- Université Paris-Saclay, INRAE, AgroParisTech, Institut Jean-Pierre Bourgin (IJPB), Versailles, France
Plant development and reproduction are complex processes during which an individual fulfills its life cycle, starting from germination and the elaboration of new organs and growth, leading to the formation of reproductive structures and ultimately terminating in the production of the next generation. These mechanisms are the result of a long evolutionary history that has led to sophisticated regulatory mechanisms involving multiple levels of regulators. MicroRNAs (miRNAs) are a class of small regulatory molecules that play a pivotal role in regulatory networks by negatively controlling target genes. Since miRNA very first identification twenty years ago, they have attracted much interest for their role as essential regulators of plant development. In this review, we propose a comprehensive and critical analysis of the importance of miRNAs during plant development and reproduction. We begin by presenting the current understanding of miRNAs’ evolutionary history, biogenesis, mode of action, position in regulatory networks, and their potential as mobile molecules, exploring how these aspects contribute to their functions in plant development and reproduction. Then, we explore the genetic strategies employed to effectively analyze their roles, with an emphasis on recent advancements resulting from genome editing techniques. Next, we focus on miRNA contributions to four crucial processes: growth, organ patterning and identity, life cycle progression and reproduction. Through this analysis, the importance of miRNAs during plant development and reproduction emerges, which we finally discuss in light of the current view miRNAs’ roles during animal development.
1 Introduction
MicroRNAs (miRNAs) are small, single-stranded regulatory RNAs that repress target gene expression. It is generally assumed that they have evolved independently several times within eukaryotes (Tarver et al., 2012), though the opposite view has also been proposed (Moran et al., 2017; Tripathi et al., 2022). Since their identification in plants more than twenty years ago (Llave et al., 2002; Reinhart et al., 2002), miRNAs have been identified in many plant species and appear to form a diverse genetic tool set encoded by multiple genes. For instance, 150 high-confidence MIRNA genes coding for 26 miRNA families have been identified in maize (Zhang et al., 2009). The observation that the first identified miRNAs could target conserved transcription factors playing key roles during plant development quickly promoted these molecules as important regulators of plant development (Rhoades et al., 2002; Floyd and Bowman, 2004). This perspective was further strengthened by the observation that key players in the biogenesis and function of plant miRNAs had previously been identified during developmental genetic screens (Garcia, 2008). Whereas it is clear that miRNAs as a whole are essential for plants because strong loss-of-function mutants in miRNA maturation machinery components display embryo lethal phenotypes (Schauer et al., 2002), the role of individual miRNAs is far less understood. In fact, in plants, like in other eukaryotes the importance of miRNAs during development is still a matter of debate. In this review, we will delve into the contribution of miRNAs to plant development. Our objective is not to present an exhaustive list of their roles but rather to explore how our current understanding of their evolution, biogenesis, and integration into regulatory networks sheds light on their significance in development. We will concentrate on recent genetic analyses that have exploded over the past five years through the use of genome editing techniques. These methodologies enable the systematic knockout of all members of MIRNA gene families, allowing to genetically define both their specific and redundant roles in plant development and reproduction. The roles of miRNAs that thus emerges in plants will be compared with the roles of miRNAs during animal development.
2 The miRNA repertoire of a plant is the result of its evolutionary history.
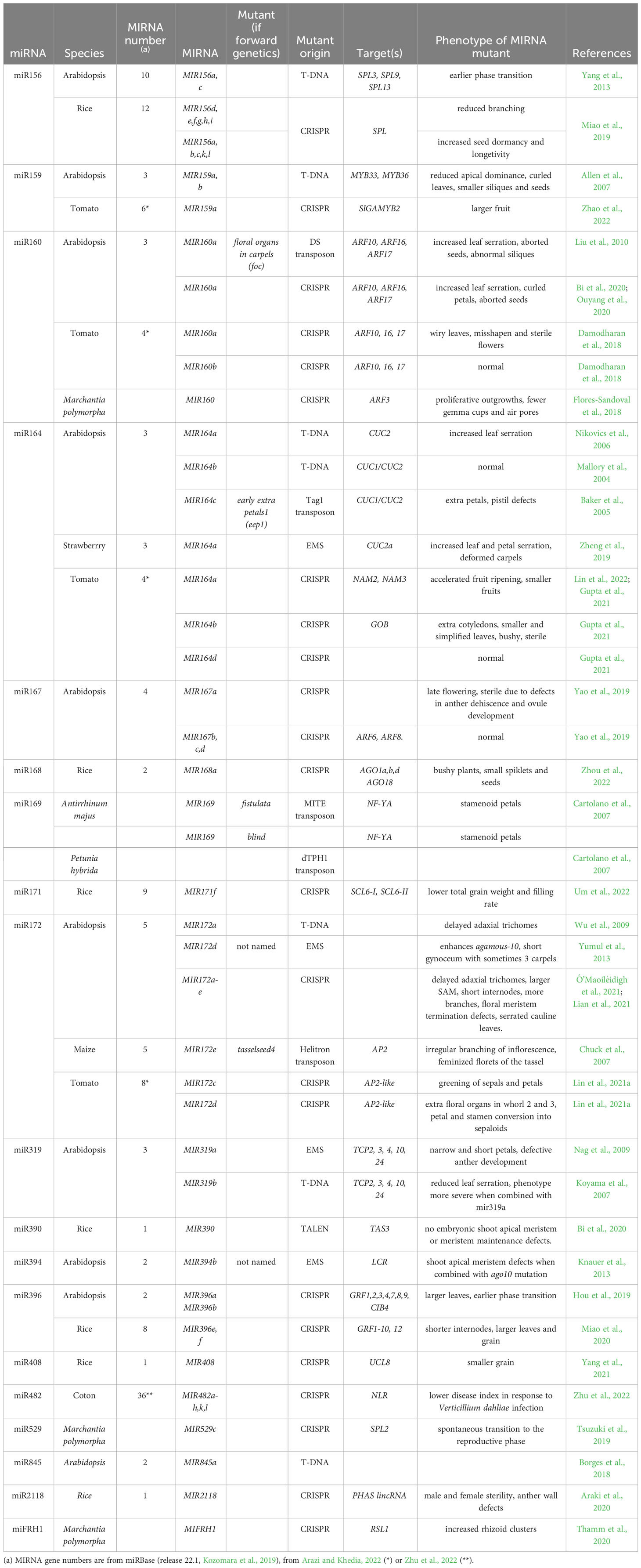
One criterion to assess the importance of miRNAs in plant development is to look at their evolutionary history and conservation. An expanding wealth of miRNA sequences is available for many species, which can be however obscured by the difficulty in distinguishing bona fide miRNA from other small RNAs (Taylor et al., 2014). A phylogenomic framework was used to attempt to reconstruct miRNA families evolution, which suggested that plant current miRNAs are in an important part inherited from the ancestral embryophyte and spermaphyte (Taylor et al., 2014). These studies showed that miRNAs root deep into the plant phylogeny, although their exact origin is still unknown as no shared miRNAs were found between the unicellular green algae Chlamydomonas reinhardtii and land plants (Nozawa et al., 2012). In addition to such ancient miRNAs, a large expansion of the miRNA families may have also occurred after the divergence of the flowering plants. To support evolution of the miRNAs, several mechanisms of miRNA gene formation have been proposed (Cui et al., 2017). As a result of such evolution, the miRNA repertoire of each plant species is formed by a continuum of ancient to younger miRNAs. The former ones are highly conserved, whereas the latter ones are specific to a genus, species, or even a particular accession (Figure 1). MIRNA genes often form families, with conserved miRNAs tending to form larger families and being on average expressed at higher levels than non-conserved ones (Figure 1; Rajagopalan et al., 2006; Fahlgren et al., 2007). An important hint pointing to the importance of miRNAs in plant development was the observation that conserved miRNAs tend to preferentially target transcription factors with well-established roles during plant development. Furthermore, this targeting was found to be evolutionary conserved (Rhoades et al., 2002; Floyd and Bowman, 2004). In contrast, targets of non-conserved miRNAs code for proteins with much more diverse biological functions (Fahlgren et al., 2007). In summary, miRNAs constitute a genetic toolkit that is both deeply conserved within the plant lineage for some of them and also comprises a highly flexible gene set which has the potential to contribute to plant morphological innovations during evolution (Rast-Somssich et al., 2015).
