- 1 Plant Science Center, RIKEN, Yokohama, Japan
- 2 Graduate School of Pharmaceutical Sciences, Chiba University, Chiba, Japan
Triterpenoid saponins are a diverse group of natural products in plants and are considered defensive compounds against pathogenic microbes and herbivores. Because of their various beneficial properties for humans, saponins are used in wide-ranging applications in addition to medicinally. Saponin biosynthesis involves three key enzymes: oxidosqualene cyclases, which construct the basic triterpenoid skeletons; cytochrome P450 monooxygenases, which mediate oxidations; and uridine diphosphate-dependent glycosyltransferases, which catalyze glycosylations. The discovery of genes committed to saponin biosynthesis is important for the stable supply and biotechnological application of these compounds. Here, we review the identified genes involved in triterpenoid biosynthesis, summarize the recent advances in the biotechnological production of useful plant terpenoids, and discuss the bioengineering of plant triterpenoids.
Introduction
Triterpenoids including steroids are a highly diverse group of natural products widely distributed in plants (Vincken et al., 2007). Plants often accumulate these compounds in their glycosylated form – saponin. Saponins comprise hydrophobic triterpenoid aglycones called sapogenin and one or more hydrophilic sugar moieties.
Biologically, plant saponins are considered defensive compounds against pathogenic microbes and herbivores (Osbourn, 1996; Kuzina et al., 2009; Szakiel et al., 2011). These saponins also have beneficial properties for humans. For example, Panax and Glycyrrhiza plants are well-known traditional herbal medicines containing saponins, ginsenosides, and glycyrrhizin, respectively, with various pharmacological effects (Shibata, 2000, 2001). Saponins have a variety of other applications as well. They show foaming ability when mixed with water, as indicated by the word sapo, meaning soap in Latin. In fact, Saponaria officinalis (common soapwort) and Quillaja saponaria (soapbark) have been used as soap. The saponins of Q. saponaria are also used as emulsifiers in cosmetics and foods. Furthermore, glycyrrhizin is used as a natural sweetener, with 150 times the sweetness of sugar.
In this article, we summarize the genes involved in triterpenoid biosynthesis identified to date and the recent advances in the bioengineering production of useful plant terpenoids; finally, we provide a perspective on the bioengineering of plant triterpenoids.
Triterpenoid Biosynthesis
Terpenoids are built up from C5 units, isopentenyl diphosphate (IPP). IPP is supplied from the cytosolic mevalonic acid (MVA) pathway and the plastidal methylerythritol phosphate (MEP) pathway. Triterpenoids and sesquiterpenoids are biosynthesized via the MVA pathway, whereas monoterpenoids, diterpenoid, and tetraterpenoids are biosynthesized via the MEP pathway. The first diversifying step in triterpenoid biosynthesis is the cyclization of 2,3-oxidosqualene catalyzed by oxidosqualene cyclase (OSC; Abe et al., 1993; Figure 1). In general, animals and fungi have only one OSC, lanosterol synthase (LAS), for sterol biosynthesis. However, higher plants have several OSCs not only for sterol biosynthesis, such as cycloartenol synthase (CAS) and LAS (Ohyama et al., 2009), but also for triterpenoid biosynthesis. The molecular diversity of OSCs enables more than 100 skeletal variations of triterpenoids in plants (Xu et al., 2004). Until now, some dozens of OSC genes from not only model plants but also crops and medicinal plants have been cloned and functionally characterized (reviewed in Kushiro and Ebizuka, 2010). For example, the Arabidopsis thaliana genome has 13 OSC genes, and the functional identification of these genes has been completed, at least, by in vitro experiments. Most of the OSCs from eudicots are phylogenetically classified into some groups, and the reaction products differ from group to group. The site-directed mutagenesis and homology modeling of plant OSCs have been carried out to investigate the reaction mechanisms regarding their product variety (reviewed in Kushiro and Ebizuka, 2010). Of OSCs from various organisms, the structure of human LAS protein was elucidated (Thoma et al., 2004).
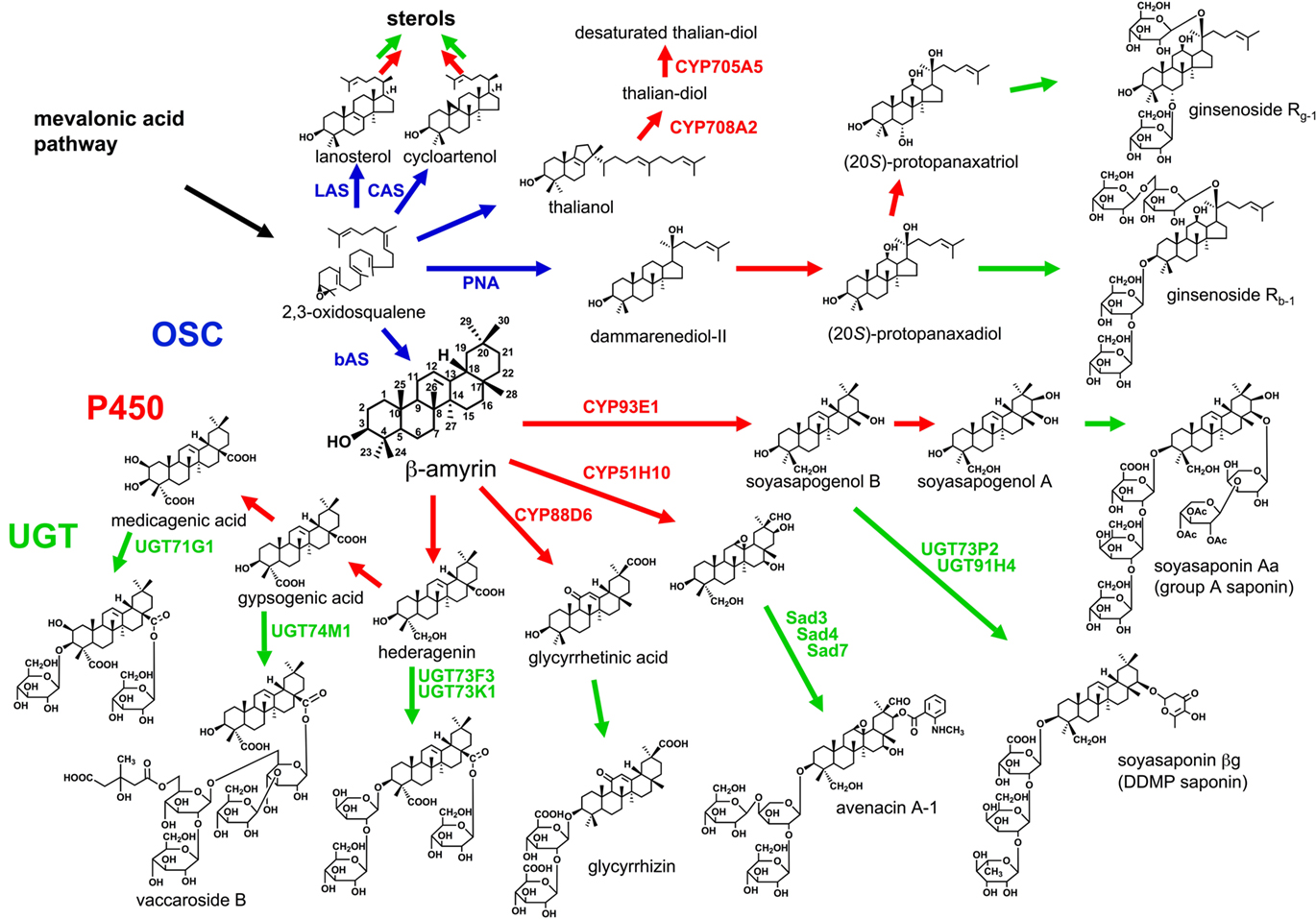
Figure 1. Triterpenoid biosynthetic pathway. After the cyclization of 2,3-oxidosqualene catalyzed by OSC, a triterpenoid undergoes various modifications including P450-catalyzed oxidation and UGT-catalyzed glycosylation. Blue arrows, OSC-catalyzed steps; red arrows, P450-catalyzed steps; green arrows, additional modifications including UGT-catalyzed steps.
