- 1Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent, Belgium
- 2Institute for Biology, Faculty of Natural Sciences, Norwegian University of Science and Technology, Trondheim, Norway
- 3Umeå Plant Science Centre (UPSC), Department of Plant Physiology, Umeå University, Umeå, Sweden
Cell walls are not just passive barriers; they are dynamic and adaptable structures that are actively remodeled in response to both internal and external cues. They are crucial in defining cellular identity, ensuring structural integrity, and mediating interactions with the environment. The concept of cell wall integrity (CWI) encompasses the mechanisms by which cells monitor and maintain their walls, ensuring proper function and response to challenges. While significant knowledge has been accumulated on CWI in certain model organisms, there remains a vast landscape of uncharted territory in others. In this review, we aim to bridge this gap, offering a comparative perspective on CWI across different evolutionary lineages, from the well-studied yeasts to the diverse world of plants. We focus especially on the green lineage –the group of green algae and land plants, hence the green wall–, but also consider some insights from organisms with radically different lifestyles and cell wall arrangements, which serves as a base to some intriguing questions about the role of CWI across evolution and environmental adaptation.
1 Introduction
Across diverse organisms, from yeasts to algae and terrestrial plants, the presence of robust extracellular matrixes, known as cell walls, is a defining feature. Beyond providing mechanical support, these walls act as dynamic interfaces, mediating interactions with the environment and playing instrumental roles in growth and development (Levin, 2005; Anderson and Kieber, 2020; Zhang et al., 2021). Their adaptability is underscored by their compositional plasticity, which allows for modifications in tune with developmental transitions, environmental shifts, and stressors.
The concept of cell wall integrity (CWI) refers to the sophisticated mechanisms that enable cells to perceive and adapt to changes in this structure, ensuring their walls remain resilient and functional (Levin, 2005; Bacete and Hamann, 2020). This adaptability is not just a passive trait; it is an active response, an essential function reflecting the cell wall’s role in safeguarding cellular identity and mediating environmental interactions. While substantial knowledge has been generated in the last years regarding CWI in specific model organisms, a vast expanse remains unexplored in others. In this mini-review, we made a concerted effort to synthesize existing knowledge on CWI, drawing parallels and distinctions across different organisms. By putting together, the well-characterized mechanisms in yeasts with the emerging insights from the plant kingdom and the intriguing recent findings in some algae, this review seeks to offer a cohesive understanding of CWI with a focus on the green lineage i.e., the green wall.
2 Defining cell wall integrity
The very notion of an “integral” cell wall is a subject of ongoing debate. What does integrity mean in the context of cell wall integrity? Is it purely structural, or does it encompass functional and dynamic aspects of the wall?
While foundational principles of CWI have been elucidated in certain organisms, such as Saccharomyces cerevisiae (Levin, 2011), the vast diversity of cell walls across different organisms introduces layers of complexity. For instance, the multicellular nature of plants brings forth challenges stemming from intercellular dynamics, where cells can exert force on their neighbors, leading to intricate mechanical interactions within tissues (Coen and Cosgrove, 2023). Similarly, unicellular organisms like algae, with their unique evolutionary histories and environmental interactions, offer distinct perspectives on CWI, further broadening the scope of our understanding (Peaucelle et al., 2012; Galindo-Trigo et al., 2016).
As we explore the complex world of CWI, it becomes clear that a comprehensive view is crucial. Analyzing each organism’s unique cell wall composition is only the beginning. We must also consider how CWI mechanisms interact with and adapt to the organism’s environment and lifestyle. This interconnectedness provides a more nuanced understanding of “integrity” in the context of cell walls and sets the stage for exploring its implications across diverse biological systems.
3 Yeast: a model system for CWI studies
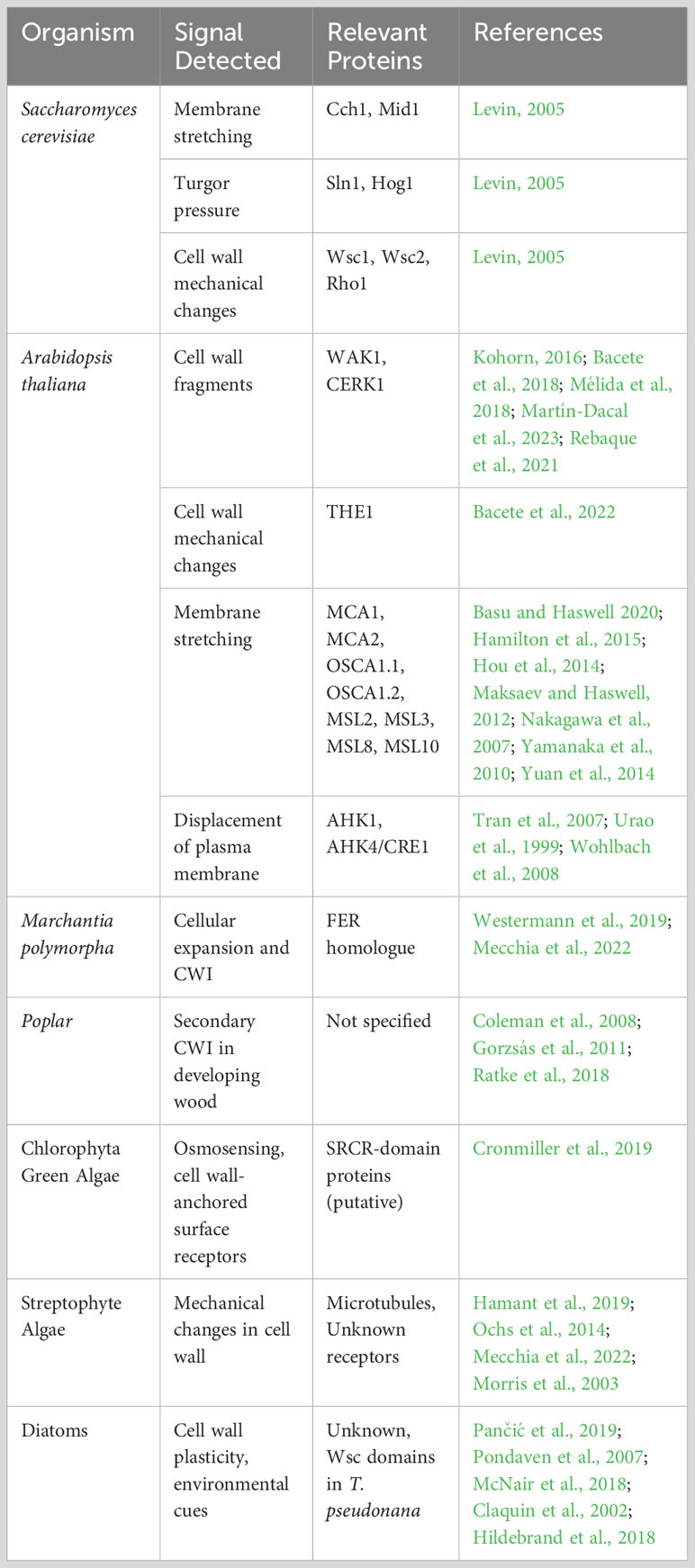
S. cerevisiae has for a long time been at the forefront of CWI studies. Its cell wall is primarily composed of glucans (glucose polymers), mannans (mannose-containing polysaccharides), and proteins. CWI is maintained through three mechanisms. First, the Cch1 and Mid1 proteins form a Ca2+ channel that detects membrane stretching, possibly from a weakened cell wall and high turgor pressure, leading to calcium ion influx and downstream signaling via calmodulin (Levin, 2005). Second, the cell-surface sensor kinase, Sln1, monitors turgor pressure. Under hyperosmotic stress, Sln1 activates the MAPK Hog1, enabling yeast cells to sense and adjust to turgor changes (Levin, 2005). Lastly, CWI sensors containing cell wall integrity and stress response component (Wsc) domains such as Wsc1 and Wsc2 detect cell wall mechanical changes. These sensors interact with the G protein Rho1 upon deformation, triggering responses essential for cell wall biosynthesis and cytoskeletal organization (Levin, 2005; Dupres et al., 2009). Central to CWI is transcriptional regulation, where activated transcription factors, especially Skn7, modulate target gene expression, guiding cellular responses to environmental or developmental cues (Kono et al., 2016). CWI pathways of other fungi share significant similarities with that of S. cerevisiae, suggesting a conserved mechanism of cell wall stress response among yeasts (Dichtl et al., 2016).
