
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Physiol. , 08 February 2024
Sec. Mitochondrial Research
Volume 15 - 2024 | https://doi.org/10.3389/fphys.2024.1338544
Mood disorders, including major depressive disorder and bipolar disorder, are highly prevalent and stand among the leading causes of disability. Despite the largely elusive nature of the molecular mechanisms underpinning these disorders, two pivotal contributors—mitochondrial dysfunctions and epigenetic alterations—have emerged as significant players in their pathogenesis. This state-of-the-art review aims to present existing data on epigenetic alterations in the mitochondrial genome in mood disorders, laying the groundwork for future research into their pathogenesis. Associations between abnormalities in mitochondrial function and mood disorders have been observed, with evidence pointing to notable changes in mitochondrial DNA (mtDNA). These changes encompass variations in copy number and oxidative damage. However, information on additional epigenetic alterations in the mitochondrial genome remains limited. Recent studies have delved into alterations in mtDNA and regulations in the mitochondrial genome, giving rise to the burgeoning field of mitochondrial epigenetics. Mitochondrial epigenetics encompasses three main categories of modifications: mtDNA methylation/hydroxymethylation, modifications of mitochondrial nucleoids, and mitochondrial RNA alterations. The epigenetic modulation of mitochondrial nucleoids, lacking histones, may impact mtDNA function. Additionally, mitochondrial RNAs, including non-coding RNAs, present a complex landscape influencing interactions between the mitochondria and the nucleus. The exploration of mitochondrial epigenetics offers valuable perspectives on how these alterations impact neurodegenerative diseases, presenting an intriguing avenue for research on mood disorders. Investigations into post-translational modifications and the role of mitochondrial non-coding RNAs hold promise to unravel the dynamics of mitoepigenetics in mood disorders, providing crucial insights for future therapeutic interventions.
Mood disorders, such as bipolar disorder (BD) and major depressive disorder (MDD), are highly prevalent and profoundly disabling conditions. BD is specifically characterized by severe lows (depression) and elevated highs (hypomania or mania), whereas MDD includes only the depressive pole. Common symptoms often observed in depression, such as psychomotor retardation, reduced engagement in daily activities, difficulty in concentrating, decreased attention span, and slower processing speed, as well as symptoms in mania, such as increased psychomotor activity, increased talkativeness, rapid speech, reduced need for sleep, racing thoughts, and distractibility, suggest that disruptions in energy-related processes through mitochondrial abnormalities may underlie mood episodes (Stork and Renshaw, 2005; Karabatsiakis et al., 2014). This connection is supported by evidence indicating a high prevalence of MDD (37%–54%) and BD (17%) in individuals with mitochondrial diseases (Kasahara and Kato, 2018).
Mitochondria, recognized as the “powerhouses of cells”, have been linked to mood disorders. Neuronal functioning requires a high amount of energy; therefore, the brain utilizes 25% of the body’s energy substrates and consumes 20% of its oxygen (Wong-Riley et al., 1989; Zhu et al., 2012). As energy derived from anaerobic glucose is insufficient to sustain the brain’s energy metabolism without high-energy sources such as glycogen, proper mitochondrial metabolism becomes crucial, generating 92% of the body’s energy through oxidative phosphorylation. Mitochondria also play direct roles in processes crucial for neural functioning, such as calcium homeostasis, apoptosis, signal conduction, and neurogenesis, all of which have significant implications in mood disorders (Rizzuto et al., 2012; Bock and Tait, 2020). Additionally, various mechanisms that actively interact with mitochondrial processes, such as oxidative stress, inflammation, neuroplasticity, neurogenesis, and stress-related processes, are associated with the pathogenesis of mood disorders (Manoli et al., 2007; Miller, 2011; Bansal and Kuhad, 2016; Midzak and Papadopoulos, 2016; Culmsee et al., 2018; Konttinen et al., 2019; Lapp et al., 2019). Additionally, studies have indicated the impact of various psychotropic agents such as antidepressants and mood stabilizers on mitochondrial function (Lundberg et al., 2020). Proteomic studies have highlighted mitochondrial pathways (Föcking et al., 2016), and animal models of depression have revealed a connection between mitochondrial dysfunction and depression-like behaviors (Kolar et al., 2021). This highlights the current evidence, emphasizing the pivotal role of mitochondria as a vital research area in mood disorders (Manji et al., 2012; Morava and Kozicz, 2013; Allen et al., 2018; Kuffner et al., 2020).
The heritability of MDD has been estimated to be approximately 40%, and for BD is estimated to be approximately 70%, with first-degree relatives of individuals with mood disorders facing an elevated risk of mood disorders 2–3 times higher for depression and up to 10 times higher for BD (Sullivan et al., 2000; Penner-Goeke and Binder, 2019). Despite its high heritability, a specific gene that strongly contributes to mood disorders remains unidentified, and genome-wide association studies explain only a fraction of the genetic diversity in these disorders (Menke et al., 2012; Malhi and Mann, 2018). One of the major challenges to comprehending the progression of mood disorders is the lack of a single genetic, biological, or psychosocial explanation. Instead, these disorders are thought to result from the complex interplay of multiple factors. Individuals with mood disorders manifest symptoms that arise from a complex interplay of genetic and environmental factors, including childhood trauma, migration, loneliness, poverty, work stress, violence, and even air pollution (Zeng et al., 2019). The interplay between genetic and environmental factors in mood disorders is shown by epigenetic alterations (Penner-Goeke and Binder, 2019; Legrand et al., 2021), which play a role in mood disorders and influence the effects of antidepressants and mood-stabilizing agents (Menke et al., 2012; Mikhed et al., 2015; Saavedra et al., 2016).
Mitochondria are semi-autonomous organelles that contain their own, circular, maternally inherited genomes. While traditional epigenetic studies have primarily concentrated on the nuclear genome, recent research underscores that mtDNA undergoes epigenetic changes, leading to the emergence of new fields of mitochondrial epigenetics and mitoepigenetics (Ghosh et al., 2015; Cavalcante et al., 2020). Given the crucial role of mitochondrial dysfunction and epigenetic alterations in the pathogenesis of mood disorders, changes in the epigenetics of the mitochondrial genome have gained significance. In this review, we aim to compile and present existing data regarding epigenetic alterations in the mitochondrial genome in mood disorders, thereby establishing a foundation for future research on the origins of these conditions.
Mitochondrial DNA (mtDNA) encodes 13 polypeptides, 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and a non-coding region known as the displacement loop (D-loop). Unlike nuclear DNA, mtDNA is in the matrix of the mitochondria rather than the cell nucleus. It lacks histones but is organized into clusters coated with nuclear counterpart proteins, called nucleoids. Research in mtDNA modifications remains limited and mainly focuses on mtDNA alterations, reporting changes in both mtDNA copy numbers and oxidation levels (Wang et al., 2018; Bodenstein et al., 2019; Czarny et al., 2020b; Chung et al., 2022). On the other hand, mitochondrial epigenetics explores changes in mtDNA and the mitochondrial genome (Manev, 2014), whereas mitoepigenetics encompasses a broader scope, incorporating interactions between mtDNA and nuclear DNA.