After an OSC constructs the basic triterpenoid skeleton, the skeleton is modified to a hydrophobic aglycone called sapogenin. The first modification is oxidation catalyzed by cytochrome P450 monooxygenase (P450), and this step enables further modifications such O-glycosylation. P450 is highly diverse and catalyzes several kinds of chemical reactions committed to the secondary metabolism (Kahn and Durst, 2000).
Glycosylation is essential for saponin biosynthesis. Glycosylation increases the water solubility and changes the biological activity of triterpenoid. Uridine diphosphate (UDP)-dependent glycosyltransferases (UGTs) recognize a wide range of natural products as acceptor molecules.
P450 species and UGTs belong to multigene families and are the key factors for explosive diversification of other natural products in plants. In the case of reported P450 species in saponin biosynthesis, those CYP families vary respecting not only the carbon skeletons of the triterpenoid substrates but also the target positions of the reactions. The diversity of these enzymes makes identification of the genes for saponin biosynthesis difficult. The genes involved in triterpenoid biosynthesis identified in plants to date are presented as follows.
Triterpenoids in Arabidopsis thaliana
The first model plant A. thaliana has total 13 OSCs, 246 P450 species (Werck-Reichhart et al., 2002) and 112 UGTs (Paquette et al., 2003). The protein encoded by At5g48010, an OSC, was identified as thalianol synthase (Fazio et al., 2004). However, no tricyclic triterpenoid including thalianol had been reported in Brassicales at that time. After that thalianol was detected at about 0.4% of total sterols in the root of A. thaliana, CYP708A2 and CYP705A5 were identified as P450 species in thalianol metabolism (Field and Osbourn, 2008). Although the details of variety and content of saponins in A. thaliana have not been clarified, Warnecke et al. (1997) reported one UGT for sterol glycosylation.
Soyasaponins in Glycine max
Glycine max mainly accumulates β-amyrin-derived oleanane-type triterpenoid saponins termed soyasaponins (Kitagawa et al., 1982, 1988; Burrows et al., 1987; Taniyama et al., 1988; Shiraiwa et al., 1991a,b; Kudou et al., 1992, 1993; Tsukamoto et al., 1993; Kikuchi et al., 1999). Soyasaponins are divided into two groups: 2,3-dihydro-2,5-dihydroxy-6-methyl-4H-pyran-4-one (DDMP) saponins have soyasapogenol B as their aglycone and group A saponins have soyasapogenol A as their aglycone. Soyasapogenol B is a C-22- and C-24-hydroxylated β-amyrin, and soyasapogenol A has an additional hydroxyl group at C-21.
Although DDMP saponins and their derivatives have beneficial effects on human health, some group A saponins are unfavorable because of their astringent taste (Okubo et al., 1992). To reduce the astringent taste of soybean, transgenic soybean plants with suppressing β-amyrin synthase (bAS), an OSC, gene by RNAi silencing were generated. The sapogenol levels of the transgenic seeds were reduced to below 500 μg g−1 or about 25% of the content in wild type (Maxwell et al., 2004). Only CYP93E1 C-24 hydroxylase was identified as an oxidase in soyasaponin biosynthesis (Shibuya et al., 2006). Soyasaponin βg, the main soyasaponin in G. max, is soyasapogenol B that attaches three sugar molecules, glucuronic acid, galactose, and rhamnose, at the C-3 hydroxyl group. UGT73P2 and UGT91H4 attach the second and third sugars in the sugar chain, respectively (Shibuya et al., 2010). These UGTs were selected as G. max expressed sequence tags (ESTs) with homologous sequences in the EST database of Medicago truncatula, which also produces a soyasaponin βg intermediate called soyasaponin I (Huhman et al., 2005). The activity of the first UGT, UDP-glucuronic acid:soyasapogenol B-glucuronyl transferase, was detected in the microsomal fraction of G. max (Kurosawa et al., 2002). However, no gene encoding the UGT has been cloned. Group A saponins with a terminal acetylated sugar in the C-22 sugar chain accumulate only in seed hypocotyls (Shimoyamada et al., 1990). A gene controlling the terminal sugar variety was mapped on soybean chromosome 7 (Takada et al., 2010).
Saponins in Medicago truncatula
Medicago truncatula, a leguminous model plant, accumulates over 30 oleanane-type saponins (Huhman et al., 2005). A corresponding bAS cDNA was identified by EST database mining (Suzuki et al., 2002) and homology-based PCR (Iturbe-Ormaetxe et al., 2003). UGT73K1 and UGT71G1 were characterized as triterpenoid glycosyltransferases by integrated analysis of the transcriptome and metabolome of M. truncatula, but UGT71G1 preferred some flavonoids to triterpenoids as substrates in vitro (Achnine et al., 2005). Although the glycosylated positions by both UGTs were not clarified in vitro, in silico docking simulation of the UGT71G1 crystal structure with UDP-glucose and medicagenic acid suggested that UGT71G1 can transfer a glucose molecule to the hydroxyl group at C-3 (Shao et al., 2005). UGT73F3 was identified as a glucosyltransferase of the hederagenin C-28 carboxyl group in an ester linkage by cluster analysis of transcription patterns and genetic loss-of-function analysis (Naoumkina et al., 2010).
Glycyrrhizin in Licorice
Glycyrrhizin is an oleanane-type saponin present in the underground parts of licorice (Glycyrrhiza). For use as a medicinal herb, the Japanese pharmacopia standard requires the root or stolon with 2.5% or more glycyrrhizin content. The biosynthetic pathway of glycyrrhizin from β-amyrin involves hydroxylations at C-11 and C-30, and two steps of glucuronyl transfers to the hydroxyl group at C-3. A bAS was identified from G. glabra (Hayashi et al., 2001). Further, we identified CYP88D6 as a β-amyrin C-11 oxidase (Seki et al., 2008). For CYP88D6 cloning, we first constructed an EST library of the underground parts (Sudo et al., 2009). On the basis of the sequence similarities, we identified P450 genes and selected the candidate P450 gene expressed in glycyrrhizin-accumulating tissues. In addition, CYP93E3 was identified as a β-amyrin C-24 oxidase in the secondary metabolism of glycyrrhizin. Further investigation is currently undertaken in our group for identifying other candidate genes including another P450 responsible for C-30 oxygenation (Seki et al., submitted) and UGTs involved in the biosynthetic pathway of glycyrrhizin.
Avenacins in Avena strigosa
Avena spp. (oats) produce antimicrobial oleanane-type saponins called avenacins. Osbourn and coworkers generated saponin-deficient (sad) mutants of A. strigosa; cloned Sad1, encoding bAS, Sad2, encoding CYP51H10 β-amyrin oxidase, and Sad7, encoding serine carboxypeptidase-like acyltransferase; and investigated sad3 and sad4 mutants accumulating a monodeglucosyl avenacin (Papadopoulou et al., 1999; Haralampidis et al., 2001; Qi et al., 2006; Mylona et al., 2008; Mugford et al., 2009). Genetic analysis showed five Sad loci, Sad2, Sad3, Sad6, Sad7, and Sad8, are within 3.6 cM around the Sad1 locus, and especially three Sad genes, Sad1, Sad3, and Sad7, clearly clustered in the genome (Qi et al., 2004). A cluster of such genes was also found in the A. thaliana genome for thalianol metabolism (Field and Osbourn, 2008). These clusters interest in the evolutionary process of triterpenoid biosynthesis in plants (Osbourn, 2010).
