1 Department of Biology, Washington University in St. Louis, St. Louis, MO, USA
- 2 Department of Biological Sciences, Webster University, Webster Groves, MO, USA
In plants, exposure to temperature extremes, heavy metal-contaminated soils, drought, air pollutants, and pathogens results in the generation of reactive oxygen species that alter the intracellular redox environment, which in turn influences signaling pathways and cell fate. As part of their response to these stresses, plants produce glutathione. Glutathione acts as an anti-oxidant by quenching reactive oxygen species, and is involved in the ascorbate–glutathione cycle that eliminates damaging peroxides. Plants also use glutathione for the detoxification of xenobiotics, herbicides, air pollutants (sulfur dioxide and ozone), and toxic heavy metals. Two enzymes catalyze glutathione synthesis: glutamate–cysteine ligase, and glutathione synthetase. Glutathione is a ubiquitous protective compound in plants, but the structural and functional details of the proteins that synthesize it, as well as the potential biochemical mechanisms of their regulation, have only begun to be explored. As discussed here, the core reactions of glutathione synthesis are conserved across various organisms, but plants have diversified both the regulatory mechanisms that control its synthesis and the range of products derived from this pathway. Understanding the molecular basis of glutathione biosynthesis and its regulation will expand our knowledge of this component in the plant stress response network.
Introduction
Plants respond to environmental stresses by regulating metabolic pathways that function to counteract resulting cellular damage. In plants exposed to temperature extremes, heavy metal-contaminated soils, drought, or air pollutants, the generation of reactive oxygen species and alterations in the intracellular redox environment perturb cellular physiology (Ogawa, 2005). As part of their response to these environmental stresses, plants produce glutathione, which acts as an anti-oxidant by quenching reactive oxygen species and is involved in the ascorbate–glutathione cycle that eliminates peroxides (Noctor and Foyer, 1998; Rouhier et al., 2008). Plants use glutathione for the metabolic detoxification of a range of xenobiotics, herbicides, air pollutants (sulfur dioxide and ozone), and heavy metals (Grill et al., 1985; Rüegsegger et al., 1990; Madamanchi and Alscher, 1991; May et al., 1998a). Glutathione also plays critical roles in plant disease resistance, cell proliferation, root development, salt tolerance, and protection against chilling damage (Vernoux et al., 2000; Mittova et al., 2003; Gómez et al., 2004; Parisy et al., 2007; Vivancos et al., 2010).
The synthesis of this chemically versatile tripeptide occurs in two ATP-dependent steps (Figure 1; Meister, 1995). In the first reaction, glutamate–cysteine ligase (GCL; also known as γ-glutamylcysteine synthetase; EC 6.3.2.2) catalyzes formation of γ-glutamylcysteine from cysteine and glutamate. In the second step, glutathione synthetase (GS; EC 6.3.2.3.) catalyzes the addition of glycine to γ-glutamylcysteine to yield glutathione. As synthesized, the reduced form of glutathione provides a substrate for multiple cellular reactions that yield oxidized glutathione, i.e., two glutathione molecules linked by a disulfide bond. Dehydroascorbate reductase consumes reduced glutathione to produce ascorbate, which is then used for the detoxification of hydrogen peroxide. In this system, glutathione reductase acts to maintain a pool of reduced glutathione. The balance between the reduced and oxidized forms of glutathione is a central component in maintaining cellular redox state (Meister, 1995).
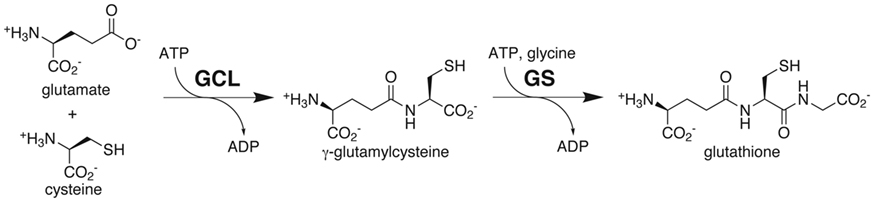
Figure 1. Glutathione biosynthesis. Substrates and products of the reactions catalyzed by glutamate-cysteine ligase (GCL) and glutathione synthetase (GS) are shown.
Although plants, like most other organisms, produce glutathione through a conserved chemical pathway, investigations of the plant GCL and GS reveal new insights on the biochemical regulatory mechanisms that control synthesis of this critical molecule and the structural basis for diversification of glutathione analogs in plants.
Oxidative Stresses and Glutathione Synthesis
Environmental and cellular conditions that cause oxidative stress increase glutathione levels in plants (May and Leaver, 1993; May et al., 1998a). Interestingly, different stresses exert different effects on the genes encoding the enzymes responsible for glutathione production in plants. For example, in response to jasmonic acid and heavy metals, transcription of GCL and GS and the levels of glutathione increase; however, expression of each gene is unaffected by treatment with either glutathione or hydrogen peroxide, even though elevated glutathione synthesis occurs (Schäfer et al., 1997, 1998; Xiang and Oliver, 1998). Similarly, exposure to cadmium increases the levels of GCL mRNA in Brassica juncea (Indian mustard) and activates transcription of both GCL and GS in Arabidopsis thaliana (thale cress; Schäfer et al., 1997, 1998; Xiang and Oliver, 1998).
