
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Physiol. , 18 December 2023
Sec. Photosynthesis and Metabolism
Volume 1 - 2023 | https://doi.org/10.3389/fphgy.2023.1330216
The primary function of proteolytic enzymes is the hydrolysis of peptide bonds. Enzymes responsible for catalyzing this reaction are commonly referred to as proteases. The vast majority of these enzymes belong to the class of hydrolases and operate in aqueous environments. However, there is a distinct group of proteases known as intramembrane proteases, which are integral membrane proteins capable of cleaving peptide bonds in the hydrophobic environment of biological membranes. Proteolysis serves several essential functions in plant cells, ranging from the degradation of damaged and unnecessary proteins to the removal of non-functional protein aggregates. It plays a role in the quality control system for proteins and even releases transcription factors from membrane proteins. Proteolytic processes are indispensable at every stage of plant development and allow for the modification of the cell’s protein composition based on developmental needs and environmental requirements. Proteases are also enzymatic components of the cell that facilitate the efficient regeneration of many key metabolic pathways, such as photosynthesis and respiration. The response of plant cells and the entire plant organism to various biotic and abiotic stresses often requires the remodeling of metabolic pathways, the regeneration of key enzymatic complexes, or changes in the protein profile. By participating in all these processes, proteases constitute a crucial element of the cellular response to environmental stresses. The aim of this work is to review the role that individual proteases play in the response of plant cells to abiotic stress factors, such as drought, salinity, cold, temperature, and light.
Proteolysis is a process in which peptide bonds within proteins are broken, resulting in the formation of shorter peptides that can then be further broken down into individual amino acids. Enzymes that catalyze these reactions are known as proteolytic enzymes, also called proteases or peptidases. These terms are often used interchangeably and refer to any enzyme belonging to the hydrolase class, whose role is to hydrolyze peptide bonds. Proteases are common enzymes throughout the whole world of living organisms from bacteria to humans as well as in the viral particles. Proteolytic enzymes can serve various functions in cells, being involved in the course of multiple metabolic pathways. They also exhibit a wide range of substrate specificity. For instance, pepsin found in the stomach of mammals, facilitating the breakdown of proteins in food, is a relatively non-specific enzyme, which is advantageous in the digestion process. The task of pepsin is to degrade as many protein chains as possible, rather than being substrate specific. In contrast, in terms of enzymatic specificity, the serine protease NS3-4A present in HCV virus particles represents an extreme example. This enzyme plays a crucial role in the virus replication process in chimpanzee hosts and is responsible for cleavage HCV polyprotein at very precisely defined place (Lin, 2006).
Plant proteases perform a variety of functions at every stage of a plant’s life. These enzymes are involved in the processes of programmed cell death, plant growth, aging, fruit, and seed ripening, and participate in the process of hydrolysis and mobilization of other proteins accumulated in seeds and cereal grains (Liu et al., 2018; Martinez et al., 2019; Tornkvist et al., 2019). In addition, they play an important role in maintaining the proper course of many metabolic processes, such as respiration and photosynthesis. Finally, proteases play key roles in plant responses to abiotic stress factors (Velasco-Arroyo et al., 2016; Velasco-Arroyo et al., 2018; Gomez-Sanchez et al., 2019; James et al., 2019) and the attack of pathogens (Misas-Villamil et al., 2016; Diaz-Mendoza et al., 2017).
The classification of proteolytic enzymes may be different and depends on the adopted criterion, which may be the structure of the active center of the enzyme, specificity of action, site of cleavage of the polypeptide chain, etc. Taking as the first classification criterion the place within the protein chain where the protease hydrolyzes it, two basic groups of proteases are distinguished. The first group in this approach are exopeptidases that cut the protein sequence at its ends. Exopeptidases can cut off the final amino acids of the protein chain both at the C-end of the protein and at its N-end, hence they are referred to as C- or N-exopeptidases. The second group are endopeptidases that catalyze the process of nucleophilic attack on the carbon atom of the peptide bond located inside the protein chain. As a result of this attack, an unstable complex is formed, the breakdown of which involves the degradation of the peptide bond and the formation of two smaller polypeptide chains (Davies, 1990).
Endopeptidases are a very broad group of proteolytic enzymes that differ, among other things, in the structure of their active site. Taking these differences into account, six main types of endopeptidases can be distinguished: aspartic, cysteine, glutamine, serine, threonine endopeptidases and metalloendoproteases. The different structure of the active center is associated with a slightly different mechanism of nucleophilic attack on the protein’s peptide bond, however, all these enzymes catalyze proteolysis reactions using a water molecule and operate only in an aquatic environment in contrast to a separate group consists of the so-called intramembrane proteases that are able to catalyze the reaction of breaking peptide bonds within biological membranes, i.e. in an extremely lipophilic environment (Wolfe and Kopan, 2004).
Oligopeptidases, which are distinguished as a subgroup of endopeptidases, are enzymes capable of cleaving short amino acid chains, but are unable to digest entire, large protein molecules. This property results from the structure of oligopeptidases themselves, whose active site is located at the end of a narrow cavity where only smaller peptides can reach. The size of the peptide that is the substrate for the protease is crucial in the process of the so-called “docking”, i.e., attaching the substrate to the active center of the protease. However, the spatial structure of the potential substrate is also important for the possibility of catalysis of the hydrolysis reaction itself. In some cases, the structure of the peptide may inhibit the activity of catalyzing enzymes (Jacchieri et al., 2009).
In addition to the properties of the potential substrate, size, spatial structure or amino acid sequence of the peptide, environmental conditions are an important factor influencing the activity of proteases. Thus, the activity of some cytosolic oligopeptidases depends on the current redox status of the environment. An example is zinc timet oligopeptidases - TOP, belonging to the M3 type of metalloproteases, present in both animal and plant cells. TOP is an enzyme whose activity is regulated by the concentration of hydrogen peroxide. Studies on the human embryonic kidney cell line HEK293 have shown that increasing the concentration of hydrogen peroxide in cells leads to an increase in the activity of TOP protease, which reaches its maximum activity at a hydrogen peroxide concentration of 1.2nM (Icimoto et al., 2017).
Plants throughout their lives are exposed to a range of unfavorable environmental factors. Abiotic stress factors include primarily drought, salinity, cold, high temperature, excessive radiation, pesticides, and heavy metals. These factors interact with plants, often leading to a series of unfavorable functional and structural changes. However, during evolution, plants have developed a range of mechanisms called adaptations that allow them to minimize or tolerate the effects of stress. Plant proteases play an extremely important role in these processes, degrading damaged proteins that hinder the efficient functioning of specific metabolic pathways. Thus, proteases contribute to the efficient regeneration of metabolic pathways or their redirection to other paths better suited to changed environmental conditions (Figure 1 and Table 1).
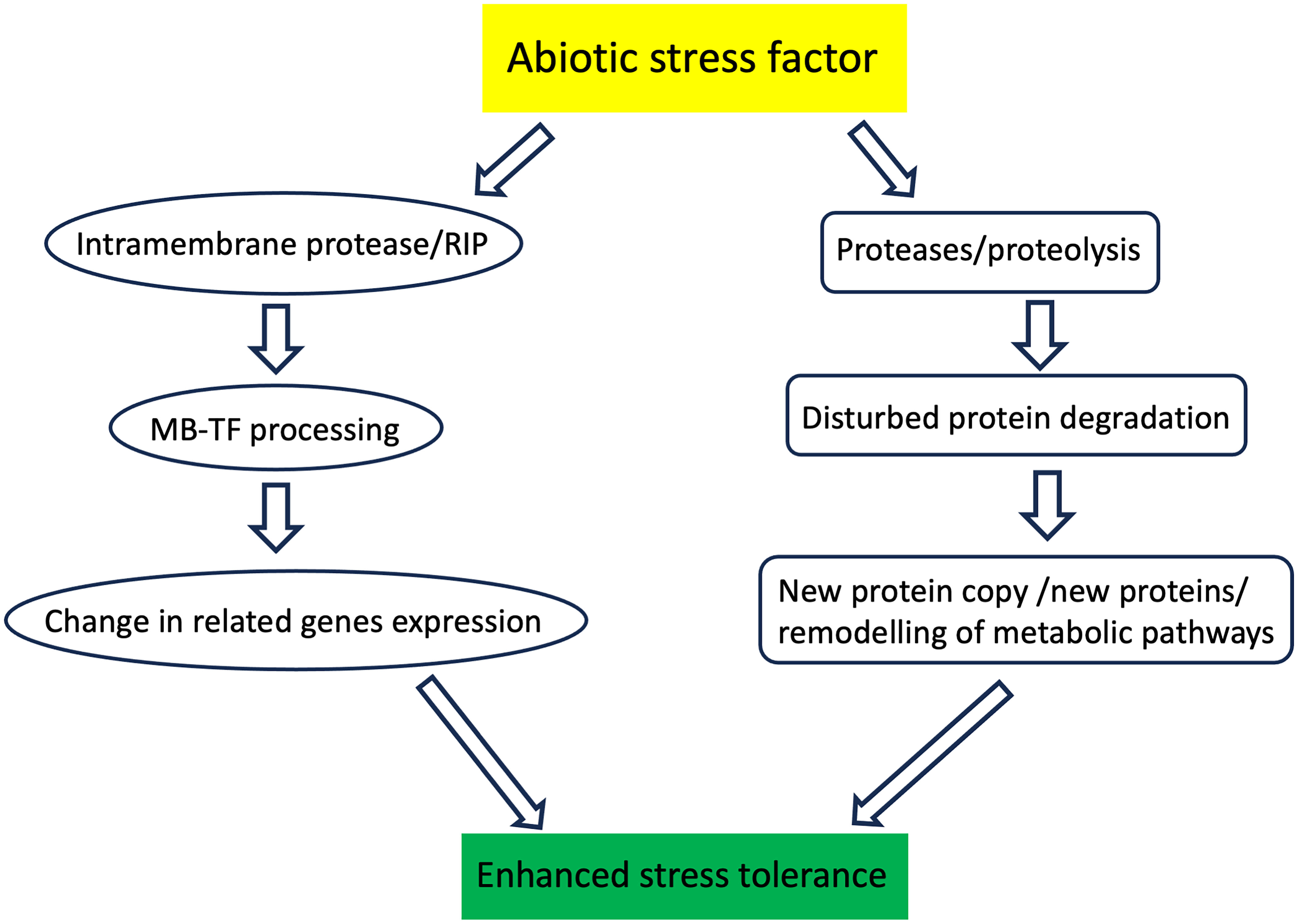
Figure 1 Schematic representation of the plant proteases action during stressful conditions. Intramembrane proteases cleave the protein transcription factor anchored in the biological membrane. This process, called regulatory intramembrane proteolysis (RIP), enables the activation of transcription factors and, ultimately, changes in the expression of related genes. The other proteases degrade proteins damaged during plant stress exposure. Degraded proteins are replaced with new functional copies. Proteases also degrade unnecessary proteins, which also enables remodeling the metabolic pathways.
