
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Physiol. , 23 January 2025
Sec. Molecular and Cellular Biology
Volume 2 - 2024 | https://doi.org/10.3389/fphgy.2024.1507833
Anthocyanins are polyphenolic compounds with antioxidant capacity, free radical scavenging power, and signaling activities in animal pathogenesis-associated pathways, thus playing an important role as nutraceuticals. Tomato fruits do not usually contain anthocyanins because their biosynthesis is switched off in these organs, but anthocyanin-enriched purple tomatoes have been produced in recent years. The varieties obtained by breeding express a functional copy of the R2R3-MYB transcription factor AN2-like, necessary to start the biosynthetic pathway, and do not produce a functional MYB-ATV repressor. The combination of these traits allows the accumulation of anthocyanins in tomatoes, strengthened under specific environmental factors such as high light intensity or low temperatures. Light starts anthocyanin synthesis and gradually extends its distribution on the fruit exocarp. The analyses carried out in the present study indicate that anthocyanin biosynthesis triggered by light is under HY5 control. However, the process is not active in mesocarp for the absence of the bHLH factor AN1, necessary to produce the MBW complex inducing the late enzymes of the biosynthetic pathway, as a consequence of insufficient expression of the R2R3-MYB gene AN2-like. This occurs since light cannot be perceived in the tissues underneath the skin because of the solar shield produced by the anthocyanins accumulated in the exocarp and for the activation of regulatory loops controlling HY5 levels. This is shown by the expression of genes involved in the production of photoreceptors and in the light signaling chain operating upstream of the anthocyanin pathway and responsible for its activation.
In recent years, increased or de novo anthocyanin accumulation in fruits and vegetables have been pursued to enrich their nutraceutical value (Martin et al., 2011; Mattoo et al., 2022). Tomato (Solanum lycopersicum L.) has been improved in some nutritional traits (Raiola et al., 2014), and the establishment of the anthocyanin biosynthetic pathway in fruits is one of them (Butelli et al., 2008; Gonzali et al., 2009).
Anthocyanins are soluble polyphenols belonging to the class of flavonoids, representing the glycosylated forms of the corresponding anthocyanidins. They are characterized by different colors, from orange/red to purple/blue, and are involved in multiple functions, from the pigmentation of flowers and fruits to the protection of plants from biotic and abiotic stresses (Landi et al., 2015; Alappat and Alappat, 2020). In tomato fruits, their presence confers a dark purple color due to the prevalence of anthocyanidins belonging to the delphinidin class (Tohge et al., 2015; Blando et al., 2019). In leaves, the major role of anthocyanins is carried out in epidermal and subepidermal cell layers, where they act as a screen to filter solar radiation harmful for the photosynthetic apparatus, particularly under conditions of high irradiance and low temperatures, that can produce free radical species and photoinhibition (Gould, 2004). In fruits, their presence is often a marker of ripening to attract seed dispersers and is controlled by developmental programs (Jaakola, 2013). However, the synthesis of anthocyanins in fruits may also be induced by light and other environmental factors to better modulate their quantity and/or quality (Jaakola, 2013; Zoratti et al., 2014). The biosynthetic process starts from the aromatic amino acid phenylalanine and proceeds with a concerted series of enzymatic reactions whose early steps are in common with other flavonoids (Winkel-Shirley, 2001). The “structural” genes encoding the enzymes of the pathway are conserved in plants (Sunil and Shetty, 2022); more variable is their regulation, being the pathway controlled by distinct factors in different organs or tissues.
Light may play a primary role in the induction of anthocyanin synthesis, and its quality is particularly important in determining the quantity of anthocyanins produced and their nature (Ma et al., 2021). Among the radiations of the solar spectrum, UV and blue light mostly affect these processes, but red light can induce anthocyanin synthesis as well (Costa Galvão and Fankhauser, 2015; Podolec and Ulm, 2018; Rai et al., 2021). For this reason, multiple photoreceptors may play important roles as mediators between light and pigment production (Zoratti et al., 2014). Higher plants perceive surrounding solar radiations through different photoreceptors, including UV-B Resistance 8 (UVR8) for UV-B/UV-A light (Rai et al., 2021), cryptochromes for UV-A/blue light, and phytochromes for red/far-red light (Costa Galvão and Fankhauser, 2015; Podolec and Ulm, 2018). Downstream, a complex signaling mechanism takes place, with the basic leucine zipper transcription factor (TF) ELONGATED HYPOCOTYL 5 (HY5) playing the role of the master switching molecule (Gangappa and Botto, 2016).
HY5 constitutes the center of a transcriptional network hub regulating the expression of hundreds of different light-responsive genes in plants, including anthocyanin regulatory and biosynthetic genes (Lee et al., 2007; Shin et al., 2013). With HY5 being destabilized in dark conditions through the proteasome machinery activated by the CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1)/SUPPRESSOR OF PHYTOCHROME A-105 (SPA) ubiquitin ligase complex (Park et al., 2017; Bian et al., 2022; Shin et al., 2007), anthocyanin synthesis can be switched off in absence of light (Albert et al., 2009; Kim et al., 2017). Also, shaded conditions and high temperatures may destabilize HY5 through the COP1/SPA complex (Sheerin and Hiltbrunner, 2017; Nieto et al., 2022), leading to reduction or suppression of anthocyanin production (Lin-Wang et al., 2011). Arabidopsis cop1 mutant seedlings can accumulate exaggerated quantities of anthocyanins in cotyledons, showing the so-called “FUSCA” phenotype, due to the stabilization of HY5 (Castle’ and Meinke, 1994; Han et al., 2020). COP10, DEETIOLATED 1 (DET1), and UV-damaged DNA-binding protein 1a (DDB1a) constitute the CDD complex, which physically interacts with COP1/SPA and the COP9 signalosome, allowing their activity (Yanagawa et al., 2004; Lau and Deng, 2012; Cañibano et al., 2021). DET1 is a chromatin-associated protein acting as a transcriptional repressor in dark conditions; its mutation, similarly to COP1 mutation, results in a stabilization of HY5 either in light or dark, with the consequent ectopic activation of HY5 targets (Cañibano et al., 2021). In tomato fruit, HY5 regulates ripening at both transcriptional and translational levels because many genes involved in carotenoid and flavonoid biosynthesis and in ethylene signaling are HY5 targets (Wang et al., 2021). Consequently, due to stabilization of HY5, DET1 mutants, known as high pigment 2 (hp2) in tomato, show enhanced phenylpropanoids, flavonoids, and carotenoids in their fruits (Mustilli et al., 1999).
The last decades have seen a significant advance in understanding the regulatory mechanisms underlying the anthocyanin production, in particular the role of the R2R3-MYB TFs which activate the early biosynthetic genes (EBGs) and of the “MBW” multiprotein complexes which mainly act on the late biosynthetic genes (LBGs) (Albert et al., 2014; Xu et al., 2015; Lloyd et al., 2017). The MBW complex is composed of R2R3-MYB, bHLH, and WDR factors, with the R2R3-MYB mainly conferring transcriptional specificity on the genomic targets via interaction with the MYB recognition elements (MREs) in their promoters (Albert et al., 2014; Xu et al., 2015). In tomato, the main R2R3-MYB protein activating the EBGs is MYB12, whereas the MBW complexes, which act in a hierarchical way (Montefiori et al., 2015), are composed by the R2R3-MYB AN2 or AN2-like (acting in vegetative tissues and fruits, respectively), the bHLH factors JAF13 and AN1, and the WDR protein AN11 (Kiferle et al., 2015; Qiu et al., 2016; Gao et al., 2018; Colanero et al., 2020; Sun et al., 2020). Repressor R2R3-MYB and R3-MYB factors can act independently or via interaction with the MBW complex to destabilize its transcriptional activations and thus inhibit or decrease the anthocyanin production (LaFountain and Yuan, 2021). Among the tomato R2R3-MYB repressors, THM27 (also known as MYB32), MYB76, and MYB72 have been recently found to be involved in inhibition of both EBGs and LBGs, even if with different specificities (Menconi et al., 2023; Suprun et al., 2023; Wu et al., 2020). MYB-ATV is a tomato CPC-type R3-MYB repressor which can sequester the bHLH proteins from the MBW complex and is transcriptionally induced by the same complex, triggering in this way a feedback repression mechanism (Colanero et al., 2018). Other tomato R3-MYB repressors are MYB-ATV-like and TRIPTYCHON (TRY) (Nukumizu et al., 2013; Cao et al., 2017). The regulation of the anthocyanin pathway is further affected by other proteins, acting under environmental or developmental control (e.g., WRKYs, SPLs, DELLAs, JAZs), which can bind the MBW complexes with positive or negative effects on their final activity (Xu et al., 2015; Lloyd et al., 2017). In anthocyanin-enriched tomato fruits, several other TFs have also been identified in recent years as positively correlated under light with anthocyanin pigmentation, some of them acting independently of HY5 (Qiu et al., 2019), and others under hormonal control (You et al., 2024).
The introgression in S. lycopersicum of the Anthocyanin fruit (Aft) or the Aubergine (Abg) alleles from Solanum chilense (Georgiev, 1972) or Solanum lycopersicoides (Rick et al., 1994), respectively, allows the activation of the synthesis of spotted anthocyanin pigmentation in the fruit exocarp (Mes et al., 2008; Menconi et al., 2024). This revealed the criticality of the fruit-specific R2R3-MYB encoding gene AN2-like, of which both Aft and Abg are functional alleles, whereas the S. lycopersicum sequence bears a splicing mutation which produces a non-functional TF (Colanero et al., 2020; Sun et al., 2020; Menconi et al., 2023, 2024). The locus atroviolacea (atv), introgressed from Solanum cheesmaniae (Rick, 1964), revealed the existence of MYB-ATV, which, when mutated, leads to derepressed anthocyanin accumulation either in fruits or in vegetative tissues (Cao et al., 2017; Colanero et al., 2018). The concomitant presence of Aft or Abg and atv leads under light to biosynthesis and enhanced accumulation of anthocyanins in the epidermal and subepidermal cell tissues of pericarp from the early stages of fruit ripening (Mes et al., 2008; Povero et al., 2011; Sun et al., 2020; Menconi et al., 2024). This generally does not occur in mesocarp, which remains green for the presence of chlorophylls in early stages and becomes red in ripe fruits when carotenoids accumulate (Mes et al., 2008; Povero et al., 2011; Menconi et al., 2024). However, the expression of a functional copy of AN2-like under the E8 fruit promoter led to tomatoes with strong anthocyanin pigmentation in all the fruit (Sun et al., 2020). The same occurred through overexpression of R2R3-MYB paralogs of AN2-like, such as AN2 (Kiferle et al., 2015; Jian et al., 2019). This suggests that anthocyanin biosynthesis is a cell autonomous process in tomato fruit and requires local presence of a suitable R2R3-MYB factor able to interact with JAF13 and AN11, respectively, the bHLH and WDR partners of the first MBW complex (MBW1), which are expressed independently of anthocyanins (Povero et al., 2011; Kiferle et al., 2015; Gao et al., 2018), to activate transcription of the bHLH gene AN1 (Montefiori et al., 2015), whose protein replaces JAF13 producing the second MBW complex (MBW2), which finally activates the expression of the LBGs.
The objective of the present study is to describe in detail the transcriptional activities, from the light stimulus downward, which in purple tomato fruit allow and not allow, respectively, anthocyanin synthesis in exocarp and internal tissues under light, by analyzing well-known regulators of the pathway as well as novel TFs recently hypothesized to be involved in the process, to understand which are the most critical steps and how such drawbacks may be overcome.