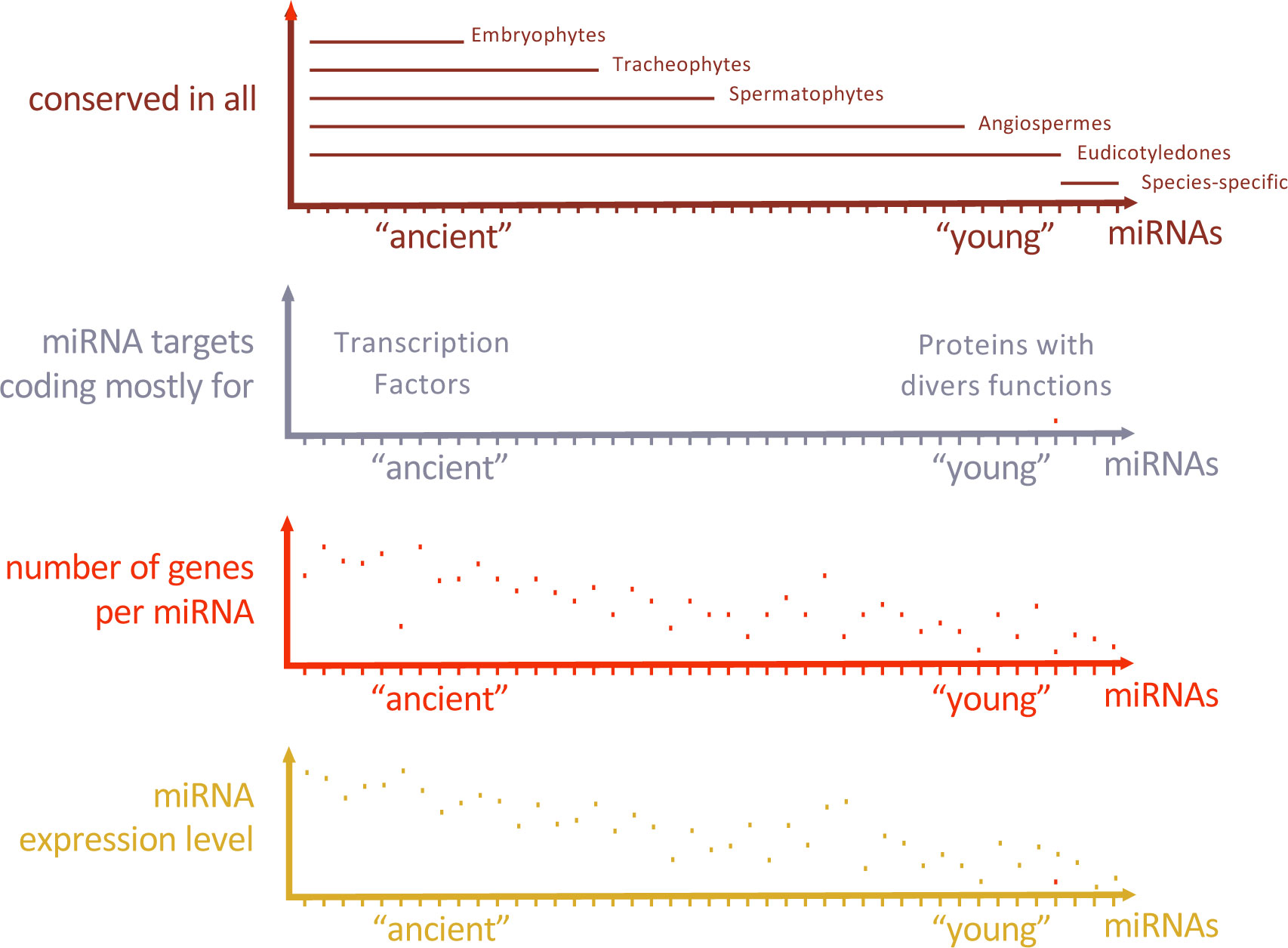
Figure 1 Characteristics of the typical miRNA repertoire of a dicotyledon. The miRNA repertoire is formed by ancient miRNAs shared with all Embryophytes and more recent ones shared with fewer lineages, some of them being specific to the species. Ancient miRNAs preferentially target genes coding for transcription factors while younger miRNAs target genes coding for proteins with much more diverse biological functions. The number of genes coding for ancient miRNA families and their expression levels tend to be higher than for younger miRNAs.
3 MiRNA biogenesis and action provide entry points to regulate their activity
In order to understand how miRNAs regulate plant development, it’s imperative to gain insight into their production, mechanisms of action, and how the modulation of these processes can influence miRNA activity. The biogenesis and action of miRNAs is a complex mechanism involving multiple actors. Some of these actors are specific to the miRNA pathway while others have roles in other biological processes. We will not detail here the growing knowledge of the mechanisms and actors at play, as very detailed reviews are available (Yu et al., 2017; Li and Yu, 2021). Briefly, miRNA genes are transcribed by the RNA polymerase II. Their transcripts are capped and polyadenylated and often spliced as they may extend over several exons. Part of the resulting pre-miRNA folds into a hairpin like structure that provides structural clues to direct a two-step maturation. First, the pre-miRNA transforms into the primary miRNA, characterized by a hairpin-like structure. Second, this primary miRNA undergoes further processing to form a small duplex structure, which consists of the miRNA itself and its complementary strand, often referred to as the miRNA* (miRNA star). This processing is mediated by the RNase DICER LIKE 1 and interacting proteins such as HYPONASTIC LEAVES 1 and SERRATE provide efficiency and precision to it. The miRNA is incorporated into the RNA INDUCED SILENCING COMPLEX (RISC) containing an ARGONAUTE (AGO) protein, often AGO1. Following the export to the cytoplasm, the miRNA containing RISC interacts with the target mRNA and triggers either its cleavage and/or inhibits its translation. MiRNA stability is also regulated: the miRNA is methylated at its 3’ end to inhibit its degradation by preventing the addition of a short U tail, and nucleases may degrade the U-tailed or 3’ non-methylated miRNA. It thus appears that the biogenesis of miRNAs and their activity is a very dynamic process with multiple levels of regulation (Meng et al., 2011). One central question that arises from this is to what extent this general pathway can be specifically modulated for one or a subgroup of miRNAs. This is far from being understood although mutations in miRNA biogenesis actors may differentially affect the miRNA population. Consequently, the extent to which regulation of miRNA biogenesis and activity is utilized as a mechanism to govern plant development remains an unanswered question. In contrast, it is clear that transcriptional regulation of MIRNA gene expression is a major determinant of miRNA activity level. Indeed, expression of MIRNA genes can be regulated by specific transcription factors (Yang et al., 2021) and chromatin remodeling resulting from histone modifications or DNA methylation (Creasey et al., 2014; Xu et al., 2016; Hou et al., 2019). Such predominance of the transcriptional regulation level for MIRNA genes allows them to be fully integrated into gene regulatory networks (GRNs).
4 MiRNA integration in developmental gene regulatory networks
Plant development requires the integrated action of regulators forming GRNs. Pioneer work in Escherichia coli identified recurrent “network motifs” as the building blocks for larger networks (Milo et al., 2002; Shen-Orr et al., 2002). MiRNAs are embedded within the architecture of these GRNs as they target a diverse range of regulators including transcription factors (TF). In many cases, miRNA-mediated gene regulatory loops are formed that may be instrumental to network (Martinez et al., 2008). However, few studies have looked at miRNA function in plants from this network perspective. Here we will discuss a few examples that have been best characterized.
Plant architecture relies on the branching pattern which is dependent on the activity of axillary meristems. Zinc finger homeodomain TFs (ZF-HD TFs) with notably HB34 have been identified as regulators of plant architecture. Indeed, HB34 positively regulates SPL10 while repressing MIR157d thus forming a feed-forward loop (FFL) (Lee et al., 2022). FFL are three-component patterns where a TF regulates the two other components that are already involved in regulatory interactions (Figure 2). The regulation of age-dependent cell death is another example for FFL including a miRNA (Kim et al., 2009). Here, ETHYLENE INSENSITIVE 2 (EIN2) is required to induce ORESARA1 (ORE1) expression which is targeted by miR164, and miR164 expression decreases with aging in an EIN2-dependent manner, forming a FFL. These two FFL examples represent coherent FFL where the sign of the indirect regulation path through the miRNA is the same as the sign of the direct regulation path (Figure 2). Work in E. coli suggests that coherent FFL generally introduce time-delay within networks. Therefore, positioning a miRNA as an intermediate component of coherent FFL in plants may contribute to network dynamics. Incoherent FFL are formed when the sign of the indirect regulation path through the miRNA and the sign of the direct regulation are different. Although less intuitive, incoherent FFL are also instrumental for developmental and environmental responses and may include a miRNA. For instance, nitrate response in the root triggers both miR393 and its target AUXIN SIGNALING F-BOX 3 (AFB3) to activate auxin responses and promote lateral root development in an incoherent FFL (Vidal et al., 2010). This mechanism might provide a rapid developmental response at first, then a way of adjusting the level of response when optimal response has been reached, allowing it to adapt to environmental fluctuations.
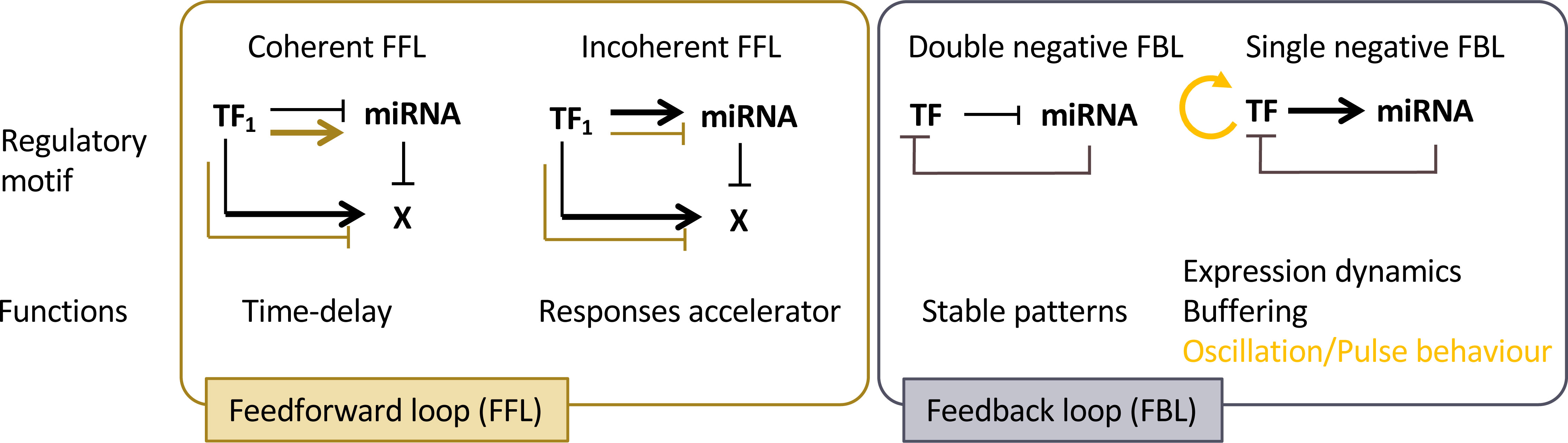
Figure 2 Regulatory motifs including miRNA are integrated into Gene Regulatory Networks. Feedforward (FBL) and Feedback loop (FBL) including a miRNA provide distinct functions which first depend on the sign of the regulatory interactions - coherent FFL when the TF regulates the gene X in the same way through both path; incoherent FFL when the TF regulates X in opposite way - then on the parameters of the regulatory motif such as expression levels and the stability of its components. For instance, oscillation or pulse behaviour may arise when an autoregulatory loop is present (orange arrow) depending on network parameters.