4 Land plants: blooming of CWI monitoring systems
Land plants, spanning from simple to complex forms, provide a rich diversity of evolutionary adaptations. The cell wall, a key element in the transition from aquatic to land environments, is no exception, with compositions and structures that vary among plant species, developmental stages, tissues, and even across a single cell wall (Knox, 2008; Somssich et al., 2016; Gigli-Bisceglia et al., 2020). The primary cell walls of land plants, predominantly composed of cellulose, hemicellulose, and pectin recently reviewed by Cosgrove (2022), have evolved to offer flexibility during cell growth. In contrast, the secondary cell walls –Recently reviewed by Zhong et al., 2019, found in specialized cells like tracheids, vessel elements, and fibers, are more rigid and provide structural support. These secondary walls are rich in cellulose, hemicellulose, and notably, lignin, which makes them rigid and resistant.
The diversity of plant cell walls is mirrored by their CWI systems, which are much more intricate than those of their unicellular counterparts (Table 1). Arabidopsis thaliana, the renowned eudicot model, has been instrumental in elucidating CWI mechanisms. The CWI in Arabidopsis is closely associated with receptor-like kinases (RLKs) and receptor-like proteins (RLPs) that perceive changes in the cell wall’s integrity by putatively binding to glycan ligands. Once bound, these RLKs initiate intricate signalling cascades that modulate a range of cellular responses, ensuring the cell wall remains robust and functional (Bacete et al., 2018). The Catharanthus roseus receptor-like kinase1-like (CrRLK1L) family –comprising 17 members in Arabidopsis– is a prominent example of RLKs involved in CWI sensing, with THESEUS1 (THE1) and FERONIA (FER) being prominent examples (Decreux and Messiaen, 2005; Kohorn, 2016; Feng et al., 2018; Gonneau et al., 2018; Bacete et al., 2022; Ortiz-Morea et al., 2022; Zhong et al., 2022). Wall-Associated Kinases (WAK) and LysM RLKs such as chitin elicitor receptor kinase 1 (CERK1) are also key elements of the receptor-mediated CWI perception (Kohorn, 2016; Bacete et al., 2018; Mélida et al., 2018; Rebaque et al., 2021; Martín-Dacal et al., 2023). Beyond the molecular interactions, the physical aspect of cell wall perception cannot be overlooked. Mechanoperception, the ability of cells to sense and respond to mechanical stimuli, is integral to CWI. The displacement between the cell wall and plasma membrane caused by mechanical forces can activate membrane-bound receptors and ion channels (Basu and Haswell, 2020; Yoshimura et al., 2021; Codjoe et al., 2022). This mechanosensory capability ensures that plants can swiftly and effectively respond to physical challenges, from turgor pressure changes to external mechanical stresses (Bacete and Hamann, 2020).
The bryophyte Marchantia polymorpha offers insights into the evolutionary transition of plants from aquatic to terrestrial habitats, with its CWI system hinting at evolutionary links with certain Arabidopsis receptors, including a FER homologue that regulates cellular expansion and cell wall integrity (Westermann et al., 2019; Mecchia et al., 2022). CrRLKs are present as well in other bryophytes such as Physcomitrium patens (Lehti-Shiu et al., 2009) although their implication in CWI sensing remains unclear. These observations give relevance to the importance of RLK-based mechanisms for plant CWI. However, no homologues for CrRLKs, WAKs, or any other relevant plant CWI RLK have been identified in fungi or algae. Conversely, the absence of Wsc sensors in plants, prevalent in other organisms, accentuates this evolutionary divergence, suggesting that plants might have evolved distinct mechanisms for CWI perception.
Poplar stands out as a model organism for studying CWI in woody plants, pivotal in understanding the intricacies of lignification and the development of secondary cell walls (Coleman et al., 2008; Gorzsás et al., 2011). However, the existence of a secondary cell wall CWI monitoring system has been questioned. Downregulation of xylan synthesis genes leads to changes in xylan content, improved lignocellulose saccharification efficiency, and stimulated growth. Shifts in cellulose orientation and cell wall thinning suggest the presence of a cell wall integrity monitoring mechanism that senses and responds to reduced xylan elongation in developing wood.(Ratke et al., 2018). However, the analysis of the stem transcriptome in Arabidopsis xylan mutants revealed minimal changes, suggesting a lack of a robust secondary CWI response (Faria-Blanc et al., 2018). Grasses, like Brachypodium distachyon, with their differential cell wall attributes (Coomey et al., 2020), can further enrich our understanding of secondary CWI and reveal both conserved and unique mechanisms distinguishing monocots from eudicots.