The tomato genotypes Aft/Aft × atv/atv (Aft/atv) and Aft/Aft × atv/atv × hp2/hp2 (Aft/atv/hp2) in the MicroTom background were used. The seeds of the line Aft/atv/hp2, containing the allele dark green of the DET1 gene (Levin et al., 2003), were donated by Prof. Peres (Sestari et al., 2014). The seeds of Aft/atv were obtained by backcrossing Aft/atv/hp2 with the cv. MicroTom and selecting for the Aft/atv genotype in the segregating F2 and F3 generations. Seeds were germinated in rock-wool plugs (Grodan) soaked in a nutritive solution (Kiferle et al., 2015). 2-week-old seedlings were transplanted in pots containing a 70:30 soil (HAWITA-Flor)/expanded clay mixture and placed in a growth chamber with 23°/20°C of day/night temperature, 12-h photoperiod, 150 µmol photons m−2 s−1, and 40% relative humidity. For both anthocyanin quantification and qPCR analysis, fruits were sampled at mature green or mature red stages with exocarp (containing also the cuticle layer) removed from the stem end and the inner parts, constituted by mesocarp and endocarp, collected without placenta and seeds. Biological replicates corresponded to fruits collected from independent plants at the same developmental stage, similar position within the plant and similar light exposition, to reduce the pigmentation variability as much as possible. The material was frozen in liquid nitrogen and stored at −80°C until use.
Anthocyanin extraction was performed starting from 50 mg of fruit material (exocarp or mesocarp + endocarp). Fruit samples were ground in 300 µl of HCl 1% (v/v) in methanol and incubated with gentle agitation overnight at 4°C. Extracts were recovered, and 200 µl of distilled water was added. 1 volume of chloroform was then added to remove chlorophylls through mixing and centrifugation (1 min at 14,000 × g). The aqueous phase containing anthocyanins was recovered, 600 µl of HCl 1% (v/v) in methanol was added, and absorption was determined spectrophotometrically. Relative anthocyanin concentrations were calculated as a difference between the absorbance read at 530 nm and 657 nm (Neff and Chory, 1998) and finally expressed as microgram petunidin-3-(p-coumaroyl rutinoside)-5-glucoside gram−1 fresh weight. Mean values were obtained from three independent replicates consisting of fruit material collected from different plants (one fruit per plant).
Total RNA was extracted from fruit exocarp or mesocarp + endocarp with the “Spectrum™ Plant Total RNA Kit” (Merck). The quality of RNA was assessed through electrophoresis on 1% agarose gels, and the quantity was measured through a μDrop™ plate on a Multiskan microplate reader (Thermo Scientific). One microgram of RNA was subjected to DNase treatment and then reverse transcribed into cDNA using the “Maxima First Strand cDNA Synthesis Kit for RT-qPCR, with dsDNase” (Thermo Fisher Scientific) and subsequently diluted with nuclease-free water (Merck) to a concentration of 5 ng/µl. Quantitative RT-PCR (qPCR) was performed with an ABI Prism 7300 Sequence Detection System (Thermo Fisher Scientific). qPCR reactions were carried out using the “PowerUp™ SYBR® Green Master Mix” (Thermo Fisher Scientific), 15 ng of cDNA template, and 300 nM forward and reverse primers (listed in Supplementary Table S1), in a final reaction volume of 10 µl. No-template control reactions were performed for each pair of primers. Amplicon dissociation curves were recorded to confirm the gene specific amplification by a single dominant peak. Elongation Factor 1-alpha (EF1A) and Abscisic Acid Stress Ripening 1 (ASR1) (Bovy et al., 2002) were used as reference genes. The relative quantitation of each individual gene expression was performed using the geometric averaging method (geNorm) (Vandesompele et al., 2002). The relative expression level was calculated as ratio between quantity of target gene and quantity of the geometric averaging of the reference genes. Values are means of four biological replicates with two technical replicates for each gene.
The analysis of the cis-acting regulatory elements contained in the 3-kb genomic region upstream of the ATG first codon of the CDS of the anthocyanin MBW regulatory genes was carried out using the database of PlantCAre (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Rombauts et al., 1999; Lescot et al., 2002) with the Sol Genomics (https://solgenomics.org) id of the genes of interest.
The expression analysis of the genes of interest in the fruit tissues of S. lycopersicum was checked by using the online tool “Tomato Expression Atlas” (https://tea.solgenomics.net/) (Pattison et al., 2015; Fernandez-Pozo et al., 2017; Shinozaki et al., 2018).
Statistical analyses were performed with GraphPad Prism 6.01 (www.graphpad.com/scientific-software/prism/). Unpaired t-test or one-way ANOVA with Tukey’s HSD post-hoc test was used to compare the means of two or more than two group samples, respectively.
To study the anthocyanin biosynthesis induced by light in anthocyanin-enriched tomato fruits, the genotypes Aft/atv and Aft/atv/hp2 were used (Sestari et al., 2014). Tomato fruit pericarp was divided into two different samples: exocarp, containing the cuticle layer, the epidermal cell layer, and the underlying thin collenchymatous tissue, and the internal parts, constituted by the intermediate parenchymatous mesocarp and by the endocarp, consisting of a cell layer covering the locular cavities (Pesaresi et al., 2014). For sake of conciseness, however, from here on, the internal layers of the pericarp will be indicated with the single word “mesocarp”. At the mature green (MG) stage, both Aft/atv and Aft/atv/hp2 fruits showed accumulation of anthocyanins in the exocarp, with a more intense and evenly distributed pigmentation on Aft/atv/hp2 fruits (Figures 1A, C, E). Instead, in both lines, anthocyanins were not present in mesocarp, as well as in the locular cavities, placenta, and developing seeds (Figures 1B, D, E).
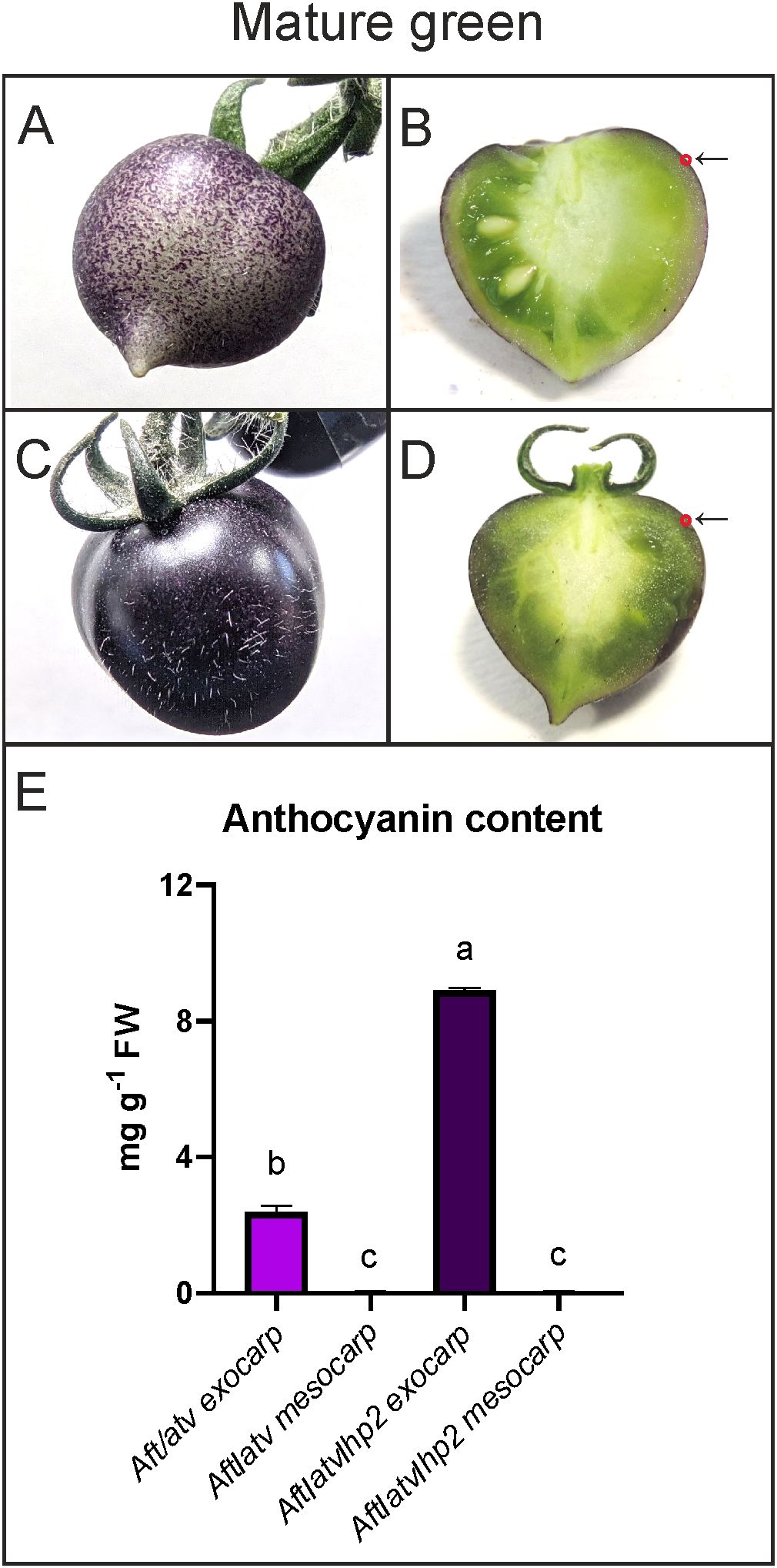
Figure 1. Phenotypes of anthocyanin-enriched tomato fruits at the mature green stage. Aft/atv (A, B) and Aft/atv/hp2 fruits (C, D), entire and in cross sections, respectively, were shown. In B and D the arrows indicate the site of accumulation of the anthocyanins in the exocarp, marked with a red circle. Anthocyanins measured in exocarp and mesocarp of Aft/atv and Aft/atv/hp2 fruits, expressed as petunidin-3-(p-coumaroyl rutinoside)-5-glucoside per gram exocarp fresh weight (FW) (E). Data are means (± SEM) of three biological replicates. One-way ANOVA with Tukey’s HSD post-hoc test was performed. Different letters indicate significant differences at p≤0.05.
The accumulation of anthocyanins was the likely consequence of activation of the biosynthetic pathway (Supplementary Figure S1): this occurred in early stages of fruit development (Supplementary Figure S2), and in MG fruits anthocyanins were already present at high levels (Figure 1E). The structural genes encoding enzymes involved in early and late reactions (Supplementary Table S2) resulted indeed expressed at MG (Figure 2A). In general, the expressions of the structural genes in Aft/atv/hp2 exocarp were higher than in Aft/atv exocarp (Figure 2A), even if the differences were not always statistically significant. In mesocarp, on the contrary, in both lines all the biosynthetic genes were not expressed (Figure 2A), thus explaining the lack of anthocyanins in this part of the fruit.
