
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 24 May 2023
Sec. Invertebrate Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1194997
 Fengshui Yang1
Fengshui Yang1 Yujie Li1,2
Yujie Li1,2 Mengyuan Gao1,2
Mengyuan Gao1,2 Qing Xia1
Qing Xia1 Qian Wang3*
Qian Wang3* Meijun Tang1
Meijun Tang1 Xiaogui Zhou1
Xiaogui Zhou1 Huawei Guo1
Huawei Guo1 Qiang Xiao1*
Qiang Xiao1* Liang Sun1*
Liang Sun1*Insect carboxylesterases (CXEs) can be expressed in multiple tissues and play crucial roles in detoxifying xenobiotic insecticides and degrading olfactory cues. Therefore, they have been considered as an important target for development of eco-friendly insect pest management strategies. Despite extensive investigation in most insect species, limited information on CXEs in sibling moth species is currently available. The Ectropis obliqua Prout and Ectropis grisescens Warren are two closely related tea geometrid species, which share the same host of tea plant but differ in geographical distribution, sex pheromone composition, and symbiotic bacteria abundance, providing an excellent mode species for studies of functional diversity of orthologous CXEs. In this study, we focused on EoblCXE14 due to its previously reported non-chemosensory organs-biased expression. First, the EoblCXE14 orthologous gene EgriCXE14 was cloned and sequence characteristics analysis showed that they share a conserved motif and phylogenetic relationship. Quantitative real-time polymerase chain reaction (qRT-PCR) was then used to compare the expression profiles between two Ectropis spp. The results showed that EoblCXE14 was predominately expressed in E. obliqua larvae, whereas EgriCXE14 was abundant in E. grisescens at multiple developmental stages. Interestingly, both orthologous CXEs were highly expressed in larval midgut, but the expression level of EoblCXE14 in E. obliqua midgut was significantly higher than that of EgriCXE14 in E. grisescens midgut. In addition, the potential effect of symbiotic bacteria Wolbachia on the CXE14 was examined. This study is the first to provide comparative expression profiles of orthologous CXE genes in two sibling geometrid moth species and the results will help further elucidate CXEs functions and identify a potential target for tea geometrid pest control.
Chemicals with ester functional groups such as species-specific pheromones, host plant odorants, and xenobiotic insecticides are important for insect physiology, biochemistry, and ecology (Xu and Turlings, 2018; Levi-Zada and Byers, 2021; Mdeni et al., 2022). Carboxylesterases (CXEs) belong to a superfamily that includes diverse functional enzymes (Oakeshott et al., 1999; Oakeshott et al., 2005). They hydrolyze esters to their corresponding acids and alcohols and are thus thought to be involved in xenobiotic detoxification, odorant degradation, and developmental regulations (Richmond et al., 1980; Leal, 2013; Godoy et al., 2021).
Insect CXEs possess highly conserved catalytic triads (Ser-Glu-His) and pentapeptide motifs (Gly-X-Ser-X-Gly). They are expressed in the midgut, fat body, and other detoxifying organs which metabolize pesticide organophosphates, pyrethroids, and carbamates in various insect species such as moths Helicoverpa armigera (Bai et al., 2019), Grapholita molesta (Li et al., 2023), Plutella xylostella (L.) (Li et al., 2021), and Spodoptera frugiperda (Carvalho et al., 2013); the aphid Aphis gossypii (Chang et al., 2010); and flies Lucilia cuprina (Heidari et al., 2005) and Drosophila melanogaster (Birner-Gruenberger et al., 2012). The overexpression of CXE genes has been reported as an important mechanism of insecticide detoxification. For instance, overexpressed CXE genes have been shown to be involved in indoxacarb resistance in Spodoptera litura (Shi et al., 2022); an overexpressed CXE (PxαE14) in the detoxification of multiple insecticides in P. xylostella (Li et al., 2022); and two overexpressed a-esterase genes mediated metabolic resistance to malathion in the oriental fruit fly, Bactrocera dorsalis (Hendel) (Wang et al., 2015). Additionally, the alternative splicing and site mutation of CXEs are considered another mechanism in insecticide detoxification. The alternative splicing of TcCCE23 with different variants enhances fenpropathrin tolerance in Tetranychus cinnabarinus (Wei et al., 2023). Two different amino acid substitutions in the ali-esterase E3 confer alternative types of organophosphorus insecticide resistance in the sheep blowfly, Lucilia cuprin (Campbell et al., 1998).
In addition to xenobiotic detoxification, some CXEs have been demonstrated to be highly expressed at the antennae, the main insect olfactory organs, and function as odorant degrading enzymes (ODEs) that play crucial roles in olfactory cue degradation. The first insect ODE (ApolPDE) was identified from the silk moth Antheraea polyphemus; it displayed male antennae-specific expression and could rapidly degrade A. polyphemus major acetate sex pheromone, (E, Z)-6, 11-hexadecadienyl acetate [E6Z11–16Ac] (Vogt and Riddiford, 1981; Ishida and Leal, 2005). Another male antennae-specific CXE gene, the encoded protein (PjapPDE) of which was identified in the Japanese beetle, Popillia japonica, could degrade the sex pheromone component (R, Z)-5-(-)-(1-decenyl) oxacyclopentan-2-one [(R)-japonilure] (Ishida and Leal, 2008). In addition, an extracellular CXE, named esterase-6 (Est-6), in D. melanogaster was reported to be highly expressed in olfactory sensilla and involved in maintaining proper temporal dynamics of cis-vaccenyl acetate (cVA) detection (Chertemps et al., 2012). Subsequently, an increasing number of insect CXEs have been identified and functionally reportedly to be involved in degradation of both plant odorants and sex pheromones (Durand et al., 2010a; Durand et al., 2010b; Durand et al., 2011; Zhang et al., 2016; He et al., 2020).
