- 1Key Laboratory of Agricultural Genetically Modified Organisms Traceability, Ministry of Agriculture and Rural Affairs, Oil Crops Research Institute of Chinese Academy of Agricultural Science/Supervision and Test Center (Wuhan) for Plant Ecological Environment Safety, Ministry of Agriculture and Rural Affairs, Wuhan, China
- 2Northern Propagation Experiment Station, Center for Science and Technology Dissemination and Industrial Development, Oil Crops Research Institute of Chinese Academy of Agricultural Science, Wuhan, China
- 3Institute of Leisure Agriculture, Jiangsu Academy of Agricultural Sciences, Nanjing, China
- 4College of Plant Protection, Henan Agricultural University, Zhengzhou, China
Mylabris sibirica is a hypermetamorphic insect whose adults feed on oilseed rape. However, due to a shortage of effective and appropriate endogenous references, studies on molecular functional genes in Mylabris sibirica, have been tremendously limited. In this study, ten internal reference genes (ACT, ARF1, AK, EF1α, GAPDH, α-TUB, RPL6, RPL13, RPS3 and RPS18) were tested and assessed under four selected treatments including adult ages, adult tissues, temperatures, and sex by RT-qPCR based on five methods (Ct value, geNorm, NormFinder, BestKeeper and RefFinder). Our findings showed that RPL6 and RPL13 were the most optimal internal reference gene combination for gene expression during various adult ages and under diverse temperatures; The combination of RPL6 and RPS18 was recommended to test gene transcription levels under different adult tissues. AK and RPL6 were the best reference genes in male and female adults. RPL6 and RPL13 were the most appropriate reference gene pair to estimate gene expression levels under four different tested backgrounds. The relative transcript levels of a uridine diphosphate (UDP)-N-acetylglucosamine-pyrophosphorylase (MsUAP), varied greatly according to normalization with the two most- and least-suited reference genes. This study will lay the basis for further molecular physiology and biochemistry studies in M. sibirica, such as development, reproduction, sex differentiation, cold and heat resistance.
Introduction
Quantitative real-time fluorescent polymerase chain reaction (qRT-PCR) is a crucial method to measure target gene transcripts (Derveaux et al., 2010; Jozefczuk and Adjaye, 2011) and microbial abundance (Ali et al., 2018a; Ali et al., 2018b; Ali et al., 2018c; Ali et al., 2019) due to its high specificity, sensitivity and accuracy. Its precision is affected by numerous biological and technical factors, such as RNA purity, PCR efficiency, inappropriate data and statistical analyses (Valasek and Repa, 2005; Bustin et al., 2009). When qRT-PCR is applied to test transcripts, it is indispensable to normalize to improve the quantitative results by combining relatively stable reference genes (Bustin et al., 2009). If poor internal genes are used, the quantitates of nucleic acid will be inaccurate (Yang et al., 2014). Hence, Suitable internal references should be verified under diverse backgrounds, including developing stages, tissues and hosts (Andersen et al., 2004; Zhang et al., 2022; Shen et al., 2022).
As usual, reference genes are housekeeping genes (HKGs), whose expression levels are stable during different physiological states or in different cells (Zhang et al., 2022). At present, the 10 most generally used references contain actin (ACT), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ribosomal protein (RPL and RPS), TATA binding protein (TATA), heat shock protein (HSP), elongation factor 1α (EF1α), tubulin (TUB), 18S ribosomal RNA (18S) and succinate dehydrogenase complex subunit A (SDHA) (Lü et al., 2018). In addition, several novel methods including bioinformatic transcripome analysis have been used for reference gene screening (Yu et al., 2020). In previously researches, one to five analysis tools were applied to evaluate gene expression stability (Lü et al., 2018). Currently, the RefFinder is the only web-based tool available for evaluating candidate housekeeping genes by integrating the four available computational programs (geNorm, NormFinder, BestKeeper, and ΔCt method) into a web-based tool (Xie et al., 2023).
Nevertheless, there is no one stable internal gene under numerous tested treatments, including developmental stages, tissues and hosts (Andersen et al., 2004; Zhang et al., 2022; Shen et al., 2022; Shen et al., 2023). To evaluate accurate gene expression levels, each candidate reference gene under diverse tested conditions must be validated (Xu et al., 2021).
Mylabris sibirica (Coleoptera, Meloidae) is a hypermetamorphic pest that leads to significant losses in oilseed rape (Brassica napus) production (Tian Yubo et al., 2021). Its adults mainly feed on flowers of B. napus (Tian Yubo et al., 2021). Until now, studies on M. sibirica have concentrated on classification (ČernÝ and Vrabec, 2019; Pan and Ren, 2020), phylogenetics (Bologna et al., 2005) and medical value (Tian Yubo et al., 2021), however, little is known on gene functions. Gene expression analyses are essential to study molecular mechanisms of development, reproduction and physiology. Nevertheless, no study evaluating M. sibirica gene expression analysis has been reported. In order to further manage M. sibirica based on novel target genes, screening for optimal reference genes is imperative.
Nowadays, many researches have reported the optimal reference genes in Coleopterans (Shi et al., 2013; Pan et al., 2015; Sang et al., 2015; Lü et al., 2018; Yang et al., 2018; Ma et al., 2021; Sellamuthu et al., 2021; Zhang et al., 2022; Sellamuthu et al., 2022). For example, the most optimal internal references have been published in Phyllotreta striolata (EF1A and VATPA) (Guo et al., 2023), Henosepilachna vigintioctomaculata (RPS18 and RPL13) (Zhang et al., 2022) and Phaedon brassicae (RPL32 and EF1α in various tissues, RPL19 and TBP across diverse developmental stages, RPL32 and RPL19 under insecticide exposure, RPL32 and TBP under thermal stress) (Ma et al., 2021). In general, at least two reference genes are used to validate expression levels in each insect species (Zhang et al., 2022).
Since top 10 most frequently used reference genes contains ACT, RPL, TUB, GAPDH, RPS, 18S, EF1α, TATA, HSP and SDHA in insects and they are the most optimal internal references in adverse experimental backgrounds (Lü et al., 2018). In this study, ten candidate references, i.e., ACT, ARF1, AK (arginine kinase), EF1α, GAPDH, α-TUB, RPL6, RPL13, RPS3 and RPS18 were accordingly selected to assess the stability of gene expression during various adult ages, among various adult tissues, under diverse temperatures, and in females and males in M. sibirica. Finally, uridine diphosphate (UDP)-N-acetylglucosamine-pyrophosphorylase (UAP) was used as the target gene to verify our findings. Our results will offer the reference foundation for further molecular mechanisms in M. sibirica.
Methods and materials
Insect collection
M. sibirica adults used in this research were collected from B. napus plants at the Northern Propagation Experiment Station of the Oil Crops Research Institute, Chinese Academy of Agricultural Sciences, located in Haidong city, Qinghai Province, China in 2023 (coordinates: 36°31′10.47″N, 101°59′7.40″E). The adults were fed in an insectary for 1 week at 25°C ± 2 °C, 16 h:8 h photoperiod and 55% ± 5% relative humidity using oilseed rape as food.
Specimens through diverse adult ages
The newly emerged adults were grouped as males and females and kept in different net cages (30 cm × 30 cm × 40 cm, hand-made). For each biological replicate, a total of five adults (three males and two females) were collected every 24 h for a continuous period of 72 h. Three biological replicates were conducted.
Samples in different adult tissues
Epidermis, foregut, midgut, hindgut, trachea and antenna were dissected from the newly-emerged adults. Ten adults (5 males and 5 females) were dissected for one biological replicate. Three biological replicates were prepared.
Specimens under various temperatures
The newly-emerged adults were reared under five temperatures (4 °C, 15 °C, 20 °C, 26 °C and 35 °C) for 8 h. Five adults (3 males and 2 females) were collected for one biological replicate. Three biological replicates were prepared.