MtDNA copy number is a measure of the quantity of mitochondrial genomes per cell and serves as a surrogate indicator of mitochondrial health. Numerous studies have investigated mtDNA copy numbers in various types of specimens, including brain slices, saliva, and blood, among individuals with mood disorders (Kakiuchi et al., 2005; Kim et al., 2011; Cai et al., 2015; Chang et al., 2015; Nicod et al., 2016; Lindqvist et al., 2018; Chung et al., 2019; Tsujii et al., 2019). In human postmortem studies, mtDNA copy number was found to be increased in the dorsolateral prefrontal cortex and decreased in the superior temporal gyrus in individuals with BD (Das et al., 2022). Wang et al. suggested that leukocyte mtDNA copy number was significantly lower in individuals with BD (even in mania or depression) than in healthy controls (Wang et al., 2018). Chung et al. found that mtDNA copy number decreased in individuals with BD type I but increased in individuals with BD type II compared to controls (Chung et al., 2022). On the other hand, Fries et al. showed that individuals with BD had elevated levels of mtDNA copy numbers compared to controls, while there was no significant difference between siblings of individuals with BD and controls (Fries et al., 2017). Despite some conflicting results, most of the literature suggests significant alterations in mtDNA copy numbers in MDD and BD (Czarny et al., 2020a).
During oxidative phosphorylation in mitochondria, approximately 5% of the oxygen consumed in the electron transport chain is transformed into reactive oxygen species (ROS), such as superoxide (O2−) and hydrogen peroxide (H2O2)). ROS can damage various components of the cell, including the mitochondrial and nuclear genomes. Owing to its proximity to oxidative phosphorylation and the absence of histone protection, mtDNA has been suggested to be three times more vulnerable to the effects of ROS than nuclear DNA (Yakes and Van Houten, 1997). To date, a very limited number of studies have focused on the oxidation of mtDNA, although increased oxidation of nuclear DNA has repeatedly been shown in mood disorders (Ceylan et al., 2018; Ahmadimanesh et al., 2019; Ceylan et al., 2020a; Czarny et al., 2020a; Kucuker et al., 2022; Çelik et al., 2023). Two clinical studies have reported increased mtDNA damage in peripheral samples of patients with unipolar depression (Chang et al., 2015; Czarny et al., 2020b). Chang et al. reported that individuals with MDD, even in remission, exhibited higher mitochondrial oxidative damage than healthy controls (Chang et al., 2015). Furthermore, Czarny et al. reported that individuals with depression showed elevated levels of mtDNA damage in peripheral blood mononuclear cells (PBMCs) compared to controls, whereas there was no alteration in mtDNA copy number (Czarny et al., 2020b). On the other hand, a post-mortem study showed a significant decrease in oxidative mtDNA damage in the brain slices of patients with BD (Bodenstein et al., 2019). In addition, a recent study showed that oxidative mtDNA damage may induce short- and long-term immune activation (Xian and Karin, 2023). While these findings may not be conclusive on their own, it can be posited that oxidatively induced mtDNA damage plays an important role in mood disorders.
Due to methodological limitations, mitochondrial epigenetic changes have not been extensively studied; however, it is known that mtDNA epigenetic changes differ from those in nuclear DNA. Three types of modifications have been identified for mitoepigenetics: 1) mtDNA methylation/hydroxymethylation, 2) modifications of mitochondrial nucleoids, 3) mitochondrial RNA modifications, and modulation of non-coding RNAs (ncRNAs) originating from nuclear DNA or mtDNA (Figure 1).
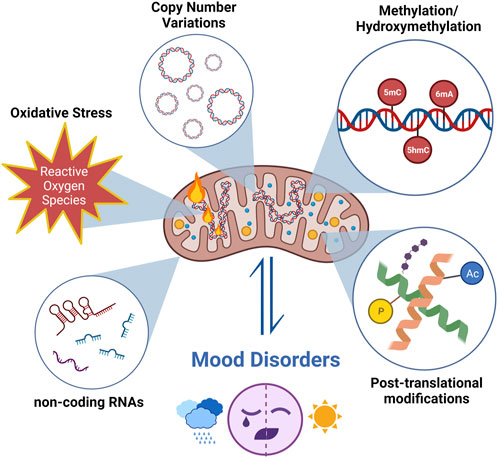
FIGURE 1. The research about mtDNA modifications focuses on changes in mtDNA copy numbers and oxidation levels, in addition to mitoepigenetic changes in mood disorders. Different types of modifications have been identified for mitoepigenetics. (1) mtDNA methylation/hydroxymethylation; (2) modifications of mitochondrial nucleoids; (3) mitochondrial RNA modifications and modulation of non-coding RNAs (ncRNA) originating from nuclear DNA or mtDNA. (Created with BioRender.com).
The most extensively studied epigenetic mechanism is DNA methylation, particularly in the nuclear DNA. DNA methylation is facilitated by a group of enzymes, known as DNA methyltransferases. These enzymes transfer a methyl group from a co-factor molecule, S-adenosyl-L-methionine, to the C5 position of cytosine and the N6 position of adenine residues, producing 5-methylcytosine (5mC) and N6-methyladenosine (6mA). Both 5mC and 6 mA are involved in either silencing or activating gene transcription. Epigenetic modification of mtDNA has been a topic of controversy and ongoing research. Although it was previously believed that mtDNA did not undergo significant epigenetic modifications, recent studies have challenged this notion (Delsite et al., 2002; Smiraglia et al., 2008; Santos, 2021). The presence of both 5mC and 5-hydroxymethylcytosine (5hmC) alterations in mtDNA suggests that epigenetic modifications may play an important role in mitochondrial functions (Shock et al., 2011; Manev et al., 2012; Bellizzi et al., 2013). A previous study showed that 6 mA in mtDNA can disrupt the DNA binding of mitochondrial transcription factor A (TFAM), thereby reducing mtDNA transcription (Hao et al., 2020). The ten-eleven translocation (TET) protein family of dioxygenases catalyzes an oxidative reaction (hydroxylation) that converts (5mC) to 5-hydroxymethylcytosine (5hmC) in an iron (II)- and α-ketoglutarate-dependent manner (Iyer et al., 2009; Tahiliani et al., 2009; Ito et al., 2010; Dzitoyeva et al., 2012) showed that both 5mC and 5hmC are present in mitochondrial DNA of the mammalian central nervous system (Dzitoyeva et al., 2012). Cytosine methylation is more widespread in eukaryotes and predominantly occurs within CpG dinucleotides located on CpG islands within the cell nucleus (Jin and Robertson, 2013; Lyko, 2018). Methylation of CpG dinucleotides occurs in certain regions, such as heavy strand promoter 1 (HSP1), heavy strand promoter 2 (HSP2), the light strand promoter (LSP), and TFAM binding sites, because mitochondrial DNA contains a low proportion of CpG islands. (Bellizzi et al., 2013; Dostal and Churchill, 2019). MtDNA methylation and hydroxymethylation have been shown that these changes have effects on neurodegenerative diseases such as Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis (Blanch et al., 2016; Stoccoro et al., 2018; Nikolac Perkovic et al., 2021; Stoccoro and Coppedè, 2021; Stoccoro et al., 2022). Several animal studies have shown TET-mediated antidepressant-like effects, but most of them are related to nuclear epigenetic regulation. One study showed that sodium butyrate exerts antidepressant-like effects, with an increase in TET1 leading to elevated 5hmC levels in the BDNF gene in the prefrontal cortex in a rat model of depression (Wei et al., 2014). DNA hydroxymethylase deficiency has been demonstrated lead to a non-negligible epigenetic alteration in response to stress and is implicated in the pathophysiology of depressive behaviors in mice (Cheng et al., 2018). Another study showed that neuroinflammation can decrease 5hmC enrichment in the brain-derived neurotrophic factor (BDNF) gene, which is an essential epigenetic element related to depression-like behaviors (Zhao et al., 2023). In addition, Scola et al. showed that lithium may have a protective effect on rotenone-induced mitochondrial complex I dysfunction and decrease the levels of mtDNA methylation and hydroxymethylation in rat primary cortical neurons (Scola et al., 2014).