Early efforts to understand the regulation of glutathione biosynthesis in plants focused on substrate availability, feedback inhibition, and transcriptional control with more recent studies investigating post-translational modifications (Yi et al., 2010a,b). Multiple experiments indicate that GCL is the rate-limiting enzyme in the plant glutathione biosynthesis (Foyer et al., 1995, 1997; Strohm et al., 1995; Noctor et al., 1996, 1997, 1998; Arisi et al., 1997). Generally, overexpression of GCL, not GS, in plants raises glutathione content by increasing flux through the pathway. Studies showing that addition of cysteine, glutamate, or glycine does not enhance glutathione synthesis also support the role of GCL as a metabolic control point in the pathway (Meyer and Fricker, 2002). Metabolic studies suggest that feedback inhibition may not be a major control feature (Meyer et al., 2001), even though glutathione inhibits both GCL and GS (Jez and Cahoon, 2004; Jez et al., 2004). Several studies describe transcriptional regulation of the pathway and report increased expression of the genes encoding GCL and GS under different stress conditions (May et al., 1998b; Schäfer et al., 1998; Xiang and Oliver, 1998). Interestingly, in Arabidopsis suspension cells transcriptional up-regulation of GCL in response to various oxidative stresses was not observed, even though both GCL activity and cellular glutathione levels increased (May et al., 1998b).
Localization of Glutathione Synthesis in Plants
The localization of GCL and GS plays a major role in the biosynthesis of glutathione. Previous studies suggest that GCL is localized to the chloroplasts and GS is localized both to chloroplasts and cytosol in plant cells (Hell and Bergmann, 1988, 1990). Recent work in Arabidopsis suggests that GCL and GS are each encoded by a single gene with alternate transcription start sites leading to either plastid-targeted or cytosolic protein (Wachter et al., 2005). Immuno-electron microscopy of Arabidopsis leaf tissue shows that GCL is localized to the chloroplast and that GS is found within chloroplasts and the cytosol (Figure 2). Likewise, assays of GCL and GS activity in the cytosol and chloroplast fractions of Arabidopsis leaf tissue show specific activities as follows: GCLcytosol – 2.2 ± 0.3 nmol min−1 mg total protein−1 (18% total activity) and GCLchloroplast – 10.0 ± 0.6 nmol min−1 mg total protein−1 (82% total activity); GScytsol – 12.4 ± 0.3 nmol min−1 mg total protein−1 (69% total activity) and GSchloroplast – 5.6 ± 0.7 nmol min−1 mg total protein−1 (31% total activity). Assays of stroma and thylakoid fractions showed no detectable GCL and GS activity in the lumen (unpublished data).
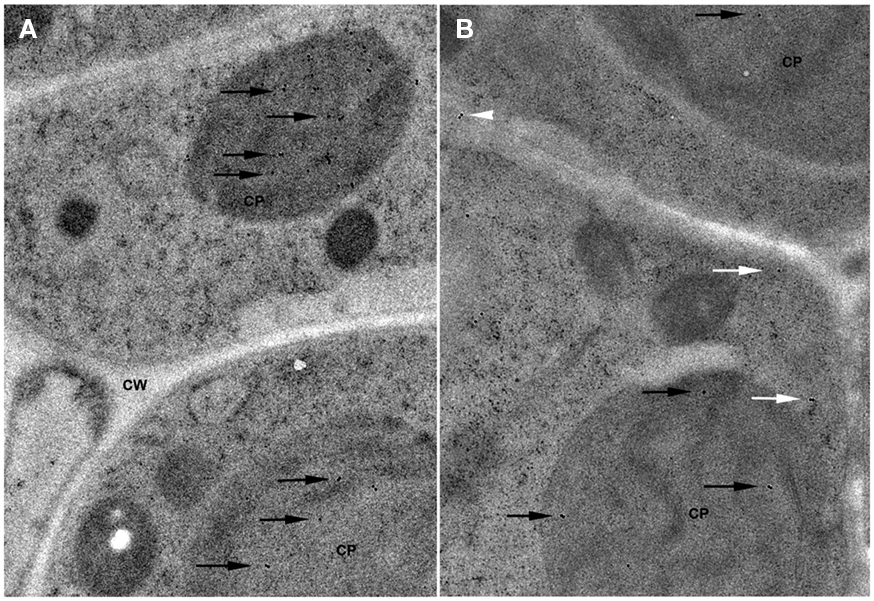
Figure 2. Localization of glutathione biosynthesis enzymes in Arabidopsis leaf cells. (A) Localization of GCL. Immunocytochemical labeling with α-GCL and gold-labeled secondary antibody is shown. Labeled GCL within the chloroplast (CP) is indicated by black arrows. Cell wall (CW) is also indicated. (B) Localization of GS. Immunocytochemical labeling with α-GS and gold-labeled secondary antibody is shown. Labeled GS within the chloroplast (CP) is indicated by black arrows and localization in the cytosol by white arrows.
Compartmentalization of glutathione biosynthesis is unique to plants (Noctor et al., 2002). The cellular localization of GCL and GS supports the need to metabolically integrate maintenance of glutathione pools in both the cytosol and plastid. Although glutathione production is regulated by GCL in the chloroplast (May et al., 1998b; Hicks et al., 2007), cytosolic pathways involved in various biotic and abiotic stress responses require glutathione. Synthesis of γ-glutamylcysteine in the chloroplast by GCL directly supports production of glutathione by plastid GS, but requires transport of the dipeptide to the cytosol for the cytosolic GS and movement of glutathione between organelles and the cytosol (Pasternak et al., 2008; Maughan et al., 2010).
Plant Glutamate–Cysteine Ligase: Structure, Function, and Regulation
Glutamate–cysteine ligase catalyzes the ATP-dependent formation of a peptide bond between glutamate and cysteine in the first reaction of glutathione synthesis. Cloning of GCL from Arabidopsis, Medicago truncatula, B. juncea, and Chorispora bungeana, revealed that the plant enzyme was unrelated by sequence to the mammalian, yeast, or bacterial enzymes (May and Leaver, 1994; Frendo et al., 1999; Hothorn et al., 2006; Wu et al., 2009). Arabidopsis mutant phenotypes for cadmium tolerance (cad2), plant disease resistance (pad2), and post-embryonic root development (rml1) are linked to the GCL gene (Cobbett et al., 1998; Vernoux et al., 2000; Parisy et al., 2007). Sequence analysis of the GCL from multiple species indicates that these enzymes group into three distinct families (Figure 3A), as follows: non-plant eukaryotes (mammals, yeast, and trypanosomes), γ-proteobacteria (Escherichia coli), and plants (Arabidopsis) and α-proteobacteria (Rhizobium; Copley and Dhillon, 2002). Comparisons within each family reveal sequence similarity, but pairwise comparisons between groups display no statistically significant relationships. Later crystallographic studies revealed that the GCL from yeast, E. coli, and plants share a common three-dimensional fold (Hibi et al., 2004; Hothorn et al., 2006; Biterova and Barycki, 2009).