Ectropis obliqua Prout and Ectropis grisescens Warren are two closely related tea geometrid species which have frequent outbreaks in tea gardens and result in serious damages to tea production (Zhang et al., 2014). At present, chemical control is still the main method against these two sibling species, which accelerates the risk of resistance development. A better understanding of the mechanism of insecticide detoxification and screening of the target genes involved in xenobiotic detoxification and odorant degradation is crucial for development of environmental friendly pest management strategies. The CXEs present potential targets due to their important roles in detoxifying various insecticides and degrading olfactory cues.
Previously, 35 EoblCXE genes have been identified in E. obliqua chemosensory organs and they displayed multiple tissues expression profiles (Sun et al., 2017). Whether these EoblCXEs in E. obliqua have their orthologs in the sibling species E. grisescens and the differences in their expression profiles are still unknown. Ectropis obliqua differs from E. grisescens in geographical distribution, sex pheromone composition, and symbiotic bacteria Wolbachia abundance (Luo et al., 2017; Li et al., 2019; Wang et al., 2020). Whether the differences in Wolbachia abundance are associated with CXE expression patterns should be investigated.
In this study, we focused on EoblCXE14, which showed a non-chemosensory organs-biased expression at the abdomen of E. obliqua adults (Sun et al., 2017). First, we cloned its orthologous gene EgriCXE14 in E. grisescens and analyzed its sequence characteristics; then we compared the expression profiles between the sibling species E. obliqua and E. grisescens and evaluated the potential effect of Wolbachia on their expression pattern. The results will help further investigation of the potential roles of CXE14 in xenobiotic detoxification.
The larvae of E. obliqua and E. grisescens was collected from Yuhang and Yueqing tea plantations in Zhejiang Province, respectively and have been reared for multiple generations in the laboratory. Identification of species to distinguish E. obliqua and E. grisescens was performed according to previous studies (Zhang et al., 2014). Larvae were fed on the fresh tea cultivar and reared under 25°C ± 1°C with 65% ± 5% humidity and L13:D11 photoperiod. The male and female pupae were identified and separated for eclosion.
The mutant population of E. grisescens, which was collected from Yizheng, Jiangsu province, and its Wolbachia was removed by using tetracycline, has been previously established and reared in our laboratory for many generations. PCR detection of the wsp gene was performed as described in previous reports (Baldo et al., 2006; Zhou et al., 2016) to ensure the removal of Wolbachia in the E. grisescens mutant with the COI gene for checking the quality of extracted DNA templates as described by Zhang et al. (2014). The rearing conditions of E. grisescens mutant were the same as those of E. obliqua and E. grisescens described above.
For the developmental expression pattern analysis of CXE14 in E. obliqua and E. grisescens, four biological replicates were collected, and each biological replicates contained the following: 200 eggs (day 0), 15 third instar larvae, 5 pupae (day 5–6), and 5 new emergence moths (day 0) of each sex. For the tissue-specific expression patterns of CXE14, 200 third instar larvae of E. obliqua, E. grisescens, and E. grisescens mutant were dissected into head, midgut, fat body, and epidermis, respectively. Four replicates were collected for each tissue sample. All of the specimens were collected and immediately stored in −80°C until use.
Total RNA was extracted using the Trizol reagent (Thermo, United States) according to the manufacturer’s protocol. Purity was assessed using NanoDrop2000 (Thermo, United States) and the quality of total RNA was detected in 1.0% agarose electrophoresis. First-strand cDNA was synthesized from 1 µg RNA using a MonScript™ RTⅢ Super Mix with dsDNase (Two-Step) (Monad, Wuhan, China).
Homology-based cloning was used to clone EgriCXE14 in E. grisescens; the specific primers (Supplementary Table S1) were designed based on its ortholog EoblCXE14 (Genbank accession No. KX015856.1) (Sun et al., 2017). PCR was performed using a MonAmp™ 2 x Taq Mix Pro (+Dye) (Monad, Wuhan, China) under the following condition: 94°C for 3 min, followed by 40 cycles at 95°C for 30 s, 65°C for 30 s, 72°C for 2 min, and a final elongation step at 72°C for 10 min. The 50 μL reaction mixture contained 25 μL MonAmpTM 2 × Taq Mix Pro(+Dye), 2 μL of each forward and reverse primers, 2 μL cDNA template, 19 μL ddH2O. The PCR product was analyzed in 1.5% agarose electrophoresis.
The PCR products were purified using a DNA Gel Extraction Kit (Axygen, Shanghai, China), and connected to pCE2 TA/Blunt-Zero vector (Vazyme, Nanjing, China) with a 5 μL reaction mixture containing 1 μL 5× TA/Blunt-Zero Cloning Mix and 4 μL purified DNA product. The product was incubated at 37°C for 30 min and then transferred into Escherichia coli DH5α (AngYuBio, Shanghai, China). Positive colonies were selected using PCR using specific primers for sequencing.
The amino acid sequences of EgriCXE14 and previously functional reported HarmCarE001a, HarmCarE001g, BmorCarE, and LcaE7 from H. armigera (Bai et al., 2019), Bombyx mori (Cui et al., 2011), and L. cuprina (Jackson et al., 2013), respectively, were aligned using ClustalW (https://www.genome.jp/tools-bin/clustalw). MEME web service was used to identify conserved motifs (https://meme-suite.org/meme/index.html). The SMART website was selected to characterize the domain in targeted protein sequences (http://smart.embl-heidelberg.de/). A neighbor-joining tree was constructed using MEGA11.0 software with p-distance amino acid substitution (Tamura et al., 2021). Bootstrapping with 1,000 replicates was used to evaluate the reliability of the tree topology. The protein accession numbers of CXEs used in phylogenetic tree construction are listed in Supplementary Table S2.
qRT-PCR was performed using Roche Light Cycle 480 (Roche, Swiss) and the specific primers of CXE14 were designed using Primer Premier 5.0 (Supplementary Table S1). The glyceraldehyde-3-phosphate dehydrogenase EoblGAPDH (Genbank accession No. KT991373) and its specific primers previously used for qPCR (EoblGAPDH-F: TATCTCTCTGAACGACAACTT; EoblGAPDH-R: TTGGTCTGGATGTACTTGAT) (Yan et al., 2020) were used to normalize the target gene CXE14 expression and correct the sample-to-sample variation. Each reaction was conducted in a 10 μL reaction mixture containing 5 μL TB Green Premix Ex Taq II, 0.4 μL of each primer, 1 μL cDNA template (approximately 200 ng), and 3.2 μL sterilized H2O. The qRT-PCR parameters were as follows: 95°C for 30 s, followed by 40 cycles at 95 °C for 5 s, and 60°C for 30 s. Then, fluorescence was measured using a 55°C–95°C melting curve to detect a specific-peak for individual gene. Each reaction was performed in three technical replicates and four independent biological replicates.