Peptides produced as a result of protease activity can also regulate the production of reactive oxygen species (Möller and Sweetlove, 2010) and thus participate in signal transduction pathways generated by reactive oxygen species. Proteases are therefore involved in the transmission of information over long distances between different tissues and organs. Proteases are involved in this aspect, among other things, in regulatory processes associated with abscisic acid (Schaller, 2004; van der Hoorn, 2008; Kato and Sakamoto, 2010; Kidric et al., 2014; van Wijk, 2015; Diaz-Mendoza et al., 2016; Sharma and Gayen, 2021).
The regulation of various plant developmental processes, starting from seed germination and the mobilization of storage materials, and extending to the formation and development of specific cell organelles, such as plastids, requires the activity of proteases (Schaller, 2004; van der Hoorn, 2008; Kato and Sakamoto, 2010; Kidric et al., 2014; van Wijk, 2015; Diaz-Mendoza et al., 2016; Sharma and Gayen, 2021).
Furthermore, intramembrane proteases are suspected of degrading small polypeptides anchored in biological membranes, leading to the release of their fragments that can function as transcription factors or elements of polymerase-stabilizing complexes (Adamiec et al., 2017; Adamiec et al., 2018). Such activity is associated with changes in the gene expression profile. This process is so significant that it has been described as intramembrane regulatory proteolysis.
The main goal of this study is to examine the role that plant proteases play in the response of plants to abiotic stress factors, such as drought, salinity, low and high temperatures, and excessive light intensity.
Drought, much like excessive water, is considered as one of the main causes of reduced yields and plant survival, especially in the case of crops such as corn, rice, and wheat (Furlan et al., 2016).
Lack of rainfall and high temperatures that increase evaporation intensity led to a situation in which the amount of transpired water by the plant exceed its capacity to its absorption by the roots. The consequence of such a situation is the occurrence of osmotic stress within the plant (Bray, 1997). Water deficit in cells results in structural and functional changes in cell walls and cell membranes, altering their permeability and disrupting basic metabolic pathways such as photosynthesis and cellular respiration (Kumar et al., 2018; Kapoor et al., 2020). One of the most common consequences of drought in plants, as well as other stresses, is secondary oxidative stress, caused by the generation of large quantities of reactive oxygen species (ROS), which damage both proteins and nucleic acids as well as lipids (de Carvalho, 2008). Naturally, over the course of evolution, plants have developed protective mechanisms to minimize the damage caused by drought.
One of the initial responses of plants to emerging soil water deficit is the synthesis of abscisic acid (ABA), which is subsequently transported to the leaves where it induces the closure of stomata (Moloi and Ngara, 2023). This restricts water loss from the leaves. However, stomatal closure is associated with limited gas exchange, thereby inhibiting the supply of CO2 to the mesophyll cells. As a result, there is a decrease of CO2 assimilation and the photosynthetic electron transport chain, leading to the generation of increased levels of ROS that need to be scavenged by the cellular machinery; otherwise, damage to lipids, proteins and nucleic acids may occur (Wang et al., 2003; Sharma et al., 2012; Moloi and Ngara, 2023). However, ABA is not the only hormone participating in a plant’s response to drought. The literature extensively describes the involvement of other hormones such as jasmonic acid, ethylene, gibberellins, cytokinins, and auxins in the comprehensive response to drought stress (Moloi and Ngara, 2023).
Drought stress is a triggering factor for the upregulation of many genes encoding proteases belonging to various classes. Available data show that in many species, the organs in which the expression of genes encoding specific proteases increases are leaves and roots (Moloi and Ngara, 2023). In A. thaliana and in Coffea, one of the key enzymes determining the plant’s drought resistance is the aspartyl protease guard cell, ASPG1 (Yao et al., 2012; Fernandes et al., 2021). Overexpression of this protein is associated with lower stomatal density and reduced stomatal aperture, resulting in increased drought tolerance in such plants (Sebastian et al., 2020). The increased drought resistance effect in A. thaliana is also observed in plants overexpressing the LON4 protein (Li et al., 2010), which is believed to play a significant role in maintaining mitochondrial homeostasis under drought conditions. LON4 is an ATP-dependent serine protease located in mitochondria. The nuclear gene encoding this protease shows increased expression under drought conditions (Pinti et al., 2016; Sharma and Gayen, 2021). It has long been known that the early response of A. thaliana to dehydration also includes the induction of the expression of ClpA and ClpB proteins (Kiyosue et al., 1993). These proteins are part of the larger Clp protease complex, which recognizes and degrades misfolded or damaged proteins. ClpA is a chaperone-like protein, which is responsible for substrate recognition, while ClpB is described as a disaggregase, facilitating the disaggregation of previously misfolded or aggregated proteins.
In the leaves of drought tolerant varieties of Barley and Wheat, an increase in the content of ClpP proteases is observed (Ashoub et al., 2013; Chmielewska et al., 2016), as well as ClpD1 protease in the case of Rice (Wu et al., 2016). ClpD1 plays an important role in the degradation of damaged proteins in plastids (Kato and Sakamoto, 2010; Ali and Baek, 2020; Moloi and Ngara, 2023), and it is encoded by the nuclear so-called gene OsClpD1, which is induced under drought conditions (Wu et al., 2016). In the leaves of drought-resistant Barley varieties, an increase in the expression of the metalloprotease FtsH1 was also observed (Wang et al., 2015). FtsH1 is a part of the chloroplast heterocomplex FtsH “Type A,” along with Ftsh5. This complex participates in the repair of damaged polypeptide PsbA (D1 protein), a component of the photosystem II reaction center (Yu et al., 2004). FtsH heterocomplex in A. thaliana has been found to be responsible for the degradation of the desiccation related damaged of photosystem II (PSII) polypeptides - Lhcb1 and Lhcb2 (Luciński and Jackowski, 2013).
Drought tolerance in Wheat is also associated with the cysteine protease SUMO (small ubiquitin-like modifier), an enzyme commonly found in plants and yeast (Le Roux et al., 2019). Additionally, in the leaves of drought-resistant Wheat varieties, increased expression of alpha-type proteasome protein (Faghani et al., 2015), as well as type II metacaspases and leucine aminopeptidases, is noted (Ford et al., 2011). In the case of leucine aminopeptidase, it is observed that its levels do not return to their previous levels after drought stress subsides, but they remain increased (Ford et al., 2011). Since leucine aminopeptidase participates in the activation of some proteins and metabolic turnover, some authors suggest that its increased level upon rehydration may be the result of an important role of this peptidase in the processes of changing the protein composition of cells regenerating after drought stress (Moloi and Ngara, 2023).
Salinity is one of the most significant environmental factors responsible for a substantial decrease in the chemical potential of soil solution water. This can lead to situations where plants lose the ability to efficiently take up water from the soil. High soil salinity results in changes in the structure and permeability of cell membranes, leading to disruptions in ion balance within cells (Shah et al., 2017). Furthermore, deficiencies in mineral nutrients and secondary oxidative stress occur due to the excessive generation of ROS, resulting in damage to nucleic acids and overproduction of oxidized proteins. This salinity-induced genotoxicity is also associated with a decrease in photosynthetic efficiency and disturbances in the mitochondrial electron transport chain (Sharma and Gayen, 2021).
