
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 04 May 2023
Sec. Clinical and Translational Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1184025
This article is part of the Research TopicModel Organisms and Experimental Models: Opportunities and Challenges in Clinical and Translational PhysiologyView all 6 articles
Introduction: Polycystic kidney disease (PKD) is a condition where fluid filled cysts form on the kidney which leads to overall renal failure. Zebrafish has been recently adapted to study polycystic kidney disease, because of its powerful embryology and genetics. However, there are concerns on the conservation of this lower vertebrate in modeling polycystic kidney disease.
Methods: Here, we aim to assess the molecular conservation of zebrafish by searching homologues polycystic kidney disease genes and carrying transcriptome studies in this animal.
Results and Discussion: We found that out of 82 human cystic kidney disease genes, 81 have corresponding zebrafish homologs. While 75 of the genes have a single homologue, only 6 of these genes have two homologs. Comparison of the expression level of the transcripts enabled us to identify one homolog over the other homolog with >70% predominance, which would be prioritized for future experimental studies. Prompted by sexual dimorphism in human and rodent kidneys, we studied transcriptome between different sexes and noted significant differences in male vs. female zebrafish, indicating that sex dimorphism also occurs in zebrafish. Comparison between zebrafish and mouse identified 10% shared genes and 38% shared signaling pathways. String analysis revealed a cluster of genes differentially expressed in male vs. female zebrafish kidneys. In summary, this report demonstrated remarkable molecular conservation, supporting zebrafish as a useful animal model for cystic kidney disease.
Polycystic kidney disease (PKD) is a condition where fluid filled cysts form on kidneys. PKDs can be separated into two distinct groups, either Autosomal Dominant PKD (ADPKD) or Autosomal Recessive PKD (ARPKD) (Harris and Torres, 2009). ADPKD is the common form of PKDs and typically affects one out of 1,000 people across various genetic backgrounds with a reported 10 million people affected globally (Bergmann et al., 2018). ADPKD patients have microcysts since childhood and massively enlarged cysts and kidney function decline around the age of 50, many of them eventually develop renal failure and require dialysis or a kidney transplant. By contrast, ARPKD is a rarer and more severe disease than ADPKD (Torres and Harris, 2014; Bergmann et al., 2018). Because proteins encoded by PKD associated genes localize in the primary cilium, in addition to other cellular places, or affect cilia-mediated signaling, PKD is largely considered as a ciliopathy (Gerdes et al., 2009; Hildebrandt et al., 2011; Hu and Harris, 2020). Renal cysts are also commonly presented in other types of ciliopathies, such as Bardet-Biedl Syndrome, Senior Loken Syndrome, Joubert Syndrome, Meckel-Gruber Syndrome, and Nephronophthisis, which are collectively referred as syndromic forms of PKD (Fliegauf et al., 2007; McConnachie et al., 2021). At this time, tolvaptan, a vasopressin V2 receptor antagonist that lowers the intracellular cAMP level, is the only FDA-approved drug to slow ADPKD progression (Chebib et al., 2018; Zhou and Torres, 2023). Thus, it is imperative to investigate potential therapies going forward.
While rodent models have been predominantly used to study PKD, there have been recent advances in utilizing zebrafish (Danio rerio) to study PKD as well. Zebrafish belongs to the teleosts that account for nearly half of the extant vertebrate species (Volff, 2005). The zebrafish pronephric kidney is comprised of a pair of tubules that are fused at a glomerulus. It has conserved nephrotic structure and morphogenesis processes comparable to the mammalian metanephric kidney (Drummond et al., 1998; Wingert et al., 2007). Zebrafish has become an attractive model for studying various ciliopathies because it provides a convenient in vivo platform for analyzing the structure, number, and length of cilia (Song et al., 2016). Additionally, zebrafish develop cysts when genes associated with human PKDs are mutated such as pkd1, pkd2 and hnf1b (Sun and Hopkins, 2001; Sun et al., 2004; Sullivan-Brown et al., 2008; Mangos et al., 2010; Zhu et al., 2017). The adult zebrafish mesonephric kidney has a largely conserved branching morphogenesis and segment composition as mammals (McCampbell et al., 2014; Diep et al., 2015). Adult zebrafish mutants of tmem67, whose human homolog TMEM67 is a major causative gene for Meckel-Gruber Syndrome (Smith et al., 2006; Leightner et al., 2013), have been shown to develop progressive renal cysts (Zhu et al., 2021).
Despite these attributes of the zebrafish model to cystic kidney diseases, there are several concerns on the conservation of the fish model. First, while mammals have primary cilia in the kidney, cilia in the zebrafish kidney are motile and cilia motility is closely associated with pronephric cyst formation (Kramer-Zucker et al., 2005; Becker-Heck et al., 2011). Second, unlike mammalian kidneys that are metanephros, adult zebrafish kidneys are mesonephros (Davidson, 2011). Third, it has been estimated that approximately 70% genes overlap between humans and zebrafish, however, the percentage of human PKD associated genes with zebrafish homologs is unknown (Howe et al., 2013). Fourth, given that approximately 30% zebrafish genome is duplicated early in the evolution of teleost fish, >100 million years ago, genetic analysis of genes with multiple homologs could be complicated (Woods et al., 2000; Howe et al., 2013). While zebrafish has proven to be an efficient model for genetic studies of disease causative genes, particularly useful for the identification of genetic modifiers for diseases such as cardiomyopathy (Ding et al., 2011; Ding et al., 2016; Ma et al., 2020), these concerns have hindered using zebrafish for genetic studies of PKD. Thus, there is an urgent need of assessing whether zebrafish is a conserved animal model for PKD research.
