- 1School of Pharmacy, Shandong University of Traditional Chinese Medicine, Jinan, China
- 2Institute of Industrial Crops, Shandong Academy of Agricultural Sciences, Jinan, China
- 3Chongqing Jiulongpo District Agro-Tech Extension and Service Station, Chongqing, China
Introduction: Semiaphis heraclei is an important economic pest affecting Caprifoliaceae and Apiaceae plants, and chemical control is still the main effective control method in the field. Afidopyropen is a new type of pyridine cyclopropyl insecticide, which can effectively control piercing-sucking mouthparts pests and is suitable for pest resistance management. However, the detoxification mechanism of S. heraclei to afidopyropen is still poorly cleared.
Methods: The insecticidal activity of afidopyropen against S. heraclei and the enzyme activity assay and synergism bioassay were evaluated. The detoxification enzyme genes were obtained by transcriptome and validated by quantitative real-time PCR (RT-qPCR). Furthermore, RNA interference was used to study the functions of detoxification enzyme genes.
Results: The activities of cytochrome P450 monooxygenases (P450s) and carboxylesterases (CarEs) were significantly increased under afidopyropen treatment. The toxicity of afidopyropen against S. heraclei was significantly increased after application the inhibitors of piperonyl butoxide and triphenyl phosphate. Sixteen P450 genes and three CarE genes were identified in the transcriptome of S. heraclei. The RT-qPCR results showed that eleven P450 genes and two CarE genes were significantly upregulated under afidopyropen treatment, and the expression of CYP6DA2 and CarFE4 was upregulated by more than 2.5 times. The expression pattern of CYP6DA2 and CarFE4 was further analyzed in different developmental stages of S. heraclei and knockdown of CYP6DA2 and CarFE4 significantly increased the susceptibility of S. heraclei to afidopyropen.
Conclusion: The results of this study uncover the key functions of CYP6DA2 and CarFE4 in the detoxification mechanism of S. heraclei to afidopyropen, and provide a theoretical basis for the scientific use of afidopyropen in the field.
1 Introduction
The Semiaphis heraclei, also known as celery aphid, is a Hemiptera insect widely distributed in the Far East and the Indian Subcontinent. The main hosts are Caprifoliaceae plants from Japan, China and East Siberia. The secondary hosts are Apiaceae plants from East and Southeast Asia, India and Pakistan, Algiers and Hawaii (Song et al., 2023). The S. heraclei will congregate on fresh stems, leaves, flower buds, and other parts of the host plant, where they feed on the plant by sucking the sap through their mouthparts. A significant infestation of S. heraclei can result in the curling of leaves, which impairs photosynthesis and ultimately leads to the yellowing or even wilting of the leaves. Additionally, they can affect the bud stage, which further impacts the growth of the plant and the content of active ingredients. In addition, the honeydew of S. heraclei can induce soot disease (Massimino Cocuzza, 2022). Moreover, the outbreak period of S. heraclei and harvest period of host plants such as honeysuckle are basically the same, which causes serious yield loss or even no yield to the honeysuckle, celery and other herbal vegetables (Song et al., 2023).
In order to reduce the impact of agricultural pests on field production, a series of field control methods are usually used, including physical and biological control. However, chemical insecticide control is still the most dependent method because of its quick effect and simple operation (Gregg et al., 2018; Mubeen et al., 2023). In recent years, as the use of pesticides increases, problems such as pesticide residues and environmental pollution become more serious. This poses a major risk to human health and to the safety of the food supply and has become a public health issue of great concern (He et al., 2023). In particular, fields require multiple applications of chemical insecticides to ensure crop and herb yields. This can easily lead to problems of excessive pesticide residues. In order to guarantee the safety of food and medication, the use of highly effective insecticides that are less harmful to natural enemies and the environment is key to the scientific control and management of pests in the field.
Afidopyropen is a derivative of pyripyropene A produced by filamentous fungus Penicillium coprobium. It is currently used worldwide as an alternative to traditional insecticides. It is particularly effective against aphids such as Myzus persicae, Aphis gossypii and Aphis craccivora (Goto et al., 2019). Afidopyropen quickly interferes with the feeding direction of insects by targeting transient receptor potential vanilloid (TRPV) cation channels. This weakens the ability of insects to coordinate movement, sense sound, maintain position and feed on plants, and ultimately lead to starvation and death (Jeschke, 2021; Yin et al., 2024). Although its insecticidal spectrum is narrow, it has high selectivity and insecticidal efficacy against target pests, low toxicity to natural enemies such as bees, seven-star lady beetles and mammals, and short persistence in crops and environment. In addition, afidopyropen can penetrate from treated to untreated leaves. This significantly improves insecticidal efficacy and reduces the amount of insecticide used (Horikoshi et al., 2022; Liu X. et al., 2024). These benefits are indicated that afidopyropen is a valuable sustainable agricultural green insecticide and an optimal agent for aphid control.
Exogenous detoxification and metabolism are the main physiological processes after insecticide exposure (Lu et al., 2021). The detoxification of insecticides in insects can be divided into three phases. Cytochrome P450 monooxygenases (P450s) and carboxylesterases (CarEs) are directly involved in the metabolism of exogenous substances as enzymes of phase I reaction (Li P. R. et al., 2023). Glutathione S-transferases (GSTs) are involved in the phase II reaction, via catalytic reaction to increase the water solubility of exogenous substances. In phase III reaction, water-soluble compounds are transferred and excreted out of the cell (Yang B. et al., 2020; Aioub and Ashour, 2023).
In this study, we evaluated the insecticidal activity of afidopyropen against the S. heraclei. Enzyme activity assay and synergism bioassay showed that P450s and CarEs participated in the detoxification of afidopyropen. The differentially expressed genes of P450s and CarEs were determined via transcriptome sequencing and their relative transcript levels were validated by quantitative real-time PCR (RT-qPCR). Furthermore, the functions of CYP6DA2 and CarFE4 in the susceptibility of S. heraclei to afidopyropen were determined by RNA interference (RNAi). The results of this study will lay a foundation for further research on S. heraclei and provide a theoretical basis for the field control of aphid and the rational use of afidopyropen.
2 Materials and methods
2.1 Insects and chemical reagents
The S. heraclei population was collected from Pingyi (35.60°N, 117.75°E; Linyi, China) in 2024. The S. heraclei was reared with celery in the laboratory at 25°C ± 1°C with 70%–80% relative humidity and 16 h light: 8 h dark photoperiod.
Afidopyropen (94.00%) was purchased from Shanghai Yuanye Biotechnology Co., Ltd (Shanghai, China). Acetone, dimethyl sulfoxide (DMSO), piperonyl butoxide (PBO), triphenyl phosphate (TPP) and diethyl maleate (DEM) were purchased from Merck KGaA (Darmstadt, Germany).
2.2 Bioassay
The bioassay was carried out by the topical application method via MICRO2T SMARTouchTM device (SMARTouchTM Controller for UMP3/Nanoliter 2010; World Precision Instruments, Inc.). Afidopyropen was dissolved in DMSO and then diluted with acetone to generate five concentrations. The third-instar nymphs of S. heraclei were anesthetized by ice and a droplet solution of 50 nL was applied topically to the dorsal plate. The nymphs treated with acetone were as the control. The treated insects were fed with celery at 25°C ± 1°C with 70%–80% relative humidity and 16 h light: 8 h dark photoperiod. Each treatment was repeated three times and contained thirty insects. Mortality was checked after 48 h.
2.3 Sample collection
The third-instar nymphs of S. heraclei treated with afidopyropen at the dose of median lethal dose (LD50), and the survival nymphs were collected at 48 h. The nymphs treated with acetone were as the control. The insects were immediately frozen in liquid nitrogen and then stored at −80°C for enzyme activity assay, transcriptome sequencing and quantitative validation. Each treatment was repeated three times and each contained thirty insects.
Besides, the first-to fourth-instar nymphs and the apterous adults of S. heraclei were collected for instar expression profile analysis. The insects were immediately frozen in liquid nitrogen and then preserved at −80°C. Each sample was repeated three times and contained thirty insects.
2.4 Enzyme activity assay
The protein concentration was determined by BCA protein quantitative/concentration determination kit provided by Beijing Labgic Technology Co. Ltd. (Beijing, China). The activities of P450, CarE and GST were determined by MFO activity assay kit, CarE activity assay kit and GST activity assay kit produced by Suzhou Comin Biotechnology Co., Ltd. (Suzhou, China).