Collections in males and females
The newly-emerged male and female adults were kept in an insectary at 26 °C for 5 days. Five males and females were collected respectively for one biological replicate. Three biological replicates were prepared.
Total RNA extraction and cDNA synthesis
Total RNAs were extracted by TRIzol reagent (YiFeiXue Tech, Nanjing, China) following the manufacturer’s instructions. The RNA integrity was assessed by electrophoresis on a 1.5% agarose gel. The purity and concentration of the total RNA samples were determined using a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies, Rockland, DE, United States), with 260/280 and 260/230 ratios ranging from 1.9 to 2.1. The HiScript III RT SuperMix (containing gDNA wiper, Vazyme Biotech Co.,Ltd., Nanjing, China)was used to synthesize cDNA. The reaction mixtures were incubated at 37°C for 15 min, followed by 85°C for 5 s. The resulting cDNA samples were diluted 5-fold for the pursuant PCR and RT-qPCR.
Selection and identification of candidate references
Ten HKG sequences (actin, ACT; α-tubulin, α-TUB;ADP ribosylation factor 1, ARF1; elongation factor 1α, EF1α; arginine kinase, AK; ribosomal proteins RPL6, RPL13, RPS3 and RPS18; glyceraldehyde-3-phosphate dehydrogenase, GAPDH) were selected by BioEdit software based on the transcriptome data of M. sibirica (unpublished data). The resulting HKG information was presented in Supplementary Table S1.
The primers of Reverse transcriptase PCR (RT-PCR) were designed by Primer Premier 5.0 and performed according to the previously described method (Shen et al., 2022). The resultant sequences were submitted to GenBank. The accession numbers were located in Supplementary Table S1.
Quantitative real-time PCR (qRT-PCR)
The primers of qRT-PCR were designed by NCBI Primer-BLAST, and were listed in Table 1. The qRT-PCR reactions were conducted using ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd.) and CFX96 Real-Time System (Bio-Rad Laboratories, Hercules, CA, United States). The reaction mixture consisted of a 20 μL reaction volume including 1 μL cDNA template, 0.4 μL of forward primer (10 μM), 0.4 μL of reverse primer (10 μM), 10 μL of 2× ChamQ Universal SYBR qPCR Master Mix and 8.2 μL of RNase Free water. The qRT-PCR reaction conditions consisted of the following steps: an initial step of 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 5 s, and subsequent annealing at 60°C for 34 s, followed by one cycle of 95 °C for 15 s, 60 °C for 60 s and 95 °C for 1 s. The specificity of PCR amplicons were assessed by a melting curve analysis, analyzing by the BioRadCFXManager and gel electrophoresis. All experiments were repeated in triplicate. Amplification efficiencies (E) were calculated using a 5-fold dilution series of template via the following equation: E = (10[−1/slope] − 1) × 100%.
Stability determination of candidate reference genes
Uridine diphosphate (UDP)-N-acetylglucosamine-pyrophosphorylase (UAP) of M. sibirica was used to verify the stability of candidate reference genes (GenBank: OR838722). The primer sequence of the target gene was as follows:
Forward: ATTATTGATGGCCGGTGGTC
Reverse: ACCATTTAAACCGGTCTTTTGTT
Based on the stability (RPL6 and RPS18) and instability (AK and GAPDH) of primary reference genes, the relative levels of MsUAP in different adult tissues were computed by the 2−ΔΔCT method and from three replicates. One-way analysis of variance was used to assess significance in UAP expression levels among various adult tissues (SPSS, Chicago, IL, United States).
Data processing
The raw Ct values were obtained using the BioRadCFXManager. The stability of candidate HKGs were measured using the ΔCt method (Silver et al., 2006), geNorm (Vandesompele et al., 2002), Normfinder (Andersen et al., 2004) and BestKeeper (Pfaffl et al., 2004). Furthermore, the optimal number of reference genes for gene expression normalization was determined by pairwise variation (Vn/n+1) using the GeNorm program Typically, a Vn/n+1 value below the threshold of 0.15 indicates that the starting n reference genes are enough for normalizing target gene expression. Lastly, the overall ranking of each experimental background was assessed based on RefFinder (Xie et al., 2012; Xie et al., 2023).
Results
Selection of candidate reference genes
Ten HKG genes, i.e., ACT, ARF1, AK, EF1α, GAPDH, α-TUB, RPL6, RPL13, RPS3 and RPS18 in M. sibirica were selected. The obtained sequences were submitted to GenBank; the accession numbers were shown in Supplementary Table S1.
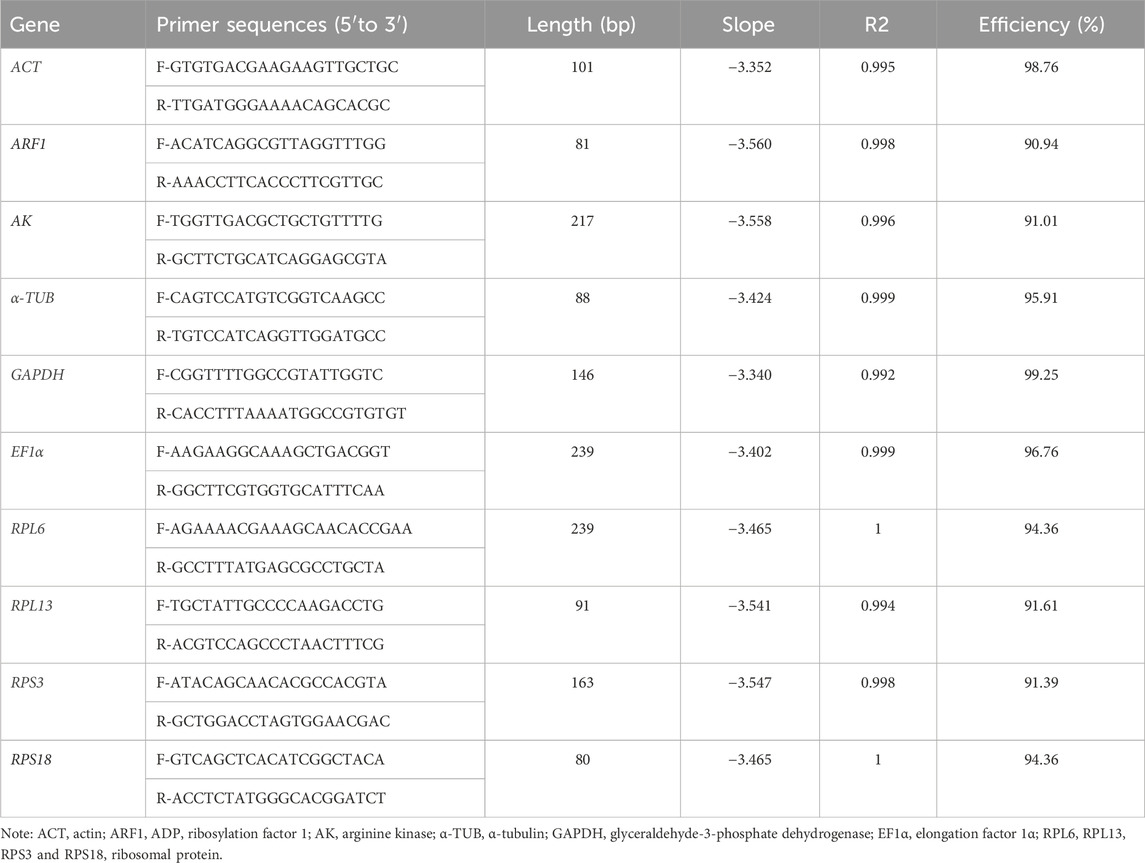
The products obtained from qRT-PCR were validated through sequencing. The primer specificity for qRT-PCR was confirmed through melting curve analysis. As expected, slopes of all primer pairs were less than −3.0, and regression coefficient (R2) and efficacy values ranged from 0.992-1 and 90.94%–99.25%, respectively (Table 1). Our findings demonstrated that efficiency met the required standards for traditional qRT-PCR (Bustin et al., 2009).