In the kidney, sexual dimorphism in gene expression patterns, transporters, responses to injury, as well as PKD progression has been reported (Stewart, 1994; Menezes et al., 2016; Veiras et al., 2017; Laouari et al., 2022; Neugarten and Golestaneh, 2022). In humans, male kidneys are usually larger than female kidneys, suggesting differing developmental dynamics (Coggins et al., 1998; Cheong et al., 2007; Kalucki et al., 2020). Renal function decreases with age, however, older male patients with chronic kidney disease usually are more prone to kidney failure, particularly as there are disruptions in the nitric oxide system (Eriksen and Ingebretsen, 2006; Baylis, 2009). Comparing with age-matched female patients, male patients with ADPKD consistently manifest more severe phenotypes, including lower estimated glomerular filtration rate and larger height-adjusted total kidney volume, and earlier onset of End-Stage Renal Disease (Neugarten et al., 2000; Reed et al., 2008). Similar to humans, sexual dimorphism has been also reported in animal models, including Han:SPRD rats and Pkd1 mutant mice (Nagao et al., 2005; Stringer et al., 2005; Menezes et al., 2016). Consistent with the phenotypic changes, differences in total RNA transcripts (transcriptome) have been reported in genes and pathways between male and female mouse kidneys (Menezes et al., 2016; Terabayashi et al., 2020). In zebrafish, the observation of renal cysts mainly in male tmem67 mutants suggests sexual dimorphism in the kidney, however, whether male and female fish have distinct transcriptome remains unknown (Zhu et al., 2021).
In this manuscript, we decided to take two approaches to assess conservation of zebrafish for PKD study. First, we assessed molecular conservation by querying how many cystic kidney disease genes have their corresponding zebrafish homologs. We found that out of 82 genes that cause renal cysts, zebrafish have homologs for 81 of these genes. Second, we compared gene expression between male and female fish kidneys and noted obvious differences. Among the differentially expressed signaling pathways, we noted a remarkable 37.9% similarities to a previously established rodent PKD model (Menezes et al., 2016). Together, our studies indicate zebrafish a valuable model for studying PKD.
Experimental design was summarized in a flowchart (Supplementary Figure S1).
Zebrafish were maintained under a 14-h light and 10-h dark cycle at 28.5°C as previously described (Shih et al., 2015). Adult WIK fish were obtained by allowing the embryos that were derived from in-house breeding to grow to 12 months of age and used in accordance with the policies of the Mayo Clinic Institutional Animal Care and Use Committee.
PKD associated genes were selected from the review by McConnachie et al. (2021). Protein sequences were found on ZFIN (The Zebrafish Information Network) (https://www.zfin.org). In most cases, the ENSEMBL (European Bioinformatics Institute and the Wellcome Trust Sanger Institute) gene ID was provided. When ENSEMBL did not have records of this gene, we gave the ZFIN gene ID. To calculate the percent identity of a gene, we searched https://www.uniprot.org/, aligned the Uniprot IDs of the zebrafish and human genes, and calculated the percent identity. To compare relative abundance of genes with multiple homologs, we added the Reads Per Kilobase of transcript per Million reads mapped (RPKM, a measure of relative RNA molar concentration of a transcript in a sample) values of each homolog together for the total abundance as previously described (Shih et al., 2015). We then divided the single value RPKM of a specific homolog to the total RPKM of the multiple homologs in order to get a percentage.
Kidneys of 12-month-old male and female fish were isolated as previously described (Ding et al., 2022). Briefly, zebrafish were anesthetized via tricaine, and kidneys were harvested as needed. A total of 10 sets of kidneys were harvested, including 5 sets of male and female kidneys, respectively. The kidneys were submerged in TRIZOL (Sigma) and metal beads were added to help dissociate the tissue. After dissociation in TRIZOL, chloroform was added to separate out the aqueous layer. This aqueous layer was then precipitated in isopropanol to isolate the RNA, before reconstituted in RNAase free water. RNA samples were considered good quality with an RNA Integrity Number (RIN) greater than seven.
RNA libraries were generated, and RNA sequencing was conducted at the Mayo Clinic Medical Genome Facility, a core laboratory offering research sequencing. The paired-end RNA reads were processed through RapMap, a custom pipeline developed by the Mayo Clinic Bioinformatics core sequencing analyses of species with small genomes. RapMap utilizes Kallisto (Bray et al., 2016) for pseudoalignment and transcript quantifications, with bootstrapping enabled to n = 100 (Bray et al., 2016). Bootstrapping is enabled while running Kallisto quantification to allow for quantifying transcript abundance. The most recently available zebrafish genome, version GRCz11, Ensembl gtf version GRCz11.101 was used for alignment. In total, 222,675 genes were identified mapping to this reference genome. RapMap also utilizes Tximport (Srivastava et al., 2016) for summarizing transcript abundances for downstream analysis and MultiQC (Ewels et al., 2016) for overall quality control. Samples with over 75% of reads mapping to the genome were considered high quality. Raw and processed files were submitted to the NCBI GEO under accession number GSE2248592. Additionally, mouse RNA-seq data was downloaded from a previous study (Geo Omnibus number GSE72554) (Menezes et al., 2016).
Using the resulting length ScaledTPM (TPM scaled up to library size) values, differentially expressed (DE) genes (FDR <0.05, log2fold change > |1.5|) were assessed using the bioinformatic package edgeR (Robinson et al., 2010). Canonical pathway analysis was performed with the DE genes using Ingenuity pathway analysis (IPA) software (QIAGEN) (Kramer et al., 2014). The top 10 canonical pathways in each species were identified and compared. Individual genes within each pathway were examined further. Additionally, we uploaded our DE genes into the STRING database, a database of known and predicted protein-protein interactions (string-db.org) (Szklarczyk et al., 2021), to identify relationships among our DE genes. The STRING network was generated and visualized at the highest confidence (0.9) level and disconnected nodes or clusters with fewer than 3 genes were removed. IPA analysis was performed on genes in the resulting network.
We picked the top 10 most differentially expressed genes that were found via STRING network analysis, which included the top 5 upregulated and top 5 downregulated genes as indicated in section 2.5. The Superscript III First-Strand Synthesis System (Invitrogen) was used to generate cDNA from 500 ng of RNA that was isolated in section 2.4. Once cDNA had been successfully generated, quantitative PCR (qPCR) was carried out using QuantStudio 7 Pro (Thermo Fischer) QPCR apparatus in 96-well qPCR plates (Roche Diagnostics Corp). The expression of the genes was normalized using the expression level of the housekeeping gene gapdh and its corresponding –∆∆Ct (cycle threshold) values. Nine RNA samples were analyzed in triplicate. Primer sequences for each gene can be found in Supplementary Table S3. Statistical analysis was conducted via vassarstats (http://vassarstats.net/). Statistically significant DEGs between male and female zebrafish were indicated by two tailed t-test with a minimum significance of p < 0.05.