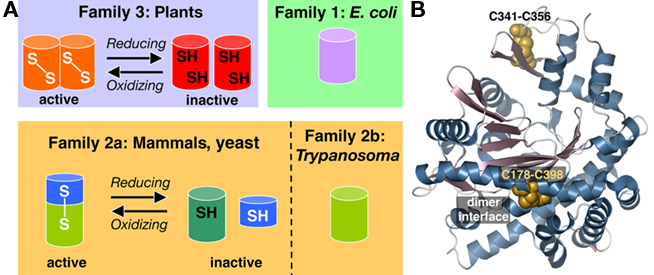
Figure 3. Overview of glutamate–cysteine ligase. (A) Oligomeric organization and redox regulation of the three types of GCL are shown. (B) Structure of the B. juncea GCL showing the positions of the two disulfide bonds (gold space-filling models). Secondary structure elements are colored as follows: α-helices in blue and β-strands in rose.
The best-studied GCL are those from non-plant eukaryotes and consist of a catalytic heavy subunit (Mr~70 kDa) and a regulatory light subunit (Mr~30 kDa; Seelig et al., 1984; Fraser et al., 2002). The heavy subunit catalyzes formation of γ-glutamylcysteine, whereas the light subunit increases affinity for glutamate and decreases the inhibition by glutathione (Seelig et al., 1984; Huang et al., 1993; Fraser et al., 2002). Formation of intermolecular disulfide bonds mediates association of the two subunits (Seelig et al., 1984; Fraser et al., 2002, 2003). Thus, these GCL respond to changes in cellular redox environment to control γ-glutamylcysteine production; however, this model does not extend to all non-plant eukaryotes. The GCL from Trypanosoma brucei and the mammalian catalytic subunit share 45% amino acid identity, but the T. brucei GCL functions as a monomer (Lueder and Phillips, 1996). Similarly, the E. coli GCL also functions as a monomeric protein (Hibi et al., 2004).
Biochemical studies of the GCL from plants revealed that these proteins are regulated by redox environment through a mechanism that differs from the heterodimeric enzymes (Jez et al., 2004; Hothorn et al., 2006; Hicks et al., 2007; Gromes et al., 2008). Kinetic analysis of Arabidopsis GCL showed that the enzyme was inactivated by buthionine sulfoximine, a potent inhibitor of mammalian GCL, and used a random ter-reactant kinetic mechanism with a preferred order of binding for catalysis (Jez et al., 2004). Although no studies examining the chemical mechanism have been published, structural similarity between the plant and yeast GCL suggests a shared reaction (Biterova and Barycki, 2009). In the yeast enzyme, an active site Mg2+, orients the glutamate γ-carboxylate for attack on the γ-phosphate of ATP, which is positioned by a second Mg2+ and an active site lysine. The resulting acyl-phosphate intermediate undergoes nucleophilic attack by the α-amino group of cysteine, which may be activated by an active site glutamate. Stabilization of the transition state in each reaction step may be provided by a conserved arginine that facilitates peptide bond formation.
Mechanistic studies of the plant GCL demonstrated that reversible disulfide bond formation alters catalytic activity with the oxidized protein more active than the reduced protein (Jez et al., 2004). Control of GCL by redox state offers a simple post-translational control mechanism of glutathione biosynthesis in plants. Glutathione maintains intracellular redox balance, and the redox regulation of GCL provides a control switch for glutathione production. Under oxidizing conditions, the demand for glutathione increases and GCL is activated. As the concentration of glutathione increases, cellular environment shifts to a more reduced potential and GCL activity decreases. Direct control of the rate-limiting enzyme in glutathione production provides a post-translational switch for responding to intracellular oxidative signals.
Subsequent crystallographic and functional studies established the molecular basis for this thiol-based redox-switch. Although regulation by redox environment occurs in the heterodimeric non-plant eukaryotic GCL, the plant GCL function as homodimeric enzymes with two intermolecular (not intramolecular) disulfide bonds (Hothorn et al., 2006; Hicks et al., 2007). The three-dimensional structure of the B. juncea GCL shows the locations of these two disulfide bonds (Figure 3B): one disulfide bond (Cys178–Cys398) is located at the interface between monomers in the dimer and the second disulfide bond (Cys341–Cys356) anchors a β-hairpin at the active site entrance (Hothorn et al., 2006). Active oxidized plant GCL functions as a dimer, but reducing conditions disrupt Cys178–Cys398 to alter the dimer interface and shift the protein to the less active monomeric form (Hothorn et al., 2006). Site-directed mutagenesis of the first disulfide bond (Cys186–Cys406 in A. thaliana GCL and Cys178–Cys398 in B. juncea GCL) abolishes the redox response and locks GCL into a monomeric form (Hothorn et al., 2006; Hicks et al., 2007). The regulatory disulfide bond of the plant GCL has a midpoint potential comparable with other known redox-responsive plant proteins (Hicks et al., 2007).
The in vitro change in GCL structure also occurs in vivo. Treatment of Arabidopsis seedlings with 5 mM H2O2 increased GCL activity and resulted in a shift from the less active reduced form to the more active oxidized form, as determined by activity assays and immunoblot analysis (Hicks et al., 2007). Similar changes in oxidation state of GCL were also observed with other oxidative stress treatments, including cadmium exposure, inhibition of glutathione synthesis by buthionine sulfoximine, or the generation of reaction oxygen species via menadione (Hicks et al., 2007).