Stability of the ten HKGs
Samples were collected from three adult ages (the first to third days of the newly-emerged adults), six adult tissues (foregut, midgut, hindgut, epidermis, trachea and tentacle), five temperature treatments (4 °C, 15 °C, 20 °C, 26 °C and 35 °C) and both sexes (5 days after emerging). We discovered that the mRNA levels of ten HKGs were abundant at various adult ages, during diverse adult tissues, under different temperatures and both sexes using qRT-PCR,.
The all cycle threshold values (Ct) under diverse tested backgrounds were presented (Figure 1). The ΔCt method assesses the genes stability based on genes average of STDEV (Silver et al., 2006). During diverse adult ages, the expression fluctuations of RPL6 and AK were smaller, whereas ACT and GAPDH were higher (Figure 1A). Under diverse tissues, except for AK, the expression variations were small in ten HKGs (Figure 1B). Under various temperatures, the expression difference of RPL13 and ARF1 was smaller (Figure 1C). In males and females, the expression variations of AK and EF1α were small in ten HKGs (Figure 1D).
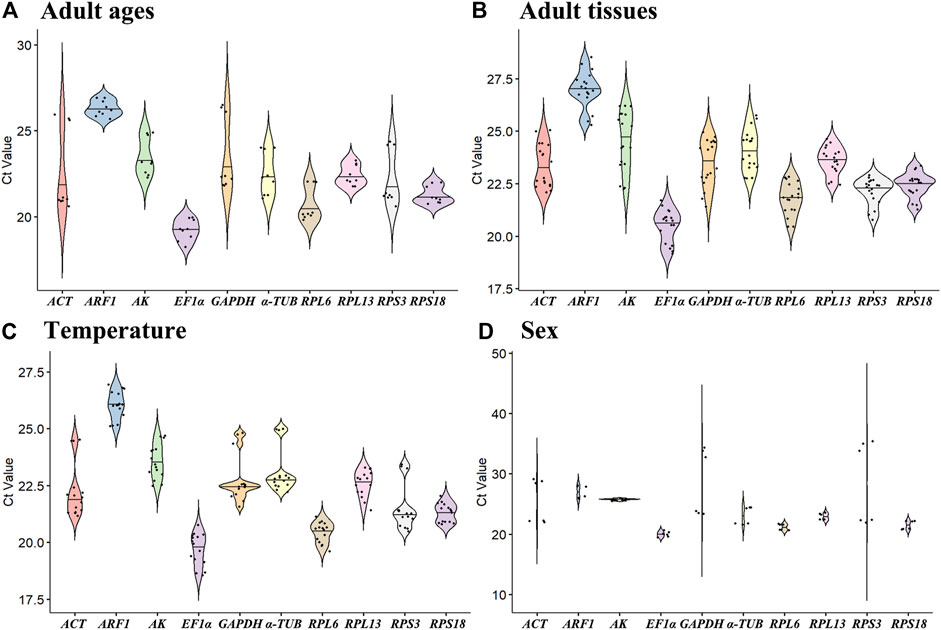
Figure 1. Expression levels of ten house-keeping genes in Mylabris sibirica. The mean Ct values for 10 candidate reference genes are shown in four different experiments: (A) adult ages, (B) adult tissues, (C) temperature, (D) sex. Mean Ct values for the eight candidate reference genes are presented in box plots, where each box indicates the 25th and 75th percentiles, and the line across the box represents the median. Abbreviation: ACT, actin; ARF1, ADP ribosylation factor 1; AK, arginine kinase; α-TUB, α-tubulin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; EF1α, elongation factor 1α; RPL6, RPL13, RPS3 and RPS18, ribosomal protein. The abbreviations are exactly the same as Figure 2 through Figure 6.
Stability of the ten HKGs under adult ages
The geNorm statistical analysis assesses the gene stability by M-values (the average expression stability) and V-values (pairwise variation). These results showed that EF1α and RPL13 were the most stable internal genes during various adult ages, with M-values below 0.25. In contrast, the most unstable genes were ACT and GAPDH (Figure 2A; Table 2). Pairwise variation analysis showed that all values were below 0.15, demonstrating that two reference genes were needed for the gene expression analysis under adult ages (Figure 2B).
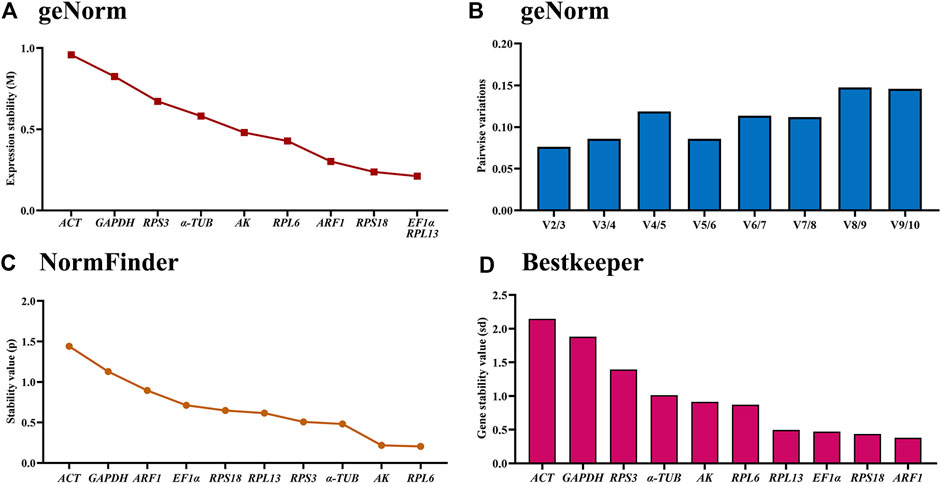
Figure 2. Stability of the ten house-keeping genes in Mylabris sibirica during diverse adult ages. All stages of Mylabris sibirica adults were sampled (collected on the first to third days of the newly-emerged adults). The expression stability rankings are determined by geNorm, NormFinder and BestKeeper.
