
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 10 May 2023
Sec. Aquatic Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1189375
This article is part of the Research Topic Endocrine Regulation and Physiological Adaptation of Stress Response in Aquatic Organisms View all 18 articles
Sepia esculenta is a cephalopod widely distributed in the Western Pacific Ocean, and there has been growing research interest due to its high economic and nutritional value. The limited anti-stress capacity of larvae renders challenges for their adaptation to high ambient temperatures. Exposure to high temperatures produces intense stress responses, thereby affecting survival, metabolism, immunity, and other life activities. Notably, the molecular mechanisms by which larval cuttlefish cope with high temperatures are not well understood. As such, in the present study, transcriptome sequencing of S. esculenta larvae was performed and 1,927 differentially expressed genes (DEGs) were identified. DEGs were subjected to functional enrichment analyses using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. The top 20 terms of biological processes in GO and 20 high-temperature stress-related pathways in KEGG functional enrichment analysis were identified. A protein-protein interaction network was constructed to investigate the interaction between temperature stress-related genes. A total of 30 key genes with a high degree of participation in KEGG signaling pathways or protein-protein interactions were identified and subsequently validated using quantitative RT-PCR. Through a comprehensive analysis of the protein-protein interaction network and KEGG signaling pathway, the functions of three hub genes (HSP90AA1, PSMD6, and PSMA5), which belong to the heat shock protein family and proteasome, were explored. The present results can facilitate further understanding of the mechanism of high temperature resistance in invertebrates and provide a reference for the S. esculenta industry in the context of global warming.
The rapid acceleration of global industrialization has resulted in a significant release of carbon dioxide into the atmosphere, which has led to global warming and subsequent oceanic warming (Nick et al., 2013; Al-Ghussain, 2019; Liu et al., 2020). High temperature will have a strong negative impact on the reproduction and development of marine biological physiological functions (Jones et al., 2013; Li et al., 2016). As an example, prolonged exposure of fish (Vargas-Chacoff et al., 2019), shellfish (Li et al., 2016; Martinez et al., 2018), mollusks (Trigg et al., 2020), and other marine animals to high-temperature marine environments will increase the burden of maintaining physiological activities, and while decreasing their ability to cope with other environmental changes, ultimately affecting their healthy growth. Additionally, high seawater temperatures can lead to stress responses in marine organisms (Dalvi et al., 2017; Samaras et al., 2018). For instance, rising seawater temperatures caused by global warming could impair the physiological function of Crassostrea virginica by disrupting the pro-oxidation-antioxidant system (Rahman and Rahman, 2021). When Piscium are exposed to high temperatures, the catecholaminergic (norepinephrine and dopaminergic) system is altered, which affects the synthesis, release, and metabolism of neurotransmitters (Alfonso et al., 2021).
Sepia esculenta is a commercially significant species belonging to the cuttlefish family, found primarily in the northern seas of China, and possessing high economic value (Guo et al., 2021). In actual aquaculture production, high-temperature seawater will significantly affect the immunity, metabolism, reproduction, and other life activities of S. esculenta, which is undoubtedly a significant challenge for S. esculenta factory farming (Bian et al., 2018).
In recent years, there has been considerable progress in high-throughput transcriptome sequencing technology, and biological analysis through high-throughput transcriptome sequencing has become more accurate (Morozova et al., 2009; Qian et al., 2014; Bao et al., 2022a; Bao et al., 2022b). To illustrate, through such technology, the expression of heat stress genes in Pinctada fucata have been found to be significantly reduced after being stimulated by multiple high temperatures, indicating that P. fucata is able to gradually adapt to the impact of high temperatures on their life activities (Li et al., 2016). The larvae possess a relatively weak capacity to endure environmental stress when in their initial phase of growth and development. Thus, elevated seawater temperature significantly impairs the routine life activities of marine larvae (Ginger et al., 2013; Kaplan et al., 2013). In the present study, transcriptome sequencing was performed to explore the mechanism of high-temperature stress in S. esculenta larvae.
In the present, high-throughput transcriptome sequencing technology was used to sequence the S. esculenta larvae exposed to high temperatures for 0, 4, and 24 h, and the data obtained were mapped to the reference genome of S. esculenta. Subsequently, differentially expressed genes (DEGs) were examined using cluster heatmap analysis and subjected to enrichment analyses of GO and KEGG. The protein-protein interaction (PPI) network was constructed by means of the selected DEGs. A combination of the KEGG signaling pathway and the protein-protein interaction network to was innovatively used explore the expression of key genes and families in S. esculenta after high-temperature stress. Finally, the expression patterns of key DEGs were verified using quantitative RT-PCR (qRT-PCR). The results provide a reference for further exploration of the temperature stress mechanism of S. esculenta, and can be beneficial for the factory farming of S. esculenta in terms of facilitating understanding of the negative effects of global warming.
In the present experiment, adult S. esculenta samples (weight = 351.87 ± 12.68 g, mantle length = 14.82 ± 0.21 mL) were collected from the marine region near Qingdao, and were held for a period of time in the pool for temporary breeding to ensure the adaptation of adults to the environment. An attachment net was placed in the pool to collect eggs. The temperature at the time of temporary breeding was 21.5°C ± 1°C. The eggs are oval, 7 ± 1 mm in diameter and translucent, which were typically collected every day and placed in perforated plastic pots. Pots were placed in another pool with flowing seawater (dissolved oxygen = 5.5 mg/L, pH = 8.2, and salinity = 30.5 ± 0.3) and continuous oxygenation, and the water temperature and other indicators were the same as those of the parent pool.
In two square 120 L buckets, each bucket was filled with 100 L seawater. The new hatched larvae were transferred to the experimental bucket half an hour after the incubation was completed. The water temperature before the larvae were transferred to the experimental bucket was 21°C ± 1°C. According to prior research, the seawater temperature of the experimental group was set to 28°C, and the temperature of the seawater in the control group was set to 23°C. During the experiment, one hundred larvae of S. esculenta were put into each bucket, and samples were taken at 4 and 24 h. Samples at the 0 h time point were obtained individually. All of the samples were stored in cryovials in liquid nitrogen.
RNA extraction, library construction, and sequencing were supported by Beijing Novogene Company. RNA was extracted from the whole larvae using standard extraction methods, followed by strict quality control with the Agilent 2100 bioanalyzer of RNA samples (Masotti and Preckel, 2006). The kit used for library construction was NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina®. When creating the library, nine larvae were collected from each group at every time point, and three samples were pooled randomly together to form a total of three biological replicates (C_0h_1, C_0h_2, C_0h_3, C_4h_1, C_4h_2, C_4h_3, C_24h_1, C_24h_2, C_24h_3, T_4h_1, T_4h_2, T_4h_3, T_24h_1, T_24h_2, and T_24h_3). S. esculenta larvae were sequenced using Illumina NovaSeq 6000 (Illumina, United States).
In the present study, the structure and function of unigenes were annotated into several databases, including NR, SwissProt, KEGG, GO, Interpro, and PFAM. DEGs were screened out using the DESeq2 package for R as a negative binomial distribution model. First of all, data were imported for building the dds model, and then the DESeq function was used to estimate the dispersion of the samples. Afterwards the difference in gene expressions was analyzed by this package. DEGs with |Log2 Fold Change| ≥ 0 (Love et al., 2014) and p-value ≤0.05 were screened out.
DAVID v6.8 (Jiao et al., 2012) was used for GO and KEGG enrichment analyses. DEGs were enriched into GO terms and KEGG signaling pathways to understand the response mechanism of S. esculenta after high-temperature exposure.
STRING v11.5 (Szklarczyk et al., 2019) was used to construct a PPI network. Genes that play significant roles in response to high-temperature stress in S. esculenta larvae were then selected based on the number of PPI and KEGG signaling pathways.
Primer Premier 5.0 (Ren et al., 2004) was used to design gene-specific primers (Table 1). In addition, qRT-PCR was performed to validate 30 genes that exhibit significant involvement in response to high-temperature stress in S. esculenta. The β-actin gene was used as a reference gene for qRT-PCR.
High-throughput sequencing technology was used to sequence the samples of S. esculenta larvae. On average, 87.68% of the high-quality reads were able to be aligned to the reference genome, and the percentage of high-quality reads with a Q30 score was 93.04% (Table 2). Such data indicate that the sequencing quality of all samples was adequate for subsequent analysis.