Conservation of the disulfide linkage that mediates the monomer/dimer transition of GCL occurs across the plant kingdom (Gromes et al., 2008). Interestingly, a disulfide bond is also found in the GCL from α-proteobacteria (Hibi et al., 2004; Gromes et al., 2008). Analysis of the redox response of the GCL from tobacco (Nicotiana tabacum), Agrobacterium tumefaciens (a-proteobacteria), and Xanthomonas campestris (γ-proteobacteria) showed that the two proteobacterial GCL remain monomeric during redox changes, even though the disulfide is retained. Although redox changes did not alter enzyme activity in vitro, it is possible that formation of the disulfide in the GCL from proteobacteria may alter the stability and/or half-life of the protein in vivo; however, this requires further investigation. Examination of the GCL from these different organisms suggests that the interplay between the redox-sensitive thiol-switch and dimerization of the plant enzymes occurred later in evolution and may be related to compartmentalization of glutathione synthesis in the chloroplast (Gromes et al., 2008).
The sub-organellar localization of GCL raises questions about redox regulation of glutathione biosynthesis in plants. In vitro and in vivo experiments indicate that GCL is redox-regulated and is more active as the oxidized dimer (Jez et al., 2004; Hothorn et al., 2006; Hicks et al., 2007; Gromes et al., 2008). Within the chloroplast, GCL activity appears localized to the stroma, which is typically considered a reducing environment, not to the lumen, which is oxidizing (Rouhier et al., 2008; Meyer et al., 2009). Many metabolic enzymes in the stroma are active as reduced proteins and less active as oxidized (Rouhier et al., 2008; Meyer et al., 2009); however, the opposite situation occurs with GCL.
The molecular mechanism involved in maintaining the redox state of GCL is currently unclear. Although the thioredoxin and glutaredoxin systems are both found in the chloroplast stroma (Rouhier et al., 2008; Meyer et al., 2009), the physical properties of GCL suggest that the glutaredoxin system may be important for modulating activity. The in vitro redox-midpoint of GCL (Hicks et al., 2007) is comparable to the redox potential of glutathione pair and to the cellular redox potential observed in plant cells (Meyer et al., 2001). The redox behavior of GCL suggests a possible link to glutaredoxins as a mechanism for maintaining a reduced pool of less active protein; however, additional work is required to establish this mechanism. Alternatively, oxidation and activation of GCL may not require additional molecular systems, but may occur by direct interaction with glutathione to mediate redox changes that modulate activity.
Plant Glutathione Synthetases: Structure, Function, and Product Diversity
In the second step of glutathione synthesis, GS catalyzes the ATP-dependent addition of glycine to γ-glutamylcysteine (Figure 1). GS from bacteria and eukaryotes form two distinct families that share no amino acid sequence homology. Structural and functional characterization of GS from bacteria demonstrates that this enzyme functions as a tetramer (Yamaguchi et al., 1993), whereas the mammalian, yeast, and plant GS are active as dimers (Polekhina et al., 1999; Gogos and Shapiro, 2002; Jez and Cahoon, 2004). The GS from Arabidopsis, wheat (Triticum aestivum), maize (Zea mays), and various legumes share ~40% amino acid sequence identity with the human and yeast homologs (Rawlins et al., 1995; Ullmann et al., 1996; Wang and Oliver, 1996; Moran et al., 2000; Matamoros et al., 2003; Skipsey et al., 2005).
Early mutagenesis studies of the Arabidopsis GS identified a putative glycine-rich loop as important for activity (Wang and Oliver, 1997a,b), which was identified in later structural studies as forming part of the active site (Galant et al., 2009). Steady-state kinetic analysis showed that Arabidopsis GS uses a random Ter-reactant mechanism with binding of either ATP or γ-glutamylcysteine preferred first and addition of glycine last (Jez and Cahoon, 2004). Extensive site-directed mutagenesis, ligand binding analysis, determination of pH–rate profiles, and solvent deuterium isotope effects established the reaction mechanism for Arabidopsis GS and for the eukaryotic homologs (Herrera et al., 2007). In the first part of the reaction, formation of an electrophilic acyl-phosphate intermediate occurs by transfer of the γ-phosphate of ATP to γ-glutamylcysteine. A group of acidic residues coordinates two Mg2+ to ATP and orienting the γ-phosphate group in the active site. The guanidyl group of Arg132 helps stabilize the pentavalent transition state that yields the acyl-phosphate intermediate. Next, the amino group of glycine acts as a nucleophile to attack this intermediate. The side-chain guanidyl group of Arg454 interacts with the carboxylate moiety of glycine to orient the substrate for attack on the acyl-phosphate intermediate (Herrera et al., 2007). Although these mechanistic studies elucidate the catalytic role of key active site residues, the basis of substrate specificity for the nucleophilic amino acid in the plant GS is less well understood.
Unlike other organisms, some plants synthesize glutathione analogs in which β-alanine, serine, or glutamate replaces glycine in the tripeptide (Figure 4A). Multiple legumes use β-alanine instead of glycine to synthesize homoglutathione (Klapheck et al., 1995; Matamoros et al., 1999; Moran et al., 2000). Hydroxymethyl–glutathione (R = serine) has been isolated from rice and Agrostis (a grass species; Klapheck et al., 1994). In maize, the synthesis of glutathione-like peptides with glutamate replacing glycine is exclusively induced by exposure to cadmium (Meuwly et al., 1995).