mtDNA replication and transcription are impacted by altered methylation in the D-loop, a non-coding region of mtDNA (Shock et al., 2011; Liu et al., 2016). Alterations or mutations in the D-loop region can affect mitochondrial function, contributing to various diseases, such as neurodegenerative disorders and mood disorders (Blanch et al., 2016; Stoccoro et al., 2018; Chung et al., 2019). Blanch et al. reported elevated levels of methylation in the D-loop region of mtDNA in the entorhinal cortex of brain samples from patients with Alzheimer’s disease (Blanch et al., 2016). Stoccoro et al. showed that patients with amyotrophic lateral sclerosis had significantly lower methylation levels in the D-loop region of mtDNA (Stoccoro et al., 2018). A previous study presented that the methylation status of the D-loop region did not differ significantly between individuals with MDD and controls (Chung et al., 2019); however, individuals with MDD displayed increased mtDNA copy number and reduced DNA methylation levels within the PGC1α promoter (Chung et al., 2019). In contrast, Ceylan et al. showed increased methylation in the D-loop region in MDD compared to that in BD and healthy controls (Ceylan et al., 2023).
Nuclear DNA has several post-translational changes of histone proteins (such as methylation, acetylation, phosphorylation, ubiquitination, sumoylation, and poly ADP-ribosylation (parylation)) (Kouzarides, 2007). Unlike nuclear DNA, mitochondrial DNA lacks histones. Mitochondrial nucleoids are distinct entities located within the mitochondria that house mtDNA and associated proteins. These nucleoids play a vital role in the organization and maintenance of the mitochondrial genome and participate in tasks such as mtDNA packaging, maintenance, and the regulation of gene expression (Kukat et al., 2015). Mitochondrial transcription factor A (TFAM), the major component of the mitochondrial nucleoids, mitochondrial single-stranded binding protein, mitochondrial RNA polymerase, and mitochondrial DNA polymerase gamma (POLG) are some of the most well-known nucleoids, and they can be post-translationally modified via acetylation, O-linked glycosylation, and phosphorylation (Sharma et al., 2019). Therefore, epigenetic changes occurring in nucleoid proteins play a crucial role in the regulation of mtDNA gene expression. POLG is responsible for replicating and repairing mitochondrial DNA and is involved in various processes that are critical for maintaining mitochondrial genome integrity. Reportedly, rapid cycling patients with BD exhibit significant downregulation of POLG expression compared with controls, regardless of illness episodes (Munkholm et al., 2015). However, our study group presented unchanged levels of POLG in both euthymic patients and depressive individuals with BD, as well as in individuals with MDD (Yılmaz et al., 2022). Neuron-specific DNA methylation analysis of neurons isolated from certain brain areas that expressed the proofreading-deficient POLG1 transgenic mice showed that mutant mice displayed depression-like and bipolar disorder-like behavioral abnormalities (Kasahara et al., 2016; Sugawara et al., 2022). On the other hand, TFAM, a nuclear-encoded transcription factor, is involved in regulating the replication and transcription of mtDNA, interacts with the DNA to facilitate condensation into nucleoids and binding to the D-loop (Choi et al., 2005), and serves as an instance of a mitochondrial protein subject to post-translational modifications (Suarez et al., 2008; Lu et al., 2013; King et al., 2018). A study showed that mitochondrial poly ADP-ribose polymerase 1 (PARP1) induces NAD + -dependent mitochondrial nucleoid poly ADP-ribosylation, influencing the recruitment of TFAM to stimulate mtDNA transcription and indicating epigenetic nucleoid reorganization (Lee et al., 2022). The expression level of TFAM has been demonstrated to be altered in various cancers, including breast cancer, lung cancer, and melanoma (Fan et al., 2017; Araujo et al., 2018; Lin et al., 2018). However, no data has indicated an association between TFAM and mood disorders. Since mitochondrial epigenetics is a newly recognized research area, no study to date has investigated the posttranslational modifications of nucleoids in mood disorders.
Mitochondrial RNA modifications and modulation of ncRNAs: There are various types of mitochondrial RNAs, including 2 rRNAs, 22 tRNAs, 13 mRNAs, and numerous ncRNAs, found in the mitochondrial matrix. Post-transcriptional mechanisms play a crucial role in governing the translation, maturation, stability, and assembly of mitochondrial RNAs. Modifications of mitochondrial mRNAs, rRNAs, and tRNAs have been linked to various mitochondrial diseases (Dong et al., 2020). N1-methyladenosine and N6-methyladenosine have been identified as post-transcriptional modifications of mitochondrial RNAs (Wang et al., 2017; Zhang and Jia, 2018). Alterations in the methylation levels of mitochondrial tRNAs have been observed in tumor tissues (Hodgkinson et al., 2014). However, the potential roles of mitochondrial RNA modifications in the pathophysiology of mood disorders have not been elucidated.
RNA transcripts that cannot be translated into proteins are referred to as non-coding RNAs (ncRNAs). ncRNAs, encompassing both small and long ncRNAs, play crucial roles in biological processes such as developmental pathways, cellular responses, and regulatory functions in transcription, and contribute to post-transcriptional processes by regulating gene expression through epigenetic mechanisms. They also play crucial roles in mitochondrial function (Kobayashi et al., 2023). Mitochondria-derived ncRNAs can be found both inside and outside the mitochondria, regulating communication between the mitochondria and nucleus through anterograde and retrograde signals. ncRNAs inside the mitochondria can be transcribed from either the nuclear or mitochondrial genome, and certain mitochondrial ncRNAs are transcribed from the mitochondrial genome before being transported to the cell nuclei or cytosol (Liang et al., 2021). MitomiRs are promising targets for future research in mood disorders.
MicroRNAs (miRNA) are the most abundant ncRNAs and play roles in regulating protein synthesis and controlling gene expression. Numerous studies have provided evidence for the significant role of miRNAs in various psychiatric disorders, including MDD, BD, and schizophrenia (Maffioletti et al., 2016; Cao and Zhen, 2018; Ceylan et al., 2020b; Zhang et al., 2020). MtDNA also encodes miRNAs that are involved in the regulation of mitochondrial gene expression (Sripada et al., 2012). miRNAs localized in mitochondria, whether transported from the nucleus or transcribed in the mitochondria, are known as mitomiRs (Rencelj et al., 2021). Most mitomiRs are encoded in the genome, and a small portion of the mitochondrial genome encodes these mitomiRs (Shinde and Bhadra, 2015). MitomiRs have been demonstrated to be important regulators of mitochondrial function (Rencelj et al., 2021). The regulation of mitochondria by mitomiRs influences the development of numerous diseases caused by mitochondrial dysfunction, contributing to the pathophysiology of conditions such as cardiovascular, metabolic, and neurodegenerative diseases as well as cancers (Dong et al., 2020; Wagner et al., 2022).
Circulating RNAs (circRNAs) are ncRNAs that are involved in depolarization, plasticity, neuronal activity, and synaptic transmission. A recent study showed that 55 circRNAs were altered in individuals with schizophrenia and BD compared to healthy controls, and 71% of these circRNAs were downregulated (Mahmoudi et al., 2021). Mitochondria-encoded circRNAs have also been identified. Four major types of circRNAs have been shown to have an impact on the function of mitochondria, including mecciND1, and mecciND5-mc-COX2-circRNA SCAR (Liu and Shan, 2021).