5 Green algae: unicellular proxies to plant-esque cell wall integrity
The green lineage is split into two significant subgroups: Chlorophyta, containing most green algae species (Cronmiller et al., 2019); and Streptophyta, consisting of the land plants (Embryophyta) and streptophyte algae. Plant terrestrialization required numerous cellular adaptations, which allowed land plant ancestors to withstand novel stress conditions, such as high UV radiation and desiccation (de Vries and Archibald, 2018). These included changes in the cell wall composition and organization, and probably novel mechanisms to monitor and modify cell wall structure accordingly.
Chlorophyta green algae have very diverse cell wall compositions, from cellulose-pectin complexes to hydroxyproline-rich glycoproteins (Domozych et al., 2012). Chlamydomonas reinhardtii has emerged as a model system for this subgroup. Its cell wall, primarily composed of hydroxyproline-rich glycoproteins, is crucial for environmental protection. In an interesting work by Cronmiller et al. (2019). C. reinhardtii was shown to respond to gametolysin by rapidly activating over 100 cell wall-related genes. This activation is partly driven by osmosensing channels and cell wall-anchored surface receptors, whereas no homologues for CrRLK1Ls, WAKs, or BR-signaling components for cell wall integrity were identified. Instead, the authors suggest that C. reinhardtii employs scavenger-receptor cysteine-rich (SRCR)-domain proteins as surface receptors. These SRCR proteins, including five TRP-type channels, are implicated in calcium-mediated signaling (Cronmiller et al., 2019). Intriguingly, Wsc domains, essential for CWI in yeast, can be found in the sequenced genomes of some other unicellular–Dunaliella tertiolecta and Tetradesmus obliquus– and colonial –Gonium pectorale– Chlorophyta green algae (Hanschen et al., 2016; Carreres et al., 2017; Malerba et al., 2020).
Streptophyte algae, also known as charophytes, are the closest relatives to land plants. They can be split into the basal-branching Klebsormidiophyceae, Chlorokybophyceae, and Mesostigmatophyceae (KCM) and the higher-branching Zygnematophyceae, Coleochaetophyceae, and Charophyceae (ZCC) group (de Vries et al., 2016). ZCC group are the closest land plant relatives with cell walls comprised of polysaccharides present in land plants, unlike the KCM group (Sørensen at al., 2011). Similarities in cell wall composition between land plants and charophytes are additionally reflected by the presence of canonical cell wall biosynthetic enzymes and structural proteins in the genomes of these algae (Feng et al., 2023). Therefore, exploring CWI mechanisms in members of ZCC group holds great promise for understanding the evolution CWI processes in land plants. Surprisingly, no FERONIA homologs were found in the genomes of charophytes (Mecchia et al., 2022), indicating that these organisms utilize alternative CWI systems. Proliferation of CrRLK1L family of receptors in land plants is more likely to be an adaptation to complex multicellular body plans (Morris et al., 2003), in which cell-to-cell communication is essential for coordinated tissue growth and development. It is plausible that in the unicellular and simple multicellular Zygnematophyceae, mechanical changes in the cell wall can be registered by alternative pathways, such as sensing by microtubules, which were proposed to act as tension sensors in plant cells (Hamant et al., 2019). Indeed, depolymerization of microtubules by oryzalin in unicellular Penium margaritaceum leads to cell swelling and abnormal cell shape (Ochs et al., 2014). It remains unclear what type of receptors, if any, are involved in CWI mechanisms in complex multicellular forms from Coleochaetophyceae and Charophyceae classes. Future research and understanding of land plant evolution is currently hampered by the lack of molecular biology tools and reliable transformation protocols for charophyte algae. With new algal genomes being published at an accelerated pace, the time has come to answer previous calls (Rensing, 2017) and establish charophytes as molecular biology model systems.
6 Diatoms: the implications of a rigid cell wall
Diatoms, which are of high ecological and biotechnological relevance, arose from several secondary endocytosis events, with their plastid derived from a eukaryotic red algae (Nelson et al., 1995; Yool and Tyrrell, 2003; Armbrust, 2009). There are estimated to be well over 100,000 diatom species, making it one of the most species-rich group of eukaryotic microorganisms (Mann and Droop, 1996). Interestingly, unlike most other photosynthetic organisms, their cell walls are not mainly composed of polysaccharides, but instead made up by two porous silica shells, called frustules, which fit together like a petri-dish (Hildebrand, 2008). This rigid cell wall is coated by an organic matrix composed of proteins, long-chain polyamines and polysaccharides, whose exact composition still remains to be studied more extensively apart from a few select examples mainly derived from the main model system for studying diatom cell wall biogenesis, Thalassiosira pseudonana (Sumper et al., 2005; Brunner et al., 2009; Richthammer et al., 2011; Kotzsch et al., 2016). Beyond the already discussed cell wall roles of providing protection and supporting the large vacuole of the cell, recent studies suggest that the frustules act as a photonic crystal, improving conditions for photosynthesis inside while providing protection from UV light (De Tommasi et al., 2018; Goessling et al., 2018). While the process of cell wall formation has been studied extensively [for a review on the topic see (Hildebrand et al., 2018)], very little is known about the plasticity of their silicified cell wall. Silica content per cell varies widely among species, but has been shown to be correlated with cell size (Conley et al., 1989). Growth-limiting conditions generally result in increased silicification rates, thus resulting in an inverse correlation between silicification and growth rates (McNair et al., 2018). Limitation of silica content, however, results in a decrease of silicification without differences in growth speed (Claquin et al., 2002). However, there are select examples for adaptation of cell wall thickness in response to grazing: The diatom Thalassiosira weissflogii has been shown to increase silicification upon incubation with grazers without changes in growth rates (Pančić et al., 2019). Similarly, Skeletonema marinoi, a chain-forming diatom, strongly reduces chain lengths upon incubation with copepods, successfully reducing grazing rates (Pondaven et al., 2007). Thus, there are examples of cell wall plasticity in diatoms, suggesting a CWI perception system, but very little is known about how these environmental cues get translated into modifications of the silica cell wall and/or its organic matrix. And it is intriguing to think on a CWI system that perceives the physicochemical integrity of a rigid wall. Like in other unicellular algae, Wsc domains have been identified in sequenced genomes such as that of T. pseudonana, but their function is unknown.