MiRNA are also integrated within networks through feedback loop motifs (FBL). These particular network motifs including miRNA are called composite FBL (Figure 2). As miRNAs exert negative regulatory effects on their targets, only two kinds of FBL exist: Single Negative (SN) - when the TF positively regulates a miRNA-, and Double Negative (DN) - when the TF negatively regulates the miRNA.
DN composite FBL results in complementary patterns that may allow spatialization of key developmental processes by maintaining terminal differentiated state (Johnston et al., 2005). In Arabidopsis flower development, specification between perianth and reproductive organs relies on such DN-FBL where the APETALA2 TF (AP2) activity is restricted to sepals and petals primordia by the action of miR172 in center of the floral meristem; LEUNING and SEUSS co-repressors restrict miR172 expression pattern in a AP2-dependent manner thus forming a composite FBL specifying flower whorls (Grigorova et al., 2011).
SN composite FBL may have various functions depending on the level of expression, the stability and/or additional regulatory interactions of its components. miRNA-TF feedback network motifs are over-represented in C. elegans and may confer robustness or oscillatory target gene expression patterns (Martinez et al., 2008). The nature of SN composite FBL implies that the TF activates the expression of the miRNA which is negatively regulating it in the same expression domain. If this regulation happens with a time delay, then the expression dynamic of the TF may be impacted. If TF autoregulatory activation occurs, then it can give rise to oscillatory or pulse behaviour depending on the parameters of the network motif. In some cases, the same cells express at the same time the TF and the miRNA. In this particular case, the function of the regulatory FBL could be to buffer TF expression to produce reproducible output. In plants, several TF and their inhibitory miRNA regulators are co-expressed in the same tissue such as miR160-ARF10/16/17, miR164-CUC1/2, miR156-SPL9/15 (Baker et al., 2005; Nikovics et al., 2006; Wang et al., 2008; Dai et al., 2021), but whether they are forming SN composite FBL and how these modules fine-tune gene expression remain to be determined.
5 MicroRNAs as mobile signals
Regulation of development involves coordination of basic cellular processes such as proliferation and differentiation at the organ or tissue levels. It also involves coordination across plant tissues or organs at the whole plant level. Communication is central for both processes and growing evidence enlightens how miRNAs may contribute to this. Indeed, like other small RNAs, miRNAs can be mobile at different distances: from cell-to-cell, between different plant organs or between individual (of the same species or not) through direct contact or through the environment (for recent reviews see eg Chen and Rechavi, 2022; Loreti and Perata, 2022; Yan and Ham, 2022). For instance, miR394 produced in the outer layer of the shoot apical meristem moves into deeper layers to define a region competent for stem cell formation (Knauer et al., 2013). Thus, miRNA mobility may provide positional information that contribute to patterning in plants. More precisely, the effects of such miRNA gradients can be at least twofold (Figure 3). During the establishment of adaxial/abaxial leaf polarity, miR166 is expressed within the abaxial epidermis. It then diffuses toward the uppermost cell layers, playing a role in creating a sharp boundary in the expression pattern of its targets, the Class III HD-ZIP genes, which are confined to the adaxial half of the leaf primordium. (Tatematsu et al., 2015; Skopelitis et al., 2017). In the developing root, a similar gradient of miR166 is formed as a result of its expression in the endodermis and movement to the root centre. However, here it leads to an opposite, increasing gradient of the Class III HD-ZIP expression levels from the endodermis to the more inner stellar tissues (Carlsbecker et al., 2010; Miyashima et al., 2011). Thus, mobile miR166 can have two different effects on its target expression pattern and thus differentiation: while in the leaf it leads to the sharp transition of the Class III HD-ZIP levels that contributes the formation of the contrasted abaxial and adaxial domains, in the root, it generates graded Class III HD-ZIP levels contributing to the differentiation of several cell types from endodermis, pericycle, proto- to metaxylem (Figure 3A). Such small range movement is a widespread mechanism affecting many miRNAs but which remains selective for the miRNAs moving, the orientation of the movement and depends on the cell type (Skopelitis et al., 2018; Brosnan et al., 2019). This process is likely to occur through plasmodesmata, as it can be impeded by their closure via callose synthesis (Vatén et al., 2011). However, it’s worth noting that other routes of diffusion may also be feasible. miRNAs can also move at a larger distance, between organs, possibly through the flux of phloem sieve. Such long-range miRNA movement constitute systemic signals that coordinate development and physiology at the whole plant scale (Figure 3B). For instance, miR172 and miR156 have been proposed to act as long-distance, graft- transmissible signals regulating tuberization in potato (Martin et al., 2009; Bhogale et al., 2014). Movement of miR399 from shoot to roots signals the onset of inorganic phosphate deficiency and represses its target PHO2 that regulates phosphate uptake from the root environment (Lin et al., 2008). Like for the small range movement, the favored form of transport is mature miRNAs, possibly free of bound AGO proteins (Brosnan et al., 2019; Dalmadi et al., 2019; Brioudes et al., 2021).
Figure 3 miRNAs as mobile signals. (A) miRNAs can move from cell-to-cell in an organ, generating a gradient of concentration that can either lead to an opposite gradient of expression of its target or to a sharp transition between cells expressing or not expressing the target. (B) miRNAs can move throughout the organism to generate a systemic signal. (C) During the interaction of a plant with a pathogen or a parasitic plant, miRNAs can be exchanged between the two neighboring cells to silence defense or virulence genes. (D) miRNAs can be released to the soil by plants and target genes of distant plants, thus possibly generating an allelopathic signal.
MiRNAs also constitute signals exchanged between two plants or with other organisms. For instance, miRNAs are transmitted from the parasitic plant Cuscuta campestris through specialized feeding structures called haustoria to the host plant. These parasitic miRNAs target conserved host genes that restrict parasite growth. Remarkably, parasitic miRNAs are typically unusually 22 nucleotides in length and have the capacity to initiate the production of secondary siRNAs, which can lead to the amplification of silencing mechanisms. (Shahid et al., 2018). Conversely, the host may also produce miRNAs to limit pathogens action: in response to Verticillium dahliae infection, cotton increases the expression and export of specific miRNAs to the pathogen to target virulence genes (Zhang et al., 2016). Whereas these examples illustrate miRNA exchanges between species in close physical connection, miRNAs can also be exchanged between distant individuals through a shared environment (Betti et al., 2021) (Figures 3C, D), with the potential to generate allelopathic signals. Again, response to these miRNAs requires the production of secondary siRNAs, which seems therefore a conserved mechanism to amplify weak signals provided by low amounts of mobile miRNAs. Cross-individual miRNA transport is thought to occur at least in part through extracellular vesicles containing different RNA binding proteins including AGO1, which may also contribute to the selective loading of a miRNA subpopulation (He et al., 2021).
6 Genetic strategies to approach miRNA functions during plant development and reproduction.
As discussed in the preceding sections miRNAs have multiple characteristics that give them the potential to play prominent roles during plant development and reproduction. In this context, the royal road to decipher their roles is a functional approach analyzing the consequences of abnormal miRNA function through genetics. We will first critically assess the different strategies used to functionally analyze miRNAs before discussing their importance in developmental processes.
Probably the most widely used approach to establish functionally the roles of a miRNA, as it is amongst the easiest and fastest to perform, is to over express the miRNA of interest. However, it is important to note that this strategy, like any ectopic gain-of-function approach, might not provide a direct insight into the miRNA’s normal physiological function. Indeed, using a promoter that differs from the endogenous promoter, either in terms of its expression pattern or level, can result in the down-regulation of targets that are distinct from the endogenous targets of this particular MIRNA gene, but are targets of other members of the same MIRNA gene family. Hence, miRNA overexpression is rather a way to reveal the collective roles of the miRNA targets which are down-regulated by miRNA over expression. Yet, such an approach remains indicative of the possible roles of the miRNA and is at least a very good way to experimentally confirm target gene predicted in silico, which can however be blurred by feedback mechanisms (Schwab et al., 2005).
Another widely used strategy is to express a target gene that is made resistant to the miRNA by introducing silent mutations in the miRNA binding site either via transgenesis or genome-editing (Lin et al., 2021b). While this strategy is highly effective in uncovering the impact of a miRNA on the expression and function of a specific target, it does not encompass its effect on multiple targets, thus overlooking the full spectrum of the miRNA’s functions. Nevertheless, this strategy is still the most effective for bridging the gap between a miRNA and a phenotype through its effects on one or several particular targets.