Long ncRNAs (lncRNAs) are the most abundantly expressed ncRNAs in the brain and contribute to the regulation of chromatin remodeling, protein scaffolding, translation, splicing, and transcription. Studies have confirmed that they interact with epigenetic mediators and transcription factors and subsequently regulate transcription by targeting gene promoters (Qureshi et al., 2010; Kadakkuzha et al., 2015; Quinn and Chang, 2016). Changes in lncRNAs have been associated with depression (Liu et al., 2014), bipolar disorder (Sayad et al., 2019; Shirvani Farsani et al., 2020), schizophrenia (Mishra and Kumar, 2021), autism (Wilkinson and Campbell, 2013), dementia, and other neurodegenerative diseases (Riva et al., 2016). Numerous lncRNA genomic regions have been associated with depression in genome-wide association studies (Zeng et al., 2017), and single-gene polymorphisms in lncRNA genes have been reported to be associated with the risk of depression (Delacrétaz et al., 2015; Ye et al., 2017). Studies have demonstrated the differential regulation of various lncRNA expressions in peripheral blood samples of individuals with MDD or BD compared to healthy controls (Liu et al., 2014; Cui et al., 2016; Cui et al., 2017; Ghafelehbashi et al., 2017; Ye et al., 2017; Naghavi-Gargari et al., 2019; Sayad et al., 2019). In addition, a recent study denoted the increased gene expression levels of long intergenic ncRNA 173 (LINC00173) in the postmortem brain tissue of individuals with BD (Akula et al., 2014).
In recent years, mitochondria-derived ncRNAs have attracted the attention of researchers because of their critical roles in the pathophysiology of many disorders (Zhao et al., 2018; Ren et al., 2023). However, there is currently no published study in the literature that has investigated mitochondrial-encoded ncRNAs, such as miRNAs, circRNAs, and lncRNAs, in mood disorders. Examining the roles of ncRNAs in mitoepigenetics remains a central focus for further exploration.
The integration of mitoepigenetics into the broader framework of mood disorder research has the potential to enrich our understanding of the molecular underpinnings of these conditions. Currently, limited research exploring D-loop methylation and mtDNA oxidation in mood disorders suggests the need for a more detailed focus on this area of research. In addition, investigating post-translational modifications of nucleoids and their impact on mitochondrial function in mood disorders could provide crucial insights into the epigenetic regulation of mtDNA. Mitochondrial RNA, including ncRNAs such as mitomiRs, circRNAs, and lncRNAs, emerges as a dynamic component influencing mitochondrial and nuclear interactions. miRNAs, in particular, have been implicated in various psychiatric disorders, and mitochondrial-encoded mitomiRs present a novel dimension in understanding mitochondrial contributions to mood regulation. Exploring the roles of mitomiRs, circRNAs, and lncRNAs in mood disorders can deepen our understanding of the intricate regulatory networks governing the mitochondrial contributions to these conditions. Considering the multifactorial nature of mood disorders, future research should explore the interactions between mitoepigenetics and environmental factors such as stressors, trauma, and lifestyle to comprehensively understand the etiology of these conditions. Translating the insights gained from mitochondrial epigenetic research into clinical applications holds great promise. Identifying therapeutic targets related to mitoepigenetics could pave the way for innovative interventions in mood disorders.
DC: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Writing–original draft, Writing–review and editing, Data curation. HEA-Ç: Data curation, Formal Analysis, Writing–review and editing. ICA: Data curation, Investigation, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This review article has been conducted within the scope of the TÜBİTAK 1001 project with the number 122Z771. Additionally, support from Koç University has been obtained for its publication.
The authors gratefully acknowledge the use of the services and facilities of the Koç University Research Center for Translational Medicine (KUTTAM), funded by the Presidency of Turkey, Head of Strategy and Budget.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmadimanesh M., Abbaszadegan M. R., Morshedi Rad D., Moallem S. A., Mohammadpour A. H., Ghahremani M. H., et al. (2019). Effects of selective serotonin reuptake inhibitors on DNA damage in patients with depression. J. Psychopharmacol. 33, 1364–1376. doi:10.1177/0269881119874461
Akula N., Barb J., Jiang X., Wendland J. R., Choi K. H., Sen S. K., et al. (2014). RNA-sequencing of the brain transcriptome implicates dysregulation of neuroplasticity, circadian rhythms and GTPase binding in bipolar disorder. Mol. Psychiatry 19, 1179–1185. doi:10.1038/mp.2013.170
Allen J., Romay-Tallon R., Brymer K. J., Caruncho H. J., Kalynchuk L. E. (2018). Mitochondria and mood: mitochondrial dysfunction as a key player in the manifestation of depression. Front. Neurosci. 12, 386. doi:10.3389/fnins.2018.00386
Araujo L. F., Siena A. D. D., Plaça J. R., Brotto D. B., Barros , Muys B. R., et al. (2018). Mitochondrial transcription factor A (TFAM) shapes metabolic and invasion gene signatures in melanoma. Sci. Rep. 8, 14190. doi:10.1038/s41598-018-31170-6
Bansal Y., Kuhad A. (2016). Mitochondrial dysfunction in depression. Curr. Neuropharmacol. 14, 610–618. doi:10.2174/1570159x14666160229114755
Bellizzi D., D'Aquila P., Scafone T., Giordano M., Riso V., Riccio A., et al. (2013). The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. 20, 537–547. doi:10.1093/dnares/dst029
Blanch M., Mosquera J. L., Ansoleaga B., Ferrer I., Barrachina M. (2016). Altered mitochondrial DNA methylation pattern in alzheimer disease-related pathology and in Parkinson disease. Am. J. Pathol. 186, 385–397. doi:10.1016/j.ajpath.2015.10.004
Bock F. J., Tait S. W. G. (2020). Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 21, 85–100. doi:10.1038/s41580-019-0173-8
Bodenstein D. F., Kim H. K., Brown N. C., Navaid B., Young L. T., Andreazza A. C. (2019). Mitochondrial DNA content and oxidation in bipolar disorder and its role across brain regions. NPJ Schizophr. 5, 21. doi:10.1038/s41537-019-0089-5
