- Department of Biological Sciences, University of New Orleans, New Orleans, LA, United States
Low dissolved oxygen (hypoxia) poses significant challenges to aquatic ecosystems, affecting the behavior, reproduction, and survival of aquatic organisms. Some fishes respond to hypoxia by changes in gene expression, which may be regulated by the hypoxia inducible factor (HIF) family of transcription factors. HIF abundance and activity depends upon the post-translational modification of the alpha protein subunit, although several studies indicate that HIFA mRNA abundance increases in tissues of fishes exposed to hypoxia. This study reviewed reports of laboratory exposures of adult ray-finned fishes to hypoxia and used generalized linear mixed effects models to examine the influence of HIFA gene, tissue sampled, and exposure conditions in explaining the diversity of responses seen in HIFA mRNA abundance. The frequency of hypoxia-induced increases in HIFA mRNA was poorly explained by gene, tissue, or the severity of the hypoxic exposure. Rather, the frequency of reported increases was strongly related to the extent to which studies adhered to guidelines for documenting quantitative real-time PCR methods: the frequency of hypoxia-induced increases in HIFA mRNA decreased sharply in studies with more thorough description of experimental design. Future research should (a) adhere to stringent reporting of experimental design, (b) address the relative paucity of data on HIF2A and HIF3A, and (c) determine levels of HIF alpha protein subunits. By following these recommendations, it is hoped that a more complete understanding will be gained of the role of the HIF family of transcription factors in the response of fish to hypoxia.
1 Introduction
Recent decades have seen an increase in the frequency and severity of aquatic hypoxia on a global scale (Breitburg et al., 2018), and the ability of fish to survive hypoxia requires a range of behavioral, physiological, and biochemical strategies (Chapman and McKenzie, 2009; Richards et al., 2009). Because many of these responses may depend on changes in gene expression (Nikinmaa and Rees, 2005; Shen et al., 2023), there is considerable interest in understanding the control of gene expression in fishes during exposure to low oxygen. The hypoxia-inducible transcription factors (HIFs) are evolutionary conserved central regulators of the molecular responses of animals to low oxygen (Semenza, 2012; Pamenter et al., 2020; Mandic et al., 2021). The active transcription factor is comprised of an alpha subunit (HIFα) and a beta subunit (HIFβ) (Semenza, 2012). HIFβ, also known as the aryl hydrocarbon receptor nuclear translocator (McIntosh et al., 2010), is constitutively expressed, whereas the cellular abundance and activity of HIFα are oxygen-dependent. At normal oxygen levels (normoxia), hydroxylation of specific proline residues targets HIFα for rapid proteasomal destruction (Semenza, 2012). In hypoxic conditions, however, the degradation of HIFα is blocked, leading to its accumulation. It then dimerizes with HIFβ, translocates into the nucleus, and activates gene expression (Semenza, 2012).
Signaling by HIF has been best studied in mammals, which possess three alpha subunits, HIF1α, HIF2α, and HIF3α, encoded by genes, HIF1A, HIF2A, and HIF3A, respectively (Pamenter et al., 2020). The initial phylogenetic analysis of HIFA genes in ray-finned fishes (Actinopterygii) demonstrated that they possess orthologs of the three genes found in mammals (Rytkonen et al., 2011). Subsequently, Rytokonen et al. (2013) showed that the cyprinids (carp and related species, including zebrafish and goldfish) possess teleost-specific duplicates, HIF1Aa/b, HIF2Aa/b, and HIF3Aa/b. Townley et al. (2022) supported the presence of HIF1Aa/b and HIF2Aa/b in a larger clade of fishes (Otocephala), which includes not only cyprinids, but also catfish, tetras, and herring. In addition, Townley et al. (2022) presented evidence that HIF3Aa and HIF3Ab correspond to one copy each of HIF3A and HIF4A in ray-finned fishes, which resulted from the genome duplication events at the base of vertebrate evolution. Moreover, Townley et al. (2022) showed that Salmonidae (salmon and trout) have independently evolved duplicates of HIF1Aa, HIF2Aa, and HIF3A. Thus, bony fishes display a diversity of HIFA genes that far exceeds the described diversity in other vertebrate lineages, demanding special attention when differentiating among them.
In the first report of HIF from fish, Soitamo et al. (2001) showed that HIF1α protein increases in abundance during hypoxic exposures of fish cells in culture in the absence of any change in HIF1A mRNA abundance, and that treatment of normoxic cells with an inhibitor of the proteasome recapitulated this increase. These results suggested that protein stabilization, rather than new transcription, was the mechanism of HIF1α upregulation in fish as it is in mammals. Since then, however, several studies have reported elevated mRNA levels of various HIFA genes in tissues of fish exposed to low oxygen (Mandic et al., 2021). In this mini-review, we attempt to discern if there are common patterns of HIFA mRNA responses to low oxygen among fishes. Specifically, we ask if the variation in reported increases in HIFA mRNA levels during hypoxia is related to the HIFA gene measured, tissue sampled, severity of hypoxia, or other experimental details (Bustin et al., 2009).
2 Methods
One hundred and forty articles were identified through searches in Web of Science using the following terms: “HIF (Topic) and mRNA (All fields) and fish (All fields)” or “hypoxia-inducible factor (Topic) and mRNA (All fields) and fish (All fields).” These articles were filtered to remove those on organisms other than ray-finned fishes (35 studies), or early developmental stages (embryos and larvae; 42 studies). The dataset was further filtered to include only studies using laboratory exposures with defined conditions (DO levels and exposure durations), that reported real-time qPCR data with associated statistical analyses. The final dataset consisted of 38 studies reporting on HIFA mRNA abundance in tissues of 25 species of ray-finned fishes.
