
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 25 October 2023
Sec. Aquatic Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1280553
This article is part of the Research Topic Endocrine Regulation and Physiological Adaptation of Stress Response in Aquatic Organisms View all 18 articles
 Xueshu Zhang1,2,3,4†
Xueshu Zhang1,2,3,4† Linfang Zhang1,3,4†
Linfang Zhang1,3,4† Yiran Si1,3,4
Yiran Si1,3,4 Xue Wen1,3,4
Xue Wen1,3,4 Lingling Wang1,2,3,4*
Lingling Wang1,2,3,4* Linsheng Song1,2,3,4*
Linsheng Song1,2,3,4*Ionotropic glutamate receptors (iGluRs), pivotal in mediating excitatory neurosignals within the central nervous system, are instrumental in environmental stress responses. In this investigation, 12 iGluRs identified in the Pacific oyster are herein designated as CgiGluRs, and further categorized into three distinct subfamilies based on their transmembrane domains. Cross-species evolutionary analysis unveiled a high degree of conservation in the sequence and structural attributes of these CgiGluRs. These receptors are ubiquitously distributed across various tissues, with pronounced expression in the oyster’s mantle, labial palps, and gills, underlining their integral role in the oyster’s environmental sensing mechanisms. Post the D-shaped larval stage, a marked upward trend in CgiGluRs expression was observed, denoting their critical involvement in oyster development beyond this phase. Exposure to five metals—cadmium (Cd), copper (Cu), zinc (Zn), mercury (Hg), and lead (Pb)—elicited a significant upregulation of CgGRIA4 expression, indicating a robust response to metal stress. A KEGG enrichment analysis on 142 genes, exhibiting parallel expression trends with CgGRIA4 under metal stress, suggests that CgGRIA4 could augment excitatory signal transmission by activating glutamatergic and dopaminergic synapses, thereby contributing to the metal stress response in the oyster. This inquiry not only bolsters our comprehension of the iGluRs gene family in metal stress response but also paves the way for future exploration of its cardinal role in cellular signaling and environmental adaptability.
Glutamate, the chief excitatory neurotransmitter within the nervous system, directs a plethora of physiological functions, encompassing neural remodeling (Endo et al., 2021), environmental sensing (Wen et al., 2020), and signal transduction (Qiu et al., 2020), via a diverse array of receptors. Pharmacological classifications segregate glutamate receptors into two primary categories: ionotropic (iGluRs) and metabotropic (mGluRs) receptors (Zhu and Gouaux, 2017). A substantial body of research accentuates the superior velocity of iGluRs in information transmission relative to mGluRs, underscoring their capability for rapid environmental stress responses. iGluRs, functioning as multimeric ion channels, are tasked with the swift excitatory transmission in the nervous system. Upon binding to pre-synaptically released glutamate, iGluRs transduce signals into post-synaptic neuronal excitation within milliseconds. This complex process gives rise to synaptic currents, crucial for neural regulatory functions, and modulates perception and information transmission (Moretto et al., 2018). iGluRs are further classified into N-methyl-D-aspartate (NMDA) receptors, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors, and kainate receptors (Mayer, 2016). According to the Motif structure diagram of model animals (human, mouse, zebrafish), we can see the differences among the three. Compared with NMDA-type receptors, Motif 8 and Motif 9 exist for AMPA-type receptors and KA-type receptors (Supplementary Figure S1). iGluRs subunits, dividing four modular structural domains including amino-terminal domain (ATD), ligand-binding domain (LBD), transmembrane domain (TMD), and C-terminal domain (CTD), coalesce into tetramers within their respective subclasses, forming ligand-gated ion channels (Karakas et al., 2015). The LBD contains two half-domains S1 and S2, which are closed to each other when LBD binds glutamate (Armstrong et al., 1998), and the Lig_Chan domain contains three transmembrane regions M1, M2, M3 and ion channel pore P (Kuner et al., 2003). Despite the extensive investigation of iGluRs in humans, mice, zebrafish, and other vertebrates over past decades (Herbrechter et al., 2021), due to their integral role in neuronal function, research in mollusks, particularly bivalves, is still nascent.
The rapid advancement of industry and agriculture in recent years has triggered a surge in marine pollution (Rahman et al., 2022). The environmental exposure to neurotoxic metals and metalloids, including cadmium, lead, mercury, copper, and zinc, has escalated into a global health concern, affecting millions worldwide (Liu et al., 2023). Research suggests that environmental neurotoxic metal stress can compromise neurotransmitter receptor function, thereby impinging on neural development, behavior, cognition, and precipitating neurodegeneration (Carmona et al., 2021). Existing evidence implicates Cd in directly affecting synaptic transmission mediated by AMPA receptors (Wang et al., 2008). Conversely, neurotoxic Pb exhibits significant selectivity for NMDA receptors, suggesting that the neurotoxicity of this metal is mediated by receptor-type-specific regulation (Marchetti and Gavazzo, 2003). Moreover, copper can bidirectionally modulate hippocampal neuronal synaptic activity: acute copper stimulation can impede signal transmission, but after a 3-h continuous copper stimulation, it amplifies the frequency and amplitude of AMPA currents (Peters et al., 2011). Recent research reveals that Cd downregulates NMDA receptors (GRIN2A and GRIN2B) and inhibits the activity of inhibitory glutamate receptor GluR2, while upregulating the phosphorylation of excitatory glutamate receptor GluR1, inducing functional impairment of glutamate receptors (Yang et al., 2023). Consequently, environmental neurotoxic metals can obstruct various functions of the entire nervous system via iGluRs, thereby disrupting organismal homeostasis (Pochwat et al., 2015). The chosen metals (Zn, Cu, Cd, Hg, and Pb), being prevalent marine pollutants with known iGluR interactions, are pivotal for examining environmental stress responses in Pacific oyster.
In this context, bivalves, such as the Pacific oyster (Crassostrea gigas), have emerged as a research focal point due to their unique resilience to metal pollution. Intriguingly, the oysters harbor high concentrations of metals without manifest toxicity, suggesting the evolution of sophisticated metal accumulation regulatory mechanisms (Jonathan et al., 2017). Investigations in vertebrates demonstrate the toxic effects of metals on ionotropic glutamate receptors, which can severely perturb iGluRs signal transmission (Sadiq et al., 2012). Prior research has corroborated the presence of a relatively comprehensive neuroendocrine system in oysters (Liu et al., 2018; Wang, 2022), yet reports on iGluRs and their regulation of metal ions are scant. Elucidating the mechanisms and strategies of bivalve iGluRs in response to metal stimulation holds profound implications for addressing environmental pollution and seafood safety issues.
Against this backdrop, the present study identified and systematically analyzed the iGluRs of C. gigas. Subsequently, the spatiotemporal expression spectrum of CgiGluRs genes was scrutinized using the RNA-seq dataset. Furthermore, this study probed the expression level and characteristics of CgiGluRs genes in the gills to decipher the molecular mechanisms underpinning oyster responses to heavy metal stress.
The BLASTP tool was deployed to decipher the gene sequence of iGluRs in the Pacific oyster. Amino acid sequences of iGluRs from a broad spectrum of invertebrates and vertebrates were leveraged as queries against the NCBI1 and Uniprot databases2 (UniProt Consortium, 2018). This exhaustive search spanned species from sea hare to human, inclusive of Xenopus tropicalis, Danio rerio, and Homo sapiens. The oyster transcriptome and whole genome sequences were meticulously examined to identify candidate iGluRs genes. Subsequent analyses involved predicting amino acid sequences using the ORF Finder tool3, identifying conserved structural domains via the SMART program4 (Letunic and Bork, 2018), and detecting conserved motifs using the MEME Suite5 (Nystrom and McKay, 2021), with a maximum motif limit set to 12 (prevent motif overlap and maintain analysis accuracy). All results were visualized using TBtools (Chen et al., 2020). The Compute pl/Mw tool6 (Wilkins et al., 1999) was utilized to calculate the GRAVY (Grand average of hydropathicity), theoretical isoelectric point (pI), and molecular weight (Mw) of the pore domain, while the secondary structure was predicted using Geneious7.0.67 (Kearse et al., 2012).