The relative fold gene expression levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001), and the statistical significance of the differences was analyzed using SPSS Statistics 26.0 software. The comparison of the relative expression levels of CXE14 amongst the developmental stages and tissues of E. obliqua and E. grisescens was conducted using a one-way nested analysis of variance (ANOVA), followed by the least significant difference test (LSD) at α = 0.05. The relative expression levels of CXE14 between two samples were determined using an independent-samples t-test. GraphPad Prism 9 software was used to create graphs.
The sequence of EgriCXE14 was identified from the adult head of E. grisescens using a homologous cloning method. The obtained partial-length sequence of EgriCXE14 was 1,098 bp and encoded 366 amino acids; it was deposited in GenBank (accession No. OQ701617). The sequence identity between orthologs EgriCXE14 and EoblCXE14 was 97.8%, and amino acid residue variations were found at Ala24Ser, Asp26Glu, Leu27Phe, Met33Ile, Asp136Asn, Ile153Leu, Ser183Thr, and Thr288Ile (Supplementary Figure S1). The conserved motif of EgriCXE14 and EoblCXE14 was analyzed and the results showed that two CXE14 orthologs had insect CXE conserved functional domains such as the catalytic triads (Ser-His-Glu) and pentapeptide motif (GXSXA) (Figure 1).
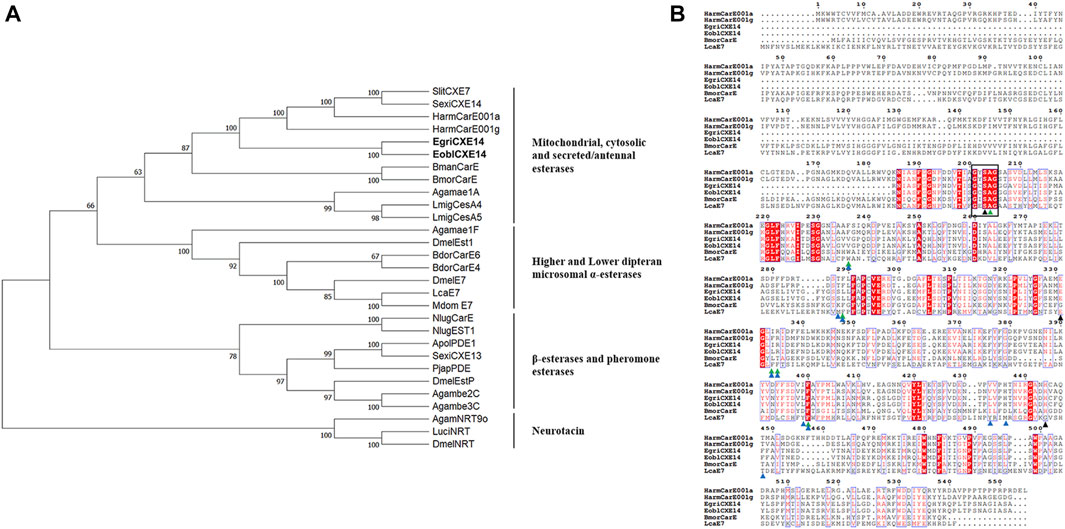
FIGURE 1. (A) Phylogenetic analyses of CXE14. CXE sequences used in this analysis are shown in Supplementary Table S2. EoblCXE14 and EgriCXE14 are shown in bold. (B) Sequence characteristics of EoblCXE14 and EgriCXE14 with previously functional reported HarmCarE001a, HarmCarE001g, BmorCarE, and LcaE7. The catalytic triad residues (Ser-His-Glu) are marked with a black triangle, the highly conserved pentapeptide residues (GXSXA) are marked with a black box; the location of key amino acids corresponding to binding pocket residues of LcaE7 and HarmCarE001g are marked with blue and green triangles, respectively.
To deduce potential roles of CXE14, the phylogenetic relationships of EgriCXE14 and EoblCXE14 with some carboxylesterases which have been previously functional reported in insecticide detoxification were analyzed. The results showed that EgriCXE14 and EoblCXE14 were clustered into the HarmCarE001g clade, a carboxylesterase reportedly associated with pyrethroids detoxification in H. armigera (Figure 1). In addition, the SMART analysis showed that EgriCXE14 and EoblCXE14 contained similar a COesterase to HarmCarE001g and HarmCarE001a (Supplementary Figure S2).
The expression profile of orthologous gene CXE14 in the two sibling species E. obliqua and E. grisescens at different developmental stages, including eggs, larvae, male and female pupae, male and female adults, were examined using qRT-PCR. The results showed that the two CXE14 orthologs were detected at all the examined developmental stages but exhibited a notably different expression pattern. In E. obliqua, EoblCXE14 was predominately expressed in larvae (Figure 2A). By contrast, EgriCXE14 was highly abundant in male adults, followed by larvae, male pupae, female adults, female pupae, and eggs (Figure 2B).