Cai N., Chang S., Li Y., Li Q., Hu J., Liang J., et al. (2015). Molecular signatures of major depression. Curr. Biol. 25, 1146–1156. doi:10.1016/j.cub.2015.03.008
Cao T., Zhen X. C. (2018). Dysregulation of miRNA and its potential therapeutic application in schizophrenia. CNS Neurosci. Ther. 24, 586–597. doi:10.1111/cns.12840
Cavalcante G. C., Magalhães L., Ribeiro-Dos-Santos Â., Vidal A. F. (2020). Mitochondrial epigenetics: non-coding RNAs as a novel layer of complexity. Int. J. Mol. Sci. 21, 1838. doi:10.3390/ijms21051838
Çelik H., Tuna G., Ceylan D., Küçükgöncü S. (2023). A comparative meta-analysis of peripheral 8-hydroxy-2'-deoxyguanosine (8-OHdG) or 8-oxo-7,8-dihydro-2'-deoxyguanosine (8-oxo-dG) levels across mood episodes in bipolar disorder. Psychoneuroendocrinology 151, 106078. doi:10.1016/j.psyneuen.2023.106078
Ceylan D., Karaçiçek B., Tufekci K. U., Aksahin I. C., Hun Şenol Ş., Genc S. (2023). Mitochondrial DNA oxidation, methylation, and copy number alterations in major and bipolar depression. Front. Psychiatry 14, 1304660. doi:10.3389/fpsyt.2023.1304660
Ceylan D., Tufekci K. U., Keskinoglu P., Genc S., Özerdem A. (2020a). Circulating exosomal microRNAs in bipolar disorder. J. Affect Disord. 262, 99–107. doi:10.1016/j.jad.2019.10.038
Ceylan D., Tuna G., Kirkali G., Tunca Z., Can G., Arat H. E., et al. (2018). Oxidatively-induced DNA damage and base excision repair in euthymic patients with bipolar disorder. DNA Repair (Amst) 65, 64–72. doi:10.1016/j.dnarep.2018.03.006
Ceylan D., Yılmaz S., Tuna G., Kant M., Er A., Ildız A., et al. (2020b). Alterations in levels of 8-Oxo-2'-deoxyguanosine and 8-Oxoguanine DNA glycosylase 1 during a current episode and after remission in unipolar and bipolar depression. Psychoneuroendocrinology 114, 104600. doi:10.1016/j.psyneuen.2020.104600
Chang C. C., Jou S. H., Lin T. T., Lai T. J., Liu C. S. (2015). Mitochondria DNA change and oxidative damage in clinically stable patients with major depressive disorder. PLoS One 10, e0125855. doi:10.1371/journal.pone.0125855
Cheng Y., Sun M., Chen L., Li Y., Lin L., Yao B., et al. (2018). Ten-eleven translocation proteins modulate the response to environmental stress in mice. Cell Rep. 25, 3194–3203. doi:10.1016/j.celrep.2018.11.061
Choi Y. S., Ryu B. K., Min H. K., Lee S. W., Pak Y. K. (2005). Analysis of proteome bound to D-loop region of mitochondrial DNA by DNA-linked affinity chromatography and reverse-phase liquid chromatography/tandem mass spectrometry. Ann. N. Y. Acad. Sci. 1042, 88–100. doi:10.1196/annals.1338.009
Chung J. K., Ahn Y. M., Kim S. A., Joo E. J. (2022). Differences in mitochondrial DNA copy number between patients with bipolar I and II disorders. J. Psychiatr. Res. 145, 325–333. doi:10.1016/j.jpsychires.2020.11.016
Chung J. K., Lee S. Y., Park M., Joo E. J., Kim S. A. (2019). Investigation of mitochondrial DNA copy number in patients with major depressive disorder. Psychiatry Res. 282, 112616. doi:10.1016/j.psychres.2019.112616
Cui X., Niu W., Kong L., He M., Jiang K., Chen S., et al. (2017). Long noncoding RNA expression in peripheral blood mononuclear cells and suicide risk in Chinese patients with major depressive disorder. Brain Behav. 7, e00711. doi:10.1002/brb3.711
Cui X., Sun X., Niu W., Kong L., He M., Zhong A., et al. (2016). Long non-coding RNA: potential diagnostic and therapeutic biomarker for major depressive disorder. Med. Sci. Monit. 22, 5240–5248. doi:10.12659/msm.899372
Culmsee C., Michels S., Scheu S., Arolt V., Dannlowski U., Alferink J. (2018). Mitochondria, microglia, and the immune system-how are they linked in affective disorders? Front. Psychiatry 9, 739. doi:10.3389/fpsyt.2018.00739
Czarny P., Bialek K., Ziolkowska S., Strycharz J., Sliwinski T. (2020a). DNA damage and repair in neuropsychiatric disorders. What do we know and what are the future perspectives? Mutagenesis 35, 79–106. doi:10.1093/mutage/gez035
Czarny P., Wigner P., Strycharz J., Swiderska E., Synowiec E., Szatkowska M., et al. (2020b). Mitochondrial DNA copy number, damage, repair and degradation in depressive disorder. World J. Biol. Psychiatry 21, 91–101. doi:10.1080/15622975.2019.1588993
Das S. C., Hjelm B. E., Rollins B. L., Sequeira A., Morgan L., Omidsalar A. A., et al. (2022). Mitochondria DNA copy number, mitochondria DNA total somatic deletions, Complex I activity, synapse number, and synaptic mitochondria number are altered in schizophrenia and bipolar disorder. Transl. Psychiatry 12, 353. doi:10.1038/s41398-022-02127-1
Delacrétaz A., Preisig M., Vandenberghe F., Saigi Morgui N., Quteineh L., Choong E., et al. (2015). Influence of MCHR2 and MCHR2-AS1 genetic polymorphisms on body mass index in psychiatric patients and in population-based subjects with present or past atypical depression. PLoS One 10, e0139155. doi:10.1371/journal.pone.0139155
Delsite R., Kachhap S., Anbazhagan R., Gabrielson E., Singh K. K. (2002). Nuclear genes involved in mitochondria-to-nucleus communication in breast cancer cells. Mol. Cancer 1, 6. doi:10.1186/1476-4598-1-6
Dong Z., Pu L., Cui H. (2020). Mitoepigenetics and its emerging roles in cancer. Front. Cell Dev. Biol. 8, 4. doi:10.3389/fcell.2020.00004
Dostal V., Churchill M. E. A. (2019). Cytosine methylation of mitochondrial DNA at CpG sequences impacts transcription factor A DNA binding and transcription. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 598–607. doi:10.1016/j.bbagrm.2019.01.006
Dzitoyeva S., Chen H., Manev H. (2012). Effect of aging on 5-hydroxymethylcytosine in brain mitochondria. Neurobiol. Aging 33, 2881–2891. doi:10.1016/j.neurobiolaging.2012.02.006
Fan X., Zhou S., Zheng M., Deng X., Yi Y., Huang T. (2017). MiR-199a-3p enhances breast cancer cell sensitivity to cisplatin by downregulating TFAM (TFAM). Biomed. Pharmacother. 88, 507–514. doi:10.1016/j.biopha.2017.01.058
Föcking M., Dicker P., Lopez L. M., Hryniewiecka M., Wynne K., English J. A., et al. (2016). Proteomic analysis of the postsynaptic density implicates synaptic function and energy pathways in bipolar disorder. Transl. Psychiatry 6, e959. doi:10.1038/tp.2016.224
Fries G. R., Bauer I. E., Scaini G., Wu M. J., Kazimi I. F., Valvassori S. S., et al. (2017). Accelerated epigenetic aging and mitochondrial DNA copy number in bipolar disorder. Transl. Psychiatry 7, 1283. doi:10.1038/s41398-017-0048-8