For the phylogenetic analysis, iGluRs proteins from C. gigas and other selected species, including the invertebrates and vertebrates, were selected. The iGluRs amino acid sequences from selected species were retrieved from the NCBI and Uniprot databases (Supplementary Table S1). Multiple sequence alignment was executed using AliView software (Larsson, 2014), followed by the construction of an evolutionary tree based on the maximum likelihood method via PhyML (v3.0) software8 (Guindon et al., 2010). The tree was subsequently refined using FigTree (v1.4.4) software9. Chromosomal locations and sizes of the iGluRs genes in the oyster were derived from the oyster genome data (cgigas_uk_roslin_v1) (Peñaloza et al., 2021), analyzed through TBtools.
Expression analysis was conducted using the RPKM (Reads Per Kilobase Million) values of each iGluRs gene from the publicly available RNA-seq dataset of the oyster. This dataset spans various developmental stages and adult tissues. Expression patterns of these genes across different stages and tissues were visualized using a heatmap generated by TBtools.
In investigating the transcriptional dynamics of iGluRs in oysters under heavy metal exposure conditions, we utilized an RNA-Seq dataset (Zhang et al., 2012), encompassing exposure data for Zn, Cu, Cd, Hg, and Pb. Specifically, oysters were exposed to one of the five metals (Zinc 1 mg/L, Cadmium 100 μg/L, Copper 100 μg/L, Lead 500 μg/L, Mercury 20 μg/L), with a control group subjected to seawater treatment. The concentrations of these metals were non-lethal, and no fatalities occurred during the experiment. The sampling time points were at 12 h and 9 days post-exposure. The original RNA-Seq data (Project number: PRJNA146329) were obtained from the NCBI database. Subsequently, these data were aligned to the oyster genome utilizing HISAT2 (v2.0.5) with default parameters. Gene expression levels were then estimated employing the Fragments Per Kilobase Million (FPKM) method. Temporal trends of gene expression under different metal exposures were analyzed and clustered using the Mfuzz R package (Kumar and Futschik, 2007) in R (version 4.2.3). A Venn diagram depicting the common expression trends of CgGRIA4 under five metal exposures was constructed using jvenn10 (Bardou et al., 2014).
Following the Venn diagram analysis, KEGG enrichment analysis was performed on all intersecting treatments using the R package clusterProfiler (Yu et al., 2012). The enrichKEGG function was used to identify enriched KEGG pathways among the genes listed in the Venn diagram, with a p-value <0.05 set as the threshold for significance. To further elucidate the response mechanism of CgGRIA4 to heavy metals, significantly enriched pathways (p < 0.05) involving the CgGRIA4 gene were screened. A network diagram of these pathways was constructed using the KEGG network tool of OmicShare Tools11. Enrichment pathways and gene information are detailed in Supplementary Table S2. Collectively, through KEGG enrichment analysis and pathway network diagramming, we aim to gain a deeper understanding of the expression pattern of CgGRIA4 under different metal exposures and its role in biological processes.
To elucidate the genomic landscape of the oyster, a comprehensive analysis of the transcriptome and genome databases was undertaken, leading to the discovery of 12 iGluRs genes. These genes, detailed in Table 2, were classified into three distinct subfamilies based on sequence homology and domain architecture: AMPA receptors, NMDA receptors, and kainate receptors (Table 1). The open reading frames (ORFs) of CgiGluRs spanned from 2,385 to 3,675, encoding between 794 and 1,224 amino acids. GRIA2 was found to be the most complex, comprising 19 exons and 18 introns (Supplementary Figure S2; Table 2). The predicted molecular weights of CgiGluRs ranged from 89.49 to 138.79 kDa, with predicted isoelectric points (pI) between 5.78 and 8.81. The secondary structure of the proteins encoded by iGluRs suggested a composition of 29–53 alpha helices, 44 to 74 beta strands, 58 to 93 coils, and 52 to 99 turns (Table 2). The amino acid consistency between CgiGluRs and iGluRs of other invertebrates ranged from 27.25% to 90.46%, and it ranged from 23.96% to 50.76% with vertebrate iGluRs (Table 3).
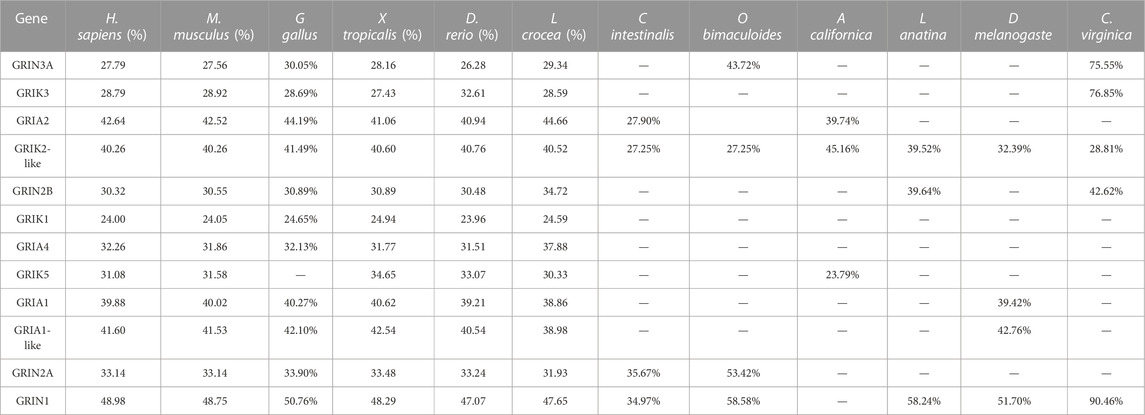
TABLE 3. Percentage of Identity(I) of C.gigas iGluRs with selected iGluRs proteins in other species.
A phylogenetic tree was constructed for CgiGluRs, and subsequent analysis of domain information and gene base sequence was conducted (Figure 1). All CgiGluRs were found to possess a Pfam Lig_Chan domain centrally, which belongs to the TMD module. (Figure 1C). The N-terminus of the CgNMDA subfamily was found to feature a PBP_type1 superfamily domain, which belongs to the LBD module. Certain CgiGluRs also contained specific structural regions, such as the Cam_bdg_C0 domain at the C-terminus of CgGRIN1, which is the key with NMDA-type receptors that allow calcium ions to pass through (Figure 1C). Twelve conserved motifs were identified in CgiGluRs, with CgiGluRs sharing eight common motifs (1–3, 5–9; Figure 1B). CgGRIN contained a unique motif, motif 11. All proteins, except for CgGRIN3A and CgGRIN1, possess motif 4 (Figure 1B). CgGRIN2B and CgGRIK3 lack motifs 10 and 12, CgGRIN2A and CgGRIN3A lack motif 10, and CgGRIK1 lacks motif 12 (Figure 1B).
