- 1College of Horticulture and Plant Protection, Henan University of Science and Technology, Luoyang, Henan Province, China
- 2Institute of Plant Protection, Henan Academy of Agricultural Sciences, Zhengzhou, Henan Province, China
Moth mouthparts, consisting of labial palps and proboscis, not only are the feeding device but also are chemosensory organs for the detection of chemical signals from surrounding environment. Up to now, the chemosensory systems in the mouthpart of moths are largely unknown. Here, we performed systematic analyses of the mouthpart transcriptome of adult Spodoptera frugiperda (Lepidoptera: Noctuidae), a notorious pest that spreads worldwide. A total of 48 chemoreceptors, including 29 odorant receptors (ORs), 9 gustatory receptors (GRs), and 10 ionotropic receptors (IRs), were annotated. Further phylogenetic analyses with these genes and homologs from other insect species determined that specific genes, including ORco, carbon dioxide receptors, pheromone receptor, IR co-receptors, and sugar receptors, were transcribed in the mouthpart of S. frugiperda adults. Subsequently, expression profiling in different chemosensory tissues demonstrated that the annotated ORs and IRs were mainly expressed in S. frugiperda antennae, but one IR was also highly expressed in the mouthparts. In comparison, SfruGRs were mainly expressed in the mouthparts, but 3 GRs were also highly expressed in the antennae or the legs. Further comparison of the mouthpart-biased chemoreceptors using RT-qPCR revealed that the expression of these genes varied significantly between labial palps and proboscises. This study provides the first large-scale description of chemoreceptors in the mouthpart of adult S. frugiperda and provides a foundation for further functional studies of chemoreceptors in the mouthpart of S. frugiperda as well as of other moth species.
Introduction
Insect head is the primary center for the communication with surrounding environment. Most insects possess two cephalic sensory organs, i.e., antenna and mouthpart. These two organs play vital roles during the life cycle of insects, such as locating host plants, feeding, and searching mates (Hansson and Stensmyr, 2011; Leal, 2013). Antennae are known to be the most crucial sensory organ of insects, they can perceive various external stimulation, such as odorants, flavors, carbon dioxide, and humidity (Dahanukar et al., 2005). In comparison, mouthparts mainly act as the device for feeding, and they were also demonstrated to have the function in chemosensation (Stocker, 1994; Kaissling, 1996; Krenn, 2010).
In Lepidoptera, most adults have a siphoning mouthpart including a retractable proboscis and a pair of labial palps. Proboscises act as the feeding device, and they consist of two elongated maxillae galeae and a hollow straw, which functions as a system for sucking liquid substances. A variety of chemosensory sensilla are distributed on the surface of the mouthpart. For example, more than 1,200 sensilla were spread on the labial-palp pit organ in Mythimna separata (Dong et al., 2014). Three major sensilla types (styloconica, chaetica, and basiconica) were determined on the proboscis of Helicoverpa armigera (Guo et al., 2018), and similar findings were reported in Athetis lepigone (Hu et al., 2021).
Chemosensory sensilla are porous hair-like structures innervated by the dendrites of chemosensory neurons (CSNs). Three chemoreceptor families, including gustatory receptors (GRs), odorant receptors (ORs), and ionotropic receptors (IRs), are expressed on the dendrite membrane of CSNs (Kaissling, 1996). These receptors can selectively identify chemical molecules and play a central role in the determination of the detection spectrum of chemosensory sensilla (Robertson, 2018; Guo et al., 2022). It is well known that ORs mediate the perception of odorant cues, whereas GRs are responsible for the detection of taste cues. According to the ligand properties, ORs can be classified into pheromone receptor (PRs) and ordinary receptors (Yang and Wang, 2020), whereas GRs can be divided into bitter, sugar, and CO2 receptors (Jones et al., 2007; Sato et al., 2011; Miyamoto et al., 2012; Fujii et al., 2015; Xu et al., 2016). Insects IRs were extensively researched in Drosophila melanogaster, and their functions mainly include olfaction, gustation, thermosensation, and hygrosensation (Chen et al., 2015; Enjin et al., 2016; Knecht et al., 2016). With the development of research techniques, IRs in moths have also been extensively studied in recent years (Zhang J. et al., 2019; Tang et al., 2020; Hou et al., 2022). Members in IR family are further divided into antennal IRs that exist in most insect species and divergent IRs of which numbers and gene types are different among species (Benton et al., 2009; Liu et al., 2018).
Spodoptera frugiperda, also called fall armyworm, is a notorious pest with more than 300 host plant species. It preferred to feed on Poaceae Barnhart species such as rice, maize, and sugarcane (Sparks, 1979). S. frugiperda is native to Americas. As the adults can undertake seasonal migrations covering long distances, it has spread to Africa and Asia in last 5 years (Goergen et al., 2016; Cock et al., 2017; Wu et al., 2019). S. frugiperda invaded Yunnan province of China in 2019 (Zhang L. et al., 2019), and has become an important agricultural pest across China in recent years (Sun et al., 2021). Adults of most Lepidopteran species (including S. frugiperda) adopt flower-visiting strategies, and floral nectar is crucial for them to sustain growth and reproduction (Krenn, 2010). Studies using anatomical, neurophysiological, and molecular approaches showed that flower evaluation by Manduca sexta is governed by specific CSNs housed in the sensilla on the moth’s proboscis (Haverkamp et al., 2016). However, identities of chemosensory receptors in the mouthpart and their functions in the feeding are still enigmatic for most Lepidoptera.
To reveal potential chemosensory systems of the mouthpart in S. frugiperda, we systematically investigated the chemoreceptors using Illumina sequencing. The candidate GR, OR, and IR genes were annotated and phylogenetic relationships between these genes and homologs from other insect species were analyzed. Lastly, expression levels of candidate genes in different chemosensory tissues were investigated by RT-qPCR. This work contributes to further functional researches on chemoreceptors in the mouthpart of S. frugiperda as well as of other moth species.
Materials and methods
Insect rearing
S. frugiperda larvae were collected from Baoshan, Yunnan Province, China. The larvae were reared with artificial diet, and moths were fed with 15% (V: V) honey water (Guo et al., 2022). Twenty moths were kept in a plastic bucket (25 cm in diameter, 30 cm in length, the opening side is wrapped with a piece of medical gauze) for mating in a sex ratio of 1: 1. The gauze was collected daily and kept in valve bags until the larvae hatched out. Successive generations were maintained in an incubator under 16 h L: 8 h D cycle at 25°C ± 1°C and 60% relative humidity. For transcriptome sequencing, mouthparts were separately collected from the 2- to 3-day old virgin male and female moths and then kept in −80°C freezer until they were used.
Transcriptome sequencing, assembly, and gene annotation
Total RNA was isolated from the sample (one biological replicate) using Trizol reagent (Invitrogen, Carlsbad, CA, United States). Concentration of the RNA was measured with an ND-2000 spectrophotometer (Nanodrop, Wilmington, DE, United States). Genomic DNA mixed in the total RNA was eliminated by DNase I (RQ1, Promega, Madison, WI, United States). mRNA was then isolated with Oligo (dT) from 5 µg of the total RNA by Dynabeads mRNA purification kit (Invitrogen, United States). RNA-seq libraries were constructed following the protocol of Illumina’s library. The prepared libraries were then sequenced on the Illumina HiSeq 2000 platform (Illumina, United States) at Sangon Biotech.