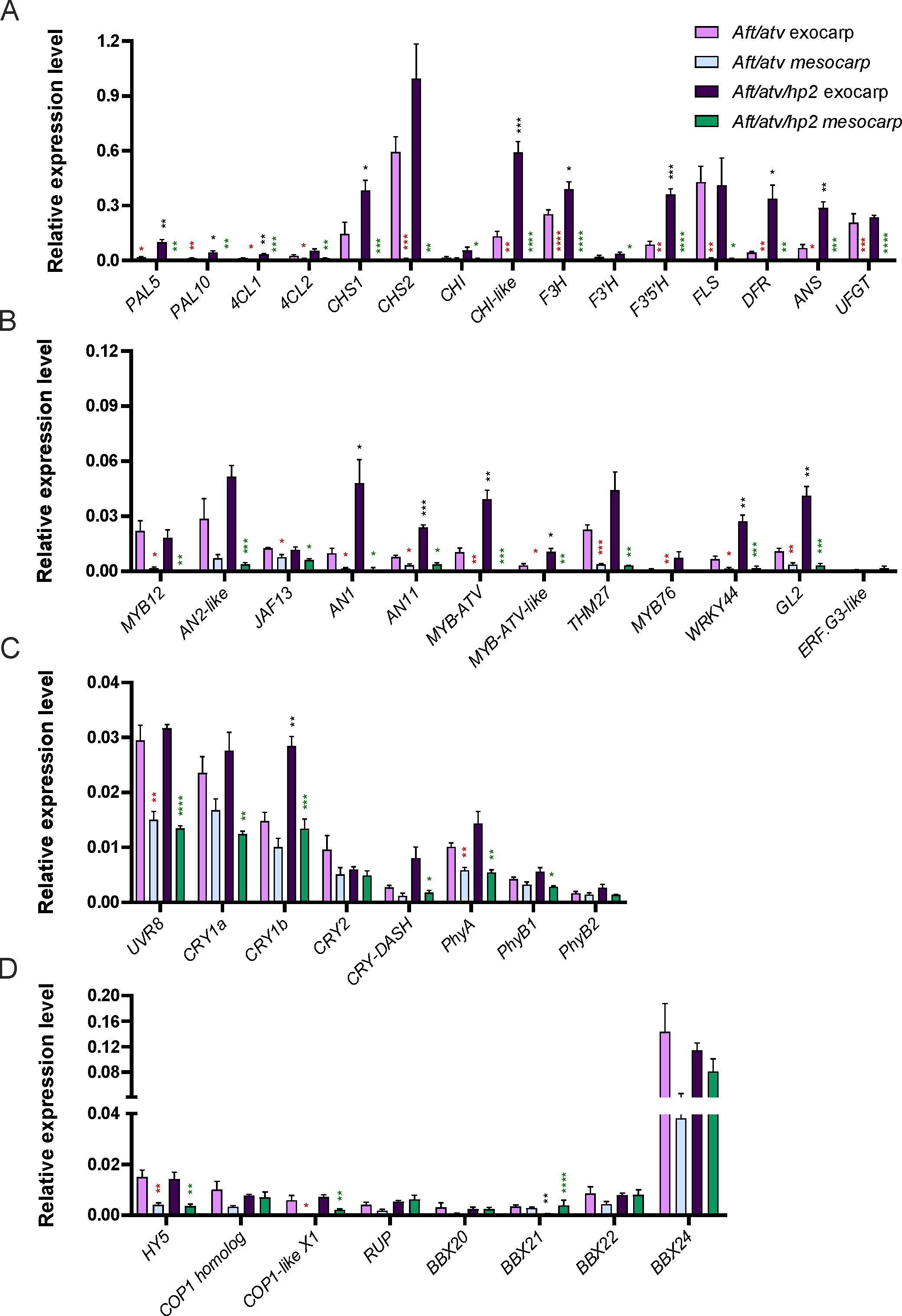
Figure 2. Relative expression levels of structural genes (A), regulatory genes (B), apoproteins of photoreceptors encoding genes (C) and light signaling genes (D) affecting or involved in the anthocyanin biosynthetic pathway, measured by qPCR in Aft/atv and Aft/atv/hp2 tomato fruit exocarp and fruit mesocarp at the mature green stage. Data are means of four biological replicates ± SEM. For each gene expression analysis, t-test was performed between exocarp and mesocarp of Aft/atv fruits (red asterisks), exocarp and mesocarp of Aft/atv/hp2 fruits (green asterisks), and Aft/atv exocarp and Aft/atv/hp2 exocarp (black asterisks). Statistical significance is reported in function of the number of asterisks: *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001.
The activator regulatory genes (Supplementary Table S2), either MYB12 or the genes encoding the MBW factors, were transcribed in the exocarps of both lines and much less or even not in the relative mesocarps, with the only exception of JAF13 which showed similar expressions in the two parts of the pericarp (Figure 2B). Excluding MYB12 and JAF13, which were expressed at similar levels in the exocarps of the two lines, the other MBW regulatory genes were expressed in Aft/atv/hp2 exocarp more than in Aft/atv (Figure 2B).
The genes encoding the repressor factors MYB-ATV, MYB-ATV-like, THM27, and MYB76 (Supplementary Table S2) were all more expressed in exocarps than in mesocarps and in Aft/atv/hp2 exocarp more than in Aft/atv exocarp. A similar trend was shown by WRKY44 and GL2. On the contrary, ERF.G3-like was not expressed in MG fruits (Figure 2B).
Genes encoding photoreceptors or their apoproteins were analyzed to highlight possible variations in the expression patterns between exocarp and mesocarp. All the genes under study, including UVR8, cryptochrome 1a (CRY1a), cryptochrome 1b (CRY1b), cryptochrome 2 (CRY2), cryptochrome DASH (CRY-DASH), phytochrome A (PhyA), phytochrome B1 (PhyB1), and phytochrome B2 (PhyB2) (Supplementary Table S2), resulted to be expressed in both Aft/atv and Aft/atv/hp2 fruits at MG, in either exocarps or mesocarps, with transcription levels in mesocarp generally lower than in exocarp (Figure 2C).
A common element downstream of the photoreceptors is HY5, but also other signaling factors, such as the B-BOX containing proteins (BBXs), may play important roles in relation to anthocyanin synthesis (Lee et al., 2007; Binkert et al., 2014; Xu, 2020; Wang et al., 2021; Menconi, 2024). The expressions of different genes known to code for key factors of the light signaling pathway (Supplementary Table S2) were analyzed. At MG, the expressions of HY5 in exocarps were similar in Aft/atv/hp2 and Aft/atv fruits, and potential HY5 target genes (Binkert et al., 2014; Burko et al., 2020), such as COP1 homolog, COP1-like isoform X1, and REPRESSOR OF UV-B PHOTOMORPHOGENESIS (RUP), also showed similar transcription levels in exocarp in the two genotypes (Figure 2D). While in Aft/atv fruits the transcription of all these genes was higher in exocarp than in mesocarp, in Aft/atv/hp2 fruits this occurred for HY5 and COP1-like isoform X1, whereas COP1 homolog and RUP showed the same expressions in exocarp and mesocarp (Figure 2D). Relative to the BBX encoding genes, they showed overall similar expression levels in the two lines with no big differences between exocarp and mesocarp, except for BBX21 which appeared downregulated in Aft/atv/hp2 exocarp compared with the other samples (Figure 2D).
At the mature red (MR) stage, the fruits of the two lines showed uniform anthocyanin pigmentation of the exocarp (Figures 3A, C), whereas the internal parts appeared homogeneously red for the presence of lycopene and absence of anthocyanins (Figures 3B, D, E). The Aft/atv/hp2 fruits still contained more anthocyanins in the exocarp than the other genotype (Figure 3E), but the quantity of pigments accumulated in the exocarp from MG to MR resulted higher in the Aft/atv line (Supplementary Figure S3).
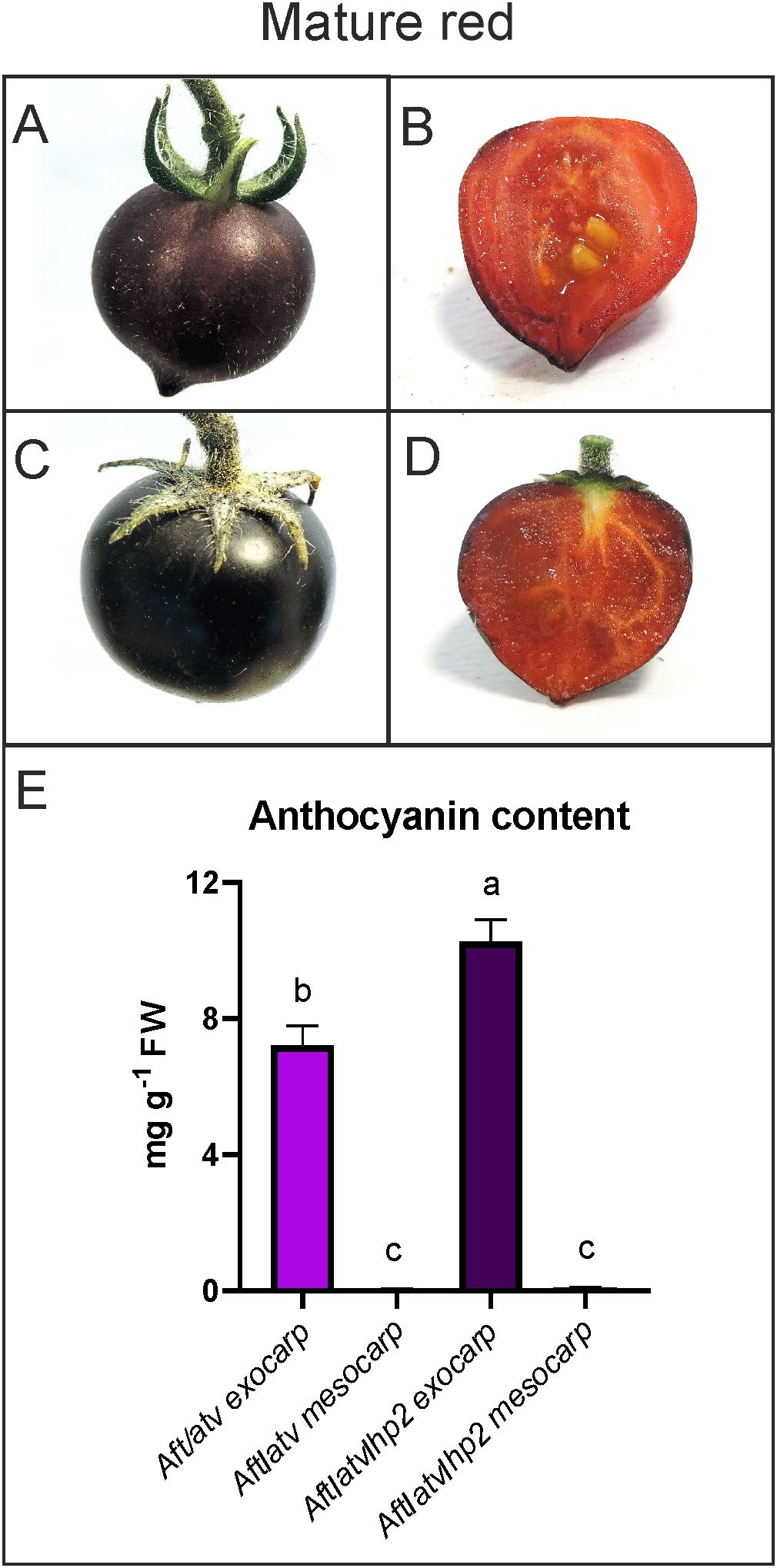
Figure 3. Phenotypes of anthocyanin-enriched tomato fruits at the mature red stage. Aft/atv (A, B) and Aft/atv/hp2 fruits (C, D), entire and in cross sections, respectively, were shown. Anthocyanins measured in exocarp and mesocarp of Aft/atv and Aft/atv/hp2 fruits, expressed as petunidin-3-(p-coumaroyl rutinoside)-5-glucoside per gram exocarp fresh weight (FW) (E). Data are means (± SEM) of three biological replicates. One-way ANOVA with Tukey’s HSD post-hoc test was performed. Different letters indicate significant differences at p≤0.05.