To test molecular conservation of the zebrafish, we queried the zebrafish genome to identify homologs of known cystic kidney disease related genes. Based on inherited pattern and phenotypes in patients, we decided on 82 genes that were separated into multiple categories including ADPKD, ARPKD, and disorders with significant presence of kidney cysts (Reiter and Leroux, 2017; McConnachie et al., 2021). Many of these gene encode proteins in renal cilia, underscoring the importance of cilia in PKD pathogenesis. We compared gene orthologs of these human genes to the ZFIN database (http://zfin.org) and Ensembl (http://www.ensembl.org/index.html) to determine if zebrafish homologs exist for these genes (Supplementary Table S1). We were able to identify zebrafish homologues for 81 of the 82 genes, indicating a 99% overlap of expression across species (Figure 1). At the protein level, the identities of each of these genes relative to their human counterparts ranged from 30% to 95% with an average of around 59% (Supplementary Table S1). All ADPKD-associated genes, including major causative gene PKD1 and PKD2 as well as genes recently linked to ADPKD such as IFT140, DNAJB11, GANAB, and ALG9, have zebrafish homologs (Supplementary Table S1) (Porath et al., 2016; Cornec-Le Gall et al., 2018; Besse et al., 2019; Senum et al., 2022). However, we were unable to find a zebrafish homolog of ARPKD causative gene PKHD1, even after searching the National Center for Biotechnology Information database using BLASTX (Figure 1 and Supplementary Table S1) (Ward et al., 2002). The closest existing homolog in zebrafish is pkhd1l (PKHD1 like), with an identity of 52% relative to its direct human homolog. Moreover, 75 of these 81 genes have a single homolog, while only 6 of these 81 genes have two separate homologs. These genes were pkd1, ganab, arl3, tmem237, glis2, and cpps1 (Figure 1 and Supplementary Table S1).
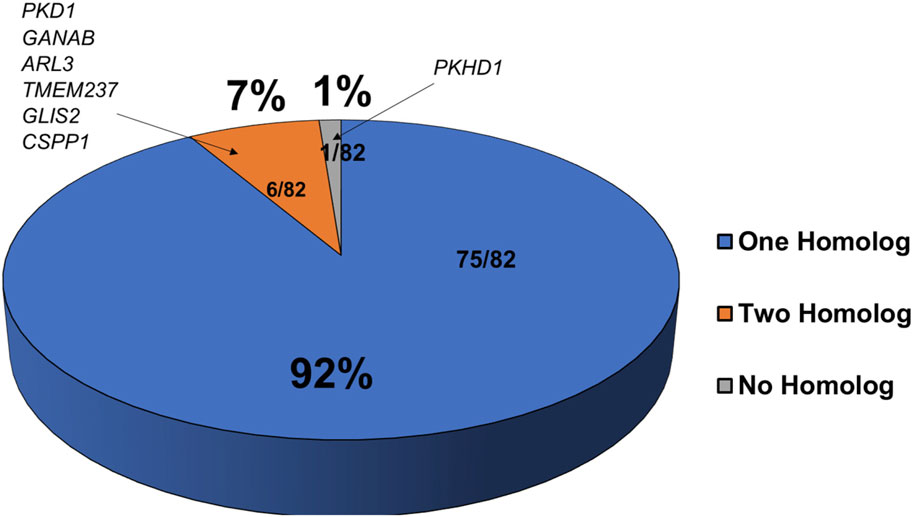
FIGURE 1. Majority of cystic kidney disease associated genes have corresponding homologs in zebrafish. Shown is a pie chart representing the number of zebrafish homologs exist for known human PKD associated genes. Genes with one zebrafish homolog are in blue, those with two homologs are in orange, and those with no known homologs are in grey.
We then defined gene expression of 12-month-old zebrafish kidney via carrying RNA sequencing (RNAseq) analysis. Both male and female fish samples were extracted with 5 sets of kidneys of each sex. We used an RPKM value of 0.3 as our cutoff for expression abundance. The average number of total reads per sample was 45,149,820 with an average of 35,128,575 reads mapped to a gene. We were able to map an average of 77% of these reads to the reference genome, enabling us to analyze expressional changes of 22,676 genes. Based on their expression abundance in zebrafish kidney transcriptomes, 81 cystic kidney disease-associated genes can be broadly divided into four categories, high, medium, low, and undetectable (Supplementary Table S2), referred as Group 1, Group II, Group III, and Group IV, respectively. Genes that comprised of Group I (with RPKM values greater than 1,000) were ganaba, ganabb, dnajb11, armc9, and ift74 in both male and female kidneys, poc1b and xpnpep3 in male kidneys, and cep290 in female kidneys (Supplementary Table S2). Genes that comprised of group II was the largest category of genes (with RPKM values between 100 and 999). Genes in group III (with RPKM values between 1 and 99) included many BBS adjacent genes (bbs1, bbs4, bbs5, bbs9, bbs10, and ttc8). The expression of the following genes in group IV had their RPKM values below threshold: c8orf37, ift74, cep164, cplane1, cep120, katnip, anks6, and cep83 (Supplementary Table S2). Although these genes had extremely low to zero expression levels in 12 months zebrafish kidney, this data cannot be interpreted as excluding these genes as kidney-expression genes. For example, it is possible that these genes are activated in the kidney during development. Indeed, cep83 is expressed in the pronephric kidney (Joo et al., 2013); ift74, cep164, cep120, and anks6 might also be expressed in the embryonic kidney since knockdown of these genes results in pronephric cysts (Chaki et al., 2012; Hoff et al., 2013; Shaheen et al., 2015; Lindstrand et al., 2016).
In our previous bioinformatic analysis of dilated cardiomyopathy genes in zebrafish, we demonstrated that comparison of expression levels of multiple zebrafish homologues can help to prioritize one homologue over the other (Shih et al., 2015). It is hypothesized that the greater the abundance, the more important this homolog is for function. To quantify the relative abundance, we took the raw RPKM value for each individual homologue and divided that by the total sum of both RPKM values to get a percentage. For the 6 genes with 2 homologs, we recommended that one homolog, based on the percentage, would be prioritized for future studies (Table 1). For example, pkd1a is more prevalent than pkd1b with pkd1a having over 90% abundance. This is consistent with our recent study showing that pkd1a is the major homolog of PKD1 in zebrafish while pkd1b plays a redundant role (Zhu et al., 2017). We also identified the more predominant homolog for the other remaining genes: ganabb, arl3b, tmem237b, glis2a, and cpps1b as compared to their other homolog (Table 1).