7 Beyond the wall: perception of extracellular matrix in other eukaryotes
The perception and composition of extracellular structures in eukaryotes is not restricted to cell walls and CWI. Extracellular matrixes (ECMs) are diverse and serve various functions, from structural support to cellular communication, similar to cell walls.
So far, we have discussed eukaryotes from Plantae –plants and green algae–, Chromalveolates –diatoms and Phaeophytes (brown algae) and Unikonts (yeast) (Figure 1). Plantae have cell walls rich in carbohydrates like cellulose, whereas certain Chromalveolates possess cell walls composed of materials like silica, and fungi in the Unikonts group have chitin-based cell walls. In contrast, animals, also part of the Unikonts, do not have cell walls but possess an ECM mainly composed of proteins like collagen (Frantz et al., 2010). This ECM serves multiple functions, including providing structural support and mediating cellular interactions through various types of receptors (Hynes, 2009). The perception of the ECM is often mediated by integrin receptors, which bind to specific ligands in the ECM and initiate cellular signalling pathways (Friedland et al., 2009). Interestingly, animals share with fungi and plants the presence of carbohydrate-binding proteins collectively known as lectins. This heterogeneous group of proteins plays crucial roles in important processes, from immune responses to cell adhesion (De Coninck and Van Damme, 2021; Radhakrishnan et al., 2023). Rhizaria and Excavates, two other eukaryotic groups, generally lack traditional cell walls but may possess unique extracellular structures. For instance, some Rhizaria have elaborate shells made of organic material or silica (Nakamura et al., 2018), while certain Excavates have a pellicle, a tough but flexible outer covering that provides shape and protection (Cavalier-Smith, 2010). The vital role of these extracellular structures for survival likely means these organisms have ways to sense and adapt to changes in their outer layers. However, the specific mechanisms remain a mystery.
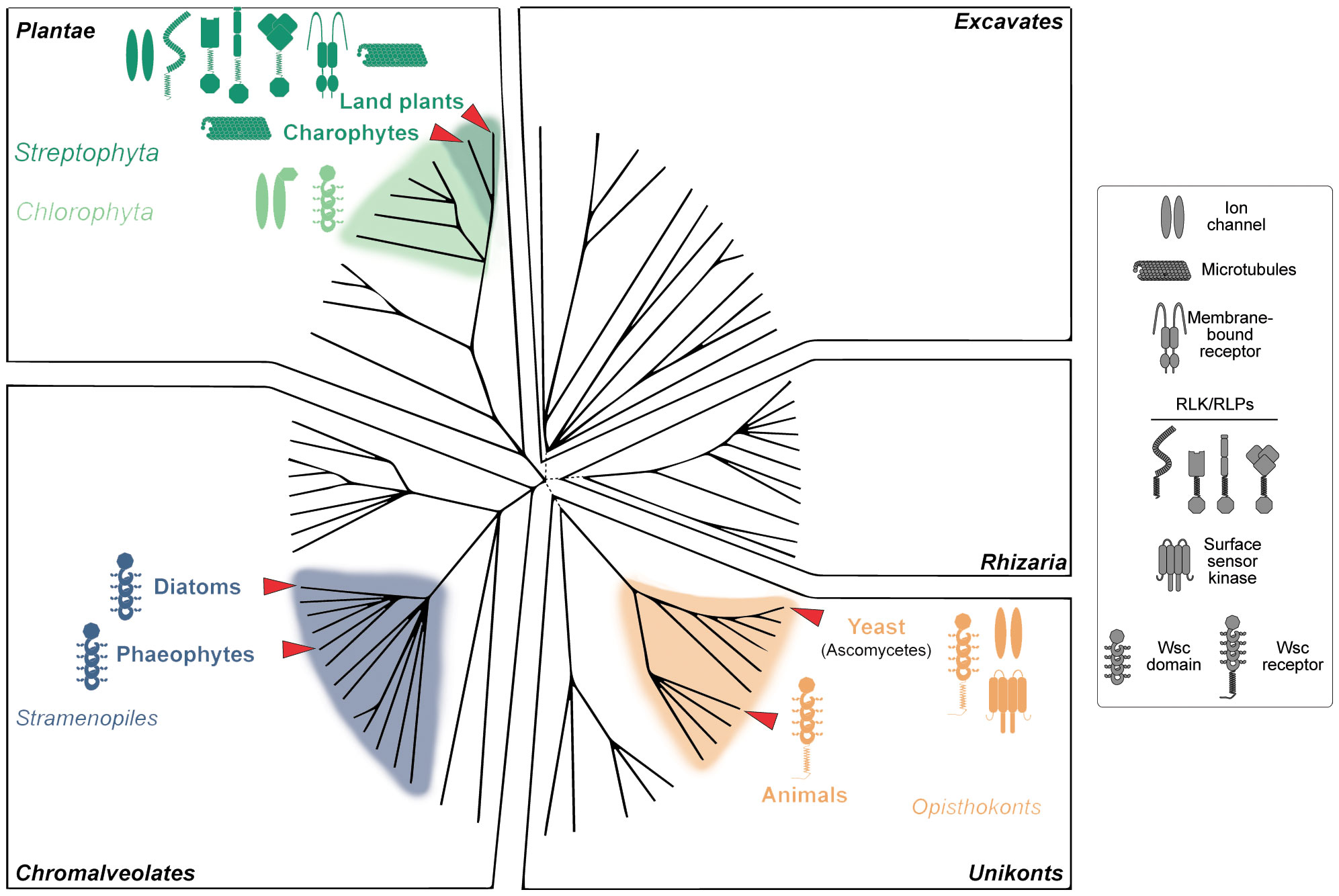
Figure 1 Distribution of cell wall integrity (CWI) sensors across the tree of life. Organisms with cell walls are distributed among various of the biological groups within the domain of Eukaryota. Plantae have cell walls rich in carbohydrates like cellulose, whereas certain Chromalveolates possess cell walls composed of materials like silica, and fungi in the Unikonts group have chitin-based cell walls. In contrast, animals, also part of the Unikonts, do not have cell walls but possess an extracellular matrix composed mainly of proteins like collagen, which serves to provide structural support and mediate cellular interactions. Rhizaria and Excavates generally lack traditional cell walls but may have other forms of extracellular structures for protection and support. Land plants, with complex cell walls and immobile cells bound together throughout the apoplast, have the most complex CWI system, comprised of different specialized receptors including receptor-like kinases (RLKs) and receptor-like proteins (RLPs), ion channels, and membrane-bound mechanoreceptors. In contrast, yeast have simpler systems, in a higher extend based on cell wall integrity and stress response component (Wsc) domain-containing receptors. Intriguingly, Wsc domains are widespread among Eukaryota, including animals, diatoms and brown algae (Phaeophytes). Based on the Eukaryotic tree of life created by Keeling et al. (2005).