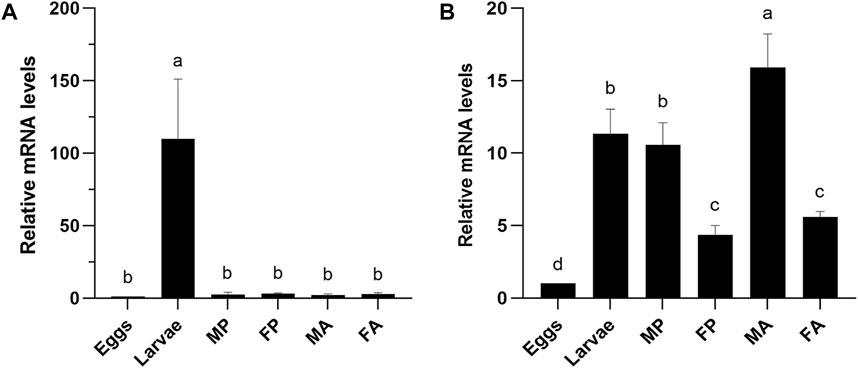
FIGURE 2. The expression profiles of CXE14 at different developmental stages of E. obliqua (A) and E. grisescens (B). MP, male pupae; FP, female pupae; MA, male adults; FA, female adults; The error bars represent the standard deviation (SD), different letters above each bar denote significant differences (one-way ANOVA followed by LSD test, p < 0.05).
The expression levels of CXE14 orthologs in different tissues of E. obliqua and E. grisescens larvae were studied. The qRT-PCR results showed that the two orthologs EoblCXE14 and EgriCXE14 had a conserved tissue-specific expression pattern; the expression levels of EoblCXE14 and EgriCXE14 were significant higher in the midgut than those in the heads, fat body, and epidermis (Figure 3).
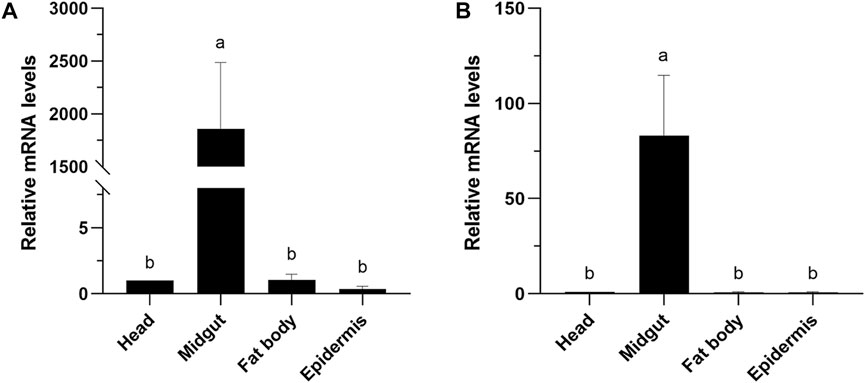
FIGURE 3. Tissue-specific expression patterns of CXE14 in E. obliqua (A) and E. grisescens (B) larvae. The error bars represent the standard deviation (SD); different letters above each bar denote significant differences (one-way ANOVA followed by LSD test, p < 0.05).
To further elucidate the functional differences of CXE14 orthologs in sibling species, we compared the expression levels of EoblCXE14 and EgriCXE14 in the midgut of E. obliqua and E. grisescens using qRT-PCR. The results showed that the expression level of EoblCXE14 in the E. obliqua midgut was higher (approximately 3.8-fold) than that of EgriCXE14 in the E. grisescens midgut (Figure 4).
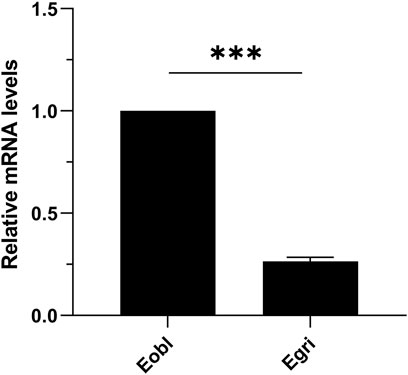
FIGURE 4. The comparison of CXE14 expression levels in the midgut of E. obliqua and E. grisescens. Eobl, E. obliqua; Egri, E. grisescens; the error bars represent the standard deviation (SD), *** indicates significant differences in CXE14 expression between E. obliqua and E. grisescens (independent-samples t-test, p < 0.001).
The mutant population of E. grisescens with Wolbachia removed using tetracycline has been previously constructed in our laboratory. PCR specific for the wsp gene was performed and the results showed a clear wsp gene band in E. grisescens, but not in the E. grisescens mutant, indicating a successful removal of Wolbachia from E. grisescens larvae (Supplementary Figure S3).
Tissue-specific expression pattern of EgriCXE14 in the E. grisescens larval mutant was examined using qRT-PCR. The results showed that EgriCXE14 in the E. grisescens mutant was highly expressed in the midgut (Figure 5).
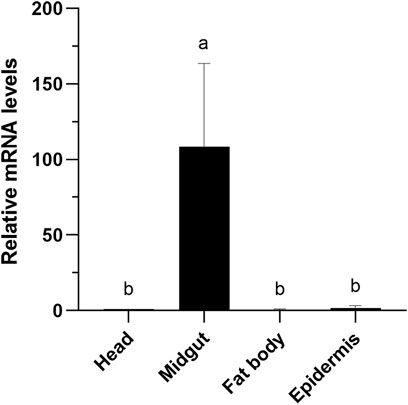
FIGURE 5. Tissue-specific expression patterns of CXE14 in E. grisescens larvae mutant. The error bars represent the standard deviation (SD); different letters above each bar denote significant differences (one-way ANOVA followed by LSD test, p < 0.05).
To deduce the potential effect of Wolbachia on the expression difference of EgriCXE14, we compared the expression levels of EgriCXE14 in larval midgut between E. grisescens and E. grisescens mutant. The qRT-PCR results showed that the expression level of EgriCXE14 in E. grisescens mutant was not significantly different from that in E. grisescens (Figure 6).