A total of 32,138 genes were annotated to different databases, and most genes were annotated to the NR database, reaching 73.67% (Table 3). At the same time, a large number of genes were annotated in the SwissProt and KEGG databases.
The volcano plot shows that 991 DEGs were identified, of which 498 DEGs were upregulated and 493 were downregulated in the 4 h exposure sample. In the 24 h exposure sample, 1,064 DEGs were identified among which, 470 DEGs were upregulated, and 594 DEGs were downregulated (Figure 1).
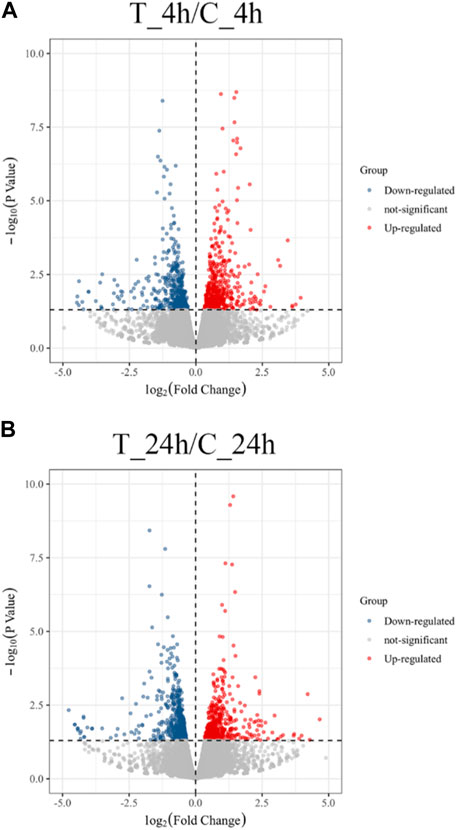
FIGURE 1. Volcano plot of the distribution expression of DEGs. (A) DEGs expression distributions between T_4h/C_4h. Red dots represent upregulated DEGs; blue dots are downregulated DEGs; grey dots indicate non-significantly genes. (B) DEGs expression distributions between T_24h/C_24h. Each dot represents a gene.
A total of 1,927 genes were differentially expressed, of which 128 genes were co-expressed at both 4 and 24 h (Figure 2).
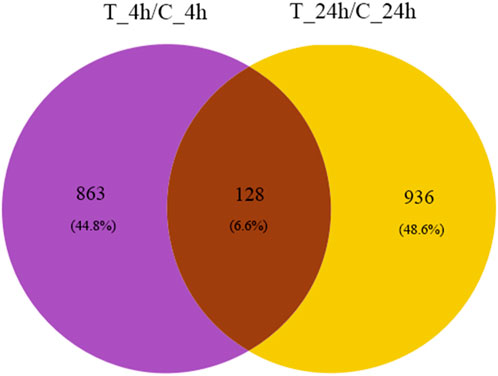
FIGURE 2. Venn diagram of DEGs. The purple and yellow regions represent the number of genes significantly expressed at 4 and 24 h after exposure, respectively. The number of genes co-expressed at 4 and 24 h is represented by brown areas.
The cluster heatmap (Figure 3) shows that the blank control group (C_0h), the 4 h control group (C_4h), and the 24 h control group (C_24h) had basically the same expression pattern. Compared with the 4 h control group (C_4h), the gene expression patterns in the 4 h experimental group (T_4h) were significantly different. A different trend was observed in the two groups (C_24h) (T_24h) at 24 h.
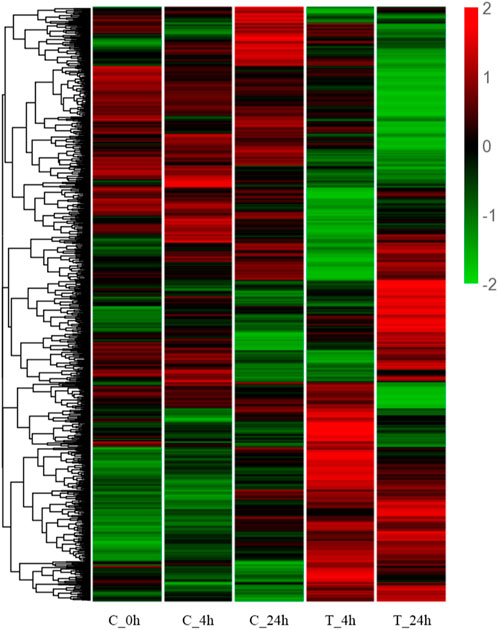
FIGURE 3. Hierarchical clustering heatmap of DEGs. The Y-axis represents each gene, and the X-axis represents the different treatment groups. Colors ranging from green to red indicating low to high expression levels.
Through GO functional enrichment analysis, DEGs were enriched into three categories: biological process, cellular component, and molecular function. The top 20 terms of the biological process and the top 10 terms of the cellular component and molecular function were selected (p-value ≤ 0.05) (Figure 4). A large number of the top 20 terms in the biological process were associated with high-temperature stress. The KEGG enrichment analysis results indicate that a large number of DEGs were enriched in multiple level-2 KEGG signaling pathways (Figure 5). Further, 20 level-3 KEGG signaling pathways were significantly enriched after exposure to high temperatures (Table 4).
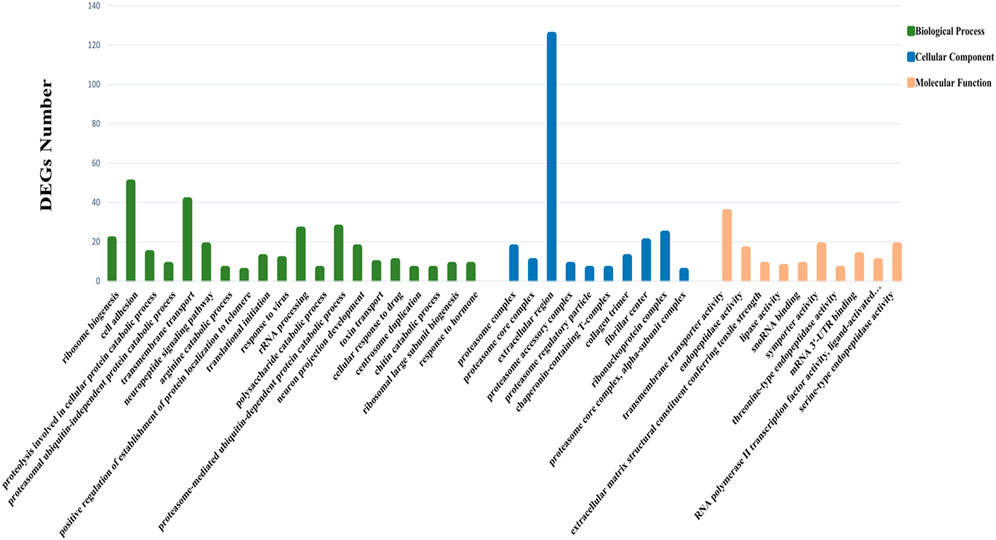
FIGURE 4. GO enrichment analysis of DEGs. The Y-axis represents the number of DEGs enriched to this term; the X-axis stands for the specific terms based on GO.
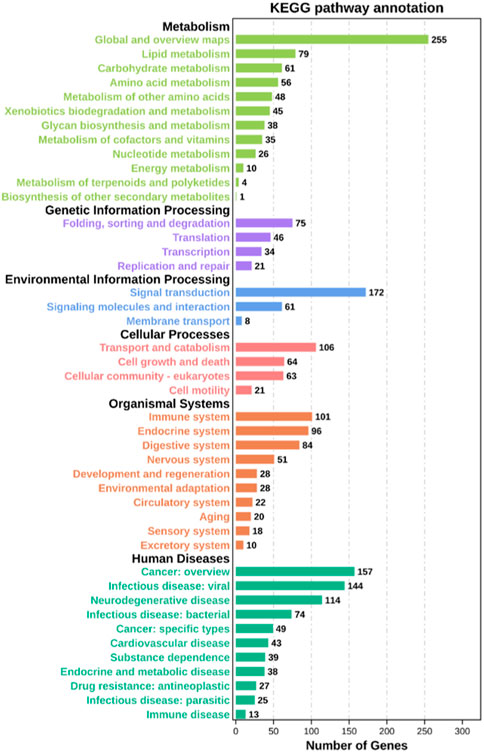
FIGURE 5. Level-2 KEGG signaling pathway annotation. The Y-axis indicates level-2 KEGG classes; the X-axis represents the corresponding number of DEGs.