De novo assembly was performed as we previously described (Sun et al., 2022). Raw reads were processed to remove low quality sequences, adaptors, and reads with microbes (Bolger et al., 2014). The results were verified with The FastQC package. The clean reads were then de novo assembled to obtain contigs using Trinity v 2.4.0. As the genome data of S. frugiperda had been made public (Gouin et al., 2017), a genome-based mapping assembling strategy was also combined to improve the quality of data analysis results.
We used BlastX tool (E-value < 1e-5) to search the Nr, Swiss-Prot, KEGG, and COG databases to retrieve putative GR, OR, and IR transcripts from the contigs. ORFs (open reading frames) of the transcripts encoding putative SfruGRs, SfruORs, and SfruIRs were predicted with ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder), the obtained ORFs were then manually checked through comparing by BlastX. Transcripts Per Kilobase of exon model per Million mapped reads (TPM) values of candidate transcripts were calculated with the RSEM package to evaluate the expression abundance in male or female mouthparts of S. frugiperda.
Phylogenetic analysis of GRs, ORs, and IRs
Phylogenetic trees of candidate chemoreceptors (SfruGRs, SfruORs, and SfruIRs) were built using the neighbor-joining method in MEGA 11 and then edited with FigTree (v1.4.2). The evolutionary distance was calculated with the Jones-Taylor-Thornton matrix-based method (Jones et al., 1992). Node support was evaluated with a bootstrap method of 1,000 replicates. The amino acid sequences of GR, OR, and IR genes used in the phylogenetic analysis are listed in (Supplementary Table S1).
Expression analysis
Real-time quantitative PCR (RT-qPCR) was conducted to compare the expression levels of candidate chemoreceptors in different tissues of S. frugiperda. Male and female antennae, mouthparts, legs, proboscises, and labial palps were collected separately from 50 to 100 individuals. Total RNA of different samples was extracted following the Trizol reagent (Invitrogen, Carlsbad, CA, United States) manual. First strand cDNA was then synthesized using reverse transcriptase (M-MLV, Promega, WI, United States). The cDNA could be directly used in RT-qPCR reaction.
RT-qPCR was performed on Roche LightCycler 480 (F. Hoffmann-La Roche Ltd., Basel, Switzerland). The solution for each reaction (total volume 20 µL) includes a 10-µL of SYBR Premix Ex TaqII (TaKaRa, Dalian, China), 2.5 ng of template cDNA, 0.4 mM of primer (forward or reverse), and right amount of sterilized deionized H2O. The reaction condition is: 1 cycle of 95°C for 30 s; 40 cycles of 95°C for 5 s, 60°C for 30 s, and 72°C for 30 s; 1 cycle of 95°C for 45 s, 55°C for 1 min. Reactions for different samples were performed with three biological replicates, and each replicate was performed in triplicate. Expression levels of tested genes in different tissues were calculated using the 2−ΔΔCT method (Schmittgen and Livak, 2008), we use β-actin as the internal control. CT values for the reactions of the β-actin gene range from 17.2 to 18.2, this indicates it is consistent across different tissues and suitable for sample normalization. The primer efficiency was evaluated by standard amplification curves constructed with 5-fold dilutions of cDNA samples. The efficiency percentage and R2 values were validated within the acceptable range. Ten PCR products were randomly selected and sequenced to ensure that the target genes were amplified. Primers used in the RT-qPCR were designed with Primer Premier v 6.0 (Supplementary Table S2).
Data analysis
RT-qPCR data were analyzed with ANOVA followed by Tukey’s test (p < 0.05). The results are presented as means ± (standard error), data are averages of three replicates. The figures were constructed with GraphPad Prism v 6.0.
Results
Transcriptome sequencing, assembly, and identification of candidate chemoreceptors
In the current study, we sequenced the transcriptome of S. frugiperda mouthpart using the Illumina HiSeq 2000 platform. Clean reads from female and male samples were then combined and generated an assembly of 119,928 unigenes with an N50 length of 785 bp and a mean length of 568 bp. Based on the sequence length distribution, 14,295 (11.91%) of the unigenes were ≥1000 bp (Supplementary Table S3).
According to the sequence analysis, we identified a total of 29 ORs in the transcriptome of S. frugiperda mouthpart. Twenty-seven of these genes possess putative complete ORFs. To keep uniformity, candidate genes in this study, if possible, were named referring to the previously reported sequences of S. frugiperda, or numbered following the names of the best hit genes in other lepidopteran species. Among the 29 ORs, 8 (SfruOR2/6/18/23/49a/67a/67c/85c) had not been annotated in previous genome or transcriptome studies of this species (Gouin et al., 2017; Sun et al., 2022).
A total of 9 GRs were identified in the transcriptome of S. frugiperda mouthpart. Except for SfruGR6, all the other annotated GRs possess full length ORFs (Supplementary Tables S4, S5). Among the 9 GRs, SfruGR1/2/3/9 had been previously reported in the head transcriptome of S. frugiperda larvae (Sun et al., 2022), while the other 5 GRs (SfruGR4/5/6/7/8) had not been reported in previous studies of this species. Moreover, we annotated 10 IRs in the mouthpart of S. frugiperda. All of these candidate IRs have complete ORFs (Supplementary Tables S4, S5). Based on the Blastx result, SfruIR8a had not been reported in previous studies of S. frugiperda. Detailed information for the candidate genes in this study, including names, accession numbers, BlastX best hits, and sequences are listed in Supplementary Tables S4, S5.
Phylogenetic analysis of candidate chemoreceptors
To reason the putative functions of candidate chemoreceptors, phylogenetic relationships were analyzed based on alignments with homologs from other insect species. According to the neighbor-joining tree of ORs from S. frugiperda (this study), H. armigera, and Bombyx mori, SfruORco was clustered in the ORco branch. Notably, we identified one putative “classic lepidopteran PR” (SfruOR6) in the transcriptome of S. frugiperda mouthpart. The other 27 SfruORs were scattered in various “ordinary OR” branches (Figure 1).