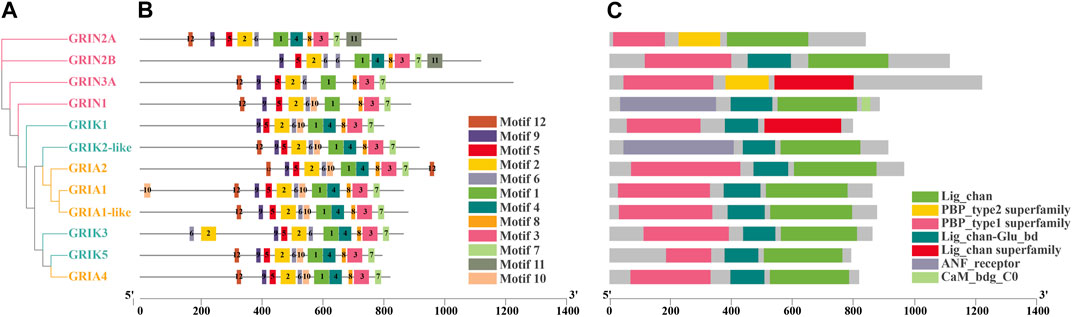
FIGURE 1. Phylogenetic relationships, protein motifs, and gene structures of 12 CgiGluRs genes. (A) Phylogenetic tree of CgiGluRs. Protein sequences were aligned using AliView, and the phylogenetic tree was constructed applying the maximum likelihood method. (B) Protein motifs of CgiGluRs. Conserved motifs (1–12) are depicted by different colored boxes, with non-conserved sequences represented by black lines. Motifs were visualized using Tbtools. (C) Gene structures: Lig_Chan domains, PBP_type1 superfamily domains, Lig_chan superfamily domains, ANF_receptor domains, and PBP_Type_2 superfamily domains are represented by green, yellow, pink, blue, and red boxes respectively.
A chromosome map of CgiGluRs was constructed based on the oyster genome sequence (Figure 2A). All 12 identified CgiGluRs were found to be located on the oyster chromosomes, primarily on chromosomes 1, 5, 7, and 10. Chr 7 hosts the majority (5 CgiGluRs) of CgiGluRs genes, while Chr 5 contains only two. Most CgiGluRs genes are found on Chr7 and Chr10 (9 out of 12, 75%), suggesting that the number of CgiGluRs genes is not related to chromosome size (Figure 2A).
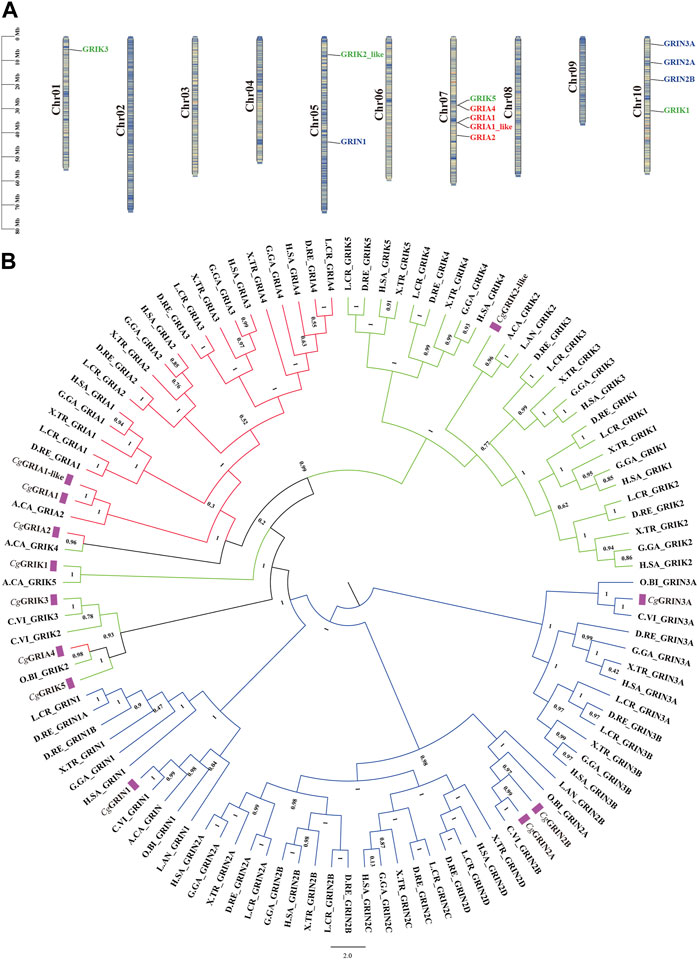
FIGURE 2. Phylogenetic Analysis and Chromosomal Localization of CgiGluRs. (A) Chromosomal distribution of the 12 CgiGluRs genes, along with the dispersion of duplicate gene pairs. Each gene is positioned on a chromosome according to its physical location, with the chromosome number (Chr01-Chr10) indicated on the left. (B) A multispecies phylogenetic tree, derived from the protein sequences of iGluRs from C. gigas and other selected species, was constructed using the maximum likelihood method and supported by 1,000 bootstrapped pseudoreplicates. CgiGluRs are marked in purple. Branches representing different subfamilies are highlighted in distinct colors (CgNMDAR: blue, CgAMPAR: red, CgKAR: green). Species abbreviations are as follows: Cg, Crassostrea gigas; H.SA, Homo sapiens; G.GA, Gallus gallus; X. TR, Xenopus tropicalis; D.RE, Danio rerio; O.BI, Octopus bimaculoides; A.CA, Aplysia californica; L. AN, Lingula anatine; L. CR, Larimichthys crocea; C.VI, Crassostrea virginica.
In this study, a phylogenetic tree was constructed by comparing the full-length amino acid sequences of CgiGluRs and those from other species (Figure 2B; Table 4). The results reveal that the CgiGluRs family can be divided into two main branches: NMDA and non-NMDA receptors, which are further subdivided into three subfamilies, namely, GRIA, GRIK, and GRIN (Figure 2B). The classification of each subfamily is based on genetic similarity. Within each subfamily, the iGluRs members of vertebrates and invertebrates form independent branches. Notably, the genes of the American oyster and the oyster share the closest evolutionary relationship within the same iGluRs subfamily (Figure 2B). In the specific construction of the phylogenetic tree, the red branch represents the GRIA subfamily. Among them, CgGRIA1, CgGRIA1-like, and A.CA GRIA2 form a branch. CgGRIA2 and A.CA GRIK4 form a branch. CgGRIA4, O.BI GRIK2, and CgGRIK5 form a branch, and then form a branch with CgGRIK3, C.VI GRIK2, and C.VI GRIK3 forms a branch. The green branch represents the GRIK subfamily. In this subfamily, CgGRIK1 forms a branch with the A.CA GRIK5, while CgGRIK2-like forms a branch with A.CA GRIK2, L. AN GRIK2. The blue branch represents the GRIN subfamily, which contains four CgGRIN genes. It is worth noting that the number of three subfamilies including GRIA, GRIK, and GRIN has significantly increased in vertebrates, indicating that these iGluRs subfamilies have been continuously expanded during evolution (Figure 2B).
RNA-seq datasets from different developmental stages and adult tissues of the oyster were used to detect the spatiotemporal expression spectrum of CgiGluRs (Figure 3). The expression patterns of CgiGluRs can be divided into two groups across different developmental stages (Figure 3A). The first group consists of 9 CgiGluRs that are highly expressed after D-shaped larvae, and these CgiGluRs have higher expression levels during the Pediveliger period than during the Later umbo larva period and Spat period. The second group consists of 3 CgiGluRs that are highly expressed before D-shaped larvae, and these CgiGluRs have different expression patterns throughout the development of the oyster.