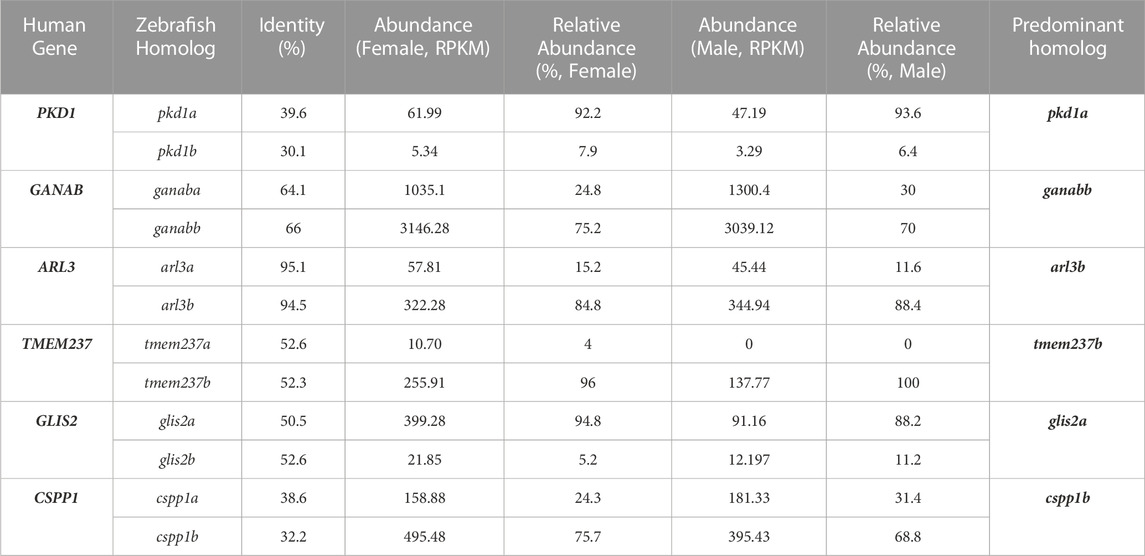
TABLE 1. Predominant homolog of zebrafish genes with multiple homologs can be predicted based on their relative abundance in kidney.
We next wanted to determine if there was sexual dimorphism at the transcriptomic level in zebrafish kidneys. Indeed, when comparing transcriptome of zebrafish kidneys of different sex, we found that transcripts in male and female kidneys form distinct clusters, with those in male fish circled in blue and those in female fish circled in red, as indicated by PCA analysis (Figure 2A). Using EdgeR, we identified a total of 2,422 DEGs between sexes (FDR <0.05, log2fold change of > |1.5|). Genes marked as green are downregulated, genes marked as red are upregulated, and genes marked as black are not differentially regulated (Figure 2B). Unbiased clustering heatmaps show distinct expression differences in genes between male and female kidneys, i.e., many genes with lower expression levels in males (shown in blue) have higher expression levels in females (shown in red), and vice versa (Figure 2C). To query what pathways these DEGs are involved, we used Metascape and found that the biggest differences were in the nuclear receptors pathway and pathways of transporting small molecules (Figure 2D). Other pathways of note include cell response to cytokines, kinases, xenobiotics, and extracellular components. These results indicate that genetic differences do exist between male and female zebrafish kidneys.
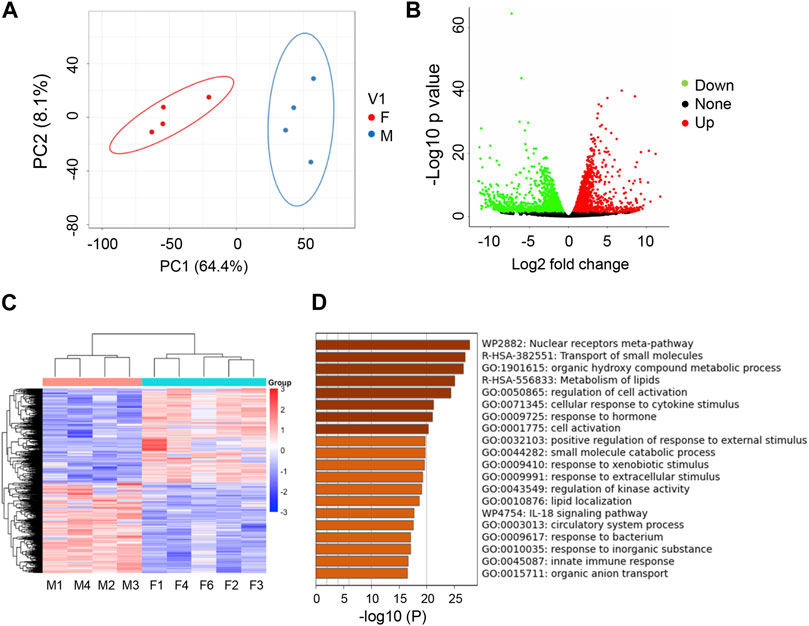
FIGURE 2. Transcriptome studies uncover sexual dimorphism in zebrafish kidneys. (A) PCA analysis of RNAseq data for male (M) vs. female (F) kidneys (4 replicates per group). (B) Volcano Plot of M vs. F kidneys (4 replicates per group). Significantly differentially expressed genes (−1.5 < log2FC > 1.5, FDR <5%) are highlighted green (downregulated) or red (upregulated). (C) Heatmap of total genes in M v F kidneys. (D) Metascape analysis of top differentiated pathways.
To further test the conservation of the zebrafish model, we sought to compare transcriptomes that define sex dimorphism in zebrafish and in mouse. In the original report, the cutoff for mouse genes was a significance of <0.05, with three to four replicates (Menezes et al., 2016). Using an FDR<5%, we identified 2,422 and 2,351 genes in zebrafish and mouse, respectively. Comparing these zebrafish and mice genes, we observed an overlap of 493 common DEGs, which is 10.2% of the total DEGs (Figure 3A). To determine if there are shared signaling pathways between zebrafish and the mouse studies, we performed ingenuity pathway analysis (IPA) on both the zebrafish and mouse genes independently. Focusing on the significant canonical pathways in each species, there were 173 shared pathways, which represent 37.9% of total significant pathways (Figure 3B). We graphed the gene expression within the top pathways to understand the expression differences of these shared pathways between species (Figure 3C; Table 2). While the direction of gene expression is similar between species, the gene expression difference between male and female was greater in zebrafish than mouse, suggesting that sexual dimorphism-associated kidney phenotypes might be more easily studied in fish than mouse. To obtain a balanced view and to determine the nature of this phenomenon, we also evaluated the different signaling pathways between zebrafish and mouse. We plotted the change in activation profile by examining the IPA canonical pathway Z-scores and comparisons were made for pathways that had an absolute Z score difference greater than 2.5 between the two species (Supplementary Figures S2A, 2B). We observed 26 pathways that fit this criterion, which include but are not limited to autophagy, PI3K-AKT signaling, and endothelin signaling amongst others. Together, these results indicate that despite some differences, molecular pathways underlying sex dimorphism are shared between zebrafish and mouse, lending credence to the zebrafish as a viable animal model to study PKD.