Another widely-used strategy based on transgenics to get access to the function of a miRNA is to express a decoy target to partially inactivate the miRNA. These decoy targets are RNA molecules that contain one or several stretches highly complementary to the miRNA that, however, have at the cleavage site a mismatch loop preventing cleavage while still allowing the binding of the miRNA. Therefore, they compete for the binding of miRNAs to endogenous targets, sequester them and lead to their degradation. A first generation of such decoy mRNAs called MIMICs was based on the long non-protein coding RNA IPS1 (INDUCED BY PHOSPHATE STARVATION 1) and contains one miRNA binding site (Franco-Zorrilla et al., 2007; Todesco et al., 2010), while a second generation called Short Tandem Target MIMICs (STTMs) contains two binding sites separated by a spacer (Yan et al., 2012). Other artificially-engineered RNAs (miRNA sponges) or endogenous RNAs (circular RNAs or long non coding RNAs) may also sequester miRNAs through their binding sites (Reichel et al., 2015; Tang et al., 2021; Zhou et al., 2021; Liu et al., 2023). These approaches are easy to set up when stable plant transformation is feasible, while transient transformation using a virus-based delivery system can be used too (Yan et al., 2014). Collections of MIMICS and STTMs lines are available for Arabidopsis, tomato, rice and maize (Todesco et al., 2010; Peng et al., 2018). Such approaches go with the advantages of generating pseudo-allelic series in which different levels of miRNA inactivation occur, are transferable to different species in the case of conserved miRNAs and allow targeting in a dominant fashion multiple members of a MIRNA gene family. The reverse of the coin is that the contribution of individual gene members of the MIRNA family cannot be investigated. Also, the efficiency of such strategies depends on the approach used and most importantly on the targeted miRNA, and remains unpredictable (Reichel et al., 2015). In addition, miRNA down-regulation is only partial. For instance, a 65% reduction of miR164 was observed in STTM tomato seedlings, while a 90% reduction was observed in the pericarp of developing tomato fruits when only MIR164a, one of the 4 MIR164 genes was inactivated (Gupta et al., 2021). In addition to being incomplete, the level of miRNA inactivation may vary within a plant both temporally or spatially (for instance due to variation of the activity of the promoter driving the MIMIC expression). Hence, while MIMIC and STTM approaches are highly valuable for assessing the general functions of a miRNA in plant development, they do exhibit limitations when it comes to elucidating the specific roles of individual MIRNA genes or uncovering their complete range of functions.
All the methods discussed above have been developed to overcome the initial difficulty to identify mutant lines impaired in miRNA production. Such a difficulty results first from the fact that MIRNA genes form families, sometimes with numerous members and that their roles are often at least partially redundant. Therefore, mutants for each member have to be identified and higher order mutants may have to be generated. In addition, finding miRNA insertion (T-DNA or transposon) mutants by chance can be unsuccessful as mature miRNA and their pri-miRNA precursors are small. Alternatively, point mutations generated by chemical mutagenesis need to affect the mature miRNA as point mutations elsewhere in the pri-miRNA do not necessarily impact its maturation into miRNA (Mateos et al., 2010). Nevertheless, mutants in MIRNA genes were identified shortly after the identification of plant miRNAs as the genetic basis of developmental mutants previously characterized, thus providing the first indications that miRNAs contribute to developmental processes (Baker et al., 2005; Chuck et al., 2007; Liu et al., 2010). However, with the exception of the MIR164 and MIR159 families in Arabidopsis (Sieber et al., 2007; Allen et al., 2010), no comprehensive analysis of all members of a MIRNA gene family could be performed due to the lack of mutants. The reports that MIRNA gene mutation could be triggered by CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated proteins)- based genome editing technologies (Jacobs et al., 2015; Zhao et al., 2016) was a game changer in this field and was rapidly applied to study the developmental roles of MIRNA genes (Table 1) (for a more comprehensive review about CRISPR applications on MIRNA genes see Deng et al., 2022). In a few years, genome editing of MIRNAs has come to a maturity. Methods to design the strategy and identify mutants have been improved. For instance, ways to test the efficiency of guide RNAs in protoplasts before stable transformation have been developed (Zhou et al., 2017). Double targeting to induce large deletions at the MIRNA locus is often used as single indels resulting from single targeting do not always affect miRNA functions (Zhou et al., 2017; Lukan et al., 2022). Nevertheless, quantifying precisely the effects of the induced mutations on miRNA function (a combined effect of MIRNA expression level, maturation, stability, target recognition and inhibition) remains a very challenging task. One pragmatic way to test it is to compare the effects of wild-type and mutant MIRNA gene overexpression (eg Ó’Maoiléidigh et al., 2021; Lian et al., 2021). Such MIRNA mutation via genome editing can be used for model species and crops including polyploids (Lukan et al., 2022; Zhu et al., 2022). This system provides a high versatility: depending on the specificities of the used guides, it can either target a single MIRNA gene of a multigene family to provide the opportunity to generate individual mutants for each member of a MIRNA family and combine them progressively via crossing (for instance Lian et al., 2021 constructed 24 combinations for 5 different MIR172 genes) or target multiple genes simultaneously using multiple specific guides (Zhu et al., 2022).
7 From specialization to full genetic redundancy of MIRNA genes
The identification of MIRNA genes as the genetic basis of mutants initially identified for their developmental defects unambiguously showed that some individual MIRNA genes have specific roles that are not masked by genetic redundancy. For instance, the early extra petal mutation leading to more petals was shown to result from the reduced expression of MIR164c, one of the three MIR164 Arabidopsis genes (Baker et al., 2005). Such a link between MIRNA genes and developmental phenotypes was not limited to the model species Arabidopsis as in maize tasselseed4 mutants have irregular branching in the inflorescence and feminization of the tassel due to mutations in a MIR172 gene (Chuck et al., 2007), while a mutation of a MIR169 gene leads to petal conversion into staminoid structures in both Antirrhinum majus and Petunia hybrida (Cartolano et al., 2007). However, a more systematic analysis of the roles and interactions between different MIRNA gene members remains limited to species with dense T-DNA or transposon mutant collections such as Arabidopsis or for which genome editing was well-developed (mostly rice and tomato). The discovery of mutants in multiple MIRNA genes within the same family has revealed that they can have roles that range from specific involvement in certain organs or developmental processes to exhibiting quantitative additive effects or even complete genetic redundancy. We will illustrate these findings below.
In line with the forward genetic approach described above, reverse genetics also identified individual MIRNA genes with important roles in specific developmental contexts. As an illustration, a strong specialization was shown for MIR164 genes in Arabidopsis and tomato. In parallel to MIR164c that determines Arabidopsis petal number via the regulation of the CUC1 target (Baker et al., 2005), MIR164a regulates leaf serration via its effect on the CUC2 target (Nikovics et al., 2006). This also suggests that specialization of some MIRNA gene members could go along with a preferential regulation of some targets in a specific developmental context. In tomato, MIR164b is the main regulator of meristem-to-organ and organ-to-organ boundary specification, stem internode elongation, and flower abscission zone development, while MIR164a is required for division and maturation in tomato fruits (Gupta et al., 2021). Such specialization of the MIRNA genes can be due to their specific expression patterns: MIR164a is specifically expressed in the leaves in domains overlapping with CUC2 (Nikovics et al., 2006) while MIR164c is the only MIR164 gene to be expressed in the floral meristem where the petals form (Sieber et al., 2007). In addition to differences in gene expression levels or patterns, differences in miRNA maturation can also contribute to variations in the importance of individual MIRNA genes of the same family, as illustrated by Arabidopsis MIR167. Within the four MIR167 members, MIR167a has the major role as its mutation leads to a strong phenotype including anther dehiscence and ovule development defects (Yao et al., 2019). In contrast, the mir167bcd triple mutant is very similar to wild type. Such differences in the developmental contribution of the MIR167 members may be due to a less efficient maturation of the MIR167b,c,d precursors compared to MIR167a as only MIR167a has strong effects when ectopically expressed while MIR167b,c,d have no or weak effects (Wu et al., 2006).
The combination of multiple mutations has shown that many MIRNA genes have redundant roles, with sometimes quantitative effects. Even in the case of a strong specialization, a certain level of genetic redundancy is observed. In tomato, MIR164a and MIR164b that have specific functions have also redundant roles as the double mir164ab mutant has very strong seedling defects not observed in any of the single mutants (Gupta et al., 2021). At the more extreme case, full functional redundancy between MIR159a and MIR159b results from similar expression patterns of these two genes and a similar mature miRNA sequence. As a result, each of the single mutant shows a wild-type phenotype while the mir159ab double mutant has hyponastic leaves, stunted growth, reduced apical dominance, reduced fertility and seed set (Allen et al., 2007). More generally, redundancy has been shown between different members of MIR156, MIR164, MIR167, MIR172, MIR396 in Arabidopsis (Sieber et al., 2007; Yang et al., 2013; Hou et al., 2019; Ó’Maoiléidigh et al., 2021; Lian et al., 2021), MIR172 in tomato (Lin et al., 2021a) and seems to be the general rule, at least to a certain level. In some instances, it has been reported that MIRNA genes control a specific character in a quantitative way. An emblematic illustration is shown by Ó’Maoiléidigh et al. (2021) who observed that the number of Arabidopsis leaves produced before flowering gradually increases when mutations of the five MIR172 genes are stacked one by one from the single mir172a mutant to the quintuple mutant. As for MIR159a and MIR159b genes, the basis of this redundancy is an overlap of MIRNA gene expression and similar effects of the miRNAs produced. An unexplored question related to MIRNA gene redundancy is the fine role of the different miRNA isoforms. Based on predictions, their efficiency towards their targets may be different and in the most extreme case, they may have different targets.
In the following sections, our objective is to discuss the roles of miRNAs during plant development and reproduction. Rather than aiming to provide an exhaustive list of roles, we will critically assess their significance in four key processes. This assessment will be primarily based on the analysis of MIRNA mutants, but we will also explore their connections with other regulators and their effects on target genes.
8 miRNAs, quantitative regulators of plant growth
Growth, when it operates at a global scale, acts as the driving force behind plants continual development. However, when growth is differential and precisely regulated at a local level, it becomes the mechanism through which morphogenesis, the development of new forms, takes place. From a more mechanistic point of view it combines different cellular processes such as cell proliferation and cell expansion that are regulated by multiple plant hormones. MiRNAs contribute to the regulation of growth at these two scales.
