- 1Aging Institute of UPMC, University of Pittsburgh School of Medicine, Pittsburgh, PA, United States
- 2Pittsburgh Liver Research Center, University of Pittsburgh, Pittsburgh, PA, United States
- 3Division of Endocrinology and Metabolism, Department of Medicine, University of Pittsburgh School of Medicine, Pittsburgh, PA, United States
Repetitive variations, such as oscillation, are ubiquitous in biology. In this mini review, we present a general summary of the ∼24 h circadian clock and provide a fundamental overview of another biological timekeeper that maintains ∼12 h oscillations. This ∼12 h oscillator is proposed to function independently of the circadian clock to regulate ultradian biological rhythms relevant to both protein homeostasis and liver health. Recent studies exploring these ∼12 h rhythms in humans are discussed, followed by our proposal that mammary gland physiology represents a promising area for further research. We conclude by highlighting potential translational applications in ∼12 h ultradian chronobiology.
Introduction
Rhythms in organismal behavior, such as the sleep-wake cycle, and cellular-level oscillations are well-documented (McClung, 2007; Goldbeter, 2008). Among biological oscillations, circadian rhythms are the most well-characterized. These evolutionarily conserved timekeepers, found across various biological domains, regulate ∼24 hour (∼24 h) cycles that align closely with the Earth’s self-rotation (Pittendrigh, 1960; Loudon et al., 2000; Loudon, 2012). Besides circadian rhythms, there are other biological oscillations, such as infradian and ultradian rhythms, with periods longer or shorter than a day, respectively.
This mini review begins with an overview of the circadian clock, followed by a concise introduction to ∼12 hour (∼12 h) ultradian rhythms that play key roles in protein homeostasis and liver health. We then explore recent research on human ∼12 h ultradian chronobiology and present a rationale for studying ∼12 h gene expression rhythms in the mammary gland as a promising new research avenue. The comprehensive study of the circadian clock has already led to novel therapeutic strategies, advancing the field of chronotherapy (Festus et al., 2024). We propose that a deeper understanding of the molecular mechanisms governing ultradian rhythms, particularly the ∼12 h oscillator, could pave the way for new chronotherapies and reveal pharmacological targets to treat a variety of human diseases.
The circadian clock is essential for organismal health
Circadian clocks can be entrained by environmental cues such as light, temperature, and food (entrainment), persist in their absence (free running), and maintain a ∼24 h period across a wide range of temperatures (temperature compensation) (Roenneberg and Merrow, 2005; Kidd et al., 2015; Mofatteh et al., 2021). The foundational study by Konopka and Benzer in 1971, titled “Clock Mutants of Drosophila melanogaster,” was the first to reveal how the molecular clock regulates circadian rhythms. They showed that mutations in the circadian gene period disrupt the circadian rhythms of eclosion and locomotion in fruit flies (Konopka and Benzer, 1971). In 1988, Ralph and Menaker demonstrated that a mutation in the tau gene alters the circadian rhythm of locomotor activity in the golden hamster, marking the first genetic link to circadian rhythms in a mammalian model (Ralph and Menaker, 1988).
The discovery of the Clock mutant in mice, which disrupts the circadian rhythm of wheel-running activity (Vitaterna et al., 1994), led to the identification of the CLOCK protein. Further research identified its binding partner, BMAL1, another core circadian clock transcription factor (Hogenesch et al., 1997; Ikeda and Nomura, 1997; Gekakis et al., 1998; Bunger et al., 2000). The BMAL1/CLOCK heterodimer regulates the transcription of two other circadian genes: Period and Cryptochrome (Gekakis et al., 1998; Jin et al., 1999; Kume et al., 1999). These genes encode the PER and CRY proteins, which together inhibit the transcriptional activity of CLOCK and BMAL1, thereby forming a negative feedback loop (Kume et al., 1999; Shearman et al., 2000; Partch et al., 2014). The transcriptional-translational feedback loop (TTFL) serves as the central mechanism driving circadian oscillations in output genes, which in turn regulate vital biological processes such as the sleep-wake and fast-feeding cycles, and many others (Roenneberg and Merrow, 2005; Takahashi, 2017; Rijo-Ferreira and Takahashi, 2019).
Our comprehensive understanding of circadian chronobiology has firmly established the connection between these ∼24 h rhythms and overall health. Research now shows that aligning feeding times (such as time-restricted feeding) with circadian rhythms can extend lifespan and promote healthy aging in mice (Acosta-Rodriguez et al., 2022). Conversely, disruptions in circadian rhythms have been causally linked to numerous negative health outcomes (Kettner et al., 2015; Cai et al., 2019; Fishbein et al., 2021; Huang et al., 2021; Huang et al., 2022; Sato and Sato, 2023; Van Drunen and Eckel-Mahan, 2023; Huang et al., 2024). These insights raise an important question: how do biological rhythms with periods other than ∼24 h influence mammalian health?
Our research group is particularly interested in ultradian rhythms, especially those cycling with a ∼12 h period, and their role in maintaining mammalian organismal health. Exploring how these faster biological oscillations influence health could reveal new opportunities for therapeutic interventions, much like the advancements made through circadian research. In the following sections, we will delve into recent discoveries on these fascinating ∼12 h ultradian rhythms.
The ∼12 h oscillator is essential for maintaining (ER) proteostasis
Endoplasmic reticulum (ER) protein homeostasis (proteostasis) is vital for maintaining a healthy secreted proteome (Plate and Wiseman, 2017). Newly synthesized proteins enter the ER, where they are properly folded and assembled for secretion. The unfolded protein response (UPR) plays a critical role in managing ER proteostasis. When misfolded proteins accumulate in the ER lumen, the UPR is activated through three ER membrane proteins: ATF6, IRE1α, and PERK. These proteins can sense ER stress and quickly initiate signaling cascades to either restore proteostasis or, if the stress is too severe, trigger apoptosis (Walter and Ron, 2011; Hetz et al., 2015).
Of these, the IRE1α branch of the UPR is the most evolutionarily conserved (Karagöz et al., 2017). Under normal conditions, the ER chaperone BiP binds to IRE1α in the ER lumen, keeping it inactive as a monomer. When unfolded proteins accumulate, they compete with IRE1α for BiP binding, freeing IRE1α and allowing it to activate through oligomerization and autophosphorylation. This activation enables the endoribonuclease domains of IRE1α to splice Xbp1 mRNA in a non-canonical manner, producing the spliced form Xbp1s by releasing a 26-nucleotide intron. Xbp1s mRNA is translated into the ∼50 kD transcription factor XBP1s that moves to the nucleus and initiates a transcriptional response aimed at restoring proteostasis (Ron and Walter, 2007; Karagöz et al., 2019). Importantly, XBP1s also plays a central role in regulating the ∼12 h oscillator, as demonstrated below.