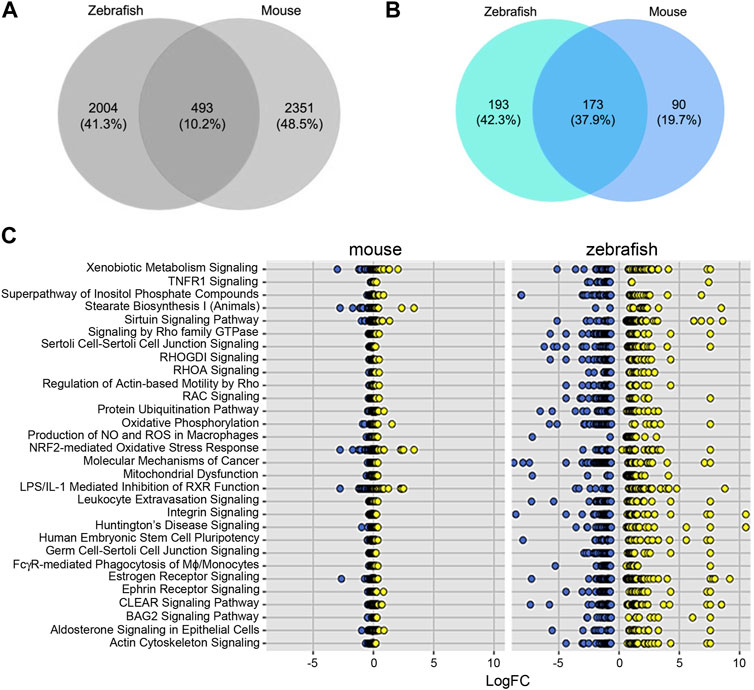
FIGURE 3. Differentially expressed genes and pathways underlying sex dimorphism are shared between zebrafish and mouse kidneys. (A) Venn diagram of shared genes between zebrafish and mouse. (B) Venn diagram of shared pathways between zebrafish and mouse. (C) Plot of log2fold change of shared IPA.
To delve deeper on genes that is important for sex dimorphism in zebrafish, we analyzed DEGs in the following shared signaling pathway between zebrafish and mouse: Coordinated Lysosomal Expression and Regulation (CLEAR) signaling, Sirtuin and Xenobiotic Metabolism. Highlighting the CLEAR pathway, we noted 59 key genes with distinct expression patterns. Heatmaps of these 59 CLEAR genes noted clear distinctions of unique genes up or downregulated in male vs. female fish (Supplementary Figure S3A). The same could be said for the genes in the Sirtuin heatmap and the xenobiotic pathway heatmap (Supplementary Figures S3B, 3C). Genes that had the greatest fold expression for the CLEAR pathway include rras2, tgfa, ntrk3a, tcirg1b, and vegefab. Sirtuin genes with the greatest fold change included nqo1, pck1, pfkfb3, arg2, and g6pd. Xenobiotic genes that were found to have the greatest fold change include nqo1, rras2, abcc2, il1b, and ugt8 (Supplementary Figure S3D). These genes in the specific pathways act as potential targets for future experimental studies to decipher underlying mechanism of sex dimorphism.
As an alternative way to discover candidate genes for sex dimorphism in zebrafish kidney, we used STRING analysis to identify gene clusters (Figure 4A). We were able to identify a cluster of around 150 genes closely associated to one another, many of which had large associations to one another based on the number of nodes (Figures 4B, C). The heatmap of the 150 network genes shows a clear distinction between male and female fish kidneys (Figure 4D). IPA analysis of this cluster of genes revealed enrichment of pathways involved in TNF-related weak inducer of apoptosis (TWEAK) signaling, NOD1/2 signaling, and cell cycle regulation (Figure 4E).
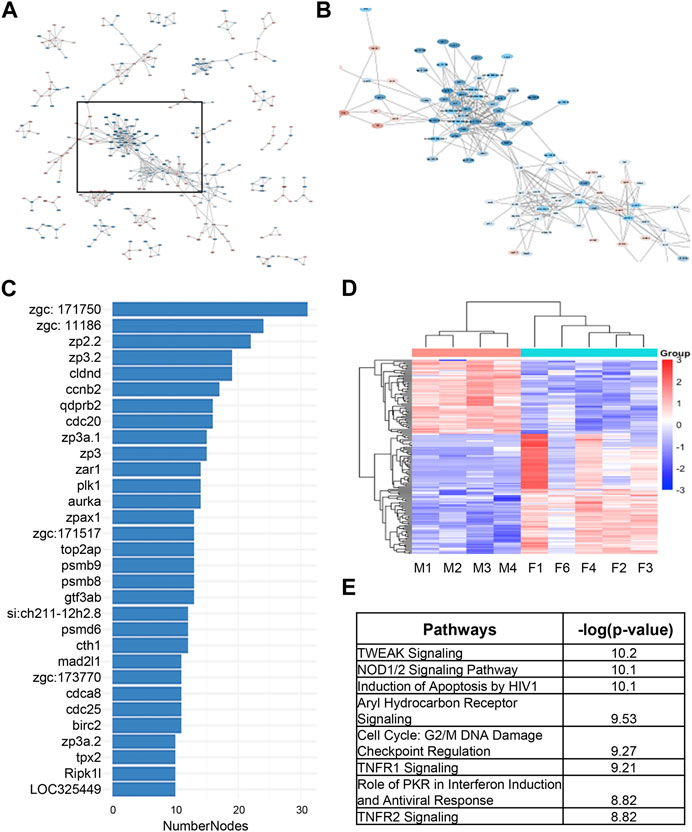
FIGURE 4. STRING analysis of differentially expressed genes underlying sex dimorphism in zebrafish kidneys. (A) String analysis of all DEGs in male vs. female zebrafish kidneys. Confidence score was set to 0.9. (B) Zoomed in area of clustered genes from (A). (C) Number of nodes in string cluster. (D) Heatmap of STRING Cluster genes in male and female zebrafish kidneys in (B). (E) IPA analysis of STRING Cluster genes in (B).