In Arabidopsis, miR159 targets seven genes encoding MYB transcription factors that contribute to the regulation of the expression of genes in response to gibberellins (GA). MiR159 acts by clearing the expression of two of them, MYB33 and MYB65, from vegetative tissues and limits their expression to anthers and seeds. Ectopic expression of MYB33 and MYB65 in vegetative tissues of the mir159ab double mutant leads to a reduced growth due to a reduced cell proliferation which is not totally compensated by an increased cell expansion (Allen et al., 2007; Alonso-Peral et al., 2010). Therefore, miR159 appears to act as a molecular switch in Arabidopsis to shut-off its targets in inappropriate tissues. In tomato, miR159 has a role during fruit growth as a mutation of MIR159a leads to larger, less elongated fruits. Here miR159, by targeting the SlGAMYB2 gene coding a transcription factor that represses the GA biosynthetic gene SlGA3ox2 indirectly promotes GA biosynthesis (Zhao et al., 2022). Because miR159 expression is induced by GAs (Achard et al., 2004), this forms a positive feedforward loop that may contribute to fruit size regulation. Thus, miR159 acts as a modulator of growth, either at the whole plant level as in Arabidopsis or the organ level like in tomato.
In Arabidopsis, differential growth at the leaf margin leads to the formation of small serrations. The balance between co-expressed MIR164a and CUC2 in the sinus of the serration determines the size of the serration, possibly by controlling their growth speed (Nikovics et al., 2006; Kawamura et al., 2010; Bilsborough et al., 2011; Maugarny-Calès et al., 2019). As a consequence, if miR164 function is compromised, Arabidopsis leaves form overdeveloped serrations, a phenotype that is also observed in strawberry (Zheng et al., 2019). Thus, by fine tuning growth, miRNAs control overall plant and organ size and also contribute to the fine regulation of plant organ shape.
9 miRNAs, regulators of plant organ patterning and identity
Morphogenesis involves the formation of new organs according to a precise spatial and temporal pattern, while also ensuring that they acquire their correct identity. As discussed above, MIR164c is required to regulate petal number in Arabidopsis (Baker et al., 2005), while in tomato MIR172d is limiting petal and stamen numbers (Lin et al., 2021a). MiRNAs have also been shown to contribute to patterning at the cellular scale as shown for the rhizoids in the liverwort Marchantia polymorpha. Rhizoids are single cell outgrowth that develop from a layer of epidermal cells and tend to be spaced and form only unfrequently linear clusters. Rhizoid formation is promoted by the basic loop helix loop transcription factor ROOT HAIR DEFECTIVE SIX-LIKE1 (RSL1) which activates the expression of its negative regulator the miRNA FEW RHIZOIDS1 (FRH1). This negative feedback loop contributes to patterning the rhizoid by generating a lateral inhibition mechanism (Thamm et al., 2020). Whether the role of FRH1 in mediating lateral inhibition involves diffusion of the miRNA it-self from the rhizoid precursor cell where it is expressed or diffusion of a downstream signal is not known yet.
Several examples also illustrate the roles of miRNAs in the regulation of organ identity. For instance, in Antirrhinum majus and Petunia hybrida mutant for MIR169, the acquisition of petal identity is compromised, resulting in an increase in stamen identity. This is due to elevated and slight expansion of the C-function expression (Cartolano et al., 2007). In tomato, petals and stamens are converted into sepaloid organs when MIR172d is reduced, suggesting that the AP2 genes that are consequently upregulated may both repress C-function genes to the most inner part of the floral meristem and repress B-function genes (Lin et al., 2021a). These two examples indicate that MIRNAs may contribute to provide positional information necessary for the correct acquisition of organ identity during flower development. A more extreme case of organ identity is shown by the role of MIR172e/Tasselseed4 (ts4) in maize (Chuck et al., 2007). In ts4 mutants, the typically male flowers in the tassel fail to undergo pistil abortion, resulting in feminization and seed formation. A similar phenotype results from dominant ts6 mutations. Ts4 encodes a member of the MIR172 family, while the ts6 mutation lies in the miR172-binding site of an AP2 gene. Compromised miR172 action does not have any discernible effect on the mRNA level or cleavage of its target but leads to the ectopic presence of the target protein in the pistil that fails to abort. Therefore, miR172 limits AP2/TS6 presence in male flowers to abort pistil development and acts mostly by repressing its translation, a mechanism also occurring in Arabidopsis (Aukerman and Sakai, 2003; Chen, 2004).
10 miRNAs, regulators of plant life cycle
Phase transition from juvenile to adult is important for the plant life cycle as it allows the plant to acquire its reproductive potential. MiR156 plays pivotal roles during this transition, maintaining the juvenile phase by repressing specific SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) activities. Indeed, SPL transcription factors have specific roles during vegetative development with SPL9 promoting adult state (Schwarz et al., 2008). Another miRNA, miR172 is acting downstream of miR156 to promote adult state (Wu et al., 2009) by repressing the activity of APETALA2-like transcription factors such as TARGET OF EAT 1 (TOE1) and TOE2 (Aukerman and Sakai, 2003; Jung et al., 2007; Wu et al., 2009). SPL9 is likely to induce MIR172b and MIR172c but not MIR172a to coordinate juvenile to adult transition (Wu et al., 2009; Lian et al., 2021). Thus, sequentially operating MIRNAs ensure the juvenile to adult transition, miR156 and miR172 having opposite effects on phase transition.
Floral transition is crucial for plant reproductive success; hence it needs to be tightly controlled to ensure flowering at the right time. Once again miR172 plays an important role here. Specific combinations of MIR172 genes responding to different endogenous or exogenous signals (such age, temperature, photoperiod) promote the transition to flowering (Lian et al., 2021). For instance, low ambient temperature modulates flowering by controlling the expression of MIR172c and MIR172d at both transcriptional and post-transcriptional levels (Jung et al., 2007). Additionally, specific SPL genes are acting as intermediate between the environmental or endogenous factor and MIR172 gene activation, therefore forming a complex network of partially interdependent regulation (Lian et al., 2021). As individual MIR172 genes integrate multiple signals, the contribution of MIR156-dependent SPL regulation remains to be determined.
MiR172 is at the core of phase transition in plants suggesting that it is required for plant growth and development. The analysis of mir172 multiple CRISPR mutants tells a different story as mir172 quintuple mutant is viable and can even produce large amounts of seeds when grown outdoors (Lian et al., 2021). This is showing that, although miR172 is required for SAM formation and control meristem size through a AP2/ARF3 module (Jung et al., 2007; Ó’Maoiléidigh et al., 2021), it is not absolutely required for seed-to-seed life cycle in Arabidopsis and that additional miR172-independent pathways exist.
11 miRNAs, essential factors for plant reproduction
To fulfill their developmental cycle, plants need to sustain continuous growth by maintaining an active pool of stem cells in meristems and develop male and female reproductive structures. miRNAs are essential for all these events. In rice, mutants for the unique MIR390 gene show a shootless phenotype and could therefore not be maintained as homozygous mutants (Bi et al., 2020). Mutation of MIR394 in Arabidopsis severely enhances the shoot apical meristem defects of ago10-1 (Knauer et al., 2013). Two families of miRNAs, miR160 and miR167 that modulate auxin responses via the targeting of different ARFs are required for proper ovule, stamen or seed development in Arabidopsis and tomato (Liu et al., 2010; Damodharan et al., 2018; Yao et al., 2019; Bi et al., 2020). For these two families, the disruption of a single member is sufficient to induce a strong reproductive defect. In addition, mutations of other miRNA genes such as MIR164b in tomato, MIR159a or b, MIR319a in Arabidopsis and MIR2118 in rice reduce fertility (Allen et al., 2007; Nag et al., 2009; Araki et al., 2020; Gupta et al., 2021).
Beside controlling morphogenesis, miRNAs also have an important contribution to genome homeostasis. Multiple miRNAs, including conserved miRNAs that target developmental genes such as miR159 or miR172, also target different transposons and trigger the production of epigenetically activated small interfering RNAs (easiRNAs), which also target transposons. This pathway is particularly active in mutants such as decreased dna methylation 1 (ddm1). Here, transposons are transcriptionally derepressed and the easiRNA pathway is required for normal plant fertility (Creasey et al., 2014). The production of easiRNAs is also triggered in a dose dependent manner by MIR845b which is specifically expressed in pollen. More precisely, easiRNAs produced in the vegetative nucleus move to the neighboring germ cell (Borges et al., 2018). A T-DNA insertion in MIR845b reduces miR845 and easiRNA accumulation and leads to a reduction of the triploid block which drives a reproductive barrier between species with different chromosome numbers. Therefore, the contribution of miRNAs contribution could extend from their role in the reproduction of individual plants to a broader role in plant evolution and speciation.
12 The role of microRNAs during plant versus animal development.
Similar to plants, animal miRNAs are organized into families, with some families being highly conserved while having at the same time a high dynamic in the birth, evolution and death of new genes (Niwa and Slack, 2007; França et al., 2016). In contrast to plant miRNAs that mostly bind to unique sequences with a high complementarity to produce strong impact on gene expression, animal miRNAs often have multiple binding sites and their action on gene expression is more limited. In fact, the action of animal miRNAs is cooperative, meaning that multiple binding sites are required to have a strong effect on gene expression. Furthermore, more limited complementarity is sufficient between an animal miRNA and its target, which has the important consequence of increasing the number of genes that are targeted by each miRNA. Therefore, essential differences exist between plant and animal miRNAs on their downstream gene target network and their quantitative effects on target expression. Functional analysis on animal miRNA gene families have led to the idea that only a few of them are essential for development (Alvarez-Saavedra and Horvitz, 2010), a view that however could be revisited with more thorough genetic analysis. Systematic analysis of animal miRNAs and their target expression studies have led to the proposal of a bimodal role of miRNAs during development (Alberti and Cochella, 2017; Avital et al., 2018). On one hand, miRNAs expressed during early embryogenesis would preferentially have a strong impact on their target expression, leading often to complementary patterns between the miRNA and its targets. These miRNAs often regulate basic mechanisms such as proliferation, apoptosis or cell signaling. On the other hand, another group of is expressed at later stages, during the process of differentiation, and exhibits cell-type-specific patterns of expression. These miRNAs have a more limited effect on the expression of their targets and may contribute to robustness of their expression (Ebert and Sharp, 2012; Cassidy et al., 2013). As discussed above, plant miRNAs may have similar effects on their target genes, being essential contributors of their expression pattern or fine regulators of their levels. Therefore, plant and animal miRNAs are likely integrated in regulatory networks of similar architectures that allow them to have similar molecular outcome despite differences in their mode of action or the number of targets. However, because plant development is a continuous process that result in an organism having at one moment organs at different developmental stages, the separation between miRNAs regulating early patterning and growth and those involved in later differentiation can not be done on a temporal basis as in animals but rather on a spatial basis. Therefore, the apparent differences in miRNA roles observed during plant and animal development are more likely to result from the distinct logic of development that exists in these organisms rather than reflecting true fundamental differences in miRNA function.