Studies by Hughes et al. (2009) and Cretenet et al. (2010) were among the first to link ∼12 h rhythms to ER proteostasis (Hughes et al., 2009; Cretenet et al., 2010). Hughes et al. (2009) showed ∼12 h rhythmic expression of selective ER proteostasis genes including BiP in the liver of mice that are fed ad libitum under constant darkness conditions, while Cretenet et al. (2010) further demonstrated ∼12 h oscillations in the IRE1α branch of the UPR in mouse liver, including ∼12 h rhythms of phosphorylated IRE1α and nuclear XBP1s levels (Cretenet et al., 2010). Their work also showed that the loss of Cry1 and Cry2 genes disrupted this ∼12 h rhythmicity. By contrast, later studies found that hundreds of ∼12 h hepatic transcripts including Bip and Xbp1s persisted even without BMAL1, the central circadian clock regulator (Yang et al., 2016; Zhu et al., 2017; Zhu and Liu, 2023). This discrepancy in how disrupting different components of the circadian clock affects ∼12 h rhythms may stem from the non-circadian clock-regulating functions of the CRY1/CRY2 proteins (Wong et al., 2022; Zhu and Liu, 2023).
Zhu et al. (2017) subsequently identified XBP1s as a key transcriptional regulator of ∼12 h rhythms of gene expression via directly binding to the promoter regions of many ER proteostasis genes, with prominent examples such as BiP, Eif2ak3 and Sec23b (Zhu et al., 2017; Zhu et al., 2018). Eif2ak3 encodes PERK, one of the sensors of ER stress that triggers the integrated stress response to attenuate global translation (Pakos-Zebrucka et al., 2016; Costa-Mattioli and Walter, 2020). Sec23b plays a role in exporting proteins from the ER for secretion (Tao et al., 2012). By performing high temporal resolution hepatic transcriptome profiling in both wild-type and XBP1 liver-specific knockout mice, Pan et al. (2020) demonstrated that XBP1s liver-specific ablation minimally affects the hepatic circadian transcriptome but greatly disrupts the ∼12 h oscillating gene program (Pan et al., 2020). Hepatic XBP1s ChIP-Seq revealed direct ∼12 h rhythmic XBP1s chromatin recruitment to the promoter regions of hundreds of genes (Pan et al., 2020). XBP1s-dependent hepatic ∼12 h cycling genes are strongly enriched in the proteostasis pathways, including ribosome biogenesis, protein processing in the ER and Golgi, protein folding, and protein export (Pan et al., 2020; Meng et al., 2020). Lastly, XBP1s-dependent cell-autonomous ∼12 h oscillations of proteostasis gene expression were further identified in mouse embryonic fibroblasts (Zhu et al., 2017; Pan et al., 2020).
Together, these studies indicate that ∼12 h ultradian rhythms operate through mechanisms distinct from circadian timekeeping and instead involve a dedicated “∼12 h oscillator”. These results further establish XBP1s as a central transcriptional regulator of the ∼12 h oscillator, playing a critical role in proteostasis. However, it is premature to conclude that the ∼12 h oscillator operates entirely independently of the circadian clock. The circadian clock regulates feeding behavior and cellular metabolism (Vollmers et al., 2009; Page et al., 2020; Schrader et al., 2024). Since metabolic cues are known to entrain the ∼12 h oscillator (Zhu et al., 2017), disruptions in circadian rhythms could indirectly influence ∼12 h ultradian rhythms via altered behaviors and metabolism.
Nuclear speckles are integral components of the ∼12 h oscillator and essential for (ER) proteostasis
In mice, besides proteostasis genes, mRNA metabolism genes also exhibit ∼12 h oscillations across various tissues (Zhu, 2020; Zhu and Liu, 2023), but the mechanism linking mRNA metabolism with proteostasis dynamics remains unclear. Our research group aims to uncover this connection by studying nuclear speckles—biomolecular condensates that regulate aspects of mRNA metabolism, including transcription, mRNA splicing, and RNA export (Spector and Lamond, 2011; Liao and Regev, 2021; Hirose et al., 2023; Bhat et al., 2024).
Nuclear speckles contain RNA-protein complexes called spliceosomes that are essential for RNA processing (Girard et al., 2012). Notably, the Gene Ontology (GO) term “spliceosome” is just as enriched in the XBP1s-dependent hepatic ∼12 h transcriptome as GO terms related to proteostasis (Pan et al., 2020). These speckles are believed to form via liquid-liquid phase separation (LLPS), where proteins rich in intrinsically disordered regions, such as SRRM2 and SON, create a scaffold that facilitates the assembly of other proteins (including splicing factors) and RNAs (such as the long non-coding RNA Malat1), forming a heterogeneous condensate with wide-ranging viscoelastic properties (Sharma et al., 2010; Sharma et al., 2011; Fei et al., 2017; Ilik et al., 2020; Ilik and Aktas, 2022). Components of nuclear speckles continuously exchange between the dense phase (the speckle itself) and the dilute phase (the surrounding nucleoplasm), resulting in irregular shapes and dynamic morphologies (Banani et al., 2017; Ilik and Aktas, 2022; Hirose et al., 2023). The proximity of nuclear speckles to genes influences transcription, with closer speckles often associated with higher transcriptional activity (Kim et al., 2020; Alexander et al., 2021; Bhat et al., 2024). This indicates that the shape and size of nuclear speckles can influence the cellular transcriptome, as larger speckles with greater surface area are likely to interact with more chromatin, potentially enhancing the expression of nearby genes (Dion et al., 2022).
Our group has linked the LLPS dynamics of nuclear speckles to the expression of proteostasis genes by uncovering an XBP1s-SON regulatory axis. This axis controls ∼12 h rhythms in both nuclear speckle morphology (Figure 1A) and their interactions with chromatin (Dion et al., 2022). The expression level of SON, a key scaffolding protein in nuclear speckles, significantly influences their LLPS dynamics, which in turn affects the transcription of proteostasis genes (Dion et al., 2022). Elevated SON levels increase the diffuseness and surface area of nuclear speckles, enhancing their interaction with chromatin, amplifying the expression of proteostasis genes (including Xbp1), and reducing protein aggregation (Dion et al., 2022). Conversely, reducing SON expression has the opposite effect, leading to smaller speckles with decreased chromatin interaction and lower proteostasis gene expression (Dion et al., 2022). Notably, Son is a direct transcriptional target of XBP1s, establishing a direct link between nuclear speckle dynamics and the transcriptional regulation of proteostasis (Dion et al., 2022).