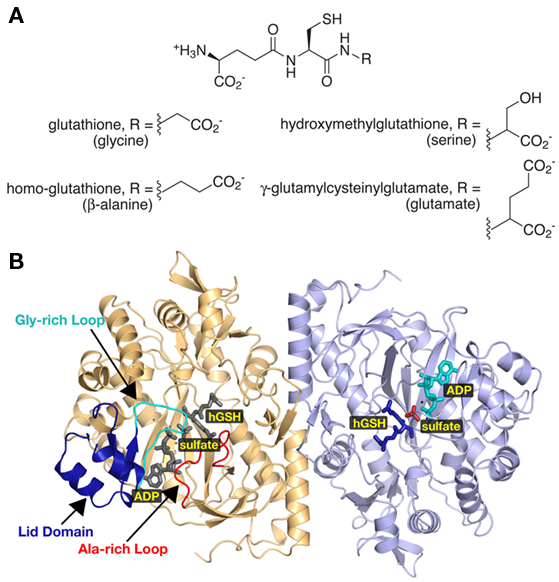
Figure 4. Diversity in Plant Glutathione Synthetases. (A) The chemical structures of glutathione analogs synthesized by various plants are shown. All share the core γ-glutamylcysteine structure with modifications to the third amino acid position as indicated. (B) Ribbon diagram of the homoglutathione (hGS) dimer (Galant et al., 2009). Monomers are colored gold and blue, respectively. The lid domain (dark blue), glycine-rich loop (cyan), and the alanine-rich loop (red) are highlighted in the gold monomer. Positions of bound ADP, sulfate, and homoglutathione are highlighted in the blue monomer with corresponding ligands colored gray in the gold monomer.
Of these alternate glutathione analogs, the most is known about homoglutathione. To date, homoglutathione is found in fourteen different legumes (pea, alfalfa, soybean, bean, mungbean, lentil, chickpea, sweet pea, cowpea, Italian clover, red clover, blue fenugreek, sweet clover, and runner bean); however, two additional legumes (broad bean and lupine) appear to lack this glutathione analog in all assayed tissue types (Matamoros et al., 1999). Additionally, cowpea and pea have homoglutathione in roots and nodules, but not in leaves (Moran et al., 2000). In soybean, leaves and seeds contain 50- to 200-fold and 135-fold more homoglutathione than glutathione, respectively (Matamoros et al., 1999). Soybean nodules contain ~4-fold more homoglutathione than glutathione, while the roots contain nearly 80-fold more homoglutathione (Matamoros et al., 1999). Physiologically, the exact role of homoglutathione is unclear. A likely possibility is that homoglutathione replaces glutathione as the dominant cellular redox-buffer in tissues associated with nitrogen fixation, as homoglutathione is required for proper development of nodules (Frendo et al., 2005).
For biosynthesis of glutathione and homoglutathione share a common first reaction, i.e., the synthesis of γ-glutamylcysteine, but the chemical diversity results from altered specificity for the nucleophilic amino acid in the second reaction of the pathway. The soybean homoglutathione synthetase (hGS) is related by ~70% sequence identity to the soybean GS. Because the soybean genome appears to have undergone two rounds of genome duplication, hGS likely arose from GS by divergent evolution after the first duplication event (Frendo et. al, 2001). In Arabidopsis, there is a single gene encoding for GS, whereas, the soybean genome contains two GS and two hGS genes, with each pair sharing 87 and 93% identity, respectively.
Recent crystallographic studies of soybean hGS demonstrate the critical role of an active site loop in determining the glycine versus β-alanine substrate specificity of GS and hGS (Figure 4; Galant et al., 2009). Overall, the three-dimensional structure of hGS resembles that of the eukaryotic GS (Polekhina et al., 1999; Gogos and Shapiro, 2002). Likewise, the amino acids forming the ATP, Mg2+, and γ-glutamylcysteine binding sites of hGS are nearly invariant with those of human, yeast, and Arabidopsis GS. Three structural features – a lid domain, a glycine-rich loop, and an alanine-rich loop – define the active site of hGS (Figure 4B). The lid domain and glycine-rich loop are critical for ATP binding, which triggers major shifts in their conformations (Galant et al., 2009). The alanine-rich loop contacts either the terminal glycine of glutathione in the GS structures or the terminal β-alanine in the hGS structure (Polekhina et al., 1999; Gogos and Shapiro, 2002; Galant et al., 2009). Interestingly, comparison of the GS and hGS structures revealed differences in the sequence of this loop.
In the structures of human and yeast GS, two alanine residues form van der Waals contacts with the glycine moiety of glutathione (Polekhina et al., 1999; Gogos and Shapiro, 2002). The corresponding residues in soybean hGS are a leucine and a proline. In the known GS, the AA-motif is conserved, whereas, the LP-motif is found in the known hGS from various legumes. Mutagenesis of the LP-motif of soybean hGS to the AA-motif reduced catalytic efficiency with β-alanine by 10-fold, but improved specificity for glycine by ~1,000-fold (Galant et al., 2009).
These structure/function studies established the basis of glutathione versus homoglutathione specificity, but it remains unclear how other glutathione analogs are formed in different plants. Skipsey et al. (2005) cloned and characterized five GS-like proteins from soybean, wheat, and corn noting that the soybean enzyme was an hGS and that all the other enzymes functioned as GS. Interestingly, their analysis demonstrated that different GS-like enzymes accepted various alternate nucleophiles, including d-serine, non-natural amino acids, and alpha-amino alcohols, to yield a range of glutathione variants, but did not produce either serine- or glutamate-substituted analogs. Although altered specificity in the GS reaction mechanism offers a direct route to analog synthesis, it is possible that other enzymes catalyze additional reactions that modify glutathione into the specialized peptides, like hydroxymethyl–glutathione.
Summary
Glutathione biosynthesis plays a key role in maintaining redox balance and homeostasis in plants. The core reactions of glutathione synthesis are conserved across various organisms, but plants have diversified both the regulatory mechanisms that control its synthesis and the range of products derived from this pathway.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge support from the National Science Foundation (MCB-0904215 to Joseph M. Jez), an American Society of Plant Biologists Pioneer Hi-Bred Graduate Research Fellowship (Ashley Galant), and an NSF Research Opportunity Award Supplement (Mary L. Preuss).