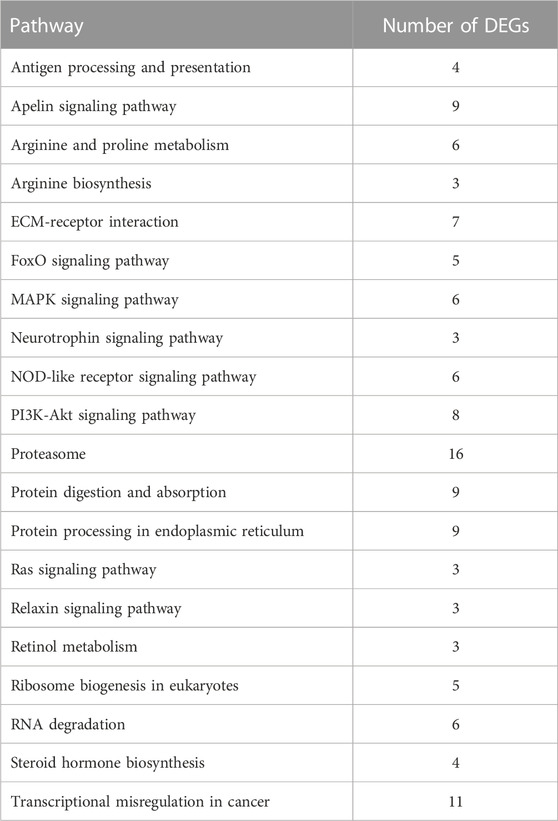
TABLE 4. Twenty significantly enriched KEGG signaling pathways related to high temperature exposure.
Among the 20 significantly enriched KEGG signaling pathways after high-temperature exposure, a total of 97 DEGs were enriched. Such genes that are involved in multiple high-temperature stress-related signaling pathways may be a significant factor in the process of resisting high-temperature stress in S. esculenta.
In the present study, 97 DEGs (Supplementary Table S1) enriched in the KEGG pathways identified after high-temperature exposure were used to construct a PPI network (Figure 6), thereby facilitating identification of key genes after high-temperature exposure.
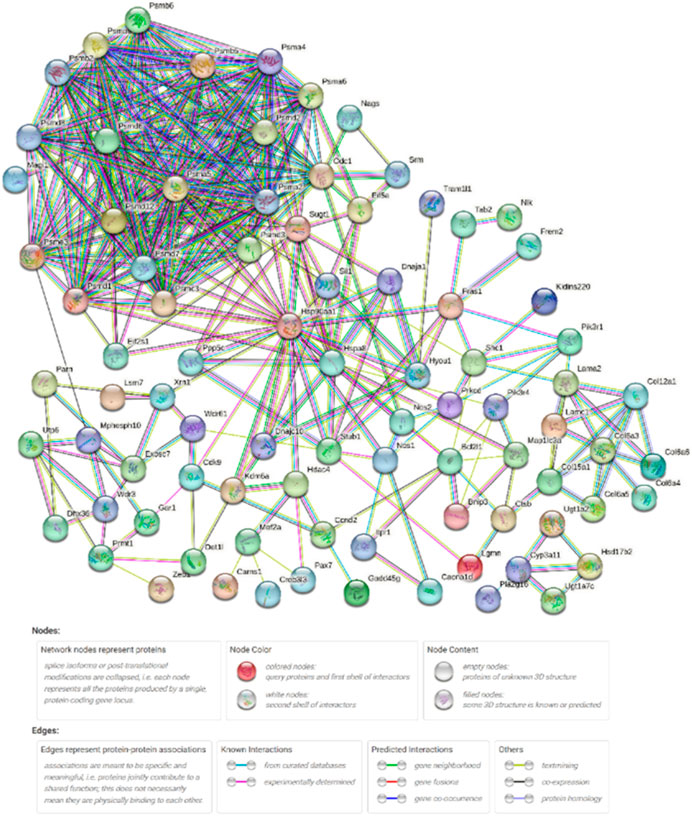
FIGURE 6. High-temperature stress-related PPI network. Network nodes represent proteins. Legends represent relationships between nodes.
Table 5 shows the network parameters. The average node degree was 6.96, the clustering coefficient was 0.574 and the PPI enrichment p-value was 1.0E-16. The parameters indicate significant interactions between the above DEGs.
After exposing S. esculenta larvae to high temperature, there was a significant level of interaction observed among the DEGs. The number of protein interactions was used as the main reference factor, combined with the number of KEGG signaling pathway participation to select the key genes. Finally, 30 key genes were identified. Table 6 displays the 30 identified key DEGs, along with their corresponding numbers of PPI and KEGG signaling pathways.
The 30 key DEGs could be further divided into five categories based on their families and functions, including the PI3K-Akt signaling pathway, heat shock protein family, proteasome family, collagen family, and other genes regulating high temperature stress.
The accuracy of the expression of 30 key genes was verified using qRT-PCR. The expression patterns of genes obtained from qRT-PCR were consistent with those obtained from RNA-Seq analysis, suggesting that the RNA-Seq results are reliable (Figure 7).
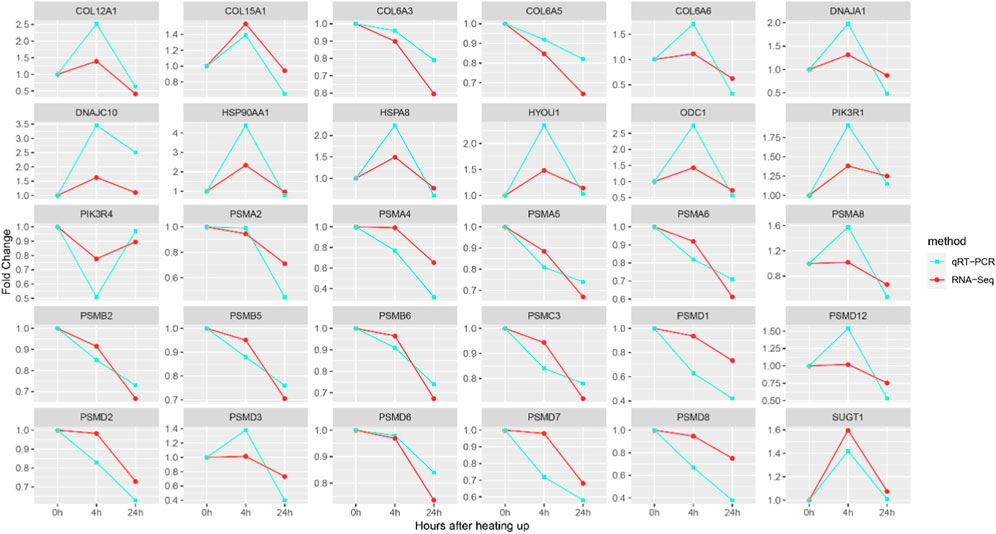
FIGURE 7. qRT-PCR and RNA-Seq results of key DEGs. The Y-axis represents the fold change and the X-axis represents the time of high temperature exposure.
In the present study, 1,927 DEGs that play significant roles in the resistance to high-temperature stress in the S. esculenta were screened. Volcano plot analysis showed that more genes appeared differentially expressed as the duration of high-temperature exposure increased. Cluster heatmap analysis showed that a large number of DEGs showed different expression patterns at 4 h (T_4h) and 24 h (T_24h). A total of 1,927 DEGs were used for GO and KEGG functional enrichment analysis. The majority of the top 20 terms identified in the cluster of biological processes, as revealed by the GO enrichment analysis, were associated with the response to high-temperature stress. The significantly enriched transmembrane transport identified through GO enrichment analysis could enhance material exchange between organelles and effectively alleviate cell stimulation (Schendzielorz et al., 2018; Jones et al., 2019; Zhong and Zhao, 2019; Ming et al., 2020). Proteasome ubiquitination, a process that helps to remove damaged proteins to reduce cell damage caused by external stimulus, was downregulated at 24 h (Shringarpure et al., 2001; Myers et al., 2018). The described biological processes may be significant factors in larval stress following high temperature exposure. In addition, among the 20 significantly enriched KEGG signaling pathways, the PI3K-Akt signaling pathway and MAPK signaling pathway play a crucial role in mollusk immunity (Song et al., 2005; Kharchenko et al., 2010; Vergadi et al., 2017; Wang et al., 2019; Qiao et al., 2021; Bao et al., 2022a). The immune response of S. esculenta larvae might be activated to protect against the injury caused by high temperature exposure. The results of GO and KEGG functional enrichment analyses show that the larvae of S. esculenta might have an intense stress response after being stimulated by high-temperature seawater. The analysis of GO terms and KEGG pathways can facilitate comprehensive understanding of the response mechanisms of S. esculenta larvae to temperature stress, which is beneficial for actual artificial breeding.