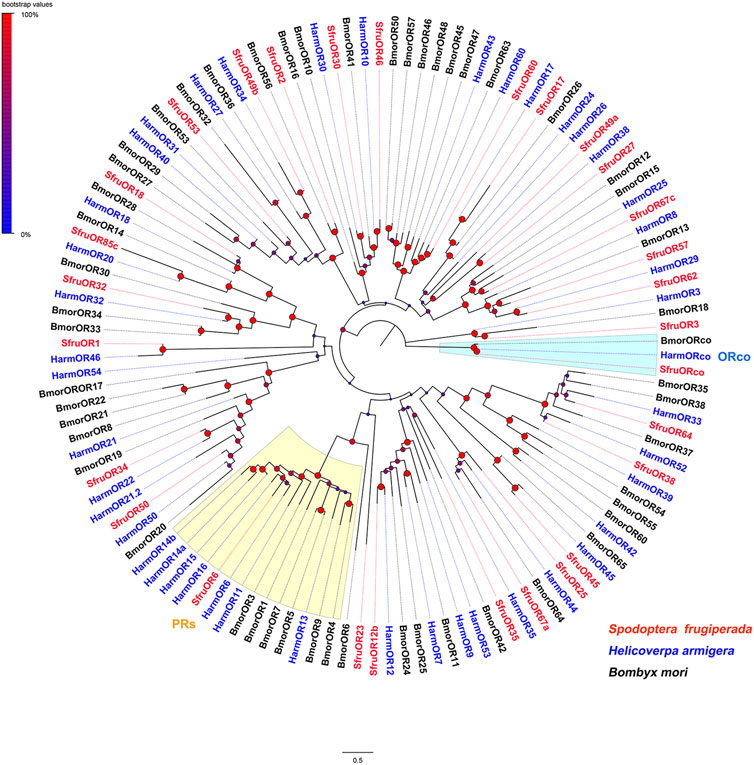
FIGURE 1. Neighbor-joining tree constructed with candidate ORs from Spodoptera frugiperda and homologs from H. armigera and B. mori. The tree was rooted by the ORco orthologs. Node support was estimated with 1000 bootstrap replicates, and bootstrap values were displayed with circles based on the scale indicated at the top left. The scale bar at the bottom indicates the branch length in proportion to amino acid substitutions per site.
A phylogenetic tree built with GRs from S. frugiperda, B. mori, and H. armigera showed that SfruGR1/2/3 were grouped with the CO2 receptors BmorGR1/2/3 and HarmGR1/2/3. And SfruGR9 clustered with the fructose receptors BmorGR9 and HarmGR9. Other five SfruGRs (SfruGR4/5/6/7/8) clustered in the “sugar-taste receptors” clade (Figure 2).
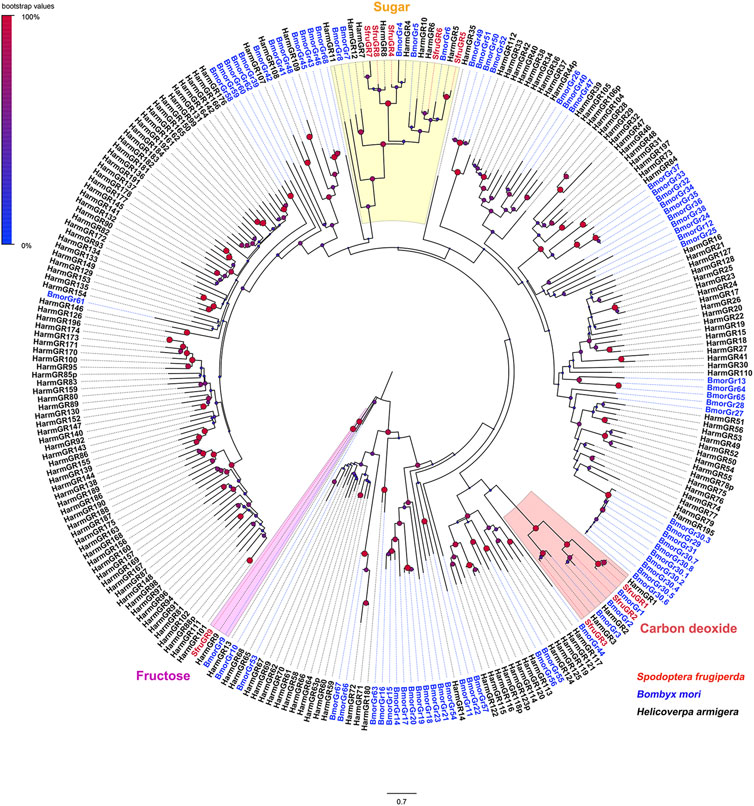
FIGURE 2. Neighbor-joining tree constructed with candidate GRs from S. frugiperda and homologs from H. armigera and B. mori. The tree was rooted by the GR9 orthologs. Node support was estimated with 1000 bootstrap replicates, and bootstrap values were displayed with circles based on the scale indicated at the top left. The scale bar at the bottom indicates the branch length in proportion to amino acid substitutions per site.
Evolutionary relationships between the candidate IRs and IRs from D. melanogaster and H. armigera showed that four putative IR co-receptors (SfruIR8a/25a/76b/93a) clustered in the co-receptor lineages of IR8a, IR25a, IR76b, and IR93a, respectively. The other 6 IRs (SfruIR21a/60a/64a/75a/75d/75p) belong to the “antennal IR” clades (Figure 3).
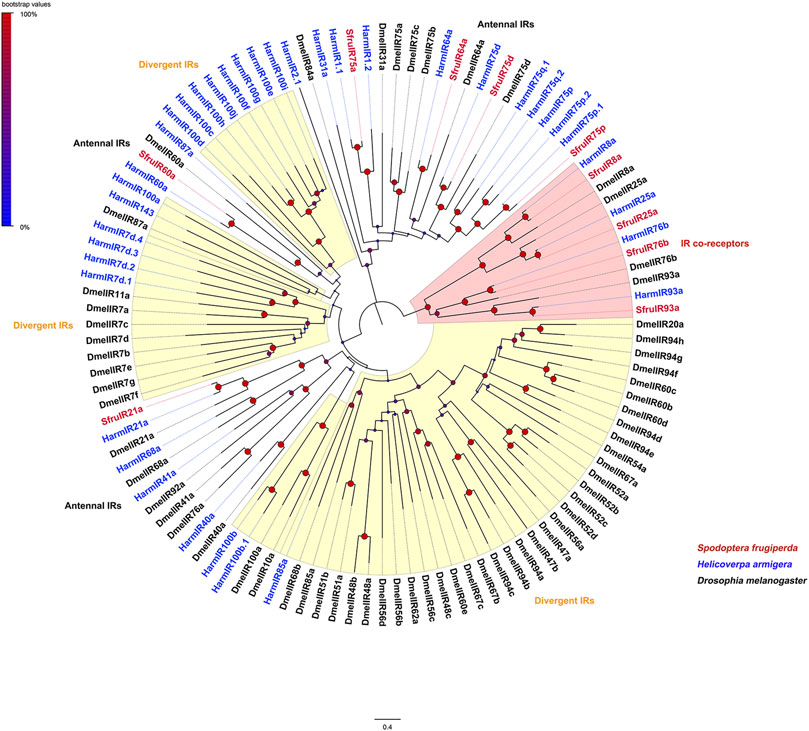
FIGURE 3. Neighbor-joining tree constructed with candidate IRs from S. frugiperda and homologs from B. mori and D. melanogaster. The tree was rooted by the IR-co receptor orthologs. Node support was estimated with 1000 bootstrap replicates, and bootstrap values were displayed with circles based on the scale indicated at the top left. The scale bar at the bottom indicates the branch length in proportion to amino acid substitutions per site.
TPM analysis
The expression abundance of candidate chemoreceptors in male and female mouthpart was then normalized across the transcriptome using the TPM method. For ORs, despite the inconsistency of expression levels between males and females, SfruORco was the highest transcribed gene (3.89/5.08 TPM, female/male, same below) among all the annotated ORs. Another candidate OR, SfruOR30, showed the second highest levels as that indicated by the TPM values (3.02/3.18). The other 27 ORs showed relative low expression levels in male and female mouthpart (the mean TPM value ≤ 1) (Figure 4A).