References
Arisi, A. C., Noctor, G., Foyer, C. H., and Jouanin, L. (1997). Modulation of the thiol contents in poplars over-expressing enzymes involved in glutathione synthesis. Planta 202, 357–359.
Biterova, E. I., and Barycki, J. J. (2009). Mechanistic details of glutathione biosynthesis revealed by crystal structures of Saccharomyces cerevisiae glutamate-cysteine ligase. J. Biol. Chem. 284, 32700–32708.
Cobbett, C. S., May, M. J., Howden, R., and Rolls, B. (1998). The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thaliana is deficient in gamma-glutamylcysteine synthetase. Plant J. 16, 73–78.
Copley, S. D., and Dhillon, J. K. (2002). Lateral gene transfer and parallel evolution in the history of glutathione biosynthesis genes. Genome Biol. 3, 25.1–16.
Foyer, C. H., Lopez-Delgado, H., Dat, J. F., and Scott, I. M. (1997). Hydrogen peroxide- and glutathione-associated mechanisms of acclimatory stress tolerance and signalling. Physiol. Plant. 100, 241–254.
Foyer, C. H., Souriau, N., Perret, S., Lelandais, M., Kunert, K. J., Pruvost, C., and Jouanin, L. (1995). Overexpression of glutathione reductase but not glutathione synthetase leads to increases in antioxidant capacity and resistance to photoinhibition in poplar tress. Plant Physiol. 109, 1047–1057.
Fraser, J. A., Kansagra, P., Kotecki, C., Saunders, R. D., and McLellan, L. I. (2003). The modifier subunit of Drosophila glutamate-cysteine ligase regulates catalytic activity by covalent and noncovalent interactions and influences glutathione homeostasis in vivo. J. Biol. Chem. 278, 46369–46377.
Fraser, J. A., Saunders, R. D., and McLellan, L. I. (2002). Drosophila melanogaster glutamate-cysteine ligase activity is regulated by a modifier subunit with a mechanism of action similar to that of the mammalian form. J. Biol. Chem. 277, 1158–1165.
Frendo, P., Harrison, J., Norman, C., Hernández-Jiménez, M. J., Van de Sype, G., Gilabert, A., and Puppo, A. (2005). Glutathione and homoglutathione play a critical role in the nodulation process of Medicago truncatula. Mol. Plant Microbe Interact. 18, 254–259.
Frendo, P., Jiménez, M. J., Mathieu, C., Duret, L., Gallesi, D., Van de Sype, G., Hérouart, D., and Puppo, A. (2001). A Medicago truncatula homoglutathione synthetase is derived from glutathione synthetase by gene duplication. Plant Physiol. 126, 706–715.
Frendo, P., Mathieu, C., Van de Sype, G., Hérouart, D., and Puppo, A. (1999). Characterisation of a cDNA encoding gamma-glutamylcysteine synthetase in Medicago truncatula. Free Radic. Res. 31, S213–S218.
Galant, A., Arkus, K. A., Zubieta, C., Cahoon, R. E., and Jez, J. M. (2009). Structural basis for evolution of product diversity in soybean glutathione biosynthesis. Plant Cell 21, 3450–3458.
Gogos, A., and Shapiro, L. (2002). Large conformational changes in the catalytic cycle of glutathione synthase. Structure 10, 1669–1676.
Gómez, L. D., Vanacker, H., Buchner, P., Noctor, G., and Foyer, C. H. (2004). Intercellular distribution of glutathione synthesis in maize leaves and its response to short-term chilling. Plant Physiol. 134, 1662–1671.
Grill, E., Winnacker, E. L., and Zenk, M. H. (1985). Phytochelatins: the principal heavy-metal complexing peptides of higher plants. Science 230, 674–676.
Gromes, R., Hothorn, M., Lenherr, E. D., Rybin, V., Scheffzek, K., and Rausch, T. (2008). The redox switch of gamma-glutamylcysteine ligase via a reversible monomer-dimer transition is a mechanism unique to plants. Plant J. 54, 1063–1075.
Hell, R., and Bergmann, L. (1988). Glutathione synthetase in tobacco suspension cultures: catalytic properties and localization. Physiol. Plant. 72, 70–76.
Hell, R., and Bergmann, L. (1990). Gamma-glutamylcysteine synthetase in higher plants: catalytic properties and subcellular localization. Planta 180, 603–612.
Herrera, K., Cahoon, R. E., Kumaran, S., and Jez, J. M. (2007). Reaction mechanism of glutathione synthetase from Arabidopsis thaliana: site-directed mutagenesis of active site residues. J. Biol. Chem. 282, 17157–17165.
Hibi, T., Nii, H., Nakatsu, T., Kimura, A., Kato, H., Hiratake, J., and Oda, J. (2004). Crystal structure of g-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis. Proc. Natl. Acad. Sci. U.S.A. 101, 15052–15057.
Hicks, L. M., Cahoon, R. E., Bonner, E. R., Rivard, R. S., Sheffield, J., and Jez, J. M. (2007). Thiol-based regulation of redox-active glutamate-cysteine ligase from Arabidopsis thaliana. Plant Cell 19, 2653–2661.
Hothorn, M., Wachter, A., Gromes, R., Stuwe, T., Rausch, T., and Scheffzek, K. (2006). Structural basis for the redox control of plant glutamate cysteine ligase. J. Biol. Chem. 281, 27557–27565.
Huang, C. S., Chang, L. S., Anderson, M. E., and Meister, A. (1993). Catalytic and regulatory properties of the heavy subunit of rat kidney γ-glutamylcysteine synthetase. J. Biol. Chem. 268, 19675–19680.
Jez, J. M., and Cahoon, R. E. (2004). Kinetic mechanism of glutathione synthetase from Arabidopsis thaliana. J. Biol. Chem. 279, 42726–42731.
Jez, J. M., Cahoon, R. E., and Chen, S. (2004). Arabidopsis thaliana glutamate-cysteine ligase: functional properties, kinetic mechanism, and regulation of activity. J. Biol. Chem. 279, 33463–33479.