We extracted information on HIFA gene(s) analyzed, tissues sampled, DO concentration, duration of exposure, temperature, and salinity (Supplementary Data Sheet S1). The identity of each HIFA gene was verified by searching author-reported accession numbers or primer sequences against all ray-finned fishes using Basic Local Alignment Search Tool (Sayers et al., 2022). We follow the nomenclature of Townley et al. (2022) in distinguishing among HIF1A, HIF2A, HIF3A, and HIF4A, as well as between teleost-specific duplicates HIF1Aa/b and HIF2Aa/b. To compare the severity of hypoxic exposures across studies, we calculated the cumulative oxygen deficit (COD) for each sampling interval in each study (Nelson and Lipkey, 2015). COD was determined as the product of the duration of the hypoxic exposure (h) and the difference in DO concentration (mg L-1) between the air-saturated concentration and the reported concentration. The air-saturated DO concentration was determined for the temperature and salinity given in each study, assuming barometric pressure was 101.3 kPa. For example, in an experiment conducted in full strength sea water (salinity 36) at 25°C, when the air-saturated DO concentration is 6.73 mg L-1, a 4-h exposure at 1.0 mg L-1 corresponds to a COD of 23 h mg L-1, whereas a 7-day exposure at 2.4 mg L-1 corresponds to a COD of 727 h mg L-1. The change in HIFA mRNA abundance at every sampling interval was coded as a binary variable. When the authors reported a statistically significant increase in mRNA abundance compared to normoxia, this sampling interval was scored as “1”, whereas, sampling intervals showing no significant difference, or a significant decrease, were scored as “0”. Finally, each study’s qPCR methodology was evaluated for the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) (Bustin et al., 2009). We determined how many of the 34 criteria deemed to be essential when reporting qPCR experiments were stated in the original citations (Supplementary Data Sheet S2).
We used generalized linear mixed-effects models (Silk et al., 2020) to evaluate the influence of gene, tissue, COD, temperature, salinity, and MIQE reporting on whether HIFA mRNA abundance was reported to increase during hypoxia. Study was included as a random factor to allow for multiple, potentially non-independent, samples from a given study. We limited our analyses of the tissue-specificity of the hypoxia response in HIFA mRNA abundance to those tissues that had been reported in three or more studies because mixed effects models are less reliable at low values of the random factor (Silk et al., 2020). The resulting outputs reflect the predicted frequency of reporting an increase in HIFA mRNA (from 0 to 1) for a given fixed factor. We used R packages lme4 (Bates et al., 2015) and emmeans (Lenth et al., 2018) and R version 3.6.1 (R Studio Team, 2022).
3 Results and discussion
3.1 Description of dataset
The final dataset consisted of 38 studies reporting on HIFA mRNA abundance in 25 species of ray-finned fishes exposed to laboratory hypoxia (Table 1). Across all studies, there were 413 discrete sampling intervals, representing different fish species, HIFA genes, tissues sampled, and severity of hypoxia (DO level and duration). HIFA mRNA abundance was significantly higher in tissues of hypoxic fish compared to normoxic controls in 182 cases (44%), unchanged in 214 cases (52%), and significantly lower in 17 cases (4%).
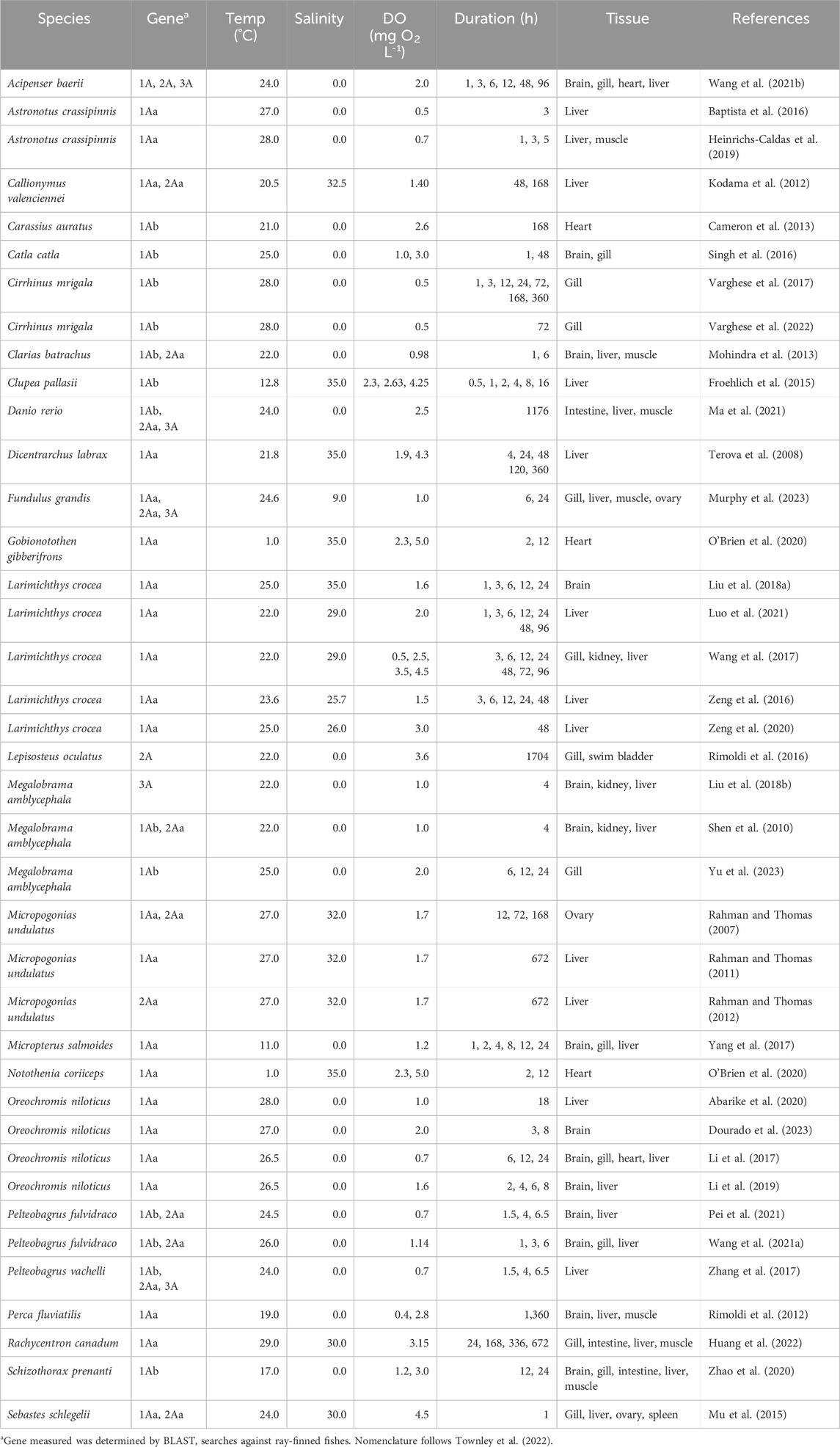
Table 1. Summary of studies measuring HIFA mRNA by qPCR of tissues from adult ray-finned fishes exposed to laboratory hypoxia.