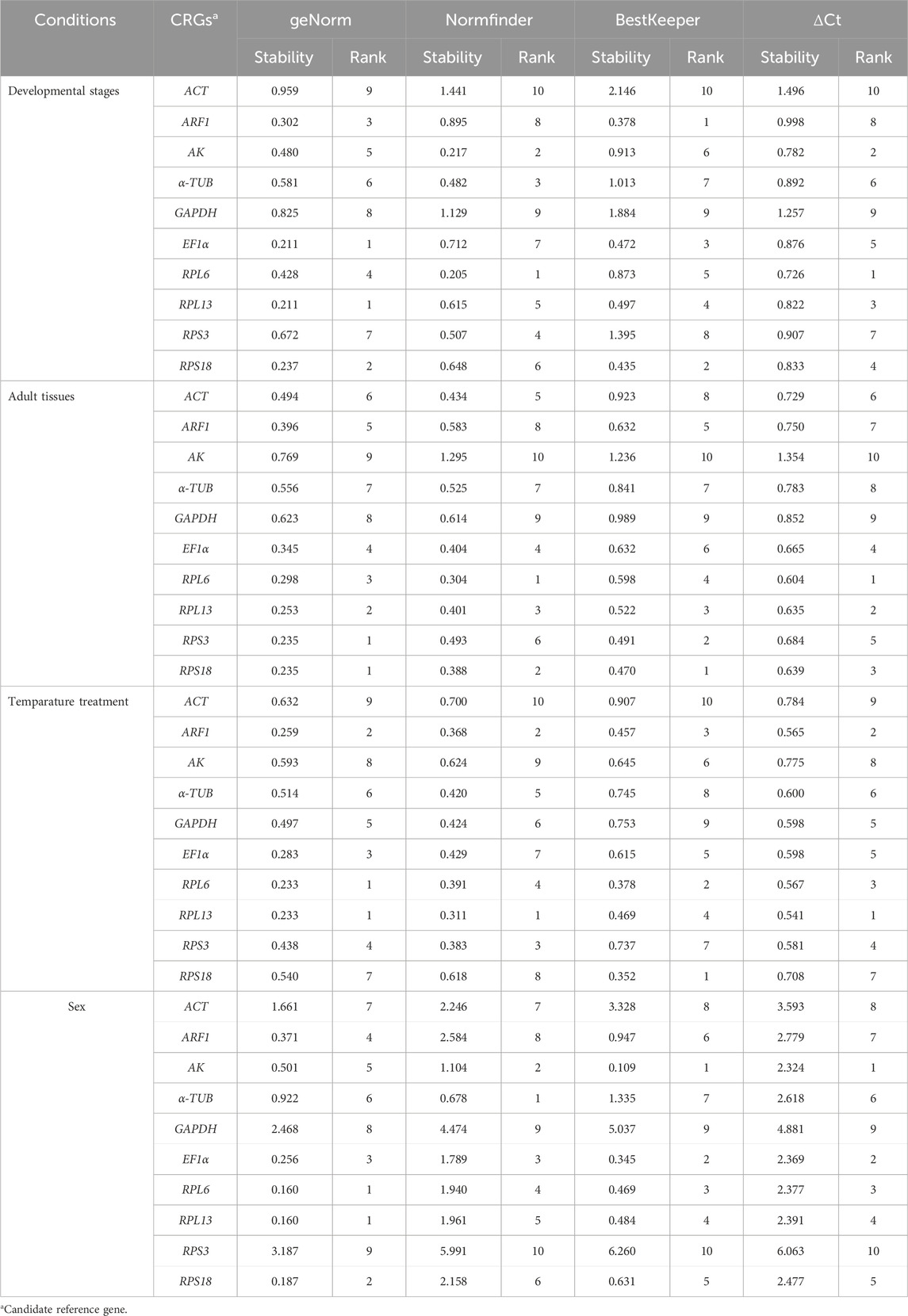
Table 2. Expression stability of the candidate reference genes under different experimental conditions.
Based on the NormFinder algorithm, the stable rankings of ten internal genes from the most stable to the least were RPL6, AK, α-TUB, RPS3, RPL13, RPS18, EF1α, ARF1, RPL27, GAPDH and ACT (Figure 2C; Table 2). Except for GAPDH and ACT, the p values of other genes were had less than 1 (Figure 2C; Table 2).
According to the BestKeeper analysis, ARF1, RPS18, EF1α, RPL13, RPL6, AK were the most stable, whose standard deviation (SD) values from raw Ct values were 0.378, 0.435, 0.472, 0.497, 0.873 and 0.913, respectively. In constrast, the SD values of α-TUB, RPS3, GAPDH and ACT were more than 1.0 (Figure 2D; Table 2).
The stability of the ten HKGs were compared and ordered by an online tool RefFinder: RPL6>RPL13>RPS18>AK>ARF1>EF1α>α-TUB>RPS3>GAPDH>ACT (Figure 6A). Thus, RPL6 and RPL13 are the optimal internal gene pair to calculate gene expression during various adult ages (Table 3).
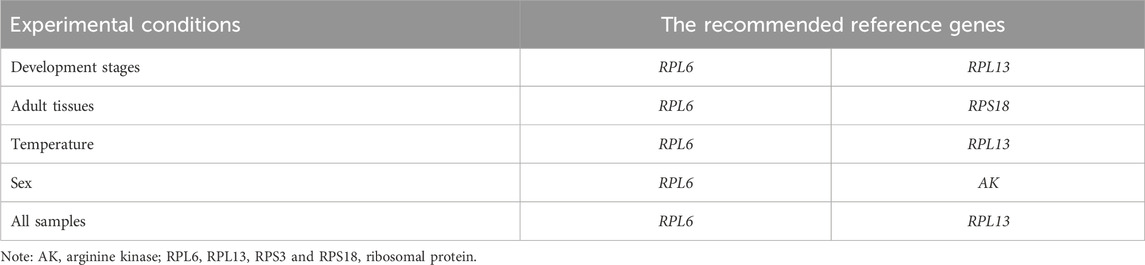
Table 3. A list of the recommended reference genes in M. sibirica for different experimental conditions.
Stability of the ten HKGs during different adult tissues
According to the geNorm data, the reference genes rankings ranging from the most stable to the least stable were RPS3, RPS18, RPL13, RPL6, EF1α, ARF1, ACT, α-TUB, GAPDH and AK (Figure 3A; Table 2). Pairwise variation analysis showed that all values were less than 0.15, indicating that two reference genes from different tissues were needed for the gene expression analysis (Figure 3B).
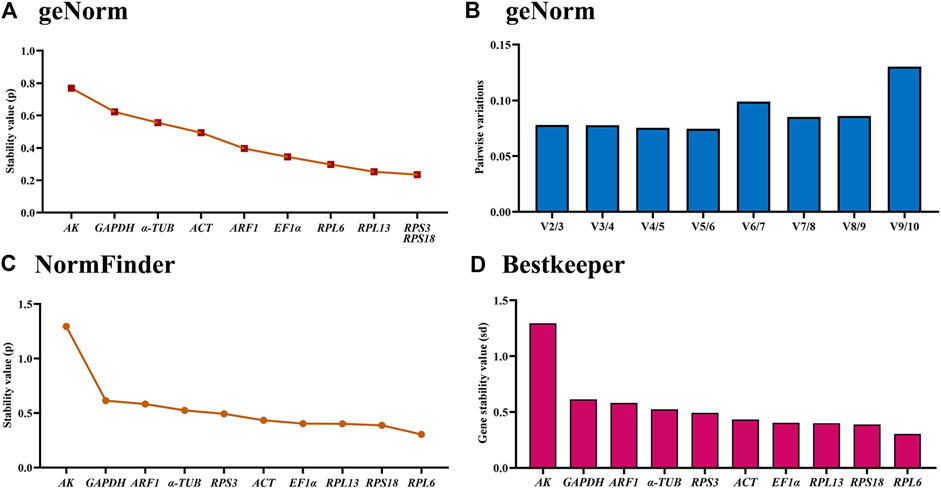
Figure 3. Stabilities of the ten house-keeping genes in Mylabris sibirica among various adult tissues. Foregut, midgut, hindgut, epidermis, trachea and antenna were dissected from the newly-emerged adults. The expression stability rankings are determined by geNorm, NormFinder and BestKeeper.
Based on the NormFinder result, the reference genes rankings were as follows: RPL6>RPS18>RPL13>EF1α>ACT>RPS3>α-TUB>ARF1>GAPDH>AK (Figure 3C; Table 2). Again, except for AK, the p values of other reference genes were below 1.0 (Figure 3C; Table 2).
The BestKeeper analysis showed that RPL6, were the most stable, with the SD values of 0.604 (Figure 3D; Table 2). Again, except for AK, the SD values of other reference genes were below 1.0 (Figure 3D; Table 2).
RefFinder provided a thorough ranking order ranging from the most stable to the least stable:RPS18>RPL6>RPL13>RPS3>EF1α>ARF1>ACT>α-TUB>GAPDH>AK (Figure 6B). Thus, RPL6 and RPS18 are the most suitable internal gene pair to measure gene expression under different adult tissues (Table 3).
Expression stability of the ten HKGs under various temperature treaments
The geNorm analysis showed that RPL6, RPL13, ARF1 and EF1α were the most stable internal genes under different temperatures, whose M-values were 0.233, 0.233, 0.259 and 0.283, respectively (Figure 4A; Table 2). Moreover, pairwise variation analysis showed that all values were less than 0.15, indicating that two different internal genes are needed for testing gene expression during various temperature treaments (Figure 4B).