Klapheck, S., Fliegner, W., and Zimmer, I. (1994). Hydroxymethyl-phytochelatins [(γ-glutamylcysteine)n-serine] are metal-induced peptides of the Poaceae. Plant Physiol. 104, 1325–1332.
Klapheck, S., Schlunz, S., and Bergmann, L. (1995). Synthesis of phytochelatins and homo-phytochelatins in Pisum sativum L. Plant Physiol. 107, 515–521.
Lueder, D. V., and Phillips, M. A. (1996). Characterization of Trypanosoma brucei γ-glutamylcysteine synthetase, an essential enzyme in the biosynthesis of trypanothione. J. Biol. Chem. 271, 17485–17490.
Madamanchi, N. R., and Alscher, N. G. (1991). Metabolic basis for differences in sensitivity of 2 pea cultivars to sulfur-dioxide. Plant Physiol. 97, 88–93.
Matamoros, M. A., Clemente, M. R., Sato, S., Asamizu, E., Tabata, S., Ramos, J., Moran, J. F., Stiller, J., Gresshoff, P. M., and Becana, M. (2003). Molecular analysis of the pathway for the synthesis of thiol tripeptides in the model legume Lotus japonicus. Mol. Plant Microbe Interact. 16, 1039–1046.
Matamoros, M. A., Moran, J. F., Iturbe-Ormaetxe, I., Rubio, M. C., and Becana, M. (1999). Glutathione and homoglutathione synthesis in legume root nodules. Plant Physiol. 121, 879–888.
Maughan, S. C., Pasternak, M., Cairns, N., Kiddle, G., Brach, T., Jarvis, R., Haas, F., Nieuwland, J., Lim, B., Müller, C., Salcedo-Sora, E., Kruse, C., Orsel, M., Hell, R., Miller, A. J., Bray, P., Foyer, C. H., Murray, J. A., Meyer, A. J., and Cobbett, C. S. (2010). Plant homologs of the Plasmodium falciparum chloroquine-resistance transporter, PfCRT, are required for glutathione homeostasis and stress responses. Proc. Natl. Acad. Sci. U.S.A. 107, 2331–2336.
May, M. J., and Leaver, C. J. (1993). Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 103, 621–627.
May, M. J., and Leaver, C. J. (1994). Arabidopsis thaliana γ-glutamylcysteine synthetase is structurally unrelated to mammalian, yeast, and Escherichia coli homologs. Proc. Natl. Acad. Sci. U.S.A. 91, 10059–10063.
May, M. J., Vernoux, T., Leaver, C., Van Montagu, M., and Inze, D. (1998a). Glutathione homeostasis in plants: implications for environmental sensing and plant development. J. Exp. Bot. 49, 649–667.
May, M. J., Vernoux, T., Sanchez-Fernandez, R., Van Montagu, M., and Inze, D. (1998b). Evidence for post-transcriptional activation of γ-glutamylcysteine synthetase during plant stress responses. Proc. Natl. Acad. Sci. U.S.A. 95, 12049–12054.
Meuwly, P., Thibault, P., Schwan, A. L., and Rauser, W. E. (1995). Three families of thiol peptides are induced by cadmium in maize. Plant J. 7, 391–400.
Meyer, A. J., and Fricker, M. D. (2002). Control of demand-driven biosynthesis of glutathione in green Arabidopsis suspension culture cells. Plant Physiol. 130, 1927–1937.
Meyer, A. J., May, M. J., and Fricker, M. D. (2001). Quantitative in vivo measurement of glutathione in Arabidopsis cells. Plant J. 27, 67–78.
Meyer, Y., Buchanan, B. B., Vignols, F., and Reichheld, J. P. (2009). Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu. Rev. Genet. 43, 335–367.
Mittova, V., Theodoulou, F. L., Kiddle, G., Gómez, L., Volokita, M., Tal, M., Foyer, C. H., and Guy, M. (2003). Coordinate induction of glutathione biosynthesis and glutathione-metabolizing enzymes is correlated with salt tolerance in tomato. FEBS Lett. 554, 417–421.
Moran, J. F., Iturbe-Ormaetxe, I., Matamoros, M. A., Rubio, M. C., Clemente, M. R., Brewin, N. J., and Becana, M. (2000). Glutathione and homoglutathione synthetases of legume nodules. Cloning, expression, and subcellular localization. Plant Physiol. 124, 1381–1392.
Noctor, G., Arisi, A. C., Jouanin, L., Kunert, K. J., Rennenberg, H., and Foyer, C. H. (1998). Glutathione: biosynthesis, metabolism, and relationship to stress tolerance explored in transformed plants. J. Exp. Bot. 49, 623–647.
Noctor, G., Arisi, A. C., Jouanin, L., Valadier, M. H., Roux, Y., and Foyer, C. H. (1997). The role of glycine in determining the rate of glutathione synthesis in poplar. Physiol. Plant 100, 255–263.
Noctor, G., and Foyer, C. H. (1998). Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 249–279.
Noctor, G., Gomez, L., Vanacker, H., and Foyer, C. H. (2002). Interactions between biosynthesis, compartmentation and transport in the control of glutathione homeostasis and signalling. J. Exp. Bot. 53, 1283–1304.
Noctor, G., Strohm, M., Jouanin, L., Kunert, K. J., Foyer, C. H., and Rennenberg, H. (1996). Synthesis of glutathione in leaves of transgenic poplar overexpressing γ-glutamylcysteine synthetase. Plant Physiol. 112, 1071–1078.
Ogawa, K. (2005). Glutathione-associated regulation of plant growth and stress responses. Antioxid. Redox Signal. 7, 973–981.
Parisy, V., Poinssot, B., Owsianowski, L., Buchala, A., Glazebrook, J., and Mauch, F. (2007). Identification of PAD2 as a gamma-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 49, 159–172.