Vaccarosides in Saponaria vaccaria
The seeds of S. vaccaria, used in traditional Chinese medicine, accumulate oleanane-type saponins called vaccarosides. The aglycone of vaccaroside B is a C-23 and C-28 carboxylated β-amyrin, gypsogenic acid. A bAS cDNA was cloned by homology-based PCR (Meesapyodsuk et al., 2007). UGT74M1, with sequence similarity to other plant ester-forming glucosyltransferases, was cloned from the developing seed EST library and identified as an UDP-glucosyltransferase to C-28 of gypsogenic acid in an ester linkage (Meesapyodsuk et al., 2007).
Ginsenosides in Panax ginseng
Panax ginseng is a famous medicinal plant in Asia. The main pharmacologically active compounds in the ginseng are saponins called ginsenosides (Shibata, 2001). Major ginsenosides have a dammarane skeleton constructed by an OSC, dammarenediol-II synthase (PNA). Ginsenoside R0, a minor ginsenoside, is derived only from β-amyrin. Ebizuka and coworkers identified CAS, two bAS, LAS, and PNA cDNAs from hairy root cultures of P. ginseng in the ginsenoside-accumulating period (Kushiro et al., 1998a,b; Suzuki et al., 2006; Tansakul et al., 2006). Han et al. (2006) also identified PNA from the flower-accumulated dammarane-type saponins. The genes encoding P450 and UGTs committed to ginsenoside biosynthesis would be identifiable from such resources in the near future.
Biotechnological Production of Useful Plant Terpenoids
Although almost all production of glycyrrhizin depends on the collection of wild licorice, its harvesting is restricted to prevent exhaustion and desertification in the main producing country, China. Similarly, ginseng requires 4–5 years of careful cultivation and prevention of injury by continuous cropping. Such problems occur not only for saponins but also for other natural plant products. To ensure their stable supply, environment-friendly, and lower-cost alternatives such as biotechnological production are necessary. In the following, we describe the recent advances in the biotechnological production of useful terpenoids.
Artemisinin
Artemisinin, a sesquiterpenoid originally sourced from Artemisia annua, is used in combination therapy for malaria. The cost of therapy is too high for people in low-income countries where malaria is prevalent, and total synthesis of artemisinin (Schmid and Hofheinz, 1983) is not easy at low cost. Semi-synthesis of artemisinin from artemisinic acid (Roth and Acton, 1989) derived by fermentation could be an alternative lower-cost supply method (White, 2008). Ro et al. (2006) genetically modified yeast to increased productivity of a sesquiterpenoid biosynthesis starter, farnesyl pyrophosphate (FPP), and expressed cDNAs encoding an amorphadiene synthase (ADS), CYP71AV1, and a cytochrome P450 reductase (CPR) from A. annua in the yeast. The recombinant yeast produced a large amount of artemisinic acid (115 mg l−1). Subsequent improvement in the fermentation process increased the artemisinic acid titer to 2.5 g l−1 (Lenihan et al., 2008). On the other hand, an engineered Escherichia coli-integrated yeast MEV pathway to supply a large amount of FPP (Martin et al., 2003), modified CYP71AV1 at the N-terminus, and CPR produced 105 mg l−1 of artemisinic acid (Chang et al., 2007). Further improvement in the MEV pathway achieved an amorphadiene titer of 27.4 g l−1 (Tsuruta et al., 2009).
Taxol (Paclitaxel)
Taxol (paclitaxel) is a diterpenoid used against numerous cancers. Originally, Taxol was isolated from the bark of pacific yew (Taxus brevifolia) at low content (Wani et al., 1971). The complex structure of this drug limits its commercial chemical synthesis (Holton et al., 1994a,b; Nicolaou et al., 1994). Therefore, semi-synthesis from more accessible biosynthetic intermediates such 10-deacetylbaccatin III and production in Taxus plant cell cultures have been developed as alternative supply methods (Kingston, 2007), which still depend on plant sources. For further improvement in productivity and reduction in the therapeutic cost, biosynthetic production has been attempted. Introduction of several biosynthetic enzyme genes for Taxol in yeast resulted in the production of only trace amounts of the first hydroxylated intermediate, taxadien-5α-ol (Dejong et al., 2006).
On the other hand, Ajikumar et al. (2010) presented an optimization termed multivariate modular pathway engineering for taxadiene production in E. coli. They divided the taxadiene-producing pathway into two modules at the isopentenyl pyrophosphate step and searched for the optimal balance of the expression strength of each module for taxadiene production. The optimization enabled over 1 g l−1 of taxadiene production in E. coli, and subsequent expression of a chimeric protein from CYP725A4, a taxadien-5α-hydroxylase, and Taxus CPR resulted in 58 mg l−1 of taxadien-5α-ol production. The researchers (Ajikumar et al., 2010) indicated that re-optimization including the chimeric protein would improve the taxadien-5α-ol productivity.
Carotenoids
Carotenoids are well-known tetraterpenoid pigments in plants and microorganisms. They are used not only as natural colorants in food and feed but also in nutraceutical, cosmetic, and pharmaceutical products because of their antioxidant property. Vitamin A is converted from some carotenoids collectively called provitamin A in human body. Deficiency of vitamin A causes blindness and mortality due to weakening of the immune system in children of the developing world. Therefore, carotenoids have received much attention as metabolic engineering targets (reviewed in Das et al., 2007; Misawa, 2010). The engineered host organisms vary from microorganisms to plants. Here are two examples of the hosts – one for a plant (rice) and one for microorganism (E. coli). “Golden rice” is one of the most successful metabolically engineered plants (Ye et al., 2000). To increase provitamin A intake from rice, a provitamin A biosynthetic pathway was constructed in the endosperm of rice. Although Golden rice with psy encoding phytoene synthase from daffodil and crtI encoding phytoene desaturase from the bacterium Erwinia uredovora produced provitamin A, the content was not enough to meet the recommended daily allowance for children even in the regions where rice is a staple food. Therefore, Golden rice 2 was developed (Paine et al., 2005). The replacement of psy from daffodil by psy from maize improved the content adequately (>30 μg g−1).
Carotenoids are also popular targets in metabolically engineered microbial production. Alper et al. (2005) combined systematic and combinatorial gene knockout target identification methods in E. coli to increase the productivity of a well-known carotenoid pigment in tomato, lycopene. First, strain genotypes were systematically designed with gene knockouts reported to improve productivity. These strains then underwent deletions of unknown genes by a combinatorial transposon-based search. After the combination, the strain produced a high amount of lycopene (23 mg g−1 dry cell weight). Recently, an engineered E. coli with the MVA pathway genes from Enterococcus faecalis and Streptococcus pneumoniae and β-carotene biosynthesis genes produced β-carotene of 465 mg l−1 (Yoon et al., 2009). Furthermore, improvement in the culture condition of the recombinant E. coli increased the β-carotene titer to 663 mg l−1 (Kim et al., 2009).