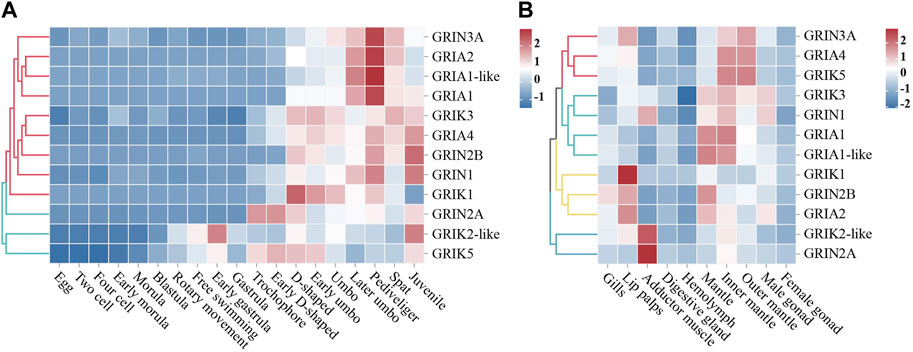
FIGURE 3. Spatiotemporal Expression Patterns of CgiGluRs. (A) A heat map depicting the expression profiles of CgiGluRs genes across various developmental stages, quantified by Log10RPKM. The stages included are egg, two-cell, four-cell, early morula, morula, blastula, rotary movement, free swimming, early gastrula, gastrula, trochophore, early D-shaped, D-shaped, early umbo, umbo, later umbo, pediveliger, spat, and juvenile. Based on the expression patterns of CgiGluRs, two primary clusters are delineated, represented in red and blue colors. (B) A heat map presenting the tissue-specific expression profiles of CgiGluRs genes in adult oysters, quantified by Log10RPKM. The tissues represented include gills, lip palps, adductor muscle, digestive gland, hemolymph, mantle, inner margin of mantle, outer margin of mantle, female gonad, and male gonad. Based on the expression patterns of CgiGluRs are grouped into four clusters, denoted by red, cyan, yellow, and blue.
In adult oyster tissues, CgiGluRs expression patterns are categorized into four distinct groups as illustrated in Figure 3B. GRIN3A, GRIA4, and GRIK5 from the first group predominantly exhibit expression in the mantle and its edge. The second group, which includes CgGRIK3, CgGRIN1, CgGRIA1, and CgGRIA1-like, primarily shows expression in the inner edge of the mantle. The labial palps are the main expression site for the third group, containing CgGRIK1, CgGRIN2B, and CgGRIA2. The adductor muscle expresses the fourth group, represented by CgGRIK2-like and CgGRIN2A. Additionally, peak expressions of CgiGluRs in adult oysters are found in neural tissues associated with environmental perception, encompassing areas like the labial palps, adductor muscle, and mantle edges.
To detect the expression pattern of CgiGluRs in response to heavy metal stress, the RNA-seq dataset of oyster gills under the stress of five heavy metals Zn, Cu, Cd, Hg, Pb were analyzed (Figure 4). The expression levels of GRIA1 and GRIA1-like mRNA decreased after short-term exposure to the five heavy metals, but under long-term exposure to Cu, the expression of these two CgiGluRs returned to normal level (Figures 4A–E). In addition, long-term exposure to Cu and Cd inhibited the expression of CgGRIA2 (Figures 4A, B). Under short-term exposure to Cu, Cd, Hg, and Pb, the expression of CgGRIA4 and CgGRIK5 was upregulated (Figures 4A–D). The expression levels of CgGRIA4 and CgGRIK5 were upregulated under long-term exposure to Zn (Figure 4E). Collectively, the results highlight the pronounced responsiveness of CgGRIA4 and CgGRIK5 to heavy metal perturbations.

FIGURE 4. Expression of CgiGluRs in Response to Metal Exposure. (A) Changes in relative gene expression (expressed as fold change relative to control) in oysters exposed to cadmium (Cd). (B) Relative gene expression alterations (expressed as fold change relative to control) in oysters in response to copper (Cu) exposure. (C) Modulations in relative gene expression (expressed as fold change relative to control) in oysters following Mercury (Hg) exposure. (D) Adjustments in relative gene expression (expressed as fold change relative to control) in oysters subjected to lead (Pb) exposure. (E) Changes in relative gene expression (expressed as fold change relative to control) in oysters upon zinc (Zn) exposure. (F) A Venn diagram presenting the genes demonstrating similar expression trends to GRIA4 under the stimulation of the five metals.
The analysis prioritized CgGRIA4 over GRIK5 due to the established association of AMPA-type receptors, to which GRIA4 belongs, with calcium ion permeation critical in metal stress response. To better understand GRIA4’s role in metal exposure, a detailed analysis following exposure to five metals identified 142 genes with similar expression trends (Supplementary Figure S3; Figure 4F). Through KEGG enrichment analysis of these genes, we found that pathways related to neural signal transmission, such as Glutamatergic synapse (ko04724), Dopaminergic synapse (ko04728), and Neuroactive ligand-receptor interaction (ko04080), were significantly enriched (Figure 5A). In addition, some antioxidant-related metabolic pathways, such as Vitamin B6 metabolism (ko00750), Vitamin digestion and absorption (ko04977), and Selenocompound metabolism (ko00450), were also significantly enriched (Figure 5A). To reveal the mechanism of CgGRIA4 in metal stress response more deeply, we drew a KEGG network map of the genes in the 142 genes that share the same pathway with CgGRIA4 (Figure 5B). In the KEGG network map, CgGRIA4 mainly participates in the activation of Glutamatergic synapse and Dopaminergic synapse, and CgGRIA4 mainly affects two neurodegenerative disease-related pathways, Spinocerebellar ataxia (ko05017), and Huntington disease (ko05016), and the activation of these two pathways is closely related to calcium ion homeostasis imbalance (Figure 5) (Begum et al., 2018; Zhou et al., 2018).
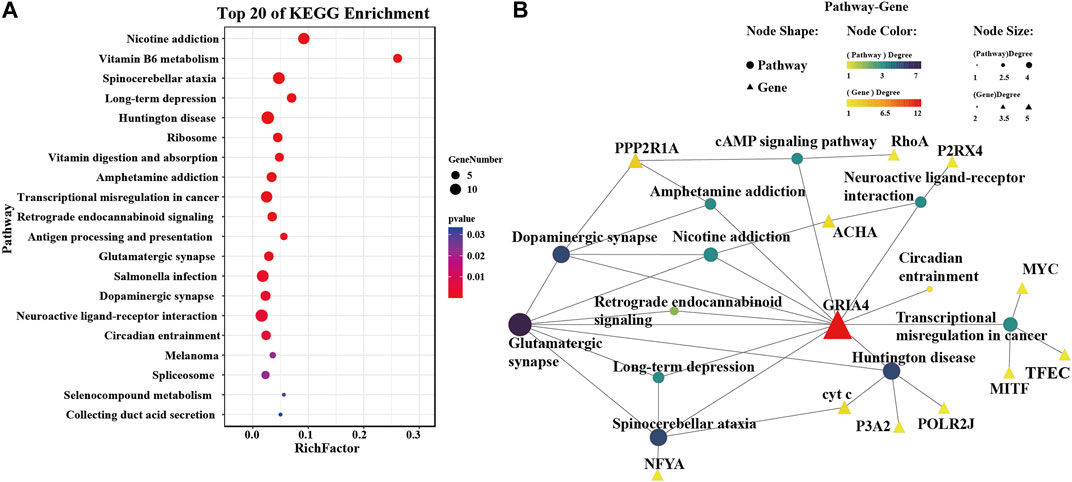
FIGURE 5. KEGG enrichment analysis of genes expressing similarly to CgGRIA4. (A) The top 20 enriched KEGG terms identified among the 142 genes depicted in Figure 4F. (B) A network interaction model constructed from the KEGG enrichment analysis performed on the 142 intersecting genes presented in Figure 4F. This analysis enabled the identification of KEGG pathways significantly engaged by CgGRIA4, and these pathways are depicted in the model to demonstrate their interconnections.
Ionotropic Glutamate Receptors (iGluRs) are a key type of ion channel widely distributed across the animal kingdom. Upon activation, they transmit signals of sodium, potassium, or calcium ions, participating in various sensory processes (Manookin et al., 2008; Sánchez-Alcañiz et al., 2018) and playing a crucial role in physiological processes such as neuroplasticity (Budreck et al., 2013), learning and memory, cell life cycle, and immune defense. In this study, a complete set of iGluRs family genes was identified in the genome of the oyster C. gigas, and their protein structure, phylogenetic relationships, expression patterns during developmental stages and in the adult tissues under heavy metal stress were analyzed. The results provide a new perspective for a deeper understanding of the molecular evolution and functional diversity of the iGluRs channel family.