Finally, to confirm the results from RNAseq analysis, we performed qPCR on the genes identified in this cluster. Consistent with the RNAseq data (Figure 5A), ahr1a, agxtb, hsp90aa1.2, cbsb, and ahcyl2a were found to be significantly upregulated in male kidney relative to female kidney (Figure 5B); tnfrsf9a, nfkbiaa, casp3b, ccnb3, and ilb1 were significantly upregulated in female fish relative to male fish (Figure 5C). These results confirmed the fidelity of our RNAseq analysis for genome-wide gene expression studies.

FIGURE 5. Sexually dimorphic genes from RNAseq analysis are validated by qPCR. (A) Table of top 10 genes and fold changes from RNAseq analysis. (B) qPCR validation of genes that were downregulated in male kidneys. (C) qPCR validation of genes that were upregulated in male kidneys. Four kidneys from male and female zebrafish were scored, respectively. *: p < 0.05.
The validity of zebrafish as a model for studying PKD in the past has come under scrutiny due to differences to mammalian models (Kramer-Zucker et al., 2005; Davidson, 2011; Howe et al., 2013). In the present manuscript, we provided the following two strong evidence to support the conservation of zebrafish for PKD studies. First, we demonstrated 99% conservation of 82 cystogenic genes, including 6 ADPKD-associated genes (PKD1, PKD2, IFT140, DNAJB11, ALG9, and GANAB), 1 ARPKD causative gene (DZIP1L), and 75 renal ciliopathy genes (Porath et al., 2016; Lu et al., 2017; Cornec-Le Gall et al., 2018; Besse et al., 2019; McConnachie et al., 2021; Senum et al., 2022). The only gene that did not have a zebrafish homolog was PKHD1, the major ARPKD gene that encodes for the Fibrocystin (Ward et al., 2002). To date there is not a known direct homolog of this gene found in zebrafish. Second, out of 81 genes, 75 have a single fish homolog, while only 6 of these genes (7.4%) have two homologs. Through abundance analysis we were able to identify which isoform is the more predominant form and should be focused upon, and thus addressed a common criticism that multiple homologs for a mammalian gene can potentially confound genetic studies in zebrafish. Our analysis indicated that renal cystic disease genes are much conserved in zebrafish than approximately 70% conservation of all genes and less expanded than the average 30% duplication (Howe et al., 2013). Importantly, our work would facilitate future experimental studies to further validate the conservation of zebrafish, via systematically generating genetic models for PKD of different etiology. When there are two or more zebrafish homologs for a certain human gene, we recommend prioritizing the predominant isoform. This notion is supported by our previous studies of pkd1a and pkd1b mutants. We showed that pkd1a fish develop severe phenotypes and pkd1b fish are generally normal (Zhu et al., 2017). For GANAB, Tmem237, Cspp1, Arl3, and Gis2 that all have 2 zebrafish homologs, the predominant homolog was also predicted. It remains to be experimentally tested whether the predominant isoform would play more important roles in cyst development and progression.
Sexual dimorphism of kidney genes has previously been reported in mice, particularly in genes that regulate fatty acid oxidation (Menezes et al., 2016). IPA analysis on this previously published data set also revealed other key differences in signaling pathways outside of fatty acid oxidation. Here, we identified 173 total pathways that are shared between zebrafish and mice, resulting in 37% shared pathways. These pathways include xenobiotic metabolism, CLEAR signaling, and sirtuin biosynthesis, amongst others. Through comparison we also showed that there is more dynamic gene change expression in zebrafish relative to mouse. These data indicated that conservation between fish and mouse is high, justifying the zebrafish PKD model as a viable model for future studies.
One of the pathways that is affected across different model systems is the CLEAR signaling pathway. Of note, some of the genes with the greatest fold change in expression were rras2, tgfα, ntrk3a, tcirg1b, and vegefα. Of these genes, serum VEGFα serves as a biomarker for ADPKD progression (Reed et al., 2011; Leierer et al., 2021); Tgfα is upregulated in the cystic epithelia of PKD patients and transgenic overexpression of Tgfα promotes renal cyst formation (Gattone et al., 1996; Lee et al., 1998). TFEB is the key transcriptional regulator of genes that harbors the CLEAR motif, and TFEB has been implicated in cystic kidney disease (Sardiello et al., 2009; Calcagni et al., 2016; Shillingford and Shayman, 2023). The xenobiotic pathway is involved in the breakdown of drugs/foreign compounds. While there have not been reports establishing the link of Xenobiotic metabolism in PKD specifically, sexually dimorphic expression of xenobiotic genes has been observed in rats (Kwekel et al., 2013). Sirt1, a member of the Sirtuin pathway, has been implicated in ADPKD, as double knockouts of Sirt1 and Pkd1 resulted in prevention of cyst formation (Zhou et al., 2013). Future studies will be needed to demonstrate whether male vs. female zebrafish manifest different severity in modeling PKD of different etiology, as we have already noted in the tmem67 model, in which renal cysts were mainly developed in males (Zhu et al., 2021). Several top pathways and top genes that have been uncovered here would be a great starting point to pinpoint underlying mechanism that is accountable for such sex dimorphism.
STRING/Hub analysis provided an alternative strategy to uncover a specific cluster of genes that could be important for the sex dimorphism. Some of the genes have been implicated in autophagy, ciliopathy, and kidney injury, which all play roles in PKD. For instance, Hsp90aa1 is involved in autophagy (Macri et al., 2015). AGXT has been linked to ciliopathy (Gee et al., 2014). NFKBIA is implicated in kidney injury through DNA damage/senescence pathways (Kolesnichenko et al., 2021). Further investigation of these genes could be conducted in the future to reveal their functions in cystogenesis.