Determination of P450 enzyme activity: according to the manufacturer’s instructions, approximately 0.1 g of the sample was taken and 1 mL of the extraction reagent was added and homogenized on ice, resulting in the production of a crude enzyme extract. The crude enzyme solution was centrifuged at 12,000 g for 20 min at 4°C. The supernatant was collected and the absorbance value of P450 was determined at 400 nm using a microplate reader with p-nitrophenol as the substrate. The production of 1 nmol of p-nitrophenol per milligram of histone per minute was defined as one unit of enzyme activity (Li Z. et al., 2023).
Determination of CarE enzyme activity: the crude enzyme solution was centrifuged at 12,000 g for 30 min at 4°C. The supernatant was collected and the absorbance value of CarE was determined at 450 nm using a microplate reader with α-naphthyl acetate as the substrate. Each milligram of histone is defined as 1 enzyme activity unit per minute increase in the catalytic absorbance value in the 37°C reaction system (Patra and Hath, 2023).
Determination of GST enzyme activity: the crude enzyme solution was centrifuged at 8,000 g for 10 min at 4°C. The supernatant was collected and the absorbance value of GST was determined at 340 nm using a microplate reader with 1-chloro2,4-dinitrobenzene (CDNB) as the substrate. At 37°C, each milligram of protein catalyses the binding of 1 nmol/L of CDNB to GSH as 1 unit of enzyme activity per minut (Cui et al., 2018; Hu et al., 2022).
BCA protein concentration assay: according to the manufacturer’s instructions, the BCA working solution was prepared. 20 μL of BCA protein standard solution (5 mg/mL) was diluted to 100 μL with PBS solution (0.01 mol L−1, pH 7.2) to give a final concentration of 1 mg/mL. The BCA standard assay solution was prepared in accordance with the instructions provided. The requisite volume of the sample to be measured was then added to the microtiter plate, and the solution was brought to a volume of 20 μL with PBS. A volume of 200 µL of the BCA working solution was added to the microtiter plate, which was then mixed thoroughly and left at 37°C for 30 min. The absorbance value at 562 nm was determined and documented, with the light absorption value of the BCA-free sample serving as the control. The protein concentration in the samples was calculated by constructing a standard curve with A562 as the vertical coordinate and BSA content as the horizontal coordinate.
2.5 Synergism bioassay
One hour before the application of afidopyropen, 2 μg of PBO, TPP and DEM in 50 nL acetone was applied topically to the dorsal plate of third-instar nymphs of S. heraclei. The nymphs treated with acetone were as the control. Then, the nymphs were anesthetized by ice and treated with afidopyropen at the dose of LD50 (Ding et al., 2013; Zhang et al., 2016; Yang B. et al., 2020). The treated insects were fed with celery at 25°C ± 1°C with 70%–80% relative humidity and 16 h light: 8 h dark photoperiod. Each treatment was repeated three times and contained thirty insects. Mortality was checked after 48 h.
2.6 RNA isolation, library construction and sequencing
Total RNA of the sample was extracted using TRIzol reagent (Invitrogen, United States) according to the manufacturer’s procedure. RNA purity, concentration and integrity were measured using a 5,300 Bioanalyzer (Agilent, United States) and an ND-2000 (NanoDrop Technolog, United States). Six S. heraclei RNA samples from the control group and the afidopyropen treatment group were taken for cDNA library construction and sequenced in BGI (Shenzhen, China).
2.7 Transcriptome assembly and function annotation
Before proceeding with the Denovo assembly, removed the reads containing joints (joint contamination) from the original reads and removed the reads with unknown base N content greater than 1%. Low-quality reads (those with a mass value of less than 15 bases accounting for more than 40% of the total base number of the reads were defined as low-quality reads) were removed to obtain clean reads. High-quality clean reads were de novo assembled using Trinity, followed by clustering of transcripts to de-redundancy using CD-HIT to obtain unigene. The assembled unigene was combined with Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), non-redundancy (Nr) and Nucleotide Sequence Database (Nt), EuKaryotic Orthologous Groups (KOG), Protein family (Pfam), Clusters of Orthologous Groups (COG) and Swiss-Prot database were blasted to obtain annotation information and functional classification of unigene with e-value <10−5 (Altschul et al., 1990; Nawrocki et al., 2009; Buchfink et al., 2015). We compared clean reads to the genome sequence using Bowtie2, and then calculated the gene expression levels of each sample using RSEM (a software package for calculating gene expression levels of RNA-seq reads and transcription subtypes). The Pearson correlation coefficient between each two samples was calculated using cor function in R software. Principal component (PCA) analysis was performed using princomp function in R software. DESeq software was used to detect deg between the control group and the experimental group (Love et al., 2014).
2.8 GO and KEGG analysis
GO can be divided into three functional categories: molecular function, cellular component and biological process. Classify according to the difference of gene detection function, at the same time, use R-phyper function to enrich and analyze in the software, calculate p-value, and then take the function of FDR correction p-value usually ≤0.05 as significant enrichment. According to KEGG annotation results and official classification, we classified the differentially expressed genes into biological pathways. At the same time, we used the phyper function in R software for enrichment analysis and calculated the p-value. The p-value was then FDR corrected, and in general, functions with q value ≤0.05 were considered significantly enriched.
2.9 Identification of P450 and CarE genes and RT-qPCR
The P450 and CarE genes were searched according to annotation information in the transcriptome of S. heraclei. The repetitive sequences were removed, and the amino acid sequences of the remaining P450 and CarE genes were BLAST compared with the Nr database of NCBI (National Center for Biotechnology Information) to remove misannotated amino acid sequences.
Reverse transcription from 1 μg total RNA to cDNA was performed using the StarScript III All-in-one RT kit with gDNA Remover (Genstar, China). The RT-qPCR was detected via LineGene 9,600 Plus real-time PCR detection system (BIOER, China) with 2×RealStar Fast SYBR qPCR Mix (Low ROX) (Genstar, China). β-Tubulin was used as the endogenous control, and the 2−ΔΔCT method was used to calculate the relative expression (Livak and Schmittgen, 2001; Ma et al., 2016). The RT-qPCR reaction consisted of three independent technical and biological replicates. Primers were listed in Supplementary Table S1.
2.10 RNA interference
The T7 high yield transcription kit (Invitrogen, United States) was used to synthesize the double-stranded RNA (dsRNA) of CYP6DA2, CarFE4 and enhanced green fluorescent protein (EGFP). The primers were listed in Supplementary Table S1. The dsRNA of CYP6DA2, CarFE4 and EGFP was added to artificial diet (sterile sucrose solution, 0.5 M) at a concentration of 150 ng/μL. The third-instar nymphs of S. heraclei were transferred onto the artificial diet containing dsRNA of CYP6DA2 and CarFE4 for rearing 2 days. The surviving insects were collected for detection of interference efficiency. The third-instar nymphs of S. heraclei feeding on artificial diet containing dsRNA of EGFP were as a control. After dsRNA feeding 1 day, the third-instar nymphs of S. heraclei were treated with afidopyropen at the dose of LD50 (Liu Y. X. et al., 2024). The mortality was checked in 48 h. Each treatment was repeated three times and contained thirty insects.
2.11 Statistical analysis
The statistical analysis was performed via SPSS 20.0 software (IBM Corporation). The Student’s t-test and Tukey’s multiple comparison test were used to compare the differences between treatments. Data were shown as mean ± standard error (SE). The p < 0.05 and p < 0.01 were considered statistically significant and very significant differences, respectively.
3 Results
3.1 Toxicity of afidopyropen to S. heraclei
After 48 h treatment, the LD50 value of afidopyropen was 0.050 ng/pest (Table 1). In further experiments, the LD50 dose of afidopyropen was used to evaluate its effect on S. heraclei.