Our comprehensive genomic screening revealed the presence of 12 CgiGluRs family genes in oysters. These genes are widely distributed across the three iGluRs subfamilies: NMDA receptors (NMDAR), AMPA receptors (AMPAR), and kainate receptors (KAR). By contrast, the iGluRs family gene combination in vertebrates, such as humans (Hansen et al., 2021), is more diverse, encompassing 7 NMDARs, 4 AMPARs, and 5 KARs. In vertebrate evolution, the expansion of KAR and AMPAR genes is notably more significant than in mollusks, as evidenced by the pronounced difference in the neural system. This expansion is likely an adaptation to the more complex requirements of neural signal transmission. In the CgNMDAR subfamily of oysters, a singular CgGRIN1 with the CaM_bdg_C0 domain has been discerned. Contrarily, zebrafish exhibit an array of GRIN copies, each embedded with the CaM_bdg_C0 domain, pivotal for the regulation of Ca2+ influx (Cox et al., 2005). Such distinctions underscore the potential evolutionary adaptation of calcium ion mediation in tandem with the intricacies of the neural system. In summary, the genomic variations in iGluRs family genes between oysters and vertebrates underscore the evolutionary intricacies and adaptive nature of neural systems, especially in calcium ion mediation, to environmental complexity.
In both vertebrate and invertebrate species, proteins of the iGluRs class, including those in the CgiGluRs family, consistently exhibit four distinct transmembrane structural domains. Intriguingly, comparative analyses reveal no significant divergence in these domains across the various iGluRs subfamilies. Within vertebrates, these subfamilies are systematically designated based on their specific affinities for synthetic agonists, namely, AMPA, NMDA, and kainate (Hansen et al., 2021). In CgGRIN1, the NMDA receptor GRIN1 subunit calmodulin binding domain C0 domain (CaM_bdg_C0) was found. This is a necessary subunit that allows Ca2+ to pass through, constant with the role of GRIN1 as an essential subunit of the NMDA receptor and its mediating function of Ca2+ channels (Ganor and Levite, 2014). There was an ANF_receptor domain identified in CgGRIN2A and CgGRIK2-like. The rest of the CgiGluRs possess the PBP_Type_2 superfamily domain. These domains form the structural basis for the C-terminal structural domain (CTD) of CgiGluRs to recognize extracellular signals. In CgiGluRs, the CTD length varies between different iGluRs subfamilies. Compared with CgAMPAR and CgKAR, CgNMDA receptors, excluding GRIN1, have a longer CTD. Studies have shown that the CTD of GluN2 is the longest (Dravid et al., 2010). This diversity of subunits in the CTD is thought to play specialized and complex roles in neurons. The above results indicate that CgiGluRs share similar domain structure with their homologues from other species, and the structural differences between members of the CgiGluRs family may directly reflect their functional diversity.
Phylogenetic analysis showcases a primary bifurcation of the CgiGluRs family into NMDA-type and non-NMDA-type receptors, aligning with prior research (Stroebel and Paoletti, 2021). CgGRIA4, CgGRIK3, and CgGRIK5 cluster with other invertebrate iGluRs proteins like the GRIK2 and GRIK3 of the Portuguese oyster, and the GRIK2 of the California double sheath, highlighting a close phylogenetic relationship. This relationship is likely fostered by structural similarities between AMPA-type and KA-type receptors, and is further supported by the molecular secretion complexity of glutamatergic synapses, illustrating an intricate evolutionary interplay. A comparative study reveals a significant phylogenetic link between the iGluRs families of the Pacific oyster and the California Sea Hare (Aplysia californica), with bootstrap analyses supporting this relationship. This underlines the conservation of the iGluRs family across species and its key role in environmental adaptability. NMDAR subfamily members are found from bacteria to mammals, suggesting it as the most ancestral lineage, followed by KAR and AMPAR (Chen et al., 1999). The iGluRs family attains functional diversity via subunit combinations and RNA editing, vital for environmental adaptation. The divergence into NMDA and non-NMDA types might reflect environmental pressures, with each type potentially offering different adaptive advantages in response to varying environmental conditions such as temperature and salinity changes (Busnardo et al., 2016; Stroebel and Paoletti, 2021). This insight offers a refined perspective on the role of iGluRs in neural signal transmission and environmental adaptability.
Prior research underscores the crucial role of iGluRs in embryonic development, with AMPA (3, 4) and Kainate (3, 4, 5) receptor abnormalities affecting mouse blastocyst development (Spirkova et al., 2022). While in vertebrates like mice, iGluRs function primarily as excitatory neurotransmitters, in bivalves, they serve different functional roles, illuminating the functional divergence across phylogenetically distant taxa. This study reveals an increase in iGluRs expression correlating with oyster larvae development (egg average RPKM = 1.5, juvenile average RPKM = 137.9), indicating CgiGluRs’ involvement in this process. Notably, iGluRs expression escalates during the pediveliger period (average RPKM = 133.7), a critical stage where the eyespot and the foot develop. Recent findings suggest the eyespot has photoreceptive abilities, and the foot engages in sensory perception and locomotion (Vogeler et al., 2016; Zhang et al., 2021). Vertebrate iGluRs are pivotal in signal transduction for environmental cue perception (Levitz et al., 2016; van Giesen and Garrity, 2017). The pronounced iGluRs expression during the pediveliger stage highlights their essential role in pediveligers’ environmental perception. Particularly, CgGRIA4 expression peaks in this phase (RPKM = 416), aligning with its AMPA-type counterparts. The functional assembly of AMPA-type receptors as either homomeric or heteromeric tetramers (Hanada, 2020) suggests the dominance of CgGRIA4 in steering the perceptual processes of pediveligers.
iGluRs are central to neural systems, mediating complex cerebral functions including neural transmission and memory (Sachser et al., 2017; Shahin et al., 2018; Hayashi, 2021). Our data from adult oysters show receptor-specific expression profiles. Elevated expression of CgGRIA4 in the mantle suggests its role in sensory and environmental perception, aligning with GRIA4’s known role in vertebrate synaptic transmission (Sagata et al., 2010). Enhanced CgGRIK1 expression in the labial palps hints at its potential role in alimentary or environmental detection, mirroring the sensory function of GRIK1 (Englund et al., 2021). Notably, pronounced CgGRIN2A expression in the adductor muscle indicates possible implications in shell dynamics, resonating with Zhao et al. (2023) identification of GRIN2A as a neural excitability modulator. Our findings highlight the critical roles of CgiGluRs in oyster physiology, emphasizing their evolutionary conservation and parallels with vertebrate neural systems. Additionally, we propose that oyster iGluRs might detect metal concentration changes, with certain metal ions potentially interacting with specific iGluRs domains, thus altering channel dynamics, and influencing neural or other physiological responses to environmental stress.
iGluRs are instrumental in mitigating the neurotoxic effects of heavy metals (Slotkin and Seidler, 2009). Our analyses delineate the nuanced responses of CgiGluRs to specific metal challenges. Intriguingly, CgGRIA1 expression is reduced under acute exposure to five metals (Zn, Cu, Cd, Hg, and Pb), yet demonstrates resilience during extended Cu and Pb challenges. This pattern suggests that CgGRIA1 might bolster cellular robustness by dynamically modulating its expression in response to metal-induced stress. Studies in vertebrates emphasize the neuroprotective advantages of GRIA1 downregulation. Furthermore, shifts in the GRIA1 to GRIA2 ratio are postulated to modulate the calcium permeability of CP-AMPAR (Li et al., 2023). In our dataset, CgGRIA2 shows a contrasting expression pattern under copper and lead exposure. Given GRIA2’s pivotal role in dictating AMPAR calcium permeability, it is plausible that CgAMPAR receptors adapt to metal stress by fine-tuning calcium homeostasis. We observed a marked upregulation of CgGRIA4 under metal stress. As a subtype of the AMPA receptor, GRIA4 is integral to rapid synaptic signaling (Song and Huganir, 2002). The pronounced expression of CgGRIA4 intimates an adaptive strategy in oysters, potentially fortifying cellular defenses against metal-induced stress. However, while the majority of AMPA receptors typically have low calcium permeability, certain stressors, such as metals, might amplify this characteristic, risking neuronal integrity. The enhanced expression of CgGRIA4, albeit potentially beneficial, could also precipitate calcium dysregulation and subsequent neurotoxicity if unchecked (Kim and von Gersdorff, 2016; Yang et al., 2023).