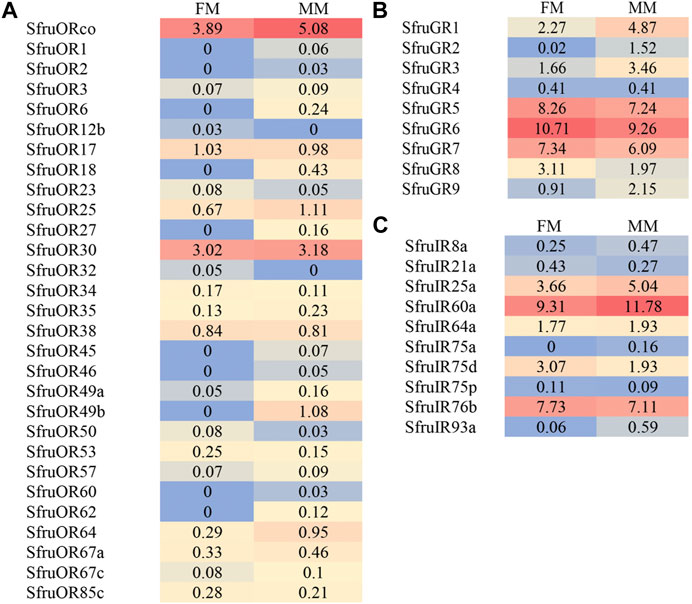
FIGURE 4. Heat-plot of TPM values for candidate chemoreceptors in female mouthpart (FM) and male mouthparts (MM). (A). SfruORs, (B). SfruGRs, (C). SfruGRs. The TPM value of each OR gene is indicated in each box.
Based on the TPM values, transcript levels of SfruGR6 in the mouthpart were the highest among all the annotated GRs, with its TPM values slightly higher in female mouthparts (10.71 TPM) than in the male ones (9.26 TPM). Other two sugar-taste receptors, SfruGR5 and SfruGR7, were also highly expressed in the mouthpart of S. frugiperda, with their TPM values of 8.26/7.24 and 7.34/6.09, respectively. In comparison, the three putative CO2 receptors had modest TPM values (2.27/4.87 for GR1, 0.02/1.52 for GR2, and 1.66/3.46 for GR3) in the mouthpart of S. frugiperda (Figure 4B).
TPM analysis showed that SfruIR60a had higher transcript levels (9.31/11.18 TPM) in the mouthpart than the other SfruIRs. SfruIR76b was also abundantly transcribed in the mouthpart (7.73/7.11 TPM). In contrast, SfruIR75a and SfruIR93a showed low TPM values of 0/0.16 (for SfruIR75a) and 0.06/0.59 (for SfruIR93a) (Figure 4C).
Expression patterns of candidate chemoreceptors
We performed RT-qPCR to analyze expression patterns of candidate chemoreceptors in different chemosensory tissues of adult S. frugiperda. Expression levels of the 48 chemoreceptors in female and male mouthparts were basically consistent with their TPM values in these two tissues.
As shown in Figure 5, all of the OR genes were mainly expressed in antennae. Among which, 10 ORs (SfruOR3/18/34/45/46/53/64/67a/67c/85c) were more expressed in female antennae than in male ones, whereas 2 ORs (SfruOR1/6) were more expressed in male antennae than in female ones, especially the putative pheromone receptor SfruOR6 which was predominantly detected in the male antennae of S. frugiperda.
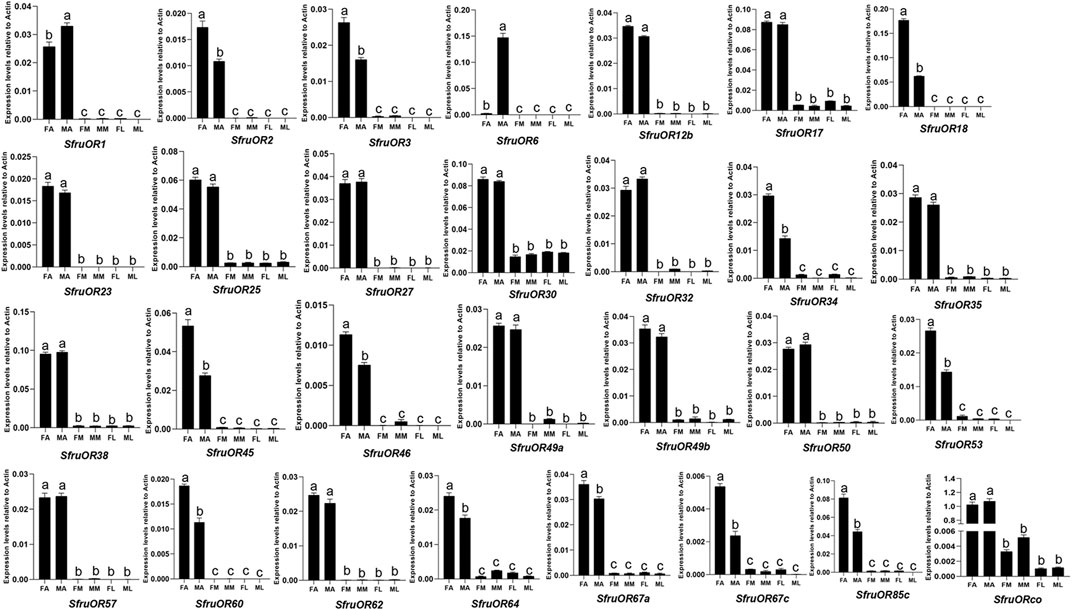
FIGURE 5. Expression patterns of candidate ORs in different chemosensory tissues of adult S. frugiperda. RT-qPCR analysis was conducted for SfruOR genes in female antennae (FA), male antennae (MA), female mouthparts (FM), male mouthparts (MM), female legs (FL), and male legs (ML). Different letters indicate significant difference based on a one-way ANOVA followed by Tukey’s multiple comparison test. Error bars show the standard errors of the means (+SE), p < 0.05, n = 3.
Based on RT-qPCR results, SfruGR1 was mainly expressed in mouthparts and legs. SfruGR7 and SfruGR8 were mainly expressed in antennae and mouthparts. In comparison, SfruGR4 was more expressed in antennae than in mouthparts and legs, and with its levels higher in female antennae than in male ones. Notably, although SfruGR9 was highly expressed in all of the tested tissues, its levels in female legs were significantly higher than that in the other tissues. The other 4 GRs, SfruGR2/3/5/6, were mainly expressed in the mouthparts. Among which, SfruGR3 was more expressed in male mouthparts than in female ones (Figure 6).