13 Conclusion
Our current knowledge unequivocally indicates that numerous miRNAs are a pillar of plant development and reproduction. Their inactivation leads to plants that are unable to carry out critical steps such as meristem maintenance, production of reproductive organs or genome homeostasis. Notably, the requirement for miRNAs in plant development is not strictly linked to gene family size, as some single genes are essential, while certain larger families appear to have milder contributions. Despite important progress in the last years, our understanding of miRNAs’ roles remains incomplete. Within a MIRNA gene family, functional data are often missing for one or several members, and some entire families are yet to be fully explored. Furthermore, such functional analysis tends to concentrate on conserved miRNAs, neglecting for the moment more recent miRNAs that could have functions more limited to some developmental particularities in a few species. In this context, genome editing has proven to be an indispensable tool to go deeper into functional analysis and gain further insights into the functions of miRNAs in plant development and reproduction.
Beside such broad-scale functional analysis, a more comprehensive understanding of the roles of miRNAs requires a deeper comprehension of their position and effects on GRN. This is a question shared with other genetic regulators and includes the key issues of the regulation of their expression. However, due to the unique characteristics of miRNAs, such as their biogenesis and mode of action, it also raises specific questions that are exclusive to these small RNA molecules, such as their quantitative effect on target expression and more generally on GRN dynamics.
Author contributions
PL: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. NA: Conceptualization, Visualization, Writing – original draft, Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. The IJPB benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) PL declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Achard P., Herr A., Baulcombe D. C., Harberd N. P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131, 3357–3365. doi: 10.1242/dev.01206
Alberti C., Cochella L. (2017). A framework for understanding the roles of miRNAs in animal development. Dev 144, 2548–2559. doi: 10.1242/dev.146613
Allen R. S., Li J., Alonso-Peral M. M., White R. G., Gubler F., Millar A. A. (2010). MicroR159 regulation of most conserved targets in Arabidopsis has negligible phenotypic effects. Silence 1, 1–18. doi: 10.1186/1758-907X-1-18
Allen R. S., Li J., Stahle M. I., Dubroué A., Gubler F., Millar A. A. (2007). Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl. Acad. Sci. U. S. A. 104, 16371–16376. doi: 10.1073/pnas.0707653104
Alonso-Peral M. M., Li J., Li Y., Allen R. S., Schnippenkoetter W., Ohms S., et al. (2010). The MicroRNA159-Regulated GAMYB-like genes inhibit growth and promote programmed Cell Death in Arabidopsis. Plant Physiol. 154, 757–771. doi: 10.1104/pp.110.160630
Alvarez-Saavedra E., Horvitz H. R. (2010). Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20, 367–373. doi: 10.1016/j.cub.2009.12.051
Araki S., Le N. T., Koizumi K., Villar-Briones A., Nonomura K. I., Endo M., et al. (2020). miR2118-dependent U-rich phasiRNA production in rice anther wall development. Nat. Commun. 11, 1–2. doi: 10.1038/s41467-020-16637-3
Arazi T., Khedia J. (2022). Tomato microRNAs and their functions. Int. J. Mol. Sci. 23, 1–20. doi: 10.3390/ijms231911979
Aukerman M. J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741. doi: 10.1105/tpc.016238
Avital G., França G. S., Yanai I. (2018). Bimodal evolutionary developmental miRNA program in animal embryogenesis. Mol. Biol. Evol. 35, 646–654. doi: 10.1093/molbev/msx316
Baker C. C., Sieber P., Wellmer F., Meyerowitz E. M. (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 15, 303–315. doi: 10.1016/j.cub.2005.02.017
Betti F., Ladera-Carmona M. J., Weits D. A., Ferri G., Iacopino S., Novi G., et al. (2021). Exogenous miRNAs induce post-transcriptional gene silencing in plants. Nat. Plants 7, 1379–1388. doi: 10.1038/s41477-021-01005-w
Bhogale S., Mahajan A. S., Natarajan B., Rajabhoj M., Thulasiram H. V., Banerjee A. K. (2014). MicroRNA156: A potential graft-transmissible microrna that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 164, 1011–1027. doi: 10.1104/pp.113.230714
Bi H., Fei Q., Li R., Liu B., Xia R., Char S. N., et al. (2020). Disruption of miRNA sequences by TALENs and CRISPR/Cas9 induces varied lengths of miRNA production. Plant Biotechnol. J. 18, 1526–1536. doi: 10.1111/pbi.13315
Bilsborough G. D., Runions A., Barkoulas M., Jenkins H. W., Hasson A., Galinha C., et al. (2011). Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. 108, 3424–3429. doi: 10.1073/pnas.1015162108
Borges F., Parent J. S., Van Ex F., Wolff P., Martínez G., Köhler C., et al. (2018). Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat. Genet. 50, 186–192. doi: 10.1038/s41588-017-0032-5
Brioudes F., Jay F., Sarazin A., Grentzinger T., Devers E. A., Voinnet O. (2021). HASTY, the Arabidopsis EXPORTIN5 ortholog, regulates cell-to-cell and vascular microRNA movement. EMBO J. 40, 1–22. doi: 10.15252/embj.2020107455
Brosnan C. A., Sarazin A., Lim P., Bologna N. G., Hirsch-Hoffmann M., Voinnet O. (2019). Genome-scale, single-cell-type resolution of micro RNA activities within a whole plant organ. EMBO J. 38, 1–19. doi: 10.15252/embj.2018100754
Carlsbecker A., Lee J. Y., Roberts C. J., Dettmer J., Lehesranta S., Zhou J., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321. doi: 10.1038/nature08977
Cartolano M., Castillo R., Efremova N., Kuckenberg M., Zethof J., Gerats T., et al. (2007). A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat. Genet. 39, 901–905. doi: 10.1038/ng2056
Cassidy J. J., Jha A. R., Posadas D. M., Giri R., Venken K. J. T., Ji J., et al. (2013). MiR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell 155, 1556–1567. doi: 10.1016/j.cell.2013.10.057
Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. doi: 10.1126/science.1088060
Chen X., Rechavi O. (2022). Plant and animal small RNA communications between cells and organisms. Nat. Rev. Mol. Cell Biol. 23, 185–203. doi: 10.1038/s41580-021-00425-y
Chuck G., Meeley R., Irish E., Sakai H., Hake S. (2007). The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Genet. 39, 1517–1521. doi: 10.1038/ng.2007.20
Creasey K. M., Zhai J., Borges F., Van Ex F., Regulski M., Meyers B. C., et al. (2014). MiRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508, 411–415. doi: 10.1038/nature13069
Cui J., You C., Chen X. (2017). The evolution of microRNAs in plants. Curr. Opin. Plant Biol. 35, 61–67. doi: 10.1016/j.pbi.2016.11.006
Dai X., Lu Q., Wang J., Wang L., Xiang F., Liu Z. (2021). MiR160 and its target genes ARF10, ARF16 and ARF17 modulate hypocotyl elongation in a light, BRZ, or PAC-dependent manner in Arabidopsis: miR160 promotes hypocotyl elongation. Plant Sci. 303, 110686. doi: 10.1016/j.plantsci.2020.110686
Dalmadi A., Gyula P., Balint J., Szittya G., Havelda Z. (2019). AGO-unbound cytosolic pool of mature miRNAs in plant cells reveals a novel regulatory step at AGO1 loading. Nucleic Acids Res. 47, 9803–9817. doi: 10.1093/nar/gkz690
Damodharan S., Corem S., Gupta S. K., Arazi T. (2018). Tuning of SlARF10A dosage by sly-miR160a is critical for auxin-mediated compound leaf and flower development. Plant J. 96, 855–868. doi: 10.1111/tpj.14073
Deng F., Zeng F., Shen Q., Abbas A., Cheng J., Jiang W., et al. (2022). Molecular evolution and functional modification of plant miRNAs with CRISPR. Trends Plant Sci. 27, 890–907. doi: 10.1016/j.tplants.2022.01.009
Ebert M. S., Sharp P. A. (2012). Roles for MicroRNAs in conferring robustness to biological processes. Cell 149, 515–524. doi: 10.1016/j.cell.2012.04.005
Fahlgren N., Howell M. D., Kasschau K. D., Chapman E. J., Sullivan C. M., Cumbie J. S., et al. (2007). High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PloS One 2, e219. doi: 10.1371/journal.pone.0000219
Flores-Sandoval E., Eklund D. M., Hong S. F., Alvarez J. P., Fisher T. J., Lampugnani E. R., et al. (2018). Class C ARFs evolved before the origin of land plants and antagonize differentiation and developmental transitions in Marchantia polymorpha. New Phytol. 218, 1612–1630. doi: 10.1111/nph.15090
Floyd S. K., Bowman J. L. (2004). Ancient microRNA target sequences in plants. Nature 428, 485–486. doi: 10.1038/428485a
França G. S., Vibranovski M. D., Galante P. A. F. (2016). Host gene constraints and genomic context impact the expression and evolution of human microRNAs. Nat. Commun. 7, 11438. doi: 10.1038/ncomms11438