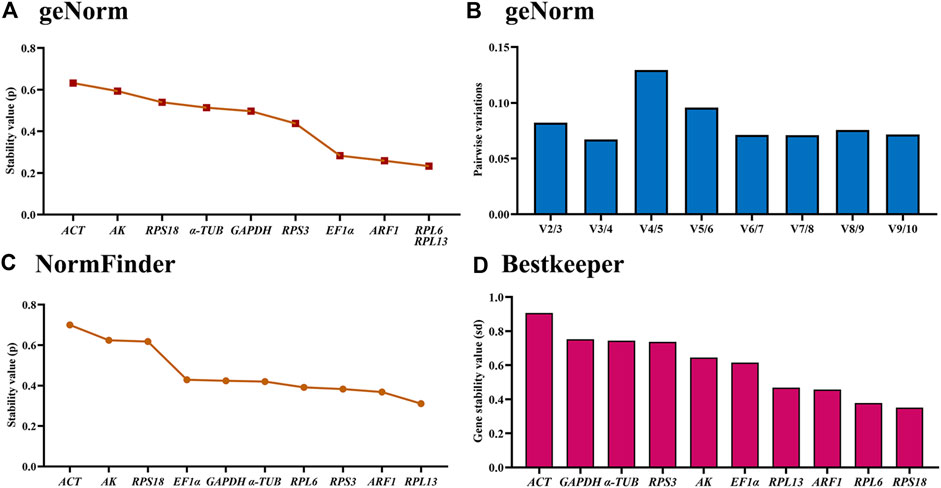
Figure 4. Stability of the ten house-keeping genes in Mylabris sibirica under different temperatures. The newly-emerged adults reared under five temperatures (4°C, 15°C, 20°C, 26 °C and 35 °C) were collected. The expression stability rankings are determined by geNorm, NormFinder and BestKeeper.
The NormFinder data uncovered that the steady rankings were RPL13, ARF1, RPS3, RPL6, α-TUB, GAPDH, EF1α, RPS18, AK and ACT (Figure 4C; Table 2).
BestKeeper data showed that RPS18, RPL6, ARF1 and RPL13 were the most optimal genes, whose SD values were 0.352, 0.378, 0.457 and 0.469, respectively (Figure 4D; Table 2). Again, these genes showed values less than 0.5, suggesting their stabilities were similar.
Based on the RefFinder data, the stability orders were as follows: RPL13>RPL6>ARF1>RPS3>RPS18>EF1α>GAPDH>α-TUB >AK>ACT (Figure 6C). Thus, RPL6 and RPL13 are the best internal gene pair for assessing mRNA levels of genes under various temperature treaments (Table 3).
Stability of the ten HKGs in males and females
Based on the geNorm algorithm results, RPL6, RPL13, RPS18, EF1α and ARF1 were the most stable, with the M-values less than 0.5 (Figure 5A; Table 2). In addition, pairwise variation data manifested that the V2/3 to V5/6 values were less than 0.15, indicating that two different internal genes are suitable for evaluating gene expression levels in males and females (Figure 5B).
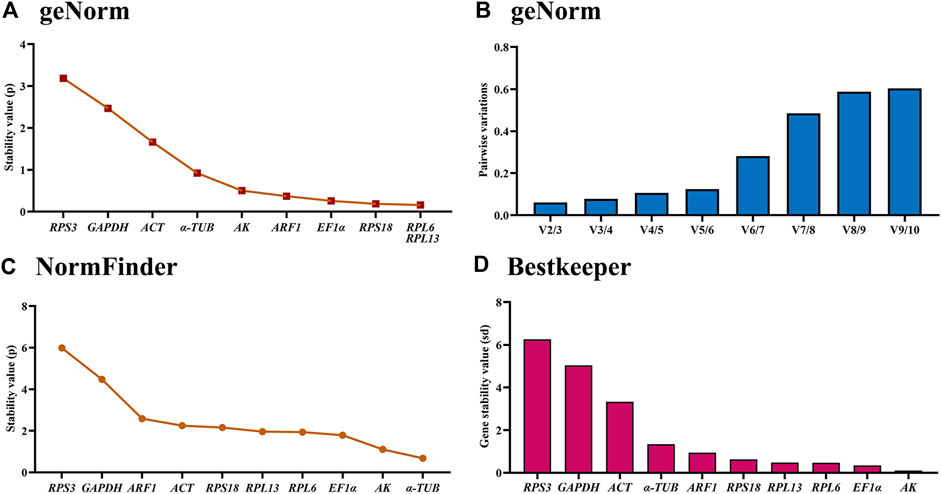
Figure 5. Stabilities of the ten house-keeping genes in Mylabris sibirica in males and females. The male and female adults were collected at day 5 after emerging. The expression stability rankings are determined by geNorm, NormFinder and BestKeeper.
Based on the NormFinder data, the stability rankings were as follows: α-TUB>AK>EF1α>RPL6>RPL13>RPS18>ACT>ARF1> GAPDH>RPS3 (Figure 5C; Table 2). Again, except for α-TUB, the p values of other reference genes were above 1.0 (Figure 5C; Table 2).
The BestKeeper result demonstrated that AK, EF1α, RPL6 and RPL13 were the most optimal genes, whose SD values were 0.109, 0.345, 0.469 and 0.484, respectively (Figure 4D; Table 2). Again, values of these genes were less than 0.5, showing their similar stabilities.
The RefFinder showed a comprehensive stability ranking: AK>RPL6>EF1α>RPL13>α-TUB>RPS18>ARF1>ACT>GAPDH>RPS3 (Figure 6D). Therefore, AK and RPL6 are considered as the most optimal internal gene pair (Table 3).

Figure 6. Stabilities of the ten house-keeping genes in Mylabris sibirica in different samples. The stability of the reference genes as calculated by the Geomean method of RefFinder. A lower Geomean of ranking value denotes more stable expression. (A) Adult ages, (B) adult tissues, (C) temperature, (D) sex, (E) all samples.
When combining the four diverse experimental conditions together, the RefFinder data showed a overall stability ranking: RPL13>RPL6>RPS18>EF1α>α-TUB>ARF1>AK>ACT>GAPDH>RPS3 (Figure 6E). In summary, RPL6 and RPL13 can be best internal genes for gene expression and microbial abundance determination in M. sibirica (Table 3).
Validation of the selected reference genes
To assess the stability of the candidate reference genes, the relative expression level of UAP was assessed in the epidermis, foregut, midgut, hindgut, trachea and antenna. The following reference genes were used to normalize: RPL6 + RPS18 (the most stable reference gene), and AK and GAPDH (the least stable reference gene). The highest accumulated mRNA level of MsUAP was detected in the midgut, hindgut and trachea, followed by those in the epidermis, the lowest level was found in the foregut and antenna. However, AK and GAPDH was used as reference genes, and MsUAP was highly expressed in the trachea, lowly expressed in the epidermis (Figure 7).
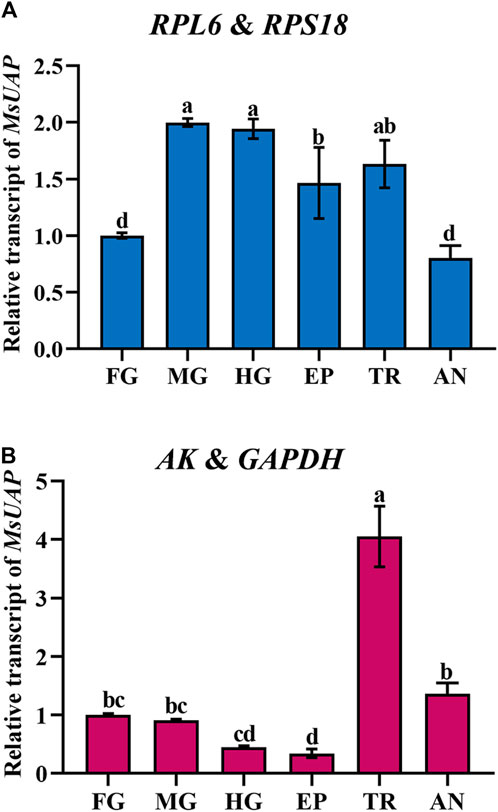
Figure 7. Relative gene expression of UAP in different adult tissues of Mylabris sibirica. The relative gene expression level of UAP in the foregut, midgut, hindgut, epidermis, trachea and antenna was normalized to the best stable [(A) RPL6 and RPS18] and least stable [(B) AK and GAPDH] reference genes, respectively. The values are means +SE. Different letters indicate significant differences in gene expression among different tissues (p < 0.05).