At the transcriptional level, comparing MR with MG, a clear attenuation was visible in the expression of most of the genes analyzed. In the exocarp of the Aft/atv fruits, the reduction of the biosynthetic pathway activity was particularly strong (Figure 4A), and evident was also the decrease in the expressions of the regulatory genes MYB12, AN2-like, and JAF13 among the activators, and MYB-ATV, MYB-ATV-like, and THM27 among the repressors (Figure 4B). In Aft/atv/hp2 exocarp, MYB12 and JAF13 expressions decreased at MR compared with MG, as well as THM27, but the other regulatory MBW genes continued to be expressed at high levels (Figure 4B). Interestingly, the regulatory gene ERF.G3-like, whose expression in MG fruits was not detected, resulted highly transcribed in MR fruits, in both mesocarps and exocarps, particularly in the Aft/atv/hp2 fruit (Figure 4B).
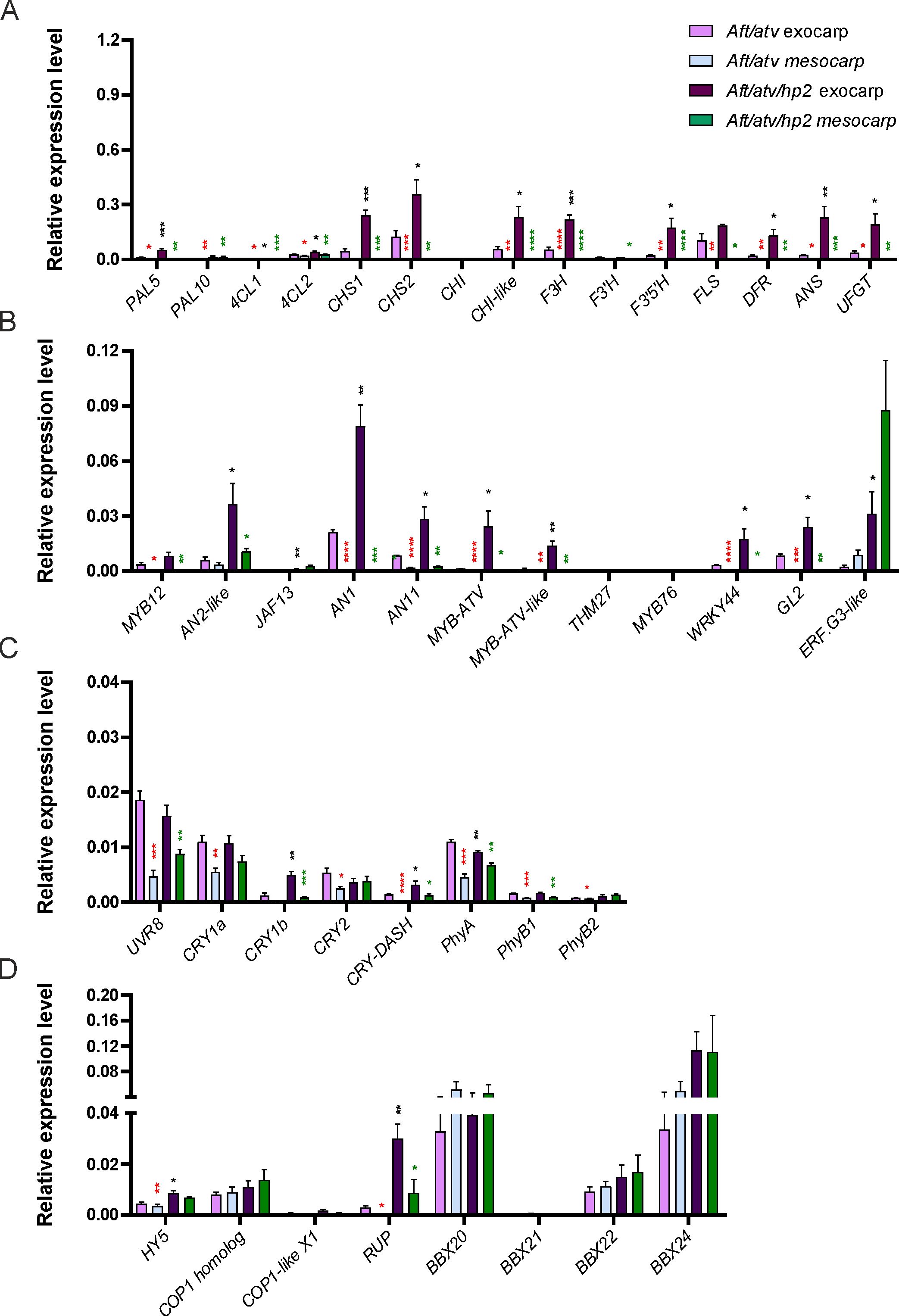
Figure 4. Relative expression levels of structural genes (A), regulatory genes (B), apoproteins of photoreceptors encoding genes (C), and light signaling genes (D) affecting or involved in the anthocyanin biosynthetic pathway, measured by qPCR in Aft/atv and Aft/atv/hp2 tomato fruit exocarp and fruit mesocarp at the mature red stage. Data are means of four biological replicates ± SEM. For each gene expression analysis, t-test was performed between exocarp and mesocarp of Aft/atv fruits (red asterisks), exocarp and mesocarp of Aft/atv/hp2 fruits (green asterisks), and Aft/atv exocarp and Aft/atv/hp2 exocarp (black asterisks). Statistical significance is reported in function of the number of asterisks: *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001.
The transcription of the genes encoding photoreceptor proteins appeared overall reduced in MR fruits compared with MG, still maintaining the same relative differences between the two genotypes and between exocarp and mesocarp previous observed (Figure 4C). The reduction of mRNA levels compared with MG was particularly evident for the gene UVR8, CRY1a, CRY1b, and PhyB1.
Finally, as for the light signaling genes, most of them resulted to be still expressed in MR fruits but with some differences compared with MG. First of all, HY5 was less transcribed in the exocarps of both lines than at MG and showed a higher expression in Aft/atv/hp2 fruits than in Aft/atv (Figure 4D). The mRNA levels of COP1 homolog slightly increased in the fruit mesocarp in both lines, whereas COP1-like isoform X1 (whose expression was already low in the mesocarps of MG fruits) resulted to be not expressed also in the exocarp of the Aft/atv MR fruits, and very low levels of its mRNA were detected only in the exocarp of the Aft/atv/hp2 fruits (Figure 4D). However, the strongest variations in gene expression between the two developmental stages interested RUP, whose expression strongly increased in Aft/atv/hp2 fruits, particularly in the exocarp, and BBX21, whose mRNA dropped at barely detectable levels in both genotypes. On the contrary, BBX20 and BBX22 expressions increased in all the fruit tissues of both lines, whereas BBX24 remained well expressed in all the samples, but its levels decreased in the Aft/atv exocarp compared with MG (Figure 4D).
Being the regulatory genes encoding the components of the MBW complexes transcriptionally activated by TFs acting upstream, an analysis on their promoter sequences was performed and different classes of cis-acting responsive elements were identified (Figure 5). In the light-mediated activation of the anthocyanin pathway, the most important classes of cis-acting sequences are the light-responsive elements (LREs), which were indeed identified in the genomic regions upstream of the CDS of all the regulatory genes. Among them, specific HY5 binding sites, belonging to both G-Box and other ACE-Box classes, were found. Several MYB responsive elements were also present and MREs specifically involved in light responsiveness were identified.
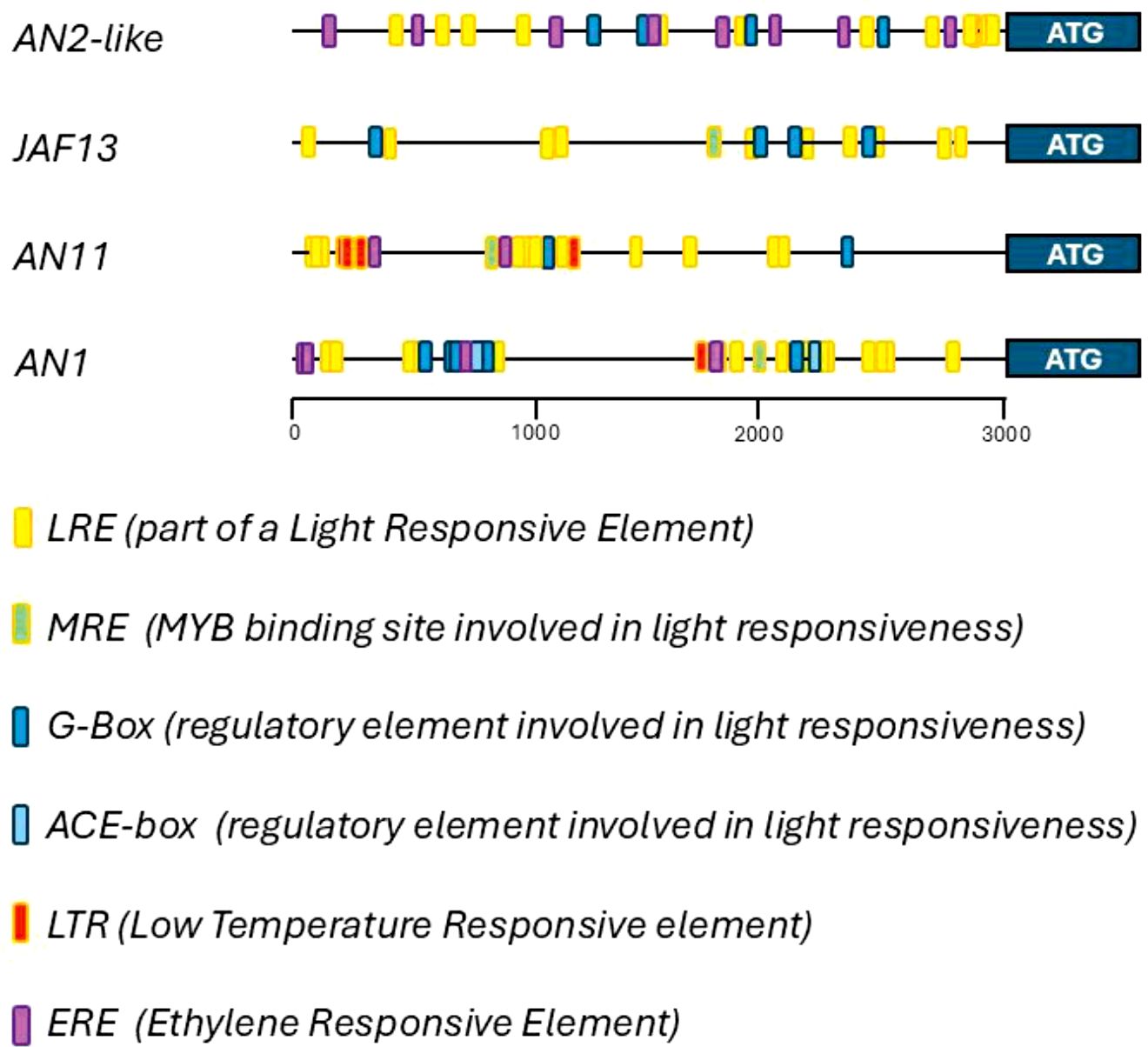
Figure 5. Cis-acting regulatory elements in the promoters of the genes involved in the production of the tomato MBW complexes. Light-responsive elements (LRE) (yellow), MYB regulatory elements (MRE) involved in light responsiveness (green), G-Box regulatory elements (blue), other ACE-box cis-acting regulatory elements involved in light responsiveness (light blue), low-temperature responsive (LTR) elements (red), and ethylene-responsive elements (EREs) (purple) identified in the 3-kb regions upstream of the ATG first codon in the genomic sequences of the regulatory genes AN2-like, JAF13, AN11, and AN1. The analysis of the regulatory elements was carried out using the database of “Plant CARE” web site.