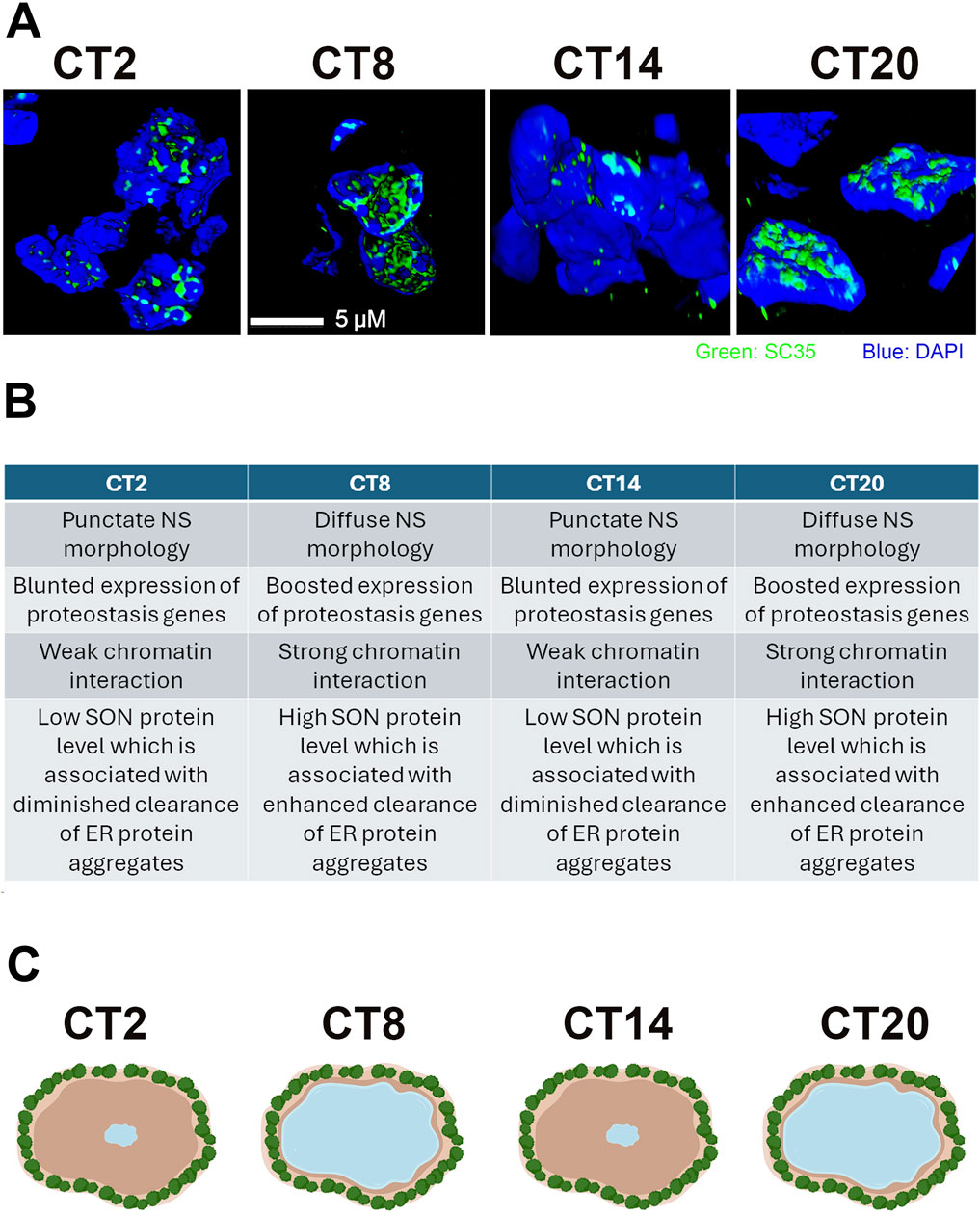
Figure 1. Ultradian biological rhythms of nuclear speckle liquid-liquid phase separation dynamics and proteostasis. (A) Nuclear speckle morphology (indicated by SC35 signal) in mouse liver at different timepoints. Normal nuclear speckle morphologies alternate between punctate (CT2 and CT14) and diffuse (CT8 and CT20). Panel taken from Figure 1A of Dion et al., 2022, © The Authors, some rights reserved; exclusive licensee AAAS. Distributed under a Creative Commons Attribution License 4.0 (CC BY) https://creativecommons.org/licenses/by/4.0/. (B) Characteristics associated with different nuclear speckle morphologies (nuclear speckle (NS), liquid-liquid phase separation (LLPS)). (C) Our lake analogy shows full and drying lakes representing normally occurring nuclear speckle morphologies and green shrubs which represent chromatin. The changes in the water’s distance from the green shrubs could be understood as how nuclear speckles' proximity to chromatin normally changes over time. Image created with BioRender.com.
Under physiological conditions, SON expression, nuclear speckle LLPS dynamics, chromatin interactions, and proteostasis gene expression all exhibit XBP1s-dependent ∼12 h rhythms (Figures 1A,B) (Dion et al., 2022). These insights led us to develop the “lake analogy”: when SON levels are high, nuclear speckles become large and diffuse, akin to a full lake, with strong chromatin interactions. In contrast, when SON levels are low, the speckles are smaller and more spherical, resembling a drying lake, with weaker chromatin interactions (Figure 1C) (Dion et al., 2022). Our exploration of ∼12 h ultradian chronobiology has deepened our understanding of the molecular mechanisms underlying proteostasis and identified nuclear speckles as a new therapeutic target for proteinopathies (Dion et al., 2024).
The ∼12 h oscillator regulates liver health via lipid remodeling
Liver disease is a growing problem affecting diverse populations (Loomba and Sanyal, 2013; Byrne and Targher, 2015; Kardashian et al., 2023; Younossi et al., 2023). Non-alcoholic fatty liver disease (NAFLD) (or “metabolic dysfunction-associated fatty liver disease” (MAFLD) (Chen et al., 2024)) is associated with dysfunctional ER proteostasis (Flessa et al., 2022). While the loss of either UPR or ER quality control components results in hepatic steatosis (also known as “fatty liver”) in mice, maintaining or activating ER quality control mechanism protects against NAFLD (Rutkowski et al., 2008; Yamamoto et al., 2010). For instance, IRE1α maintains lipid balance during ER stress (Zhang et al., 2011) and XBP1s reduces the production of lipids in the livers of both obese and insulin-resistant mice (Herrema et al., 2016). XBP1s-selective pharmacological activation of IRE1α also improves liver function in obese mice (Madhavan et al., 2022). The importance of the ∼12 h oscillator’s regulator XBP1s to hepatic function suggests a link between ∼12 h ultradian rhythms and liver health.
In addition to regulating mRNA metabolism and proteostasis, XBP1s also plays key roles in lipid metabolism (Moncan et al., 2021). Recent studies have shown that activating XBP1s and other UPR pathways can protect against hepatic steatosis by modulating membrane lipid composition (Rutkowski et al., 2008; Yamamoto et al., 2010; Zhang et al., 2011; Herrema et al., 2016). For instance, during diet-induced ER stress, activation of the Lysophosphatidylcholine Acyltransferase 3 (Lpcat3) gene, which promotes the incorporation of polyunsaturated fatty acids into ER membrane phospholipids, helps maintain ER membrane fluidity, reducing both hepatic inflammation and ER stress (Rong et al., 2013; Zhu et al., 2017). Notably, Lpcat3 mRNA and levels of 2-Lysophosphatidylcholine species (LPCAT3 catalyzes the conjugation of 2-Lysophosphatidylcholine with unsaturated Acyl-CoA to form phosphatidylcholine) exhibit strong ∼12 h rhythms in the mouse liver, along with rhythmic expression of fatty acid-modifying enzymes like Scd1 and Elovl6 (Zhu et al., 2017; Zhu et al., 2018; Meng et al., 2020). These rhythmic changes in lipid composition impact the fluidity of cellular membranes, affecting signal transduction across lipid bilayers and potentially influencing systemic metabolism—a connection that remains to be fully explored. In mice with liver-specific XBP1 deletion, the ∼12 h rhythm of Lpcat3 expression is disrupted, leading to lower levels of polyunsaturated phospholipids, reduced membrane fluidity, and impaired lipid metabolism (Meng et al., 2020). This disruption accelerates the development of NAFLD and liver aging, while also contributing to glucose intolerance and hyperinsulinemia (Meng et al., 2020).