The growth and development of organisms, and metabolism, immunity, and other physiological functions are inseparable from the participation of proteins, which are the basis of all life activities (Ahmad et al., 2021; Chandhini et al., 2021; Li et al., 2021). The analysis of the interaction between proteins facilitates further understanding of the resisting mechanism of high-temperature stress in S. esculenta larvae. In the present study, a PPI network was constructed using 97 high-temperature stress-related DEGs in 20 significantly enriched KEGG signaling pathways. The results of the network show that there were strong interactions between proteins. For instance, HSP90AA1 (Heat shock protein 90 alpha family class A member 1), PSMD6 (Proteasome 26S subunit, non-ATPase 6), PSMA5 (Proteasome 20S subunit alpha 5), and ODC1 (Ornithine decarboxylase 1) interacted with over 20 proteins. Therefore, the suggestion of the present authors is that the aforementioned proteins may play central roles in the mechanism of high-temperature stress in S. esculenta larvae. Further investigation is necessary to elucidate the mechanism of action of the identified key genes involved in the response to high-temperature stress in S. esculenta larvae.
In the present study, transcriptome sequencing technology was used to analyze the stress reaction of S. esculenta larvae in high-temperature seawater, and the results can facilitate further understanding of to further understand the stress mechanism of high-temperature resistance in larvae. Finally, 30 key genes with a high number of interactions or a high number of KEGG signaling pathway involvement were identified.
The PI3K-Akt signaling pathway plays a central role in immune regulation by controlling the proliferation, differentiation, and migration of immune cells (Song et al., 2005; Vergadi et al., 2017; Bao et al., 2022a). Previous studies have shown that the PI3K-Akt signaling pathway in mollusks is involved in and regulates immune processes (Canesi et al., 2002; Fukao and Koyasu, 2003; Troutman et al., 2012). For example, the PI3K-Akt signaling pathway can regulate phagocytosis during the immune response elicited by organismal stimulation. Further, the pathway can induce an immune defense response and control the apoptosis and growth of immune cells following environmental stimulation (Sun et al., 2016; Yu et al., 2019; Wang et al., 2020). In the present study, two genes including PIK3R1 (Phosphoinositide-3-kinase regulatory subunit 1) and PIK3R4 (Phosphoinositide-3-kinase regulatory subunit 4) were identified. As previously reported, environmental stimuli can down-regulate the expression of PIK3R1, which in turn activates the mitochondrial apoptosis and death receptor pathways (Yin et al., 2020). PIK3R4 expression promotes the formation of autolysosomes for protein degradation (Gámez-Díaz et al., 2022). The results suggest that the PI3K-Akt signaling pathway may play a significant role in the resistance of S. esculenta larvae to high-temperature stress. We speculated that the PI3K-Akt signaling pathway may induce the activation of immune signaling factors to induce immune defense in larvae when stimulated by high temperature. At the same time, the expressions levels of PIK3R1 and PIK3R4 enriched in this pathway were significantly downregulated, which may also induce the apoptotic pathway to alleviate the damage of high-temperature exposure.
Heat shock proteins are a class of chaperone proteins that is widely present in various organisms and is expressed in large quantities in response to high temperature stimulation (Brokordt et al., 2009; Zuehlke et al., 2015; Wu et al., 2017). The main function of heat shock proteins is to assist in the refolding of misfolded proteins, as well as to remove damaged amino acid chains that cannot be properly folded and to degrade damaged proteins. Such actions contribute to maintaining protein homeostasis within organisms (Sghaier et al., 2004; Brun et al., 2008; Kumar et al., 2022). At the same time, HSPs can also improve the resistance to stress in the organism. Previous studies have indicated that the energy metabolism and antioxidant capacity of Ruditapes philippinarum can be improved after a brief high-temperature treatment (Zhang et al., 2023). In the present study, HSP90AA1, HSPA8 [Heat shock protein family A (HSP70) member 8], DNAJC10 [DNAJ heat shock protein family (HSP40) member C10], and DNAJA1 [DNAJ heat shock protein family (HSP40) member A1] in the HSPs family had strong interactions with other proteins, and were significantly upregulated in 4 h after high-temperature stress. HSP90AA1 had the highest number of PPI. Previous studies have linked HSP90AA1 to protein trafficking, transcriptional regulation of gene expression, and epigenetic processes (Csermely et al., 1998; Wegele et al., 2004). HSP90AA1 is sensitive to temperature and can respond quickly to high-temperature stimulation, being a soluble protein of the HSPs family, which mainly exists in the cytoplasm (Pearl and Prodromou, 2006; Swirplies et al., 2019). Similar to the functions of other HSPs family genes, HSP90AA1 can bind to hydrophobic fragments of the proteasome, thereby facilitating generation and removal of damaged proteins (Kuckelkorn et al., 2000; Gouy and Delmotte, 2008; Hayashi and Kamikawa, 2011; Reeg et al., 2016). However, differing from other chaperone proteins, HSP90AA1 has a high binding specificity (Lawless et al., 2013). HSPA8, with the participation of ATP, binds to the hydrophobic fragment of the newly generated protein to help the protein fold correctly (Stricher et al., 2013). Deletion of the gene causes selective tissue deformities during embryonic development (Wang et al., 2022). DNAJC10 and DNAJA1 belong to small molecule HSPs (HSP40s), which can bind to HSP70s through the J domain to enhance the interactions between HSP70s and substrates (Qiu et al., 2006; Liu et al., 2020). HSP40s are involved in immune defense when the organism is stimulated (Yan et al., 2021). Figure 8 shows that the genes of HSPs were significantly upregulated after 4 h of exposure to high temperature, while the genes of the HSPs were significantly downregulated after 24 h of exposure. Such findings could be attributed to prolonged exposure to high temperatures causing damage to the larval stress resistance system, leading to inadequate expression of HSPs. Based on the described results, macromolecular HSPs, HSP90AA1 and HSPA8, may protect against protein damage caused by high temperatures by binding to newly generated proteins to help them fold correctly. Small molecule HSPs, DNAJC10 and DNAJA1, may combine with HSP70s to help the protein fold correctly and improve the heat resistance of the organism. At the same time, the anti-stress mechanism of S. esculenta larvae was activated by HSPs families to alleviate the damage caused by high temperature stimulation. Extended exposure to high temperature can surpass the threshold of the anti-stress system, resulting in a decrease or impairment of the system.
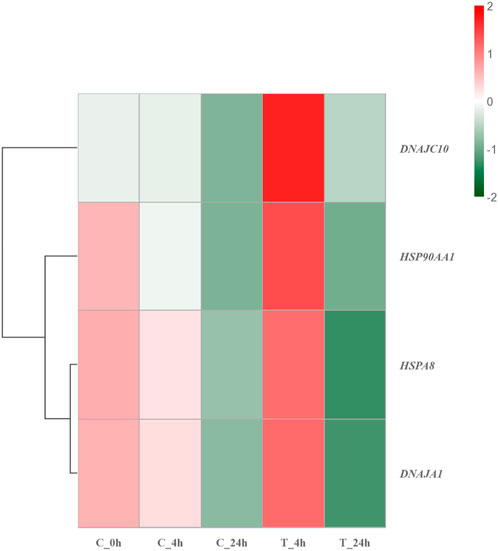
FIGURE 8. Hierarchical clustering heatmap of HSP family genes. Colors ranging from green to red indicating low to high expression levels.