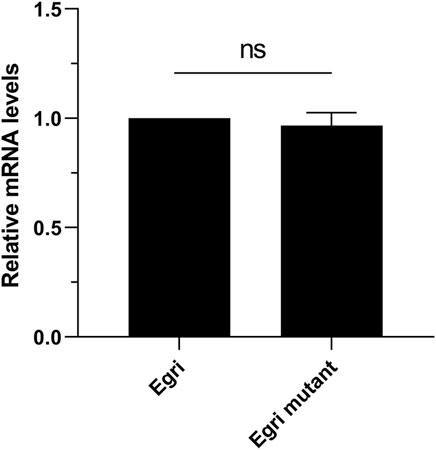
FIGURE 6. The potential effect of Wolbachia on the expression level of CXE14 in the midgut of E. grisescens. Egri, E. grisescens; Egri mutant, E. grisescens without Wolbachia; The error bars represent the standard deviation (SD), ns indicates no significant difference in CXE14 expression level between E. grisescens and E. grisescens mutant (independent-samples t-test, p > 0.05).
Insect CXEs have been widely demonstrated to be involved in pesticide detoxification and pheromone and plant odorant degradation. Therefore, they represent a major target for development of eco-friendly pest management strategies. This study reports a pair of CXE orthologous genes, EoblCXE14 and EgriCXE14, which share a conserved motif and phylogenetic relationship but have different expression profiles in two closely related tea geometrid species, E. obliqua and E. grisescens.
Insect CXEs have been identified in most of the lepidopteran moth species, such as Spodoptera littoralis (Durand et al., 2010b), S. litura (Zhang et al., 2016), Carposina sasakii (Li and Zhang, 2022), Hyphantria cunea (Ye et al., 2021), and P. xylostella (Xie et al., 2017; Li et al., 2021; Wang et al., 2021), which represent important pests of agricultural food crops, cotton, fruit trees, and vegetables. By contrast, the identification of CXEs in tea plant pest, especially in the sibling species E. obliqua and E. grisescens, have not been well explored, although we have previously identified and characterized 35 CXEs in tea geometrid moth E. obliqua (Sun et al., 2017). In this study, we cloned a CXE gene from E. grisescens named EgriCXE14 which is the orthologs of EoblCXE14. Although it was not the full-length sequence, EgriCXE14 contained the main functional domain of insect CXEs including the highly conserved pentapeptide residues (GXSXA) and the Ser-His-Glu catalytic triad residues. In addition, some amino acid residues such as Arg, Phe, and His, which are considered key residues in the binding pocket associated with insecticide binding of H. armigera HarmCarE001g (Bai et al., 2019) or L. cuprina LcaE7 (Jackson et al., 2013), are also conserved in both EoblCXE14 and EgriCXE14 (Figure 1), indicating potential roles of these two CXE genes in detoxifying insecticides.
The phylogenetic analyses and larval tissue-specific expression patterns observed in this study support the above-mentioned speculation. EgriCXE14 and EoblCXE14 were found to cluster with HarmCarE001g, a carboxylesterase which has been demonstrated to detoxify pyrethroids in H. armigera (Bai et al., 2019) (Figure 1). Furthermore, qRT-PCR results showed that both EgriCXE14 and EoblCXE14 were highly expressed in the midgut, a detoxification metabolic organ of E. obliqua and E. grisescens larvae (Figure 3). Indeed, many CXEs of different moth species are highly detected in the midgut and play crucial roles in insecticide detoxification. The CXEs genes (CarE) in G. molesta have higher expression in the detoxification metabolic organs such as the midgut and fat body but lower expression in the head and cuticle, and are involved in the tolerance of insecticides (Li et al., 2023). In the diamondback moth, P. xylostella, PxEst-6 is highly expressed in the midgut and cuticles of the third instar larvae and is associated with pyrethroid insecticides metabolization (Li et al., 2021), whereas PxαE14 is predominantly expressed in the midgut and Malpighian tubule of larvae and contributes to detoxification of multiple insecticides (Li et al., 2022). CXE genes (SlituCOE) of S. litura are expressed in the midgut, Malpighian tubule, and fat body, and are related to insecticide indoxacarb resistance (Shi et al., 2022). Further functional studies of enzyme activities and RNAi knockdown are needed to test this hypothesis in two closely related tea geometrid Ectropis spp.
Despite the predominant expression in the larval midgut, the expression profiles of EoblCXE14 and EgriCXE14 are significantly different. The investigation of expression profiles at different developmental stages showed that EoblCXE14 was larvae-enriched in E. obliqua but EgriCXE14 was ubiquitously expressed at different developmental stages of E. grisescens (Figure 2). These results indicate that EoblCXE14 likely plays roles in E. obliqua larvae; however, EgriCXE14 probably functions in E. grisescens at multiple developmental stages. A possible role of CXE14 in degrading ester odorants cannot be excluded, because several CXEs in insect species reportedly contribute to degradation of insect pheromones and plant volatiles (Ishida and Leal, 2005; Durand et al., 2011; Chertemps et al., 2012; He et al., 2020).
The expressional comparison in the larval midgut of the two Ectropis spp. showed that the expression level of EoblCXE14 was significant higher than that of EgriCXE14 (Figure 4). Previous studies proposed that E. obliqua differs from E. grisescens in symbiotic bacteria composition, particularly the Wolbachia (Zhou et al., 2016; Wang et al., 2020); specifically, E. grisescens has a significant higher abundance of Wolbachia than E. obliqua. Wolbachia has been shown to affect the expression of genes in insect (Pfarr et al., 2008; Cai et al., 2021; Sun et al., 2022). Therefore, in this study, we tested whether Wolbachia was correlated with different expression levels of CXE14 in the midgut of the two Ectropis spp. The qRT-PCR results showed that EgriCXE14 in E. grisescens mutant, in which Wolbachia was removed using tetracycline, was also highly expressed in the midgut (Figure 5), and no significant differences in EgriCXE14 expression levels between E. grisescens and E. grisescens mutant were found (Figure 6). These results suggest that Wolbachia may not be related to EgriCXE14 expression in the E. grisescens midgut, and may not contribute to the different expression level in the midgut of these two sibling species. These findings are not consistent with our previous reports that Wolbachia could be correlated with the expression of a chemosensory gene CSP8 in E. obliqua and E. grisescens (Yan et al., 2022). Notably, E. grisescens mutant collected from the Yizheng, Jiangsu province, differs from the E. grisescens from Yueqing, Zhejiang province; therefore, we cannot rule out the possible effect of geographical population on the aforementioned findings. Further expression variation of target genes in different geographical population, and between wild (with Wolbachia) and mutant (without Wolbachia) E. grisescens from the same geographical region need to be evaluated.