In a follow-up study, Meng et al. (2022) characterized SRC-3 (Ncoa3) as a transcriptional co-activator of XBP1s essential for hepatic ∼12 h rhythms of gene expression and proper metabolic function (Meng et al., 2022). Considering the loss of ∼12 h hepatic rhythms of gene expression preceded the manifestation of steatosis (Meng et al., 2020), disruption of the hepatic ∼12 h oscillator is suggested to drive, rather than be a consequence of, NAFLD. Chronobiological therapies that maintain ∼12 h ultradian rhythmicity could prevent or slow the progression of NAFLD. One future research direction could be administering the XBP1s-selective IRE1α activating compound IXA4 (Grandjean et al., 2020; Madhavan et al., 2022) at regular intervals to possibly synchronize/reinforce the ∼12 h oscillator. This approach could be applied to different mouse models of NAFLD to see if pharmacologically boosting the ∼12 h oscillator could slow or prevent liver disease progression.
∼12 h rhythms exist in humans
Previous research has identified ∼12 h oscillations in human physiological metrics, suggesting the existence of a ∼12 h oscillator in humans (Broughton and Mullington, 1992; Wan et al., 1992; Hayashi et al., 2002; Otsuka et al., 2016; Otsuka et al., 2022; Otsuka et al., 2023a; Otsuka et al., 2023b). As previously discussed, the ∼12 h oscillator plays a crucial role in regulating proteostasis in mice, and disruptions in proteostasis are also strongly linked to human neurodegenerative diseases such as Alzheimer’s, Parkinson’s, and Huntington’s, as well as psychiatric disorders like schizophrenia (SZN) (Hetz and Saxena, 2017). For example, altered expression of XBP1s in the brain was observed in Alzheimer’s and Huntington’s diseases (Hetz and Mollereau, 2014), while dysfunction of the IRE1α component of the UPR has been linked to SZN (Kim et al., 2021).
To explore whether ∼12 h rhythms exist in the human brain and their potential connection to psychiatric disorders, Scott et al. (2023) conducted a post hoc analysis of RNA-seq data from human brain samples, using time-of-death as a proxy for circadian time. The analysis included both control subjects and individuals with SZN (Scott et al., 2023). In the dorsolateral prefrontal cortex—a region critical for cognitive function—Scott et al. (2023) identified ∼12 h rhythms of gene expression in control subjects. These rhythms peaked at sleep/wake transitions (around 9 AM and 9 PM) and at static times (around 3 AM and 3 PM) (Scott et al., 2023). Intriguingly, in subjects with SZN, genes associated with the UPR and neuronal structural maintenance lost their ∼12 h rhythmic expressions (Scott et al., 2023). Additionally, genes involved in mitochondrial function and protein translation, which normally peak at sleep/wake transitions in control subjects, peak at static times in SZN subjects (Scott et al., 2023). These findings suggest that pharmacological realignment of ∼12 h gene expression rhythms might help alleviate some symptoms of schizophrenia. This approach aligns with existing strategies that target the circadian clock as a therapeutic option for circadian disruptions (Rasmussen et al., 2022).
In a separate study, Zhu et al. (2024) provided direct evidence of ∼12 h ultradian rhythms in humans through prospective temporal transcriptome profiling of peripheral white blood cells from three healthy male subjects (Zhu et al., 2024). This study identified robust ∼12 h transcriptional rhythms, particularly those implicated in RNA and protein metabolism, with striking homology to the circatidal gene programs previously found in marine species like Cnidarians (Zhu et al., 2024). In addition, Zhu et al. (2024). uncovered ∼12 h rhythms of intron retention in genes involved in MHC class I antigen presentation, which were synchronized with mRNA splicing gene expression in each individual (Zhu et al., 2024). These findings suggest that human ∼12 h biological rhythms have a primordial evolutionary origin and may have significant implications for human health and disease beyond neurological disorders and metabolic syndromes.
Mammary gland physiology as a future direction
The synthesis and secretory demands of lactation are associated with an abundance of Golgi and ER in alveolar epithelial cells (Anderson et al., 2007; Hannan et al., 2023). Xbp1 gene expression increases in the pregnant murine mammary gland (Tsuchiya et al., 2017), and knockout of Xbp1 in the mammary epithelium caused ER stress during lactation and impeded milk production (Hasegawa et al., 2015; Davis et al., 2016). The transcriptional co-activator SRC-3 is also essential for proper mammary gland development (Xu et al., 2000). These studies show that previously identified aspects of the ∼12 h oscillator are relevant to mammary gland physiology.
Maningat et al. (2009) completed a temporal analysis of human milk fat globule (hMFG) gene expression to study the cycling transcriptome of human mammary epithelial cells (Maningat et al., 2009). They uncovered a circadian transcriptional program in the hMFG which prompted our post hoc analysis of their published gene expression data. We used RAIN (Thaben and Westermark, 2014) to test for ultradian oscillations of gene expression and uncovered a distribution of cycling genes with ultradian and circadian periods among the participants (Maningat et al., 2009) (Figure 2). Based on these findings, we propose that there is strong justification for a study profiling the temporal transcriptome in murine mammary glands, both with and without functional XBP1s activity. Such research could help identify chronotherapeutic targets that address barriers to healthy lactation, ultimately benefiting mothers and their infants (Rollins et al., 2016; Wang and Scherer, 2019).

Figure 2. Post hoc analysis of the human milk fat globule temporal transcriptome. Data from the published study are available through the NCBI Gene Expression Omnibus, identifier GSE12669 (Maningat et al., 2009). (A) Total cycling genes with p values less than 0.05 for corresponding periods as determined with RAIN (Thaben and Westermark, 2014). (B) Temporal expression profiles of individual genes previously shown to have ultradian rhythms (Zhu et al., 2017; Pan et al., 2020).
Closing remarks
Our understanding of ∼12 h biological rhythms in humans, though largely descriptive at this point, suggests translational studies are an appropriate future direction. Pharmacological adjustment of circadian rhythms is a proposed therapy to address the disruption of the circadian clock caused by jetlag (Ruan et al., 2021). This suggests that manipulating other biological timekeepers may also benefit human health. The loss of ∼12 h rhythmicity preceding NAFLD progression (Meng et al., 2020) and the misalignment of ∼12 h rhythms in the dorsolateral prefrontal cortex of individuals with SZN (Scott et al., 2023) are two previously discussed examples in which synchronizing or realigning ∼12 h ultradian rhythms could prove as effective therapies. Perhaps inducing low levels of ER stress—which synchronizes the ∼12 h oscillator (Zhu et al., 2017)—to reset ultradian biological rhythms could be an effective chronotherapy to slow NAFLD progression or address some symptoms of SZN. Furthermore, the development of compounds that specifically activate the UPR—such as IXA4 mentioned previously (Grandjean et al., 2020; Madhavan et al., 2022)—could prove to be convenient therapies to manipulate ultradian rhythms.