The proteasome is a large cylindrical protein complex that primarily degrades unwanted or damaged proteins by cleaving peptide bonds (Morozov et al., 2019; Račková and Csekes, 2020). Such function is the primary mechanism through which cells regulate the concentrations of specific proteins and remove misfolded proteins (Shin et al., 2020; Wang et al., 2020). The components of the proteasome family include 20s core particles, 19s regulatory particles, and 11s regulatory particles (Raynes et al., 2016). Previous studies have reported significant upregulation of genes encoding proteasome and antioxidant enzymes in Chlamys opercularis when exposed to the toxin domoic acid (Ventoso et al., 2019). Similarly, genes involved in the ubiquitin-proteasome pathway were found to be significantly upregulated after R. philippinarum infection with Vibrio anguillarum (Lin et al., 2022). Among the identified key DEGs in the present study, PSMCs and PSMDs belong to the 26s proteasome, which consists of a 20s core granule and two 19s regulatory granules. PSMD6, a member of the 26S proteasome, had the highest number of PPI within the top three. During the degradation of proteins damaged by high-temperature stimulation by the 26S proteasome, the small molecule protein ubiquitin is required to covalently link the degraded proteins in a process called ubiquitination (Saeki, 2017; Shin et al., 2020). The ubiquitinated protein is then recognized by the 19S regulatory particles, which requires the participation of ATP, and the degradation of the protein is conducted by the β subunit in the 20S core particle (Saeki, 2017; Shin et al., 2020; Bhat et al., 2022). When the high-temperature stress response occurs, heat shock proteins are abundantly expressed. HSPs can bind to the hydrophobic regions of misfolded proteins to guide the conjugation of ubiquitin to misfolded proteins and improve the degradation efficiency of proteasomes (Kuckelkorn et al., 2000; Reeg et al., 2016). The PSMAs and PSMBs identified in the genes belong to the 20S proteasome, which contains only one 20S core particle. PSMA5 belongs to the category of 20S proteasome and is one of the top three genes in the number of PPI. The 20S complex can also act alone to degrade damaged proteins when the organism is acutely stimulated (Shringarpure et al., 2001; Myers et al., 2018; Abi Habib et al., 2020; Sahu et al., 2021). The proteasome plays a central role in removing erroneous proteins and maintaining cell stability in S. esculenta larvae when stimulated by high temperatures. In contrast to the expression trend of HSPs family genes, genes of the proteasome family were consistently downregulated with increasing exposure time after high-temperature exposure (Figure 9). In consideration of the fact that the main function of proteasomes is to degrade erroneous proteins, we hypothesized that proteasomes should operate at normal temperature rather than excessive temperature (28°C). Exposure to high temperatures can lead to a malfunction of the proteasome-ubiquitin system. Meanwhile, prolonged exposure to high-temperature can cause more serious damage to the anti-stress system of S. esculenta larvae, which in turn leads to the reduction of gene expression.
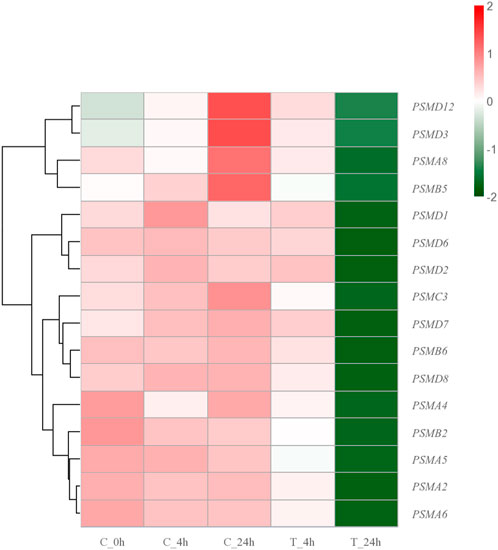
FIGURE 9. Hierarchical clustering heatmap of proteasome family genes. The colors ranging from green to red indicate low to high expression levels.
Collagen is a structural protein containing a triple helical domain that plays a central role in maintaining cellular organization (Ricard-Blum and Ruggiero, 2005; Ricard-Blum, 2011). Collagen also has other vital roles, such as being involved in cell adhesion, chemotaxis and migration, as well as regulating wound healing and tissue remodeling (Brown and Timpl, 1995; Myllyharju and Kivirikko, 2004). Previous research has suggested that HSPs play a role in regulating collagen synthesis induced by transforming growth factor-β, possibly through modulation of Smad 2/3 phosphorylation (Lee et al., 2016). Additionally, HSPs have the ability to bind to newly synthesized collagen and assist in proper folding, resulting in the formation of a stable triple helix structure (Nagata, 1996; Mala and Rose, 2010). In the present study, the associated genes enriched to encode the proteasome included COL6A3 (Collagen type VI alpha 3 chain), COL12A1 (Collagen type XII alpha 1 chain), COL15A1 (Collagen type XV alpha 1 chain), COL6A5 (Collagen type VI alpha 5 chain), and COL6A6 (Collagen type VI alpha 6 chain). The expression levels of such genes were significantly upregulated after 4 and 24 h of exposure. The results suggest that collagen is a significant factor in the resistance of S. esculenta larvae to high temperature stress. It is hypothesized that collagen is involved in tissue repair and facilitate repair and regeneration of cells after high temperature stress.
ODC1, SUGT1 (SGT1 homolog, MIS12 kinetochore complex assembly cochaperone), and HYOU1 (Hypoxia upregulated 1) were also identified as genes that have significant roles in the stress resistance mechanism of S. esculenta larvae after high-temperature exposure. The significant downregulation of ODC1 can mitigate the apoptosis and damage resulting from exposure to high temperatures in the organism (Jeffries et al., 2012; Jiang et al., 2018). Additionally, SUGT1 can activate the NOD-like receptor family to eliminate damaged proteins, thereby counteracting the negative impacts of high-temperature stress (da Silva Correia et al., 2007; Hong and Hahn, 2016). The expression product of the HYOU1 gene can activate the PI3K-AKT signaling pathway to promote cell growth and migration (Li et al., 2019; Wang et al., 2022). Such genes were all significantly downregulated, indicating that excessive temperature would destroy the anti-stress system of S. esculenta larvae and reduce the expression of proteins.
In the present study, transcriptome sequencing technology was used to preliminarily analyze the mechanism of high-temperature stress in S. esculenta larvae. Through the analysis of functional enrichment and protein-protein interaction networks, a significant number of DEGs that play crucial roles in response to stress were identified. The expression trends of key genes in the heat shock protein family and proteasome family indicated that prolonged high-temperature exposure would disrupt the larval stress system. In conclusion, high-temperature exposure could significantly affect the S. esculenta larvae stress system, and even prolonged high temperature could cause severe damage to the stress system. The obtained results offer valuable insights to investigate the mechanism underlying the stress response of cephalopods upon exposure to high temperatures.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA947123.
The animal study was reviewed and approved by the protocols of the Institutional Animal Care and Use Committee of the Ludong University (protocol number LDU-IRB20210308NXY) and the China Government Principles for the Utilization and Care of Invertebrate Animals Used in Testing, Research, and Training (State Science and Technology Commission of the People’s Republic of China for No. 2, 31 October 1988. http://www.gov.cn/gongbao/content/2011/content_1860757.htm).