3.2 Effect of afidopyropen on the detoxification enzymes of S. heraclei
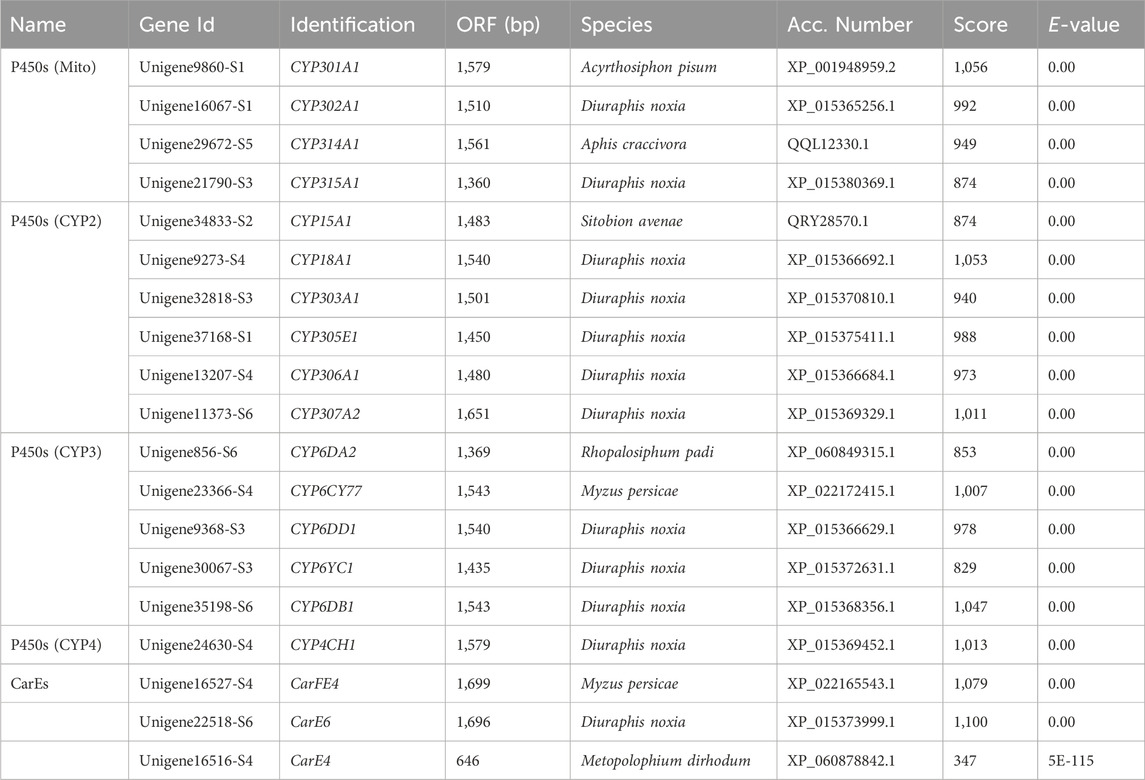
The relative level of P450 activity in S. heraclei was remarkably increased by 2.60-fold under afidopyropen treatment at 48 h compared to the control (Figure 1A). The relative level of CarE activity was remarkably increased by 4.86-fold under afidopyropen treatment at 48 h compared to the control (Figure 1B). The relative level of GST activity was significantly decreased under afidopyropen treatment at 48 h compared to the control (Figure 1C).
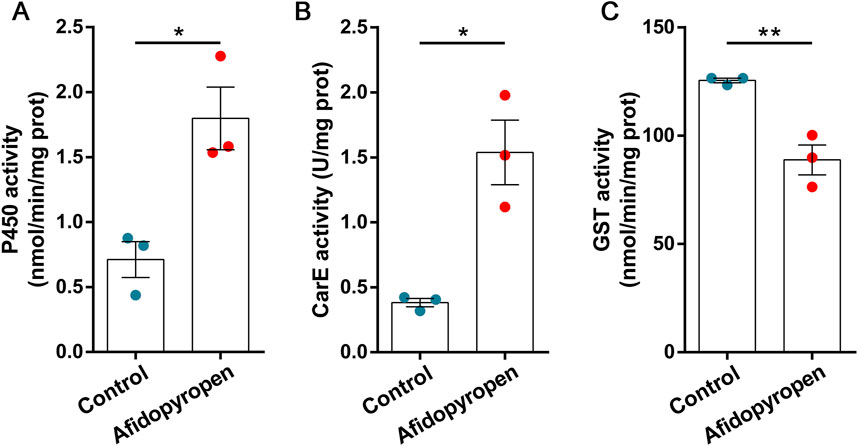
Figure 1. Detoxification enzyme activity in S. heraclei after afidopyropen treatment. (A) MFO activity after treatment of afidopyropen. (B) CarE activity after treatment of afidopyropen. (C) GST activity after treatment of afidopyropen. The asterisk (*) and asterisks (**) indicate significant differences with p < 0.05 and p < 0.01 compared to control group.
3.3 Synergistic effects of PBO, TPP and DEM on the mortality of S. heraclei
After PBO treatment, the mortality of S. heraclei was significantly increased by 1.41-fold under afidopyropen treatment compared to the control (Figure 2A). After TPP treatment, the mortality of S. heraclei was significantly increased by 1.24-fold under afidopyropen treatment compared to the control (Figure 2B). After DEM treatment, there was no significant synergistic effect on the mortality of S. heraclei treated with afidopyropen compared to the control (Figure 2C).
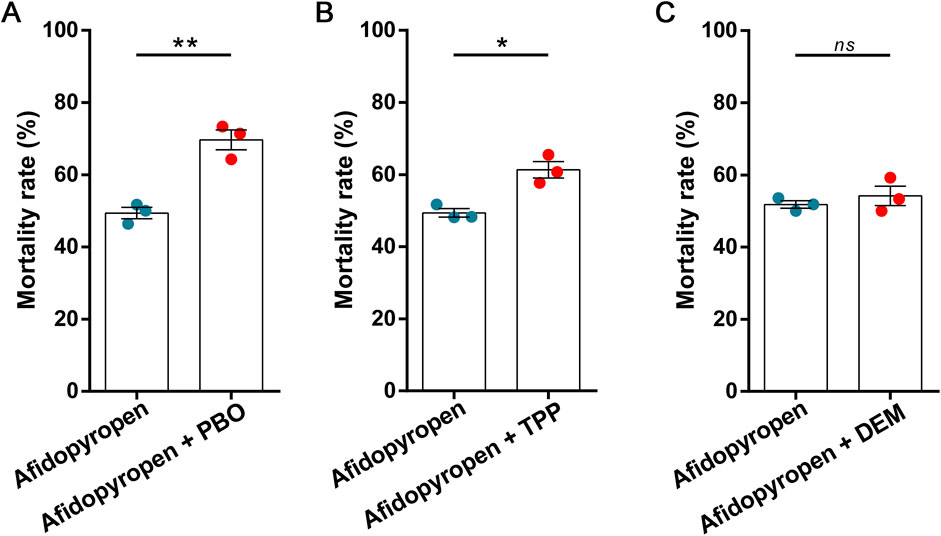
Figure 2. Effects of synergists on mortality of S. heraclei under afidopyropen treatment. (A) The synergism of PBO on S. heraclei mortality after treatment of afidopyropen. (B) The synergism of TPP on S. heraclei mortality after treatment of afidopyropen. (C) The synergism of DEM on S. heraclei mortality after treatment of afidopyropen. The asterisk (*) and asterisks (**) indicate significant differences with p < 0.05 and p < 0.01 compared to afidopyropen group. The ns represents no significant difference.
3.4 Overall analysis of transcriptome data
A total of six samples were sequenced, yielding an average of 21, 925, 000 clean reads. The clean reads ratio was greater than 95.00%. The data quality evaluation showed that the Q20 values of the six samples were greater than 96.00%, and the Q30 values were greater than 89.50% (Supplementary Table S2), indicating good data quality. The clean reads were finally assembled into 62,844 unigenes, with an average length of 1,486 bp, N50 length 2,829 bp, N70 length 1,804 bp, and GC percentage counting range from 33.13% to 33.56% (Supplementary Table S3).
The number of unigenes in the range of sequence length from 200 nt to 300 nt was the largest (11,818), and the number of unigenes with sequence length ≥3,000 nt was the second (9,618) (Supplementary Figure S1A). In order to ensure the rationality and reliability of samples and the repeatability of biological experiment, correlation analysis and principal component analysis were performed on six samples before differential gene analysis. The correlation coefficients of the six samples were all above 96.80%, indicating that the gene expression levels of the six samples were very similar (Supplementary Figure S1B). The principal component analysis demonstrated that the percentage value of PC1 was 98.71% and PC2 was 1.04%. This indicates that the repeatability of intra-group samples was good, the data similarity was high, the discrimination between groups was good, and there were no outlier samples (Supplementary Figure S1C).
3.5 Functional annotation and analysis of differentially expressed genes (DEGs)
Through the Blastx homology search, a total of 12,600 unigenes matched entries in the NR database. Most of them shared homology with Hemiptera, such as Diuraphis noxia (37.14%), Acyrthosiphon pisum (26.73%), M. persicae (13.82%), Aphis glycines (8.60%) and Aphis craccivora (7.11%) (Figure 3A). To identify DEGs associated with afidopyropen treatment, DESeq method was used to analyze the DEGs between control and treatment group samples, and a total of 11,144 DEGs were detected, of which 5,887 were upregulated and 5,257 were downregulated (Figure 3B). A heat map of differential gene expression showed a clear visual comparison of DEGs under afidopyropen treatment (Figure 3C).