Within the neural framework, iGluRs play an indispensable role in preserving neuronal health and orchestrating functional dynamics. Our research highlights the sensitivity of CgGRIA4 to heavy metal stress. We observed genes with expression patterns that mirror CgGRIA4, primarily associated with Glutamatergic and Dopaminergic synapses. GRIA4, recognized for its prompt responsiveness to glutamate, is pivotal in facilitating efficient neurotransmission (Tritsch and Sabatini, 2012). Parallelly, dopaminergic modulation has been documented to sculpt the functional dynamics and membrane transport of AMPA receptors. This interplay suggests that CgGRIA4 might channel neurotransmission via the Glutamatergic pathway, with its signal intensity potentially under the regulatory purview of the Dopaminergic signaling axis. Additionally, our data suggest a potential role for CgGRIA4 in pathways related to neurodegenerative conditions. The activation of these pathways appears to be intertwined with perturbations in calcium homeostasis (Wakazono et al., 2023), underscoring the prospective role of CgGRIA4 in bolstering cellular defenses against heavy metal stress through judicious calcium regulation. Additionally, our analysis reveals a pronounced enrichment in antioxidant metabolic pathways, notably those pivoting around Vitamin B6 and selenium derivatives (Binte Hossain et al., 2018; Ko et al., 2022). This enrichment suggests a strategic role for CgGRIA4 in mitigating oxidative duress stemming from heavy metal exposure, echoing seminal research that underscores the neuroprotective virtues of Vitamin B6 and selenium compounds in countering oxidative stress. This suggests a strategic role for CgGRIA4 in mitigating oxidative stress from heavy metal exposure, possibly aiding in oysters’ environmental adaptability to varying conditions like different water temperatures, salinity levels, or pollution levels.
This research illuminates the pervasive distribution of iGluRs in oysters, emphasizing their central importance in physiological functions. A comprehensive set of iGluRs family genes has been identified within the genome, with these genes demonstrating varied expression patterns across developmental stages, within adult tissues, and under the duress of heavy metal stress. Importantly, our findings indicate that CgGRIA4 can actively respond to heavy metal stress, potentially aiding cells in resisting such stress by engaging in neural signal transmission and antioxidant stress response. However, the specific modulation of these signaling pathways by CgGRIA4, and the question of whether its upregulation might precipitate calcium overload and neurotoxicity, still necessitates further exploration. These findings provide a fresh vantage point for a more profound understanding of the mechanisms of neurotoxicity.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA146329.
The animal study was approved by the Laboratory Animal Ethics Committee of Dalian Ocean University. The study was conducted in accordance with the local legislation and institutional requirements.
XZ: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. LZ: Data curation, Formal Analysis, Investigation, Software, Visualization, Writing–original draft. YS: Visualization, Writing–original draft, Data curation, Investigation, Methodology, Software. XW: Visualization, Writing–original draft, Formal Analysis. LW: Data curation, Funding acquisition, Methodology, Project administration, Resources, Writing–review and editing. LS: Data curation, Funding acquisition, Methodology, Project administration, Resources, Writing–review and editing.
The authors declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by grants from National Natural Science Foundation of China (41961124009), National key R&D Program of China (2018YFD0900606), earmarked fund (CARS-49) from Modern Agro-industry Technology Research System, the fund for Outstanding Talents and Innovative Team of Agricultural Scientific Research in MARA, Distinguished Professor in Liaoning (to LW, XLYC1902012), the innovation team of Aquaculture Environment Safety from Liaoning Province (LT202009), and Dalian High Level Talent Innovation Support Program (2022RG14), and Talented Scholars in Dalian Ocean University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1280553/full#supplementary-material
SUPPLEMENTARY TABLE S1 | Statistical information on the amino acid sequences of the iGluRs of the selected species (search databases: NCBI, Uniprot).
SUPPLEMENTARY TABLE S2 | Statistical table of KEGG-enriched gene and pathway information.
SUPPLEMENTARY FIGURE S1 | Phylogenetic relationships and protein motifs of vertebrates iGluRs genes (Homo sapiens, Mus musculus, Danio rerio). (A) Phylogenetic tree of vertebrates iGluRs (Homo sapiens, Mus musculus, Danio rerio). Protein sequences were aligned using AliView, and the phylogenetic tree was constructed applying the maximum likelihood method. (B) Protein motifs of vertebrates iGluRs (Homo sapiens, Mus musculus, Danio rerio). Conserved motifs (1–12) are depicted by different colored boxes, with non-conserved sequences represented by black lines. Motifs were visualized using Tbtools.
SUPPLEMENTARY FIGURE S2 | Phylogenetic tree and gene structure of CgiGluRs. (A) Phylogenetic tree of CgiGluRs. (B) Black lines, green and yellow boxes indicate the structure of the untranslated region (UTR), Intervening region, and expressed region.
SUPPLEMENTARY FIGURE S3 | Temporal trend and clustering of gene expression under different metal stimulation were analyzed using the Mfuzz package. A to E correspond to different time patterns of protein expression under the stimulation of five metals (Cd, Cu, Hg, Pb, and Zn). The x axis represents three stimulation duration, while the y axis represents log2-transformed, normalized intensity ratios in each stage.