Despite significant progress in the field of ultradian chronobiology, there is still much more to be learned. We encourage others to uncover more of the molecular clockwork regulating ∼12 h rhythms. Such discoveries promise to benefit human health, given the recent establishment of ∼12 h transcriptional programs among different human tissues. Our understanding of the ∼12 h oscillator is biased toward males in both mice and humans and present studies focus heavily on the liver. We firmly believe that future studies across both sexes and of different tissue types are essential to understanding the full translatable impact of the ∼12 h oscillator on human health.
Author contributions
WD: Writing–original draft, Writing–review and editing. BZ: Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. WD was supported by fellowship F31 AG080998 through the NIH and BZ was supported by grants DP2 GM140924 and R21 AG071893 through the NIH and grants from both the Richard King Mellon Foundation and the American Federation for Aging Research/Hevolution Foundation.
Acknowledgments
Figures were created using Adobe Photoshop 2024, BioRender.com, GraphPad Prism, Microsoft Office PowerPoint, and RStudio (Wickham et al., 2019; RStudio Team 2021).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acosta-Rodriguez V., Rijo-Ferreira F., Izumo M., Xu P., Wight-Carter M., Green C. B., et al. (2022). Circadian alignment of early onset caloric restriction promotes longevity in male C57BL/6J mice. Science 376 (6598), 1192–1202. doi:10.1126/science.abk0297
Alexander K. A., Coté A., Nguyen S. C., Zhang L., Gholamalamdari O., Agudelo-Garcia P., et al. (2021). p53 mediates target gene association with nuclear speckles for amplified RNA expression. Mol. Cell 81 (8), 1666–1681.e6. doi:10.1016/j.molcel.2021.03.006
Anderson S. M., Rudolph M. C., McManaman J. L., Neville M. C. (2007). Key stages in mammary gland development. Secretory activation in the mammary gland: it’s not just about milk protein synthesis. Breast Cancer Res. 9 (204), 204–214. doi:10.1186/bcr1653
Banani S. F., Lee H. O., Hyman A. A., Rosen M. K. (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18 (5), 285–298. doi:10.1038/nrm.2017.7
Bhat P., Chow A., Emert B., Ettlin O., Quinodoz S. A., Strehle M., et al. (2024). Genome organization around nuclear speckles drives mRNA splicing efficiency. Nature 629 (8014), 1165–1173. doi:10.1038/s41586-024-07429-6
Broughton R., Mullington J. (1992). Circasemidian sleep propensity and the phase-amplitude maintenance model of human sleep/wake regulation. J. Sleep. Res. 1, 93–98. doi:10.1111/j.1365-2869.1992.tb00017.x
Bunger M. K., Wilsbacher L. D., Moran S. M., Clendenin C., Radcliffe L. A., Hogenesch J. B., et al. (2000). Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103 (7), 1009–1017. doi:10.1016/S0092-8674(00)00205-1
Byrne C. D., Targher G. (2015). NAFLD: a multisystem disease. J. Hepatol. 62 (1), S47–S64. doi:10.1016/j.jhep.2014.12.012
Cai C., Vandermeer B., Khurana R., Nerenberg K., Featherstone R., Sebastianski M., et al. (2019). The impact of occupational shift work and working hours during pregnancy on health outcomes: a systematic review and meta-analysis. Am. J. Obstet. Gynecol. 221 (6), 563–576. doi:10.1016/j.ajog.2019.06.051
Chen L., Tao X., Zeng M., Mi Y., Xu L. (2024). Clinical and histological features under different nomenclatures of fatty liver disease: NAFLD, MAFLD, MASLD and MetALD. J. Hepatol. 80 (2), e64–e66. doi:10.1016/j.jhep.2023.08.021
Costa-Mattioli M., Walter P. (2020). The integrated stress response: from mechanism to disease. Science 368 (6489), eaat5314. doi:10.1126/science.aat5314
Cretenet G., Le Clech M., Gachon F. (2010). Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 11 (1), 47–57. doi:10.1016/j.cmet.2009.11.002
Davis K. R., Giesy S. L., Long Q., Krumm C. S., Harvatine K. J., Boisclair Y. R. (2016). XBP1 regulates the biosynthetic capacity of the mammary gland during lactation by controlling epithelial expansion and endoplasmic reticulum formation. Endocrinology 157 (1), 417–428. doi:10.1210/en.2015-1676
Dion W., Ballance H., Lee J., Pan Y., Irfan S., Edwards C., et al. (2022). Four-dimensional nuclear speckle phase separation dynamics regulate proteostasis. Sci. Adv. 8 (1), eabl4150. doi:10.1126/sciadv.abl4150
Dion W., Tao Y., Chambers M., Zhao S., Arbuckle R. K., Sun M., et al. (2024). Nuclear speckle rejuvenation alleviates proteinopathies at the expense of YAP1. bioRxiv 2024 (04.18), 590103. doi:10.1101/2024.04.18.590103
Fei J., Jadaliha M., Harmon T. S., Li I. T. S., Hua B., Hao Q., et al. (2017). Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J. Cell. Sci. 130 (24), 4180–4192. doi:10.1242/jcs.206854
Festus I. D., Spilberg J., Young M. E., Cain S., Khoshnevis S., Smolensky M. H., et al. (2024). Pioneering new frontiers in circadian medicine chronotherapies for cardiovascular health. Trends Endocrinol. Metab. 35 (7), 607–623. doi:10.1016/j.tem.2024.02.011
Fishbein A. B., Knutson K. L., Zee P. C. (2021). Circadian disruption and human health. J. Clin. Invest. 131 (19), e148286. doi:10.1172/JCI148286
Flessa C. M., Kyrou I., Nasiri-Ansari N., Kaltsas G., Kassi E., Randeva H. S. (2022). Endoplasmic reticulum stress in nonalcoholic (metabolic associated) fatty liver disease (NAFLD/MAFLD). J. Cell. Biochem. 123 (10), 1585–1606. doi:10.1002/jcb.30247
Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., et al. (1998). Role of the CLOCK protein in the mammalian circadian mechanism. Science 280 (5369), 1564–1569. doi:10.1126/science.280.5369.1564