Pasternak, M., Lim, B., Wirtz, M., Hell, R., Cobbett, C. S., and Meyer, A. J. (2008). Restricting glutathione biosynthesis to the cytosol is sufficient for normal plant development. Plant J. 53, 999–1012.
Polekhina, G., Board, P. G., Gali, R. R., Rossjohn, J., and Parker, M. W. (1999). Molecular basis of glutathione synthetase deficiency and a rare gene permutation event. EMBO J. 18, 3204–3213.
Rawlins, M. R., Leaver, C. J., and May, M. J. (1995). Characterisation of an Arabidopsis thaliana cDNA encoding glutathione synthetase. FEBS Lett. 376, 81–86.
Rouhier, N., Lemaire, S. D., and Jacquot, J. P. (2008). The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 59, 143–166.
Rüegsegger, A., Schmutz, D., and Brunold, C. (1990). Regulation of glutathione synthesis by cadmium in Pisum sativum L. Plant Physiol. 93, 1579–1584.
Schäfer, H. J., Greiner, S., Rausch, T., and Haag-Kerwer, A. (1997). In seedlings of the heavy metal accumulator Brassica juncea Cu2+ differentially affects transcript amounts for gamma-glutamylcysteine synthetase (gamma-ECS) and metallothionein (MT2). FEBS Lett. 404, 216–220.
Schäfer, H. J., Haag-Kerwer, A., and Rausch, T. (1998). cDNA cloning and expression analysis of genes encoding GSH synthesis in roots of the heavy-metal accumulator Brassica juncea: evidence for Cd-induction of a putative mitochondrial γ-glutamylcysteine synthetase isoform. Plant Mol. Biol. 37, 87–97.
Seelig, G. F., Simondsen, R. P., and Meister, A. (1984). Reversible dissociation of γ-glutamylcysteine synthetase into two subunits. J. Biol. Chem. 259, 9345–9347.
Skipsey, M., Davis, B. G., and Edwards, R. D. (2005). Diversification in substrate usage by glutathione synthetases from soya bean (Glycine max), wheat (Triticum aestivum) and maize (Zea mays). Biochem. J. 391, 567–574.
Strohm, M., Jouanin, L., Kunert, K. J., Pruvost, C., Polle, A., Foyer, C. H., and Rennenburg, H. (1995). Regulation of glutathione synthesis in leaves of transgenic poplar overexpressing glutathione synthetase. Plant J. 7, 141–145.
Ullmann, P., Gondet, L., Potier, S., and Bach, T. J. (1996). Cloning of Arabidopsis thaliana glutathione synthetase (GSH2) by functional complementation of a yeast gsh2 mutant. Eur. J. Biochem. 236, 662–669.
Vernoux, T., Wilson, R. C., Seeley, K. A., Reichheld, J. P., Muroy, S., Brown, S., Maughan, S. C., Cobbett, C. S., Van Montagu, M., Inzé, D., May, M. J., and Sung, Z. R. (2000). The root meristemless1/cadmium sensitive2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12, 97–110.
Vivancos, P. D., Dong, Y., Ziegler, K., Markovic, J., Pallardo, F. V., Pellny, T. K., Verrier, P. J., and Foyer, C. H. (2010). Recruitment of glutathione into the nucleus during cell proliferation adjusts whole-cell redox homeostasis in Arabidopsis thaliana and lowers the oxidative defence shield. Plant J. 64, 825–838.
Wachter, A., Wolf, S., Steininger, H., Bogs, J., and Rausch, T. (2005). Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J. 41, 15–30.
Wang, C. L., and Oliver, D. J. (1996). Cloning of the cDNA and genomic clones for glutathione synthetase from Arabidopsis thaliana and complementation of a gsh2 mutant in fission yeast. Plant Mol. Biol. 31, 1093–1104.
Wang, C. L., and Oliver, D. J. (1997a). Identification of a putative flexible loop in Arabidopsis glutathione synthetase. Biochem. J. 322, 241–244.
Wang, C. L., and Oliver, D. J. (1997b). Glutathione synthetase: similarities of the proteins from Schizosaccharomyces pombe and Arabidopsis thaliana. Biochem. J. 326, 563–566.
Wu, J., Qu, T., Chen, S., Zhao, Z., and An, L. (2009). Molecular cloning and characterization of a gamma-glutamylcysteine synthetase gene from Chorispora bungeana. Protoplasma 235, 27–36.
Xiang, C., and Oliver, D. J. (1998). Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. Plant Cell 10, 1539–1550.
Yamaguchi, H., Kato, H., Hata, Y., Nishioka, T., Kimura, A., Oda, J., and Katsube, Y. (1993). Three-dimensional structure of the glutathione synthetase from Escherichia coli B at 2.0 Å resolution. J. Mol. Biol. 229, 1083–1100.
Yi, H., Galant, A., Ravilious, G. E., Preuss, M. L., and Jez, J. M. (2010a). Sensing sulfur conditions: simple to complex biochemical regulatory mechanisms in plant thiol metabolism. Mol. Plant 3, 269–279.
Keywords: glutathione, Arabidopsis, metabolism, redox regulation, biosynthesis
Citation: Galant A, Preuss ML, Cameron JC and Jez JM (2011) Plant glutathione biosynthesis: diversity in biochemical regulation and reaction products. Front. Plant Sci. 2:45. doi: 10.3389/fpls.2011.00045
Received: 02 August 2011;
Paper pending published: 15 August 2011;
Accepted: 16 August 2011;
Published online: 05 September 2011.
Edited by:
Ikuro Abe, The University of Tokyo, JapanCopyright: © 2011 Galant, Preuss, Cameron and Jez. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Joseph M. Jez, Department of Biology, Washington University in St. Louis, One Brookings Drive, Campus Box 1137, St. Louis, MO 63130, USA. e-mail:jjez@biology2.wustl.edu
 Ashley Galant1
Ashley Galant1