Plants have evolved a range of adaptations to minimize the negative effects of excessive salinity. The plant’s response includes both enzymatic and non-enzymatic reactions. Accumulation of proline, sugars, and other osmotically active compounds is one of the elements that lower the water potential of the cell sap and extend the cell’s ability to take up water in unfavorable osmotic conditions.
Since salinity induces a strong synthesis of ROS in plant cells, there is a need for the degradation of oxidized proteins. Cysteine proteases play a significant role in these processes (van der Hoorn, 2008), especially proteins RD19 and RD21 belonging to the papain-like cysteine protease family. A strong induction of the expression of genes encoding them has been observed under salinity stress conditions (Koizumi et al., 1993). It is suggested that changes in the osmotic potential of cells are the direct stimulus for the alteration in the expression levels of genes RD19 and RD21. The levels of RD19 and RD21 did not change significantly under low or high-temperature conditions or in the presence of ABA (Koizumi et al., 1993). Another cysteine protease whose levels significantly increase in response to high salt concentrations in the environment is Cyp15 from Pisum sativum. The activity of this protease is linked to changes in the levels of various mRNA molecules under salt stress conditions (Jones and Mullet, 1995).
A separate class of proteases involved in the response to high salinity is serine proteases. It has been shown that under conditions of high salinity in the cyanobacterium species Halothece sp., there is a significant increase in the expression of the gene encoding the protease HtrA2 (Deg). This enzyme is responsible for the degradation of misfolded proteins (Patipong et al., 2020). Protease HtrA2 has an optimum pH of 8.0 and remains active under conditions of high salt concentration and high temperature. This protein was detected in both the soluble protein fraction and within the cell membrane and thylakoid membranes. Exposure of cells to high salt stress and high-temperature stress caused a significant change in the subcellular localization of HtrA2. Under such conditions, the protein mostly localized to the cell membrane (Patipong et al., 2020). In higher plants, within the thylakoid membranes of chloroplasts, there are also cysteine, ATP-independent proteases like Deg (Huesgen et al., 2005; Huesgen et al., 2006; Sun et al., 2007; Chi et al., 2012; Schuhmann et al., 2012, Butenko et al., 2018). One of them is Deg2, which is involved in the degradation of the Lhcb6 polypeptide under salt stress conditions (Luciński et al., 2011). The Lhcb6 polypeptide is part of the minor antenna complexes of PSII and plays an important role in the formation of PSII-LHCII supercomplexes. The absence of Lhcb6 is associated with the preferential formation of C2S2-type supercomplexes and an increase in non-photochemical quenching of chlorophyll a fluorescence (Illikova et al., 2021). Therefore, protease Deg2 may play a significant role in maintaining the proper composition of PSII-LHCII supercomplexes and the efficiency of photosynthesis under salt stress conditions.
The stress associated with low temperatures is most commonly considered as chilling (for temperature ranges from 0°C to slightly below 15°C) and freezing (below 0°C). Prolonged exposure of plants to low temperatures results in the inhibition or slowing down of development, a decrease in growth rate, and a drop in productivity. Low temperatures significantly affect the efficiency of basic metabolic pathways such as respiration and photosynthesis. Therefore, the ability to cope with low temperatures is crucial for most plants living on Earth. The photosynthetic conversion of light into energy stored in chemical bonds and CO2 assimilation are processes that are highly temperature dependent. Low temperatures reduce the efficiency of reactions within PSII and decrease the synthesis of the large subunit of Rubisco, the enzyme directly responsible for CO2 assimilation in the Calvin-Benson cycle (Sharma and Gayen, 2021). Such changes necessarily imply a decrease in photosynthesis efficiency.
There are several proteases located in the chloroplasts, which mostly belong to FtsH, Deg, and Clp and play a significant role in a range of processes related to the quality control of chloroplast proteins and in maintaining chloroplast homeostasis (van Wijk, 2015). Some of these proteases also have importance in plant acclimatization to low temperatures. Under low-temperature conditions, a decrease in the content of polypeptides of the main energy antenna of PSII (LHCII), namely Lhcb1 and Lhcb2, has been observed. The degradation of these proteins in low-temperature conditions is attributed to the heterocomplex AtFtsH, located in the thylakoid membranes (Luciński and Jackowski, 2013). It is also possible that the inner membrane protease S2P2, located within chloroplasts, plays a role in shaping cold resistance in A. thaliana (Wang et al., 2016). S2P2 is a metalloprotease belonging to the class of site-2-proteases (S2P), which are known as enzymes that release transcription factors from membranes. Therefore, it can be imagined that the involvement of S2P2 in shaping cold resistance is more related to shaping the expression profiles of certain genes rather than the direct degradation of proteins damaged by the action of a stress factor.
High temperature has a significant impact on the development, growth, flowering, and productivity of plants. It also alters the rate and efficiency of metabolic pathways. Throughout evolution, plants have developed various mechanisms that allow them to rapidly respond to temperature changes in their environment and, consequently, adapt quickly to changing temperatures. Therefore, the temperature values that are harmful to plants are largely species-specific (Sharma and Gayen, 2021). Plants often respond to the presence of temperature stress factors through changes in the composition of membrane lipids, increased activity of stress-related transcription factors, alterations in the course and regulation of metabolic pathways, and various detoxification pathways (Kai and Iba, 2014). Proteins are molecules that undergo significant changes due to temperature stress. Dangerous phenomena for plants include protein denaturation, improper folding, and, consequently, changes in spatial structure. Protein denaturation itself is an irreversible process in which proteins lose their biological activity. Improper folding can lead to the formation of aggregates, preventing proper cell function. The consequences of such a state can be equally severe. In plant cells, protein aggregates can cause endoplasmic reticulum dysfunction, known as ER stress (Li and Sun, 2021). To prevent such situations or mitigate their consequences, cells have a range of structures and mechanisms. Firstly, lysosomes and proteasomes should be mentioned, whose primary role is the degradation of damaged proteins. Additionally, plant cells contain special chaperone proteins, known as heat shock proteins (HSP; Burgos et al., 2020).
Proteases also play a significant role in the response of plant cells to temperature stress. In the green tissues of leaves with constitutive expression, Clp proteases are involved (Sharma and Gayen, 2021). These serine proteases, ATP-dependent, play a crucial role in the functioning and development of plastids by removing improperly folded, aggregated, or otherwise unwanted proteins. The plant Clp system consists of a tetradecameric proteolytic core with catalytically active ClpP subunits and inactive ClpR subunits, hexameric ATP-dependent chaperone proteins, and adaptor proteins (Nishimura and van Wijk, 2015). ClpB, ClpC, and ClpD subunits function as chaperone proteins (Lee et al., 2007). In A. thaliana cells, these proteins are essential for plant acclimatization to high temperatures. ClpB is constitutively expressed in chloroplasts and mitochondria in A. thaliana (Lee et al., 2007) and P. lunatus (Keeler et al., 2000). On the other hand, strong upregulation of the ClpB gene was observed in O. sativa cells in response to high temperatures. Moreover, A. thaliana mutants lacking the ClpB protein grew significantly slower than the wild-type form, and under high-temperature conditions, the absence of this protein in mutants led to premature plant death (Lee et al., 2007).
The FtsH metalloproteases are crucial for the functioning of chloroplasts and mitochondria under high-temperature conditions. These proteases are a fundamental component that determines a plant’s ability to survive in high-temperature conditions (Langer, 2000). In chloroplasts, the FtsH11 protease is essential for the functioning of photosystem I (PSI) and PSII under high-temperature conditions. A. thaliana mutants lacking FtsH11 showed significantly reduced efficiency in PSI and PSII reactions (Chen et al., 2018). Chlorophyll a fluorescence analyses indicate that the presence of FtsH11 is necessary for regulating the response of photosystems to high-temperature stress (Chen et al., 2018).
The mitochondrial protease FtsH4 and OMA1 are crucial for A. thaliana’s acclimatization to moderate, prolonged temperature stress (Maziak et al., 2021). Mutants lacking FtsH4 or OMA1 displayed a range of phenotypic changes when grown at 30°C, although their phenotype remained unchanged compared to wild-type plants when grown at 22°C (Smakowska et al., 2016; Migdal et al., 2017). Phenotypic changes observed in ftsh4 mutants included delayed germination, reduced rosette size, and shorter root length. Similar changes in rosette size and root length were observed in oma1 mutants (Smakowska et al., 2016; Migdal et al., 2017; Maziak et al., 2021). These phenotypic effects are associated with the accumulation of protein aggregates composed of small heat shock proteins (sHSPs). The protease and chaperone activities of FtsH4 and the protease activity of OMA1 are crucial for protecting mitochondria from the formation of these aggregates (Maziak et al., 2021). Additionally, the FtsH4 protease plays a significant role in regulating the developmental processes of A. thaliana by mediating a peroxidase-dependent interplay between hydrogen peroxide and auxin homeostasis (Zhang et al., 2014).
For the proper functioning of plants under elevated temperature conditions, another mitochondrial protease, Deg10, is also essential. The absence of this protein in A. thaliana mutants was associated with disruptions in root elongation, reduced seed production, and changes in the content of proteins that build components of the mitochondrial respiratory chain (Huber et al., 2019).