Girard C., Will C. L., Peng J., Makarov E. M., Kastner B., Lemm I., et al. (2012). Post-transcriptional spliceosome are retained in nuclear speckles until splicing completion. Nat. Commun. 3, 994. doi:10.1038/ncomms1998
Goldbeter A. (2008). Biological rhythms: clocks for all times. Curr. Biol. 18 (17), R751–R753. doi:10.1016/j.cub.2008.06.044
Grandjean J. M. D., Madhavan A., Cech L., Seguinot B. O., Paxman R. J., Smith E., et al. (2020). Pharmacologic IRE1/XBP1s activation confers targeted ER proteostasis reprogramming. Nat. Chem. Biol. 16 (10), 1052–1061. doi:10.1038/s41589-020-0584-z
Hannan F. M., Elajnaf T., Vandenberg L. N., Kennedy S. H., Thakker R. V. (2023). Hormonal regulation of mammary gland development and lactation. Nat. Rev. Endocrinol. 19 (1), 46–61. doi:10.1038/s41574-022-00742-y
Hasegawa D., Calvo V., Avivar-Valderas A., Lade A., Chou H. I., Lee Y. A., et al. (2015). Epithelial Xbp1 is required for cellular proliferation and differentiation during mammary gland development. Mol. Cell. Biol. 35 (9), 1543–1556. doi:10.1128/Mcb.00136-15
Hayashi M., Morikawa T., Hori T. (2002). Circasemidian 12 h cycle of slow wave sleep under constant darkness. Clin. Neurophysiol. 113 (9), 1505–1516. doi:10.1016/s1388-2457(02)00168-2
Herrema H., Zhou Y., Zhang D., Lee J., Hernandez M. A. S., Shulman G. I., et al. (2016). XBP1s is an anti-lipogenic protein. J. Biol. Chem. 291 (33), 17394–17404. doi:10.1074/jbc.M116.728949
Hetz C., Chevet E., Oakes S. A. (2015). Proteostasis control by the unfolded protein response. Nat. Cell. Biol. 17 (7), 829–838. doi:10.1038/ncb3184
Hetz C., Mollereau B. (2014). Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat. Rev. Neurosci. 15 (4), 233–249. doi:10.1038/nrn3689
Hetz C., Saxena S. (2017). ER stress and the unfolded protein response in neurodegeneration. Nat. Rev. Neurol. 13 (8), 477–491. doi:10.1038/nrneurol.2017.99
Hirose T., Ninomiya K., Nakagawa S., Yamazaki T. (2023). A guide to membraneless organelles and their various roles in gene regulation. Nat. Rev. Mol. Cell Biol. 24 (4), 288–304. doi:10.1038/s41580-022-00558-8
Hogenesch J. B., Chan W. K., Jackiw V. H., Brown R. C., Gu Y. Z., Pray-Grant M., et al. (1997). Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J. Biol. Chem. 272 (13), 8581–8593. doi:10.1074/jbc.272.13.8581
Huang S., Jiao X., Lu D., Pei X., Qi D., Li Z. (2021). Light cycle phase advance as a model for jet lag reprograms the circadian rhythms of murine extraorbital lacrimal glands. Ocul. Surf. 20, 95–114. doi:10.1016/j.jtos.2021.02.001
Huang S., Si H., Liu J., Qi D., Pei X., Lu D., et al. (2022). Sleep loss causes dysfunction in murine extraorbital lacrimal glands. Invest. Ophthalmol. Vis. Sci. 63 (6), 19. doi:10.1167/iovs.63.6.19
Huang S., Zhang W., Xuan S., Si H., Huang D., Ba M., et al. (2024). Chronic sleep deprivation impairs retinal circadian transcriptome and visual function. Exp. Eye Res. 243, 109907. doi:10.1016/j.exer.2024.109907
Hughes M. E., DiTacchio L., Hayes K. R., Vollmers C., Pulivarthy S., Baggs J. E., et al. (2009). Harmonics of circadian gene transcription in mammals. PLoS Genet. 5 (4), e1000442. doi:10.1371/journal.pgen.1000442
Ikeda M., Nomura M. (1997). cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS protein (BMAL1) and identification of alternatively spliced variants with alternative translation initiation site usage. Biochem. Biophys. Res. Commun. 233 (1), 258–264. doi:10.1006/bbrc.1997.6371
Ilik İ. A., Aktaş T. (2022). Nuclear speckles: dynamic hubs of gene expression regulation. FEBS J. 289 (22), 7234–7245. doi:10.1111/febs.16117
Ilik İ. A., Malszycki M., Lübke A. K., Schade C., Meierhofer D., Aktaş T. (2020). SON and SRRM2 are essential for nuclear speckle formation. Elife 9, e60579. doi:10.7554/eLife.60579
Jin X., Shearman L. P., Weaver D. R., Zylka M. J., de Vries G. J., Reppert S. M. (1999). A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96 (1), 57–68. doi:10.1016/s0092-8674(00)80959-9
Karagöz G. E., Acosta-Alvear D., Nguyen H. T., Lee C. P., Chu F., Walter P. (2017). An unfolded protein-induced conformational switch activates mammalian IRE1. Elife 6, e30700. doi:10.7554/eLife.30700
Karagöz G. E., Acosta-Alvear D., Walter P. (2019). The unfolded protein response: detecting and responding to fluctuations in the protein-folding capacity of the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 11 (9), a033886. doi:10.1101/cshperspect.a033886
Kardashian A., Serper M., Terrault N., Nephew L. D. (2023). Health disparities in chronic liver disease. Hepatology 77 (4), 1382–1403. doi:10.1002/hep.32743
Kettner N. M., Mayo S. A., Hua J., Lee C., Moore D. D., Fu L. (2015). Circadian dysfunction induces leptin resistance in mice. Cell Metab. 22 (3), 448–459. doi:10.1016/j.cmet.2015.06.005
Kidd P. B., Young M. W., Siggia E. D. (2015). Temperature compensation and temperature sensation in the circadian clock. Proc. Natl. Acad. Sci. U. S. A. 112 (46), E6284–E6292. doi:10.1073/pnas.1511215112
Kim J., Venkata N. C., Hernandez Gonzalez G. A., Khanna N., Belmont A. S. (2020). Gene expression amplification by nuclear speckle association. J. Cell Biol. 219 (1), e201904046. doi:10.1083/jcb.201904046
Kim P., Scott M. R., Meador-Woodruff J. H. (2021). Dysregulation of the unfolded protein response (UPR) in the dorsolateral prefrontal cortex in elderly patients with schizophrenia. Mol. Psychiatry 26 (4), 1321–1331. doi:10.1038/s41380-019-0537-7
Konopka R. J., Benzer S. (1971). Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 68 (9), 2112–2116. doi:10.1073/pnas.68.9.2112
Kume K., Zylka M. J., Sriram S., Shearman L. P., Weaver D. R., Jin X., et al. (1999). mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98 (2), 193–205. doi:10.1016/s0092-8674(00)81014-4