Discussion
Quantitative real-time PCR (qRT-PCR) is a popular method for assessing gene expression (Derveaux et al., 2010; Jozefczuk and Adjaye, 2011) and microbial abundance determination (Ali et al., 2018a; Ali et al., 2018b; Ali et al., 2018c; Ali et al., 2019). Optimal reference genes are critical greatly in eliminating heterogeneity in diverse datasets and improving the quantitative results (Bustin et al., 2009). An ideal internal gene are abundantly expressed under diverse experimental treatments (Derveaux et al., 2010). However, not all internal genes remains suitable in various species (Zhang et al., 2022). Therefore, selecting suitable references must be conducted before qRT-PCR. Studies on internal genes have been reported in many insect species (Shakeel et al., 2018), such as Aphidoletes aphidimyza (Shen et al., 2023), Nilaparvata lugens (Zhang et al., 2022), Phthorimaea operculella (Shen et al., 2022), H. vigintioctomaculata (Zhang et al., 2022) and P. striolata (Guo et al., 2023). As an oilseed rape pest, evaluating optimal reference genes are helpful in studying molecular mechanisms in M. sibirica. As we all knows, the current research is the first report on reference genes assessment in M. sibirica.
In the current text, the stability of ten HKGs for M. sibirica (ACT, ARF1, AK, EF1α, GAPDH, α-TUB, RPL6, RPL13, RPS3 and RPS18) were investigated under four experiment treaments (adult ages, adult tissues, temperature and sex), with five methods (Ct value, geNorm, NormFinder, BestKeeper and RefFinder).
Our findings indicated that the best reference gene pairs were RPL6 and RPL13 during different adult ages (Figures 1, 2, 6; Table 2) and under diverse temperatures (Figures 1, 4, 6; Table 2), RPL6 and RPS18 in various adult tissues (Figures 1, 3, 6; Table 2). Moreover, RPL6 and RPL13 were the most stable reference gene pair in tested backgrounds (Figure 6). These results also demonstrated that ribosomal protein genes were the most optimal internal genes to meassur target gene expression in M. sibirica. As we all know, ribosomal proteins play an important part in ribosome assembly. They combine with four ribosomal RNAs (rRNAs) to form the ribosomal subunits, which have an important function cellular protein translation (Landry-Voyer et al., 2023). Consistent with our findings, reference genes involving ribosomal protein genes have been selected frequently for expression analysis in various insects during the last 10 years (Zhang et al., 2022; Shen et al., 2022). They are selected as reference genes in Coleopterans Ips sexdentatus (RPS3) (Sellamuthu et al., 2021), Tribolium castaneum (RPS6, RPL13a, RPS3 and RPL18) (Toutges et al., 2010), L. decemlineata (RP18 and RP4) (Shi et al., 2013), H. vigintioctopunctata (RPL13 and RPS18) (Lü et al., 2018), H. vigintioctomaculata (RPS18 and RPL13) (Zhang et al., 2022) and P. brassicae (RPL32 and RPL19) (Ma et al., 2021), Lepidopteran species Helicoverpa armigera (RPS15 and RPL27) (Zhang et al., 2015), Plutella xylostella (RPS13 and RPS23) (Fu et al., 2013) and P. operculella (RPL13) (Shen et al., 2022), Thysanopterans Frankliniella occidentalis (RPL32) (Zheng et al., 2014), Dipterans Chlorops oryzae (RPS15) (Tian et al., 2019) and A. aphidimyza (RPL8 and RPS3) (Shen et al., 2023), Hymenopterans such as Apis mellifera (RPS5 and RPS18) (Jeon et al., 2020) and Anastatus japonicus Ashmead (RPL13 and RPS6) (Liu et al., 2022), Hemipterans N. lugens (RPL5, RPS8 and RPL14) (Zhang et al., 2022), Aphis glycines (RPS9) (Bansal et al., 2012), Dichelops melacanthus (RPL9 and RPS23) (Pinheiro et al., 2020), Diaphorina citri (RPL7) (Bassan et al., 2017) and Rhopalosiphum padi (RPL13, RPS6 and RPS18) (Li et al., 2021), and Orthopteran Locusta migratoria (RPL32) (Yang et al., 2014), as well as Acari Brevipalpus yothersi (RPL13 and RPL32) (Rogerio et al., 2019).
In male and female adults, the most reliable reference genes were AK and RPL6 (Figures 1, 5, 6; Table 2). As we discussed in the previous paragraph, RPL6 was expressed abundantly in males and females. Arginine kinase (AK) has a prominent function in invertebrate energy metabolism, which catalyzes the reversible phosphorylation of l-arginine to make up phosphoarginine (Shi et al., 2012). Similar to our results, the arginine kinase gene has been selected as the most suitable gene for expression assessment in Xylosandrus germanus (Patwa et al., 2021), Spodoptera frugiperda (Han et al., 2021) and Spodoptera litura (Lu et al., 2013).
Moreover, The BestKeeper results demonstrated the SD values of ACT, α-TUB and GAPDH were above 1.0 (Table 2), suggesting that the three reference genes were unsuitable as internal genes for RT-qPCR.
Actin (ACT) is extremely abundant in eukaryotes, and has an vital function in cellular activities, including cell motility and the regulation of transcription cell secretion (Dominguez and Holmes, 2011). At present, many publictions have verified that the expression level of ACT is less steady in various insects, such as P. operculella (RPL13) (Shen et al., 2022), H. vigintioctopunctata (RPL13 and RPS18) (Lü et al., 2018) and Hippodamia convergens (Yang et al., 2016).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a key enzyme participating in glycolysis and glucose metabolism (Zhao et al., 2022). Similar to our results, the instability of GAPDH expression has reported in Ophraella communa (Zhang et al., 2020) P. brassicae (Ma et al., 2021) and Colaphellus bowringi (Tan et al., 2015).
Microtubule α-tubulin (α-TUB), interacts with many microtubule-associated proteins to conduct a variety of cellular functions, such as intracellular transport and cell division (Hammond et al., 2008). Currently, many studies indicated that the stability of GAPDH expression was greatly unsteady in P. operculella (Shen et al., 2022), H. vigintioctomaculata (Zhang et al., 2022), N. lugens (RPL5, RPS8 and RPL14) (Zhang et al., 2022) and A. aphidimyza (Shen et al., 2023), In brief, our results recommend RPL6 and RPL13 as the most stable internal gene pair under four tested backgrounds (Figure 6; Table 3). To further validate the reference genes in M. sibirica, we assessed the relative expression level of UAP in different adult tissues. Our findings revealed that UAP expression pattern was inconsistent in the different adult tissues when normalized to the two best- and least-stable reference genes (Figure 7). These results showed that the unreasonable use of reference genes may lead to inaccurate results for target genes. Therefore, it is crucial to choose and validate the best reference genes to ensure the accuracy of gene expression. Our study would provide a foundation for further gene molecular functions and microbial abundance determination in M. sibirica.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because the study is about pest, not involving animal ethics.