Deg proteases also play a significant role in the functioning of chloroplasts and their photosynthetic apparatus. Under high-temperature conditions, damage can occur to, among others, the polypeptide Lhcb6, which is subsequently intensively degraded. Experiments conducted using deg2 mutants have determined that the degradation of Lhcb6 under temperature stress, as well as under salt stress, is carried out by the Deg2 protease (Luciński et al., 2011).
Light is a crucial factor that determines the functioning of plants and the existence of life on our planet. Light energy is converted into chemical energy, which is used in biochemical reactions in the process of photosynthesis. Simultaneously, photosynthesis is the primary source of oxygen in the Earth’s atmosphere. Excessive light intensity can lead to the phenomenon of photoinhibition, and prolonged light stress can result in damage to key proteins that make up the photosynthetic complexes in thylakoid membranes (i.e., Haußühl et al., 2001; Luciński and Jackowski, 2006; Sharma and Gayen, 2021). As a result, there is a limitation or inhibition of the light reactions of photosynthesis and a decrease in the efficiency of CO2 assimilation. Chloroplast proteases play a role in plant acclimatization to high light intensities. They are responsible for the degradation of light-damaged proteins, thereby enabling the regeneration of entire photosynthetic complexes.
The protein most susceptible to photodamage, especially, is the polypeptide PsbA (D1), which, along with PsbD (D2), forms the core of the PSII reaction center (Baena-Gonzales and Aro, 2002; Luciński and Jackowski, 2006; Sun et al., 2007). Damage to PsbA leads to the inactivation of the entire PSII complex and necessitates its repair by replacing the damaged PsbA with a new, functional copy (Baena-Gonzales and Aro, 2002). The degradation of photodamaged PsbA is a multi-step process involving proteases from different classes. It is believed that proteases Deg and FtsH play a crucial role in this process. Deg1 and Deg2 proteases perform the initial proteolytic cleavage within the hydrophilic loops connecting the individual transmembrane domains of the PsbA protein (Chassin et al., 2002; Kapri-Perdes et al., 2007). There are also reports in the literature suggesting that the Deg5/Deg8 protease complex is also involved in this process (Sun et al., 2007). Likely, the protease Deg1 cleaves the CD loop, while the Deg5/Deg8 complex cleaves the CD loop of photodamaged PsbA (Kapri-Perdes et al., 2007). Furthermore, high light intensity stimulates the activation of the Deg1 protease by inducing the transformation of an inactive monomer into an active proteolytic hexamer (Kley et al., 2011). All Deg proteases perform peptide bond hydrolysis in an ATP-independent manner. The degradation products generated by Deg proteases are subsequently digested in an ATP-dependent manner by the FtsH protease (Sakamoto et al., 2002; Sakamoto et al., 2003; Kato and Sakamoto, 2018). It has been shown that the degradation of the 23 kDa fragment of PsbA is mediated by the FtsH1 protease (Lindahl et al., 2000), although other experiments indicate the involvement of FtsH2 and FtsH5 proteases, which are part of the FtsH heterocomplex (Sakamoto et al., 2002, 2003, Kato and Sakamoto, 2018). In the case of Synechocystis spp. cells, the degradation of D1 is attributed to the FtsH2/FtsH3 heterocomplex (Krynicka et al., 2015).
Photodamaged PsbA is not the sole proteolytic target of chloroplast proteases under high light stress conditions. Polypeptides Lhcb1 and Lhcb2, damaged under light stress conditions, can be degraded by the chloroplast heterocomplex FtsH (Luciński and Jackowski, 2013). Protease Deg1 is also suspected of degrading other PSII polypeptides, specifically PsbS and PsbO. Furthermore, based on in vitro experimental results, it is attributed the ability to degrade the polypeptide Lhcb4 and Lhcb5, which constitute the polypeptide moiety of CP29 and CP24, respectively (Chassin et al., 2002; Kapri-Perdes et al., 2007; Zienkiewicz et al., 2011).
On the other hand, protease Deg2 is responsible for the degradation of polypeptide Lhcb6, similarly under temperature and salt stress conditions (Luciński et al., 2011).
The chloroplast subunit of the Clp complex with chaperone activity, ClpB3, is also upregulated under high light stress conditions (Adamiec et al., 2011). It has been demonstrated that the gene encoding ClpB3 contains specific cis-regulatory elements to which transcription factor PAP1 can bind. It is suggested that PAP1 acts as an intermediary in the induction of CLPB3 gene expression under stress conditions (Adamiec et al., 2011).
High light intensity also enhances the expression of the gene encoding the pseudo-protease EGY3 (similar to what occurs under temperature stress) and the protease EGY2. However, despite demonstrated proteolytic activity of EGY2 (Chen et al., 2012), its substrates are generally not known. However, there are speculations that such a substrate may be the chloroplast transcription factor pTAC16, which is able to interact with chloroplast RNA polymerase (Adamiec et al., 2018 and unpublish data). It is known that its absence significantly alters the levels of PsbA, PsbD, and PsbC polypeptides by affecting the expression of the genes that encode them (Adamiec et al., 2018). Therefore, EGY2 may be another protein with protease activity that plays a significant role in a plant’s response to light stress.
Intramembrane proteases are a relatively recently discovered, remarkable group of proteases. What distinguishes these enzymes from other proteases is their ability to hydrolyze the peptide bond within the lipid bilayer of the biological membrane (Chen et al., 2012; Adam, 2013; Adamiec et al., 2017). Despite the uniqueness of this process, it occurs widely in practically all living organisms. Intramembrane proteases are classified into several different families, including site-2-proteases, rhomboid proteases, presenilins, and signal peptide peptidases. These enzymes play a significant role in plant developmental processes and in the course of many metabolic pathways, being involved in protein quality control, cell adhesion, membrane remodeling, and cell signaling (Adam, 2013; Schneider and Glickman, 2013; Adamiec et al., 2017).
It is often believed that the primary function of these proteases is to cleave the polypeptide chain, releasing membrane-anchored transcription factors. This activity ultimately leads to changes in the expression profile of certain genes, a process referred to as regulated intramembrane proteolysis (RIP) (Figure 1) (Adam, 2013; Schneider and Glickman, 2013, Adamiec et al., 2018; Adamiec et al., 2021). The first substrate described within the RIP framework was the endoplasmic reticulum (ER)-associated transcription factor bZIP28 (Liu et al., 2007a; Tajima et al., 2008; Adam, 2013; Sun et al., 2015; De Backer et al., 2022). It has been demonstrated that the release of bZIP28 from the ER membrane to the cytoplasm allows it to migrate to the cell nucleus, where it induces the expression of genes related to the plant’s response to salt stress (Liu et al., 2007b; Tajima et al., 2008). It is possible that the protease directly responsible for releasing bZIP28 from the ER is S1P or S2P (Liu et al., 2007a; Tajima et al., 2008). Another transcription factor, whose release from the ER membrane is associated with the response to stress is bZIP60 (Iwata and Koizumi, 2005). Accumulation in the cell nucleus of the soluble domain of this protein is linked to increased transcription of BIP proteins, which belong to the HSP70 family and are typical representatives of the plant’s ER stress response (Iwata et al., 2008). S1P and S2P proteases are once again involved in the release of the bZIP60 factor from ER membranes. Intramembrane proteases are also suspected of releasing the transcription factors belonging to the NAC family: NTL1, NTL7 and NTL8 from endoplasmic reticulum membranes and NTL6 from the plasma membrane. NTL8 is associated with the regulation of flowering processes in A. thaliana under saline and low temperature conditions (Kim et al., 2007). NT1 and NTL7 is activated under oxidative stress conditions conditions (De Backer et al., 2022), while NTL6 and NTL8 are related to salinity and low temperature (Kim et al., 2007, Seo , 2008, 2010) (Table 2).
Despite the still limited knowledge about the role of intramembrane proteases, some of them are suspected to be involved in the response of plant cells to various stress factors. In this context, examples include the chloroplasts’ rhomboid protease RBL10, which may be involved in photoprotective mechanisms in response to low temperatures (Thompson et al., 2012). On the other hand, the plasma membrane-localized RBL14 seems to be involved in the response to elevated temperature (Lee et al., 2015). Rhomboid proteases are also suspected to be involved in the response to H2O2-mediated stress signaling by releasing the ANAC017 transcription factor from ER membranes (Ng et al., 2013) (Table 2).