Other environmental factors may induce anthocyanin biosynthesis besides light, and cold temperatures are known to be able to stimulate anthocyanin production in tomato leaves (Kiferle et al., 2015). Low temperature-responsive (LTR) elements were in fact present in the promoters of AN11 and AN1 genes, but not in those of AN2-like and JAF13 (Figure 5).
Finally, several hormonal responsive-elements were found, and ethylene-responsive elements (EREs), which may link anthocyanin biosynthesis with ethylene production occurring during fruit ripening, were identified in the promoters of AN2-like, AN11, and AN1 (Figure 5).
When the Aft and atv alleles are present in the tomato genome, under adequate light intensities the flavonoid biosynthetic pathway produces anthocyanins in the fruit exocarp. In the experimental setup used in this work, light activated the anthocyanin synthesis in immature fruits (Supplementary Figure S2), and the quantity of pigments increased in exocarp till MR (Supplementary Figure S3). As expected, both EBGs and LBGs were expressed in the exocarp, likely induced, respectively, by MYB12 (Adato et al., 2009; Ballester et al., 2010) and by the MBW complexes (Montefiori et al., 2015), whose genes were all expressed at MG (Figures 2A, B). From MG on, some activators of the pathway tended to reduce their expression in Aft/atv exocarp, and this was followed by a general reduction in the expression of the biosynthetic genes (Figures 4A, B). In Aft/atv fruit mesocarp, anthocyanins were not produced at both MG and MR for the lack of expression of the structural genes, accompanied by reduced or null transcription of the regulatory factors (Figures 2A, B, 4A, B). The mutation hp2 increased the anthocyanin accumulation in Aft/atv fruit exocarp, as a consequence of higher activation of most regulatory and structural genes at both MG and MR but, remarkably, did not affect anthocyanin synthesis in the fruit mesocarp (Figures 2A, B, 4A, B). Interestingly, not only were the genes analyzed expressed in Aft/atv/hp2 exocarp generally more than in Aft/atv exocarp (Supplementary Figure S4, columns A and D) but also the ratios of gene expression between mesocarp and exocarp resulted lower in Aft/atv/hp2 fruits than in Aft/atv fruits for many of them (Supplementary Figure S4, columns B, C, E, F).
The DET1/hp2 mutation, impairing the COP1/SPA ubiquitin ligase activity, stabilizes HY5 and induces its targets, thus conferring hypersensitivity to light (Mustilli et al., 1999; Lau and Deng, 2012; Cañibano et al., 2021). The stabilization of HY5 in the exocarp of Aft/atv/hp2 fruits was proved by the increased expression at MR of some of its known targets, such as COP1 homolog and COP1-like isoform X1 (Burko et al., 2020; Menconi, 2024), RUP (Binkert et al., 2014; Zhang et al., 2021), and the same HY5 gene (Figure 4D), which is prone of autoactivation (Binkert et al., 2014). Likewise, the increased transcription of almost all the genes encoding the regulatory factors and the biosynthetic enzymes in Aft/atv/hp2 exocarp compared with Aft/atv exocarp indicated that the light-dependent regulation of the anthocyanin pathway in purple tomato fruit is mediated by HY5.
The transcription of the MBW genes AN2-like and AN1 was specifically induced in Aft/atv fruits by light, being null their expressions in wild-type fruits (Supplementary Figure S5). Conversely, the other MBW genes JAF13 and AN11 are expressed also in wild-type fruits independently from the presence of anthocyanins (Supplementary Figure S6). The transcriptions of AN2-like, AN1, and AN11 resulted upregulated in Aft/atv/hp2 fruits and their promoters showed the presence of G-box and ACE-box sequences, putative cis-acting HY5 binding sites (Shin et al., 2013; Binkert et al., 2014; Zoratti et al., 2014) (Figure 5): thus, they might be directly targeted by HY5.
The induction by light of AN2-like switched on the hierarchical regulatory chain which produced the MBW1 complex through interaction of AN2-like with JAF13 and AN11, constitutively present (Supplementary Figure S6). AN1 contains in its promoter both LREs and MREs involved in light responsiveness (Figure 5). Similarly to the light-mediated induction of other genes of the flavonoid pathway, which require MREs as part of the light-responsive units (Hartmann et al., 2005), it is possible that the activation of AN1 requires the concerted action of the MBW1 complex, binding its promoter through AN2-like, with HY5. The same mechanism might allow transcription of the structural genes, being activated, respectively, by MYB12 and by the MBW2 complex, but also directly bound in their promoters by HY5 (Wang et al., 2021): as a confirmation, both EBGs and LBGs resulted to be more expressed in Aft/atv/hp2 than in Aft/atv exocarp (Figure 2A). Furthermore, feedback repression mechanisms (LaFountain and Yuan, 2021), carried out by negative regulators of the pathway, tended to break the accumulation of anthocyanins, once activated. MYB-ATV was expressed more than the other repressors, but, being not functional in both genotypes for the presence of the atv allele (Cao et al., 2017; Colanero et al., 2018), its role may have been replaced by MYB-ATV-like and THM27, which showed a similar pattern (Figure 2B).
With ripening, the transcription of the anthocyanin structural genes decreased in Aft/atv fruits, highlighting, besides the action of the repressors, an overall reduced activation exerted by light on the metabolic pathway under study, proved by the lower expression of HY5 and of some activator genes (Figures 4A, B, D). In Aft/atv/hp2 exocarp, on the other hand, the reduced expression of HY5 at MR may have been counterbalanced by its stabilization at the protein level, since some of its targets, including most anthocyanin regulatory genes, still showed high transcription (Figure 4B). Nevertheless, also in this genotype many structural genes at MR were expressed less in exocarp than at MG: since many negative regulators were expressed in Aft/atv/hp2 more than in Aft/atv exocarp, stronger feedback repression mechanisms may have contributed to slow down the pathway.
WRKY44 and GL2, similarly to the other regulators of the process, were expressed almost exclusively in the exocarps (Figures 2B, 4B), and, differently from earlier hypotheses (Qiu et al., 2019), they also appeared to be under HY5 control, being significantly more expressed in Aft/atv/hp2 fruits. WRKY TFs involved in anthocyanidin and proanthocyanidin synthesis and homologs of Arabidopsis TTG2, which regulates the seed coat tannin accumulation (Gonzalez et al., 2016), have been identified in different species: they could interact with the MBW complex to increase its activity toward specific targets (Lloyd et al., 2017; Amato et al., 2019). The homolog of WRKY44 in kiwifruit, for example, has been recently shown to activate the promoters of F3′H and F3′5′H (Supplementary Figure S1), regulating important branch points of the pathway (Peng et al., 2020). WRKY44, like AN2-like and AN1, is not expressed in wild-type tomato fruit (Supplementary Figure S5); thus, its transcription in anthocyanin-enriched tomatoes suggests a possible role as an additional activator of the pathway. GL2 is a leucine-zipper TF still not well characterized in tomato and homolog to Arabidopsis GL2, which is involved in trichome development and inhibition of anthocyanin synthesis (Chen and Wang, 2019). Actually, some MG Aft/atv/hp2 fruits showed slightly longer trichomes on their skin (Figure 1C): therefore, tomato GL2 might play functions close to its Arabidopsis counterpart, also inhibiting the anthocyanin pathway.
ERF.G3-like behaved differently from all the other TFs analyzed. Its expression was absent in MG fruits and strongly increased with ripening, particularly in Aft/atv/hp2 fruits (Figure 4B). Recent studies have indicated that ERF.G3-like is transcriptionally activated by the master ripening regulator RIN and particularly expressed in tomato mesocarp (You et al., 2024); it has been also associated with activation of ethylene synthesis and expression of some flavonoid EBGs (Li et al., 2020). In our system, we observed activation of ERF.G3-like expression in MR mesocarps, besides exocarps, but without concomitant activation of the EBGs (Figures 4A, B). Ethylene has been recently found to inhibit anthocyanin synthesis in Aft/atv tomatoes by specifically repressing the transcription of AN2-like and of several structural genes (Xu et al., 2022). Remarkably, multiple EREs were found in the promoter of AN2-like, as well as in AN1 and AN11 (Figure 5): as a consequence, in Aft/atv fruits a possible repressor activity of ethylene on AN2-like expression at MR, possibly mediated by ERF.G3-like, could not be excluded. The higher expression of this gene in Aft/atv/hp2 fruits may also indicate that ERF.G3-like is a target of HY5, as other ethylene signaling genes (Wang et al., 2021).
The anthocyanin biosynthetic pathway may be controlled by the light signaling factors not only at the transcriptional level but also through physical interactions of these factors with the MBW complexes or single components of them, altering their activities. In particular, degradation of AN2-like, whose homologs in other species can be direct targets of the COP1/SPA system (Li et al., 2012; Maier et al., 2013), may not be excluded in Aft/atv fruits, where both COP1 homolog and COP1-like X1 isoform resulted expressed, particularly in mesocarp where also the expression of AN2-like was lower. However, degradation of AN2-like mediated by the COP1/SPA system could not occur in Aft/atv/hp2 fruits due to the mutation of DET1 which impaired COP1 activity (Lau and Deng, 2012; Cañibano et al., 2021), but these fruits did not show pigmentation in the mesocarp, as well as the Aft/atv fruits. Other mechanisms must therefore be hypothesized.
RUP and the BBXs may have indirectly affected the anthocyanin pathway by altering the HY5 levels (Xu, 2020; Zhang et al., 2021). RUP expression was very similar in exocarp in the two lines at MG, and then it strongly increased at MR, but only in Aft/atv/hp2 fruit (Figure 4D). RUP is a UV-B light signaling inhibitor, whose expression can be induced by UV light via UVR8 and by HY5 (Zhang et al., 2021), and is produced to revert the active UVR8 monomer to the inactive homodimer (Heijde and Ulm, 2013), contributing, in this way, to also control HY5 expression. In lettuce, an inhibitory action on the anthocyanin pathway has been recently demonstrated (Yamashita et al., 2023). With the quality of light not changed from MG to MR stages, the strong increase of RUP expression in Aft/atv/hp2 fruits at MR may have been induced only by the stabilization of HY5, thus creating an inhibitory loop on further HY5 expression and anthocyanin production, counteracting, at least in part, the positive effects on the pathway of the stabilization of HY5.
The BBX encoding genes showed expression levels not very different in the two genotypes (Figures 2D, 4D), with only a slight tendency, statistically not significant, of higher mRNA levels in Aft/atv/hp2 MR fruits. Recently, in tomato, the redundant roles of BBX20 and BBX21 in photomorphogenesis have been hypothesized, and the complex produced by BBX20 or BBX21 with HY5 under UV-B has resulted able to activate the expression of HY5. However, HY5 protein, in turn, would outcompete BBX20 and BBX21 for binding to its promoter, thus producing an autoregulatory negative feedback loop attenuating its transcription (Yang et al., 2022). In our system, BBX20 and BBX21 were both transcribed at MG, and at MR the expression of BBX20 increased a lot: this could imply a higher inhibitory loop on HY5 transcription in MR, thus contributing to reduce the HY5 expression. In Arabidopsis thaliana, BBX22 and BBX24 act in both cases in association with HY5, but the first is an activator and the second an inhibitor of the anthocyanin pathway (Jiang et al., 2012; Job et al., 2018; Liu et al., 2022b). The expression patterns of these two genes in MG and MR fruits were overall quite similar in the two lines: thus, they would not seem to be targets of HY5 at the transcriptional level. Furthermore, they were expressed in both fruit exocarps and mesocarps; then, a specific role in the pigmentation patterns of the fruits cannot be inferred by these experimental data.