Liao S. E., Regev O. (2021). Splicing at the phase-separated nuclear speckle interface: a model. Nucleic Acids Res. 49 (2), 636–645. doi:10.1093/nar/gkaa1209
Loomba R., Sanyal A. J. (2013). The global NAFLD epidemic. Nat. Rev. Gastroenterol. Hepatol. 10 (11), 686–690. doi:10.1038/nrgastro.2013.171
Loudon A. S., Semikhodskii A. G., Crosthwaite S. K. (2000). A brief history of circadian time. Trends Genet. 16 (11), 477–481. doi:10.1016/s0168-9525(00)02122-3
Loudon A. S. I. (2012). Circadian biology: a 2.5 billion year old clock. Curr. Biol. 22 (14), R570–R571. doi:10.1016/j.cub.2012.06.023
Madhavan A., Kok B. P., Rius B., Grandjean J. M. D., Alabi A., Albert V., et al. (2022). Pharmacologic IRE1/XBP1s activation promotes systemic adaptive remodeling in obesity. Nat. Commun. 13 (1), 608. doi:10.1038/s41467-022-28271-2
Maningat P. D., Sen P., Rijnkels M., Sunehag A. L., Hadsell D. L., Bray M., et al. (2009). Gene expression in the human mammary epithelium during lactation: the milk fat globule transcriptome. Physiol. Genomics 37 (1), 12–22. doi:10.1152/physiolgenomics.90341.2008
McClung C. A. (2007). Circadian genes, rhythms and the biology of mood disorders. Pharmacol. Ther. 114 (2), 222–232. doi:10.1016/j.pharmthera.2007.02.003
Meng H., Gonzales N. M., Jung S. Y., Lu Y., Putluri N., Zhu B., et al. (2022). Defining the mammalian coactivation of hepatic 12-h clock and lipid metabolism. Cell Rep. 38 (10), 110491. doi:10.1016/j.celrep.2022.110491
Meng H., Gonzales N. M., Lonard D. M., Putluri N., Zhu B., Dacso C. C., et al. (2020). XBP1 links the 12-hour clock to NAFLD and regulation of membrane fluidity and lipid homeostasis. Nat. Commun. 11 (1), 6215. doi:10.1038/s41467-020-20028-z
Mofatteh M., Echegaray-Iturra F., Alamban A., Dalla Ricca F., Bakshi A., Aydogan M. G. (2021). Autonomous clocks that regulate organelle biogenesis, cytoskeletal organization, and intracellular dynamics. Elife 10, e72104. doi:10.7554/eLife.72104
Moncan M., Mnich K., Blomme A., Almanza A., Samali A., Gorman A. M. (2021). Regulation of lipid metabolism by the unfolded protein response. J. Cell. Mol. Med. 25 (3), 1359–1370. doi:10.1111/jcmm.16255
Otsuka K., Cornelissen G., Furukawa S., Kubo Y., Hayashi M., Shibata K., et al. (2016). Long-term exposure to space’s microgravity alters the time structure of heart rate variability of astronauts. Heliyon 2 (12), e00211. doi:10.1016/j.heliyon.2016.e00211
Otsuka K., Cornelissen G., Furukawa S., Shibata K., Kubo Y., Mizuno K., et al. (2022). Unconscious mind activates central cardiovascular network and promotes adaptation to microgravity possibly anti-aging during 1-year-long spaceflight. Sci. Rep. 12 (1), 11862. doi:10.1038/s41598-022-14858-8
Otsuka K., Cornelissen G., Kubo Y., Shibata K., Mizuno K., Aiba T., et al. (2023a). Methods for assessing change in brain plasticity at night and psychological resilience during daytime between repeated long-duration space missions. Sci. Rep. 13 (1), 10909. doi:10.1038/s41598-023-36389-6
Otsuka K., Murakami S., Okajima K., Shibata K., Kubo Y., Gubin D. G., et al. (2023b). Appropriate circadian-circasemidian coupling protects blood pressure from morning surge and promotes human resilience and wellbeing. Clin. Interv. Aging 18, 755–769. doi:10.2147/CIA.S398957
Page A. J., Christie S., Symonds E., Li H. (2020). Circadian regulation of appetite and time restricted feeding. Physiol. Behav. 220, 112873. doi:10.1016/j.physbeh.2020.112873
Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A. M. (2016). The integrated stress response. EMBO Rep. 17 (10), 1374–1395. doi:10.15252/embr.201642195
Pan Y., Ballance H., Meng H., Gonzalez N., Kim S. M., Abdurehman L., et al. (2020). 12-h clock regulation of genetic information flow by XBP1s. PLoS Biol. 18 (1), e3000580. doi:10.1371/journal.pbio.3000580
Partch C. L., Green C. B., Takahashi J. S. (2014). Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 24 (2), 90–99. doi:10.1016/j.tcb.2013.07.002
Pittendrigh C. S. (1960). Circadian rhythms and the circadian organization of living systems. Cold Spring Harb. Symp. Quant. Biol. 25, 159–184. doi:10.1101/sqb.1960.025.01.015
Plate L., Wiseman R. L. (2017). Regulating secretory proteostasis through the unfolded protein response: from function to therapy. Trends Cell Biol. 27 (10), 722–737. doi:10.1016/j.tcb.2017.05.006
Ralph M. R., Menaker M. (1988). A mutation of the circadian system in golden hamsters. Science 241 (4870), 1225–1227. doi:10.1126/science.3413487
Rasmussen E. S., Takahashi J. S., Green C. B. (2022). Time to target the circadian clock for drug discovery. Trends biochem. Sci. 47 (9), 745–758. doi:10.1016/j.tibs.2022.04.009
Rijo-Ferreira F., Takahashi J. S. (2019). Genomics of circadian rhythms in health and disease. Genome Med. 11 (1), 82. doi:10.1186/s13073-019-0704-0
Roenneberg T., Merrow M. (2005). Circadian clocks - the fall and rise of physiology. Nat. Rev. Mol. Cell Biol. 6 (12), 965–971. doi:10.1038/nrm1766
Rollins N. C., Bhandari N., Hajeebhoy N., Horton S., Lutter C. K., Martines J. C., et al. (2016). Why invest, and what it will take to improve breastfeeding practices? Lancet 387 (10017), 491–504. doi:10.1016/S0140-6736(15)01044-2
Ron D., Walter P. (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8 (7), 519–529. doi:10.1038/nrm2199
Rong X., Albert C. J., Hong C., Duerr M. A., Chamberlain B. T., Tarling E. J., et al. (2013). LXRs regulate ER stress and inflammation through dynamic modulation of membrane phospholipid composition. Cell Metab. 18 (5), 685–697. doi:10.1016/j.cmet.2013.10.002
RStudio Team (2021). RStudio. Boston, MA: Integrated Development for R. RStudio, PBC. Available at: http://www.rstudio.com/.