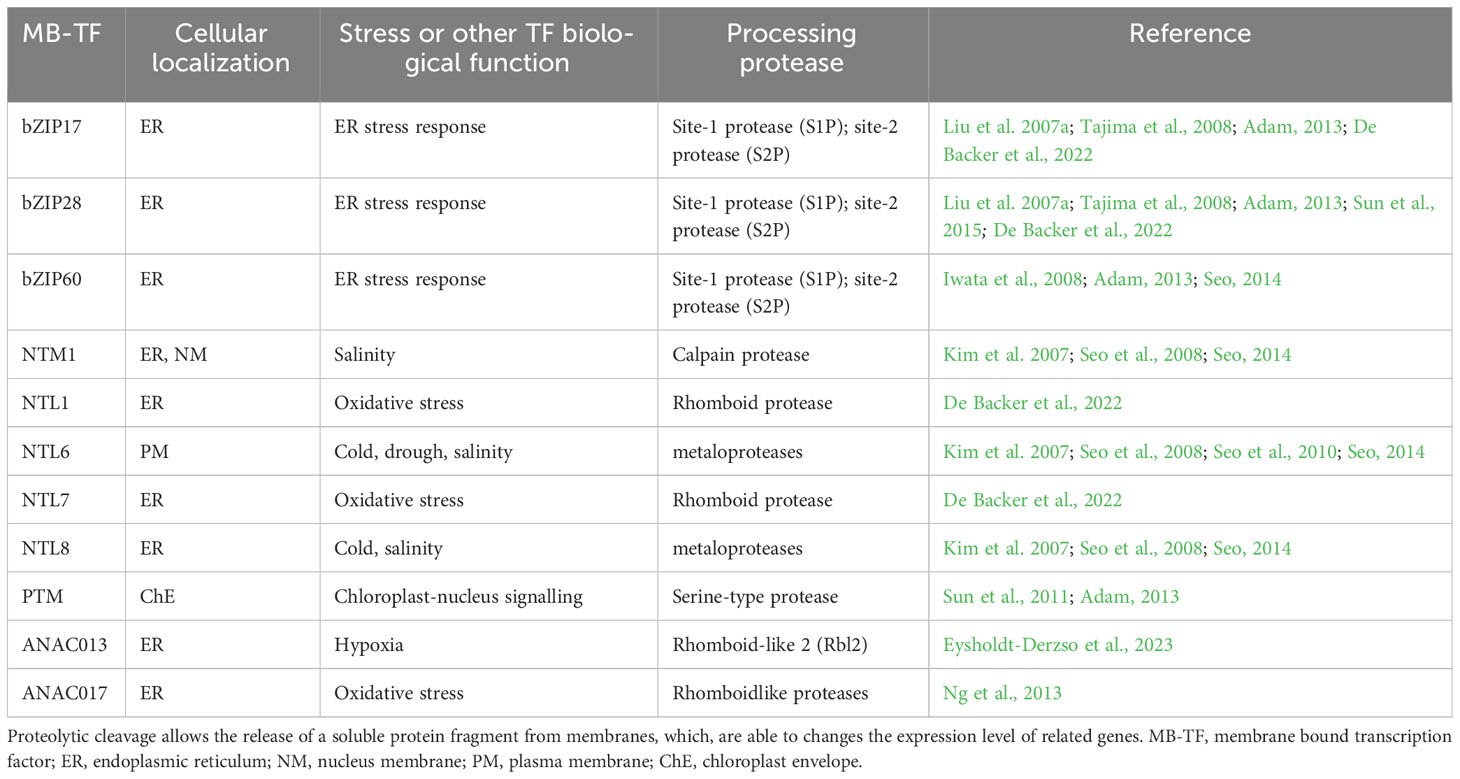
Table 2 Membrane bound transcription factors that are substrates for intramembrane proteases in different stress conditions.
Within chloroplasts, intramembrane proteases belonging to the S2P family, namely EGY1 and EGY2, may also influence the functioning of these organelles under light stress conditions. Both proteins are located within the thylakoid membranes (Chen et al., 2005; Chen et al., 2012). It has been shown that Arabidopsis thaliana mutants lacking EGY1 or EGY2 exhibit changes in the stoichiometry of proteins building the PSII core and LHCII antennas, resulting in increased sensitivity to photoinhibition (Adamiec et al., 2018; Adamiec et al., 2022). There are also suggestions that EGY2 may affect the expression of chloroplast genes PSBA, PSBC, and PSBD by releasing the protein factor pTAC16 from the thylakoid membranes. On the other hand, the protein EGY3, considered a pseudoprotease due to the lack of proteolytic activity, seems to play a significant role under salt stress conditions, influencing the amount of H2O2 produced in chloroplasts by stabilizing copper/zinc superoxide dismutase 2 (CSD2; Zhuang et al., 2021).
Programmed Cell Death (PCD) is an essential part of the plant life cycle, occurring throughout the development of a plant, from embryogenesis to the death of the entire plant (Stael et al., 2019). PCD in plants can be executed through various pathways, including autophagy, apoptosis, hypersensitive reactions, and autolytic (vacuolar) PCD. Inducing PCD is required for many growth and development processes, such as the development of aleurone cells and grain in cereals, tissues that store seeds, the differentiation of female gametophytes, leaf abscission, pollen separation, the aging of the entire plant, and many more (Mondal et al., 2021; Basak and Kundu, 2022).
Several groups of proteolytic enzymes are involved in the process of PCD, including metacaspases (Fagundes et al., 2015). Metacaspases are structural homologs of caspases, which are enzymes found in animals. Unlike caspases, metacaspases do not show substrate specificity for aspartate (Asp); instead, they cleave substrates just after arginine (Arg) or lysine (Lys) residues (Vercammen et al., 2004). Two main types of metacaspases are recognized (Fagundes et al., 2015). Type I metacaspases have a proline-rich N-terminal prodomain and a C-terminal metacaspase domain with a zinc finger motif. Type II metacaspases lack a prodomain at the N-terminus but have a connecting region between the large (p20) and small (p10) caspase-like catalytic domains. Most metacaspases feature a catalytic dyad His-Cys in their active site, with the Cys residue acting as a nucleophile for peptide bond hydrolysis of substrates (Vercammen et al., 2004). In Arabidopsis thaliana, there are nine metacaspases, with AtMC1, AtMC2, and AtMC3 exhibiting characteristics of type I metacaspases, while enzymes from AtMC4 to AtMC9 display type II metacaspase features.
AtMC1 and AtMC2 antagonistically regulate the PCD process in A. thaliana. AtMC1 promotes programmed cell death, while AtMC2 acts as an inhibitor of AtMC1-dependent PCD (Coll et al., 2014). Vacuolar processing enzymes (VPEs) are activated in the acidic environment provided by the vacuole, as well as during the hypersensitive response (HR) and abiotic stresses. Thus, AtMC1 activates hydrogen peroxide-dependent cell death and initiates the hypersensitivity response (Coll et al., 2010; Coll et al., 2014).
The CEP proteases are another group involved in PCD in plants. CEP1 is a protease responsible for PCD in the tapetum of anthers, which is essential for pollen grain development in angiosperms (Zhang et al., 2014). Premature or interrupted cell death of tapetal cells disrupts the nutrient supply to pollen microspores, leading to plant infertility (Ku et al., 2003). CEP1 initially appears as a proenzyme in precursor vesicles of the protease and is then transported to the vacuole, where it matures into an active enzyme. CEP1 influences nuclear degeneration, the formation of tapetal secretory structures, and pollen development (Zhang et al., 2014). Additionally, CEP1 is involved in xylem maturation in A. thaliana, regulating secondary wall thickening (Han et al., 2019).
Proteases in plant cells are a crucial part of the enzymatic machinery, contributing to a plant’s ability to acclimatize to changing environmental conditions. These enzymes are responsible not only for the degradation of damaged or unnecessary proteins within the cell but also for preventing the formation of non-functional protein aggregates or breaking them down. A well-functioning cellular proteolytic system is a condition that enables the cell to change its protein composition and redirect its metabolism to new pathways that are better suited to the altered environmental conditions. Additionally, proteases can play a significant role in processes aimed at removing unfavorable metabolites from the cell. Proteolytic enzymes are also essential components of many signaling pathways and can influence changes in the gene expression profile.
Despite many years of research into various aspects of proteolysis in plant cells, many aspects remain unclear. In particular, intramembrane proteases, commonly believed to be involved in intramembrane regulatory proteolysis processes, appear to be poorly understood. This process leads to the release of transcription factors from membranes, resulting in changes in gene expression. Intramembrane proteases are enzymes that operate in the extremely hydrophobic environment of the lipid bilayer of biological membranes. This characteristic alone makes them extraordinary enzymes, the full significance of which is not yet completely understood. A thorough understanding of the mechanisms of protease action and the processes they are involved in will undoubtedly bring us many tangible benefits in the future.