Figure 3. Differential gene expression analysis of S. heraclei in response to afidopyropen treatment. (A) Species distributions of top blastx matches to Nr database. (B) Number of up- and downregulated DEGs and volcano plot of DEGs in afidopyropen exposure. (C) Heatmap of DEGs between afidopyropen and control.
To further elucidate the biological functions and metabolic pathways of 11,144 DEGs, the biological processes and metabolic pathways of these unigenes were classified using the GO database and KEGG database. According to the GO analysis results, a total of 23 biological processes, 14 cell components and 9 molecular function catalogues were gathered. In biological processes, most DEGs were involved in “cellular process” (3,120), “metabolic process” (2,513), “biological regulation” (1,938) and “biological regulation” (1,785). In the cell component, the number of “cells” was 3,350, the number of “cell parts” was 3,350 and the number of “organelle” was 2,762. In terms of the molecular function catalogs, most GO terms were involved in “binding” (2,803) and “catalytic activity” (1,908) (Supplementary Figure S2A). These data indicated that many biological processes, organelles, and molecular functions were involved in the response of S. heraclei to afidopyropen. KEGG pathway analysis, according to the results of these unigenes involved five metabolic pathways, including Cellular the Processes, Environmental Information Processing, Genetic Information Processing, Metabolism and Organismal Systems. In the end, a total of 1,605 unigenes related to metabolism, including Amino acid metabolism, Biosynthesis of other secondary metabolites, Carbohydrate metabolism, Energy metabolism, Global and overview maps, Glycan biosynthesis and metabolism, Lipid metabolism, Metabolism of cofactors and vitamins, Metabolism of other amino acids, Metabolism of terpenoids and polyketides, Nucleotide metabolism, and Xenobiotics biodegradation and metabolism (Supplementary Figure S1B). Analysis of the top 20 KEGG pathways showed that the metabolism was significantly enriched in seven pathways including “Biosynthesis of amino acids”, “Selenocompound metabolism”, “Nitrogen metabolism”, Citrate cycle (TCA cycle)”, “Tyrosine metabolism”, “Ether lipid metabolism” and “2-Oxocarboxylic acid metabolism”. The genetic information processing was enriched in five pathways including “Ribosome”, “Nucleocytoplasmic transport”, “Spliceosome”, “Proteasome” and “Ubiquitin mediated proteolysis”. The cellular processes pathway was enriched in four pathways including “Apoptosis”, “Apoptosis–fly”, “Mitophagy–animal” and “Autophagy–animal”. The organismal systems pathway was enriched in three pathways including “Antigen processing and presentation”, “Renin secretion” and “Fat digestion and absorption”. The environmental information processing was enriched in one pathway including “Hedgehog signaling pathway–fly” (Supplementary Table S4).
3.6 Identification and screening of differentially expressed P450 and CarE genes
A total of sixteen P450 genes were identified in the transcriptome of S. heraclei, including four in Mito clade, six in CYP2 clade, five in CYP3 clade and one in CYP4 clade. In addition, three CarE genes were identified, including CarFE4, CarE6 and CarE4 (Table 2).
To identify potential P450 and CarE genes which involved in afidopyropen susceptibility, we examined the expression induction of sixteen P450 genes and three CarE genes using RT-qPCR after afidopyropen treatment. After afidopyropen treatment, eleven P450 genes were significantly upregulated, including one in Mito clade, six in CYP2 clade, three in CYP3 clade and one in CYP4 clade, among which CYP6DA2 in CYP3 clade was the most significant (Figure 4). Two CarE genes CarFE4 and CarE6 were significantly upregulated, of which CarFE4 was the most significant (Figure 4).
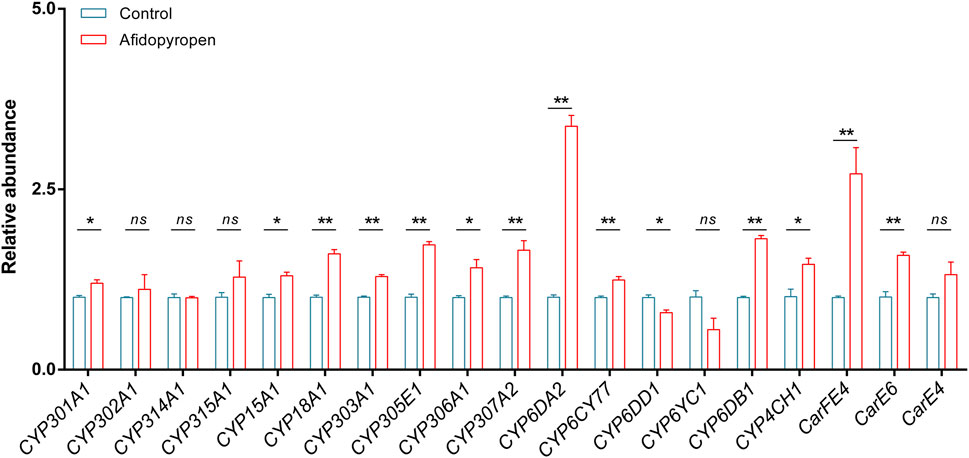
Figure 4. Screening of differentially expressed P450 and CarE genes by RT-qPCR analysis. The asterisk (*) and asterisks (**) indicate significant differences with p < 0.05 and p < 0.01 compared to control group. The ns represents no significant difference.
In the transcriptome of S. heraclei, the expression of up-regulated P450 genes included CYP18A1, CYP306A1, CYP307A2, CYP6DA2, and CYP6CY77, and the expression of downregulated P450 genes included CYP301A1 and CYP303A1 (Supplementary Table S5). There was no difference in the transcriptome for the CarE genes.
3.7 CYP6DA2 and CarFE4 affect the susceptibility of S. heraclei to afidopyropen
To further explore the functions of CYP6DA2 and CarFE4, we examined the expression of these two genes in S. heraclei at different developmental stages after treatment with afidopyropen. The results demonstrated that CYP6DA2 and CarFE4 were expressed in all five stages, from the first-instar larvae to the adult stage, following treatment with the drug. The highest expression of CYP6DA2 was observed in the adult stage, followed by the fourth-instar larvae stage. The expression of CYP6DA2 was observed to be relatively low in the second and third-instar larval stages, with the lowest expression observed in the second-instar stage (Figure 5A). The highest levels of CarFE4 expression were observed in the adult stage, followed by the first and fourth-instar stages. Conversely, relatively low levels of CarFE4 expression were noted in the second and third-instar stages, with the lowest levels observed in the second-instar stage (Figure 5B). To verify the role of CYP6DA2 and CarFE4 in the sensitivity of S. heraclei to afidopyropen, we disrupted these two genes, and the results showed that the relative abundance of CYP6DA2 and CarFE4 (Figures 5C, D) genes was significantly reduced in the disrupted S. heraclei compared with the dsEGFP group. Further bioassay results indicated that downregulation of CYP6DA2 and CarFE4 (Figures 5E, F) expression significantly increased the mortality of S. heraclei after afidopyropen treatment.