Although Aft/atv/hp2 fruits showed an exaggerated photomorphogenic phenotype under light, they did not synthesize anthocyanins underneath the exocarp, like the Aft/atv fruits. Thus, in both lines, a factor necessary to inducing the pathway should have been missed. HY5 expression in mesocarp was lower than in exocarp and remained quite stable from MG to MR in both lines. The transcription of HY5 is mainly activated by UV light through the UVR8/COP1 complex (Chen et al., 2022), whereas its protein stability is controlled by red and blue light photoreceptors via COP1 removal (Texteira, 2020; Bianchetti et al., 2022; Yan et al., 2023), less effective in the presence of the DET1/hp2 mutation. In our system, all the genes encoding photoreceptors or their apoproteins were expressed at MG; then, with ripening, a global reduction in their expressions was observed in both lines (Figures 2C, 4C), and this could have in parallel affected HY5 levels.
In each genotype, at both MG and MR, most of the photoreceptor genes were less expressed in mesocarp than in exocarp (Figures 2C, 4C). If the anthocyanins already present in the exocarp had absorbed specific radiations more than others, the same should have been perceived less in the underneath tissues. UV-A and UV-B are the wavelengths generally mainly filtered by anthocyanins, but also blue photons can be effectively absorbed (Landi et al., 2021). The artificial lightening system used (Supplementary Figure S7) had a spectrum very similar to sunlight; thus, the anthocyanins in the fruit exocarps should have mainly filtered UV and blue light, which are the radiations perceived by UVR8 and cryptochromes: interestingly, the relative encoding genes resulted the more dampened in mesocarp. The reduction of UVR8 expression in mesocarp may have particularly contributed to inhibit the early steps of the pathway, since flavonoid genes, including MYB12, CHS1, and CHS2, are mainly induced in tomato by UV via UVR8 (Liu et al., 2020). Blue light has been involved in inducing anthocyanin accumulation in strawberry, pepper, and blueberry fruits (Kadomura-Ishikawa et al., 2013; Liu et al., 2022a; Wei et al., 2023), and in tomato flavonoids, chlorophylls and carotenoids are strongly affected by CRY1 and CRY2 activities (Giliberto et al., 2005; Fantini et al., 2019). A reduction in UV perception by UVR8 and in blue-light perception by cryptochromes may have thus strongly affected anthocyanin synthesis in mesocarps through reduction of HY5 gene transcription and HY5 protein stability, and this should have been more severe where anthocyanin concentrations in exocarps were higher, that was in Aft/atv/hp2 fruits. Confirming that, all the photoreceptor genes reduced their expressions from exocarp to mesocarp more in Aft/atv/hp2 fruits than in Aft/atv (Supplementary Figure S4, columns B, C, E, F): a light hypersensitivity, such as the one shown by the hp2 mutants, was thus not more permissive since the higher anthocyanins levels accumulated in the exocarp produced an even thicker shield for light penetration inside the fruit, neutralizing the stabilization of HY5 due to the failure of COP1-mediated turnover.
The lack of anthocyanins in mesocarp might thus indicate that AN2-like, insufficiently induced by HY5, could not have reached a threshold necessary to produce the MBW1 complex in the amounts needed to activate AN1 transcription. The same direct activation exerted by HY5 and other light-responsive factors on the transcription of AN1 and AN11 might have been lost or highly reduced in mesocarp because of the filter exerted by the anthocyanin light screen. As a consequence, AN1 expression resulted negligible in mesocarp (Figures 2B, 4B) and thus ineffective to produce the MBW2 complex necessary to activate the expression of the LBGs: this would have finally impaired the synthesis of anthocyanins inside the fruit.
To circumvent this bottleneck, an adequate increase of the transcription rate of AN2-like or AN1 in mesocarp would be necessary. Low temperature may represent a trigger for anthocyanin synthesis in fruits of several species (Zhang et al., 2011; Gao-Takai et al., 2019; Xue et al., 2021; Dai et al., 2022), increasing the expression of R2R3-MYB or bHLH TF-encoding genes and/or improving the binding ability of bHLH and MYB inside the MBW complexes (Sheerin and Hiltbrunner, 2017). Furthermore, HY5 levels under light are positively regulated by low temperature, both transcriptionally, via a CBF- and ABA-independent pathway, and posttranslationally, via protein stabilization through nuclear depletion of COP1 (Catalá et al., 2011). Previous studies indicated that cold could activate both AN2 and AN1 expressions in tomato plants (Kiferle et al., 2015), and, as observed in the present work, it may act on the R2R3-MYB gene transcription through HY5 but also directly on AN1 and AN11 expression being several LTR elements present in their promoters (Figure 5). Expositions to low temperature may therefore effectively integrate the light stimulus, finally leading to purple tomatoes containing anthocyanins in all the pericarp.
In conclusion, the analyses carried out in the present study, whose main results are summarized in Figure 6, indicated that i) the anthocyanin biosynthetic pathway could be activated by light in the exocarp of Aft/atv fruits, reflecting the role of flavonoids in tomato fruit photoprotection; ii) between photoreceptors and anthocyanin production, a signaling cascade based on HY5 activated, either directly or indirectly, anthocyanin regulatory and biosynthetic genes; iii) a combinatorial effect of light-mediated signals with AN2-like may have been necessary to guide AN1 expression as well as transcription of many structural genes; iv) the light penetrating inside the fruit was qualitatively/quantitatively different from radiations incident on the skin, as a result of the anthocyanins accumulated in the exocarp which shaded the inner fruit tissues; v) HY5 expression in mesocarp was low because of the scarce activation exerted by the photoreceptors on its expression and protein stability, and inhibitory loops on its transcription produced by BBX factors and RUP proteins may not be excluded; and vi) in the absence of other inducing factors, e.g., low temperatures, the anthocyanin regulatory genes were very poorly transcribed in tomato mesocarp, starting from the R2R3-MYB activators MYB12 and AN2-like till the bHLH gene AN1, whose absence is the primary cause of the silence of both EBGs and LBGs.
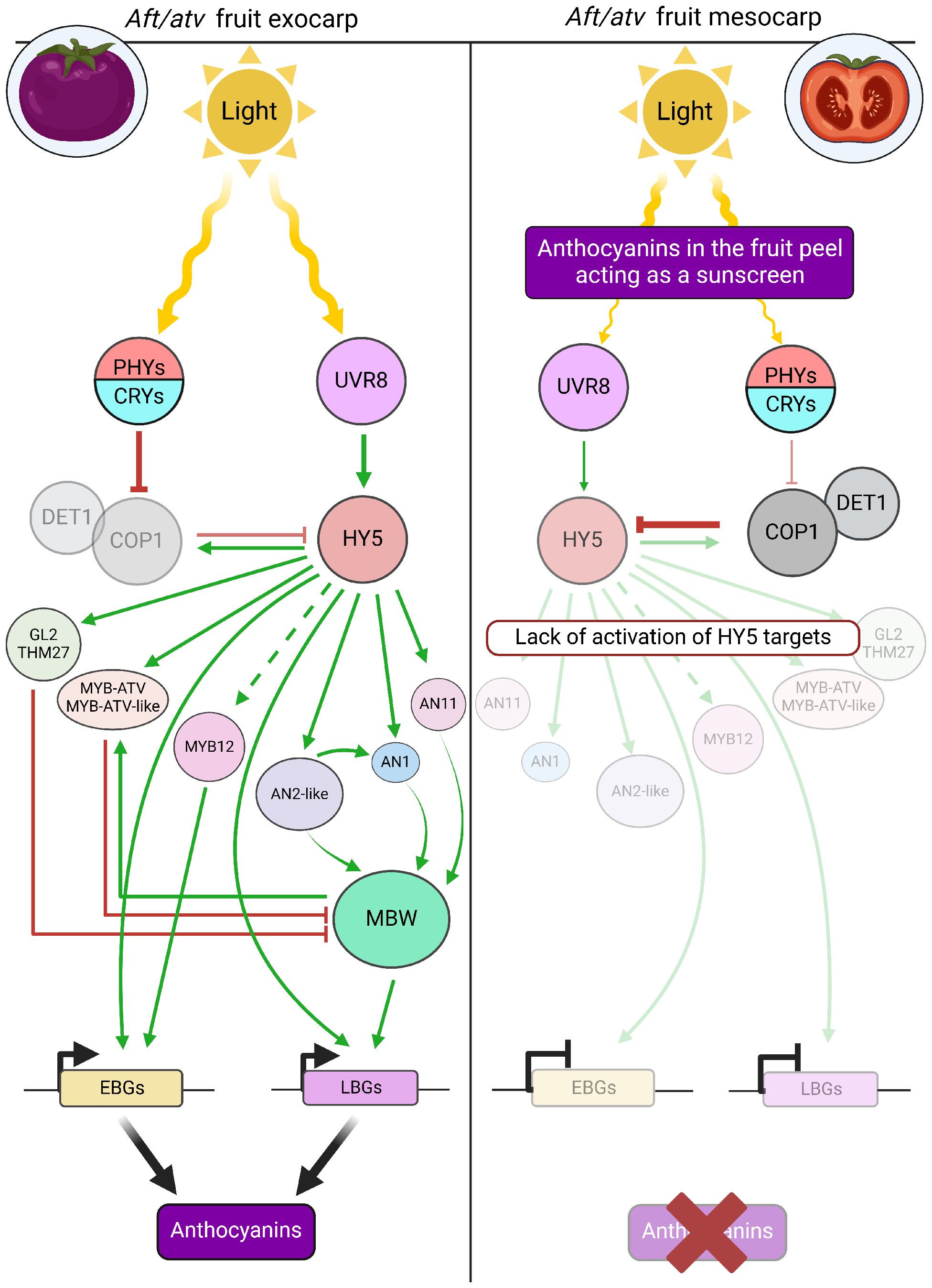
Figure 6. Final scheme summarizing the main elements which under light regulate anthocyanin pigmentation in the exocarp (left) and mesocarp (right) of Aft/atv tomato fruits.
The data presented in the study are deposited in the Gene Expression Omnibus repository, accession number GSE282571.
SG: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing. JM: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. PP: Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Scuola Superiore Sant'Anna, Pisa, Italy.