Thirty-four studies described the effects of hypoxic exposure on HIF1A mRNA levels, 13 described changes in HIF2A mRNA, and five described changes in HIF3A (Supplementary Figure S1A). All but two studies were on species from lineages arising after the teleost-specific genome duplication (Hughes et al., 2018), meaning that certain species were predicted to have teleost-specific “a” and “b” duplicates (Rytkonen et al., 2013; Townley et al., 2022). The exceptions were Wang X. et al. (2021), who reported on HIF1A, HIF2A, and HIF3A in Siberian sturgeon (Acipenser baerii), and Rimoldi et al. (2016), who reported on HIF2A from spotted gar (Lepiosteus oculatus).
Apart from these reports on basal taxa, 21 studies examined changes in HIF1Aa in Neoteleostei (more derived ray-finned fishes) and 13 examined changes in HIF1Ab in Otocephala. For HIF2A, all studies reported data for HIF2Aa, even though HIF2Ab is found in most fish lineages (Rytkonen et al., 2013; Townley et al., 2022). The lack of data on HIF2Ab is a critical gap in knowledge, considering that recent work suggests that sequence variation in this duplicate is associated with variation in hypoxia tolerance among ray-finned fishes (Babin et al., 2024). For HIF3A, only a single teleost-specific duplicate has been retained in ray-finned fishes (Townley et al., 2022), and we did not distinguish between “a” and “b” duplicates. No studies were found on HIF4A.
3.2 Effects of gene and teleost-specific duplicate
Prior to assessing differences in HIF1A, HIF2A, and HIF3A, we compared the frequency of reported hypoxia-dependent increases in mRNA abundance for HIF1Aa and HIF1Ab (excluding data for ancestral HIF1A from Siberian sturgeon). No significant difference was found in the frequency of reported increases in the levels of HIF1Aa and HIF1Ab mRNA (p = 0.160) (Supplementary Table S1). Thus, data were combined for teleost-specific duplicates, along with ancestral forms, of each gene. Hereafter, HIF1A, HIF2A, and HIF3A refer to data from all forms of each gene.
The effect of HIFA gene on the frequency of reported hypoxia-induced increases in mRNA abundance approached statistical significance (Supplementary Table S2). The frequency of increased HIF1A mRNA tended to be higher than that for HIF2A (p = 0.0525), but lower than that for HIF3A (p = 0.0566) (Figure 1A). Law et al. (2006) used Northern blot analysis and found higher HIF3A mRNA levels in several tissues of the grass carp (Ctenopharyngodon idella) under hypoxia when HIF1A mRNA levels were largely unaffected in the same tissues. These observations, combined with the widespread expression of HIF3A in tissues of normoxic fish (Townley et al., 2022), suggest that more attention should be given to this understudied gene (Duan, 2016).
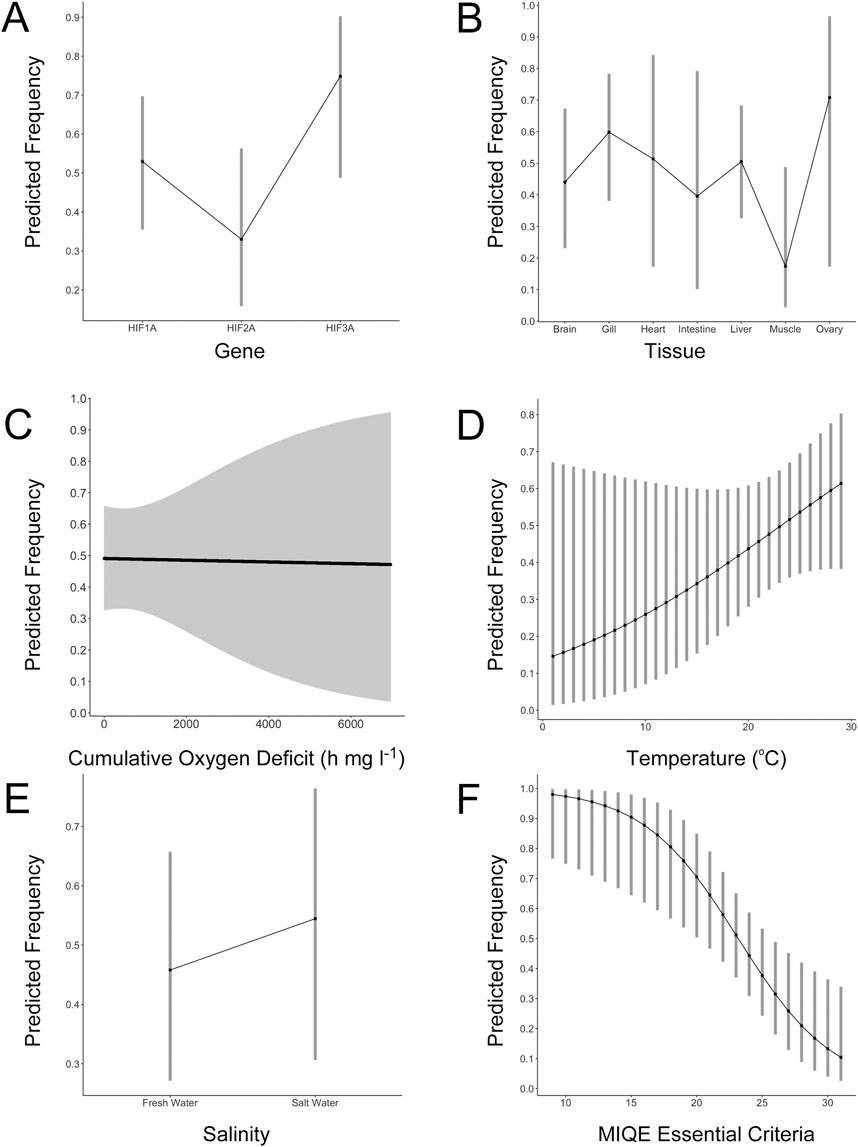
Figure 1. Results of generalized linear mixed effects modeling of factors influencing the abundance of HIFA mRNA during hypoxic exposure of adult ray-finned fishes. (A) The effects of gene (HIF1A, HIF2A, and HIF3A) on the predicted frequency of increased HIFA mRNA abundance. (B) The effects of tissue on the predicted frequency of increased HIF1A mRNA abundance. (C) The effects of cumulative oxygen deficit on the predicted frequency of increased HIF1A mRNA abundance. (D) The effects of experimental temperature on the predicted frequency of increased HIF1A mRNA abundance. (E) The effects of experimental salinity on the predicted frequency of increased HIF1A mRNA abundance. (F) The effects of methods reporting (number of MIQE essential criteria) on the predicted frequency of increased HIF1A mRNA abundance. For all panels, the y-axis represents the model-predicted frequency of increased HIFA mRNA abundance during hypoxic exposure and the error bars represent the 95% confidence intervals of the estimates. See Supplementary Tables S2–S5 for complete model results.