8 Discussion
As we explore the complexities of CWI, the importance of investigating less-studied organisms and systems becomes increasingly clear. These could offer fresh perspectives and expand our understanding of CWI across a range of walled Eukaryote (Figure 1). One captivating aspect of CWI is the variety of cues that activate its monitoring systems. While we have made significant progress in understanding chemical cues, such as interactions between ligands and receptors, the role of physical cues like mechanical stress remains relatively uncharted territory. In animals, integrins serve as vital sensory molecules, converting chemical and physical cues from ECM into signals that guide cell behavior. They achieve this by responding to the ligand (proteins and glycoproteins) density and chemical affinity between receptors and ligands in the ECM, initiating processes like integrin clustering which in turn translate these cues into biochemical signals (Paszek et al., 2009). To date, no similar system has been identified in plants, where chemical and mechanical CWI cues have traditionally been studied separately (Bacete and Hamann, 2020).
How have different organisms tailored their CWI mechanisms to meet their unique environmental challenges and life cycles? The lack of certain CWI receptors in fungi and algae, compared to plants, suggests an evolutionary divergence that deserves further study. Understanding these evolutionary intricacies could provide insights into why some CWI mechanisms are conserved across species while others are not. The unique challenges and life cycles of plants–composed of immobile cells connected to each other through their cell walls, in a dry environment and under the pressure of gravity– raise compelling questions about how they perceive and respond to mechanical stimuli affecting their cell walls. All in all, the plant cell walls of different cells are bound together forming the apoplast, and thus CWI alterations could be an efficient mechanism to transfer signals across the plant, but this would require specific systems different from those in motile cells. Given that plants can respond to CWI challenges with both death and growth, could the complexity of CWI mechanisms be linked to the complexity of the organism itself? In contrast, for unicellular organisms like yeasts, CWI is often a life-or-death issue, either due to severe damage or the death of a similar organism nearby. Charophytes live in water, which involves a very different context for mechanoperception and self-perception. This could be a reason for the lack of plant-like CWI receptors in these organisms. Interestingly, carbohydrate-binding Wsc domains are widespread in numerous unicellular organisms from yeast to green algae, although with unknown functions and in many cases lacking a transmembrane domain which would be essential for intracellular signal transduction. These domains are present even in mammals, where they are in some cases linked to diseases associated with a misperception of the extracellular matrix such as polycystic kidney disease (Weston et al., 2003). However, to date, there have not been identified in any member of the Streptophyta subgroup. Brown algae such as Ectocarpus siliculosus with more than 100 Wsc-containing proteins seem the only example of multicellular photosynthetic organisms with the possibility of a putative Wsc-based CWI monitoring system (Cock et al., 2010). The question of when and why this system was replaced by other CWI monitoring systems can shed light on the origins of multicellularity, the transition to land and the loss of cell motility.
Emerging technologies offer promising directions for CWI research. Non-invasive live-imaging methods are crucial for capturing real-time data on plant cell wall dynamics. Synthetic biology enables engineering cells with customized cell walls. Computational models are needed to integrate data across different organisms and cell types. These models could simulate how various factors affect CWI. Machine learning algorithms could predict how changes in one factor affect overall CWI. Nanotechnology can provide real-time insights into CWI through nano-sensors, providing immediate data crucial in such a dynamic process.
Author contributions
NV: Writing – original draft, Writing – review & editing. MS: Writing – original draft, Writing – review & editing. LB: Conceptualization, Writing – original draft, Writing – review & editing, Visualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Research Council of Norway through grants 300550 to M.S. and 334633 to LB and by Research Foundation-Flanders through grant 12R7819N to NV. LB was supported also by the Science and Technology Faculty and the Plant Physiology department of UmU and by the Umeå Plant Science Centre (UPSC), which receives funding from the Knut and Alice Wallenberg Foundation and the Swedish Governmental Agency for Innovation Systems (VINNOVA).
Acknowledgments
We extend our gratitude to colleagues and mentors for their invaluable insights, and to reviewers for their constructive feedback. Our gratitude also goes to the editorial team for their support. Due to space constraints, we regretfully could not cite all deserving works and apologize for any omissions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anderson C. T., Kieber J. J. (2020). Dynamic construction, perception, and remodeling of plant cell walls. Annu. Rev. Plant Biol. 71, 39–69. doi: 10.1146/annurev-arplant-081519-035846
Bacete L., Hamann T. (2020). The role of mechanoperception in plant cell wall integrity maintenance. Plants (Basel Switzerland 9, 574. doi: 10.3390/plants9050574
Bacete L., Mélida H., Miedes E., Molina A. (2018). Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J. 93, 614–636. doi: 10.1111/tpj.13807
Bacete L., Schulz J., Engelsdorf T., Bartosova Z., Vaahtera L., Yan G., et al. (2022). THESEUS1 modulates cell wall stiffness and abscisic acid production in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 119, e2119258119. doi: 10.1073/pnas.2119258119
Basu D., Haswell E. S. (2020). The mechanosensitive ion channel MSL10 potentiates responses to cell swelling in arabidopsis seedlings. Curr. Biol. 30, 2716–2728.e6. doi: 10.1016/j.cub.2020.05.015
Brunner E., Richthammer P., Ehrlich H., Paasch S., Simon P., Ueberlein S., et al. (2009). Chitin-based organic networks: an integral part of cell wall biosilica in the diatom thalassiosira pseudonana. Angew Chem. Int. Ed Engl. 48, 9724–9727.