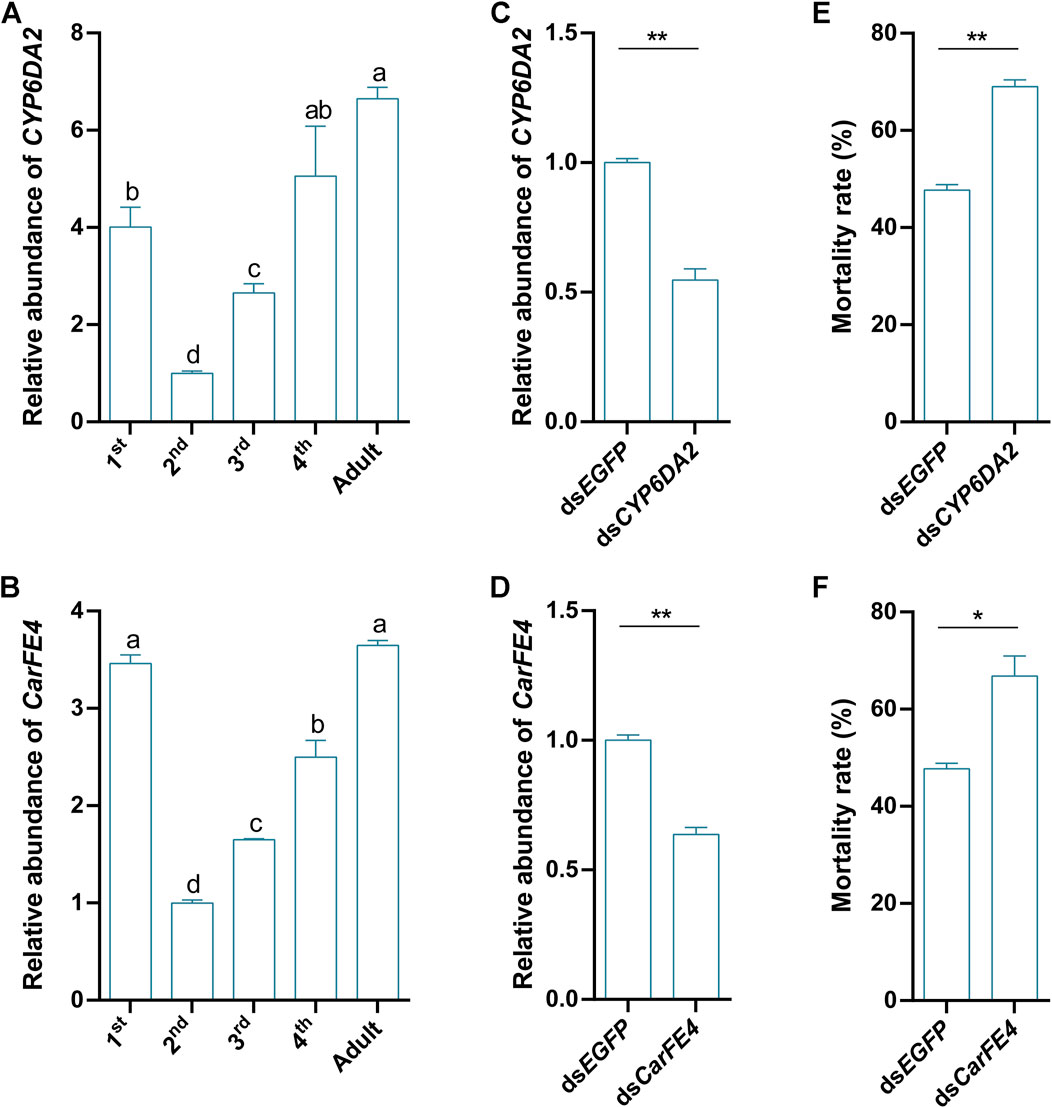
Figure 5. Functional analysis of CYP6DA2 and CarFE4 by RNAi and their expression patterns in S. heraclei. (A) The expression patterns of CYP6DA2 at different developmental stages. (B) The expression patterns of CarFE4 at different developmental stages. (C) Relative abundance of CYP6DA2 in S. heraclei at 2 days after feeding dsRNA. (D) Relative abundance of CarFE4 in S. heraclei at 2 days after feeding dsRNA. (E) Mortality rate of S. heraclei fed dsCYP6DA2 treated with LD50 concentrations of afidopyropen for 48 h. (F) Mortality rate of S. heraclei fed dsCarFE4 treated with LD50 concentrations of afidopyropen for 48 h. The asterisk (*) and asterisks (**) indicate significant differences with p < 0.05 and p < 0.01 compared to control group. The letters a, b, c, and d present significant differences (p < 0.05) of the expression.
4 Discussion
At present, chemical control is an important mean to manage aphids, which is simple and fast, but with the repeated use of chemical insecticides, insecticide residues and insecticide resistance have gradually become a major challenge to be solved (Silva et al., 2012; Field et al., 2017). Afidopyropen is a highly targeted and environmentally friendly insecticide with no cross-resistance to other insecticides (Ding et al., 2024), and is an important tool for integrated field management or resistance management of piercing-sucking mouthparts pests such as aphids, whitefly and leafhoppers. Understanding the detoxification mechanism of afidopyropen by pests is very important for the scientific use of afidopyropen in the field.
Metabolic resistance is one of the main mechanisms of insecticide resistance in field pests (Hemingway, 2000). The enhanced activity of detoxifying enzymes is an important line of defense against insecticides, and the main enzymes involved are P450, CarE and GST (Paula et al., 2020). Previous studies have shown that single insecticide could not induce the activity of all detoxification enzymes, such as P450 and CarE were involved in the detoxification process of A. gossypii to many insecticides, while GST was selectively resistant to different insecticides (Shi et al., 2023), and the activity of detoxification enzyme of Bemisia tabaci was different in different host plants (Guo et al., 2014). In addition, study has shown that the enhancement of P450 and CarE activity was closely related to the multiple resistance of Laodelphax striatellus (Gao et al., 2008). At the same time, due to the different metabolic pathways of P450 and CarE in insects and mammals, the toxicity was different, which also reflected the selective function of insecticides (Liu X. et al., 2024). The results of this study showed that after afidopyropen treatment, the activities of P450 and CarE were significantly increased, while the activity of GST was decreased. We speculated that the P450 and CarE enzymes might be involved in the detoxification process of S. heraclei to afidopyropen, and the direct relationship and detoxification mechanism between them were verified by synergism bioassay, but we could not deny the function of GST, which acted as a detoxification enzyme in phase II, played a late role in the detoxification process. For example, the activity of GST in Nilaparvata lugens was detected only 72 h after insecticide treatment (Yang B. et al., 2020). The decrease in GST activity, may be due to the fact that GST does not possess selective resistance to afidopyropen, whereas aphids, when attacked by insecticides, adjust their resources in order to survive by prioritizing the assurance of other, more critical physiological processes, with a corresponding decrease in the investment in non-emergency defense mechanisms such as GST (King and MacRae, 2015). Furthermore, it is possible that excessive oxidative stress induced by insecticides leads to a decrease in the stability of GST proteins (Hao et al., 2021; Dong et al., 2024). Chemicals and host plants affected the activity of detoxification enzymes. Different enzymes involved in the detoxification process reflected the multiplicity of insect resistance mechanisms, and the joint action of P450 and CarE might also reflect the selective insecticidal characteristics of afidopyropen. Furthermore, the metabolism of enzymes affected the efficacy of insecticides (Khan, 2020; Shakya et al., 2022), and the expression of regulatory enzymes could restore or maintain the sensitivity of insects to insecticides to a certain extent.
Transcriptome is a technology for analyzing transcripts to reveal the regulation of gene expression and to annotate its biological function (Jones et al., 2024). In the transcriptome data of this study, we found seventeen P450 genes and three CarE genes. The number of P450 and CarE genes is relatively low. This may be related to the nature of exogenous substances (Katsavou et al., 2022), the true sociality of insect species (Zhang et al., 2023), the adaptability of host plants and survival environments (Li Y. et al., 2020; Lu et al., 2023), and even the influence of sequencing techniques on transcriptional data (Yang Y. X. et al., 2020). P450 genes are divided into several families, among which CYP3 and CYP4 families are related to the detoxification metabolism of insecticides. CYP3 family is even more important genes involved in the detoxification metabolism that are unique to insects (Feyereisen, 2012; Han et al., 2022). Quantitative detection of P450 and CarE genes showed that thirteen genes were up-regulated after afidopyropen treatment, of which one-third belonged to CYP3 and CYP4 family, and the expression of CYP6DA2 and CarFE4 was upregulated by more than 2.5-fold. CYP6DA2 was significantly expressed in A. gossypii (Peng et al., 2016) under insecticides exposure, which was consistent with our results. CarFE4 was involved in the resistance mechanism of insects and would respond to external adverse factors (Ma et al., 2018). Under the selection of insecticides, M. persicae with low CarFE4 gene level were gradually replaced by M. persicae with high level of CarE4 gene (Field and Foster, 2002). At the same time, the amplification of CarFE4 gene made M. persicae resistant to a variety of insecticides in the field (Tang et al., 2017). Based on this, we have reason to speculate that CYP6DA2 and CarFE4 play an important role in the insecticide detoxification in insects.