Author contributions
C-HS: Writing–original draft, Conceptualization, Data curation, Formal Analysis, Methodology. MT: Writing–original draft, Data curation, Formal Analysis, Methodology. X-FL: Data curation, Methodology, Writing–original draft. LZ: Methodology, Writing–original draft. WL: Methodology, Writing–original draft. PD: Formal Analysis, Writing–original draft. QZ: Formal Analysis, Writing–original draft. GW: Conceptualization, Funding acquisition, Writing–original draft. X-HY: Conceptualization, Funding acquisition, Methodology, Writing–original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Major Special Project of Breeding for Genetically Modified Organisms in China (No. 2016ZX08012-005) and the Innovation Project of Chinese Academy of Agricultural Sciences (No. 2060302-049-091).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2024.1345836/full#supplementary-material
References
Ali H., Muhammad A., Bala N. S., Wang G., Chen Z., Peng Z., et al. (2018a). Genomic evaluations of Wolbachia and mtDNA in the population of coconut hispine beetle, Brontispa longissima (Coleoptera: Chrysomelidae). Mol. Phylogenet Evol. 127, 1000–1009. doi:10.1016/j.ympev.2018.07.003
Ali H., Muhammad A., Islam S. U., Islam W., Hou Y. (2018b). A novel bacterial symbiont association in the hispid beetle, Octodonta nipae (Coleoptera: Chrysomelidae), their dynamics and phylogeny. Microb. Pathog. 118, 378–386. doi:10.1016/j.micpath.2018.03.046
Ali H., Muhammad A., Sanda N. B., Huang Y., Hou Y. (2019). Pyrosequencing uncovers a shift in bacterial communities across life stages of Octodonta nipae (Coleoptera: Chrysomelidae). Front. Microbiol. 10, 466. doi:10.3389/fmicb.2019.00466
Ali H., Muhammad A., Sanda Bala N., Hou Y. (2018c). The endosymbiotic Wolbachia and host COI gene enables to distinguish between two invasive palm pests; coconut leaf beetle, Brontispa longissima and hispid leaf beetle, Octodonta nipae. J. Econ. Entomol. 111 (6), 2894–2902. doi:10.1093/jee/toy233
Andersen C. L., Jensen J. L., Ørntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64 (15), 5245–5250. doi:10.1158/0008-5472.Can-04-0496
Bansal R., Mamidala P., Mian M. A., Mittapalli O., Michel A. P. (2012). Validation of reference genes for gene expression studies in Aphis glycines (Hemiptera: Aphididae). J. Econ. Entomol. 105 (4), 1432–1438. doi:10.1603/ec12095
Bassan M. M., Angelotti-Mendonc A. J., Alves G. R., Yamamoto P. T., Moura O. F. F. A. A. (2017). Selection of reference genes for expression studies in Diaphorina citri (Hemiptera: Liviidae). J. Econ. Entomol. 110 (6), 2623–2629. doi:10.1093/jee/tox253
Bologna M. A., D'Inzillo B., Cervelli M., Oliverio M., Mariottini P. (2005). Molecular phylogenetic studies of the Mylabrini blister beetles (Coleoptera, Meloidae). Mol. Phylogenet Evol. 37 (1), 306–311. doi:10.1016/j.ympev.2005.03.034
Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 (4), 611–622. doi:10.1373/clinchem.2008.112797
ČernÝ L., Vrabec V. (2019). Mylabris (Mylabris) snizeki sp. nov. from Jordan, with a key to the Jordanian species of the nominotypical subgenus (Coleoptera: Meloidae). Zootaxa 4555 (1), 146–150. doi:10.11646/zootaxa.4555.1.13
Derveaux S., Vandesompele J., Hellemans J. (2010). How to do successful gene expression analysis using real-time PCR. Methods 50 (4), 227–230. doi:10.1016/j.ymeth.2009.11.001
Dominguez R., Holmes K. C. (2011). Actin structure and function. Annu. Rev. Biophys. 40, 169–186. doi:10.1146/annurev-biophys-042910-155359
Fu W., Xie W., Zhang Z., Wang S., Wu Q., Liu Y., et al. (2013). Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 9 (8), 792–802. doi:10.7150/ijbs.5862
Guo M., Gao R., Nanda S., Li Y., Guo C., Zhou X., et al. (2023). RNAi assays in the striped flea beetle (Phyllotreta striolata) suggest Psγ-COPI and PsArf1COPI as potential molecular targets for pest control. Pestic. Biochem. Physiol. 193, 105428. doi:10.1016/j.pestbp.2023.105428
Hammond J. W., Cai D., Verhey K. J. (2008). Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 20 (1), 71–76. doi:10.1016/j.ceb.2007.11.010
Han S., Qin Q., Wang D., Zhou Y., He Y. (2021). Selection and evaluation of reference genes for qRT-PCR in Spodoptera frugiperda (Lepidoptera: Noctuidae). Insects 12 (10), 902. doi:10.3390/insects12100902
Jeon J. H., Moon K., Kim Y., Kim Y. H. (2020). Reference gene selection for qRT-PCR analysis of season- and tissue-specific gene expression profiles in the honey bee Apis mellifera. Sci. Rep. 10 (1), 13935. doi:10.1038/s41598-020-70965-4
Jozefczuk J., Adjaye J. (2011). Quantitative real-time PCR-based analysis of gene expression. Methods Enzymol. 500, 99–109. doi:10.1016/b978-0-12-385118-5.00006-2
Landry-Voyer A. M., Mir Hassani Z., Avino M., Bachand F. (2023). Ribosomal protein uS5 and friends: protein-protein interactions involved in ribosome assembly and beyond. Biomolecules 13 (5), 853. doi:10.3390/biom13050853
Li M., Li X., Wang C., Li Q., Zhu S., Zhang Y., et al. (2021). Selection and validation of reference genes for qRT-PCR analysis of Rhopalosiphum padi (Hemiptera: Aphididae). Front. Physiol. 12, 663338. doi:10.3389/fphys.2021.663338
Liu Z., Xiao J., Xia Y., Wu Q., Zhao C., Li D. (2022). Selection and validation of reference genes for RT-qPCR-based analyses of Anastatus japonicus Ashmead (Hymenoptera: Helicopteridae). Front. Physiol. 13, 1046204. doi:10.3389/fphys.2022.1046204
Lü J., Yang C., Zhang Y., Pan H. (2018). Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: a systematic review. Front. Physiol. 9, 1560. doi:10.3389/fphys.2018.01560
Lu Y., Yuan M., Gao X., Kang T., Zhan S., Wan H., et al. (2013). Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS One 8 (7), e68059. doi:10.1371/journal.pone.0068059
Ma L., Jiang T., Liu X., Xiao H., Peng Y., Zhang W. (2021). Evaluation of candidate reference genes for gene expression analysis in the brassica leaf beetle, Phaedon brassicae (Coleoptera: Chrysomelidae). PLoS One 16 (6), e0251920. doi:10.1371/journal.pone.0251920
Pan H., Yang X., Siegfried B. D., Zhou X. (2015). A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae). PLoS One 10 (4), e0125868. doi:10.1371/journal.pone.0125868
Pan Z., Ren G. D. (2020). New synonyms, combinations and status in the Chinese species of the family Meloidae Gyllenhal, 1810 (Coleoptera: Tenebrionoidea) with additional faunistic records. Zootaxa 4820 (2), zootaxa.4820.2.3. zootaxa.4820.4822.4823. doi:10.11646/zootaxa.4820.2.3
Patwa N., Ranger C. M., Lehenberger M., Biedermann P. H., Reding M. E. (2021). Stability of nuclear and mitochondrial reference genes in selected tissues of the ambrosia beetle Xylosandrus germanus. Insects 12 (12), 1125. doi:10.3390/insects12121125
Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26 (6), 509–515. doi:10.1023/b:bile.0000019559.84305.47
Pinheiro D. H., Moreira R. O., Leite N. A., Redoan A. C., Xavier A. D. S., Barros B. A., et al. (2020). Suitable reference genes for RT-qPCR analysis in Dichelops melacanthus (Hemiptera: Pentatomidae). Mol. Biol. Rep. 47 (7), 4989–5000. doi:10.1007/s11033-020-05550-z