Ruan W., Yuan X., Eltzschig H. K. (2021). Circadian rhythm as a therapeutic target. Nat. Rev. Drug Discov. 20 (4), 287–307. doi:10.1038/s41573-020-00109-w
Rutkowski D. T., Wu J., Back S. H., Callaghan M. U., Ferris S. P., Iqbal J., et al. (2008). UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev. Cell 15 (6), 829–840. doi:10.1016/j.devcel.2008.10.015
Sato T., Sato S. (2023). Circadian regulation of metabolism: commitment to health and diseases. Endocrinology 164 (7), bqad086. doi:10.1210/endocr/bqad086
Schrader L. A., Ronnekleiv-Kelly S. M., Hogenesch J. B., Bradfield C. A., Malecki K. M. C. (2024). Circadian disruption, clock genes, and metabolic health. J. Clin. Invest. 134 (14), e170998. doi:10.1172/JCI170998
Scott M. R., Zong W., Ketchesin K. D., Seney M. L., Tseng G. C., Zhu B., et al. (2023). Twelve-hour rhythms in transcript expression within the human dorsolateral prefrontal cortex are altered in schizophrenia. PLoS Biol. 21 (1), e3001688. doi:10.1371/journal.pbio.3001688
Sharma A., Markey M., Torres- Muñoz K., Varia S., Kadakia M., Bubulya A., et al. (2011). Son maintains accurate splicing for a subset of human pre-mRNAs. J. Cell Sci. 124 (Pt 24), 4286–4298. doi:10.1242/jcs.092239
Sharma A., Takata H., Shibahara K., Bubulya A., Bubulya P. A. (2010). Son is essential for nuclear speckle organization and cell cycle progression. Mol. Biol. Cell 21 (4), 650–663. doi:10.1091/mbc.e09-02-0126
Shearman L. P., Sriram S., Weaver D. R., Maywood E. S., Chaves I., Zheng B., et al. (2000). Interacting molecular loops in the mammalian circadian clock. Science 288 (5468), 1013–1019. doi:10.1126/science.288.5468.1013
Spector D. L., Lamond A. I. (2011). Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3 (2), a000646. doi:10.1101/cshperspect.a000646
Takahashi J. S. (2017). Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18 (3), 164–179. doi:10.1038/nrg.2016.150
Tao J., Zhu M., Wang H., Afelik S., Vasievich M. P., Chen X. W., et al. (2012). SEC23B is required for the maintenance of murine professional secretory tissues. Proc. Natl. Acad. Sci. U. S. A. 109 (29), E2001–E2009. doi:10.1073/pnas.1209207109
Thaben P. F., Westermark P. O. (2014). Detecting rhythms in time series with RAIN. J. Biol. Rhythms 29 (6), 391–400. doi:10.1177/0748730414553029
Tsuchiya M., Koizumi Y., Hayashi S., Hanaoka M., Tokutake Y., Yonekura S. (2017). The role of unfolded protein response in differentiation of mammary epithelial cells. Biochem. Biophys. Res. Commun. 484 (4), 903–908. doi:10.1016/j.bbrc.2017.02.042
Van Drunen R., Eckel-Mahan K. (2023). Circadian rhythms as modulators of brain health during development and throughout aging. Front. Neural Circuits 16, 1059229. doi:10.3389/fncir.2022.1059229
Vitaterna M. H., King D. P., Chang A. M., Kornhauser J. M., Lowrey P. L., McDonald J. D., et al. (1994). Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264 (5159), 719–725. doi:10.1126/science.8171325
Vollmers C., Gill S., DiTacchio L., Pulivarthy S. R., Le H. D., Panda S. (2009). Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl. Acad. Sci. U. S. A. 106 (50), 21453–21458. doi:10.1073/pnas.0909591106
Walter P., Ron D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334 (6059), 1081–1086. doi:10.1126/science.1209038
Wan C., Wang Z., Cornélissen G., Halberg F. (1992). Age, gender and circadian or circasemidian blood pressure and heart rate variation of children. Chronobiologia 19 (3-4), 121–129.
Wang Q. A., Scherer P. E. (2019). Remodeling of murine mammary adipose tissue during pregnancy, lactation, and involution. J. Mammary Gland. Biol. Neoplasia 24 (3), 207–212. doi:10.1007/s10911-019-09434-2
Wickham H., Averick M., Bryan J., Chang W., McGowan L. D. A., François R., et al. (2019). Welcome to the tidyverse. J. Open Source Softw. 4 (43), 1686. doi:10.21105/joss.01686
Wong D. C. S., Seinkmane E., Zeng A., Stangherlin A., Rzechorzek N. M., Beale A. D., et al. (2022). CRYPTOCHROMES promote daily protein homeostasis. EMBO J. 41 (1), e108883. doi:10.15252/embj.2021108883
Xu J., Liao L., Ning G., Yoshida-Komiya H., Deng C., O’Malley B. W. (2000). The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc. Natl. Acad. Sci. U. S. A. 97 (12), 6379–6384. doi:10.1073/pnas.120166297
Yamamoto K., Takahara K., Oyadomari S., Okada T., Sato T., Harada A., et al. (2010). Induction of liver steatosis and lipid droplet formation in ATF6alpha-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol. Biol. Cell 21 (17), 2975–2986. doi:10.1091/mbc.E09-02-0133
Yang G., Chen L., Grant G. R., Paschos G., Song W. L., Musiek E. S., et al. (2016). Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci. Transl. Med. 8 (324), 324ra16. doi:10.1126/scitranslmed.aad3305
Younossi Z. M., Golabi P., Paik J. M., Henry A., Van Dongen C., Henry L. (2023). The global epidemiology of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH): a systematic review. Hepatology 77 (4), 1335–1347. doi:10.1097/HEP.0000000000000004
Zhang K., Wang S., Malhotra J., Hassler J. R., Back S. H., Wang G., et al. (2011). The unfolded protein response transducer IRE1α prevents ER stress-induced hepatic steatosis. EMBO J. 30 (7), 1357–1375. doi:10.1038/emboj.2011.52
Zhu B. (2020). Decoding the function and regulation of the mammalian 12h-clock. J. Mol. Cell Biol. 12 (10), 752–758. doi:10.1093/jmcb/mjaa021
Zhu B., Dacso C. C., O’Malley B. W. (2018). Unveiling “Musica Universalis” of the cell: a brief history of biological 12-hour rhythms. J. Endocr. Soc. 2 (7), 727–752. doi:10.1210/js.2018-00113
Zhu B., Liu S. (2023). Preservation of ∼12-h ultradian rhythms of gene expression of mRNA and protein metabolism in the absence of canonical circadian clock. Front. Physiol. 14, 1195001. doi:10.3389/fphys.2023.1195001
Zhu B., Liu S., David N. L., Dion W., Doshi N. K., Siegel L. B., et al. (2024). Evidence for ∼12-h ultradian gene programs in humans. NPJ Biol. Timing Sleep. 1 (1), 4. doi:10.1038/s44323-024-00005-1
Keywords: circadian rhythm, ultradian rhythm, proteostasis, NAFLD/MAFLD, nuclear speckles, XBP1s, unfolded protein response
Citation: Dion W and Zhu B (2024) Basic research and opportunities for translational advancement in the field of mammalian ∼12-hour ultradian chronobiology. Front. Physiol. 15:1497836. doi: 10.3389/fphys.2024.1497836
Received: 17 September 2024; Accepted: 28 October 2024;
Published: 20 November 2024.
Edited by:
Jingling Jin, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Baharan Fekry, University of Texas Health Science Center at Houston, United StatesShenzhen Huang, People’s Hospital of Zhengzhou University, China
Copyright © 2024 Dion and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bokai Zhu, YnpodUBwaXR0LmVkdQ==
 William Dion
William Dion Bokai Zhu
Bokai Zhu