ZL and JY designed and supervised the study. YW, XB, TY, WW, XX, and XL prepared the samples. YW, XB, XX, and XL analyzed all sequencing data. YW wrote the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This research was supported by the earmarked fund for CARS-49, and the Natural Science Foundation of Shandong Province (No. ZR2019BC052).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1189375/full#supplementary-material
Abi Habib, J., De Plaen, E., Stroobant, V., Zivkovic, D., Bousquet, M. P., Guillaume, B., et al. (2020). Efficiency of the four proteasome subtypes to degrade ubiquitinated or oxidized proteins. Sci. Rep. 10 (1), 15765. doi:10.1038/s41598-020-71550-5
Ahmad, I., Ahmed, I., Fatma, S., and Peres, H. (2021). Role of branched-chain amino acids on growth, physiology and metabolism of different fish species: A review. Aquacult. Nutr. 27, 1270–1289. doi:10.1111/anu.13267
Al-Ghussain, L. (2019). Global warming: Review on driving forces and mitigation. Environ. Prog. Sustain. Energy. 38, 13–21. doi:10.1002/ep.13041
Alfonso, S., Gesto, M., and Sadoul, B. (2021). Temperature increase and its effects on fish stress physiology in the context of global warming. J. Fish. Biol. 98 (6), 1496–1508. doi:10.1111/jfb.14599
Bao, X., Wang, W., Chen, X., Feng, Y., Xu, X., Sun, G., et al. (2022a). Exploration of immune response mechanisms in cadmium and copper co-exposed juvenile golden cuttlefish (Sepia esculenta) based on transcriptome profiling. Front. Immunol. 13, 963931. doi:10.3389/fimmu.2022.963931
Bao, X., Wang, W., Yuan, T., Li, Y., Chen, X., Liu, X., et al. (2022b). Transcriptome profiling based on larvae at different time points after hatching provides a core set of gene resource for understanding the immune response mechanisms of the egg-protecting behavior against Vibrio anguillarum infection in Amphioctopus fangsiao. Fish. Shellfish Immunol. 124, 430–441. doi:10.1016/j.fsi.2022.04.030
Bhat, S. A., Vasi, Z., Adhikari, R., Gudur, A., Ali, A., Jiang, L., et al. (2022). Ubiquitin proteasome system in immune regulation and therapeutics. Curr. Opin. Pharmacol. 67, 102310. doi:10.1016/j.coph.2022.102310
Bian, L., Liu, C., Chen, S., Zhao, F., Ge, J., and Tan, J. (2018). Transcriptome analysis of gene expression patterns during embryonic development in golden cuttlefish (Sepia esculenta). Genes Genomics 40 (3), 253–263. doi:10.1007/s13258-017-0588-6
Brokordt, K., Leiva, N., Jeno, K., Martínez, G., and Winkler, F. (2009). Effect of allozyme heterozygosity on basal and induced levels of heat shock protein (Hsp70), in juvenile Concholepas concholepas (Mollusca). J. Exp. Mar. Biol. Ecol. 370 (1-2), 18–26. doi:10.1016/j.jembe.2008.11.007
Brown, J. C., and Timpl, R. (1995). The collagen superfamily. Int. Arch. Allergy Immunol. 107 (4), 484–490. doi:10.1159/000237090
Brun, N. T., Bricelj, V. M., MacRae, T. H., and Ross, N. W. (2008). Heat shock protein responses in thermally stressed bay scallops, Argopecten irradians, and sea scallops, Placopecten magellanicus. J. Exp. Mar. Biol. Ecol. 358 (2), 151–162. doi:10.1016/j.jembe.2008.02.006
Canesi, L., Gallo, G., Gavioli, M., and Pruzzo, C. (2002). Bacteria-hemocyte interactions and phagocytosis in marine bivalves. Microsc. Res. Tech. 57 (6), 469–476. doi:10.1002/jemt.10100
Chandhini, S., Trumboo, B., Jose, S., Varghese, T., Rajesh, M., and Kumar, V. J. R. (2021). Insulin-like growth factor signalling and its significance as a biomarker in fish and shellfish research. Fish. Physiol. Biochem. 47 (4), 1011–1031. doi:10.1007/s10695-021-00961-6
Csermely, P., Schnaider, T., Soti, C., Prohászka, Z., and Nardai, G. (1998). The 90-kDa molecular chaperone family: Structure, function, and clinical applications. A comprehensive review. Pharmacol. Ther. 79 (2), 129–168. doi:10.1016/s0163-7258(98)00013-8
da Silva Correia, J., Miranda, Y., Leonard, N., and Ulevitch, R. (2007). SGT1 is essential for Nod1 activation. Proc. Natl. Acad. Sci. U. S. A. 104 (16), 6764–6769. doi:10.1073/pnas.0610926104
Dalvi, R. S., Das, T., Debnath, D., Yengkokpam, S., Baruah, K., Tiwari, L. R., et al. (2017). Metabolic and cellular stress responses of catfish, Horabagrus brachysoma (Gunther) acclimated to increasing temperatures. J. Therm. Biol. 65, 32–40. doi:10.1016/j.jtherbio.2017.02.003
Fukao, T., and Koyasu, S. (2003). PI3K and negative regulation of TLR signaling. Trends Immunol. 24 (7), 358–363. doi:10.1016/s1471-4906(03)00139-x
Gámez-Díaz, L., Ligeon, L., Sindram, E., Deau, M., Sanchez-Martin, P., Nestel, S., et al. (2022). LRBA balances antigen presentation and T-cell responses by facilitating autophagy through the binding to PIK3R4 and FYCO1. Biorxiv. doi:10.1101/2022.10.17.512524
Ginger, K. W., Vera, C. B., R, D., Dennis, C. K., Adela, L. J., Yu, Z., et al. (2013). Larval and post-larval stages of Pacific oyster (Crassostrea gigas) are resistant to elevated CO2. PLoS One 8 (5), e64147. doi:10.1371/journal.pone.0064147
Gouy, M., and Delmotte, S. (2008). Remote access to ACNUC nucleotide and protein sequence databases at PBIL. Biochimie 90 (4), 555–562. doi:10.1016/j.biochi.2007.07.003
Guo, H., Zhang, D., Wang, L., Li, W., He, P., Näslund, J., et al. (2021). Sperm competition in golden cuttlefish Sepia esculenta: The impact of mating order and male size. Aquaculture 530, 735929. doi:10.1016/j.aquaculture.2020.735929
Hayashi, K., and Kamikawa, Y. (2011). HSP90 is crucial for regulation of LAT expression in activated T cells. Mol. Immunol. 48 (6-7), 941–946. doi:10.1016/j.molimm.2010.12.014
Hong, T. J., and Hahn, J. S. (2016). Application of SGT1-Hsp90 chaperone complex for soluble expression of NOD1 LRR domain in E. coli. Biochem. Biophys. Res. Commun. 478 (4), 1647–1652. doi:10.1016/j.bbrc.2016.08.174
Jeffries, K. M., Hinch, S. G., Sierocinski, T., Clark, T. D., Eliason, E. J., Donaldson, M. R., et al. (2012). Consequences of high temperatures and premature mortality on the transcriptome and blood physiology of wild adult sockeye salmon (Oncorhynchus nerka). Ecol. Evol. 2 (7), 1747–1764. doi:10.1002/ece3.274
Jiang, F., Gao, Y., Dong, C., and Xiong, S. (2018). ODC1 inhibits the inflammatory response and ROS-induced apoptosis in macrophages. Biochem. Biophys. Res. Commun. 504 (4), 734–741. doi:10.1016/j.bbrc.2018.09.023
Jiao, X., Sherman, B. T., Huang da, W., Stephens, R., Baseler, M. W., Lane, H. C., et al. (2012). DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 28 (13), 1805–1806. doi:10.1093/bioinformatics/bts251
Jones, H. R., Johnson, K. M., and Kelly, M. W. (2019). Synergistic effects of temperature and salinity on the gene expression and physiology of Crassostrea virginica. Integr. Comp. Biol. 59 (2), 306–319. doi:10.1093/icb/icz035
Jones, M. C., Dye, S. R., Fernandes, J. A., Frölicher, T. L., Pinnegar, J. K., Warren, R., et al. (2013). Predicting the impact of climate change on threatened species in UK waters. PLoS One 8 (1), e54216. doi:10.1371/journal.pone.0054216
Kaplan, M. B., Mooney, T. A., McCorkle, D. C., and Cohen, A. L. (2013). Adverse effects of ocean acidification on early development of squid (Doryteuthis pealeii). PLoS One 8 (5), e63714. doi:10.1371/journal.pone.0063714
Kharchenko, O. A., Grinkevich, V. V., Vorobiova, O. V., and Grinkevich, L. N. (2010). Learning-induced lateralized activation of the MAPK/ERK cascade in identified neurons of the food-aversion network in the mollusk Helix lucorum. Neurobiol. Learn. Mem. 94 (2), 158–166. doi:10.1016/j.nlm.2010.05.002
Kuckelkorn, U., Knuehl, C., Boes-Fabian, B., Drung, I., and Kloetzel, P. M. (2000). The effect of heat shock on 20S/26S proteasomes. Biol. Chem. 381 (9-10), 1017–1023. doi:10.1515/BC.2000.125