At the same time, the number of genes obtained from the transcriptome is so large that it is impossible to analyze its function alone, so in the course of the experiment, genes with similar functions are grouped together to form pathways, so as to realize the correlation between function and phenotype (Li et al., 2022; Kakino et al., 2023). The GO and KEGG annotations in this study showed some of the most representative biological functions and information processing pathways. The analysis of these results revealed that the pathways related to higher concentration of differentially expressed genes, mainly included carbohydrate metabolism, energy metabolism, lipid metabolism, amino acid metabolism and so on. These metabolic pathways played an important role in the daily actions, growth and development of insects (Jiang et al., 2020; Xu et al., 2022). After afidopyropen treatment, the genes in these metabolic pathways were enriched, and it was speculated that S. heraclei achieved metabolic detoxification of insecticides through these pathways. A large amount of energy will be generated in the process of Metabolism to provide enough power for life activities, and a large amount of reactive oxygen species (ROS) will be generated at the same time. Previous studies have reported that ROS can regulate detoxification metabolism related genes through CncC pathway to enhance the resistance of Bactrocera dorsalis to β-cypermethrin (Zeng et al., 2024). In addition, signal transduction was also a relatively enriched pathway of differentially expressed genes, which regulated various physiological activities throughout the insect’s life and was an extremely important mechanism in the organism (Meng et al., 2019), and we speculated that it also regulated the detoxification process of S. heraclei to afidopyropen.
In order to further verify the functions of CYP6DA2 and CarFE4, we need to silence these two genes by RNAi technology (Zhou et al., 2021), and then observe the effect of afidopyropen on S. heraclei. Because of its simplicity and good control effect, RNAi has been widely used in gene function identification. For example, in the experiment of resistance of A. gossypii to plant defense substances, the mortality of A. gossypii was significantly increased by silencing related genes by RNAi (Ma et al., 2017). The main methods of RNAi administration are microinjection and artificial feeding. Although the artificial feeding method has its own determinations, such as the inability to accurately determine the feeding habits of insects (Jain et al., 2020), many Hemiptera insects have successfully studied the function of key genes, such as Sitobion avenae (Fan et al., 2015), M. persicae (Tariq et al., 2019), B. tabaci (Vyas et al., 2017), N. lugens (Chen et al., 2010) and so on, so the feasibility of feeding is also very high. The interference results of this study showed that the silencing of CYP6DA2 and CarFE4 could significantly increase the susceptibility of S. heraclei to afidopyropen. The results suggested that CYP6DA2 and CarFE4 were indeed involved in the regulation of the detoxification process of S. heraclei and played a key role in reducing the susceptibility of insecticides.
In order to determine the critical period of the control effect of afidopyropen on S. heraclei, the expression of CYP6DA2 and CarFE4 in S. heraclei at different developmental stages was detected. It has been proved that the cuticle of insects played an important role in resisting insecticides (Yan et al., 2022), and the microbiota of insects also protected insects at different developmental stages in the face of external stimuli (Huang et al., 2019). The activity of insect detoxification enzymes increased with the increase of insect development (Durak et al., 2021), which meant that with the gradual development of insects, their susceptibility to insecticides would gradually decrease. The reason for choosing the third instar nymph for most of the insect experiments is that at this time, the insects are physiologically more mature and more sensitive to environmental changes, making it easier to observe the effects of specific treatments on the insect (Kim and Roberts, 2012; Durak et al., 2021; Pers and Hansen, 2021). The body size is moderate, which is convenient for experimental operation, and it begins to show more obvious activity behavior, which is convenient for observing its behavioral pattern. Meanwhile, in the entomological study of Hemiptera, the third-instar nymph stage has been widely studied and experienced (Fan et al., 2020). The results showed that the expression of CYP6DA2 and CarFE4 related to detoxification was the lowest in the second- and third-instar nymphs and the highest in adults, indicating that the activity of detoxification enzyme in the second- and third-instar nymphs was low, at this time, S. heraclei was weak in detoxification ability to insecticides, and the control effects in these two periods might be better than those in other periods.
5 Conclusion
In conclusion, the results of this study can lay a foundation for further exploring the detoxification mechanism of S. heraclei on afidopyropen. It will also provide a theoretical basis for the rational use of afidopyropen to control aphids in the field.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE272830: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE272830.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
XF: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Validation, Writing–original draft, Writing–review and editing. CX: Data curation, Investigation, Methodology, Writing–review and editing. XW: Data curation, Investigation, Writing–review and editing. AW: Data curation, Investigation, Writing–review and editing. YaZ: Data curation, Investigation, Writing–review and editing. YY: Data curation, Funding acquisition, Investigation, Writing–review and editing. YZg: Data curation, Investigation, Writing–review and editing. YZu: Conceptualization, Data curation, Funding acquisition, Investigation, Writing–review and editing. MZ: Conceptualization, Data curation, Investigation, Writing–review and editing. CS: Conceptualization, Data curation, Funding acquisition, Investigation, Writing–review and editing. JZ: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by Technology Innovation Guidance Plan of Shandong Province, China (2024LYXZ005), Innovation Fund Project for Outstanding postgraduates in Shandong University of Traditional Chinese Medicine (YYCXJJ-202429), Key Research and Development Plan of Shandong Province (2022TZXD0034), Shandong Province Modern Agricultural Technology System Chinese Herbal Medicine Industrial Innovation Team (SDAIT-20) and Agricultural Scientific and Technological Innovation Project of Shandong Academy of Agricultural Sciences (CXGC 2024F03, CXGC 2024D03).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2024.1478869/full#supplementary-material
References
Aioub A. A. A., Ashour M.-B. A. (2023). Editorial for the special issue “detoxification mechanisms in insects.”. Toxics 11, 691. doi:10.3390/toxics11080691
Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi:10.1016/S0022-2836(05)80360-2
Buchfink B., Xie C., Huson D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi:10.1038/nmeth.3176
Chen J., Zhang D., Yao Q., Zhang J., Dong X., Tian H., et al. (2010). Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 19, 777–786. doi:10.1111/j.1365-2583.2010.01038.x
Cui L., Yuan H., Wang Q., Wang Q., Rui C. (2018). Sublethal effects of the novel cis-nitromethylene neonicotinoid cycloxaprid on the cotton aphid Aphis gossypii Glover (Hemiptera: aphididae). Sci. Rep. 8, 8915. doi:10.1038/s41598-018-27035-7