In conclusion, this study characterized a pair of CXEs orthologs, EoblCXE14 and EgriCXE14, in two sibling geometrid mode species, E. obliqua and E. grisescens. The results revealed that EoblCXE14 and EgriCXE14 share a conserved motif and phylogenetic relationship but differ in expression profiles at different developmental stages and expression levels in the larval midgut. EoblCXE14 was predominately expressed in the E. obliqua larvae, whereas EgriCXE14 was abundant in E. grisescens at multiple developmental stages. The expression level of EoblCXE14 in the E. obliqua midgut was significantly higher than that of EgriCXE14 in the E. grisescens midgut. In addition, potential effects of Wolbachia on the CXE14 expression levels were examined. This study provides the first report on comparative expression profiles of orthologous CXE genes in two sibling geometrid moth species and could help identify potential targets for controlling tea geometrid.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
LS conceived and designed the experiments. FY, YL, MG and Qing Xia performed the experiments. FY and YL analyzed the data. Qiang Xiao provided insect population especially the mutant population of E. grisescens with Wolbachia removed. FY, LS and QW prepared and edited the manuscript. MT, HG and XZ reviewed the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by National Natural Science Foundation of China (31871977, 32102200), Zhejiang Provincial Natural Science Foundation of China under Grant No. LQ21C140001 and Central Public-Interest Scientific Institution Basal Research Fund (1610212020001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1194997/full#supplementary-material
Bai, L.-S., Zhao, C.-X., Xu, J.-J., Feng, C., Li, Y.-Q., Dong, Y.-L., et al. (2019). Identification and biochemical characterization of carboxylesterase 001G associated with insecticide detoxification in Helicoverpa armigera. Pestic. Biochem. Phys. 157, 69–79. doi:10.1016/j.pestbp.2019.03.009
Baldo, L., Dunning Hotopp, J. C., Jolley, K. A., Bordenstein, S. R., Biber, S. A., Choudhury, R. R., et al. (2006). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 72 (11), 7098–7110. doi:10.1128/aem.00731-06
Birner-Gruenberger, R., Bickmeyer, I., Lange, J., Hehlert, P., Hermetter, A., Kollroser, M., et al. (2012). Functional fat body proteomics and gene targeting reveal in vivo functions of Drosophila melanogaster α-Esterase-7. Insect Biochem. Mol. Biol. 42 (3), 220–229. doi:10.1016/j.ibmb.2011.12.004
Cai, T., Zhang, Y., Liu, Y., Deng, X., He, S., Li, J., et al. (2021). Wolbachia enhances expression of NlCYP4CE1 in Nilaparvata lugens in response to imidacloprid stress. Insect Sci. 28 (2), 355–362. doi:10.1111/1744-7917.12834
Campbell, P. M., Newcomb, R. D., Russell, R. J., and Oakeshott, J. G. (1998). Two different amino acid substitutions in the ali-esterase, E3, confer alternative types of organophosphorus insecticide resistance in the sheep blowfly, Lucilia cuprina. Insect Biochem. Mol. Biol. 28 (3), 139–150. doi:10.1016/s0965-1748(97)00109-4
Carvalho, R. A., Omoto, C., Field, L. M., Williamson, M. S., and Bass, C. (2013). Investigating the molecular mechanisms of organophosphate and pyrethroid resistance in the fall armyworm Spodoptera frugiperda. PLoS One 8 (4), e62268. doi:10.1371/journal.pone.0062268
Chang, J., Cao, C. W., and Gao, X. W. (2010). The effect of pretreatment with S,S,S-tributyl phosphorotrithioate on deltamethrin resistance and carboxylesterase activity in Aphis gossypii (Glover) (Homoptera: Aphididae). Pestic. Biochem. Phys. 98 (2), 296–299. doi:10.1016/j.pestbp.2010.06.021
Chertemps, T., François, A., Durand, N., Rosell, G., Dekker, T., Lucas, P., et al. (2012). A carboxylesterase, Esterase-6, modulates sensory physiological and behavioral response dynamics to pheromone in Drosophila. BMC Biol. 10, 56. doi:10.1186/1741-7007-10-56
Cui, F., Lin, Z., Wang, H., Liu, S., Chang, H., Reeck, G., et al. (2011). Two single mutations commonly cause qualitative change of nonspecific carboxylesterases in insects. Insect Biochem. Mol. Biol. 41 (1), 1–8. doi:10.1016/j.ibmb.2010.09.004
Durand, N., Carot-Sans, G., Bozzolan, F., Rosell, G., Siaussat, D., Debernard, S., et al. (2011). Degradation of pheromone and plant volatile components by a same odorant-degrading enzyme in the cotton leafworm, Spodoptera littoralis. PLoS One 6 (12), e29147. doi:10.1371/journal.pone.0029147