Kumar, V., Roy, S., Behera, B. K., and Das, B. K. (2022). Heat shock proteins (hsps) in cellular homeostasis: A promising tool for health management in Crustacean aquaculture. Life (Basel). 12 (11), 1777. doi:10.3390/life12111777
Lawless, N., Blacklock, K., Berrigan, E., and Verkhivker, G. (2013). Structural bioinformatics and protein docking analysis of the molecular chaperone-kinase interactions: Towards allosteric inhibition of protein kinases by targeting the hsp90-cdc37 chaperone machinery. Pharm. (Basel) 6 (11), 1407–1428. doi:10.3390/ph6111407
Lee, S. B., Lim, A. R., Rah, D. K., Kim, K. S., and Min, H. J. (2016). Modulation of heat shock protein 90 affects TGF-β-induced collagen synthesis in human dermal fibroblast cells. Tissue Cell 48 (6), 616–623. doi:10.1016/j.tice.2016.09.002
Li, S., Liu, C., Huang, J., Liu, Y., Zhang, S., Zheng, G., et al. (2016). Transcriptome and biomineralization responses of the pearl oyster Pinctada fucata to elevated CO2 and temperature. Sci. Rep. 6, 18943. doi:10.1038/srep18943
Li, X., Zhang, N. X., Ye, H. Y., Song, P. P., Chang, W., Chen, L., et al. (2019). HYOU1 promotes cell growth and metastasis via activating PI3K/AKT signaling in epithelial ovarian cancer and predicts poor prognosis. Eur. Rev. Med. Pharmacol. Sci. 23 (10), 4126–4135. doi:10.26355/eurrev_201901_17914
Li, Z., Bao, X., Liu, X., Li, Y., Cui, M., Liu, X., et al. (2021). Transcriptome profiling based on protein-protein interaction networks provides a set of core genes for understanding the immune response mechanisms of the egg-protecting behavior in Octopus ocellatus. Fish. Shellfish Immunol. 117, 113–123. doi:10.1016/j.fsi.2021.07.020
Lin, Z., Nie, H., Zhang, Y., Yin, Z., and Yan, X. (2022). Genome-wide identification and analysis of HECT E3 ubiquitin ligase gene family in Ruditapes philippinarum and their involvement in the response to heat stress and Vibrio anguillarum infection. Comp. Biochem. Physiol. Part D. Genomics Proteomics. 43, 101012. doi:10.1016/j.cbd.2022.101012
Liu, L., Lin, L., Ma, Z., Wang, G., and Wu, M. (2020). iTRAQ-based quantitative proteomic analysis of Sargassum fusiformein response to high temperature stress. Aquacult. Res. 52 (1), 185–195. doi:10.1111/are.14880
Liu, Q., Liang, C., and Zhou, L. (2020). Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci. 29 (2), 378–390. doi:10.1002/pro.3725
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
Mala, J. G., and Rose, C. (2010). Interactions of heat shock protein 47 with collagen and the stress response: An unconventional chaperone model? Life Sci. 87 (19-22), 579–586. doi:10.1016/j.lfs.2010.09.024
Martinez, M., Mangano, M. C., Maricchiolo, G., Genovese, L., Mazzola, A., and Sarà, G. (2018). Measuring the effects of temperature rise on Mediterranean shellfish aquaculture. Ecol. Indic. 88, 71–78. doi:10.1016/j.ecolind.2018.01.002
Masotti, A., and Preckel, T. (2006). Analysis of small RNAs with the agilent 2100 bioanalyzer. Nat. Methods. 3, 658. doi:10.1038/nmeth908
Ming, Z., Pang, Y., and Liu, J. (2020). Mechanical deformation mediated transmembrane transport. Macromol. Rapid Commun. 41 (2), e1900518. doi:10.1002/marc.201900518
Morozov, A. V., Burov, A. V., Astakhova, T. M., Spasskaya, D. S., Margulis, B. A., and Karpov, V. L. (2019). Dynamics of the functional activity and expression of proteasome subunits during cellular adaptation to heat shock. Mol. Biol. Mosk. 53 (4), 638–647. doi:10.1134/S0026898419040086
Morozova, O., Hirst, M., and Marra, M. A. (2009). Applications of new sequencing technologies for transcriptome analysis. Annu. Rev. Genomics Hum. Genet. 10, 135–151. doi:10.1146/annurev-genom-082908-145957
Myers, N., Olender, T., Savidor, A., Levin, Y., Reuven, N., and Shaul, Y. (2018). The disordered landscape of the 20S proteasome substrates reveals tight association with phase separated granules. Proteomics 18 (21-22), e1800076. doi:10.1002/pmic.201800076
Myllyharju, J., and Kivirikko, K. I. (2004). Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 20 (1), 33–43. doi:10.1016/j.tig.2003.11.004
Nagata, K. (1996). Hsp47: A collagen-specific molecular chaperone. Trends biochem. Sci. 21 (1), 22–26. doi:10.1016/0968-0004(96)80881-4
Nick, F. M., Vieli, A., Andersen, M. L., Joughin, I., Payne, A., Edwards, T. L., et al. (2013). Future sea-level rise from Greenland’s main outlet glaciers in a warming climate. Nature 497 (7448), 235–238. doi:10.1038/nature12068
Pearl, L. H., and Prodromou, C. (2006). Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 75, 271–294. doi:10.1146/annurev.biochem.75.103004.142738
Qian, X., Ba, Y., Zhuang, Q., and Zhong, G. (2014). RNA-Seq technology and its application in fish transcriptomics. OMICS 18 (2), 98–110. doi:10.1089/omi.2013.0110
Qiao, Y., Yan, W., He, J., Liu, X., Zhang, Q., and Wang, X. (2021). Identification, evolution and expression analyses of mapk gene family in Japanese flounder (Paralichthys olivaceus) provide insight into its divergent functions on biotic and abiotic stresses response. Aquat. Toxicol. 241, 106005. doi:10.1016/j.aquatox.2021.106005
Qiu, X. B., Shao, Y. M., Miao, S., and Wang, L. (2006). The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol. Life Sci. 63 (22), 2560–2570. doi:10.1007/s00018-006-6192-6
Račková, L., and Csekes, E. (2020). Proteasome biology: Chemistry and bioengineering insights. Polym. (Basel) 12 (12), 2909. doi:10.3390/polym12122909
Rahman, M. S., and Rahman, M. S. (2021). Effects of elevated temperature on prooxidant-antioxidant homeostasis and redox status in the American oyster: Signaling pathways of cellular apoptosis during heat stress. Environ. Res. 196, 110428. doi:10.1016/j.envres.2020.110428
Raynes, R., Pomatto, L. C., and Davies, K. J. (2016). Degradation of oxidized proteins by the proteasome: Distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Mol. Asp. Med. 50, 41–55. doi:10.1016/j.mam.2016.05.001
Reeg, S., Jung, T., Castro, J. P., Davies, K. J. A., Henze, A., and Grune, T. (2016). The molecular chaperone Hsp70 promotes the proteolytic removal of oxidatively damaged proteins by the proteasome. Free Radic. Biol. Med. 99, 153–166. doi:10.1016/j.freeradbiomed.2016.08.002
Ren, L., Zhu, B., Zhang, Y., Wang, H., Li, C., Su, Y., et al. (2004). The research of applying primer premier 5.0 to design PCR primer. J. Jinzhou. Med. Coll. 25 (6), 43–46. doi:10.3969/j.issn.1674-0424.2004.06.015
Ricard-Blum, S., and Ruggiero, F. (2005). The collagen superfamily: From the extracellular matrix to the cell membrane. Pathol. Biol. Paris. 53 (7), 430–442. doi:10.1016/j.patbio.2004.12.024
Ricard-Blum, S. (2011). The collagen family. Cold Spring Harb. Perspect. Biol. 3 (1), a004978. doi:10.1101/cshperspect.a004978
Saeki, Y. (2017). Ubiquitin recognition by the proteasome. J. Biochem. 161 (2), 113–124. doi:10.1093/jb/mvw091
Sahu, I., Mali, S. M., Sulkshane, P., Xu, C., Rozenberg, A., Morag, R., et al. (2021). The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag. Nat. Commun. 12 (1), 6173. doi:10.1038/s41467-021-26427-0
Samaras, A., Papandroulakis, N., Lika, K., and Pavlidis, M. (2018). Water temperature modifies the acute stress response of European sea bass, Dicentrarchus labrax L. (1758). J. Therm. Biol. 78, 84–91. doi:10.1016/j.jtherbio.2018.09.006
Schendzielorz, A. B., Bragoszewski, P., Naumenko, N., Gomkale, R., Schulz, C., Guiard, B., et al. (2018). Motor recruitment to the TIM23 channel's lateral gate restricts polypeptide release into the inner membrane. Nat. Commun. 9 (1), 4028. doi:10.1038/s41467-018-06492-8
Sghaier, H., Le Ai, T. H., Horiike, T., and Shinozawa, T. (2004). Molecular chaperones: Proposal of a systematic computer-oriented nomenclature and construction of a centralized database. Silico. Biol. 4 (3), 311–322.