Bioengineering of Plant Triterpenoids
In plants, overexpression of 3-hydroxy-3-methylgulutaryl-CoA reductase (HMGR), FPP synthase (FPS), and squalene synthase in the isoprenoid pathway has been attempted to increase triterpenoid productivity (Chappell et al., 1995; Schaller et al., 1995; Harker et al., 2003; Lee et al., 2004; Seo et al., 2005; Hey et al., 2006; Muñoz-Bertomeu et al., 2007; Lu et al., 2008; Kim et al., 2010). Despite the improved productivities per unit weight, the transgenic plants sometimes showed growth inhibition (Masferrer et al., 2002; Manzano et al., 2004; Shim et al., 2010), probably caused by metabolic imbalances. In fact, co-overexpression of FPS and the HMGR catalytic domain alleviated the growth inhibition caused by the individual overexpression of FPS or the HMGR catalytic domain (Manzano et al., 2004). Further elucidation of isoprenoid biosynthetic mechanisms is required to improve triterpenoid productivity in plants.
Furthermore, future challenging targets to elucidate for bioengineering of plant triterpenoids should be the regulatory mechanisms of the biosynthetic gene expressions and the accumulation mechanisms of triterpenoids. Saponins are frequently accumulated in specific tissues and organs. Glycyrrhizin and ginsenosides are accumulated in xylems of roots of licorice and ginseng, respectively (Shan et al., 2001; Fukuda et al., 2006). Genes for saponin biosynthesis also express in specific tissues and organs. In avena, Sad genes are expressed in the root epidermal cells accumulating avenacin A-1 (Haralampidis et al., 2001; Qi et al., 2006; Mylona et al., 2008; Mugford et al., 2009). In addition, metabolomic and transcriptomic analyses showed good correlations between expressions of the biosynthetic genes and the accumulations (Matsuda et al., 2010). In fact, recent successes for identification of saponin biosynthetic genes are based on such correlation analysis as described above. Those observations indicate that saponin productions are regulated most likely at the transcription level and thus implying the existence of specific transcription factor(s) for saponin biosynthesis. The engineering of transcription factor is a promising way to modify the biosynthetic pathway in addition to an introduction of multiple biosynthetic enzyme genes (Borevitz et al., 2000; Hirai et al., 2007; Gonzalez et al., 2008). The discovery of novel structural genes involved in the pathway can be achieved by an analysis of overexpressing lines of a transcription factor (Tohge et al., 2005; Luo et al., 2007). Furthermore, the control by the transcription factor would be a useful reference to improve the productivity by the optimization of the expression levels of multiple pathway genes introduced in a heterologous host.
At the subcellular level, saponins are accumulated in vacuoles (Kesselmeier and Urban, 1983; Mylona et al., 2008). However OSCs, P450s and some UGTs for saponin biosynthesis are known as microsomal enzymes (Hayashi et al., 1996; Kurosawa et al., 2002). These facts suggest the presence of a vacuolar transporter of saponin. The transporter is also a target of engineering to improve the accumulation of the target compounds. So far, as a plant terpenoid transporter, an ATP-binding cassette transporter, NpPDR1, involved in secretion of an antifungal diterpenoid, sclareol, in tobacco plant was reported (Jasiński et al., 2001). A homolog of human sterol carrier protein-2 in A. thaliana was identified as a lipid transfer protein (Edqvist et al., 2004). There is, however, no report to identify a triterpenoid transporter in a plant to date. To narrow down the transcription factor and transporter candidates in plant secondary metabolism, omics analyses are also useful strategies (Goossens et al., 2003; Hirai et al., 2007; Morita et al., 2009; Sawada et al., 2009).
Noteworthily, almost all OSCs, catalyzing the first triterpenoid diversification step, cloned from plants have been heterologously expressed in yeast for functional identification (reviewed in Kushiro and Ebizuka, 2010), because OSCs are membrane binding-type proteins and require eukaryotic developed intracellular membrane systems for their heterologous expression. Furthermore, yeast has a sterol biosynthetic pathway for producing the membrane constituent, and this pathway could be converted to a useful triterpenoid-producing pathway.
Previously, we developed a recombinant yeast system for the mechanistic investigation of P450 species in glycyrrhizin biosynthesis (Seki et al., 2008). To supply β-amyrin as the CYP88D6 substrate endogenously, a bAS was constitutively expressed. After the accumulation of β-amyrin in the yeast, CYP88D6 was co-expressed with a CPR as the redox partner. The final yields of 11-oxo-β-amyrin and 11α-hydroxy-β-amyrin at 2 days of culture after the induction were approximately 1.6 and 0.2 mg l−1, respectively. Although we employed Lotus bAS and CPR at that time, G. uralensis bAS and CPR would be more suitable to improve the production.
Genetic engineering in yeast sterol biosynthesis to enhance the availability of β-amyrin may also improve the production of 11-oxo-β-amyrin. Kirby et al. (2008) isolated a bAS from A. annua and achieved 6 mg l−1 of β-amyrin production in yeast expressing an N-terminal-truncated HMGR and restricting the expression of a native OSC, LAS. A large amount of squalene accumulated in yeast means that the yeast can produce even more β-amyrin. In addition to the efforts to improve the common isoprenoid pathway, as already described, enhancement of the catalytic properties of enzymes in triterpenoid biosynthesis should be effective. In Solanaceae, CYP71Ds are involved in sesquiterpenoid phytoalexin biosynthesis. Protein engineering of CYP71Ds based on sequence alignment analysis with phylogenetically related P450 species and homology modeling successfully enhanced the catalytic efficiencies of the enzymes (Takahashi et al., 2007). In Glycyrrhiza, not only glycyrrhizin-producing species but also non-producing species can produce other oleanane-type triterpenoid saponins (Hayashi et al., 2000). The saponin diversity in Glycyrrhiza spp. could be derived from the variation in homologous P450 species and UGTs in saponin biosynthesis. Evaluation of the differences in these enzymes would be useful to improve their activities.
Although the number of identified genes has increased in the last decade, yet there is no saponin biosynthetic pathway of which all genes encoding the proteins involved in the biosynthetic steps have been identified. In fact only one CYP88D6 has been identified in glycyrrhizin biosynthetic pathway that requires two P450 species and two UGTs. The recent transcriptomic and metabolomic approaches have accelerated the elucidation of plant secondary metabolisms (Ziegler et al., 2006; Hirai et al., 2007; Yonekura-Sakakibara et al., 2007, 2008; Liscombe et al., 2009; Okazaki et al., 2009; Matsuda et al., 2010; Saito and Matsuda, 2010), and some saponin biosynthetic genes were identified by such strategies (Achnine et al., 2005; Seki et al., 2008; Naoumkina et al., 2010). Introduction of the current advanced DNA sequence technology in the omics strategies should enhance gene discovery (Li et al., 2010; Sun et al., 2010). In addition to the efforts to discover the proteins for the lacking biosynthetic steps, understanding the regulatory mechanisms of the expression of biosynthetic genes and the accumulation mechanisms of triterpenoids in plant and microbial hosts should enable further promising application for the production of useful triterpenoids.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN). The authors thank Dr. Hikaru Seki (Osaka University) for his comments on the manuscript.
References
Abe, I., Rohmer, M., and Prestwich, G. D. (1993). Enzymatic cyclization of squalene and oxidosqualene to sterols and triterpenes. Chem. Rev. 93, 2189–2206.
Achnine, L., Huhman, D. V., Farag, M. A., Sumner, L. W., Blount, J. W., and Dixon, R. A. (2005). Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. 41, 875–887.