3.3 Effects of tissue
Measurements of mRNA abundance were not uniformly distributed among tissues (Supplementary Figure S1B). Across all HIFA genes, the most well-documented tissue was liver, followed by gill, brain, skeletal muscle, heart, kidney, ovary, intestine, spleen, testes, and swim bladder. For HIF1A, the frequency of hypoxia-induced increases in mRNA was highly variable and not significantly different across tissues (Figure 1B; Supplementary Table S3), although there tended to be fewer reports of increased HIF1A mRNA in skeletal muscle (p = 0.113). For HIF2A, there was a trend toward a higher frequency of reported effects of hypoxia in gill (p = 0.0782; Supplementary Table S4). Interestingly, HIF2A is more highly expressed in gill than in other tissues even under normoxia (Townley et al., 2022) and it may play a role in oxygen-sensing in fishes (Pan et al., 2022). For HIF3A, only one tissue (liver) was measured in at least three studies, precluding analysis of the effects of tissue on mRNA changes during hypoxia.
3.4 Effects of exposure conditions
We evaluated the effects of exposure conditions (COD, temperature, and salinity) on the frequency of reported increases in HIF1A mRNA, pooling measurements from all tissues (Supplementary Table S5). Surprisingly, COD, which reflects both the duration and the depth of hypoxia (Nelson and Lipkey, 2015), had no effect on HIF1A abundance (p = 0.918; Figure 1C). The only pattern was that the variability in responses increased as a function of COD. Because we pooled all tissues for this analysis, it is possible that tissue-specific responses were obscured (e.g., increasing mRNA levels over time versus decreasing over time in different tissues). Consequently, we repeated the analysis on the two most-studied tissues, liver and gill; COD was not statistically related to the frequency of increased abundance of liver HIF1A (p = 0.786) or gill HIF1A (p = 0.472). For liver HIF1A, some studies reported higher mRNA levels with increasing time of exposure, as one might expect (Froehlich et al., 2015; Heinrichs-Caldas et al., 2019). However, there were more studies where HIF1A was not different from controls at any time point (Li et al., 2017; Murphy et al., 2023; Wang X. et al., 2021; Yang et al., 2017) or it was higher at all time points (Huang et al., 2022; Luo et al., 2021). Thus, there was no consistent pattern across all studies due to the duration or severity of hypoxia.
We also evaluated temperature and salinity as potential factors related to the increase in HIFA mRNA abundance. We observed a non-significant trend toward a greater frequency of increased HIF1A mRNA at increasing temperature (p = 0.114; Figure 1D). Using Northern blot analyses, Rissanen et al. (2006) presented evidence of an interaction between temperature and hypoxia exposure on levels of HIF1A mRNA in Crucian carp, although this effect differed among tissues. When data from freshwater species were grouped and compared to data pooled from estuarine and marine species, there was no effect of salinity on the frequency of elevated HIF1A mRNA (p = 0.394; Figure 1E). Like the effects of temperature, though, direct tests of the effects of salinity on HIFA mRNA abundance in single study are scarce. COD, temperature, and salinity were unrelated to the frequency of reported increases in HIF2A or HIF3A mRNA (Supplementary Tables S6, S7; Supplementary Figure S2).
3.5 Effects of methods reporting
We determined whether the number of MIQE criteria deemed essential for ensuring data uniformity, comparability, and reliability (Bustin et al., 2009) reported in each study was related to the frequency of elevated mRNA abundance during hypoxia (pooling tissues and sampling intervals for a given HIFA transcript). For HIF1A, there was a strong, non-linear, negative relationship between the number of essential MIQE criteria and the frequency of reported increases in mRNA abundance (p = 0.0032; Figure 1F; Supplementary Table S8). Studies that reported more details were less likely to find increased HIF1A mRNA compared to those reporting fewer of these details. Similar results were generated for HIF2A (p = 0.127; Supplementary Figure S3A; Supplementary Table S9) and HIF3A (p = 0.0547; Supplementary Figure S3B; Supplementary Table S10), although they were not individually significant due to a much smaller number of observations.
Some of the MIQE criteria that were under-reported in the literature include assessment of RNA integrity prior to reverse transcription, linearity of calibration curves, and justification of reference genes. The last is especially important because many of the “housekeeping” genes used to standardize the levels of a gene of interest can be influenced by hypoxia (e.g., GAPDH). If the abundance of a reference gene is lower during hypoxia, then levels of HIFA mRNA will be overestimated. In addition, given the above-mentioned diversity of HIFA paralogs among fishes, the targets of pPCR must be unambiguously identified. Unfortunately, no consensus exists on the nomenclature of teleost-specific duplicates of HIFA, which has led to the different names for a given gene (e.g., HIF2Aa) or uninformative names (e.g., HIF1-like). We adhered to the nomenclature of Townley et al. (2022) and Babin et al. (2024), which are the most complete phylogenetic surveys of HIFA in ray-finned fishes at the present time.