Through validation of molecular conservation of PKD associated genes, the present study justified future experimental efforts to establish zebrafish as a faithful animal model for this disease. The identification of all zebrafish homologs of cystic kidney disease genes laid the foundation for systematic generation of the corresponding genetic models. Phenotyping of these future models in adult zebrafish is feasible, as exemplified by our recent study of tmem67 mutants (Zhu et al., 2021). It is anticipated that comparison among these models could uncover both common and unique pathological events among PKD models with different etiology, which would justify the use of this lower vertebrate model for developing genotype-guided therapies. Another stringent criterion to test the fidelity of this vertebrate model would be to assess whether replacing a wild type gene with a disease-causing variant that was found in human patients (knock in) can recapitulate PKD-like phenotypes in zebrafish. If established, zebrafish could offer an efficient in vivo platform for evaluating large number of PKD1 or PKD2 variants that are discovered clinically with unknown significance.
The major driving force for establishing cystic kidney disease in a simpler but more efficient zebrafish model is the potential to conduct genetic screens. As exemplified by our recent studies in cardiomyopathy, insertional mutagenesis-based forward genetic screens can be carried out in zebrafish for systematic identification of genetic modifiers, which could lead to the identification of novel therapeutic targets such as dnajb6 and rxra (Ding et al., 2011; Ding et al., 2016; Ma et al., 2020). F0-based genetic assays, which do not involve the necessity of generating stable mutants, have also been recently implemented in zebrafish for rapid assessment of modifying effects of candidate genes, such as those suggested from transcriptome studies (Ata et al., 2018; Ding et al., 2022). Both the forward genetic screen and F0-based screen could be used to search genetic modifiers of PKD. Therefore, zebrafish models have the potential to accelerate the discovery of underlying mechanisms of PKD, as well as the development of mechanism-based therapies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by Laboratory Animal Ethics Committee of Mayo Clinic.
The authors confirm their contribution to the paper as follows: study conception and design: MK, PZ, XX, and XL; data collection: MK and PZ; analysis and interpretation of results: MK and CM; draft manuscript preparation: MK, CM, XX, and XL. CM, XX, and XL approved the final version of the manuscript.
This work was supported by NIH T32HL07111 (MK), Mayo Foundation (XX), and Mayo PKD Center Zell PKD Research Innovation Fund (XL).
We appreciate Beninio Gore and Kashia Strategy for taking care of the zebrafish facility.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1184025/full#supplementary-material
Ata, H., Ekstrom, T. L., Martinez-Galvez, G., Mann, C. M., Dvornikov, A. V., Schaefbauer, K. J., et al. (2018). Robust activation of microhomology-mediated end joining for precision gene editing applications. PLoS Genet. 14, e1007652. doi:10.1371/journal.pgen.1007652
Baylis, C. (2009). Sexual dimorphism in the aging kidney: Differences in the nitric oxide system. Nat. Rev. Nephrol. 5, 384–396. doi:10.1038/nrneph.2009.90
Becker-Heck, A., Zohn, I. E., Okabe, N., Pollock, A., Lenhart, K. B., Sullivan-Brown, J., et al. (2011). The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nat. Genet. 43, 79–84. doi:10.1038/ng.727
Bergmann, C., Guay-Woodford, L. M., Harris, P. C., Horie, S., Peters, D. J. M., and Torres, V. E. (2018). Polycystic kidney disease. Nat. Rev. Dis. Prim. 4, 50. doi:10.1038/s41572-018-0047-y
Besse, W., Chang, A. R., Luo, J. Z., Triffo, W. J., Moore, B. S., Gulati, A., et al. (2019). ALG9 mutation carriers develop kidney and liver cysts. J. Am. Soc. Nephrol. 30, 2091–2102. doi:10.1681/ASN.2019030298
Bray, N. L., Pimentel, H., Melsted, P., and Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. doi:10.1038/nbt.3519
Calcagni, A., Kors, L., Verschuren, E., de Cegli, R., Zampelli, N., Nusco, E., et al. (2016). Modelling TFE renal cell carcinoma in mice reveals a critical role of WNT signaling. Elife 5, e17047. doi:10.7554/eLife.17047
Chaki, M., Airik, R., Ghosh, A. K., Giles, R. H., Chen, R., Slaats, G. G., et al. (2012). Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell 150, 533–548. doi:10.1016/j.cell.2012.06.028
Chebib, F. T., Perrone, R. D., Chapman, A. B., Dahl, N. K., Harris, P. C., Mrug, M., et al. (2018). A practical guide for treatment of rapidly progressive ADPKD with tolvaptan. J. Am. Soc. Nephrol. 29, 2458–2470. doi:10.1681/ASN.2018060590
Cheong, B., Muthupillai, R., Rubin, M. F., and Flamm, S. D. (2007). Normal values for renal length and volume as measured by magnetic resonance imaging. Clin. J. Am. Soc. Nephrol. 2, 38–45. doi:10.2215/CJN.00930306
Coggins, C. H., Breyer Lewis, J., Caggiula, A. W., Castaldo, L. S., Klahr, S., and Wang, S. R. (1998). Differences between women and men with chronic renal disease. Nephrol. Dial. Transpl. 13, 1430–1437. doi:10.1093/ndt/13.6.1430
Cornec-Le Gall, E., Olson, R. J., Besse, W., Heyer, C. M., Gainullin, V. G., Smith, J. M., et al. (2018). Monoallelic mutations to DNAJB11 cause atypical autosomal-dominant polycystic kidney disease. Am. J. Hum. Genet. 102, 832–844. doi:10.1016/j.ajhg.2018.03.013
Davidson, A. J. (2011). Uncharted waters: Nephrogenesis and renal regeneration in fish and mammals. Pediatr. Nephrol. 26, 1435–1443. doi:10.1007/s00467-011-1795-z
Diep, C. Q., Peng, Z., Ukah, T. K., Kelly, P. M., Daigle, R. V., and Davidson, A. J. (2015). Development of the zebrafish mesonephros. Genesis 53, 257–269. doi:10.1002/dvg.22846