Rogerio L. A., Galdeano D. M., Arena G. D., Nunes M. A., Machado M. A., Novelli V. M. (2019). Reference genes for gene expression studies by RT-qPCR in Brevipalpus yothersi (Acari: Tenuipalpidae), the mite vector of citrus leprosis virus. Sci. Rep. 9 (1), 6536. doi:10.1038/s41598-019-42993-2
Sang W., He L., Wang X. P., Zhu-Salzman K., Lei C. L. (2015). Evaluation of reference genes for RT-qPCR in Tribolium castaneum (Coleoptera: Tenebrionidae) under UVB stress. Environ. Entomol. 44 (2), 418–425. doi:10.1093/ee/nvv010
Sellamuthu G., Amin S., Bílý J., Synek J., Modlinger R., Sen M. K., et al. (2021). Reference gene selection for normalizing gene expression in Ips sexdentatus (Coleoptera: Curculionidae: Scolytinae) under different experimental conditions. Front. Physiol. 12, 752768. doi:10.3389/fphys.2021.752768
Sellamuthu G., Bílý J., Joga M. R., Synek J., Roy A. (2022). Identifying optimal reference genes for gene expression studies in Eurasian spruce bark beetle, Ips typographus (Coleoptera: Curculionidae: Scolytinae). Sci. Rep. 12 (1), 4671. doi:10.1038/s41598-022-08434-3
Shakeel M., Rodriguez A., Tahir U. B., Jin F. (2018). Gene expression studies of reference genes for quantitative real-time PCR: an overview in insects. Biotechnol. Lett. 40 (2), 227–236. doi:10.1007/s10529-017-2465-4
Shen C. H., Peng L. J., Zhang Y. X., Zeng H. R., Yu H. F., Jin L., et al. (2022). Reference genes for expression analyses by qRT-PCR in Phthorimaea operculella (Lepidoptera: Gelechiidae). Insects 13 (2), 140. doi:10.3390/insects13020140
Shen X. X., Zhang G. Q., Zhao Y. X., Zhu X. X., Yu X. F., Yang M. F., et al. (2023). Selection and validation of optimal reference genes for RT-qPCR analyses in Aphidoletes aphidimyza Rondani (Diptera: Cecidomyiidae). Front. Physiol. 14, 1277942. doi:10.3389/fphys.2023.1277942
Shi X., Wang L., Zhou Z., Yang C., Gao Y., Wang L., et al. (2012). The arginine kinase in Zhikong scallop Chlamys farreri is involved in immunomodulation. Dev. Comp. Immunol. 37 (2), 270–278. doi:10.1016/j.dci.2012.03.008
Shi X. Q., Guo W. C., Wan P. J., Zhou L. T., Ren X. L., Ahmat T., et al. (2013). Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say). BMC Res. Notes 6, 93. doi:10.1186/1756-0500-6-93
Silver N., Best S., Jiang J., Thein S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7, 33. doi:10.1186/1471-2199-7-33
Tan Q. Q., Zhu L., Li Y., Liu W., Ma W. H., Lei C. L., et al. (2015). A de novo transcriptome and valid reference genes for quantitative real-time PCR in Colaphellus bowringi. PLoS One 10 (2), e0118693. doi:10.1371/journal.pone.0118693
Tian P., Qiu L., Zhou A., Chen G., He H., Ding W., et al. (2019). Evaluation of appropriate reference genes for investigating gene expression in Chlorops oryzae (Diptera: Chloropidae). J. Econ. Entomol. 112 (5), 2207–2214. doi:10.1093/jee/toz142
Tian Yubo R. Y., Liu H., Mao Y. (2021). Distribution characteristics of the main medicinal insects of Meloidae in the Hasi mountain, Gansu province. Sichuan J. Zoology 40 (4), 383–393.
Toutges M. J., Hartzer K., Lord J., Oppert B. (2010). Evaluation of reference genes for quantitative polymerase chain reaction across life cycle stages and tissue types of Tribolium castaneum. J. Agric. Food Chem. 58 (16), 8948–8951. doi:10.1021/jf101603j
Valasek M. A., Repa J. J. (2005). The power of real-time PCR. Adv. Physiol. Educ. 29 (3), 151–159. doi:10.1152/advan.00019.2005
Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3 (7), Research0034. doi:10.1186/gb-2002-3-7-research0034
Xie F., Wang J., Zhang B. (2023). RefFinder: a web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genomics 23 (2), 125. doi:10.1007/s10142-023-01055-7
Xie F., Xiao P., Chen D., Xu L., Zhang B. (2012). miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80, 75–84. doi:10.1007/s11103-012-9885-2
Xu J., Welker D. L., James R. R. (2021). Variation in expression of reference genes across life stages of a bee, Megachile rotundata. Insects 12 (1), 36. doi:10.3390/insects12010036
Yang C., Preisser E. L., Zhang H., Liu Y., Dai L., Pan H., et al. (2016). Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front. Plant Sci. 7, 1672. doi:10.3389/fpls.2016.01672
Yang Q., Li Z., Cao J., Zhang S., Zhang H., Wu X., et al. (2014). Selection and assessment of reference genes for quantitative PCR normalization in migratory locust Locusta migratoria (Orthoptera: Acrididae). PLoS One 9 (6), e98164. doi:10.1371/journal.pone.0098164
Yang X., Pan H., Yuan L., Zhou X. (2018). Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 8 (1), 2689. doi:10.1038/s41598-018-20612-w
Yu J., Su Y., Sun J., Liu J., Li Z., Zhang B. (2020). Selection of stable reference genes for gene expression analysis in sweet potato (Ipomoea batatas L.). Mol. Cell Probes 53, 101610. doi:10.1016/j.mcp.2020.101610
Zhang S., An S., Li Z., Wu F., Yang Q., Liu Y., et al. (2015). Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 555 (2), 393–402. doi:10.1016/j.gene.2014.11.038
Zhang Y., Chen J., Chen G., Ma C., Chen H., Gao X., et al. (2020). Identification and validation of reference genes for quantitative gene expression analysis in Ophraella communa. Front. Physiol. 11, 355. doi:10.3389/fphys.2020.00355
Zhang Y., Zeng L., Wei Y., Zhang M., Pan W., Sword G. A., et al. (2022b). Reliable reference genes for gene expression analyses under the hypomagnetic field in a migratory insect. Front. Physiol. 13, 954228. doi:10.3389/fphys.2022.954228
Zhang Y.-X., Tan Q., Shen C.-H., Wu J.-J., Wu Y.-K., Li W.-Z., et al. (2022a). Reference gene selection for transcriptional profiling by RT-qPCR in the 28-spotted larger potato ladybird. J. Asia-Pac Entomol. 25 (2), 101900. doi:10.1016/j.aspen.2022.101900
Zhao W., Zhang B., Geng Z., Chang Y., Wei J., An S. (2022). The uncommon function and mechanism of the common enzyme glyceraldehyde-3-phosphate dehydrogenase in the metamorphosis of Helicoverpa armigera. Front. Bioeng. Biotechnol. 10, 1042867. doi:10.3389/fbioe.2022.1042867
Keywords: Mylabris sibirica, reference gene, RT-qPCR, RefFinder, geNorm
Citation: Shen C-H, Tang M, Li X-F, Zhu L, Li W, Deng P, Zhai Q, Wu G and Yan X-H (2024) Evaluation of reference genes for quantitative expression analysis in Mylabris sibirica (Coleoptera, Meloidae). Front. Physiol. 15:1345836. doi: 10.3389/fphys.2024.1345836
Received: 20 December 2023; Accepted: 25 March 2024;
Published: 08 April 2024.
Edited by:
Natraj Krishnan, Mississippi State University, United StatesReviewed by:
Habib Ali, Khwaja Fareed University of Engineering and Information Technology (KFUEIT), PakistanKang He, Zhejiang University, China
Copyright © 2024 Shen, Tang, Li, Zhu, Li, Deng, Zhai, Wu and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Wu, d3VnYW5nQGNhYXMuY24=; Xiao-Hong Yan, eWFueGlhb2hvbmdAY2Fhcy5jbg==
 Chen-Hui Shen1
Chen-Hui Shen1 Gang Wu
Gang Wu Xiao-Hong Yan
Xiao-Hong Yan