Carreres B. M., de Jaeger L., Springer J., Barbosa M. J., Breuer G., van den End E. J., et al. (2017). Draft genome sequence of the oleaginous green alga tetradesmus obliquus UTEX 393. Genome Announc 5, e01449–e01416. doi: 10.1128/genomeA.01449-16
Cavalier-Smith T. (2010). Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol. Lett. 6, 342–345. doi: 10.1098/rsbl.2009.0948
Claquin P., Martin-Jézéquel V., Kromkamp J. C., Veldhuis M. J. W., Kraay G. W. (2002). Uncoupling of silicon compared with carbon and nitrogen metabolisms and the role of the cell cycle in continuous cultures of thalassiosira pseudonana (Bacillariophyceae) under light, nitrogen, and phosphorus Control1. J. Phycology 38, 922–930.
Cock J. M., Sterck L., Rouzé P., Scornet D., Allen A. E., Amoutzias G., et al. (2010). The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465, 617–621. doi: 10.1038/nature09016
Codjoe J. M., Miller K., Haswell E. S. (2022). Plant cell mechanobiology: Greater than the sum of its parts. Plant Cell 34, 129–145. doi: 10.1093/plcell/koab230
Coen E., Cosgrove D. J. (2023). The mechanics of plant morphogenesis. Science 379, eade8055. doi: 10.1126/science.ade8055
Coleman H. D., Samuels A. L., Guy R. D., Mansfield S. D. (2008). Perturbed lignification impacts tree growth in hybrid poplar—A function of sink strength, vascular integrity, and photosynthetic assimilation. Plant Physiol. 148, 1229–1237. doi: 10.1104/pp.108.125500
Conley D. J., Kilham S. S., Theriot E. (1989). Differences in silica content between marine and freshwater diatoms. Limnol. Oceanogr. 34, 205–212.
Coomey J. H., Sibout R., Hazen S. P. (2020). Grass secondary cell walls, Brachypodium distachyon as a model for discovery. New Phytol. 227, 1649–1667. doi: 10.1111/nph.16603
Cosgrove D. J. (2022). Building an extensible cell wall. Plant Physiol. 189, 1246–1277. doi: 10.1093/plphys/kiac184
Cronmiller E., Toor D., Shao N. C., Kariyawasam T., Wang M. H., Lee J.-H. (2019). Cell wall integrity signaling regulates cell wall-related gene expression in Chlamydomonas reinhardtii. Sci. Rep. 9, 12204. doi: 10.1038/s41598-019-48523-4
De Coninck T., Van Damme E. J. M. (2021). Review: The multiple roles of plant lectins. Plant Science 313, pp.111096. doi: 10.1016/j.plantsci.2021.111096
Decreux A., Messiaen J. (2005). Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 46, 268–278. doi: 10.1093/pcp/pci026
De Tommasi E., Congestri R., Dardano P., De Luca A. C., Manago S., Rea I., et al. (2018). UV-shielding and wavelength conversion by centric diatom nanopatterned frustules. Sci. Rep. 8, 16285.
de Vries J., Archibald J. M. (2018). Plant evolution: landmarks on the path to terrestrial life. New Phytol. 217 (4), 1428–1434.
de Vries J., Stanton A., Archibald J. M., Gould S. B. (2016). Streptophyte terrestrialization in light of plastid evolution. Trends Plant Sci. 21 (6), 467–476.
Dichtl K., Samantaray S., Wagener J. (2016). Cell wall integrity signalling in human pathogenic fungi: CWI signalling in pathogenic fungi. Cell. Microbiol. 18, 1228–1238. doi: 10.1111/cmi.12612
Domozych D. S., Ciancia M., Fangel J. U., Mikkelsen M. D., Ulvskov P., Willats W. G. T. (2012). The cell walls of green algae: A journey through evolution and diversity. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00082
Dupres V., Alsteens D., Wilk S., Hansen B., Heinisch J. J., Dufrene Y. F. (2009). The yeast Wsc1 cell surface sensor behaves like a nanospring in vivo. Nat. Chem. Biol. 5, 857–862. doi: 10.1038/nchembio.220
Faria-Blanc N., Mortimer J. C., Dupree P. (2018). A transcriptomic analysis of xylan mutants does not support the existence of a secondary cell wall integrity system in arabidopsis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00384
Feng W., Kita D., Peaucelle A., Cartwright H. N., Doan V., Duan Q., et al. (2018). The FERONIA Receptor Kinase Maintains Cell-Wall Integrity during Salt Stress through Ca(2+) Signaling. Curr. Biol. 28, 666–675 e5. doi: 10.1016/j.cub.2018.01.023
Feng X., Zheng J., Irisarri I., Yu H., Zheng B., Ali Z., et al. (2023). Chromosome-level genomes of multicellular algal sisters to land plants illuminate signaling network evolution. bioRxiv.
Frantz C., Stewart K. M., Weaver V. M. (2010). The extracellular matrix at a glance. J. Cell Sci. 123, 4195–4200. doi: 10.1242/jcs.023820
Friedland J. C., Lee M. H., Boettiger D. (2009). Mechanically activated integrin switch controls α 5 β 1 function. Science 323, 642–644. doi: 10.1126/science.1168441
Galindo-Trigo S., Gray J. E., Smith L. M. (2016). Conserved roles of crRLK1L receptor-like kinases in cell expansion and reproduction from algae to angiosperms. Front. Plant Sci. 07. doi: 10.3389/fpls.2016.01269
Gigli-Bisceglia N., Engelsdorf T., Hamann T. (2020). Plant cell wall integrity maintenance in model plants and crop species-relevant cell wall components and underlying guiding principles. Cell. Mol. Life Sci. 77, 2049–2077. doi: 10.1007/s00018-019-03388-8
Goessling J. W., Su Y., Cartaxana P., Maibohm C., Rickelt L. F., Trampe E. C. L., et al. (2018). Structure-based optics of centric diatom frustules: modulation of the in vivo light field for efficient diatom photosynthesis. New Phytol. 219, 122–134.