We acknowledge Prof. Lázaro Eustáquio Pereira Peres and his group working at Universidade de São Paulo, Brazil, for kindly providing us with seeds of Aft/atv/hp2 line in MT background.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphgy.2024.1507833/full#supplementary-material
Adato A., Mandel T., Mintz-Oron S., Venger I., Levy D., Yativ M., et al. (2009). Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PloS Genet. 5, e1000777. doi: 10.1371/journal.pgen.1000777
Alappat B., Alappat J. (2020). Anthocyanin pigments: beyond aesthetics. Molecules 25, 5500. doi: 10.3390/molecules25235500
Albert N. W., Davies K. M., Lewis D. H., Zhang H., Montefiori M., Brendolise C., et al. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26, 962–980. doi: 10.1105/tpc.113.122069
Albert N. W., Lewis D. H., Zhang H., Irving L. J., Jameson P. E., Davies K. M. (2009). Light-induced vegetative anthocyanin pigmentation in Petunia. J. Exp. Bot. 60, 2191–2202. doi: 10.1093/jxb/erp097
Amato A., Cavallini E., Walker A. R., Pezzotti M., Bliek M., Quattrocchio F., et al. (2019). The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. Plant J. 99, 1220–1241. doi: 10.1111/tpj.14419
Ballester A.-R., Molthoff J., de Vos R., Lintel Hekkert B., Orzaez D., Fernández-Moreno J.-P., et al. (2010). Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol. 152, 71–84. doi: 10.1104/pp.109.147322
Bian Y., Chu L., Lin H., Qi Y., Fang Z., Xu D. (2022). PIFs- and COP1-HY5-mediated temperature signaling in higher plants. Stress Biol. 2, 35. doi: 10.1007/s44154-022-00059-w
Bianchetti R., Bellora N., de Haro L. A., Zuccarelli R., Rosado D., Freschi L., et al. (2022). Phytochrome-mediated light perception affects fruit development and ripening through epigenetic mechanisms. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.870974
Binkert M., Kozma-Bognár L., Terecskei K., De Veylder L., Nagy F., Ulm R. (2014). UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26, 4200–4213. doi: 10.1105/tpc.114.130716
Blando F., Berland H., Maiorano G., Durante M., Mazzucato A., Picarella M. E., et al. (2019). Nutraceutical characterization of anthocyanin-rich fruits produced by “Sun Black” tomato line. Front. Nutr. 6. doi: 10.3389/fnut.2019.00133
Bovy A., de Vos R., Kemper M., Schijlen E., Almenar Pertejo M., Muir S., et al. (2002). High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14, 2509–2526. doi: 10.1105/tpc.004218
Burko Y., Seluzicki A., Zander M., Pedmale U. V., Ecker J. R., Chory J. (2020). Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism. Plant Cell 32, 967–983. doi: 10.1105/tpc.19.00772
Butelli E., Titta L., Giorgio M., Mock H. P., Matros A., Peterek S., et al. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308. doi: 10.1038/nbt.1506
Cañibano E., Bourbousse C., García-León M., Garnelo Gómez B., Wolff L., García-Baudino C., et al. (2021). DET1-mediated COP1 regulation avoids HY5 activity over second-site gene targets to tune plant photomorphogenesis. Mol. Plant 14, 963–982. doi: 10.1016/j.molp.2021.03.009
Cao X., Qiu Z., Wang X., Van Giang T., Liu X., Wang J., et al. (2017). A putative R3 MYB repressor is the candidate gene underlying atroviolacium, a locus for anthocyanin pigmentation in tomato fruit. J. Exp. Bot. 68, 5745–5758. doi: 10.1093/jxb/erx382
Castle’ L. A., Meinke D. W. (1994). A FUSCA gene of arabidopsis encodes a nove1 protein essential for plant development. Plant Cell 6, 25–41. doi: 10.1105/tpc.6.1.25
Catalá R., Medina J., Salinas J. (2011). Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 16475–16480. doi: 10.1073/pnas.1107161108
Chen Z., Dong Y., Huang X. (2022). Plant responses to UV-B radiation: signaling, acclimation and stress tolerance. Stress Biol. 2, 51. doi: 10.1007/s44154-022-00076-9
Chen S., Wang S. (2019). GLABRA2, A common regulator for epidermal cell fate determination and anthocyanin biosynthesis in arabidopsis. Int. J. Mol. Sci. 20, 4997. doi: 10.3390/ijms20204997
Colanero S., Perata P., Gonzali S. (2018). The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00830
Colanero S., Tagliani A., Perata P., Gonzali S. (2020). Alternative splicing in the Anthocyanin fruit gene encoding an R2R3 MYB transcription factor affects anthocyanin biosynthesis in tomato fruits. Plant Commun. 1, 100006. doi: 10.1016/j.xplc.2019.100006
Costa Galvão V., Fankhauser C. (2015). Sensing the light environment in plants: photoreceptors and early signaling steps. Curr. Opin. Neurobiol. 34, 46–53. doi: 10.1016/j.conb.2015.01.013
Dai Y., Zhang L., Sun X., Li F., Zhang S., Zhang H., et al. (2022). Transcriptome analysis reveals anthocyanin regulation in Chinese cabbage (Brassica rapa L.) at low temperatures. Sci. Rep. 12, 6308. doi: 10.1038/s41598-022-10106-1
Fantini E., Sulli M., Zhang L., Aprea G., Jiménez-Gómez J. M., Bendahmane A., et al. (2019). Pivotal roles of cryptochromes 1a and 2 in tomato development and physiology. Plant Physiol. 179, 732–748. doi: 10.1104/pp.18.00793
Fernandez-Pozo N., Zheng Y., Snyder S. I., Nicolas P., Shinozaki Y., Fei Z., et al. (2017). The tomato expression atlas. Bioinformatics 33, 2397–2398. doi: 10.1093/bioinformatics/btx190
Gangappa S. N., Botto J. F. (2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant 9, 1353–1365. doi: 10.1016/j.molp.2016.07.002
Gao Y., Liu J., Chen Y., Tang H., Wang Y., He Y., et al. (2018). Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins. Hortic. Res. 5, 27. doi: 10.1038/s41438-018-0032-3
Gao-Takai M., Katayama-Ikegami A., Matsuda K., Shindo H., Uemae S., Oyaizu M. (2019). A low temperature promotes anthocyanin biosynthesis but does not accelerate endogenous abscisic acid accumulation in red-skinned grapes. Plant Sci. 283, 165–176. doi: 10.1016/j.plantsci.2019.01.015
Giliberto L., Perrotta G., Pallara P., Weller J. L., Fraser P. D., Bramley P. M., et al. (2005). Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol. 137, 199–208. doi: 10.1104/pp.104.051987
Gonzalez A., Brown M., Hatlestad G., Akhavan N., Smith T., Hembd A., et al. (2016). TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev. Biol. 419, 54–63. doi: 10.1016/j.ydbio.2016.03.031
Gonzali S., Mazzucato A., Perata P. (2009). Purple as a tomato: towards high anthocyanin tomatoes. Trends Plant Sci. 14, 237–241. doi: 10.1016/j.tplants.2009.02.001
Gould K. S. (2004). Nature’s swiss army knife: the diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 2004, 314–320. doi: 10.1155/S1110724304406147
Han X., Huang X., Deng X. W. (2020). The photomorphogenic central repressor COP1: conservation and functional diversification during evolution. Plant Commun. 1, 100044. doi: 10.1016/j.xplc.2020.100044
Hartmann U., Sagasser M., Mehrtens F., Stracke R., Weisshaar B. (2005). Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 57, 155–171. doi: 10.1007/s11103-004-6910-0
Heijde M., Ulm R. (2013). Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc. Natl. Acad. Sci. U.S.A. 110, 1113–1118. doi: 10.1073/pnas.1214237110
Jaakola L. (2013). New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18, 477–483. doi: 10.1016/j.tplants.2013.06.003
Jian W., Cao H., Yuan S., Liu Y., Lu J., Lu W., et al. (2019). SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 6, 22. doi: 10.1038/s41438-018-0098-y
Jiang L., Wang Y., Li Q.-F., Björn L. O., He J.-X., Li S.-S. (2012). Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res. 22, 1046–1057. doi: 10.1038/cr.2012.34
Job N., Yadukrishnan P., Bursch K., Datta S., Johansson H. (2018). Two B-Box Proteins Regulate Photomorphogenesis by Oppositely Modulating HY5 through their Diverse C-Terminal Domains. Plant Physiol. 176, 2963–2976. doi: 10.1104/pp.17.00856
Kadomura-Ishikawa Y., Miyawaki K., Noji S., Takahashi A. (2013). Phototropin 2 is involved in blue light-induced anthocyanin accumulation in Fragaria x ananassa fruits. J. Plant Res. 126, 847–857. doi: 10.1007/s10265-013-0582-2
Kiferle C., Fantini E., Bassolino L., Povero G., Spelt C., Buti S., et al. (2015). Tomato R2R3-MYB proteins SlANT1 and SlAN2: Same protein activity, different roles. PloS One 10, e0136365. doi: 10.1371/journal.pone.0136365
Kim S., Hwang G., Lee S., Zhu J.-Y., Paik I., Nguyen T. T., et al. (2017). High ambient temperature represses anthocyanin biosynthesis through degradation of HY5. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01787
LaFountain A. M., Yuan Y. W. (2021). Repressors of anthocyanin biosynthesis. New Phytol. 231, 933–949. doi: 10.1111/nph.17397
Landi M., Agati G., Fini A., Guidi L., Sebastiani F., Tattini M. (2021). Unveiling the shade nature of cyanic leaves: A view from the “blue absorbing side” of anthocyanins. Plant Cell Environ. 44, 1119–1129. doi: 10.1111/pce.13818
Landi M., Tattini M., Gould K. S. (2015). Multiple functional roles of anthocyanins in plant-environment interactions. Environ. Exp. Bot. 119, 4–17. doi: 10.1016/j.envexpbot.2015.05.012
Lau O. S., Deng X. W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17, 584–593. doi: 10.1016/j.tplants.2012.05.004
Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., et al. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749. doi: 10.1105/tpc.106.047688
Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Levin I., Frankel P., Gilboa N., Tanny S., Lalazar A. (2003). The tomato dark green mutation is a novel allele of the tomato homolog of the DEETIOLATED1 gene. Theor. Appl. Genet. 106, 454–460. doi: 10.1007/s00122-002-1080-4
Li Y., Chen Y., Zhou L., You S., Deng H., Chen Y., et al. (2020). MicroTom metabolic network: rewiring tomato metabolic regulatory network throughout the growth cycle. Mol. Plant 13, 1203–1218. doi: 10.1016/j.molp.2020.06.005
Li Y.-Y., Mao K., Zhao C., Zhao X.-Y., Zhang H.-L., Shu H.-R., et al. (2012). MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160, 1011–1022. doi: 10.1104/pp.112.199703
Lin-Wang K., Micheletti D., Palmer J., Volz R., Lozano L., Espley R., et al. (2011). High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 34, 1176–1190. doi: 10.1111/j.1365-3040.2011.02316.x
Liu Y., Schouten R. E., Tikunov Y., Liu X., Visser R. G. F., Tan F., et al. (2022a). Blue light increases anthocyanin content and delays fruit ripening in purple pepper fruit. Postharvest Biol. Technol. 192, 112024. doi: 10.1016/j.postharvbio.2022.112024
Liu Y., Ye Y., Wang Y., Jiang L., Yue M., Tang L., et al. (2022b). B-box transcription factor faBBX22 promotes light-induced anthocyanin accumulation in strawberry (Fragaria × ananassa). Int. J. Mol. Sci. 23, 7757. doi: 10.3390/ijms23147757
Liu X., Zhang Q., Yang G., Zhang C., Dong H., Liu Y., et al. (2020). Pivotal roles of Tomato photoreceptor SlUVR8 in seedling development and UV-B stress tolerance. Biochem. Biophys. Res. Commun. 522, 177–183. doi: 10.1016/j.bbrc.2019.11.073
Lloyd A., Brockman A., Aguirre L., Campbell A., Bean A., Cantero A., et al. (2017). Advances in the MYB-bHLH-WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58, 1431–1441. doi: 10.1093/pcp/pcx075
Ma Y., Ma X., Gao X., Wu W., Zhou B. (2021). Light induced regulation pathway of anthocyanin biosynthesis in plants. Int. J. Mol. Sci. 22, 11116. doi: 10.3390/ijms222011116
Maier A., Schrader A., Kokkelink L., Falke C., Welter B., Iniesto E., et al. (2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 74, 638–651. doi: 10.1111/tpj.12153
Martin C., Butelli E., Petroni K., Tonelli C. (2011). How can research on plants contribute to promoting human health? Plant Cell 23, 1685–1699. doi: 10.1105/tpc.111.083279
Mattoo A. K., Dwivedi S. L., Dutt S., Singh B., Garg M., Ortiz R. (2022). Anthocyanin-rich vegetables for human consumption-focus on potato, sweetpotato and tomato. Int. J. Mol. Sci. 23, 2634. doi: 10.3390/ijms23052634
Menconi J. (2024). From light and temperature to genomes: genetic regulation of anthocyanin biosynthesis in Solanum lycopersicum fruit. Scuola Superiore Sant’Anna, Pisa, Italy.