RL: Writing – review & editing. MA: Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adam Z. (2013). Emerging roles for diverse intramembrane proteases in plant biology. Biochim. Biophys. Acta: Biomembr. 1828, 2933–2936. doi: 10.1016/j.bbamem.2013.05.013
Adamiec M., Ciesielska M., Zalaś P., Luciński R. (2017). Arabidopsis thaliana intramembrane proteases. Acta Physol. Plant 39, 146. doi: 10.1007/s11738-017-2445-2
Adamiec M., Dobrogojski J., Wojtyla Ł., Luciński R. (2022). Stress-related expression of the chloroplast EGY3 pseudoprotease and its possible imact on chloroplasts’ proteome composition. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.965143
Adamiec M., Luciński R., Jackowski G. (2011). The irradiance dependent transcriptional regulation of AtCLPB3 expression. Plant Sci. 181, 449–456. doi: 10.1016/j.plantsci.2011.07.004
Adamiec M., Misztal L., iesielska M., Luciński R. (2021). The changes of PSII supercomplexes stoichiometry in egy1 mutants are related to chlorophyll b deficiency. Phosynthet. 59, 294–302. doi: 10.32615/ps.2021.027
Adamiec M., Misztal L., Kosicka E., Paluch-Lubawa E., Luciński R. (2018). Arabidopsis thaliana egy2 mutants display altered expression level of genes encoding crucial photosystem II proteins. J. Plant Physiol. 231, 155–167. doi: 10.1016/j.jplph.2018.09.010
Ali M. S., Baek H. K. (2020). Protective roles of cytosolic and plastidal proteasomes on abiotic stress and pathogen invasion. Plants 9, 832. doi: 10.3390/plants9070832
Ashoub A., Beckhaus T., Berberich T., Karas M., Bruggemann W. (2013). Comparative analysis of barley leaf proteome as affected by drought stress. Planta 237, 771–781. doi: 10.1007/s00425-012-1798-4
Baena-Gonzales E., Aro E. M. (2002). Biogenesis, assembly and turnover of photosystem II units. Philos. Trans. R. Soc Lond. B Biol. Sci. 357, 1451–1459. doi: 10.1098/rstb.2002.1141
Basak S., Kundu P. (2022). Plant metacaspases: Decoding their dynamics in development and disease. Rev. Plant Physiol. Biochem. 180, 50–63. doi: 10.1016/j.plaphy.2022.03.024
Bray E. A. (1997). Plant responses to water deficit. Trends Plant Sci. 2, 48–54. doi: 10.1016/S1360-1385(97)82562-9
Burgos R., Weber M., Martinez S., Lluch-Senar M., Serrano L. (2020). Protein quality control and regulated proteolysis in the genome-reduced organism Mycoplasma pneumoniae. Mol. Syst. Biol. 16, e9530. doi: 10.15252/msb.20209530
Butenko Y., Lin A., Naveh L., Kupervase M., Levin Y., Reich Z., et al. (2018). Differential roles of the thylakoid lumenal deg protease homologs in chloroplast proteostasis. Plant Physiol. 178, 1065–1080. doi: 10.1104/pp.18.00912
Chassin Y., Kapri-Pardes E., Sinvany G., Arad T., Adam Z. (2002). Expression and characterization of the thylakoid lumen protease DegP1 from Arabidopsis thaliana. Plant Physiol. 130, 857–864. doi: 10.1104/pp.007922
Chen G., Bi Y. R., Li N. (2005). EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J. 41, 364–375. doi: 10.1111/j.1365-313X.2004.02308.x
Chen J., Burke J. J., Xin Z. (2018). Chlorophyll fluorescence analysis revealed essential roles of FtsH11 protease in regulation of the adaptive responses of photosynthetic systems to high temperature. BMC Plant Biol. 18, 1–13. doi: 10.1186/s12870-018-1228-2
Chen G., Law K., Ho P., Zhang X., Li N. (2012). EGY2, a chloroplast membrane metalloprotease, plays a role in hypocotyl elongation in Arabidopsis. Mol. Biol. Rep. 39, 2147–2155. doi: 10.1007/s11033-011-0962-4
Chi W., Sun X., Zhang L. (2012). The roles of chloroplast proteases in the biogenesis and maintenance of photosystem II. Bioch Bioph. Acta Bioenerg. 1817, 239–246. doi: 10.1016/j.bbabio.2011.05.014
Chmielewska K., Rodziewicz P., Swarcewicz B. (2016). Analysis of droughtinduced proteomic and metabolomic changes in barley (Hordeum vulgare L.) leaves and roots unravels some aspects of biochemical mechanisms involved in drought tolerance. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.011082016
Coll N., Smidler A., Puigvert M., Popa C., Valls M., Dangl J. (2014). The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ 2, 1399–1408. doi: 10.1038/cdd.2014.50
Coll N. S., Vercammen D., Smidler A., Clover C., van Breusegem F., Dangl J. L., et al. (2010). Arabidopsis type I metacaspases control cell death. Science 330, 1393–1397. doi: 10.1126/science.1194980
Davies D. R. (1990). The structure and function of the aspartic proteinase. Ann. Rev. Biophys. Biophys. Chem. 19, 189–215. doi: 10.1146/annurev.bb.19.060190.001201
De Backer J., van Breusegen F., de Clercq I. (2022). Proteolytic activation of plant membrane-bound transcription factors. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.927746
de Carvalho M. H. C. (2008). Drought stress and reactive oxygen species. Plant Signal. Behav. 3, 156–165. doi: 10.4161/psb.3.3.5536
Diaz-Mendoza M., Velasco-Arroyo B., Santamaria M. E., Diaz I., Martinez M. (2017). HvPap-1 C1A protease participates differentially in the barley response to a pathogen and an herbivore. Front. Plant Sci. 12. doi: 10.3389/fpls.2017.01585
Diaz-Mendoza M., Velasco-Arroyo B., Santamaria M. E., González-Melendi P., Martinez M., Diaz I. (2016). Plant senescence and proteolysis: two processes with one destiny. Genet. Mol. Biol. 338, 329–338. doi: 10.1590/1678-4685-GMB-2016-0015
Eysholdt-Derzso E., Renzienhausen T., Frings S., Frohn S, von Bongarz K, Igisch C.P., et al. (2023). Ecdolpasmic reticulum-bound ANAC013 factor is cleaved by Rhomboid-like 2 during the initial response to hypoxia in Arabidopsis thaliana. Proc. Natl. Acad. Scie. U.S.A. 120, e2221308120. doi: 10.1073/pnas.2221308120
Fagundes D., Bohn B., Cabreira C., Leipelt F., Dias N., Bodanese-Zanettini M. H., et al. (2015). Caspases in plants: metacaspase gene family in plant stress responses. Funct. Integr. Genomics 15, 639–649. doi: 10.1007/s10142-015-0459-7
Faghani E., Gharechahi J., Komatsu S., Mirzaei M., Khavarinejad R.A., Najafi F., et al. (2015). Comparative physiology and proteomic analysis of two wheat genotypes contrasting in drought tolerance. J. Proteomics 114, 1–15. doi: 10.1016/j.jprot.2014.10.018
Fernandes I., Marques I., Paulo O. S., Batista D., Partelli F. L., Lidon F. C., et al. (2021). Understanding the impact of drought in coffea genotypes: transcriptomic analysis supports a common high resilience to moderate water deficit but a genotype dependent sensitivity to severe water deficit. Agronomy 11, 2255. doi: 10.3390/agronomy11112255
Ford K. L., Cassin A., Bacic A. (2011). Quantitative proteomic analysis of wheat cultivars with differing drought stress tolerance. Front. Plant Sci. 2. doi: 10.3389/fpls.2011.00044
Furlan A. L., Bianucci E., Castro S. (2016). “Signaling role of ROS in modulating drought stress tolerance,” in Drought Stress Tolerance in Plants, vol. 1. , 309–330. doi: 10.1007/978-3-319-28899-4_13
Gomez-Sanchez A., Gonzalez-melendi P., Santamaria M. E., Arbona V., Lopez-Gonzalvez A., Garcia A. (2019). Repression of drought-induced cysteine-protease genes alters barley leaf structure and responses to abiotic and biotic stresses. J. Exp. Bot. 70, 2143–2155. doi: 10.1093/jxb/ery410
Han J., Li H., Yin B., Zhang Y., Liu Y., Cheng Z., et al. (2019). The papain-like cysteine protease CEP1 is involved in programmed cell death and secondary wall thickening during xylem development in Arabidopsis. J. Exp. Bot. 70, 205–215. doi: 10.1093/jxb/ery356
Haußühl K., Andersson B., Adamska I. (2001). A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant Photosystem II. EMBO J. 20, 713–722. doi: 10.1093/emboj/20.4.713
Huber C. V., Jakobs B. D., Mishra L. S., Niedermaier S., Stift M., Winter G., et al. (2019). DEG10 contributes to mitochondrial proteostasis, root growth and seed yield in Arabidopsis. J. Exp. Bot. 70, 5423–5436. doi: 10.1093/jxb/erz294
Huesgen P. F., Schuhmann H., Adamska I. (2005). The family of Deg proteases in cyanobacteria and chloroplasts of higher plants. Physil. Plant 123, 413–420. doi: 10.1111/j.1399-3054.2005.00458.x
Huesgen P. F., Schuhmann H., Adamska I. (2006). Photodameged D1 protein is degraded in Arabidopsis mutants lacking the Deg2 protease. FEBS Lett. 580, 6929–6932. doi: 10.1016/j.febslet.2006.11.058
Icimoto M. Y., Ferreira J. C., Yokomizo C. H., Bim L. V., Marem A., Gilio J. M., et al. (2017). Redox modulation of thimet oligopeptidase activity by hydrogen peroxide. FEBS Open Bio. 7, 1037–1050. doi: 10.1002/2211-5463.12245
Illikova I., Ilik P., Opatikova M., Arshad R., Nosek L., Karlicky V., et al. (2021). Towards spruce-type photosystem II: consequences of the loss of light-harvesting proteins LHCB3 and LHCB6 in Arabidopsis. Plant Physiol. 187, 2691–2715. doi: 10.1093/plphys/kiab396
Iwata Y., Federoff N. V., Koizumi N. (2008). Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20, 3107–3121. doi: 10.1105/tpc.108.061002
Iwata Y., Koizumi N. (2005). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. U. S. A. 102, 5280–5285. doi: 10.1073/pnas.0408941102
Jacchieri S. G., Gomes M. D., Juliano L., Camargo A. C. M. (2009). A comparative conformational analysis of thimet oligopeptidase (EC 3.4.24.15) substrates. J. Pept. Res. 51, 452–459. doi: 10.1111/j.1399-3011.1998.tb00644.x
James M., Masclaux-Daubresse C., Marmagne A., Azzopardi M., Laine P., Goux D., et al. (2019). A new role for SAG12 cysteine protease in roots of arabidopsis thaliana. Science 9, 1998. doi: 10.3389/fpls.2018.01998
Jones J. T., Mullet J. E. (1995). A salt- and dehydration-inducibile pea gene, Cyp15a, encodes a cell-wall protein with sequence similarity to cysteine protease. Plant Mol. Biol. 28, 1055–1065. doi: 10.1007/BF00032666
Kai H., Iba K. (2014). Temperature stress in plants (Chichester: Wiley Online Library). doi: 10.1002/9780470015902.a0001320.pub2
Kapoor D., Bhardwaj S., Landi M., Sharma A., Ramakrishnan M., Sharma A. (2020). The impact of drought in plant metabolism: How to exploit tolerance mechanisms to increase crop production. Appl. Sci. 10, 5692. doi: 10.3390/app10165692
Kato Y., Sakamoto W. (2010). “New insights into the types and function of proteases in plastids,” in International review of cell and molecular biology. Ed. Jeon K. W. (San Diego, CA: Elsevier Inc), 185–218.