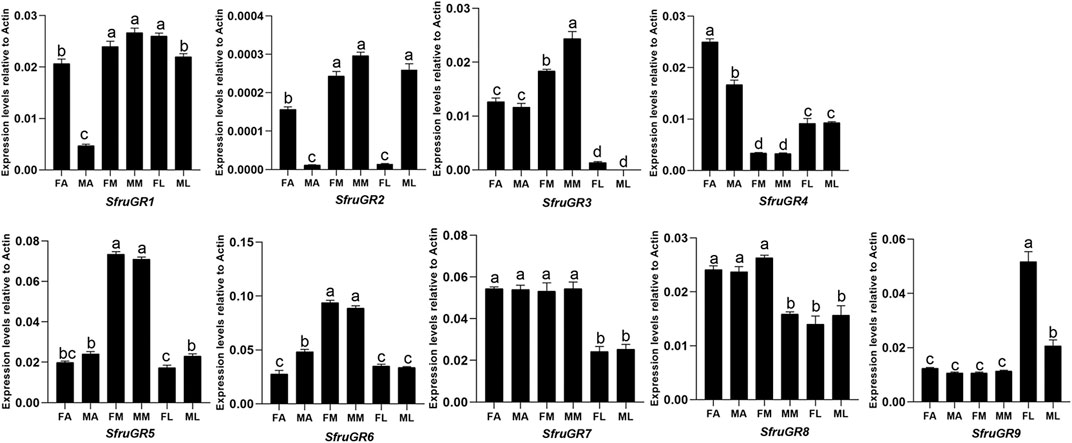
FIGURE 6. Expression patterns of candidate GRs in different chemosensory tissues of adult S. frugiperda. RT-qPCR analysis was conducted for SfruGR genes in female antennae (FA), male antennae (MA), female mouthparts (FM), male mouthparts (MM), female legs (FL), and male legs (ML). Different letters indicate significant difference based on a one-way ANOVA followed by Tukey’s multiple comparison test. Error bars show the standard errors of the means (+SE), p < 0.05, n = 3.
According to the expression profiling, all of the candidate IR genes were mainly expressed in the antennae, especially SfruIR8a, SfruIR75a, SfruIR75p, and SfruIR93a, which were almost exclusively expressed in the antennae. Moreover, the expression of SfruIR60a was also detected in the mouthparts, with comparable levels to that in the antennae and the legs (Figure 7).
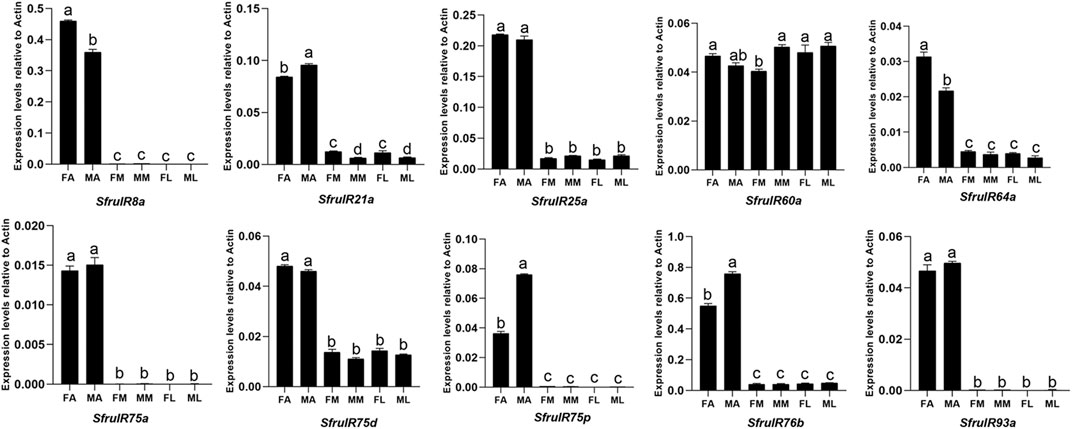
FIGURE 7. Expression patterns of candidate IRs in different chemosensory tissues of adult S. frugiperda. RT-qPCR analysis was conducted for SfruIR genes in female antennae (FA), male antennae (MA), female mouthparts (FM), male mouthparts (MM), female legs (FL), and male legs (ML). Different letters indicate significant difference based on a one-way ANOVA followed by Tukey’s multiple comparison test. Error bars show the standard errors of the means (+SE), p < 0.05, n = 3.
Expression comparison of specific chemoreceptors between proboscises and labial palps
Subsequently, we compared the expression levels of mouthpart-biased chemoreceptors between proboscises and labial palps of S. frugiperda. In total, 7 GRs (SfruGR1/2/3/5/6/7/8) and 1 IR (SfruIR60a) were selected and subjected to RT-qPCR. According to the results, 3 putative CO2 receptors (SfruGR1/2/3) were predominantly expressed in labial palps, and no significant differences between sexes were noted. In contrast, the 4 putative sugar-taste receptors (SfruGR5/6/7/8/9) and SfruIR60a were mainly expressed in proboscises. Among which, the expression of SfruGR8 was significantly higher in female proboscises than in male ones (p < 0.05). The other measured genes exhibited similar expression levels between the proboscises of both sexes (Figure 8).

FIGURE 8. Comparison of the genes displaying high expression in the mouthpart among female proboscises (FP), male proboscises (MP), female labial palps (FL), and male labial palps (ML). Different letters indicate significant difference based on a one-way ANOVA followed by Tukey’s multiple comparison test. Error bars show the standard errors of the means (+SE), p < 0.05, n = 3.
Discussion
It has long been reported that many chemosensilla are distributed on the mouthpart of moths and that the mouthpart functions in chemosensation in moths (Kent et al., 1986; Liu et al., 2014; Hu et al., 2021). However, the identity of the chemoreceptor genes in the mouthpart and their functions in feeding are still unclear for most moth species. In this study, through analyzation of the transcriptome data, we identified 29 ORs, 9 GRs, and 10 IRs in the mouthpart of S. frugiperda adults. This number is bigger than the reported 4 ORs, 7 GRs, and 6 IRs in the transcriptome of the H. armigera mouthpart (Guo et al., 2018). The high quality of the transcriptome data provides us confidence for the next step researches. Moreover, we found that the number of chemoreceptors in the adult mouthparts (29 ORs, 9 GRs, and 10 IRs) is larger than that in the larval (6th instar) ones (11 ORs, 4 GRs, and 6 IRs) (Sun et al., 2022). The quantity variation of chemoreceptors may relate to their living environment. The habitats of the larvae are relatively simple and concealed. In contrast, the adults live in open environments and their habitats are relatively complicated. Correspondingly, the S. frugiperda adults have a larger number of chemoreceptors than that in the larvae. Though such inference needs to be validated with more instances.
Insect functional ORs are dimeric complexes of specifically tuned ORs and a highly conserved OR co-receptor, ORco (Leal, 2013). Although all of the annotated SfruORs are mainly expressed in the antennae, SfruORco also shows high expression in the mouthpart of S. frugiperda. This finding indicates olfaction roles of the S. frugiperda mouthpart. Furthermore, according to the RT-qPCR results, the expression of SfruORco is significantly higher in the antennae than in the mouthparts. Such expression profile verifies the previous inference that ORco is mainly expressed in the antennae of insects (Jones, et al., 2005). Another interesting finding relates to the detection of the putative pheromone receptor SfruOR6 in the S. frugiperda mouthpart (although the expression level is very weak). A series of functional studies in heterologous system reported the best ligand of OR6 (ortholog of SfurOR6) in H. armigera and Helicoverpa assulta is Z9-16: OH (Chang et al., 2016; Yang et al., 2017). The participation of S. frugiperda mouthpart (whether or not use SfruOR6 to detect sex pheromones) in sex pheromone detection remains to be determined.