Ghafelehbashi H., Pahlevan Kakhki M., Kular L., Moghbelinejad S., Ghafelehbashi S. H. (2017). Decreased expression of IFNG-AS1, IFNG and IL-1B inflammatory genes in medicated schizophrenia and bipolar patients. Scand. J. Immunol. 86, 479–485. doi:10.1111/sji.12620
Ghosh S., Singh K. K., Sengupta S., Scaria V. (2015). Mitoepigenetics: the different shades of grey. Mitochondrion 25, 60–66. doi:10.1016/j.mito.2015.09.003
Hao Z., Wu T., Cui X., Zhu P., Tan C., Dou X., et al. (2020). N(6)-Deoxyadenosine methylation in mammalian mitochondrial DNA. Mol. Cell 78, 382–395. doi:10.1016/j.molcel.2020.02.018
Hodgkinson A., Idaghdour Y., Gbeha E., Grenier J. C., Hip-Ki E., Bruat V., et al. (2014). High-resolution genomic analysis of human mitochondrial RNA sequence variation. Science 344, 413–415. doi:10.1126/science.1251110
Ito S., D'Alessio A. C., Taranova O. V., Hong K., Sowers L. C., Zhang Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133. doi:10.1038/nature09303
Iyer L. M., Tahiliani M., Rao A., Aravind L. (2009). Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle 8, 1698–1710. doi:10.4161/cc.8.11.8580
Jin B., Robertson K. D. (2013). DNA methyltransferases, DNA damage repair, and cancer. Adv. Exp. Med. Biol. 754, 3–29. doi:10.1007/978-1-4419-9967-2_1
Kadakkuzha B. M., Liu X. A., McCrate J., Shankar G., Rizzo V., Afinogenova A., et al. (2015). Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Front. Cell Neurosci. 9, 63. doi:10.3389/fncel.2015.00063
Kakiuchi C., Ishiwata M., Kametani M., Nelson C., Iwamoto K., Kato T. (2005). Quantitative analysis of mitochondrial DNA deletions in the brains of patients with bipolar disorder and schizophrenia. Int. J. Neuropsychopharmacol. 8, 515–522. doi:10.1017/s1461145705005213
Karabatsiakis A., Böck C., Salinas-Manrique J., Kolassa S., Calzia E., Dietrich D. E., et al. (2014). Mitochondrial respiration in peripheral blood mononuclear cells correlates with depressive subsymptoms and severity of major depression. Transl. Psychiatry 4, e397. doi:10.1038/tp.2014.44
Kasahara T., Kato T. (2018). What can mitochondrial DNA analysis tell us about mood disorders? Biol. Psychiatry 83, 731–738. doi:10.1016/j.biopsych.2017.09.010
Kasahara T., Takata A., Kato T. M., Kubota-Sakashita M., Sawada T., Kakita A., et al. (2016). Depression-like episodes in mice harboring mtDNA deletions in paraventricular thalamus. Mol. Psychiatry 21, 39–48. doi:10.1038/mp.2015.156
Kim M. Y., Lee J. W., Kang H. C., Kim E., Lee D. C. (2011). Leukocyte mitochondrial DNA (mtDNA) content is associated with depression in old women. Arch. Gerontol. Geriatr. 53, e218–e221. doi:10.1016/j.archger.2010.11.019
King G. A., Hashemi Shabestari M., Taris K. H., Pandey A. K., Venkatesh S., Thilagavathi J., et al. (2018). Acetylation and phosphorylation of human TFAM regulate TFAM-DNA interactions via contrasting mechanisms. Nucleic Acids Res. 46, 3633–3642. doi:10.1093/nar/gky204
Kobayashi A., Takeiwa T., Ikeda K., Inoue S. (2023). Roles of noncoding RNAs in regulation of mitochondrial electron transport chain and oxidative phosphorylation. Int. J. Mol. Sci. 24, 9414. doi:10.3390/ijms24119414
Kolar D., Kleteckova L., Brozka H., Vales K. (2021). Mini-review: brain energy metabolism and its role in animal models of depression, bipolar disorder, schizophrenia and autism. Neurosci. Lett. 760, 136003. doi:10.1016/j.neulet.2021.136003
Konttinen H., Cabral-da-Silva M. E. C., Ohtonen S., Wojciechowski S., Shakirzyanova A., Caligola S., et al. (2019). PSEN1ΔE9, APPswe, and APOE4 confer disparate phenotypes in human iPSC-derived microglia. Stem Cell Rep. 13, 669–683. doi:10.1016/j.stemcr.2019.08.004
Kouzarides T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi:10.1016/j.cell.2007.02.005
Kucuker M. U., Ozerdem A., Ceylan D., Cabello-Arreola A., Ho A. M. C., Joseph B., et al. (2022). The role of base excision repair in major depressive disorder and bipolar disorder. J. Affect Disord. 306, 288–300. doi:10.1016/j.jad.2022.03.033
Kuffner K., Triebelhorn J., Meindl K., Benner C., Manook A., Sudria-Lopez D., et al. (2020). Major depressive disorder is associated with impaired mitochondrial function in skin fibroblasts. Cells 9, 884. doi:10.3390/cells9040884
Kukat C., Davies K. M., Wurm C. A., Spåhr H., Bonekamp N. A., Kühl I., et al. (2015). Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. U. S. A. 112, 11288–11293. doi:10.1073/pnas.1512131112
Lapp H. E., Bartlett A. A., Hunter R. G. (2019). Stress and glucocorticoid receptor regulation of mitochondrial gene expression. J. Mol. Endocrinol. 62, R121–R128. doi:10.1530/jme-18-0152
Lee J. H., Hussain M., Kim E. W., Cheng S. J., Leung A. K. L., Fakouri N. B., et al. (2022). Mitochondrial PARP1 regulates NAD(+)-dependent poly ADP-ribosylation of mitochondrial nucleoids. Exp. Mol. Med. 54, 2135–2147. doi:10.1038/s12276-022-00894-x
Legrand A., Iftimovici A., Khayachi A., Chaumette B. (2021). Epigenetics in bipolar disorder: a critical review of the literature. Psychiatr. Genet. 31, 1–12. doi:10.1097/ypg.0000000000000267
Liang H., Liu J., Su S., Zhao Q. (2021). Mitochondrial noncoding RNAs: new wine in an old bottle. RNA Biol. 18, 2168–2182. doi:10.1080/15476286.2021.1935572
Lin C. S., Liu L. T., Ou L. H., Pan S. C., Lin C. I., Wei Y. H. (2018). Role of mitochondrial function in the invasiveness of human colon cancer cells. Oncol. Rep. 39, 316–330. doi:10.3892/or.2017.6087
Lindqvist D., Wolkowitz O. M., Picard M., Ohlsson L., Bersani F. S., Fernström J., et al. (2018). Circulating cell-free mitochondrial DNA, but not leukocyte mitochondrial DNA copy number, is elevated in major depressive disorder. Neuropsychopharmacology 43, 1557–1564. doi:10.1038/s41386-017-0001-9
Liu B., Du Q., Chen L., Fu G., Li S., Fu L., et al. (2016). CpG methylation patterns of human mitochondrial DNA. Sci. Rep. 6, 23421. doi:10.1038/srep23421
Liu X., Shan G. (2021). Mitochondria encoded non-coding RNAs in cell Physiology. Front. Cell Dev. Biol. 9, 713729. doi:10.3389/fcell.2021.713729
Liu Z., Li X., Sun N., Xu Y., Meng Y., Yang C., et al. (2014). Microarray profiling and co-expression network analysis of circulating lncRNAs and mRNAs associated with major depressive disorder. PLoS One 9, e93388. doi:10.1371/journal.pone.0093388
Lu B., Lee J., Nie X., Li M., Morozov Y. I., Venkatesh S., et al. (2013). Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol. Cell 49, 121–132. doi:10.1016/j.molcel.2012.10.023