Kato Y., Sakamoto W. (2018). FtsH protease in the thylakoid membrane: physiological functions and the regulation of protease activity. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00855
Keeler S. J., Boettger C. M., Haynes J. G., Kuches K. A., Johnson M. M., Thureen D. L., et al. (2000). Acquired thermotolerance and expression of the HSP100/ClpB genes of lima bean. Plant Physiol. 123, 1121–1132. doi: 10.1104/pp.123.3.1121
Kidric M., Kos J., Sabotic J. (2014). Proteases and their endogenous inhibitors in the plant response to abiotic stress. Bot. Serbica 38, 139–158.
Kim S. Y., Kim S. G., Kim Y. S., Seo P. J., Bae M., Yoon H. K., et al. (2007). Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res. 35, 203–213. doi: 10.1093/nar/gkl1068
Kiyosue T, Yamaguchishinozaki K, Shinozaki K. (1993). cCharacterization of cdna for a dehydration-inducible gene that encodes a CLP a, B-like protein in Arabidopsis thaliana L. Biochem. Biophys. Res. Communic. 196, 1214–1220. doi: 10.1006/bbrc.1993.2381
Kley J., Schmidt B., Boyanov B., Stolt-Bergner P. C., Kirk R., Ehrmann M., et al. (2011). Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure. Nta. Struct. Mol. Biol. 18, 728–731. doi: 10.1038/nsmb.2055
Koizumi M., Yamaguchi-Shinozaki K., Tsuji H., Shinozaki K. (1993). Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene 129, 175–182. doi: 10.1016/0378-1119(93)90266-6
Krynicka V., Shao S., Nixon P. J., Komenda J. (2015). Accessibility controls selective degradation of photosystem II subunits by FtsH protease. Nat. Plants 15 (1), 15168. doi: 10.1038/NPLANTS.2015.168
Ku S., Yoon H., Suh H. S., Chung Y. Y. (2003). Male-sterility of thermosensitive genic male-sterile rice is associated with premature programmed cell death of the tapetum. Planta 217, 559–565. doi: 10.1007/s00425-003-1030-7
Kumar S., Sachdeva S., Bhat K. V., Vats S. (2018). “Plant responses to drought stress: Physiological, biochemical and molecular basis,” in Biotic and abiotic stress tolerance in plants. Ed. Vats S. (Singapore: Springer), 1–25.
Langer T. (2000). AAA proteases: cellular machines for degrading membrane proteins. Trends Biochem. Sci. 25, 247–251. doi: 10.1016/S0968-0004(99)01541-8
Lee U., Rioflorido I., Hong S. W., Larkindale J., Waters E. R., Vierling E. (2007). The Arabidopsis ClpB/Hsp100 family of proteins: chaperones for stress and chloroplast development. Plant J. 49, 115–127. doi: 10.1111/j.1365-313X.2006.02940.x
Lee T., Yang S.E., Ko Y., Hwang S., Shin J., Shim J.E., et al. (2015). AraNet v2: an improved database of cofunctional gene networks for study of Arabidopsis thaliana and 27 other non-model plant species. Nuc Acid Res 43, D996–D1002. doi: 10.1093/nar/gku1053
Le Roux M. L., Kunert K. J., van der Vyver C., Cullis C. A., Botha A. M. (2019). Expression of a small ubiquitin-like modifier protease increases drought tolerance in wheat (Triticum aestivum L.). Front. Plant Sci. 10, 266. doi: 10.3389/fpls
Li X., Mu Y., Sun X., Zhang L. (2010). Increased sensitivity to drought stress in atlon4 Arabidopsis mutant. Chin. Sci. Bull. 55, 3668–3672. doi: 10.1007/s11434-010-4166-4
Li H., Sun S. (2021). Protein aggregation in the ER: Calm behind the storm. Cells 10, 3337. doi: 10.3390/cells10123337
Lin C. (2006). “HCV NS3-4A serine protease,” in Hepatitis C Viruses: Genomes and Molecular Biology. Ed. Tan S. L. (Norfolk (UK: Horizon bioscience). Available at: https://www.ncbi.nlm.nih.gov/books/NBK1623/. Chapter 6.
Lindahl M., Spetea C., Hundal T., Oppenheim A. B., Adam Z., Andersson B. (2000). The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12, 419e431. doi: 10.1105/tpc.12.3.419
Liu H., Hu M., Wang Q., Cheng L., Zhang Z. (2018). Role of papain-like cysteine proteases in plant development. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01717
Liu J. X., Srivastava R., Che P., Howell S. H. (2007a). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19, 4111–41119. doi: 10.1105/tpc.106.050021
Liu J. X., Srivastava R., Che P., Howell S. H. (2007b). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 51, 897–909. doi: 10.1111/j.1365-313X.2007.03195.x
Luciński R., Jackowski G. (2006). The structure, functions and degradation of pigment-binding roteins of photosystem II. Acta Bioch. Pol. 53, 693–708. doi: 10.18388/abp.2006_3297
Luciński R., Jackowski G. (2013). AtFtsH heterocomplex-mediated degradation of apoproteins of the major light harvesting complex of photosystem II (LHCII) in response to stresses. J. Plant Physiol. 170, 1082–1089. doi: 10.1016/j.jplph.2013.03.008
Luciński R., Misztal L., Samardakiewicz S., Jackowski G. (2011). The thylakoid protease Deg2 is involved in stress-related degradation of the photosystem II light-harvesting protein Lhcb6 in Arabidopsis thaliana. New Phytol. 192, 74–86. doi: 10.1111/j.1469-8137.2011.03782.x
Martinez M., Gómez-Cabellos S., Giménez M. J., Barro F., Diaz I., Diaz-Mendoza M. (2019). Plant proteases: from key enzymes in germination to allies for fighting human gluten-related disorders. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00721
Maziak A., Heidorn-Czarna M., Weremczuk A., Jańska H. (2021). FTSH4 and OMA1 mitochondrial proteases reduce moderate heat stress-induced protein aggregation. Plant Physiol. 187, 769–786. doi: 10.1093/plphys/kiab296
Migdal I., Skibior-Blaszczyk R., Heidorn-Czarna M., Kolodziejczak M., Garbiec A., Janska H. (2017). AtOMA1 affects the OXPHOS system and plant growth in contrast to other newly identified ATP-independent proteases in Arabidopsis mitochondria. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01543
Misas-Villamil J. C., van der Hoorn R. A., Doehlemann G. (2016). Papainlike cysteine proteases as hubs in plant immunity. New Phytol. 212, 902–907. doi: 10.1111/nph.14117
Möller I. M., Sweetlove L. J. (2010). ROS signalling – specificity is required. Trend. Plant Sci. 15, 370–374. doi: 10.1016/j.tplants.2010.04.008
Moloi S. J., Ngara R. (2023). The roles of plant proteases and protease inhibitors in drought response: a review. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1165845
Mondal R., Antony S., Roy S., Chattopadhyay S. K. (2021). “Programmed cell death (PCD) in plant: molecular mechanism, regulation, and cellular dysfunction in response to development and stress,” in Regulation and Dysfunction of Apoptosis (IntechOpen). Ed. Tutar Y. doi: 10.5772/intechopen.97940
Ng S., Ivanova A., Duncan O., Law S. R., Van Aken O., De Clercq I., et al. (2013). A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25, 3450–3471. doi: 10.1105/tpc.113.113985
Nishimura K., van Wijk K. (2015). Organization, function and substrates of the essential Clp protease system in plastids. Biochim. Biophys. Acta Bioenerg. 1847, 915–930. doi: 10.1016/j.bbabio.2014.11.012
Patipong T., Hibino T., Kageyama H., Waditee-Sirisattha R. (2020). The evolutionarily conserved HtrA is associated with stress tolerance and protein homeostasis in the halotolerant cyanobacterium Halothece sp. PCC7418. Extremophiles 24, 377–389. doi: 10.1007/s00792-020-01162-4
Pinti M., Gibellini L., Nasi M., De Biasi S., Bortolotti C. A., Iannone A., et al. (2016). Emerging role of Lon protease as a master regulator of mitochondrial functions. Biochim. Biophys. Acta Bioenerg. 1857, 1300–1306. doi: 10.1016/j.bbabio.2016.03.025
Sakamoto W., Tamura T., Hanba-Tomita Y., Murata M., Sodmergen (2002). The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells 7, 769–780. doi: 10.1046/j.1365-2443.2002.00558.x