Ajikumar, P. K., Xiao, W. H., Tyo, K. E. J., Wang, Y., Simeon, F., Leonard, E., Mucha, O., Phon, T. H., Pfeifer, B., and Stephanopoulos, G. (2010). Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330, 70–74.
Alper, H., Miyaoku, K., and Stephanopoulos, G. (2005). Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat. Biotechnol. 23, 612–616.
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394.
Burrows, J. C., Price, K. R., and Fenwick, R. G. (1987). Soyasaponin IV, an additional monodesmosidic saponin isolated from soyabean. Phytochemistry 26, 1214–1215.
Chang, M. C. Y., Eachus, R. A., Trieu, W., Ro, D. K., and Keasling, J. D. (2007). Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat. Chem. Biol. 3, 274–277.
Chappell, J., Wolf, F., Proulx, J., Cuellar, R., and Saunders, C. (1995). Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme A reductase a rate-limiting step for isoprenoid biosynthesis in plants? Plant Physiol. 109, 1337–1343.
Das, A., Yoon, S. H., Lee, S. H., Kim, J. Y., Oh, D. K., and Kim, S. W. (2007). An update on microbial carotenoid production: application of recent metabolic engineering tools. Appl. Microbiol. Biotechnol. 77, 505–512.
Dejong, J. M., Liu, Y., Bollon, A. P., Long, R. M., Jennewein, S., Williams, D., and Croteau, R. B. (2006). Genetic engineering of taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol. Bioeng. 93, 212–224.
Edqvist, J., Rönnberg, E., Rosenquist, S., Blomqvist, K., Viitanen, L., Salminen, T. A., Nylund, M., Tuuf, J., and Mattjus, P. (2004). Plants express a lipid transfer protein with high similarity to mammalian sterol carrier protein-2. J. Biol. Chem. 279, 53544–53553.
Fazio, G. C., Xu, R., and Matsuda, S. P. T. (2004). Genome mining to identify new plant triterpenoids. J. Am. Chem. Soc. 126, 5678–5679.
Field, B., and Osbourn, A. E. (2008). Metabolic diversification-independent assembly of operon-like gene clusters in different plants. Science 320, 543–547.
Fukuda, N., Shan, S., Tanaka, H., and Shoyama, Y. (2006). New staining methodology: Eastern blotting for glycosides in the field of Kampo medicines. J. Nat. Med. 60, 21–27.
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827.
Goossens, A., Häkkinen, S. T., Laakso, I., Seppänen-Laakso, T., Biondi, S., De Sutter, V., Lammertyn, F., Nuutila, A. M., Söderlund, H., Zabeau, M., Inzé, D., and Oksman-Caldentey, K. M. (2003). A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc. Natl. Acad. Sci. U.S.A. 100, 8595–8600.
Han, J. Y., Kwon, Y. S., Yang, D. C., Jung, Y. R., and Choi, Y. E. (2006). Expression and RNA interference-induced silencing of the dammarenediol synthase gene in Panax ginseng. Plant Cell Physiol. 47, 1653–1662.
Haralampidis, K., Bryan, G., Qi, X., Papadopoulou, K., Bakht, S., Melton, R., and Osbourn, A. (2001). A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc. Natl. Acad. Sci. U.S.A. 98, 13431–13436.
Harker, M., Holmberg, N., Clayton, J. C., Gibbard, C. L., Wallace, A. D., Rawlins, S., Hellyer, S. A., Lanot, A., and Safford, R. (2003). Enhancement of seed phytosterol levels by expression of an N-terminal truncated Hevea brasiliensis (rubber tree) 3-hydroxy-3-methylglutaryl-CoA reductase. Plant Biotechnol. J. 1, 113–121.
Hayashi, H., Hosono, N., Kondo, M., Hiraoka, N., Ikeshiro, Y., Shibano, M., Kusano, G., Yamamoto, H., Tanaka, T., and Inoue, K. (2000). Phylogenetic relationship of six Glycyrrhiza species based on rbcL sequences and chemical constituents. Biol. Pharm. Bull. 23, 602–606.
Hayashi, H., Huang, P., Kirakosyan, A., Inoue, K., Hiraoka, N., Ikeshiro, Y., Kushiro, T., Shibuya, M., and Ebizuka, Y. (2001). Cloning and characterization of a cDNA encoding β-amyrin synthase involved in glycyrrhizin and soyasaponin biosyntheses in licorice. Biol. Pharm. Bull. 24, 912–916.
Hayashi, H., Nishiyama, Y., Tomizawa, N., Hiraoka, N., and Ikeshiro, Y. (1996). UDP-glucuronic acid: triterpene glucuronosyltransferase activity in cultured licorice cells. Phytochemistry 42, 665–666.
Hey, S. J., Powers, S. J., Beale, M. H., Hawkins, N. D., Ward, J. L., and Halford, N. G. (2006). Enhanced seed phytosterol accumulation through expression of a modified HMG-CoA reductase. Plant Biotechnol. J. 4, 219–229.
Hirai, M. Y., Sugiyama, K., Sawada, Y., Tohge, T., Obayashi, T., Suzuki, A., Araki, R., Sakurai, N., Suzuki, H., Aoki, K., Goda, H., Nishizawa, O. I., Shibata, D., and Saito, K. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 6478–6483.
Holton, R. A., Somoza, C., Kim, H. B., Liang, F., Biediger, R. J., Boatman, P. D., Shindo, M., Smith, C. C., Kim, S., Nadizadeh, H., Suzuki, Y., Tao, C., Vu, P., Tang, S., Zhang, P., Murthi, K. K., Gentile, L. N., and Liu, J. H. (1994a). First total synthesis of taxol. 1. Functionalization of the B ring. J. Am. Chem. Soc. 116, 1597–1598.
Holton, R. A., Kim, H. B., Somoza, C., Liang, F., Biediger, R. J., Boatman, P. D., Shindo, M., Smith, C. C., Kim, S., Nadizadeh, H., Suzuki, Y., Tao, C., Vu, P., Tang, S., Zhang, P., Murthi, K. K., Gentile, L. N., and Liu, J. H. (1994b). First total synthesis of taxol. 2. Completion of the C and D rings. J. Am. Chem. Soc. 116, 1599–1600.
Huhman, D. V., Berhow, M. A., and Sumner, L. W. (2005). Quantification of saponins in aerial and subterranean tissues of Medicago truncatula. J. Agric. Food Chem. 53, 1914–1920.
Iturbe-Ormaetxe, I., Haralampidis, K., Papadopoulou, K., and Osbourn, A. E. (2003). Molecular cloning and characterization of triterpene synthases from Medicago truncatula and Lotus japonicus. Plant Mol. Biol. 51, 731–743.
Jasiński, M., Stukkens, Y., Degand, H., Purnelle, B., Marchand-Brynaert, J., and Boutry, M. (2001). A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 13, 1095–1107.
Kahn, R. A., and Durst, F. (2000). Function and evolution of plant cytochrome P450. Recent Adv. Phytochem. 34, 151–189.
Kesselmeier, J., and Urban, B. (1983). Subcellular localization of saponins in green and etiolated leaves and green protoplasts of oat (Avena sativa L.). Protoplasma 114, 133–140
Kikuchi, A., Tsukamoto, C., Tabuchi, K., Adachi, T., and Okubo, K. (1999). Inheritance and characterization of a null allele for group A acetyl saponins found in a mutant soybean (Glycine max (L.) Merrill). Breed. Sci. 49, 167–171.
Kim, J. H., Kim, S. W., Nguyen, D. Q. A., Li, H., Kim, S. B., Seo, Y. G., Yang, J. K., Chung, I. Y., Kim, D. H., and Kim, C. J. (2009). Production of β-carotene by recombinant Escherichia coli with engineered whole mevalonate pathway in batch and fed-batch cultures. Biotechnol. Bioprocess Eng. 14, 559–564.