1https://www.ncbi.nlm.nih.gov/
3https://www.ncbi.nlm.nih.gov/orffinder/
4http://smart.embl-heidelberg.de/
5https://meme-suite.org/meme/tools/meme
6https://web.expasy.org/compute_pi/
8http://www.atgc-montpellier.fr/
9http://tree.bio.ed.ac.uk/software/Figtree/
10https://jvenn.toulouse.inrae.fr/app/example.html
Armstrong N., Sun Y., Chen G.-Q., Gouaux E. (1998). Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature 395, 913–917. doi:10.1038/27692
Bardou P., Mariette J., Escudié F., Djemiel C., Klopp C. (2014). jvenn: an interactive Venn diagram viewer. BMC Bioinforma. 15, 293. doi:10.1186/1471-2105-15-293
Begum G., Otsu M., Ahmed U., Ahmed Z., Stevens A., Fulton D. (2018). NF-Y-dependent regulation of glutamate receptor 4 expression and cell survival in cells of the oligodendrocyte lineage. Glia 66, 1896–1914. doi:10.1002/glia.23446
Binte Hossain K. F., Rahman Md. M., Sikder Md. T., Saito T., Hosokawa T., Kurasaki M. (2018). Inhibitory effects of selenium on cadmium-induced cytotoxicity in PC12 cells via regulating oxidative stress and apoptosis. Food Chem. Toxicol. 114, 180–189. doi:10.1016/j.fct.2018.02.034
Budreck E. C., Kwon O.-B., Jung J. H., Baudouin S., Thommen A., Kim H.-S., et al. (2013). Neuroligin-1 controls synaptic abundance of NMDA-type glutamate receptors through extracellular coupling. Proc. Natl. Acad. Sci. 110, 725–730. doi:10.1073/pnas.1214718110
Busnardo C., Crestani C. C., Fassini A., Resstel L. B. M., Corrêa F. M. A. (2016). NMDA and non-NMDA glutamate receptors in the paraventricular nucleus of the hypothalamus modulate different stages of hemorrhage-evoked cardiovascular responses in rats. Neuroscience 320, 149–159. doi:10.1016/j.neuroscience.2016.02.003
Carmona A., Roudeau S., Ortega R. (2021). Molecular mechanisms of environmental metal neurotoxicity: a focus on the interactions of metals with synapse structure and function. Toxics 9, 198. doi:10.3390/toxics9090198
Chen G.-Q., Cui C., Mayer M. L., Gouaux E. (1999). Functional characterization of a potassium-selective prokaryotic glutamate receptor. Nature 402, 817–821. doi:10.1038/45568
Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Cox J. A., Kucenas S., Voigt M. M. (2005). Molecular characterization and embryonic expression of the family of N-methyl-D-aspartate receptor subunit genes in the zebrafish. Dev. Dyn. 234, 756–766. doi:10.1002/dvdy.20532
Dravid S. M., Burger P. B., Prakash A., Geballe M. T., Yadav R., Le P., et al. (2010). Structural determinants of d-cycloserine efficacy at the NR1/NR2C NMDA receptors. J. Neurosci. 30, 2741–2754. doi:10.1523/JNEUROSCI.5390-09.2010
Endo Y., Zhang Y., Olumi S., Karvar M., Argawal S., Neppl R. L., et al. (2021). Exercise-induced gene expression changes in skeletal muscle of old mice. Genomics 113, 2965–2976. doi:10.1016/j.ygeno.2021.06.035
Englund J., Haikonen J., Shteinikov V., Amarilla S. P., Atanasova T., Shintyapina A., et al. (2021). Downregulation of kainate receptors regulating GABAergic transmission in amygdala after early life stress is associated with anxiety-like behavior in rodents. Transl. Psychiatry 11, 538. doi:10.1038/s41398-021-01654-7
Ganor Y., Levite M. (2014). The neurotransmitter glutamate and human T cells: glutamate receptors and glutamate-induced direct and potent effects on normal human T cells, cancerous human leukemia and lymphoma T cells, and autoimmune human T cells. J. Neural Transm. 121, 983–1006. doi:10.1007/s00702-014-1167-5
Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi:10.1093/sysbio/syq010
Hanada T. (2020). Ionotropic glutamate receptors in epilepsy: a review focusing on AMPA and NMDA receptors. Biomolecules 10, 464. doi:10.3390/biom10030464
Hansen K. B., Wollmuth L. P., Bowie D., Furukawa H., Menniti F. S., Sobolevsky A. I., et al. (2021). Structure, function, and pharmacology of glutamate receptor ion channels. Pharmacol. Rev. 73, 1469–1658. doi:10.1124/pharmrev.120.000131
Hayashi T. (2021). Post-translational palmitoylation of ionotropic glutamate receptors in excitatory synaptic functions. Br. J. Pharmacol. 178, 784–797. doi:10.1111/bph.15050
Herbrechter R., Hube N., Buchholz R., Reiner A. (2021). Splicing and editing of ionotropic glutamate receptors: a comprehensive analysis based on human RNA-Seq data. Cell. Mol. Life Sci. 78, 5605–5630. doi:10.1007/s00018-021-03865-z
Jonathan M. P., Muñoz-Sevilla N. P., Góngora-Gómez A. M., Luna Varela R. G., Sujitha S. B., Escobedo-Urías D. C., et al. (2017). Bioaccumulation of trace metals in farmed pacific oysters Crassostrea gigas from SW Gulf of California coast, Mexico. Chemosphere 187, 311–319. doi:10.1016/j.chemosphere.2017.08.098
Karakas E., Regan M. C., Furukawa H. (2015). Emerging structural insights into the function of ionotropic glutamate receptors. Trends Biochem. Sci. 40, 328–337. doi:10.1016/j.tibs.2015.04.002
Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi:10.1093/bioinformatics/bts199
Kim M.-H., von Gersdorff H. (2016). Postsynaptic plasticity triggered by Ca2+-permeable AMPA receptor activation in retinal amacrine cells. Neuron 89, 507–520. doi:10.1016/j.neuron.2015.12.028
Ko J. W., Jeon S., Kwon Y. H. (2022). Dietary vitamin B6 restriction aggravates neurodegeneration in mice fed a high-fat diet. Life Sci. 309, 121041. doi:10.1016/j.lfs.2022.121041
Kumar L., Futschik M. E. (2007). Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2, 5–7. doi:10.6026/97320630002005
Kuner T., Seeburg P. H., Robert Guy H. (2003). A common architecture for K+ channels and ionotropic glutamate receptors? Trends Neurosci. 26, 27–32. doi:10.1016/S0166-2236(02)00010-3
Larsson A. (2014). AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30, 3276–3278. doi:10.1093/bioinformatics/btu531
Letunic I., Bork P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi:10.1093/nar/gkx922
Levitz J., Popescu A. T., Reiner A., Isacoff E. Y. (2016). A toolkit for orthogonal and in vivo optical manipulation of ionotropic glutamate receptors. Front. Mol. Neurosci., 9. doi:10.3389/fnmol.2016.00002
Li Y., Liang Z., Lei S., Wu X., Yuan T., Ma K., et al. (2023). Sevoflurane preconditioning downregulates GRIA1 expression to attenuate cerebral ischemia-reperfusion-induced neuronal injury. Neurotox. Res. 41, 29–40. doi:10.1007/s12640-022-00620-5
Liu Z., Li M., Yi Q., Wang L., Song L. (2018). The neuroendocrine-immune regulation in response to environmental stress in marine bivalves. Front. Physiol. 9. doi:10.3389/fphys.2018.01456
Liu D., Shi Q., Liu C., Sun Q., Zeng X. (2023). Effects of endocrine-disrupting heavy metals on human health. Toxics 11, 322. doi:10.3390/toxics11040322
Manookin M. B., Beaudoin D. L., Ernst Z. R., Flagel L. J., Demb J. B. (2008). Disinhibition combines with excitation to extend the operating range of the OFF visual pathway in daylight. J. Neurosci. 28, 4136–4150. doi:10.1523/JNEUROSCI.4274-07.2008
Marchetti C., Gavazzo P. (2003). Subunit-dependent effects of nickel on NMDA receptor channels. Mol. Brain Res. 117, 139–144. doi:10.1016/S0169-328X(03)00293-6
Mayer M. L. (2016). Structural biology of glutamate receptor ion channel complexes. Curr. Opin. Struct. Biol. 41, 119–127. doi:10.1016/j.sbi.2016.07.002
Moretto E., Murru L., Martano G., Sassone J., Passafaro M. (2018). Glutamatergic synapses in neurodevelopmental disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry 84, 328–342. doi:10.1016/j.pnpbp.2017.09.014