3.6 Limitations of this study
One limitation of this study is that we cannot exclude the possibility that the response of HIFA mRNA to hypoxia is species-specific. We included study as a random factor in our analyses, which would largely explain the same variation as species because all but one study (O’Brien et al., 2020) measured a single species. Nevertheless, the lack of consistent responses across studies, species, tissues, and experimental conditions suggests that attempts to use HIFA mRNA as a biomarker of hypoxic exposure (e.g., Zhang et al., 2009; Thomas and Rahman, 2009; Froehlich et al., 2015) must be coupled with careful laboratory experiments that adhere to guidelines for conducting and reporting qPCR. Moreover, those results might be useful only for that species. A second limitation is that our analyses considered only author-reported significant differences, rather than the magnitude of the changes in HIFA mRNA abundance. Considering the great variability in the magnitude of those changes across studies, however, it is unlikely that including effect sizes in our analyses would have led to dramatically different conclusions. Finally, we calculated the cumulative oxygen deficit for each sampling interval to represent the degree of hypoxic exposure. Because fish species are known to differ in their hypoxia tolerance (Verberk et al., 2022), relating the DO level of the laboratory exposures to some measure of each species’ hypoxia tolerance, for example, the critical oxygen tension (Ultsch and Regan, 2019), could be a better metric of the stress experienced. Unfortunately, there is not a single measure of hypoxia tolerance that is universally accepted (Wood, 2018) or available for all the species represented in the current analysis.
4 Conclusion
Based upon our results, we conclude that increased HIFA mRNA during hypoxia is not a universal response of fish to low oxygen, but one that appears to be variable among studies and best explained by the rigor of reporting experimental methods rather than any other factor. Even allowing for species-specific increases in HIFA mRNA, several important questions remain. First, what is the mechanism of this increase? To address this question, studies should focus on the upstream regulators of HIFA transcription, including elucidating specific transcription factor binding sites in the promoter of HIFA genes (e.g., Rytkonen et al., 2013). For example, is HIFA transcription under the control of HIF, itself, or some other transcription factor? Second, does an increase in HIFA mRNA result in an increase in the corresponding HIFα protein? The abundance of mRNA does not always correlate with protein abundance due to post-transcriptional regulation, including changes in protein stability (Vogel and Marcotte, 2012). Early studies in fish cell culture (Soitamo et al., 2001) and recent studies on tissues of fish exposed to hypoxia (Murphy et al., 2023) showed that HIFα protein levels increase even when HIFA mRNA does not. Clearly, more studies need to couple measures of HIFA mRNA with measures of HIFα protein. Third, and most importantly, do changes in HIFA mRNA result in changes in gene expression and phenotypes that improve the fish’s capacity to tolerate aquatic hypoxia? With the advent of broad scale measures of mRNA abundance, it is possible to correlate levels of HIFA mRNA (or HIFα protein) and levels of target genes or pathways. Experiments such as these would enhance our understanding of HIF’s role in the responses of fish to aquatic hypoxia.
Author contributions
TM: Data curation, Formal Analysis, Investigation, Methodology, Visualization, Writing–original draft. BR: Conceptualization, Funding acquisition, Project administration, Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. BR is supported by the Greater New Orleans Foundation Endowed Chair in Aquatic Conservation.
Acknowledgments
We thank Drs. Courtney H. Babin and Anastasia Konefal for constructive feedback on this Mini Review.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2024.1496226/full#supplementary-material
SUPPLEMENTARY TABLE 1 | Tables S1-S10.
SUPPLEMENTARY IMAGE 1 | Figures S1-S3.
References
Abarike E. D., Jian J., Tang J., Cai J., Sakyi E. M., Kuebutornye F. K. A. (2020). A mixture of Chinese herbs and a commercial probiotic Bacillus species improves hemato-immunological, stress, and antioxidant parameters, and expression of HSP70 and HIF-1α mRNA to hypoxia, cold, and heat stress in Nile tilapia, Oreochromis niloticus. Aquac. Rep. 18, 100438. doi:10.1016/j.aqrep.2020.100438
Babin C. H., Leiva F. P., Verberk W. C. E. P., Rees B. B. (2024). Evolution of key oxygen-sensing genes is associated with hypoxia tolerance in fishes. Genome. Biol. Evol. 16 (9), evae183. doi:10.1093/gbe/evae183
Baptista R. B., Souza-Castro N., Almeida-Val V. M. F. (2016). Acute hypoxia up-regulates HIF-1α and VEGF mRNA levels in Amazon hypoxia-tolerant Oscar (Astronotus ocellatus). Fish Physiology Biochem. 42 (5), 1307–1318. doi:10.1007/s10695-016-0219-1
Bates D., Mächler M., Bolker B., Walker S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67 (1), 1–48. doi:10.18637/jss.v067.i01
Breitburg D., Levin L. A., Oschlies A., Grégoire M., Chavez F. P., Conley D. J., et al. (2018). Declining oxygen in the global ocean and coastal waters. Science 359 (6371), eaam7240. doi:10.1126/science.aam7240
Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 (4), 611–622. doi:10.1373/clinchem.2008.112797
Cameron J. S., DeWitt J. P., Ngo T. T., Yajnik T., Chan S., Chung E., et al. (2013). Cardiac KATP channel alterations associated with acclimation to hypoxia in goldfish (Carassius auratus L.). Comp. Biochem. Physiology Part A Mol. and Integr. Physiology 164 (4), 554–564. doi:10.1016/j.cbpa.2012.12.020
Chapman L. J., McKenzie D. (2009). Chapter 2 behavioral responses and ecological consequences. Fish. Physiol. 27, 25–77. doi:10.1016/S1546-5098(08)00002-2
Dourado P. L. R., Lima D., Mattos J. J., Bainy A. C. D., Grott S. C., Alves T. C., et al. (2023). Fipronil impairs the GABAergic brain responses of Nile Tilapia during the transition from normoxia to acute hypoxia. J. Exp. Zoology Part A Ecol. Integr. Physiology 339 (2), 138–152. doi:10.1002/jez.2662
Duan C. (2016). Hypoxia-inducible factor 3 biology: complexities and emerging themes. Am. J. Physiology-Cell Physiology 310 (4), C260–C269. doi:10.1152/ajpcell.00315.2015
Froehlich H. E., Roberts S. B., Essington T. E. (2015). Evaluating hypoxia-inducible factor-1α mRNA expression in a pelagic fish, Pacific herring Clupea pallasii, as a biomarker for hypoxia exposure. Comp. Biochem. Physiology Part A Mol. and Integr. Physiology 189, 58–66. doi:10.1016/j.cbpa.2015.07.016
Heinrichs-Caldas W., Campos D. F., Paula-Silva M. N., Almeida-Val V. M. F. (2019). Oxygen-dependent distinct expression of hif-1α gene in aerobic and anaerobic tissues of the Amazon Oscar, Astronotus crassipinnis. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 227, 31–38. doi:10.1016/j.cbpb.2018.08.011
Huang J., Amenyogbe E., Yang L., Wang Z., Chen G., Wang W., et al. (2022). Cloning and expression analysis of hif-1α and downstream genes during hypoxic stress in cobia (Rachycentron canadum). Aquac. Int. 30 (2), 803–824. doi:10.1007/s10499-021-00820-4
Hughes L. C., Ortí G., Huang Y., Sun Y., Baldwin C. C., Thompson A. W., et al. (2018). Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc. Natl. Acad. Sci. 115 (24), 6249–6254. doi:10.1073/pnas.1719358115
Kodama K., Saydur Rahman Md., Horiguchi T., Thomas P. (2012). Upregulation of hypoxia-inducible factor (HIF)-1α and HIF-2α mRNA levels in dragonet Callionymus valenciennei exposed to environmental hypoxia in Tokyo Bay. Mar. Pollut. Bull. 64 (7), 1339–1347. doi:10.1016/j.marpolbul.2012.05.002
Law S. H., Wu R. S., Ng P. K., Yu R. M., Kong R. Y. (2006). Cloning and expression analysis of two distinct HIF-alpha isoforms – gcHIF-1alpha and gcHIF-4alpha – from the hypoxia-tolerant grass carp, Ctenopharyngodon idellus. BMC Mol. Biol. 7 (1), 15. doi:10.1186/1471-2199-7-15
Lenth R., Singmann H., Love J., Buerkner P., Herve M. (2018). Package “emmeans”. R. Package Version 4.0-3. Available at: http://cran.r-project.org/package=emmeans.
Li H. L., Gu X. H., Li B. J., Chen X., Lin H. R., Xia J. H. (2017). Characterization and functional analysis of hypoxia-inducible factor HIF1α and its inhibitor HIF1αn in tilapia. PLOS ONE 12 (3), e0173478. doi:10.1371/journal.pone.0173478
Li X., Wang T., Yin S., Zhang G., Cao Q., Wen X., et al. (2019). The improved energy metabolism and blood oxygen-carrying capacity for pufferfish, Takifugu fasciatus, against acute hypoxia under the regulation of oxygen sensors. Fish Physiology Biochem. 45 (1), 323–340. doi:10.1007/s10695-018-0565-2
Liu W., Liu X., Wu C., Jiang L. (2018a). Transcriptome analysis demonstrates that long noncoding RNA is involved in the hypoxic response in Larimichthys crocea. Fish Physiology Biochem. 44 (5), 1333–1347. doi:10.1007/s10695-018-0525-x
Liu Z., Zhao X., Jiang X., Zou S. (2018b). Transcription of blunt snout bream (Megalobrama amblycephala) HIF3α and its localization in the nucleus under both normoxic and hypoxic conditions. Biochem. Biophysical Res. Commun. 500 (2), 443–449. doi:10.1016/j.bbrc.2018.04.099
Luo S.-Y., Wang J.-Q., Liu C., Gao X.-M., Zhang Y.-B., Ding J., et al. (2021). Hif-1α/Hsf1/Hsp70 signaling pathway regulates redox homeostasis and apoptosis in large yellow croaker (Larimichthys crocea) under environmental hypoxia. Ningbo, Zhejiang 315012, China 42 (6), 746–760. doi:10.24272/j.issn.2095-8137.2021.224
Ma Q., Wang X., Li L.-Y., Qiao F., Zhang M.-L., Du Z.-Y. (2021). High protein intake promotes the adaptation to chronic hypoxia in zebrafish (Danio rerio). Aquaculture 535, 736356. doi:10.1016/j.aquaculture.2021.736356
Mandic M., Joyce W., Perry S. F. (2021). The evolutionary and physiological significance of the Hif pathway in teleost fishes. J. Exp. Biol. 224 (18), jeb231936. doi:10.1242/jeb.231936
McIntosh B. E., Hogenesch J. B., Bradfield C. A. (2010). Mammalian per-arnt-sim proteins in environmental adaptation. Annu. Rev. Physiology 72 (1), 625–645. doi:10.1146/annurev-physiol-021909-135922
Mohindra V., Tripathi R. K., Singh R. K., Lal K. K. (2013). Molecular characterization and expression analysis of three hypoxia-inducible factor alpha subunits, HIF-1α, -2α and -3α in hypoxia-tolerant Indian catfish, Clarias batrachus [Linnaeus, 1758]. Mol. Biol. Rep. 40 (10), 5805–5815. doi:10.1007/s11033-013-2685-1
Mu W., Wen H., Li J., He F. (2015). HIFs genes expression and hematology indices responses to different oxygen treatments in an ovoviviparous teleost species Sebastes schlegelii. Mar. Environ. Res. 110, 142–151. doi:10.1016/j.marenvres.2015.04.008