Lundberg M., Millischer V., Backlund L., Martinsson L., Stenvinkel P., Sellgren C. M., et al. (2020). Lithium and the interplay between telomeres and mitochondria in bipolar disorder. Front. Psychiatry 11, 586083. doi:10.3389/fpsyt.2020.586083
Lyko F. (2018). The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92. doi:10.1038/nrg.2017.80
Maffioletti E., Cattaneo A., Rosso G., Maina G., Maj C., Gennarelli M., et al. (2016). Peripheral whole blood microRNA alterations in major depression and bipolar disorder. J. Affect Disord. 200, 250–258. doi:10.1016/j.jad.2016.04.021
Mahmoudi E., Green M. J., Cairns M. J. (2021). Dysregulation of circRNA expression in the peripheral blood of individuals with schizophrenia and bipolar disorder. J. Mol. Med. Berl. 99, 981–991. doi:10.1007/s00109-021-02070-6
Malhi G. S., Mann J. J. (2018). Depression. Lancet 392, 2299–2312. doi:10.1016/s0140-6736(18)31948-2
Manev H. (2014). Mitoepigenetics and neuropsychiatric disorders. Epigenetics Psychiatry 2014, 463–478. doi:10.1016/B978-0-12-417114-5.00022-X
Manev H., Dzitoyeva S., Chen H. (2012). Mitochondrial DNA: a blind spot in neuroepigenetics. Biomol. Concepts 3, 107–115. doi:10.1515/bmc-2011-0058
Manji H., Kato T., Di Prospero N. A., Ness S., Beal M. F., Krams M., et al. (2012). Impaired mitochondrial function in psychiatric disorders. Nat. Rev. Neurosci. 13, 293–307. doi:10.1038/nrn3229
Manoli I., Alesci S., Blackman M. R., Su Y. A., Rennert O. M., Chrousos G. P. (2007). Mitochondria as key components of the stress response. Trends Endocrinol. Metab. 18, 190–198. doi:10.1016/j.tem.2007.04.004
Menke A., Klengel T., Binder E. B. (2012). Epigenetics, depression and antidepressant treatment. Curr. Pharm. Des. 18, 5879–5889. doi:10.2174/138161212803523590
Midzak A., Papadopoulos V. (2016). Adrenal mitochondria and steroidogenesis: from individual proteins to functional protein assemblies. Front. Endocrinol. (Lausanne) 7, 106. doi:10.3389/fendo.2016.00106
Mikhed Y., Görlach A., Knaus U. G., Daiber A. (2015). Redox regulation of genome stability by effects on gene expression, epigenetic pathways and DNA damage/repair. Redox Biol. 5, 275–289. doi:10.1016/j.redox.2015.05.008
Miller W. L. (2011). Role of mitochondria in steroidogenesis. Endocr. Dev. 20, 1–19. doi:10.1159/000321204
Mishra P., Kumar S. (2021). Association of lncRNA with regulatory molecular factors in brain and their role in the pathophysiology of schizophrenia. Metab. Brain Dis. 36, 849–858. doi:10.1007/s11011-021-00692-w
Morava E., Kozicz T. (2013). Mitochondria and the economy of stress (mal)adaptation. Neurosci. Biobehav Rev. 37, 668–680. doi:10.1016/j.neubiorev.2013.02.005
Munkholm K., Peijs L., Vinberg M., Kessing L. V. (2015). A composite peripheral blood gene expression measure as a potential diagnostic biomarker in bipolar disorder. Transl. Psychiatry 5, e614. doi:10.1038/tp.2015.110
Naghavi-Gargari B., Zahirodin A., Ghaderian S. M. H., Shirvani-Farsani Z. (2019). Significant increasing of DISC2 long non-coding RNA expression as a potential biomarker in bipolar disorder. Neurosci. Lett. 696, 206–211. doi:10.1016/j.neulet.2018.12.044
Nicod J., Wagner S., Vonberg F., Bhomra A., Schlicht K. F., Tadic A., et al. (2016). The amount of mitochondrial DNA in blood reflects the course of a depressive episode. Biol. Psychiatry 80, e41–e42. doi:10.1016/j.biopsych.2015.12.019
Nikolac Perkovic M., Videtic Paska A., Konjevod M., Kouter K., Svob Strac D., Nedic Erjavec G., et al. (2021). Epigenetics of Alzheimer's disease. Biomolecules 11, 195. doi:10.3390/biom11020195
Penner-Goeke S., Binder E. B. (2019). Epigenetics and depression dialogues clin neurosci. Dialogues Clin. Neurosci. 21, 397–405. doi:10.31887/DCNS.2019.21.4/ebinder
Quinn J. J., Chang H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi:10.1038/nrg.2015.10
Qureshi I. A., Mattick J. S., Mehler M. F. (2010). Long non-coding RNAs in nervous system function and disease. Brain Res. 1338, 20–35. doi:10.1016/j.brainres.2010.03.110
Ren B., Guan M. X., Zhou T., Cai X., Shan G. (2023). Emerging functions of mitochondria-encoded noncoding RNAs. Trends Genet. 39, 125–139. doi:10.1016/j.tig.2022.08.004
Rencelj A., Gvozdenovic N., Cemazar M. (2021). MitomiRs: their roles in mitochondria and importance in cancer cell metabolism. Radiol. Oncol. 55, 379–392. doi:10.2478/raon-2021-0042
Riva P., Ratti A., Venturin M. (2016). The long non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr. Alzheimer Res. 13, 1219–1231. doi:10.2174/1567205013666160622112234
Rizzuto R., De Stefani D., Raffaello A., Mammucari C. (2012). Mitochondria as sensors and regulators of calcium signalling. Nat. Rev. Mol. Cell Biol. 13, 566–578. doi:10.1038/nrm3412
Saavedra K., Molina-Márquez A. M., Saavedra N., Zambrano T., Salazar L. A. (2016). Epigenetic modifications of major depressive disorder. Int. J. Mol. Sci. 17, 1279. doi:10.3390/ijms17081279
Santos J. H. (2021). Mitochondria signaling to the epigenome: a novel role for an old organelle. Free Radic. Biol. Med. 170, 59–69. doi:10.1016/j.freeradbiomed.2020.11.016
Sayad A., Taheri M., Omrani M. D., Fallah H., Kholghi Oskooei V., Ghafouri-Fard S. (2019). Peripheral expression of long non-coding RNAs in bipolar patients. J. Affect Disord. 249, 169–174. doi:10.1016/j.jad.2019.02.034
Scola G., Kim H. K., Young L. T., Salvador M., Andreazza A. C. (2014). Lithium reduces the effects of rotenone-induced complex I dysfunction on DNA methylation and hydroxymethylation in rat cortical primary neurons. Psychopharmacol. Berl. 231, 4189–4198. doi:10.1007/s00213-014-3565-7
Sharma N., Pasala M. S., Prakash A. (2019). Mitochondrial DNA: epigenetics and environment. Environ. Mol. Mutagen 60, 668–682. doi:10.1002/em.22319
Shinde S., Bhadra U. (2015). A complex genome-microRNA interplay in human mitochondria. Biomed. Res. Int. 2015, 206382. doi:10.1155/2015/206382
Shirvani Farsani Z., Zahirodin A., Ghaderian S. M. H., Shams J., Naghavi Gargari B. (2020). The role of long non-coding RNA MALAT1 in patients with bipolar disorder. Metab. Brain Dis. 35, 1077–1083. doi:10.1007/s11011-020-00580-9
Shock L. S., Thakkar P. V., Peterson E. J., Moran R. G., Taylor S. M. (2011). DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. U. S. A. 108, 3630–3635. doi:10.1073/pnas.1012311108
Smiraglia D. J., Kulawiec M., Bistulfi G. L., Gupta S. G., Singh K. K. (2008). A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer Biol. Ther. 7, 1182–1190. doi:10.4161/cbt.7.8.6215