Franco-Zorrilla J. M., Valli A., Todesco M., Mateos I., Puga M. I., Rubio-Somoza I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
Garcia D. (2008). A miRacle in plant development: Role of microRNAs in cell differentiation and patterning. Semin. Cell Dev. Biol. 19, 586–595. doi: 10.1016/j.semcdb.2008.07.013
Grigorova B., Mara C., Hollender C., Sijacic P., Chen X., Liu Z. (2011). LEUNIG and SEUSS co-repressors regulate miR172 expression in Arabidopsis flowers. Development 138, 2451–2456. doi: 10.1242/dev.058362
Gupta S. K., Vishwakarma A., Kenea H. D., Galsurker O., Cohen H., Aharoni A., et al. (2021). CRISPR/Cas9 mutants of tomato MICRORNA164 genes uncover their functional specialization in development. Plant Physiol. 187, 1636–1652. doi: 10.1093/plphys/kiab376
He B., Cai Q., Qiao L., Huang C.-Y., Wang S., Miao W., et al. (2021). RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants 7, 342–352. doi: 10.1038/s41477-021-00863-8
Hou N., Cao Y., Li F., Yuan W., Bian H., Wang J., et al. (2019). Epigenetic regulation of miR396 expression by SWR1-C and the effect of miR396 on leaf growth and developmental phase transition in Arabidopsis. J. Exp. Bot. 70, 5217–5229. doi: 10.1093/jxb/erz285
Jacobs T. B., LaFayette P. R., Schmitz R. J., Parrott W. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15, 1–10. doi: 10.1186/s12896-015-0131-2
Johnston R. J., Chang S., Etchberger J. F., Ortiz C. O., Hobert O. (2005). MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. 102, 12449–12454. doi: 10.1073/pnas.0505530102
Jung J.-H., Seo Y.-H., Seo P. J., Reyes J. L., Yun J., Chua N.-H., et al. (2007). The GIGANTEA -regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in arabidopsis. Plant Cell 19, 2736–2748. doi: 10.1105/tpc.107.054528
Kawamura E., Horiguchi G., Tsukaya H. (2010). Mechanisms of leaf tooth formation in Arabidopsis. Plant J. 62, 429–441. doi: 10.1111/j.1365-313X.2010.04156.x
Kim J. H., Woo H. R., Kim J., Lim P. O., Lee I. C., Choi S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Sci. (80-.). 323, 1053–1057. doi: 10.1126/science.1166386
Knauer S., Holt A. L., Rubio-Somoza I., Tucker E. J., Hinze A., Pisch M., et al. (2013). A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 24, 125–132. doi: 10.1016/j.devcel.2012.12.009
Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19, 473–484. doi: 10.1105/tpc.106.044792
Kozomara A., Birgaoanu M., Griffiths-Jones S. (2019). MiRBase: From microRNA sequences to function. Nucleic Acids Res. 47, D155–D162. doi: 10.1093/nar/gky1141
Lee Y. K., Kumari S., Olson A., Hauser F., Ware D. (2022). Role of a ZF-HD transcription factor in miR157-mediated feed-forward regulatory module that determines plant architecture in arabidopsis. Int. J. Mol. Sci. 23, 8665. doi: 10.3390/ijms23158665
Li M., Yu B. (2021). Recent advances in the regulation of plant miRNA biogenesis. RNA Biol. 18, 2087–2096. doi: 10.1080/15476286.2021.1899491
Lian H., Wang L., Ma N., Zhou C. M., Han L., Zhang T. Q., et al. (2021). Redundant and specific roles of individual MIR172 genes in plant development. PloS Biol. 19, 1–25. doi: 10.1371/journal.pbio.3001044
Lin S. I., Chiang S. F., Lin W. Y., Chen J. W., Tseng C. Y., Wu P. C., et al. (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147, 732–746. doi: 10.1104/pp.108.116269
Lin W., Gupta S. K., Arazi T., Spitzer-rimon B. (2021a). Mir172d is required for floral organ identity and number in tomato. Int. J. Mol. Sci. 22, 4659. doi: 10.3390/ijms22094659
Lin Y., Zhu Y., Cui Y., Chen R., Chen Z., Li G., et al. (2021b). Derepression of specific miRNA-target genes in rice using CRISPR/Cas9. J. Exp. Bot. 72, 7067–7077. doi: 10.1093/jxb/erab336
Lin D., Zhu X., Qi B., Gao Z., Tian P., Li Z., et al. (2022). SlMIR164A regulates fruit ripening and quality by controlling SlNAM2 and SlNAM3 in tomato. Plant Biotechnol. J. 20, 1456–1469. doi: 10.1111/pbi.13824
Liu X., Huang J., Wang Y., Khanna K., Xie Z., Owen H. A., et al. (2010). The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in MicroRNA160a, in organogenesis and the mechanism regulating its expression. Plant J. 62, 416–428. doi: 10.1111/j.1365-313X.2010.04164.x
Liu R., Ma Y., Guo T., Li G. (2023). Identification, biogenesis, function, and mechanism of action of circular RNAs in plants. Plant Commun. 4, 100430. doi: 10.1016/j.xplc.2022.100430
Llave C., Kasschau K. D., Rector M. A., Carrington J. C. (2002). Endogenous and silencing-associated small RNAs in plants. Plant Cell 14, 1605–1619. doi: 10.1105/tpc.003210
Loreti E., Perata P. (2022). Mobile plant microRNAs allow communication within and between organisms. New Phytol. 235, 2176–2182. doi: 10.1111/nph.18360
Lukan T., Veillet F., Križnik M., Coll A., Mahkovec Povalej T., Pogačar K., et al. (2022). CRISPR/Cas9-mediated fine-tuning of miRNA expression in tetraploid potato. Hortic. Res. 9, 1–15. doi: 10.1093/hr/uhac147
Mallory A. C., Dugas D. V., Bartel D. P., Bartel B. (2004). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14, 1035–1046. doi: 10.1016/j.cub.2004.06.022
Martin A., Adam H., Díaz-Mendoza M., Źurczak M., González-Schain N. D., Suárez-López P. (2009). Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 136, 2873–2881. doi: 10.1242/dev.031658
Martinez N. J., Ow M. C., Barrasa M. I., Hammell M., Sequerra R., Doucette-Stamm L., et al. (2008). A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 22, 2535–2549. doi: 10.1101/gad.1678608
Mateos J. L., Bologna N. G., Chorostecki U., Palatnik J. F. (2010). Identification of microRNA processing determinants by random mutagenesis of arabidopsis MIR172a precursor. Curr. Biol. 20, 49–54. doi: 10.1016/j.cub.2009.10.072
Maugarny-Calès A., Cortizo M., Adroher B., Borrega N., Gonçalves B., Brunoud G., et al. (2019). Dissecting the pathways coordinating patterning and growth by plant boundary domains. PloS Genet. 15, e1007913. doi: 10.1371/journal.pgen.1007913
Meng Y., Shao C., Wang H., Chen M. (2011). The regulatory activities of plant microRNAs: A more dynamic perspective. Plant Physiol. 157, 1583–1595. doi: 10.1104/pp.111.187088
Miao C., Wang Z., Zhang L., Yao J., Hua K., Liu X., et al. (2019). The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat. Commun. 10, 3822. doi: 10.1038/s41467-019-11830-5
Miao C., Wang D., He R., Liu S., Zhu J. K. (2020). Mutations in MIR396e and MIR396f increase grain size and modulate shoot architecture in rice. Plant Biotechnol. J. 18, 491–501. doi: 10.1111/pbi.13214
Milo R., Shen-Orr S., Itzkovitz S., Kashtan N., Chklovskii D., Alon U. (2002). Network motifs: simple building blocks of complex networks. Science 298, 824–827. doi: 10.1126/science.298.5594.824
Miyashima S., Koi S., Hashimoto T., Nakajima K. (2011). Non-cell-autonomous microRNA 165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138, 2303–2313. doi: 10.1242/dev.060491
Moran Y., Agron M., Praher D., Technau U. (2017). The evolutionary origin of plant and animal microRNAs. Nat. Ecol. Evol. 1, 27. doi: 10.1038/s41559-016-0027
Nag A., King S., Jack T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 22534–22539. doi: 10.1073/pnas.0908718106
Nikovics K., Blein T., Peaucelle A., Ishida T., Morin H., Aida M., et al. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in arabidopsis. Plant Cell 18, 2929–2945. doi: 10.1105/tpc.106.045617
Niwa R., Slack F. J. (2007). The evolution of animal microRNA function. Curr. Opin. Genet. Dev. 17, 145–150. doi: 10.1016/j.gde.2007.02.004
Nozawa M., Miura S., Nei M. (2012). Origins and evolution of microRNA genes in plant species. Genome Biol. Evol. 4, 230–239. doi: 10.1093/gbe/evs002
Ó’Maoiléidigh D. S., van Driel A. D., Singh A., Sang Q., Le Bec N., Vincent C., et al. (2021). Systematic analyses of the MIR172 family members of Arabidopsis define their distinct roles in regulation of APETALA2 during floral transition. PloS Biol. 19, e3001043. doi: 10.1371/journal.pbio.3001043
Ouyang L., Ma M., Li L. (2020). An efficient transgene-free DNA-editing system for Arabidopsis using a fluorescent marker. Biotechnol. Lett. 42, 313–318. doi: 10.1007/s10529-019-02778-z
Peng T., Qiao M., Liu H., Teotia S., Zhang Z., Zhao Y., et al. (2018). A resource for inactivation of microRNAs using short tandem target mimic technology in model and crop plants. Mol. Plant 11, 1400–1417. doi: 10.1016/j.molp.2018.09.003
Rajagopalan R., Vaucheret H., Trejo J., Bartel D. P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20, 3407–3425. doi: 10.1101/gad.1476406