The sensing of CO2 has been long documented in insects. The neuron cells involved in the detection of CO2 are located in maxillary palps (for mosquitoes) and labial palps (for Lepidoptera adults) (Kellogg, 1970; Bogner et al., 1986; Grant et al., 1995). The mechanism underlying the detection of CO2 stimuli at the molecular level was first unraveled in D. melanogaster (Jones et al., 2007; Kwon et al., 2007). Subsequently, the molecular mechanism concerning CO2 sensing was uncovered in several moth species, and 3 GR genes, GR1/GR2/GR3, are determined to be responsible for the CO2 sensing (Xu and Anderson, 2015; Ning et al., 2016). In this study, three putative CO2 receptors (SfruGR1/2/3) have been annotated in the mouthpart of S. frugiperda. RT-qPCR demonstrated they are predominantly expressed in the labial palps, which is in accordance with the findings reported in other moth species (Xu and Anderson, 2015; Ning et al., 2016). Further work is required to validate the function of SfruGR1/2/3 so as to elucidate the molecular mechanism of CO2 detection in S. frugiperda.
Sugars are vital in insect life as valuable food resources. The detection of sugars is always utilized by insects to evaluate the nutritional values of foods (Slone et al., 2007; Kent and Robertson, 2009). In the silkmoth B. mori, 5 sugar-taste receptors (BmorGR4–8) were identified (Wanner and Robertson, 2008). BmorGR8 responds to myo-inositol and epi-inositol (Zhang et al., 2011), and BmorGR9 selectively responds to D-fructose (Sato et al., 2011). Nine putative sugar-taste receptors (HarmGR4−12) were identified in H. armigera (Xu et al., 2017). Heterologous expression in sf9 cells combined with calcium imaging found that HarmGR9 responds to D-galactose, D-maltose, and D-fructose (Xu et al., 2012). While Xenopus oocytes expression and two-electrode voltage clamping reported that HarmGR9 responds specifically to D-fructose (Jiang et al., 2015). In our study, a total of 6 putative sugar-taste receptors (SfruGR4/5/6/7/8/9) were identified in the mouthpart of S. frugiperda adults. Expression analysis showed that SfruGR5/6/7/8 are predominantly expressed in the proboscis of S. frugiperda, corroborating the vital roles of adult proboscises in the feeding of sweet substances. Moreover, the expression of SfruGR8 was documented to be significantly higher in female proboscises than in male ones, indicating its roles in female-associated behaviors of S. frugiperda. According to the RT-qPCR results, SfruGR4 and SfruGR9 displayed the highest expression in female antennae and female legs, respectively. We speculate that these GRs are involved in the gustatory process in the antennae and legs of female S. frugiperda. Specific ligands of the six putative GRs in the detection of sugar tastants in S. frugiperda will be one focus of our research in the future. We did not identify members that belonged to the “bitter-taste receptors” in the transcriptome of S. frugiperda mouthpart. This may echo the previous inference that most bitter-taste receptors have low transcript levels in chemosensory tissues (Xu et al., 2016).
Although the function of insect IRs was initially limited to olfaction, recent findings extended their roles to the sensing of taste, humidity, temperature, and sound (Rytz et al., 2013; Ni et al., 2016; Pitts et al., 2017; Hou et al., 2022). In this study, we annotated a total of 10 IRs in the mouthpart of S. frugiperda. Phylogenetic analysis showed 6 (SfruIR21a/60a/64a/75a/75d/75p) of them were classified into the “antennal IRs”. Most of the annotated “antennal IRs” were highly or specifically expressed in the antennae of S. frugiperda, consistent with that reported in other species (Liu et al., 2018; Zhu et al., 2018), However, SfruIR60a was also highly expressed in proboscises and legs. Similar findings had been reported for HarmIR60a in H. armigera (Liu et al., 2018). Thus, we suggest that SfruIR60a may be involved in both olfaction and gustation in the mouthpart of S. frugiperda. Like insect ORs, a heteromeric complex is needed for functional IRs, with at least one specific IR and an IR co-receptor within a single CSN (Abuin et al., 2011; Silbering et al., 2011). In our study, 4 putative IR co-receptors (SfruIR8a/25a/76b/IR93a) were identified in the mouthpart of S. frugiperda. According to the TPM values, SfruIR76b is the highest expressed co-receptor among the four co-receptors in the mouthpart. In D. melanogaster, DmelIR76b had been demonstrated to function as the co-receptor of specific IRs that tune to amino acids (Ganguly et al., 2017). The role of SfruIR76b (whether or not sense amino acids) in S. frugiperda mouthpart remains to be elucidated. Although “divergent IRs” were reported to be the largest group in D. melanogaster (Croset et al., 2010), we did not identify putative “divergent IRs” in the current study. This may be due to the relative small number or/and low expression levels of “divergent IRs” in the mouthpart of S. frugiperda which need to be experimentally investigated in the future.
Conclusion
By analyzing the mouthpart transcriptome of adult S. frugiperda, we annotated 48 chemoreceptors. Expression pattern investigation revealed several chemoreceptors being highly expressed in the mouthpart (labial palps or proboscises). We suggest that these genes could be important in the chemosensation in S. frugiperda mouthpart. These findings give us useful information for further investigation of chemosensory mechanism in the mouthpart of S. frugiperda as well as of other moth species.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
J-FD and C-HT conceived and designed the study. B-XD reared the insects and collected the biological materials. J-FD performed the transcriptome data analysis. J-FD and Z-JH performed the molecular work. J-FD wrote the manuscript. Z-JH and C-HT revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was funded by National Key R&D Programs of China (Grant No. 2021YFD1400701) and Major Special Science and Technology Project of Henan Province (Grant No. 201300111500).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1193085/full#supplementary-material
References
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi:10.1016/j.cell.2008.12.001
Bogner, F., Boppre, M., Ernst, K.-D., and Boeckh, J. (1986). CO2 sensitive receptors on labial palps of rhodogastria moths (Lepidoptera: Arctiidae): Physiology, fine structure and central projection. J. Comp. Physiol. A 158, 741–749. doi:10.1007/BF01324818
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi:10.1093/bioinformatics/btu170
Chang, H., Guo, M., Wang, B., Liu, Y., Dong, S., and Wang, G. (2016). Sensillar expression and responses of olfactory receptors reveal different peripheral coding in two Helicoverpa species using the same pheromone components. Sci. Rep. 6, 18742. doi:10.1038/srep18742
Chen, C., Buhl, E., Xu, M., Croset, V., Rees, J. S., Lilley, K. S., et al. (2015). Drosophila ionotropic receptor 25a mediates circadian clock resetting by temperature. Nature 527, 516–520. doi:10.1038/nature16148
Cock, M. J. W., Beseh, P. K., Buddie, A. G., Cafá, G., and Crozier, J. (2017). Molecular methods to detect Spodoptera frugiperda in Ghana, and implications for monitoring the spread of invasive species in developing countries. Sci. Rep. 7, 4103. doi:10.1038/s41598-017-04238-y
Croset, V., Rytz, R., Cummins, S. F., Budd, A., Brawand, D., Kaessmann, H., et al. (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6, e1001064. doi:10.1371/journal.pgen.1001064
Dahanukar, A., Hallem, E. A., and Carlson, J. R. (2005). Insect chemoreception. Curr. Opin. Neurobiol. 15, 423–430. doi:10.1016/j.conb.2005.06.001
Dong, J. F., Liu, H., Tang, Q. B., Liu, Y., Zhao, X. C., and Wang, G. R. (2014). Morphology, type and distribution of the labial-palp pit organ and its sensilla in the oriental armyworm, Mythimna separata (Lepidoptera: Noctuidae). Acta Entomol. Sin. 57, 681–687. CNKI:SUN:KCXB.0.2014-06-008.