Murphy T. E., Harris J. C., Rees B. B. (2023). Hypoxia-inducible factor 1 alpha protein increases without changes in mRNA during acute hypoxic exposure of the Gulf killifish, Fundulus grandis. Biol. Open 12 (12), bio060167. doi:10.1242/bio.060167
Nelson J. A., Lipkey G. K. (2015). Hypoxia tolerance variance between swimming and resting striped bass Morone saxatilis. J. Fish Biol. 87 (2), 510–518. doi:10.1111/jfb.12735
Nikinmaa M., Rees B. B. (2005). Oxygen-dependent gene expression in fishes. Am. J. Physiology-Regulatory, Integr. Comp. Physiology 288 (5), R1079–R1090. doi:10.1152/ajpregu.00626.2004
O’Brien K. M., Rix A. S., Grove T. J., Sarrimanolis J., Brooking A., Roberts M., et al. (2020). Characterization of the hypoxia-inducible factor-1 pathway in hearts of Antarctic notothenioid fishes. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 250, 110505. doi:10.1016/j.cbpb.2020.110505
Pamenter M. E., Hall J. E., Tanabe Y., Simonson T. S. (2020). Cross-species insights into genomic adaptations to hypoxia. Front. Genet. 11, 743. doi:10.3389/fgene.2020.00743
Pan W., Godoy R. S., Cook D. P., Scott A. L., Nurse C. A., Jonz M. G. (2022). Single-cell transcriptomic analysis of neuroepithelial cells and other cell types of the gills of zebrafish (Danio rerio) exposed to hypoxia. Sci. Rep. 12 (1), 10144. doi:10.1038/s41598-022-13693-1
Pei X., Chu M., Tang P., Zhang H., Zhang X., Zheng X., et al. (2021). Effects of acute hypoxia and reoxygenation on oxygen sensors, respiratory metabolism, oxidative stress, and apoptosis in hybrid yellow catfish “Huangyou-1.”. Fish Physiology Biochem. 47 (5), 1429–1448. doi:10.1007/s10695-021-00989-8
Rahman Md. S., Thomas P. (2007). Molecular cloning, characterization and expression of two hypoxia-inducible factor alpha subunits, HIF-1α and HIF-2α, in a hypoxia-tolerant marine teleost, Atlantic croaker (Micropogonias undulatus). Gene 396 (2), 273–282. doi:10.1016/j.gene.2007.03.009
Rahman Md. S., Thomas P. (2011). Characterization of three IGFBP mRNAs in Atlantic croaker and their regulation during hypoxic stress: potential mechanisms of their upregulation by hypoxia. Am. J. Physiology-Endocrinology Metabolism 301 (4), E637–E648. doi:10.1152/ajpendo.00168.2011
Rahman Md. S., Thomas P. (2012). Effects of hypoxia exposure on hepatic cytochrome P450 1A (CYP1A) expression in Atlantic croaker: molecular mechanisms of CYP1A down-regulation. PLoS ONE 7 (7), e40825. doi:10.1371/journal.pone.0040825
Richards J. G. (2009). Chapter 10 metabolic and molecular responses of fish to hypoxia. Fish. Physiol. 27, 443–485. doi:10.1016/S1546-5098(08)00010-1
Rimoldi S., Terova G., Ceccuzzi P., Marelli S., Antonini M., Saroglia M. (2012). HIF-1α mRNA levels in Eurasian perch (Perca fluviatilis) exposed to acute and chronic hypoxia. Mol. Biol. Rep. 39 (4), 4009–4015. doi:10.1007/s11033-011-1181-8
Rimoldi S., Terova G., Zaccone G., Parker T., Kuciel M., Dabrowski K. (2016). The effect of hypoxia and hyperoxia on growth and expression of hypoxia-related genes and proteins in spotted gar Lepisosteus oculatus larvae and juveniles. J. Exp. Zoology Part B Mol. Dev. Evol. 326 (4), 250–267. doi:10.1002/jez.b.22680
Rissanen E., Tranberg H. K., Sollid J., Nilsson G. E., Nikinmaa M. (2006). Temperature regulates hypoxia-inducible factor-1 (HIF-1) in a poikilothermic vertebrate, crucian carp (Carassius carassius). J. Exp. Biol. 209 (6), 994–1003. doi:10.1242/jeb.02103
Rytkonen K. T., Akbarzadeh A., Miandare H. K., Kamie H., Duan C., Williams T. A., et al. (2013). Subfunctionalization of Cyprinid hypoxia-inducible factors for roles in development and oxygen sensing. Evolution 67 (3), 873–882. doi:10.1111/j.1558-5646.2012.01820.x
Rytkonen K. T., Williams T. A., Renshaw G. M., Primmer C. R., Nikinmaa M. (2011). Molecular evolution of the metazoan PHD-HIF oxygen sensing system. Mol. Biol. Evol. 28 (6), 1913–1926. doi:10.1093/molbev/msr012
Sayers E. W., Bolton E. E., Brister J. R., Canese K., Chan J., Comeau D. C., et al. (2022). Database resources of the national center for biotechnology information. Nucleic Acids Res. 50 (D1), D20–D26. doi:10.1093/nar/gkab1112
Semenza G. L. (2012). Hypoxia-inducible factors in physiology and medicine. Cell 148 (3), 399–408. doi:10.1016/j.cell.2012.01.021
Shen R.-J., Jiang X.-Y., Pu J.-W., Zou S.-M. (2010). HIF-1α and -2α genes in a hypoxia-sensitive teleost species Megalobrama amblycephala: cDNA cloning, expression and different responses to hypoxia. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 157 (3), 273–280. doi:10.1016/j.cbpb.2010.06.013
Shen Y., You W., Luo X., Lu Y., Huang M., Ke C. (2023). An overview of the mechanisms underlying hypoxia tolerance differences in aquatic animals and their inspirations for aquaculture. Rev. Fish Biol. Fish. 33, 1223–1236. doi:10.1007/s11160-023-09793-4
Silk M. J., Harrison X. A., Hodgson D. J. (2020). Perils and pitfalls of mixed-effects regression models in biology. PeerJ 8, e9522. doi:10.7717/peerj.9522
Singh S. P., Sharma J., Ahmad T., Chakrabarti R. (2016). Oxygen stress: impact on innate immune system, antioxidant defense system and expression of HIF-1α and ATPase 6 genes in Catla catla. Fish Physiology Biochem. 42 (2), 673–688. doi:10.1007/s10695-015-0168-0