Kim, O. T., Kim, S. H., Ohyama, K., Muranaka, T., Choi, Y. E., Lee, H. Y., Kim, M. Y., and Hwang, B. (2010). Upregulation of phytosterol and triterpene biosynthesis in Centella asiatica hairy roots overexpressed ginseng farnesyl diphosphate synthase. Plant Cell Rep. 29, 403–411.
Kingston, D. G. I. (2007). The shape of the things to come: structural and synthetic studies of Taxol and related compounds. Phytochemistry 68, 1844–1854.
Kirby, J., Romanini, D. W., Paradise, E. M., and Keasling, J. D. (2008). Engineering triterpene production in Saccharomyces cerevisiae β-amyrin synthase from Artemisia annua. FEBS J. 275, 1852–1859.
Kitagawa, I., Taniyama, T., Nagahama, Y., Okubo, K., Yamauchi, F., and Yoshikawa, M. (1988). Saponin and sapogenol. XLII. Structures and acetyl-soyasaponins A1, A2, and A3, astringent partially acetylated bisdesmosides of soyasapogenol A, from American soybean, the seeds of Glycine max Merrill. Chem. Pharm. Bull. 36, 2819–2828.
Kitagawa, I., Yoshikawa, M., Wang, H. K., Saito, M., Tosirisuk, V., Fujiwara, T., and Tomita, T. (1982). Revised structures of soyasapogenols A, B and E, oleanene-sapogenols from soybean. Structures of soyasaponins I, II and III. Chem. Pharm. Bull. 30, 2294–2297.
Kudou, S., Tonomura, M., Tsukamoto, C., Shimoyamada, M., Uchida, T., and Okubo, K. (1992). Isolation and structural elucidation of the major genuine soybean saponin. Biosci. Biotechnol. Biochem. 56, 142–143.
Kudou, S., Tonomura, M., Tsukamoto, C., Uchida, T., Sakabe, T., Tamura, N., and Okubo, K. (1993). Isolation and structural elucidation of DDMP-conjugated soyasaponins as genuine saponins from soybean seeds. Biosci. Biotechnol. Biochem. 57, 546–550.
Kurosawa, Y., Takahara, H., and Shiraiwa, M. (2002). UDP-glucuronic acid:soyasapogenol glucuronosyltransferase involved in saponin biosynthesis in germinating soybean seeds. Planta 215, 620–629.
Kushiro, T., and Ebizuka, Y. (2010). “Triterpenes,” in Comprehensive Natural Products II: Chemistry and Biology 1, eds L. Mander and H. W. B. Liu (Oxford: Elsevier), 673–708.
Kushiro, T., Shibuya, M., and Ebizuka, Y. (1998a). β-Amyrin synthase—Cloning of oxidosqualene cyclase that catalyzes the formation of the most popular triterpene among higher plants. Eur. J. Biochem. 256, 238–244.
Kushiro, T., Shibuya, M., and Ebizuka, Y. (1998b). “Molecular cloning of oxidosqualene cyclase cDNA from Panax ginseng: the isogene that encodes β-amyrin synthase,” in Towards Natural Medicine Research in the 21st Century, Excerpta Medica International Congress Series 1157, eds H. Ageta, N. Aimi, Y. Ebizuka, T. Fujita, and G. Honda (Amsterdam: Elsevier), 421–428.
Kuzina, V., Ekstrøm, C. T., Andersen, S. B., Nielsen, J. K., Olsen, C. E., and Bak, S. (2009). Identification of defense compounds in Barbarea vulgaris against the herbivore Phyllotreta nemorum by an ecometabolomic approach. Plant Physiol. 151, 1977–1990.
Lee, M. H., Jeong, J. H., Seo, J. W., Shin, C. G., Kim, Y. S., In, J. G., Yang, D. C., Yi, J. S., and Choi, Y. E. (2004). Enhanced triterpene and phytosterol biosynthesis in Panax ginseng overexpressing squalene synthase gene. Plant Cell Physiol. 45, 976–984.
Lenihan, J. R., Tsuruta, H., Diola, D., Renninger, N. S., and Regentin, R. (2008). Developing an industrial artemisinic acid fermentation process to support the cost-effective production of antimalarial artemisinin-based combination therapies. Biotechnol. Prog. 24, 1026–1032.
Li, Y., Luo, H. M., Sun, C., Song, J. Y., Sun, Y. Z., Wu, Q., Wang, N., Yao, H., Steinmetz, A., and Chen, S. L. (2010). EST analysis reveals putative genes involved in glycyrrhizin biosynthesis. BMC Genomics 11, 268. doi: 10.1186/1471-2164-11-268
Liscombe, D. K., Ziegler, J., Schmidt, J., Ammer, C., and Facchini, P. J. (2009). Targeted metabolite and transcript profiling for elucidating enzyme function: isolation of novel N-methyltransferases from three benzylisoquinoline alkaloid-producing species. Plant J. 60, 729–743.
Lu, H. Y., Liu, J. M., Zhang, H. C., Yin, T., and Gao, S. L. (2008). Ri-mediated transformation of Glycyrrhiza uralensis with a squalene synthase gene (GuSQS1) for production of glycyrrhizin. Plant Mol. Biol. Rep. 26, 1–11.
Luo, J., Nishiyama, Y., Fuell, C., Taguchi, G., Elliott, K., Hill, L., Tanaka, Y., Kitayama, M., Yamazaki, M., Bailey, P., Parr, A., Michael, A. J., Saito, K., and Martin, C. (2007). Convergent evolution in the BAHD family of acyl transferases; identification and characterisation of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 50, 678–695.
Manzano, D., Fernández-Busquets, X., Schaller, H., González, V., Boronat, A., Arró, M., and Ferrer, A. (2004). The metabolic imbalance underlying lesion formation in Arabidopsis thaliana overexpressing farnesyl diphosphate synthase (isoform 1S) leads to oxidative stress and is triggered by the developmental decline of endogenous HMGR activity. Planta 219, 982–989.
Martin, V. J., Pitera, D. J., Withers, S. T., Newman, J. D., and Keasling, J. D. (2003). Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21, 796–802.
Masferrer, A., Arró, M., Manzano, D., Schaller, H., Fernández-Busquets, X., Moncaleán, P., Fernández, B., Cunillera, N., Boronat, A., and Ferrer, A. (2002). Overexpression of Arabidopsis thaliana farnesyl diphosphate synthase (FPS1S) in transgenic Arabidopsis induces a cell death/senescence-like response and reduced cytokinin levels. Plant J. 30, 123–132.
Matsuda, F., Hirai, M. Y., Sasaki, E., Akiyama, K., Yonekura-Sakakibara, K., Provart, N. J., Sakurai, T., Shimada, Y., and Saito, K. (2010). AtMetExpress development: a phytochemical atlas of Arabidopsis thaliana development. Plant Physiol. 152, 566–578.
Maxwell, C. A., Restrepo-Hartwig, M. A., Hession, A. O., and McGonigle, B. (2004). “Metabolic engineering of soybean for improved flavor and health benefits,” in Recent Advances in Phytochemistry 38, ed. J. T. Romeo (Amsterdam: Elsevier), 153–176.
Meesapyodsuk, D., Balsevich, J., Reed, D. W., and Covello, P. S. (2007). Saponin biosynthesis in Saponaria vaccaria. cDNAs encoding β-amyrin synthase and a triterpene carboxylic acid glucosyltransferase. Plant Physiol. 143, 959–969.