Sakamoto W., Zaltsman A., Adam Z., Takahashi Y. (2003). Coordinated Regulation and Complex Formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, Chloroplastic FtsH Metalloproteases Involved in the Repair Cycle of Photosystem II in Arabidopsis Thylakoid Membranes. Plant Cell 15, 2843–2855. doi: 10.1105/tpc.017319
Schaller A. (2004). A cut above the rest: the regulatory function of plant proteases. Planta 220, 183–197. doi: 10.1007/s00425-004-1407-2
Schneider J. S., Glickman M. S. (2013). Function of site-2 proteases in bacteria and bacterial pathogens. Bioch Bioph Acta Biomembr. 1828, 2808–2814. doi: 10.1016/j.bbamem.2013.04.019
Schuhmann H., Huesgen P. F., Adamska I. (2012). The family of Deg/HtrA proteases in plants. BMC Plant Biol. 12, 52. doi: 10.1186/1471-2229-12-52
Sebastian D., Fernando F. D., Raúl D. G., Gabriela G. M. (2020). Overexpression of Arabidopsis aspartic protease APA1 gene confers drought tolerance. Plant Sci. 292, 110406. doi: 10.1016/J.PLANTSCI.2020.110406
Seo P. J. (2014). Recent advances in plant membrane-bound transcription factor research: mphasis on intracellular movement. J. Integr. Plant Biol. 56, 334–342. doi: 10.1111/jipb.12139
Seo P. J., Kim S. G., Park C. M. (2008). Membrane-bound transcription factors in plants. Trend Plant Sci. 13, 550–556. doi: 10.1016/j.tplants.2008.06.008
Seo P. J., Kim S.-G., Song J. S., Kim Y. S., Kim H. J., Park C. M. (2010). Proteolytic processing of an Arabidopsis membrane-bound NAC transcription factor is triggered by cold-induced changes in membrane fluidity. Biochem. J. 427, 359–367. doi: 10.1042/BJ20091762
Shah Z. H., Rehman H. M., Akhtar T., Daur I., Nawaz M. A., Ahmad M. Q., et al. (2017). Redox and ionic homeostasis regulations against oxidative, salinity and drought stress in wheat (A systems biology approach). Front. Genet. 8. doi: 10.3389/fgene.2017.00141
Sharma P., Gayen D. (2021). Plant protease as regulator and signaling molecule for enhancing environmental stress-tolerance. Plant Cell Rep. 40, 2081–2095. doi: 10.1007/s00299-021-02739-9
Sharma P., Jha A. B., Dubey R. S., Pessarakli M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 1–26. doi: 10.1155/2012/217037
Smakowska E., Skibior-Blaszczyk R., Czarna M., Kolodziejczak M., Kwasniak-Owczarek M., Parys K., et al. (2016). Lack of FTSH4 protease affects protein carbonylation, mitochondrial morphology, and phospholipid content in mitochondria of Arabidopsis: new insights into a complex interplay. Plant Physiol. 171, 2516–2535. doi: 10.1104/pp.16.00370
Stael S., Van Breusegem F., Gevaert K., Novack M. K. (2019). Plant proteases and programmed cell death. J. Exp. Bot. 70, 1991–1995. doi: 10.1093/jxb/erz126
Sun X., Feng P., Xu X., Guo H., Ma J., Chi W., et al. (2011). A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2, 477. doi: 10.1038/ncomms148
Sun L., Zhang S. S., Lu S. J., Liu J. X. (2015). Site-1 protease cleavage site is important for the ER stress-induced activation of membrane-associated transcription factor bZIP28 in Arabidopsis. Sci. China 58, 270–275. doi: 10.1007/s11427-015-4807-6
Sun X., Peng L., Guo J., Chi W., Ma J., Lu C., et al. (2007). Formation of DEG5 and DEG8 and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell 19, 1347–1361. doi: 10.1105/tpc.106.049510
Tajima H., Iwata Y., Iwano M., Takayama S., Koizumi N. (2008). Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochem. Biophys. Res. Commun. 374, 242–247. doi: 10.1016/j.bbrc.2008.07.021
Thompson E. P., Smith L. S. G., Glover B. J. (2012). An Arabidopsis rhomboid protease has roles in the chloroplast and in flower development. J. Exp. Bot. 63, 3559–3570. doi: 10.1093/jxb/ers012
Tornkvist A., Liu C., Moschou P. N. (2019). Proteolysis and nitrogen: emerging insights. J. Exp. Bot. 70, 2009–2019. doi: 10.1093/jxb/erz024
van der Hoorn A. L. (2008). Plant proteases: from phenotypes to molecular mechanisms. Ann. Rev. Plant Biol. 59, 111–223. doi: 10.1146/annurev.arplant.59.032607.092835
van Wijk K. J. (2015). Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Ann. Rev. Plant Biol. 66, 75–111. doi: 10.1146/annurev-arplant-043014-115547
Velasco-Arroyo B., Diaz-Mendoza M., Santamaria M. E., Gonzalez-Melendi P., Gomez-Sanchez A., Arnaiz A., et al. (2016). Senescence-associated genes in response to abiotic/biotic stresses. Prog. Bot. 79, 89–109. doi: 10.1007/124_2017_1
Velasco-Arroyo B., Martínez M., Díaz I., Diaz-Mendoza M. (2018). Differential response of silencing HvIcy2 barley plants against Magnaporthe oryzae infection and light deprivation. BMC Plant Biol. 18, 337. doi: 10.1186/s12870-018-1560-6
Vercammen D., van de Cotte B., De Jaeger G., Van Beeumen J., Inza D., Van Breusegem F. (2004). Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J. Biol. Chem. 279, 45329–45336. doi: 10.1074/jbc.M406329200
Wang W. H., He E. M., Guo Y., Tong Q. X., Zheng H. L. (2016). Chloroplast calcium and ROS signaling networks potentially facilitate the primed state for stomatal closure under multiple stresses. Environ. Exp. Bot. 122, 85–93. doi: 10.1016/j.envexpbot.2015.09.008
Wang W., Vinocur B., Altman A. (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14. doi: 10.1007/s00425-003-1105-5
Wang N., Zhao J., He X., Sun H., Zhang G., Wu F. (2015). Comparative proteomic analysis of drought tolerance in the two contrasting Tibetan wild genotypes and cultivated genotype. BMC Genomics 16, 1–19. doi: 10.1186/s12864-015-1657-3
Wolfe M. S., Kopan R. (2004). Intramembrane proteolysis: theme and variations. Science 305, 1119–1123. doi: 10.1126/science.1096187
Wu Y., Mirzaei M., Pascovici D., Chick J. M., Atwell B. J., Haynes P. A. (2016). Quantitative proteomic analysis of two different rice varieties reveals that drought tolerance is correlated with reduced abundance of photosynthetic machinery and increased abundance of ClpD1 protease. J. Proteomics 143, 73–82. doi: 10.1016/j.jprot.2016.05.014
Yao X., Xiong W., Ye T., Wu Y. (2012). Overexpression of the aspartic protease ASPG1 gene confers drought avoidance in Arabidopsis. J. Exp. Bot. 63, 2579–2593. doi: 10.1093/jxb/err433
Yu F., Park S., Rodermel S. R. (2004). The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J. 37, 864–876. doi: 10.1111/j.1365-313X.2003.02014.x
Zhang S., Zhang D., Yang C. (2014). AtFtsH4 perturbs the mitochondrial respiratory chain complexes and auxin homeostasis in Arabidopsis. Plant Signal. Behav. 9, e29709. doi: 10.4161/psb.29709
Zhuang Y., Wei M., Ling C., Liu Y., Amin A. K., Li P., et al. (2021). EGY3 mediates chloroplastic ROS homeostasis and promotes retrograde signaling in response to salt stress in Arabidopsis. Cell Rep. 36, 109384. doi: 10.1016/j.celrep.2021.109384
Keywords: abiotic stress factor, plant proteases, proteolysis, stress response, protein degradation
Citation: Luciński R and Adamiec M (2023) The role of plant proteases in the response of plants to abiotic stress factors. Front. Plant Physiol. 1:1330216. doi: 10.3389/fphgy.2023.1330216
Received: 30 October 2023; Accepted: 01 December 2023;
Published: 18 December 2023.
Edited by:
Harvey J. M. Hou, Alabama State University, United StatesReviewed by:
Qingfang He, University of Arkansas at Little Rock, United StatesCopyright © 2023 Luciński and Adamiec. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert Luciński, cnRsQGFtdS5lZHUucGw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.