Gonneau M., Desprez T., Martin M., Doblas V. G., Bacete L., Miart F., et al. (2018). Receptor kinase THESEUS1 is a rapid alkalinization factor 34 receptor in arabidopsis. Curr. Biol. 28, 2452–2458.e4. doi: 10.1016/j.cub.2018.05.075
Gorzsás A., Stenlund H., Persson P., Trygg J., Sundberg B. (2011). Cell-specific chemotyping and multivariate imaging by combined FT-IR microspectroscopy and orthogonal projections to latent structures (OPLS) analysis reveals the chemical landscape of secondary xylem: The chemical landscape of secondary xylem. Plant J. 66, 903–914. doi: 10.1111/j.1365-313X.2011.04542.x
Hamant O., Inoue D., Bouchez D., Dumais J., Mjolsness E. (2019). Are microtubules tension sensors? Nat. Commun. 10 (1), 2360.
Hamilton E. S., Jensen G. S., Maksaev G., Katims A., Sherp A. M., Haswell E. S. (2015). Mechanosensitive channel MSL8 regulates osmotic forces during pollen hydration and germination. Science 350, 438–441. doi: 10.1126/science.aac6014
Hanschen E. R., Marriage T. N., Ferris P. J., Hamaji T., Toyoda A., Fujiyama A., et al. (2016). The Gonium pectorale genome demonstrates co-option of cell cycle regulation during the evolution of multicellularity. Nat. Commun. 7, 11370. doi: 10.1038/ncomms11370
Hildebrand M. (2008). Diatoms, biomineralization processes, and genomics. Chem. Rev. 108, 4855–4874.
Hildebrand M., Lerch S. J. L., Shrestha R. P. (2018). Understanding diatom cell wall silicification–moving forward. Front. Mar. Sci. 5.
Hou C., Tian W., Kleist T., He K., Garcia V., Bai F., et al. (2014). DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Res. 24, 632–635. doi: 10.1038/cr.2014.14
Hynes R. O. (2009). The extracellular matrix: not just pretty fibrils. Science 326, 1216–1219. doi: 10.1126/science.1176009
Keeling P. J., Burger G., Durnford D. G., Lang B. F., Lee R. W., Pearlman R. E., et al. (2005). The tree of eukaryotes. Trends Ecol. Evol. 20, 670–676. doi: 10.1016/j.tree.2005.09.005
Knox J. (2008). Revealing the structural and functional diversity of plant cell walls. Curr. Opin. Plant Biol. 11, 308–313. doi: 10.1016/j.pbi.2008.03.001
Kohorn B. D. (2016). Cell wall-associated kinases and pectin perception. J. Exp. Bot. 67, 489–494. doi: 10.1093/jxb/erv467
Kono K., Al-Zain A., Schroeder L., Nakanishi M., Ikui A. E. (2016). Plasma membrane/cell wall perturbation activates a novel cell cycle checkpoint during G1 in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 113, 6910–6915. doi: 10.1073/pnas.1523824113
Kotzsch A., Pawolski D., Milentyev A., Shevchenko A., Scheffel A., Poulsen N., et al. (2016). Biochemical composition and assembly of biosilica-associated insoluble organic matrices from the diatom thalassiosira pseudonana. J. Biol. Chem. 291, 4982–4997.
Lehti-Shiu M. D., Zou C., Hanada K., Shiu S.-H. (2009). Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150, 12–26. doi: 10.1104/pp.108.134353
Levin D. E. (2005). Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. reviews: MMBR 69, 262–291. doi: 10.1128/MMBR.69.2.262-291.2005
Levin D. E. (2011). Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189, 1145–1175. doi: 10.1534/genetics.111.128264
Maksaev G., Haswell E. S. (2012). MscS-Like10 is a stretch-activated ion channel from arabidopsis thaliana with a preference for anions. Proc. Natl. Acad. Sci. 109, 19015–19020. doi: 10.1073/pnas.1213931109
Malerba M. E., Ghedini G., Marshall D. J. (2020). Genome size affects fitness in the eukaryotic alga dunaliella tertiolecta. Curr. Biol. 30, 3450–3456.e3. doi: 10.1016/j.cub.2020.06.033
Mann D. G., Droop S. J. M. (1996). Biodiversity, biogeography and conservation of diatoms. Hydrobiologia 336, 19–32.
Martín-Dacal M., Fernández-Calvo P., Jiménez-Sandoval P., López G., Garrido-Arandía M., Rebaque D., et al. (2023). Arabidopsis immune responses triggered by cellulose- and mixed-linked glucan-derived oligosaccharides require a group of leucine-rich repeat malectin receptor kinases. Plant J. 113, 833–850. doi: 10.1111/tpj.16088
McNair H. M., Brzezinski M. A., Krause J. W. (2018). Diatom populations in an upwelling environment decrease silica content to avoid growth limitation. Environ. Microbiol. 20, 4184–4193.
Mecchia M. A., Rövekamp M., Giraldo-Fonseca A., Meier D., Gadient P., Vogler H., et al. (2022). The single Marchantia polymorpha FERONIA homolog reveals an ancestral role in regulating cellular expansion and integrity. Development 149, dev200580. doi: 10.1242/dev.200580
Mélida H., Sopeña-Torres S., Bacete L., Garrido-Arandia M., Jordá L., López G., et al. (2018). Non-branched beta-1,3-glucan oligosaccharides trigger immune responses in Arabidopsis. Plant J. 93, 34–49. doi: 10.1111/tpj.13755
Morris E.R., Walker J. C. (2003). Receptor-like protein kinases: the keys to response. Current opinion in plant biology, 64, pp.339–342.
Nakagawa Y., Katagiri T., Shinozaki K., Qi Z., Tatsumi H., Furuichi T., et al. (2007). Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc. Natl. Acad. Sci. U.S.A. 104, 3639–3644. doi: 10.1073/pnas.0607703104
Nakamura Y., Iwata I., Hori R. S., Uchiyama N., Tuji A., Fujita M. J., et al. (2018). Elemental composition and ultrafine structure of the skeleton in shell-bearing protists-A case study of phaeodarians and radiolarians. J. Struct. Biol. 204, 45–51. doi: 10.1016/j.jsb.2018.06.008
Nelson D. M., Treguer P., Brzezinski M. A., Leynaert A., Queguiner B. (1995). Production and dissolution of biogenic silica in the ocean - revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob. Biogeochem Cycles 9, 359–372.