Misawa, N. (2010). “Carotenoids,” in Comprehensive Natural Products II: Chemistry and Biology 1, eds. L. Mander and H. W. B. Liu (Oxford: Elsevier), 733–753.
Morita, M., Shitan, N., Sawada, K., Van Montagu, M. C., Inzé, D., Rischer, H., Goossens, A., Oksman-Caldentey, K. M., Moriyama, Y., and Yazaki, K. (2009). Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc. Natl. Acad. Sci. U.S.A. 106, 2447–2452.
Mugford, S. T., Qi, X., Bakht, S., Hill, L., Wegel, E., Hughes, R. K., Papadopoulou, K., Melton, R., Philo, M., Sainsbury, F., Lomonossoff, G. P., Roy, A. D., Goss, R. J., and Osbourn, A. (2009). A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell 21, 2473–2484.
Muñoz-Bertomeu, J., Sales, E., Ros, R., Arrillaga, I., and Segura, J. (2007). Up-regulation of an N-terminal truncated 3-hydroxy-3-methylglutaryl CoA reductase enhances production of essential oils and sterols in transgenic Lavandula latifolia. Plant Biotechnol. J. 5, 746–758.
Mylona, P., Owatworakit, A., Papadopoulou, K., Jenner, H., Qin, B., Findlay, K., Hill, L., Qi, X., Bakht, S., Melton, R., and Osbourn, A. (2008). Sad3 and Sad4 are required for saponin biosynthesis and root development in oat. Plant Cell 20, 201–212.
Naoumkina, M. A., Modolo, L. V., Huhman, D. V., Urbanczyk-Wochniak, E., Tang, Y., Sumner, L. W., and Dixon, R. A. (2010). Genomic and coexpression analyses predict multiple genes involved in triterpene saponin biosynthesis in Medicago truncatula. Plant Cell 22, 850–866.
Nicolaou, K. C., Yang, Z., Liu, J. J., Ueno, H., Nantermet, P. G., Guy, R. K., Claiborne, C. F., Renaund, J., Couladouros, E. A., Paulvannan, K., and Sorensen, E. J. (1994). Total synthesis of taxol. Nature 367, 630–634.
Ohyama, K., Suzuki, M., Kikuchi, J., Saito, K., and Muranaka, T. (2009). Dual biosynthetic pathways to phytosterol via cycloartenol and lanosterol in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 725–730.
Okazaki, Y., Shimojima, M., Sawada, Y., Toyooka, K., Narisawa, T., Mochida, K., Tanaka, H., Matsuda, F., Hirai, A., Hirai, M. Y., Ohta, H., and Saito, K. (2009). Chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 21, 892–909.
Okubo, K., Iijima, M., Kobayashi, Y., Yoshikoshi, M., Uchida, T., and Kudou, S. (1992). Components responsible for the undesirable taste of soybean seeds. Biosci. Biotech. Biochem. 56, 99–103.
Osbourn, A. E. (1996). Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell 8, 1821–1831.
Osbourn, A. (2010). Gene clusters for secondary metabolic pathways: an emerging theme in plant biology. Plant Physiol. 154, 531–535.
Paine, J. A., Shipton, C. A., Chaggar, S., Howells, R. M., Kennedy, M. J., Vernon, G., Wright, S. Y., Hinchliffe, E., Adams, J. L., Silverstone, A. L., and Drake, R. (2005). Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 23, 482–487.
Papadopoulou, K., Melton, R., Leggett, M., Daniels, M. J., and Osbourn, A. E. (1999). Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. U.S.A. 96, 12923–12928.
Paquette, S., Møller, B. L., and Bak, S. (2003). On the origin of family 1 plant glycosyltransferases. Phytochemistry 62, 399–413.
Qi, X., Bakht, S., Leggett, M., Maxwell, C., Melton, R., and Osbourn, A. (2004). A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. U.S.A. 101, 8233–8238.
Qi, X., Bakht, S., Qin, B., Leggett, M., Hemmings, A., Mellon, F., Eagles, J., Werck-Reichhart, D., Schaller, H., Lesot, A., Melton, R., and Osbourn, A. (2006). A different function for a member of an ancient and highly conserved cytochrome P450 family: from essential sterols to plant defense. Proc. Natl. Acad. Sci. U.S.A. 103, 18848–18853.
Ro, D. K., Paradise, E. M., Ouellet, M., Fisher, K. J., Newman, K. L., Ndungu, J. M., Ho, K. A., Eachus, R. A., Ham, T. S., Kirby, J., Chang, M. C., Withers, S. T., Shiba, Y., Sarpong, R., and Keasling, J. D. (2006). Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943.
Roth, R. J., and Acton, N. (1989). A simple conversion of artemisinic acid into artemisinin. J. Nat. Prod. 52, 1183–1185.
Saito, K., and Matsuda, F. (2010). Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 61, 463–489.
Sawada, Y., Toyooka, K., Kuwahara, A., Sakata, A., Nagano, M., Saito, K., and Hirai, M. Y. (2009). Arabidopsis bile acid:sodium symporter family protein 5 is involved in methionine-derived glucosinolate biosynthesis. Plant Cell Physiol. 50, 1579–1586.
Schaller, H., Grausem, B., Benveniste, P., Chye, M. L., Tan, C. T., Song, Y. H., and Chua, N. H. (1995). Expression of the Hevea brasiliensis (H. B. K.) Müll. Arg. 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 in tobacco results in sterol overproduction. Plant Physiol. 109, 761–770.
Seki, H., Ohyama, K., Sawai, S., Mizutani, M., Ohnishi, T., Sudo, H., Akashi, T., Aoki, T., Saito, K., and Muranaka, T. (2008). Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc. Natl. Acad. Sci. U.S.A. 105, 14204–14209.
Seo, J. W., Jeong, J. H., Shin, C. G., Lo, S. C., Han, S. S., Yu, K. W., Harada, E., Han, J. Y., and Choi, Y. E. (2005). Overexpression of squalene synthase in Eleutherococcus senticosus increases phytosterol and triterpene accumulation. Phytochemistry 66, 869–877.
Shan, S., Tanaka, H., and Shoyama, Y. (2001). Enzyme-linked immunosorbent assay for glycyrrhizin using anti-glycyrrhizin monoclonal antibody and an eastern blotting technique for glucuronides of glycyrrhetic acid. Anal. Chem. 73, 5784–5790.
Shao, H., He, X., Achnine, L., Blount, J. W., Dixon, R. A., and Wang, X. (2005). Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17, 3141–3154.
Shibata, S. (2000). A drug over the millennia: pharmacognosy, chemistry, and pharmacology of licorice. Yakugaku Zasshi 120, 849–862.
Shibata, S. (2001). Chemistry and cancer preventing activities of ginseng saponins and some related triterpenoid compounds. J. Korean Med. Sci. 16, S28–S37.
Shibuya, M., Hoshino, M., Katsube, Y., Hayashi, H., Kushiro, T., and Ebizuka, Y. (2006). Identification of β-amyrin and sophoradiol 24-hydroxylase by expressed sequence tag mining and functional expression assay. FEBS J. 273, 948–959.
Shibuya, M., Nishimura, K., Yasuyama, N., and Ebizuka, Y. (2010). Identification and characterization of glycosyltransferases involved in the biosynthesis of soyasaponin I in Glycine max. FEBS Lett. 584, 2258–2264.