Durand, N., Carot-Sans, G., Chertemps, T., Bozzolan, F., Party, V., Renou, M., et al. (2010a). Characterization of an antennal carboxylesterase from the pest moth Spodoptera littoralis degrading a host plant odorant. PLoS One 5 (11), e15026. doi:10.1371/journal.pone.0015026
Durand, N., Carot-Sans, G., Chertemps, T., Montagné, N., Jacquin-Joly, E., Debernard, S., et al. (2010b). A diversity of putative carboxylesterases are expressed in the antennae of the noctuid moth Spodoptera littoralis. Insect Mol. Biol. 19 (1), 87–97. doi:10.1111/j.1365-2583.2009.00939.x
Godoy, R., Machuca, J., Venthur, H., Quiroz, A., and Mutis, A. (2021). An overview of antennal esterases in Lepidoptera. Front. Physiol. 12, 643281. doi:10.3389/fphys.2021.643281
He, P., Mang, D.-Z., Wang, H., Wang, M.-M., Ma, Y.-F., Wang, J., et al. (2020). Molecular characterization and functional analysis of a novel candidate of cuticle carboxylesterase in Spodoptera exigua degradating sex pheromones and plant volatile esters. Pestic. Biochem. Phys. 163, 227–234. doi:10.1016/j.pestbp.2019.11.022
Heidari, R., Devonshire, A. L., Campbell, B. E., Dorrian, S. J., Oakeshott, J. G., and Russell, R. J. (2005). Hydrolysis of pyrethroids by carboxylesterases from Lucilia cuprina and Drosophila melanogaster with active sites modified by in vitro mutagenesis. Insect Biochem. Mol. Biol. 35 (6), 597–609. doi:10.1016/j.ibmb.2005.02.018
Ishida, Y., and Leal, W. S. (2008). Chiral discrimination of the Japanese beetle sex pheromone and a behavioral antagonist by a pheromone-degrading enzyme. P. Natl. Acad. Sci. U. S. A. 105 (26), 9076–9080. doi:10.1073/pnas.0802610105
Ishida, Y., and Leal, W. S. (2005). Rapid inactivation of a moth pheromone. P. Natl. Acad. Sci. U. S. A. 102 (39), 14075–14079. doi:10.1073/pnas.0505340102
Jackson, C. J., Liu, J. W., Carr, P. D., Younus, F., Coppin, C., Meirelles, T., et al. (2013). Structure and function of an insect α-carboxylesterase (αEsterase7) associated with insecticide resistance. P. Natl. Acad. Sci. 110 (25), 10177–10182. doi:10.1073/pnas.1304097110
Leal, W. S. (2013). Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi:10.1146/annurev-ento-120811-153635
Levi-Zada, A., and Byers, J. A. (2021). Circadian rhythms of insect pheromone titer, calling, emission, and response: A review. Naturwissenschaften 108 (5), 35. doi:10.1007/s00114-021-01746-w
Li, J., Jia, Y., Zhang, D., Li, Z., Zhang, S., and Liu, X. (2023). Molecular identification of carboxylesterase genes and their potential roles in the insecticides susceptibility of Grapholita molesta. Insect Mol. Biol. 32 (3), 305–315. doi:10.1111/imb.12831
Li, J., and Zhang, L. (2022). Identification and expression patterns of candidate carboxylesterases in Carposina sasakii Matsumura (Lepidoptera: Carposinidae), an important pest of fruit trees. Bull. Entomol. Res. 112 (4), 567–573. doi:10.1017/S0007485322000244
Li, R., Zhu, B., Hu, X.-P., Shi, X.-Y., Qi, L.-L., Liang, P., et al. (2022). Overexpression of PxαE14 contributing to detoxification of multiple insecticides in Plutella xylostella (L). J. Agric. Food Chem. 70 (19), 5794–5804. doi:10.1021/acs.jafc.2c01867
Li, Y., Sun, H., Tian, Z., Li, Y., Ye, X., Li, R., et al. (2021). Identification of key residues of carboxylesterase PxEst-6 involved in pyrethroid metabolism in Plutella xylostella (L). J. Hazard. Mater. 407, 124612. doi:10.1016/j.jhazmat.2020.124612
Li, Z. Q., Cai, X. M., Luo, Z. X., Bian, L., Xin, Z. J., Liu, Y., et al. (2019). Geographical distribution of Ectropis grisescens (Lepidoptera: Geometridae) and Ectropis obliqua in China and description of an efficient identification method. J. Econ. Entomol. 112 (1), 277–283. doi:10.1093/jee/toy358
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Method Methods (San Diego, Calif. 25 (4), 402–408. doi:10.1006/meth.2001.1262
Luo, Z. X., Li, Z. Q., Cai, X. M., Bian, L., and Chen, Z. M. (2017). Evidence of premating isolation between two sibling moths: Ectropis grisescens and Ectropis obliqua (Lepidoptera: Geometridae). J. Econ. Entomol. 110 (6), 2364–2370. doi:10.1093/jee/tox216
Mdeni, N. L., Adeniji, A. O., Okoh, A. I., and Okoh, O. O. (2022). Analytical evaluation of carbamate and organophosphate pesticides in human and environmental matrices: A review. Molecules 27 (3), 618. doi:10.3390/molecules27030618
Oakeshott, J. G., Claudianos, C., Campbell, P. M., Newcomb, R. D., and Russell, R. (2005). “Biochemical genetics and genomics of insect esterases,” in Comprehensive molecular insect science. Editors L. I. Gilbert, K. Iatrou, and S. S. Gill (London: Elsevier), 309–361.