Enjin, A., Zaharieva, E. E., Frank, D. D., Mansourian, S., Suh, G. S., Gallio, M., et al. (2016). Humidity sensing in Drosophila. Curr. Biol. 26, 1352–1358. doi:10.1016/j.cub.2016.03.049
Fujii, S., Yavuz, A., Slone, J., Jagge, C., Song, X., and Amrein, H. (2015). Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr. Biol. 25, 621–627. doi:10.1016/j.cub.2014.12.058
Ganguly, A., Pang, L., Duong, V. K., Lee, A., Schoniger, H., Varady, E., et al. (2017). A molecular and cellular context-dependent role for Ir76b in detection of amino acid taste. Cell Rep. 18, 737–750. doi:10.1016/j.celrep.2016.12.071
Goergen, G., Kumar, P. L., Sankung, S. B., Togola, A., and Tamò, M. (2016). First report of outbreaks of the fall armyworm Spodoptera frugiperda (J E Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in West and Central Africa. PLoS One 11, e0165632. doi:10.1371/journal.pone.0165632
Gouin, A., Bretaudeau, A., Nam, K., Gimenez, S., Aury, J. M., Duvic, B., et al. (2017). Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci. Rep. 7, 11816. doi:10.1038/s41598-017-10461-4
Grant, A. J., Aghajanian, J. G., O'Connell, R. J., and Wigton, B. E. (1995). Electrophysiological responses of receptor neurons in mosquito maxillary palp sensilla to carbon dioxide. J. Comp. Physiol. A 177, 389–396. doi:10.1007/BF00187475
Guo, H., Gong, X. L., Li, G. C., Mo, B. T., Jiang, N. J., Huang, L. Q., et al. (2022). Functional analysis of pheromone receptor repertoire in the fall armyworm, Spodoptera frugiperda. Pest Manag. Sci. 78, 2052–2064. doi:10.1002/ps.6831
Guo, M., Chen, Q., Liu, Y., Wang, G., and Han, Z. (2018). Chemoreception of mouthparts: Sensilla morphology and discovery of chemosensory genes in proboscis and labial palps of adult Helicoverpa armigera (Lepidoptera: Noctuidae). Front. Physiol. 9, 970. doi:10.3389/fphys.2018.00970
Hansson, B. S., and Stensmyr, M. C. (2011). Evolution of insect olfaction. Neuron 72, 698–711. doi:10.1016/j.neuron.2011.11.003
Haverkamp, A., Yon, F., Keesey, I. W., Missbach, C., Koenig, C., Hansson, B. S., et al. (2016). Hawkmoths evaluate scenting flowers with the tip of their proboscis. eLife 5, e15039. doi:10.7554/eLife.15039
Hou, X. Q., Zhang, D. D., Powell, D., Wang, H. L., Andersson, M. N., and Löfstedt, C. (2022). Ionotropic receptors in the turnip moth Agrotis segetum respond to repellent medium-chain fatty acids. BMC Biol. 20, 34. doi:10.1186/s12915-022-01235-0
Hu, G. L., Zhang, C. M., Wang, Z. Q., Chen, Q. X., and Lu, J. Q. (2021). Sensilla of the antenna and proboscis of Athetis lepigone (Möschler) (Lepidoptera: Noctuidae). J. Morphol. 282, 733–745. doi:10.1002/jmor.21342
Jiang, X. J., Ning, C., Guo, H., Jia, Y. Y., Huang, L. Q., Qu, M. J., et al. (2015). Gustatory receptor tuned to D-fructose in antennal sensilla chaetica of Helicoverpa armigera. Insect biochem. Mol. Biol. 60, 39–46. doi:10.1016/j.ibmb.2015.03.002
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. doi:10.1093/bioinformatics/8.3.275
Jones, W. D., Cayirlioglu, P., Kadow, I. G., and Vosshall, L. B. (2007). Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445, 86–90. doi:10.1038/nature05466
Jones, W. D., Nguyen, T. A., Kloss, B., Lee, K. J., and Vosshall, L. B. (2005). Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Curr. Biol. 15, R119–R121. doi:10.1016/j.cub.2005.02.007
Kaissling, K. E. (1996). Peripheral mechanisms of pheromone reception in moths. Chem. Senses 21, 257–268. doi:10.1093/chemse/21.2.257
Kellogg, F. E. (1970). Water vapour and carbon dioxide receptors in Aedes aegypti. J. Insect Physiol. 16, 99–108. doi:10.1016/0022-1910(70)90117-4
Kent, K. S., Harrow, I. D., Quartararo, P., and Hildebrand, J. G. (1986). An accessory olfactory pathway in Lepidoptera: The labial pit organ and its central projections in Manduca sexta and certain other sphinx moths and silk moths. Cell Tissue Res. 245, 237–245. doi:10.1007/BF00213927
Kent, L. B., and Robertson, H. M. (2009). Evolution of the sugar receptors in insects. BMC Evol. Biol. 9, 41. doi:10.1186/1471-2148-9-41
Knecht, Z. A., Silbering, A. F., Ni, L., Klein, M., Budelli, G., Bell, R., et al. (2016). Distinct combinations of variant ionotropic glutamate receptors mediate thermosensation and hygrosensation in Drosophila. eLife 5, e17879. doi:10.7554/eLife.17879
Krenn, H. W. (2010). Feeding mechanisms of adult Lepidoptera: Structure, function, and evolution of the mouthparts. Annu. Rev. Entomol. 55, 307–327. doi:10.1146/annurev-ento-112408-085338
Kwon, J. Y., Dahanukar, A., Weiss, L. A., and Carlson, J. R. (2007). The molecular basis of CO2 reception in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 104, 3574–3578. doi:10.1073/pnas.0700079104
Leal, W. S. (2013). Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi:10.1146/annurev-ento-120811-153635
Liu, N. Y., Xu, W., Dong, S. L., Zhu, J. Y., Xu, Y. X., and Anderson, A. (2018). Genome-wide analysis of ionotropic receptor gene repertoire in Lepidoptera with an emphasis on its functions of Helicoverpa armigera. Insect biochem. Mol. Biol. 99, 37–53. doi:10.1016/j.ibmb.2018.05.005
Liu, Y. L., Guo, H., Huang, L. Q., Pelosi, P., and Wang, C. Z. (2014). Unique function of a chemosensory protein in the proboscis of two Helicoverpa species. J. Exp. Biol. 217, 1821–1826. doi:10.1242/jeb.102020
Miyamoto, T., Slone, J., Song, X., and Amrein, H. (2012). A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell 151, 1113–1125. doi:10.1016/j.cell.2012.10.024
Ni, L., Klein, M., Svec, K. V., Budelli, G., Chang, E. C., Ferrer, A. J., et al. (2016). The ionotropic receptors IR21a and IR25a mediate cool sensing in Drosophila. eLife 5, e13254. doi:10.7554/eLife.13254