Nystrom S. L., McKay D. J. (2021). Memes: a motif analysis environment in R using tools from the MEME Suite. PLoS Comput. Biol. 17, e1008991. doi:10.1371/journal.pcbi.1008991
Peñaloza C., Gutierrez A. P., Eöry L., Wang S., Guo X., Archibald A. L., et al. (2021). A chromosome-level genome assembly for the Pacific oyster Crassostrea gigas. Gigascience 10. doi:10.1093/gigascience/giab020
Peters C., Muñoz B., Sepúlveda F. J., Urrutia J., Quiroz M., Luza S., et al. (2011). Biphasic effects of copper on neurotransmission in rat hippocampal neurons. J. Neurochem. 119, 78–88. doi:10.1111/j.1471-4159.2011.07417.x
Pochwat B., Nowak G., Szewczyk B. (2015). Relationship between zinc (Zn2+) and glutamate receptors in the processes underlying neurodegeneration. Neural Plast. 2015, 1–9. doi:10.1155/2015/591563
Qiu X. M., Sun Y. Y., Ye X. Y., Li Z. G. (2020). Signaling role of glutamate in plants. Front. Plant Sci. 10. doi:10.3389/fpls.2019.01743
Rahman M. M., Hossain M. K. F. B., Afrin S., Saito T., Kurasaki M. (2022). “Effects of metals on human health and ecosystem,” in The handbook of environmental chemistry (Berlin, Heidelberg: Springer), 81–119. doi:10.1007/698_2021_825
Sachser R. M., Haubrich J., Lunardi P. S., de Oliveira Alvares L. (2017). Forgetting of what was once learned: exploring the role of postsynaptic ionotropic glutamate receptors on memory formation, maintenance, and decay. Neuropharmacology 112, 94–103. doi:10.1016/j.neuropharm.2016.07.015
Sadiq S., Ghazala Z., Chowdhury A., Büsselberg D. (2012). Metal toxicity at the synapse: presynaptic, postsynaptic, and long-term effects. J. Toxicol. 2012, 1–42. doi:10.1155/2012/132671
Sagata N., Iwaki A., Aramaki T., Takao K., Kura S., Tsuzuki T., et al. (2010). Comprehensive behavioural study of GluR4 knockout mice: implication in cognitive function. Genes. Brain Behav. 9, 899–909. doi:10.1111/j.1601-183X.2010.00629.x
Sánchez-Alcañiz J. A., Silbering A. F., Croset V., Zappia G., Sivasubramaniam A. K., Abuin L., et al. (2018). An expression atlas of variant ionotropic glutamate receptors identifies a molecular basis of carbonation sensing. Nat. Commun. 9, 4252. doi:10.1038/s41467-018-06453-1
Shahin S., Banerjee S., Swarup V., Singh S. P., Chaturvedi C. M. (2018). From the cover: 2.45-GHz microwave radiation impairs hippocampal learning and spatial memory: involvement of local stress mechanism-induced suppression of iGluR/ERK/CREB signaling. Toxicol. Sci. 161, 349–374. doi:10.1093/toxsci/kfx221
Slotkin T. A., Seidler F. J. (2009). Oxidative and excitatory mechanisms of developmental neurotoxicity: Transcriptional profiles for chlorpyrifos, diazinon, dieldrin, and divalent nickel in PC12 cells. Environ. Health Perspect. 117 (4), 587–596. doi:10.1289/ehp.0800251
Song I., Huganir R. L. (2002). Regulation of AMPA receptors during synaptic plasticity. Trends Neurosci. 25, 578–588. doi:10.1016/S0166-2236(02)02270-1
Spirkova A., Kovaříková V., Šefčíková Z., Pisko J., Kšiňanová M., Koppel J., et al. (2022). Glutamate can act as a signaling molecule in mouse preimplantation embryos. Biol. Reprod. doi:10.1093/biolre/ioac126
Stroebel D., Paoletti P. (2021). Architecture and function of NMDA receptors: an evolutionary perspective. J. Physiol. 599, 2615–2638. doi:10.1113/JP279028
Tritsch N. X., Sabatini B. L. (2012). Dopaminergic modulation of synaptic transmission in cortex and striatum. Neuron 76, 33–50. doi:10.1016/j.neuron.2012.09.023
UniProt Consortium T. (2018). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46, 2699. doi:10.1093/nar/gky092
van Giesen L., Garrity P. A. (2017). More than meets the IR: The expanding roles of variant Ionotropic Glutamate Receptors in sensing odor, taste, temperature and moisture. F1000Research 6, 1753. doi:10.12688/f1000research.12013.1
Vogeler S., Bean T. P., Lyons B. P., Galloway T. S. (2016). Dynamics of nuclear receptor gene expression during Pacific oyster development. BMC Dev. Biol. 16, 33. doi:10.1186/s12861-016-0129-6
Wakazono Y., Midorikawa R., Takamiya K. (2023). Temporal and quantitative analysis of the functional expression of Ca2+-permeable AMPA receptors during LTP. Neurosci. Res. doi:10.1016/j.neures.2023.07.002
Wang S., Hu P., Wang H., Wang M., Chen J., Tang J., et al. (2008). Effects of Cd2+ on AMPA receptor-mediated synaptic transmission in rat hippocampal CA1 area. Toxicol. Lett. 176, 215–222. doi:10.1016/j.toxlet.2007.11.008
Wang L. (2022). Regulatory mechanism of neuroendocrine system on immune response in molluscs:a review. J. Dalian Ocean Univ. 37, 363–375. doi:10.16535/j.cnki.dlhyxb.2022-140
Wen X., Chen Y.-H., Li R., Ge M.-H., Yin S.-W., Wu J.-J., et al. (2020). Signal decoding for glutamate modulating egg laying oppositely in Caenorhabditis elegans under varied environmental conditions. iScience 23, 101588. doi:10.1016/j.isci.2020.101588
Wilkins M. R., Gasteiger E., Bairoch A., Sanchez J. C., Williams K. L., Appel R. D., et al. (1999). “Protein identification and analysis tools in the ExPASy server,” in 2-D proteome analysis protocols (New Jersey: Humana Press), 531–552. doi:10.1385/1-59259-584-7:531
Yang J. Y., Wang J., Hu Y., Shen D. Y., Xiao G. L., Qin X. Y., et al. (2023). Paeoniflorin improves cognitive dysfunction, restores glutamate receptors, attenuates gliosis and maintains synaptic plasticity in cadmium-intoxicated mice. Arabian J. Chem. 16, 104406. doi:10.1016/j.arabjc.2022.104406
Yu G., Wang L. G., Han Y., He Q.-Y. (2012). clusterProfiler: an R Package for comparing biological themes among gene clusters. OMICS 16, 284–287. doi:10.1089/omi.2011.0118
Zhang G., Fang X., Guo X., Li L., Luo R., Xu F., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi:10.1038/nature11413
Zhang X., Fan C., Zhang X., Li Q., Li Y., Wang Z. (2021). Effects of light intensity and wavelength on the phototaxis of the Crassostrea gigas (♂) and Crassostrea sikamea (♀) hybrid larvae. Front. Mar. Sci. 8. doi:10.3389/fmars.2021.698874
Zhao T., Zhong R., Zhang X., Li G., Zhou C., Fang S., et al. (2023). Efavirenz restored NMDA receptor dysfunction and inhibited epileptic seizures in GluN2A/Grin2a mutant mice. Front. Neurosci. 17. doi:10.3389/fnins.2023.1086462
Zhou H., Cheng Z., Bass N., Krystal J. H., Farrer L. A., Kranzler H. R., et al. (2018). Genome-wide association study identifies glutamate ionotropic receptor GRIA4 as a risk gene for comorbid nicotine dependence and major depression. Transl. Psychiatry 8, 208. doi:10.1038/s41398-018-0258-8
Keywords: ionotropic glutamate receptors, Crassostrea gigas, metal stress, environmental sensing, physiological adaptation
Citation: Zhang X, Zhang L, Si Y, Wen X, Wang L and Song L (2023) Unveiling the functional diversity of ionotropic glutamate receptors in the Pacific oyster (Crassostrea gigas) by systematic studies. Front. Physiol. 14:1280553. doi: 10.3389/fphys.2023.1280553
Received: 20 August 2023; Accepted: 10 October 2023;
Published: 25 October 2023.
Edited by:
Yi-Feng Li, Shanghai Ocean University, ChinaReviewed by:
Chenghua Li, Ningbo University, ChinaCopyright © 2023 Zhang, Zhang, Si, Wen, Wang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingling Wang, d2FuZ2xpbmdsaW5nQGRsb3UuZWR1LmNu; Linsheng Song, bHNoc29uZ0BkbG91LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.