Menconi J., Perata P., Gonzali S. (2023). Novel R2R3 MYB transcription factors regulate anthocyanin synthesis in Aubergine tomato plants. BMC Plant Biol. 23, 148. doi: 10.1186/s12870-023-04153-7
Menconi J., Perata P., Gonzali S. (2024). In pursuit of purple: anthocyanin biosynthesis in fruits of the tomato clade. Trends Plant Sci. 29, 589–604. doi: 10.1016/j.tplants.2023.12.010
Mes P. J., Boches P., Myers J. R. (2008). Characterization of tomatoes expressing anthocyanin in the fruit. J. Am. Soc Hortic. Sci. 133, 262–269. doi: 10.21273/JASHS.133.2.262
Montefiori M., Brendolise C., Dare A. P., Lin-Wang K., Davies K. M., Hellens R. P., et al. (2015). In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J. Exp. Bot. 66, 1427–1436. doi: 10.1093/jxb/eru494
Mustilli A. C., Fenzi F., Ciliento R., Alfano F., Bowler C. (1999). Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11, 145–157. doi: 10.1105/tpc.11.2.145
Neff M.M., Chory J. (1998). Genetic Interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118, 27–35. doi: 10.1104/pp.118.1.27
Nieto C., Catalán P., Luengo L. M., Legris M., López-Salmerón V., Davière J. M., et al. (2022). COP1 dynamics integrate conflicting seasonal light and thermal cues in the control of Arabidopsis elongation. Sci. Adv. 8, eabp8412. doi: 10.1126/sciadv.abp8412
Nukumizu Y., Wada T., Tominaga-Wada R. (2013). Tomato (Solanum lycopersicum) homologs of TRIPTYCHON (SlTRY) and GLABRA3 (SlGL3) are involved in anthocyanin accumulation. Plant Signal. Behav. 8, e24575. doi: 10.4161/psb.24575
Park Y.-J., Lee H.-J., Ha J.-H., Kim J. Y., Park C.-M. (2017). COP1 conveys warm temperature information to hypocotyl thermomorphogenesis. New Phytol. 215, 269–280. doi: 10.1111/nph.14581
Pattison R. J., Csukasi F., Zheng Y., Fei Z., van der Knaap E., Catala C. (2015). Comprehensive tissue-specific transcriptome analysis reveals distinct regulatory programs during early tomato fruit development. Plant Physiol. 168, 1684–1701. doi: 10.1104/pp.15.00287
Peng Y., Thrimawithana A. H., Cooney J. M., Jensen D. J., Espley R. V., Allan A. C. (2020). The proanthocyanin-related transcription factors MYBC1 and WRKY44 regulate branch points in the kiwifruit anthocyanin pathway. Sci. Rep. 10, 14161. doi: 10.1038/s41598-020-70977-0
Pesaresi P., Mizzotti C., Colombo M., Masiero S. (2014). Genetic regulation and structural changes during tomato fruit development and ripening. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00124
Podolec R., Ulm R. (2018). Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 45, 18–25. doi: 10.1016/j.pbi.2018.04.018
Povero G., Gonzali S., Bassolino L., Mazzucato A., Perata P. (2011). Transcriptional analysis in high-anthocyanin tomatoes reveals synergistic effect of Aft and atv genes. J. Plant Physiol. 168, 270–279. doi: 10.1016/j.jplph.2010.07.022
Qiu Z., Wang X., Gao J., Guo Y., Huang Z., Du Y. (2016). The Tomato Hoff-man’s anthocyaninless gene encodes a bHLH transcription factor involved in anthocyanin biosynthesis that is developmentally regulated and induced by low temperatures. PloS One 11, e0151067. doi: 10.1371/journal.pone.0151067
Qiu Z., Wang H., Li D., Yu B., Hui Q., Yan S., et al. (2019). Identification of candidate HY5-dependent and -independent regulators of anthocyanin biosynthesis in tomato. Plant Cell Physiol. 60, 643–656. doi: 10.1093/pcp/pcy236
Rai N., Morales L. O., Aphalo P. J. (2021). Perception of solar UV radiation by plants: photoreceptors and mechanisms. Plant Physiol. 186, 1382–1396. doi: 10.1093/plphys/kiab162
Raiola A., Rigano M. M., Calafiore R., Frusciante L., Barone A. (2014). Enhancing the health-promoting effects of tomato fruit for biofortified food. Mediators Inflamm. 2014, 139873. doi: 10.1155/2014/139873
Rick C. M. (1964). Biosystematic studies on Galapagos Island tomatoes. Occas. Paper Calif. Acad. Sci. 44, 59.
Rick C. M., Cisneros P., Chetelat R. T., Deverona J. W. (1994). Abg, a gene on chromosome 10 for purple fruit derived from S. lycopersiciodes. Rep. Tomato Genet. Coop. 44, 29–30.
Rombauts S., Déhais P., Van Montagu M., Rouzé P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Sestari I., Zsögön A., Garcia Rehder G., de Lira Teixeira L., Aymoto Hassimotto N. M., Purgatto E., et al. (2014). Near-isogenic lines enhancing ascorbic acid, anthocyanin and carotenoid content in tomato (Solanum lycopersicum L. cv Micro-Tom) as a tool to produce nutrient-rich fruits. Scientia Hortic. 75, 111–120. doi: 10.1016/j.scienta.2014.06.010
Sheerin D. J., Hiltbrunner A. (2017). Molecular mechanisms and ecological function of far-red light signalling. Plant Cell Environ. 40, 2509–2529. doi: 10.1111/pce.12915
Shin D. H., Choi M. G., Kim K., Bang G., Cho M., Choi S.-B., et al. (2013). HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 587, 1543–1547. doi: 10.1016/j.febslet.2013.03.037
Shin J., Park E., Choi G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49, 981–994. doi: 10.1111/j.1365-313X.2006.03021.x
Shinozaki Y., Nicolas P., Fernandez-Pozo N., Ma Q., Evanich D. J., Shi Y., et al. (2018). High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nat. Commun. 9, 364. doi: 10.1038/s41467-017-02782-9
Sun C., Deng L., Du M., Zhao J., Chen Q., Huang T., et al. (2020). A transcriptional network promotes anthocyanin biosynthesis in tomato mesocarp. Mol. Plant 13, 42–58. doi: 10.1016/j.molp.2019.10.010
Sunil L., Shetty N. P. (2022). Biosynthesis and regulation of anthocyanin pathway genes. Appl. Microbiol. Biotechnol. 106, 1783–1798. doi: 10.1007/s00253-022-11835-z
Suprun A. R., Kiselev K. V., Dubrovina A. S. (2023). Exogenously induced silencing of four MYB transcription repressor genes and activation of anthocyanin accumulation in solanum lycopersicum. Int. J. Mol. Sci. 24, 9344. doi: 10.3390/ijms24119344
Texteira R. T. (2020). Distinct responses to light in plants. Plants (Basel) 9, 894. doi: 10.3390/plants9070894
Tohge T., Zhang Y., Peterek S., Matros A., Rallapalli G., Tandròn Y.-A., et al. (2015). Ectopic expression of snapdragon transcription factors facilitates the identification of genes encoding enzymes of anthocyanin decoration in tomato. Plant J. 83, 686–704. doi: 10.1111/tpj.12920
Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., DePaepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Wang W., Wang P., Li X., Wang Y., Tian S., Qin G. (2021). The transcription factor SlHY5 regulates the ripening of tomato fruit at both the transcriptional and translational levels. Hortic. Res. 8, 83. doi: 10.1038/s41438-021-00523-0
Wei Z., Yang H., Shi J., Duan Y., Wu W., Lyu L., et al. (2023). Effects of different light wavelengths on fruit quality and gene expression of anthocyanin biosynthesis in blueberry (Vaccinium corymbosum). Cells. 12, 1225. doi: 10.3390/cells12091225
Winkel-Shirley B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Wu M., Xu X., Hu X., Liu Y., Cao H., Chan H., et al. (2020). SlMYB72 regulates the metabolism of chlorophylls, carotenoids, and flavonoids in tomato fruit. Plant Physiol. 183, 854–868. doi: 10.1104/pp.20.00156
Xu D. (2020). COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol. 228, 1748–1753. doi: 10.1111/nph.16296
Xu W., Dubos C., Lepiniec L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Xu Y., Liu X., Huang Y., Xia Z., Lian Z., Qian L., et al. (2022). Ethylene inhibits anthocyanin biosynthesis by repressing the R2R3-MYB regulator slAN2-like in tomato. Int. J. Mol. Sci. 23, 7648. doi: 10.3390/ijms23147648
Xue X., Duan Y., Wang J., Ma F., Li P. (2021). Nighttime temperatures and sunlight intensities interact to influence anthocyanin biosynthesis and photooxidative sunburn in “Fuji” Apple. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.694954
Yamashita H., Wada K. C., Inagaki N., Fujimoto Z., Yonemaru J.-I., Itoh H. (2023). Deciphering transcriptomic signatures explaining the phenotypic plasticity of nonheading lettuce genotypes under artificial light conditions. Plant Cell Environ. 46, 3971–3985. doi: 10.1111/pce.14677
Yan J., Liu J., Yang S., Jiang C., Liu Y., Zhang N., et al. (2023). Light quality regulates plant biomass and fruit quality through a photoreceptor-dependent HY5-LHC/CYCB module in tomato. Hortic. Res. 10, uhad219. doi: 10.1093/hr/uhad219
Yanagawa Y., Sullivan J. A., Komatsu S., Gusmaroli G., Suzuki G., Yin J., et al. (2004). Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18, 2172–2181. doi: 10.1101/gad.1229504
Yang G., Zhang C., Dong H., Liu X., Guo H., Tong B., et al. (2022). Activation and negative feedback regulation of SlHY5 transcription by the SlBBX20/21-SlHY5 transcription factor module in UV-B signaling. Plant Cell 34, 2038–2055. doi: 10.1093/plcell/koac064
You S., Wu Y., Li W., Liu X., Tang Q., Huang F., et al. (2024). SlERF.G3-Like mediates a hierarchical transcriptional cascade to regulate ripening and metabolic changes in tomato fruit. Plant Biotechnol. J. 22, 165–180. doi: 10.1111/pbi.14177
Zhang C., Zhang Q., Guo H., Yu X., Liang W., Chen Y., et al. (2021). Tomato SlRUP is a negative regulator of UV-B photomorphogenesis. Mol. Hortic. 1, 8. doi: 10.1186/s43897-021-00010-z
Zhang Y., Zheng S., Liu Z., Wang L., Bi Y. (2011). Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J. Plant Physiol. 168, 367–374. doi: 10.1016/j.jplph.2010.07.025
Keywords: Solanum lycopersicum, purple tomato, anthocyanins, MBW, DEETIOLATED 1 (DET1), ELONGATED HYPOCOTYL 5 (HY5), photoreceptors, light signaling
Citation: Gonzali S, Menconi J and Perata P (2025) Transcriptional survey of the light-induced anthocyanin pathway in non-GM purple tomatoes. Front. Plant Physiol. 2:1507833. doi: 10.3389/fphgy.2024.1507833
Received: 08 October 2024; Accepted: 13 December 2024;
Published: 23 January 2025.
Edited by:
Prasenjit Saha, Planet 13 Holdings, Inc., United StatesReviewed by:
Ashutosh Mukherjee, Vivekananda College, IndiaCopyright © 2025 Gonzali, Menconi and Perata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Silvia Gonzali, cy5nb256YWxpQHNhbnRhbm5hcGlzYS5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.