Ning, C., Yang, K., Xu, M., Huang, L. Q., and Wang, C. Z. (2016). Functional validation of the carbon dioxide receptor in labial palps of Helicoverpa armigera moths. Insect biochem. Mol. Biol. 73, 12–19. doi:10.1016/j.ibmb.2016.04.002
Pitts, R. J., Derryberry, S. L., Zhang, Z., and Zwiebel, L. J. (2017). Variant ionotropic receptors in the malaria vector mosquito Anopheles gambiae tuned to amines and carboxylic acids. Sci. Rep. 7, 40297. doi:10.1038/srep40297
Robertson, H. M. (2018). Molecular evolution of the major arthropod chemoreceptor gene families. Annu. Rev. Entomol. 64, 227–242. doi:10.1146/annurev-ento-020117-043322
Rytz, R., Croset, V., and Benton, R. (2013). Ionotropic receptors (IRs): Chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect biochem. Mol. Biol. 43, 888–897. doi:10.1016/j.ibmb.2013.02.007
Sato, K., Tanaka, K., and Touhara, K. (2011). Sugar-regulated cation channel formed by an insect gustatory receptor. Proc. Natl. Acad. Sci. U. S. A. 108, 11680–11685. doi:10.1073/pnas.1019622108
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi:10.1038/nprot.2008.73
Silbering, A. F., Rytz, R., Grosjean, Y., Abuin, L., Ramdya, P., Jefferis, G. S., et al. (2011). Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 31, 13357–13375. doi:10.1523/JNEUROSCI.2360-11.2011
Slone, J., Daniels, J., and Amrein, H. (2007). Sugar receptors in Drosophila. Curr. Biol. 17, 1809–1816. doi:10.1016/j.cub.2007.09.027
Sparks, A. N. (1979). A review of the biology of the fall armyworm. Flo. Entomol. 62, 82–87. doi:10.2307/3494083
Stocker, R. F. (1994). The organization of the chemosensory system in Drosophila melanogaster: A review. Cell Tissue Res. 275, 3–26. doi:10.1007/BF00305372
Sun, X. X., Hu, C. X., Jia, H. R., Wu, Q. L., Shen, X. J., Zhao, S. Y., et al. (2021). Case study on the first immigration of fall armyworm, Spodoptera frugiperda invading into China. J. Integr. Agr. 20, 664–672. doi:10.1016/S2095-3119(19)62839-X
Sun, Y. L., Jiang, P. S., Dong, B. X., Tian, C. H., and Dong, J. F. (2022). Candidate chemosensory receptors in the antennae and maxillae of Spodoptera frugiperda (J. E. Smith) larvae. Front. Physiol. 13, 970915. doi:10.3389/fphys.2022.970915
Tang, R., Jiang, N. J., Ning, C., Li, G. C., Huang, L. Q., and Wang, C. Z. (2020). The olfactory reception of acetic acid and ionotropic receptors in the Oriental armyworm, Mythimna separata Walker. Insect biochem. Mol. Biol. 118, 103312. doi:10.1016/j.ibmb.2019.103312
Wanner, K. W., and Robertson, H. M. (2008). The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol. Biol. 17, 621–629. doi:10.1111/j.1365-2583.2008.00836.x
Wu, Q. L., Jiang, Y. Y., and Wu, K. M. (2019). Analysis of migration routes of the fall armyworm Spodoptera frugiperda (J.E. Smith) from Myanmar to China. Plant Prot. 45 (1–6), 18. doi:10.16688/j.zwbh.2019047
Xu, W., and Anderson, A. (2015). Carbon dioxide receptor genes in cotton bollworm Helicoverpa armigera. Naturwissenschaften 102, 11. doi:10.1007/s00114-015-1260-0
Xu, W., Liu, N. Y., Liao, Y. L., and Anderson, A. (2017). Molecular characterization of sugar taste receptors in the cotton bollworm Helicoverpa armigera. Genome 60, 1037–1044. doi:10.1139/gen-2017-0086
Xu, W., Papanicolaou, A., Zhang, H.-J., and Anderson, A. (2016). Expansion of a bitter taste receptor family in a polyphagous insect herbivore. Sci. Rep. 6, 23666. doi:10.1038/srep23666
Xu, W., Zhang, H. J., and Anderson, A. A. (2012). Sugar gustatory receptor identified from the foregut of cotton bollworm Helicoverpa armigera. J. Chem. Ecol. 38, 1513–1520. doi:10.1007/s10886-012-0221-8
Yang, K., Huang, L. Q., Ning, C., and Wang, C. Z. (2017). Two single-point mutations shift the ligand selectivity of a pheromone receptor between two closely related moth species. eLife 6, e29100. doi:10.7554/eLife.29100
Yang, K., and Wang, C. Z. (2020). Review of pheromone receptors in heliothine species: Expression, function, and evolution. Entomol. Exp. Appl. 169, 156–171. doi:10.1111/eea.12982
Zhang, H. J., Anderson, A. R., Trowell, S. C., Luo, A. R., Xiang, Z. H., and Xia, Q. Y. (2011). Topological and functional characterization of an insect gustatory receptor. PLoS One 6, e24111. doi:10.1371/journal.pone.0024111
Zhang, J., Bisch-Knaden, S., Fandino, R. A., Yan, S., Obiero, G. F., Grosse-Wilde, E., et al. (2019a). The olfactory coreceptor IR8a governs larval feces-mediated competition avoidance in a hawkmoth. Proc. Natl. Acad. Sci. U. S. A. 116, 21828–21833. doi:10.1073/pnas.1913485116
Zhang, L., Jin, M. H., Zhang, D. D., Jiang, Y. Y., Liu, J., Wu, K. M., et al. (2019b). Molecular identification of invasive fall armyworm Spodoptera frugiperda in Yunnan province. Plant Prot. 45, 19–24. doi:10.16688/j.zwbh.2019121
Keywords: chemoreceptor, transcriptome, mouthpart, expression profiling, Spodoptera frugiperda
Citation: Dong J-F, Hu Z-J, Dong B-X and Tian C-H (2023) A mouthpart transcriptome for Spodoptera frugiperda adults: identification of candidate chemoreceptors and investigation of expression patterns. Front. Physiol. 14:1193085. doi: 10.3389/fphys.2023.1193085
Received: 24 March 2023; Accepted: 14 April 2023;
Published: 25 April 2023.
Edited by:
Fengqi Li, Guizhou University, ChinaReviewed by:
Han Wu, Guangdong Academy of Science (CAS), ChinaRui Tang, Guangdong Academy of Science (CAS), China
Copyright © 2023 Dong, Hu, Dong and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cai-Hong Tian, Y2FpaG9uZ3RpYW5AaG5hZ3JpLm9yZy5jbg==
†These authors have contributed equally to this work
 Jun-Feng Dong
Jun-Feng Dong Zhen-Jie Hu1†
Zhen-Jie Hu1† Cai-Hong Tian
Cai-Hong Tian