Shim, J. S., Lee, O. R., Kim, Y. J., Lee, J. H., Kim, J. H., Jung, D. Y., In, J. G., Lee, B. S., and Yang, D. C. (2010). Overexpression of PgSQS1 increases ginsenoside production and negatively affects ginseng growth rate in Panax ginseng. J. Ginseng Res. 34, 98–103.
Shimoyamada, M., Kudo, S., Okubo, K., Yamauchi, F., and Harada, K. (1990). Distribution of saponin constituents in some varieties of soybean plant. Agric. Biol. Chem. 54, 77–81.
Shiraiwa, M., Kudo, S., Shimoyamada, M., Harada, K., and Okubo, K. (1991a). Composition and structure of “group A saponin” in soybean seed. Agric. Biol. Chem. 55, 315–322.
Shiraiwa, M., Harada, K., and Okubo, K. (1991b). Composition and structure of “group B saponin” in soybean seed. Agric. Biol. Chem. 55, 911–917.
Sudo, H., Seki, H., Sakurai, N., Suzuki, H., Shibata, D., Toyoda, A., Totoki, Y., Sakaki, Y., Iida, O., Shibata, T., Kojoma, M., Muranaka, T., and Saito, K. (2009). Expressed sequence tags from rhizomes of Glycyrrhiza uralensis. Plant Biotechnol. 26, 105–107.
Sun, C., Li, Y., Wu, Q., Luo, H., Sun, Y., Song, J., Lui, E. M., and Chen, S. (2010). De novo sequencing and analysis of the American ginseng root transcriptome using a GS FLX Titanium platform to discover putative genes involved in ginsenoside biosynthesis. BMC Genomics 11, 262. doi: 10.1186/1471-2164-11-262
Suzuki, H., Achnine, L., Xu, R., Matsuda, S. P. T., and Dixon, R. A. (2002). A genomics approach to the early stages of triterpene saponin biosynthesis in Medicago truncatula. Plant J. 32, 1033–1048.
Suzuki, M., Xiang, T., Ohyama, K., Seki, H., Saito, K., Muranaka, T., Hayashi, H., Katsube, Y., Kushiro, T., Shibuya, M., and Ebizuka, Y. (2006). Lanosterol synthase in dicotyledonous plants. Plant Cell Physiol. 47, 565–571.
Szakiel, A., Peęczkowski, C., and Henry, M. (2011). Influence of environmental abiotic factors on the content of saponins in plants. Phytochem. Rev. doi: 10.1007/s11101-010-9177-x
Takada, Y., Sayama, T., Kikuchi, A., Kato, S., Ttsuzaki, N., Nakamoto, Y., Suzuki, A., Tsukamoto, C., and Ishimoto, M. (2010). Genetic analysis of variation in sugar chain composition at the C-22 position of group A saponins in soybean, Glycine max (L.) Merrill. Breed. Sci. 60, 3–8.
Takahashi, S., Yeo, Y. S., Zhao, Y., O’Maille, P. E., Greenhagen, B. T., Noel, J. P., Coates, R. M., and Chappell, J. (2007). Functional characterization of premnaspirodiene oxygenase, a cytochrome P450 catalyzing regio- and stereo-specific hydroxylations of diverse sesquiterpene substrates. J. Biol. Chem. 282, 31744–31754.
Taniyama, T., Nagahama, Y., Yoshikawa, M., and Kitagawa, I. (1988). Saponin and sapogenol. XLIII. Acetyl-soyasaponins A4, A5, and A6, new astringent bisdesmosides of soyasapogenol A, from Japanese soybean, the seeds of Glycine max Merrill. Chem. Pharm. Bull. 36, 2829–2839.
Tansakul, P., Shibuya, M., Kushiro, T., and Ebizuka, Y. (2006). Dammarenediol-II synthase, the first dedicated enzyme for ginsenoside biosynthesis, in Panax ginseng. FEBS Lett. 580, 5143–5149.
Thoma, R., Schulz-Gasch, T., D’Arcy, B., Benz, J., Aebi, J., Dehmlow, H., Hennig, M., Stihle, M., and Ruf, A. (2004). Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature 432, 118–122.
Tohge, T., Nishiyama, Y., Hirai, M. Y., Yano, M., Nakajima, J.-I., Awazuhara, M., Inoue, E., Takahashi, H., Goodenowe, D. B., Kitayama, M., Noji, M., Yamazaki, M., and Saito, K. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42, 218–235.
Tsukamoto, C., Kikuchi, A., Harada, K., Kitamura, K., and Okubo, K. (1993). Genetic and chemical polymorphisms of saponins in soybean seed. Phytochemistry 34, 1351–1356.
Tsuruta, H., Paddon, C. J., Eng, D., Lenihan, J. R., Horning, T., Anthony, L. C., Regentin, R., Keasling, J. D., Renninger, N. S., and Newman, J. D. (2009). High-level production of amorpha-4,11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PLoS ONE 4, e4489. doi: 10.1371/journal.pone.0004489
Vincken, J. P., Heng, L., de Groot, A., and Gruppen, H. (2007). Saponins, classification and occurrence in the plant kingdom. Phytochemistry 68, 275–297.
Wani, M. C., Taylor, H. L., Wall, M. E., Coggon, P., and McPhail, A. T. (1971). Plant antitumor agents. VI. Isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J. Am. Chem. Soc. 93, 2325–2327.
Warnecke, D. C., Baltrusch, M., Buck, F., Wolter, F. P., and Heinz, E. (1997). UDP-glucose:sterol glucosyltransferase: cloning and functional expression in Escherichia coli. Plant Mol. Biol. 35, 597–603.
Xu, R., Fazio, G. C., and Matsuda, S. P. T. (2004). On the origins of triterpenoid skeletal diversity. Phytochemistry 65, 261–291.
Ye, X., Al-Babili, S., Klöti, A., Zhang, J., Lucca, P., Beyer, P., and Potrykus, I. (2000). Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287, 303–305.
Yonekura-Sakakibara, K., Tohge, T., Matsuda, F., Nakabayashi, R., Takayama, H., Niida, R., Takahashi, A. W., Inoue, E., and Saito, K. (2008). Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20, 2160–2176.
Yonekura-Sakakibara, K., Tohge, T., Niida, R., and Saito, K. (2007). Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression and reverse genetics. J. Biol. Chem. 282, 14932–14941.
Yoon, S. H., Lee, S. H., Das, A., Ryu, H. K., Jang, H. J., Kim, J. Y., Oh, D. K., Keasling, J. D., and Kim, S. W. (2009). Combinatorial expression of bacterial whole mevalonate pathway for the production of β-carotene in E. coli. J. Biotechnol. 140, 218–226.
Keywords: saponin, terpenoid, biotechnological production, P450, UGT
Citation: Sawai S and Saito K (2011) Triterpenoid biosynthesis and engineering in plants. Front. Plant Sci. 2:25. doi: 10.3389/fpls.2011.00025
Received: 22 March 2011;
Paper pending published: 20 April 2011;
Accepted: 16 June 2011;
Published online: 30 June 2011.
Edited by:
Frikkie C. Botha, BSES Limited, AustraliaReviewed by:
Kevin Davies, New Zealand Institute for Plant and Food Research, New ZealandLloyd W. Sumner, The Samuel Roberts Noble Foundation, USA
Copyright: © 2011 Sawai and Saito. This is an open-access article subject to an exclusive license agreement between the authors and Frontiers Media SA, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Kazuki Saito, RIKEN Plant Science Center, Suehiro-cho 1-7-22, Tsurumi-ku, Yokohama, Kanagawa 230-0045, Japan. e-mail:a3NhaXRvQHBzYy5yaWtlbi5qcA==