Oakeshott, J. G., Claudianos, C., Russell, R. J., and Robin, G. C. (1999). Carboxyl/cholinesterases: A case study of the evolution of a successful multigene family. BioEssays news Rev. Mol. Cell. Dev. Biol. 21 (12), 1031–1042. doi:10.1002/(SICI)1521-1878(199912)22:1<1031:AID-BIES7>3.0.CO;2-J
Pfarr, K. M., Heider, U., Schmetz, C., Büttner, D. W., and Hoerauf, A. (2008). The mitochondrial heat shock protein 60 (HSP60) is up-regulated in Onchocerca volvulus after the depletion of Wolbachia. Parasitology 135 (4), 529–538. doi:10.1017/s003118200700409x
Richmond, R. C., Gilbert, D. G., Sheehan, K. B., Gromko, M. H., and Butterworth, F. M. (1980). Esterase 6 and reproduction in Drosophila melanogaster. Science 207 (4438), 1483–1485. doi:10.1126/science.6767273
Shi, Y., Li, W., Zhou, Y., Liao, X., and Shi, L. (2022). Contribution of multiple overexpressed carboxylesterase genes to indoxacarb resistance in Spodoptera litura. Pest Manag. Sci. 78 (5), 1903–1914. doi:10.1002/ps.6808
Sun, L., Wang, Q., Wang, Q., Zhang, Y., Tang, M., Guo, H., et al. (2017). Identification and expression patterns of putative diversified carboxylesterases in the tea geometrid Ectropis obliqua Prout. Front. Physiol. 8, 1085. doi:10.3389/fphys.2017.01085
Sun, Z., Liu, Y., Xu, H., and Yan, C. (2022). Genome-wide identification of P450 genes in Chironomid Propsilocerus akamusi reveals candidate genes involved in gut microbiota-mediated detoxification of Chlorpyrifos. Insects 13 (9), 765. doi:10.3390/insects13090765
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi:10.1093/molbev/msab120
Vogt, R. G., and Riddiford, L. M. (1981). Pheromone binding and inactivation by moth antennae. Nature 293 (5828), 161–163. doi:10.1038/293161a0
Wang, L. L., Huang, Y., Lu, X. P., Jiang, X. Z., Smagghe, G., Feng, Z. J., et al. (2015). Overexpression of two α-esterase genes mediates metabolic resistance to malathion in the oriental fruit fly, Bactrocera dorsalis (Hendel). Insect Mol. Biol. 24 (4), 467–479. doi:10.1111/imb.12173
Wang, M. M., Long, G. J., Guo, H., Liu, X. Z., Wang, H., Dewer, Y., et al. (2021). Two carboxylesterase genes in Plutella xylostella associated with sex pheromones and plant volatiles degradation. Pest Manag. Sci. 77 (6), 2737–2746. doi:10.1002/ps.6302
Wang, Z., Li, H., Zhou, X., Tang, M., Sun, L., Zhan, S., et al. (2020). Comparative characterization of microbiota between the sibling species of tea geometrid moth Ectropis obliqua Prout and E. grisescens Warren. Bull. Entomol. Res. 110 (6), 684–693. doi:10.1017/S0007485320000164
Wei, P., Zeng, X., Han, H., Yang, Y., Zhang, Y., and He, L. (2023). Alternative splicing of a carboxyl/choline esterase gene enhances the fenpropathrin tolerance of Tetranychus cinnabarinus. Insect Sci. doi:10.1111/1744-7917.13166
Xie, M., Ren, N. N., You, Y. C., Chen, W. J., Song, Q. S., and You, M. S. (2017). Molecular characterisation of two α-esterase genes involving chlorpyrifos detoxification in the diamondback moth, Plutella xylostella. Pest Manag. Sci. 73 (6), 1204–1212. doi:10.1002/ps.4445
Xu, H., and Turlings, T. C. J. (2018). Plant volatiles as mate-finding cues for insects. Trends Plant Sci. 23 (2), 100–111. doi:10.1016/j.tplants.2017.11.004
Yan, Y., Li, Y., Wang, Q., Tang, M., Guo, H., Li, H., et al. (2022). The expression profiles of chemosensory protein 8 orthologs in two closely related tea geometrid species, Ectropis obliqua Prout and Ectropis grisescens Warren. J. Tea Sci. 42 (2), 200–210. doi:10.13305/j.cnki.jts.2022.02.007
Yan, Y., Zhang, Y., Tu, X., Wang, Q., Li, Y., Li, H., et al. (2020). Functional characterization of a binding protein for Type-II sex pheromones in the tea geometrid moth Ectropis obliqua Prout. Pestic. Biochem. Phys. 165, 104542. doi:10.1016/j.pestbp.2020.02.008
Ye, J., Mang, D., Kang, K., Chen, C., Zhang, X., Tang, Y., et al. (2021). Putative carboxylesterase gene identification and their expression patterns in Hyphantria cunea (Drury). PeerJ 9, e10919. doi:10.7717/peerj.10919
Zhang, G. H., Yuan, Z. J., Zhang, C. X., Yin, K. S., Tang, M. J., Guo, H. W., et al. (2014). Detecting deep divergence in seventeen populations of tea geometrid (Ectropis obliqua Prout) in China by COI mtDNA and cross-breeding. PLoS One 9 (6), e99373. doi:10.1371/journal.pone.0099373
Zhang, Y.-N., Li, J.-B., He, P., Sun, L., Li, Z.-Q., Fang, L.-P., et al. (2016). Molecular identification and expression patterns of carboxylesterase genes based on transcriptome analysis of the common cutworm, Spodoptera litura (Lepidoptera: Noctuidae). J. Asia-Pac. Entomol. 19 (4), 989–994. doi:10.1016/j.aspen.2016.07.020
Keywords: tea geometrid moth, sibling species, carboxylesterases, Wolbachia, expression profiles
Citation: Yang F, Li Y, Gao M, Xia Q, Wang Q, Tang M, Zhou X, Guo H, Xiao Q and Sun L (2023) Comparative expression profiles of carboxylesterase orthologous CXE14 in two closely related tea geometrid species, Ectropis obliqua Prout and Ectropis grisescens Warren. Front. Physiol. 14:1194997. doi: 10.3389/fphys.2023.1194997
Received: 27 March 2023; Accepted: 12 May 2023;
Published: 24 May 2023.
Edited by:
Wei Xu, Murdoch University, AustraliaReviewed by:
Sheng Sheng, Jiangsu University of Science and Technology, ChinaCopyright © 2023 Yang, Li, Gao, Xia, Wang, Tang, Zhou, Guo, Xiao and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Sun, c3VubGlhbmcwMUBjYWFzLmNu; Qian Wang, d2FuZ3FpYW5tYXlAMTYzLmNvbQ==; Qiang Xiao, eHF0ZWFAbWFpbC50cmljYWFzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.