Rast-Somssich M. I., Broholm S., Jenkins H., Canales C., Vlad D., Kwantes M., et al. (2015). Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta. Genes Dev. 29, 2391–2404. doi: 10.1101/gad.269050.115
Reichel M., Li Y., Li J., Millar A. A. (2015). Inhibiting plant microRNA activity: Molecular SPONGEs, target MIMICs and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol. J. 13, 915–926. doi: 10.1111/pbi.12327
Reinhart B. J., Weinstein E. G., Rhoades M. W., Bartel B., Bartel D. P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. doi: 10.1101/gad.1004402
Rhoades M. W., Reinhart B. J., Lim L. P., Burge C. B., Bartel B., bartel D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Schauer S. E., Jacobsen S. E., Meinke D. W., Ray A. (2002). DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 7, 487–491. doi: 10.1016/S1360-1385(02)02355-5
Schwab R., Palatnik J. F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517–527. doi: 10.1016/j.devcel.2005.01.018
Schwarz S., Grande A. V., Bujdoso N., Saedler H., Huijser P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67, 183–195. doi: 10.1007/s11103-008-9310-z
Shahid S., Kim G., Johnson N. R., Wafula E., Wang F., Coruh C., et al. (2018). MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553, 82–85. doi: 10.1038/nature25027
Shen-Orr S. S., Milo R., Mangan S., Alon U. (2002). Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31, 64–68. doi: 10.1038/ng881
Sieber P., Wellmer F., Gheyselinck J., Riechmann J. L., Meyerowitz E. M. (2007). Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134, 1051–1060. doi: 10.1242/dev.02817
Skopelitis D. S., Benkovics A. H., Husbands A. Y., Timmermans M. C. P. (2017). Boundary formation through a direct threshold-based readout of mobile small RNA gradients. Dev. Cell 43, 265–273.e6. doi: 10.1016/j.devcel.2017.10.003
Skopelitis D. S., Hill K., Klesen S., Marco C. F., von Born P., Chitwood D. H., et al. (2018). Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nat. Commun. 9, 3107. doi: 10.1038/s41467-018-05571-0
Tang Y., Qu Z., Lei J., He R., Adelson D. L., Zhu Y., et al. (2021). The long noncoding RNA FRILAIR regulates strawberry fruit ripening by functioning as a noncanonical target mimic. PloS Genet. 17, e1009461. doi: 10.1371/journal.pgen.1009461
Tarver J. E., Donoghue P. C. J., Peterson K. J. (2012). Do miRNAs have a deep evolutionary history? BioEssays 34, 857–866. doi: 10.1002/bies.201200055
Tatematsu K., Toyokura K., Miyashima S., Nakajima K., Okada K. (2015). A molecular mechanism that confines the activity pattern of miR165 in Arabidopsis leaf primordia. Plant J. 82, 596–608. doi: 10.1111/tpj.12834
Taylor R. S., Tarver J. E., Hiscock S. J., Donoghue P. C. J. (2014). Evolutionary history of plant microRNAs. Trends Plant Sci. 19, 175–182. doi: 10.1016/j.tplants.2013.11.008
Thamm A., Saunders T. E., Dolan L. (2020). MpFEW RHIZOIDS1 miRNA-Mediated Lateral Inhibition Controls Rhizoid Cell Patterning in Marchantia polymorpha. Curr. Biol. 30, 1905–1915.e4. doi: 10.1016/j.cub.2020.03.032
Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PloS Genet. 6, e1001031. doi: 10.1371/journal.pgen.1001031
Tripathi A. M., Admoni Y., Fridrich A., Lewandowska M., Surm J. M., Aharoni R., et al. (2022). Functional characterization of a ‘plant-like’ HYL1 homolog in the cnidarian Nematostella vectensis indicates a conserved involvement in microRNA biogenesis. Elife 11, 1–22. doi: 10.7554/eLife.69464
Tsuzuki M., Futagami K., Shimamura M., Inoue C., Kunimoto K., Oogami T., et al. (2019). An early arising role of the microRNA156/529-SPL module in reproductive development revealed by the liverwort marchantia polymorpha. Curr. Biol. 29, 3307–3314.e5. doi: 10.1016/j.cub.2019.07.084
Um T., Choi J., Park T., Chung P. J., Jung S. E., Shim J. S., et al. (2022). Rice microRNA171f/SCL6 module enhances drought tolerance by regulation of flavonoid biosynthesis genes. Plant Direct 6, 1–12. doi: 10.1002/pld3.374
Vatén A., Dettmer J., Wu S., Stierhof Y. D., Miyashima S., Yadav S. R., et al. (2011). Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 21, 1144–1155. doi: 10.1016/j.devcel.2011.10.006
Vidal E. A., Araus V., Lu C., Parry G., Green P. J., Coruzzi G. M., et al. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 107, 4477–4482. doi: 10.1073/pnas.0909571107
Wang J. W., Schwab R., Czech B., Mica E., Weigel D. (2008). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20, 1231–1243. doi: 10.1105/tpc.108.058180
Wu G., Park M. Y., Conway S. R., Wang J. W., Weigel D., Poethig R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. doi: 10.1016/j.cell.2009.06.031
Wu M. F., Tian Q., Reed J. W. (2006). Arabidopis microRNA 167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218. doi: 10.1242/dev.02602
Xu M., Hu T., Smith M. R., Poethig R. S. (2016). Epigenetic regulation of vegetative phase change in arabidopsis. Plant Cell 28, 28–41. doi: 10.1105/tpc.15.00854
Yan J., Gu Y., Jia X., Kang W., Pan S., Tang X., et al. (2012). Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24, 415–427. doi: 10.1105/tpc.111.094144
Yan F., Guo W., Wu G., Lu Y., Peng J., Zheng H., et al. (2014). A virus-based miRNA suppression (VbMS) system for miRNA loss-of-function analysis in plants. Biotechnol. J. 9, 702–708. doi: 10.1002/biot.201300523
Yan Y., Ham B. K. (2022). The mobile small RNAs: important messengers for long-distance communication in plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.928729
Yang L., Xu M., Koo Y., He J., Scott Poethig R. (2013). Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. Elife 2013, 1–15. doi: 10.7554/eLife.00260
Yang X., Zhao X., Dai Z., Ma F., Miao X., Shi Z. (2021). OsmiR396/growth regulating factor modulate rice grain size through direct regulation of embryo-specific miR408. Plant Physiol. 186, 519–533. doi: 10.1093/plphys/kiab084
Yao X., Chen J., Zhou J., Yu H., Ge C., Zhang M., et al. (2019). An essential role for mirna167 in maternal control of embryonic and seed development. Plant Physiol. 180, 453–464. doi: 10.1104/pp.19.00127
Yu Y., Jia T., Chen X. (2017). The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 216, 1002–1017. doi: 10.1111/nph.14834
Yumul R. E., Kim Y. J., Liu X., Wang R., Ding J., Xiao L., et al. (2013). POWERDRESS and diversified expression of the MIR172 gene family bolster the floral stem cell network. PloS Genet. 9, e1003218. doi: 10.1371/journal.pgen.1003218
Zhang L., Chia J.-M., Kumari S., Stein J. C., Liu Z., Narechania A., et al. (2009). A genome-wide characterization of microRNA genes in maize. PloS Genet. 5, e1000716. doi: 10.1371/journal.pgen.1000716
Zhang T., Zhao Y.-L., Zhao J.-H., Wang S., Jin Y., Chen Z.-Q., et al. (2016). Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2, 16153. doi: 10.1038/nplants.2016.153
Zhao P., Wang F., Deng Y., Zhong F., Tian P., Lin D., et al. (2022). Sly-miR159 regulates fruit morphology by modulating GA biosynthesis in tomato. Plant Biotechnol. J. 20, 833–845. doi: 10.1111/pbi.13762
Zhao Y., Zhang C., Liu W., Gao W., Liu C., Song G., et al. (2016). An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci. Rep. 6, 1–11. doi: 10.1038/srep23890
Zheng G., Wei W., Li Y., Kan L., Wang F., Zhang X., et al. (2019). Conserved and novel roles of miR164-CUC2 regulatory module in specifying leaf and floral organ morphology in strawberry. New Phytol. 224, 480–492. doi: 10.1111/nph.15982
Zhou J., Deng K., Cheng Y., Zhong Z., Tian L., Tang X., et al. (2017). CRISPR-cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01598
Zhou J., Yuan M., Zhao Y., Quan Q., Yu D., Yang H., et al. (2021). Efficient deletion of multiple circle RNA loci by CRISPR-Cas9 reveals Os06circ02797 as a putative sponge for OsMIR408 in rice. Plant Biotechnol. J. 19, 1240–1252. doi: 10.1111/pbi.13544
Zhou J., Zhang R., Jia X., Tang X., Guo Y., Yang H., et al. (2022). CRISPR-Cas9 mediated OsMIR168a knockout reveals its pleiotropy in rice. Plant Biotechnol. J. 20, 310–322. doi: 10.1111/pbi.13713
Keywords: miRNA, development, morphogenesis, growth, reproduction, robustness, gene regulatory network, mobile signal
Citation: Arnaud N and Laufs P (2023) Plant miRNA integrated functions in development and reproduction. Front. Plant Physiol. 1:1271423. doi: 10.3389/fphgy.2023.1271423
Received: 02 August 2023; Accepted: 14 September 2023;
Published: 02 October 2023.
Edited by:
Tzahi Arazi, Agricultural Research Organization (ARO), IsraelReviewed by:
Attila Molnar, University of Edinburgh, United KingdomTony Millar, Australian National University, Australia
Copyright © 2023 Arnaud and Laufs. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick Laufs, cGF0cmljay5sYXVmc0BpbnJhZS5mcg==
 Nicolas Arnaud
Nicolas Arnaud Patrick Laufs
Patrick Laufs