Soitamo A. J., Råbergh C. M. I., Gassmann M., Sistonen L., Nikinmaa M. (2001). Characterization of a hypoxia-inducible factor (HIF-1alpha) from rainbow trout. Accumulation of protein occurs at normal venous oxygen tension. J. Biol. Chem. 276 (23), 19699–19705. doi:10.1074/jbc.M009057200
Terova G., Rimoldi S., Corà S., Bernardini G., Gornati R., Saroglia M. (2008). Acute and chronic hypoxia affects HIF-1α mRNA levels in sea bass (Dicentrarchus labrax). Aquaculture 279 (1–4), 150–159. doi:10.1016/j.aquaculture.2008.03.041
Thomas P., Rahman M. D. (2009). Biomarkers of hypoxia exposure and reproductive function in Atlantic croaker: a review with some preliminary findings from the northern Gulf of Mexico hypoxic zone. J. Exp. Mar. Biol. Ecol. 381, S38–S50. doi:10.1016/j.jembe.2009.07.008
Townley I. K., Babin C. H., Murphy T. E., Summa C. M., Rees B. B. (2022). Genomic analysis of hypoxia inducible factor alpha in ray-finned fishes reveals missing Ohnologs and evidence of widespread positive selection. Sci. Rep. 12 (1), 22312. doi:10.1038/s41598-022-26876-7
Ultsch G. R., Regan M. D. (2019). The utility and determination of Pcrit in fishes. J. Exp. Biol. 222, jeb203646. doi:10.1242/jeb.203646
Varghese T., Dasgupta S., Anand G., Rejish Kumar V. J., Sahu N. P., Pal A. K., et al. (2022). Dietary arginine attenuates hypoxia-induced HIF expression, metabolic responses and oxidative stress in Indian Major Carp, Cirrhinus mrigala. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 259, 110714. doi:10.1016/j.cbpb.2022.110714
Varghese T., Pal A., Puthiyottil M., Sahu N., Dasgupta S. (2017). Physiological and molecular responses of a bottom dwelling carp, Cirrhinus mrigala to short term environmental hypoxia. Turkish J. Fish. Aquatic Sci. 18. doi:10.4194/1303-2712-v18_3_14
Verberk W. C. E. P., Sandker J. F., van de Pol I. L. E., Urbina M. A., Wilson R. W., McKenzie D. J., et al. (2022). Body mass and cell size shape the tolerance of fishes to low oxygen in a temperature-dependent manner. Glob. Change Biol. 28, 5695–5707. doi:10.1111/gcb.16319
Vogel C., Marcotte E. (2012). Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232. doi:10.1038/nrg3185
Wang M., Wu F., Xie S., Zhang L. (2021a). Acute hypoxia and reoxygenation: effect on oxidative stress and hypoxia signal transduction in the juvenile yellow catfish (Pelteobagrus fulvidraco). Aquaculture 531, 735903. doi:10.1016/j.aquaculture.2020.735903
Wang Q.-F., Shen W.-L., Hou C.-C., Liu C., Wu X.-F., Zhu J.-Q. (2017). Physiological responses and changes in gene expression in the large yellow croaker Larimichthys crocea following exposure to hypoxia. Chemosphere 169, 418–427. doi:10.1016/j.chemosphere.2016.11.099
Wang X., Xiao S., Zhang R., Liu L., Zhu H. (2021b). Physiological changes and transcriptional modulation of HIF-αs in Siberian sturgeon in response to hypoxia. Aquaculture 545, 737219. doi:10.1016/j.aquaculture.2021.737219
Wood C. M. (2018). The fallacy of the Pcrit—are there more useful alternatives? J. Exp. Biol. 221, jeb163717. doi:10.1242/jeb.163717
Yang S., Yan T., Wu H., Xiao Q., Fu H. M., Luo J., et al. (2017). Acute hypoxic stress: effect on blood parameters, antioxidant enzymes, and expression of HIF-1alpha and GLUT-1 genes in largemouth bass (Micropterus salmoides). Fish and Shellfish Immunol. 67, 449–458. doi:10.1016/j.fsi.2017.06.035
Yu X., Zhang Y., Li S., Zheng G., Zou S. (2023). Effects of hypoxia on the gill morphological structure, apoptosis and hypoxia-related gene expression in blunt snout bream (Megalobrama amblycephala). Fish Physiology Biochem. 49 (5), 939–949. doi:10.1007/s10695-023-01233-1
Zeng L., Ai C.-X., Zhang J.-S., Li W.-C. (2020). Pre-hypoxia exposure inhibited copper toxicity by improving energy metabolism, antioxidant defense and mitophagy in the liver of the large yellow croaker Larimichthys crocea. Sci. Total Environ. 708, 134961. doi:10.1016/j.scitotenv.2019.134961
Zeng L., Wang Y.-H., Ai C.-X., Zheng J.-L., Wu C.-W., Cai R. (2016). Effects of β-glucan on ROS production and energy metabolism in yellow croaker (Pseudosciaena crocea) under acute hypoxic stress. Fish Physiology Biochem. 42 (5), 1395–1405. doi:10.1007/s10695-016-0227-1
Zhang G., Zhao C., Wang Q., Gu Y., Li Z., Tao P., et al. (2017). Identification of HIF-1 signaling pathway in Pelteobagrus vachelli using RNA-Seq: effects of acute hypoxia and reoxygenation on oxygen sensors, respiratory metabolism, and hematology indices. J. Comp. Physiology B 187 (7), 931–943. doi:10.1007/s00360-017-1083-8
Zhang Z., Ju Z., Wells M. C., andWalter R. B. (2009). Genomic approaches in the identification of hypoxia biomarkers in model fish species. J. Exp. Mar. Biol. Ecol. 381, S180-S187–87. doi:10.1016/j.jembe.2009.07.021
Keywords: oxygen, gene expression, transcription factor, hypoxia inducible factor, quantitative PCR
Citation: Murphy TE and Rees BB (2024) Diverse responses of hypoxia-inducible factor alpha mRNA abundance in fish exposed to low oxygen: the importance of reporting methods. Front. Physiol. 15:1496226. doi: 10.3389/fphys.2024.1496226
Received: 13 September 2024; Accepted: 20 September 2024;
Published: 04 October 2024.
Edited by:
Guosong Zhang, Heze University, ChinaReviewed by:
D. K. Meena, Central Inland Fisheries Research Institute (ICAR), IndiaCopyright © 2024 Murphy and Rees. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bernard B. Rees, YnJlZXNAdW5vLmVkdQ==
 Taylor E. Murphy
Taylor E. Murphy Bernard B. Rees
Bernard B. Rees