Ochs J., LaRue T., Tinaz B., Yongue C., Domozych D. S. (2014). The cortical cytoskeletal network and cell-wall dynamics in the unicellular charophycean green alga penium margaritaceum. Ann. Bot. 114 (6), 1237–1249.
Ortiz-Morea F. A., Liu J., Shan L., He P. (2022). Malectin-like receptor kinases as protector deities in plant immunity. Nat. Plants 8, 27–37. doi: 10.1038/s41477-021-01028-3
Pančić M., Torres R. R., Almeda R., Kiørboe T. (2019). Silicified cell walls as a defensive trait in diatoms. Proc. Biol. Sci. 286, 20190184.
Paszek M. J., Boettiger D., Weaver V. M., Hammer D. A. (2009). Integrin clustering is driven by mechanical resistance from the glycocalyx and the substrate. PloS Comput. Biol. 5, e1000604. doi: 10.1371/journal.pcbi.1000604
Peaucelle A., Braybrook S., Höfte H. (2012). Cell wall mechanics and growth control in plants: the role of pectins revisited. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00121
Pondaven P., Gallinari M., Chollet S., Bucciarelli E., Sarthou G., Schultes S., et al. (2007). Grazing-induced changes in cell wall silicification in a marine diatom. Protist 158, 21–28.
Radhakrishnan A., Chellapandian H., Ramasamy P., Jeyachandran S. (2023). RBack2Basics: animal lectins: an insight into a highly versatile recognition protein. J Proteins Proteom. doi: 10.1007/s42485-022-00102-4
Ratke C., Terebieniec B. K., Winestrand S., Derba-Maceluch M., Grahn T., Schiffthaler B., et al. (2018). Downregulating aspen xylan biosynthetic GT43 genes in developing wood stimulates growth via reprograming of the transcriptome. New Phytol. 219, 230–245. doi: 10.1111/nph.15160
Rebaque D., Hierro I., López G., Bacete L., Vilaplana F., Dallabernardina P., et al. (2021). Cell wall-derived mixed-linked β-1,3/1,4-glucans trigger immune responses and disease resistance in plants. Plant J. 106, 601–615. doi: 10.1111/tpj.15185
Richthammer P., Bormel M., Brunner E., van Pee K. H. (2011). Biomineralization in diatoms: the role of silacidins. ChemBioChem 12, 1362–1366.
Somssich M., Khan G. A., Persson S. (2016). Cell wall heterogeneity in root development of arabidopsis. Front. Plant Sci. 07. doi: 10.3389/fpls.2016.01242
Sørensen I., Pettolino F. A., Bacic A., Ralph J., Lu F., et al. (2011). The charophycean green algae provide insights into the early origins of plant cell walls. J. Plant 68 (2), 201–211.
Sumper M., Brunner E., Lehmann G. (2005). Biomineralization in diatoms: characterization of novel polyamines associated with silica. FEBS Lett. 579, 3765–3769.
Tran L. S., Urao T., Qin F., Maruyama K., Kakimoto T., Shinozaki K., et al. (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 20623–20628. doi: 10.1073/pnas.0706547105
Urao T., Yakubov B., Satoh R., Yamaguchi-Shinozaki K., Seki M., Hirayama T., et al. (19991743). A transmembrane hybrid-type histidine kinase in arabidopsis functions as an osmosensor. Plant Cell 11. doi: 10.2307/3871051
Westermann J., Streubel S., Franck C. M., Lentz R., Dolan L., Boisson-Dernier A. (2019). An evolutionarily conserved receptor-like kinases signaling module controls cell wall integrity during tip growth. Curr. Biol. 29, 3899–3908.e3. doi: 10.1016/j.cub.2019.09.069
Weston B. S., Malhas A. N., Price R. G. (2003). Structure–function relationships of the extracellular domain of the autosomal dominant polycystic kidney disease-associated protein, polycystin-1. FEBS Lett. 538, 8–13. doi: 10.1016/S0014-5793(03)00130-3
Wohlbach D. J., Quirino B. F., Sussman M. R. (2008). Analysis of the arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 20, 1101–1117. doi: 10.1105/tpc.107.055871
Yamanaka T., Nakagawa Y., Mori K., Nakano M., Imamura T., Kataoka H., et al. (2010). MCA1 and MCA2 that mediate Ca2+ uptake have distinct and overlapping roles in arabidopsis. Plant Physiol. 152, 1284–1296. doi: 10.1104/pp.109.147371
Yool A., Tyrrell T. (2003). Role of diatoms in regulating the ocean's silicon cycle. Glob Biogeochem Cycles 17, 1103–1121.
Yoshimura K., Iida K., Iida H. (2021). MCAs in Arabidopsis are Ca2+-permeable mechanosensitive channels inherently sensitive to membrane tension. Nat. Commun. 12, 6074. doi: 10.1038/s41467-021-26363-z
Yuan F., Yang H., Xue Y., Kong D., Ye R., Li C., et al. (2014). OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in arabidopsis. Nature 514, 367–371. doi: 10.1038/nature13593
Zhang B., Gao Y., Zhang L., Zhou Y. (2021). The plant cell wall: Biosynthesis, construction, and functions. J. Integr. Plant Biol. 63, 251–272. doi: 10.1111/jipb.13055
Zhong R., Cui D., Ye Z. (2019). Secondary cell wall biosynthesis. New Phytol. 221, 1703–1723. doi: 10.1111/nph.15537
Keywords: mechanosensing, cell wall plasticity, cell wall dynamics, cell wall physicochemical properties, Streptophyta, Zygnematophyceae, Bacillariophyceae, Charophyceae
Citation: Vukašinović N, Serif M and Bacete L (2023) Cracking the green wall code: insights into cell wall integrity across organisms. Front. Plant Physiol. 1:1323899. doi: 10.3389/fphgy.2023.1323899
Received: 18 October 2023; Accepted: 30 October 2023;
Published: 27 November 2023.
Edited by:
Yohann Boutté, UMR200 Laboratoire de Biogenèse Membranaire (LBM), FranceReviewed by:
Jozef Mravec, Institute of Plant Genetics and Biotechnology (SAS), SlovakiaCopyright © 2023 Vukašinović, Serif and Bacete. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura Bacete, bGF1cmEuYmFjZXRlQHVtdS5zZQ==
 Nemanja Vukašinović
Nemanja Vukašinović Manuel Serif
Manuel Serif Laura Bacete
Laura Bacete