Ding W., Guo L., Xue Y., Wang M., Li C., Zhang R., et al. (2024). Life parameters and physiological reactions of cotton aphids Aphis gossypii (Hemiptera: aphididae) to sublethal concentrations of afidopyropen. Agron. J. 14, 258. doi:10.3390/agronomy14020258
Ding Z., Wen Y., Yang B., Zhang Y., Liu S., Liu Z., et al. (2013). Biochemical mechanisms of imidacloprid resistance in Nilaparvata lugens: over-expression of cytochrome P450 CYP6AY1. Insect Biochem. Mol. Biol. 43, 1021–1027. doi:10.1016/j.ibmb.2013.08.005
Dong W., Hou D., Hou Q., Jin H., Li F., Wu S., et al. (2024). Effects of ultraviolet light stress on protective and detoxification enzymes in insects. T 3, 0. doi:10.48130/tp-0024-0008
Durak R., Dampc J., Kula-Maximenko M., Mołoń M., Durak T. (2021). Changes in antioxidative, oxidoreductive and detoxification enzymes during development of aphids and temperature increase. Antioxidants (Basel) 10, 1181. doi:10.3390/antiox10081181
Fan J., Zhang Y., Francis F., Cheng D., Sun J., Chen J. (2015). Orco mediates olfactory behaviors and winged morph differentiation induced by alarm pheromone in the grain aphid, Sitobion avenae. Insect Biochem. Mol. Biol. 64, 16–24. doi:10.1016/j.ibmb.2015.07.006
Fan Y., Li X., Mohammed A. A. A. H., Liu Y., Gao X. (2020). miR-147b-modulated expression of vestigial regulates wing development in the bird cherry-oat aphid Rhopalosiphum padi. BMC Genomics 21, 71. doi:10.1186/s12864-020-6466-7
Feyereisen R. (2012). Insect CYP genes and P450 enzymes. Insect Biochem. Mol. Biol., 236–316. doi:10.1016/B978-0-12-384747-8.10008-X
Field L. M., Bass C., Davies T. G. E., Williamson M. S., Zhou J. J. (2017). Aphid genomics and its contribution to understanding aphids as crop pests. CABI, Aphids as crop pests, 37–49. doi:10.1079/9781780647098.0037
Field L. M., Foster S. P. (2002). Amplified esterase genes and their relationship with other insecticide resistance mechanisms in English field populations of the aphid, Myzus persicae (Sulzer). Pest Manag. Sci. 58, 889–894. doi:10.1002/ps.552
Gao B., Wu J., Huang S., Mu L., Han Z. (2008). Insecticide resistance in field populations of Laodelphax striatellus Fallén (Homoptera: Delphacidae) in China and its possible mechanisms. Int. J. Pest Manag. 54, 13–19. doi:10.1080/09670870701553303
Goto K., Horikoshi R., Mitomi M., Oyama K., Hirose T., Sunazuka T., et al. (2019). Synthesis and insecticidal efficacy of pyripyropene derivatives. Part II-Invention of afidopyropen. J. Antibiot. (Tokyo) 72, 661–681. doi:10.1038/s41429-019-0193-9
Gregg P. C., Socorro A. P. D., Landolt P. J. (2018). Advances in attract-and-kill for agricultural pests: beyond pheromones. Annu. Rev. Entomol. 63, 453–470. doi:10.1146/annurev-ento-031616-035040
Guo L., Xie W., Wang S., Wu Q., Li R., Yang N., et al. (2014). Detoxification enzymes of Bemisia tabaci B and Q: biochemical characteristics and gene expression profiles. Pest Manag. Sci. 70, 1588–1594. doi:10.1002/ps.3751
Han H., Yang Y., Hu J., Wang Y., Zhao Z., Ma R., et al. (2022). Identification and characterization of CYP6 family genes from the oriental fruit moth (grapholita molesta) and their responses to insecticides. Insects 13, 300. doi:10.3390/insects13030300
Hao L. I., Ya-ping C., Zhi-hui L. U., Jian-yang G. U. O., Ya-hong L. I., Lin-yun Z. H. U., et al. (2021). Response comparison of protective and detoxification enzymes in Spodoptera frugiperda (J. E. Smith) and Spodoptera litura (Fabricius) larvae to two insecticides. nfnyxb 52, 559–569. doi:10.3969/j.issn.2095-1191.2021.03.002
He Q., Sun Y., Yi M., Huang H. (2023). How to promote agricultural enterprises to reduce the use of pesticides and fertilizers? An evolutionary game approach. Front. Sustain. Food Syst. 7. doi:10.3389/fsufs.2023.1238683
Hemingway J. (2000). The molecular basis of two contrasting metabolic mechanisms of insecticide resistance. Insect Biochem. Mol. Biol. 30, 1009–1015. doi:10.1016/s0965-1748(00)00079-5
Horikoshi R., Goto K., Mitomi M., Oyama K., Hirose T., Sunazuka T., et al. (2022). Afidopyropen, a novel insecticide originating from microbial secondary extracts. Sci. Rep. 12, 2827. doi:10.1038/s41598-022-06729-z
Hu C., Liu J.-Y., Wang W., Mota-Sanchez D., He S., Shi Y., et al. (2022). Glutathione S-transferase genes are involved in lambda-cyhalothrin resistance in Cydia pomonella via sequestration. J. Agric. Food Chem. 70, 2265–2279. doi:10.1021/acs.jafc.2c00360
Huang H., Li H., Ren L., Cheng D. (2019). Microbial communities in different developmental stages of the oriental fruit fly, Bactrocera dorsalis, are associated with differentially expressed peptidoglycan recognition protein-encoding genes. Appl. Environ. Microbiol. 85, e00803. doi:10.1128/AEM.00803-19
Jain R. G., Robinson K. E., Fletcher S. J., Mitter N. (2020). RNAi-Based functional genomics in Hemiptera. Insects 11, 557. doi:10.3390/insects11090557
Jeschke P. (2021). Status and outlook for acaricide and insecticide discovery. Pest Manag. Sci. 77, 64–76. doi:10.1002/ps.6084
Jiang L., Peng L.-L., Cao Y.-Y., Thakur K., Hu F., Tang S.-M., et al. (2020). Transcriptome analysis reveals gene expression changes of the fat body of silkworm (Bombyx mori L.) in response to selenium treatment. Chemosphere 245, 125660. doi:10.1016/j.chemosphere.2019.125660
Jones E. F., Haldar A., Oza V. H., Lasseigne B. N. (2024). Quantifying transcriptome diversity: a review. Brief. Funct. Genomics 23, 83–94. doi:10.1093/bfgp/elad019
Kakino K., Mon H., Ebihara T., Hino M., Masuda A., Lee J. M., et al. (2023). Comprehensive transcriptome analysis in the testis of the silkworm, Bombyx mori. Insects 14, 684. doi:10.3390/insects14080684
Katsavou E., Riga M., Ioannidis P., King R., Zimmer C. T., Vontas J. (2022). Functionally characterized arthropod pest and pollinator cytochrome P450s associated with xenobiotic metabolism. Pestic. Biochem. Physiol. 181, 105005. doi:10.1016/j.pestbp.2021.105005
Khan H. A. A. (2020). Susceptibility to indoxacarb and synergism by enzyme inhibitors in laboratory and field strains of five major stored product insects in Pakistan. Chemosphere 257, 127167. doi:10.1016/j.chemosphere.2020.127167
Kim J. J., Roberts D. W. (2012). The relationship between conidial dose, moulting and insect developmental stage on the susceptibility of cotton aphid, Aphis gossypii, to conidia of Lecanicillium attenuatum, an entomopathogenic fungus. Biocontrol Sci. Techn 22, 319–331. doi:10.1080/09583157.2012.656580
King A. M., MacRae T. H. (2015). Insect heat shock proteins during stress and diapause. Annu. Rev. Entomol. 60, 59–75. doi:10.1146/annurev-ento-011613-162107
Li H., Xia X., He X., Li S., Dai L., Ye J., et al. (2022). Comparative transcriptome analysis reveals molecular insights in overwintering monochamus alternatus (Coleoptera: cerambycidae). J. Insect Sci. 22, 8. doi:10.1093/jisesa/ieac025
Li P. R., Shi Y., Ju D., Liu Y.-X., Wang W., He Y.-S., et al. (2023). Metabolic functional redundancy of the CYP9A subfamily members leads to P450-mediated lambda-cyhalothrin resistance in Cydia pomonella. Pest Manag. Sci. 79, 1452–1466. doi:10.1002/ps.7317
Li Y., Liu X., Wang N., Zhang Y., Hoffmann A. A., Guo H. (2020). Background-dependent Wolbachia-mediated insecticide resistance in Laodelphax striatellus. Environ. Microbiol. 22, 2653–2663. doi:10.1111/1462-2920.14974
Li Z., Li W., Qin W., Liu J., He Y. (2023). Ampicillin enhanced the resistance of Myzus persicae to imidacloprid and cyantraniliprole. Pest Manag. Sci. 79, 1388–1398. doi:10.1002/ps.7310
Liu X., Wang Q., Liu X., Xiao D., Liu T.-X., Liang P. (2024). Molecular mechanisms for selective action of afidopyropen to Myzus persicae and Coccinella septempunctata. Pest Manag. Sci. 80, 3893–3900. doi:10.1002/ps.8092
Liu Y. X., Hu C., Li Y. T., Gao P., Yang X.-Q. (2024). Identification of G Protein-Coupled receptors (GPCRs) associated with lambda-cyhalothrin detoxification in Cydia pomonella. J. Agric. Food Chem. 72, 363–377. doi:10.1021/acs.jafc.3c06522