Sripada L., Tomar D., Prajapati P., Singh R., Singh A. K., Singh R. (2012). Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS One 7, e44873. doi:10.1371/journal.pone.0044873
Stoccoro A., Baldacci F., Ceravolo R., Giampietri L., Tognoni G., Siciliano G., et al. (2022). Increase in mitochondrial D-loop region methylation levels in mild cognitive impairment individuals. Int. J. Mol. Sci. 23, 5393. doi:10.3390/ijms23105393
Stoccoro A., Coppedè F. (2021). Mitochondrial DNA methylation and human diseases. Int. J. Mol. Sci. 22, 4594. doi:10.3390/ijms22094594
Stoccoro A., Mosca L., Carnicelli V., Cavallari U., Lunetta C., Marocchi A., et al. (2018). Mitochondrial DNA copy number and D-loop region methylation in carriers of amyotrophic lateral sclerosis gene mutations. Epigenomics 10, 1431–1443. doi:10.2217/epi-2018-0072
Stork C., Renshaw P. F. (2005). Mitochondrial dysfunction in bipolar disorder: evidence from magnetic resonance spectroscopy research. Mol. Psychiatry 10, 900–919. doi:10.1038/sj.mp.4001711
Suarez J., Hu Y., Makino A., Fricovsky E., Wang H., Dillmann W. H. (2008). Alterations in mitochondrial function and cytosolic calcium induced by hyperglycemia are restored by mitochondrial transcription factor A in cardiomyocytes. Am. J. Physiol. Cell Physiol. 295, C1561–C1568. doi:10.1152/ajpcell.00076.2008
Sugawara H., Bundo M., Kasahara T., Nakachi Y., Ueda J., Kubota-Sakashita M., et al. (2022). Cell-type-specific DNA methylation analysis of the frontal cortices of mutant Polg1 transgenic mice with neuronal accumulation of deleted mitochondrial DNA. Mol. Brain 15, 9. doi:10.1186/s13041-021-00894-4
Sullivan P. F., Neale M. C., Kendler K. S. (2000). Genetic epidemiology of major depression: review and meta-analysis. Am. J. Psychiatry 157, 1552–1562. doi:10.1176/appi.ajp.157.10.1552
Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. doi:10.1126/science.1170116
Tsujii N., Otsuka I., Okazaki S., Yanagi M., Numata S., Yamaki N., et al. (2019). Mitochondrial DNA copy number raises the potential of left frontopolar hemodynamic response as a diagnostic marker for distinguishing bipolar disorder from major depressive disorder. Front. Psychiatry 10, 312. doi:10.3389/fpsyt.2019.00312
Wagner A., Kosnacova H., Chovanec M., Jurkovicova D. (2022). Mitochondrial genetic and epigenetic regulations in cancer: therapeutic potential. Int. J. Mol. Sci. 23, 7897. doi:10.3390/ijms23147897
Wang D., Li Z., Liu W., Zhou J., Ma X., Tang J., et al. (2018). Differential mitochondrial DNA copy number in three mood states of bipolar disorder. BMC Psychiatry 18, 149. doi:10.1186/s12888-018-1717-8
Wang Z., Tang K., Zhang D., Wan Y., Wen Y., Lu Q., et al. (2017). High-throughput m6A-seq reveals RNA m6A methylation patterns in the chloroplast and mitochondria transcriptomes of Arabidopsis thaliana. PLoS One 12, e0185612. doi:10.1371/journal.pone.0185612
Wei Y., Melas P. A., Wegener G., Mathé A. A., Lavebratt C. (2014). Antidepressant-like effect of sodium butyrate is associated with an increase in TET1 and in 5-hydroxymethylation levels in the Bdnf gene. Int. J. Neuropsychopharmacol. 18, pyu032. doi:10.1093/ijnp/pyu032
Wilkinson B., Campbell D. B. (2013). Contribution of long noncoding RNAs to autism spectrum disorder risk. Int. Rev. Neurobiol. 113, 35–59. doi:10.1016/b978-0-12-418700-9.00002-2
Wong-Riley M. T., Tripathi S. C., Trusk T. C., Hoppe D. A. (1989). Effect of retinal impulse blockage on cytochrome oxidase-rich zones in the macaque striate cortex: I. Quantitative electron-microscopic (EM) analysis of neurons. Vis. Neurosci. 2, 483–497. doi:10.1017/s0952523800012384
Xian H., Karin M. (2023). Oxidized mitochondrial DNA: a protective signal gone awry. Trends Immunol. 44, 188–200. doi:10.1016/j.it.2023.01.006
Yakes F. M., Van Houten B. (1997). Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 94, 514–519. doi:10.1073/pnas.94.2.514
Ye N., Rao S., Du T., Hu H., Liu Z., Shen Y., et al. (2017). Intergenic variants may predispose to major depression disorder through regulation of long non-coding RNA expression. Gene 601, 21–26. doi:10.1016/j.gene.2016.11.041
Yılmaz S., Akan P., Baysal K., Özerdem A., Ceylan D. (2022). DNA base excision repair genes in unipolar and bipolar depression. Neurosci. Appl. 1, 69. doi:10.1016/j.nsa.2022.100183
Zeng Y., Lin R., Liu L., Liu Y., Li Y. (2019). Ambient air pollution exposure and risk of depression: a systematic review and meta-analysis of observational studies. Psychiatry Res. 276, 69–78. doi:10.1016/j.psychres.2019.04.019
Zeng Y., Navarro P., Shirali M., Howard D. M., Adams M. J., Hall L. S., et al. (2017). Genome-wide regional heritability mapping identifies a locus within the TOX2 gene associated with major depressive disorder. Biol. Psychiatry 82, 312–321. doi:10.1016/j.biopsych.2016.12.012
Zhang C., Jia G. (2018). Reversible RNA modification N(1)-methyladenosine (m(1)A) in mRNA and tRNA. Genomics Proteomics Bioinforma. 16, 155–161. doi:10.1016/j.gpb.2018.03.003
Zhang H. P., Liu X. L., Chen J. J., Cheng K., Bai S. J., Zheng P., et al. (2020). Circulating microRNA 134 sheds light on the diagnosis of major depressive disorder. Transl. Psychiatry 10, 95. doi:10.1038/s41398-020-0773-2
Zhao T., Piao L. H., Li D. P., Xu S. H., Wang S. Y., Yuan H. B., et al. (2023). BDNF gene hydroxymethylation in hippocampus related to neuroinflammation-induced depression-like behaviors in mice. J. Affect Disord. 323, 723–730. doi:10.1016/j.jad.2022.12.035
Zhao Y., Sun L., Wang R. R., Hu J. F., Cui J. (2018). The effects of mitochondria-associated long noncoding RNAs in cancer mitochondria: new players in an old arena. Crit. Rev. Oncol. Hematol. 131, 76–82. doi:10.1016/j.critrevonc.2018.08.005
Keywords: mood disorders, bipolar disorder, major depression, epigenetics, mitochondrial dysfunction, mitoepigenetics
Citation: Ceylan D, Arat-Çelik HE and Aksahin IC (2024) Integrating mitoepigenetics into research in mood disorders: a state-of-the-art review. Front. Physiol. 15:1338544. doi: 10.3389/fphys.2024.1338544
Received: 14 November 2023; Accepted: 24 January 2024;
Published: 08 February 2024.
Edited by:
Abdülhadi Cihangir Uguz, Karamanoğlu Mehmetbey University, TürkiyeReviewed by:
Nina Dalkner, Medical University of Graz, AustriaCopyright © 2024 Ceylan, Arat-Çelik and Aksahin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Deniz Ceylan, ZGVuaXpjZXlsYW5kckBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.