Shin, J. Y., Muniyappan, S., Tran, N. N., Park, H., Lee, S. B., and Lee, B. H. (2020). Deubiquitination reactions on the proteasome for proteasome versatility. Int. J. Mol. Sci. 21 (15), 5312. doi:10.3390/ijms21155312
Shringarpure, R., Grune, T., and Davies, K. J. (2001). Protein oxidation and 20S proteasome-dependent proteolysis in mammalian cells. Cell Mol. Life Sci. 58 (10), 1442–1450. doi:10.1007/PL00000787
Song, G., Ouyang, G., and Bao, S. (2005). The activation of Akt/PKB signaling pathway and cell survival. J. Cell. Mol. Med. 9 (1), 59–71. doi:10.1111/j.1582-4934.2005.tb00337.x
Stricher, F., Macri, C., Ruff, M., and Muller, S. (2013). HSPA8/HSC70 chaperone protein: Structure, function, and chemical targeting. Autophagy 9 (12), 1937–1954. doi:10.4161/auto.26448
Sun, Y., Zhang, X., Wang, G., Lin, S., Zeng, X., Wang, Y., et al. (2016). PI3K-AKT signaling pathway is involved in hypoxia/thermal-induced immunosuppression of small abalone Haliotis diversicolor. Fish. Shellfish Immunol. 59, 492–508. doi:10.1016/j.fsi.2016.11.011
Swirplies, F., Wuertz, S., Baßmann, B., Orban, A., Schäfer, N., Brunner, R. M., et al. (2019). Identification of molecular stress indicators in pikeperch Sander lucioperca correlating with rising water temperatures. Aquaculture 501, 260–271. doi:10.1016/j.aquaculture.2018.11.043
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47 (D1), D607–D613. doi:10.1093/nar/gky1131
Trigg, S. A., Mitchell, K. R., Thompson, R. E., Eudeline, B., Vadopalas, B., Timmins-Schiffman, E. B., et al. (2020). Temporal proteomic profiling reveals insight into critical developmental processes and temperature-influenced physiological response differences in a bivalve mollusc. Bmc. Genomics. 21 (1), 723. doi:10.1186/s12864-020-07127-3
Troutman, T. D., Bazan, J. F., and Pasare, C. (2012). Toll-like receptors, signaling adapters and regulation of the pro-inflammatory response by PI3K. Cell Cycle 11 (19), 3559–3567. doi:10.4161/cc.21572
Vargas-Chacoff, L., Muñoz, J. L. P., Ocampo, D., Paschke, K., and Navarro, J. M. (2019). The effect of alterations in salinity and temperature on neuroendocrine responses of the Antarctic fish Harpagifer antarcticus. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 235, 131–137. doi:10.1016/j.cbpa.2019.05.029
Ventoso, P., Pazos, A. J., Pérez-Parallé, M. L., Blanco, J., Triviño, J. C., and Sánchez, J. L. (2019). RNA-seq transcriptome profiling of the queen scallop (Aequipecten opercularis) digestive gland after exposure to domoic acid-producing pseudo-nitzschia. Toxins (Basel). 11 (2), 97. doi:10.3390/toxins11020097
Vergadi, E., Ieronymaki, E., Lyroni, K., Vaporidi, K., and Tsatsanis, C. (2017). Akt signaling pathway in macrophage activation and M1/M2 polarization. J. Immunol. 198 (3), 1006–1014. doi:10.4049/jimmunol.1601515
Wang, C., Zhang, X., Wang, X., Zhai, Y., Li, M., Pan, J., et al. (2022). Genetic deletion of hspa8 leads to selective tissue malformations in zebrafish embryonic development. J. Cell Sci. 135 (21), jcs259734. doi:10.1242/jcs.259734
Wang, H., Pan, L., Xu, R., Si, L., and Zhang, X. (2019). The molecular mechanism of Nrf2-Keap1 signaling pathway in the antioxidant defense response induced by BaP in the scallop Chlamys farreri. Fish. Shellfish Immunol. 92, 489–499. doi:10.1016/j.fsi.2019.06.006
Wang, W., Jiang, X., Xia, F., Chen, X., Li, G., Liu, L., et al. (2022). HYOU1 promotes cell proliferation, migration, and invasion via the PI3K/AKT/FOXO1 feedback loop in bladder cancer. Mol. Biol. Rep. 50, 453–464. doi:10.1007/s11033-022-07978-x
Wang, X., Meul, T., and Meiners, S. (2020). Exploring the proteasome system: A novel concept of proteasome inhibition and regulation. Pharmacol. Ther. 211, 107526. doi:10.1016/j.pharmthera.2020.107526
Wang, Y., Zhou, S., Liu, T., Chen, M., and Zhang, X. (2020). De novo transcriptome analysis of stressed blood clam (Anadara broughtonii) and identification of genes associated with hemoglobin. Genes Genomics 42 (2), 189–202. doi:10.1007/s13258-019-00887-7
Wegele, H., Müller, L., and Buchner, J. (2004). Hsp70 and Hsp90-a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 151, 1–44. doi:10.1007/s10254-003-0021-1
Wu, J., Liu, T., Rios, Z., Mei, Q., Lin, X., and Cao, S. (2017). Heat shock proteins and cancer. Trends Pharmacol. Sci. 38 (3), 226–256. doi:10.1016/j.tips.2016.11.009
Yan, W., Qiao, Y., Qu, J., Liu, X., Zhang, Q., and Wang, X. (2021). The hsp40 gene family in Japanese flounder: Identification, phylogenetic relationships, molecular evolution analysis, and expression patterns. Front. Mar. Sci. 7. doi:10.3389/fmars.2020.596534
Yin, K., Cui, Y., Sun, T., Qi, X., Zhang, Y., and Lin, H. (2020). Antagonistic effect of selenium on lead-induced neutrophil apoptosis in chickens via miR-16-5p targeting of PiK3R1 and IGF1R. Chemosphere 246, 125794. doi:10.1016/j.chemosphere.2019.125794
Yu, J., Wang, H., Yue, X., and Liu, B. (2019). Dynamic immune and metabolism response of clam Meretrix petechialis to Vibrio challenge revealed by a time series of transcriptome analysis. Fish. Shellfish Immunol. 94, 17–26. doi:10.1016/j.fsi.2019.08.057
Zhang, Y., Nie, H., and Yan, X. (2023). Metabolomic analysis provides new insights into the heat-hardening response of Manila clam (Ruditapes philippinarum) to high temperature stress. Sci. Total Environ. 857 (2), 159430. doi:10.1016/j.scitotenv.2022.159430
Zhong, J., and Zhao, X. (2019). Transcriptomic analysis of viable but non-culturable Escherichia coli O157:H7 formation induced by low temperature. Microorganisms 7 (12), 634. doi:10.3390/microorganisms7120634
Keywords: high-temperature, stress, protein-protein interaction network, Sepia esculenta, transcriptome
Citation: Wang Y, Bao X, Wang W, Xu X, Liu X, Li Z, Yang J and Yuan T (2023) Exploration of anti-stress mechanisms in high temperature exposed juvenile golden cuttlefish (Sepia esculenta) based on transcriptome profiling. Front. Physiol. 14:1189375. doi: 10.3389/fphys.2023.1189375
Received: 19 March 2023; Accepted: 28 April 2023;
Published: 10 May 2023.
Edited by:
Yiming Li, Fishery Machinery and Instrument Research Institute, ChinaReviewed by:
Tianming Wang, Zhejiang Ocean University, ChinaCopyright © 2023 Wang, Bao, Wang, Xu, Liu, Li, Yang and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zan Li, bGl6YW5seG1AMTYzLmNvbQ==; Jianmin Yang, bGFkZGVydXBAMTI2LmNvbQ==; Tingzhu Yuan, eXVhbnRpbmd6aHVAc2luYS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.