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Lu J., Zhang H., Wang Q., Huang X. (2023). Genome-wide identification and expression pattern of cytochrome P450 genes in the social aphid Pseudoregma bambucicola. Insects 14, 212. doi:10.3390/insects14020212
Lu K., Song Y., Zeng R. (2021). The role of cytochrome P450-mediated detoxification in insect adaptation to xenobiotics. Curr. Opin. Insect Sci. 43, 103–107. doi:10.1016/j.cois.2020.11.004
Ma K., Li F., Liang P., Chen X., Liu Y., Tang Q., et al. (2017). RNA interference of Dicer-1 and Argonaute-1 increasing the sensitivity of Aphis gossypii Glover (Hemiptera: aphididae) to plant allelochemical. Pestic. Biochem. Physiol. 138, 71–75. doi:10.1016/j.pestbp.2017.03.003
Ma K.-S., Li F., Liang P.-Z., Chen X.-W., Liu Y., Gao X.-W. (2016). Identification and validation of reference genes for the normalization of gene expression data in qRT-PCR analysis in Aphis gossypii (Hemiptera: aphididae). J. Insect Sci. 16, 17. doi:10.1093/jisesa/iew003
Ma M., Jia H., Cui X., Zhai N., Wang H., Guo X., et al. (2018). Isolation of carboxylesterase (esterase FE4) from Apis cerana and its role in oxidative resistance during adverse environmental stress. Biochimie 144, 85–97. doi:10.1016/j.biochi.2017.10.022
Massimino Cocuzza G. (2022). Semiaphis heraclei (celery aphid). CABI Compendium. doi:10.1079/cabicompendium.3216353232163532
Meng X., Dong F., Qian K., Miao L., Yang X., Ge H., et al. (2019). Transcriptome analysis reveals global gene expression changes of Chilo suppressalis in response to sublethal dose of chlorantraniliprole. Chemosphere 234, 648–657. doi:10.1016/j.chemosphere.2019.06.129
Mubeen I., Fawzi Bani Mfarrej M., Razaq Z., Iqbal S., Naqvi S. A. H., Hakim F., et al. (2023). Nanopesticides in comparison with agrochemicals: outlook and future prospects for sustainable agriculture. PPB 198, 107670. doi:10.1016/j.plaphy.2023.107670
Nawrocki E. P., Kolbe D. L., Eddy S. R. (2009). Infernal 1.0: inference of RNA alignments. Bioinformatics 25, 1335–1337. doi:10.1093/bioinformatics/btp157
Patra B., Hath T. K. (2023). Resistance status and activity of detoxifying enzymes in Oligonychus coffeae (Nietner) (Acari:Tetranychidae) on tea. Crop Prot. 167, 106201. doi:10.1016/j.cropro.2023.106201
Paula D. P., Menger J., Andow D. A., Koch R. L. (2020). Diverse patterns of constitutive and inducible overexpression of detoxifying enzyme genes among resistant Aphis glycines populations. Pestic. Biochem. Phys. 164, 100–114. doi:10.1016/j.pestbp.2019.12.012
Peng T., Pan Y., Gao X., Xi J., Zhang L., Yang C., et al. (2016). Cytochrome P450 CYP6DA2 regulated by cap ‘n’collar isoform C (CncC) is associated with gossypol tolerance in Aphis gossypii Glover. Insect Mol. Biol. 25, 450–459. doi:10.1111/imb.12230
Pers D., Hansen A. K. (2021). The boom and bust of the aphid’s essential amino acid metabolism across nymphal development. G3 (Bethesda) 11, jkab115. doi:10.1093/g3journal/jkab115
Shakya M., Nandi A., Fular A., Kumar S., Bisht N., Sharma A. K., et al. (2022). Synergistic property of piperonyl butoxide, diethyl maleate, triphenyl phosphate and verapamil hydrochloride with deltamethrin and ivermectin against Rhipicephalus microplus ticks. Ticks Tick. Borne Dis. 13, 102006. doi:10.1016/j.ttbdis.2022.102006
Shi D., Wang T., Lv H., Li X., Wan H., He S., et al. (2023). Insecticide resistance monitoring and diagnostics of resistance mechanisms in cotton-melon aphid, Aphis gossypii Glover in Central China. J. Appl. Entomol. 147, 392–405. doi:10.1111/jen.13119
Silva A. X., Jander G., Samaniego H., Ramsey J. S., Figueroa C. C. (2012). Insecticide resistance mechanisms in the green peach aphid Myzus persicae (Hemiptera: aphididae) I: a transcriptomic survey. PLOS ONE 7, e36366. doi:10.1371/journal.pone.0036366
Song K.-X., Wang J.-Y., Qiao H.-L., Wei H.-S., Guo K., Xu C.-Q. (2023). Secondary rhinaria contribute to major sexual dimorphism of antennae in the aphid Semiaphis heraclei (takahashi). Insects 14, 468. doi:10.3390/insects14050468
Tang Q.-L., Ma K.-S., Hou Y.-M., Gao X.-W. (2017). Monitoring insecticide resistance and diagnostics of resistance mechanisms in the green peach aphid, Myzus persicae (Sulzer) (Hemiptera: aphididae) in China. Pestic. Biochem. Physiol. 143, 39–47. doi:10.1016/j.pestbp.2017.09.013
Tariq K., Ali A., Davies T. G. E., Naz E., Naz L., Sohail S., et al. (2019). RNA interference-mediated knockdown of voltage-gated sodium channel (MpNav) gene causes mortality in peach-potato aphid, Myzus persicae. Sci. Rep. 9, 5291. doi:10.1038/s41598-019-41832-8
Vyas M., Raza A., Ali M. Y., Ashraf M. A., Mansoor S., Shahid A. A., et al. (2017). Knock down of whitefly gut gene expression and mortality by orally delivered gut gene-specific dsRNAs. PLoS One 12, e0168921. doi:10.1371/journal.pone.0168921
Xu L., Zhao J., Xu D., Xu G., Gu Z., Xiao Z., et al. (2022). Application of transcriptomic analysis to unveil the toxicity mechanisms of fall armyworm response after exposure to sublethal chlorantraniliprole. Ecotoxicol. Environ. Saf. 230, 113145. doi:10.1016/j.ecoenv.2021.113145
Yan Z., Tong X., Xiong G., Yang W., Lu K., Yuan Y., et al. (2022). A blueprint of microstructures and stage-specific transcriptome dynamics of cuticle formation in Bombyx mori. Int. J. Mol. Sci. 23, 5155. doi:10.3390/ijms23095155
Yang B., Lin X., Yu N., Gao H., Zhang Y., Liu W., et al. (2020). Contribution of glutathione S-transferases to imidacloprid resistance in Nilaparvata lugens. J. Agric. Food Chem. 68, 15403–15408. doi:10.1021/acs.jafc.0c05763
Yang Y. X. Y. X., Lin R.-H., Li Z., Wang A.-Y., Xue C., Duan A.-L., et al. (2020). Function analysis of P450 and GST genes to imidacloprid in Aphis craccivora (koch). Front. Physiol. 11, 624287. doi:10.3389/fphys.2020.624287
Yin T., Zhang J., Liu C., Xue Y., Liu Z., Liu S., et al. (2024). Environmental-related doses of afidopyropen induced toxicity effects in earthworms (Eisenia fetida). Ecotoxicol. Environ. Saf. 274, 116223. doi:10.1016/j.ecoenv.2024.116223
Zeng T., Fu Q.-Y., Luo F.-Y., Dai J., Fu R., Qi Y.-X., et al. (2024). Lactic acid bacteria modulate the CncC pathway to enhance resistance to β-cypermethrin in the oriental fruit fly. ISME J. 18 (1), wrae058. doi:10.1093/ismejo/wrae058
Zhang H., Liu Q., Chen K., Ni X., Cheng Z., Qiao G., et al. (2023). Mother knows best: reproductive regulation of caste ratio in a social hemipteran. Insect Sci. 30, 582–586. doi:10.1111/1744-7917.13105
Zhang Y., Yang Y., Sun H., Liu Z. (2016). Metabolic imidacloprid resistance in the brown planthopper, Nilaparvata lugens, relies on multiple P450 enzymes. Insect Biochem. Mol. Biol. 79, 50–56. doi:10.1016/j.ibmb.2016.10.009
Keywords: Semiaphis heraclei, afidopyropen, detoxification mechanism, CYP6DA2, CarFE4
Citation: Fu X, Xue C, Wang X, Wang A, Zhu Y, Yang Y, Zhang Y, Zhou Y, Zhao M, Shan C and Zhang J (2024) Two detoxification enzyme genes, CYP6DA2 and CarFE4, mediate the susceptibility to afidopyropen in Semiaphis heraclei. Front. Physiol. 15:1478869. doi: 10.3389/fphys.2024.1478869
Received: 11 August 2024; Accepted: 25 November 2024;
Published: 06 December 2024.
Edited by:
Senthil-Nathan, Sengottayan, Manonmaniam Sundaranar University, IndiaReviewed by:
Xue-Qing Yang, Shenyang Agricultural University, ChinaLu Xu, Jiangsu Academy of Agricultural Sciences (JAAS), China
Copyright © 2024 Fu, Xue, Wang, Wang, Zhu, Yang, Zhang, Zhou, Zhao, Shan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Zhou, enlzYXNzMjAyMUAxNjMuY29t; Ming Zhao, c2NyY3pobUAxNjMuY29t; Chenggang Shan, c2hhbmNoZW5nZ2FuZ0AxMjYuY29t; Jianhua Zhang, emhhbmdqaWFuaHVhMTk4OTA0QDE2My5jb20=
 Xiaochen Fu
Xiaochen Fu Chao Xue2
Chao Xue2 Yuanxue Yang
Yuanxue Yang Ming Zhao
Ming Zhao Jianhua Zhang
Jianhua Zhang