Ding, Y., Long, P. A., Bos, J. M., Shih, Y. H., Ma, X., Sundsbak, R. S., et al. (2016). A modifier screen identifies DNAJB6 as a cardiomyopathy susceptibility gene. JCI Insight 1, e88797. doi:10.1172/jci.insight.88797
Ding, Y., Sun, X., Huang, W., Hoage, T., Redfield, M., Kushwaha, S., et al. (2011). Haploinsufficiency of target of rapamycin attenuates cardiomyopathies in adult zebrafish. Circ. Res. 109, 658–669. doi:10.1161/CIRCRESAHA.111.248260
Ding, Y., Wang, M., Bu, H., Li, J., Lin, X., and Xu, X. (2022). Application of an F0-based genetic assay in adult zebrafish to identify modifier genes of an inherited cardiomyopathy. Dis. Model Mech. 16, dmm049427. doi:10.1242/dmm.049427
Drummond, I. A., Majumdar, A., Hentschel, H., Elger, M., Solnica-Krezel, L., Schier, A. F., et al. (1998). Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development 125, 4655–4667. doi:10.1242/dev.125.23.4655
Eriksen, B. O., and Ingebretsen, O. C. (2006). The progression of chronic kidney disease: A 10-year population-based study of the effects of gender and age. Kidney Int. 69, 375–382. doi:10.1038/sj.ki.5000058
Ewels, P., Magnusson, M., Lundin, S., and Kaller, M. (2016). MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi:10.1093/bioinformatics/btw354
Fliegauf, M., Benzing, T., and Omran, H. (2007). When cilia go bad: Cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8, 880–893. doi:10.1038/nrm2278
Gattone, V. H., Kuenstler, K. A., Lindemann, G. W., Lu, X., Cowley, B. D., Rankin, C. A., et al. (1996). Renal expression of a transforming growth factor-alpha transgene accelerates the progression of inherited, slowly progressive polycystic kidney disease in the mouse. J. Lab. Clin. Med. 127, 214–222. doi:10.1016/s0022-2143(96)90081-5
Gee, H. Y., Otto, E. A., Hurd, T. W., Ashraf, S., Chaki, M., Cluckey, A., et al. (2014). Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal ciliopathies. Kidney Int. 85, 880–887. doi:10.1038/ki.2013.450
Gerdes, J. M., Davis, E. E., and Katsanis, N. (2009). The vertebrate primary cilium in development, homeostasis, and disease. Cell 137, 32–45. doi:10.1016/j.cell.2009.03.023
Harris, P. C., and Torres, V. E. (2009). Polycystic kidney disease. Annu. Rev. Med. 60, 321–337. doi:10.1146/annurev.med.60.101707.125712
Hildebrandt, F., Benzing, T., and Katsanis, N. (2011). Ciliopathies. N. Engl. J. Med. 364, 1533–4310. doi:10.1056/NEJMra1010172
Hoff, S., Halbritter, J., Epting, D., Frank, V., Nguyen, T. M., van Reeuwijk, J., et al. (2013). ANKS6 is a central component of a nephronophthisis module linking NEK8 to INVS and NPHP3. Nat. Genet. 45, 951–956. doi:10.1038/ng.2681
Howe, K., Clark, M. D., Torroja, C. F., Torrance, J., Berthelot, C., Muffato, M., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503. doi:10.1038/nature12111
Hu, J., and Harris, P. C. (2020). Regulation of polycystin expression, maturation and trafficking. Cell Signal 72, 109630. doi:10.1016/j.cellsig.2020.109630
Joo, K., Kim, C. G., Lee, M. S., Moon, H. Y., Lee, S. H., Kim, M. J., et al. (2013). CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc. Natl. Acad. Sci. U. S. A. 110, 5987–5992. doi:10.1073/pnas.1220927110
Kalucki, S. A., Lardi, C., Garessus, J., Kfoury, A., Grabherr, S., Burnier, M., et al. (2020). Reference values and sex differences in absolute and relative kidney size. A Swiss autopsy study. BMC Nephrol. 21, 289–1186. doi:10.1186/s12882-020-01946-y
Kolesnichenko, M., Mikuda, N., Hopken, U. E., Kargel, E., Uyar, B., Tufan, A. B., et al. (2021). Transcriptional repression of NFKBIA triggers constitutive IKK- and proteasome-independent p65/RelA activation in senescence. EMBO J. 40, e104296. doi:10.15252/embj.2019104296
Kramer, A., Green, J., Pollard, J., and Tugendreich, S. (2014). Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530. doi:10.1093/bioinformatics/btt703
Kramer-Zucker, A. G., Olale, F., Haycraft, C. J., Yoder, B. K., Schier, A. F., and Drummond, I. A. (2005). Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development 132, 1907–1921. doi:10.1242/dev.01772
Kwekel, J. C., Desai, V. G., Moland, C. L., Vijay, V., and Fuscoe, J. C. (2013). Sex differences in kidney gene expression during the life cycle of F344 rats. Biol. Sex. Differ. 4, 14–1186. doi:10.1186/2042-6410-4-14
Laouari, D., Vergnaud, P., Hirose, T., Zaidan, M., Rabant, M., Nguyen, C., et al. (2022). The sexual dimorphism of kidney growth in mice and humans. Kidney Int. 102, 78–95. doi:10.1016/j.kint.2022.02.027
Lee, D. C., Chan, K. W., and Chan, S. Y. (1998). Expression of transforming growth factor alpha and epidermal growth factor receptor in adult polycystic kidney disease. J. Urol. 159, 291–296. doi:10.1016/s0022-5347(01)64084-9
Leierer, J., Perco, P., Hofer, B., Eder, S., Dzien, A., Kerschbaum, J., et al. (2021). Coregulation analysis of mechanistic biomarkers in autosomal dominant polycystic kidney disease. Int. J. Mol. Sci. 2210, 6885. doi:10.3390/ijms22136885
Leightner, A. C., Hommerding, C. J., Peng, Y., Salisbury, J. L., Gainullin, V. G., Czarnecki, P. G., et al. (2013). The Meckel syndrome protein meckelin (TMEM67) is a key regulator of cilia function but is not required for tissue planar polarity. Hum. Mol. Genet. 22, 2024–2040. doi:10.1093/hmg/ddt054
Lindstrand, A., Frangakis, S., Carvalho, C. M., Richardson, E. B., Mcfadden, K. A., Willer, J. R., et al. (2016). Copy-number variation contributes to the mutational load of bardet-biedl syndrome. Am. J. Hum. Genet. 99, 318–336. doi:10.1016/j.ajhg.2015.04.023
Lu, H., Galeano, M. C. R., Ott, E., Kaeslin, G., Kausalya, P. J., Kramer, C., et al. (2017). Mutations in DZIP1L, which encodes a ciliary-transition-zone protein, cause autosomal recessive polycystic kidney disease. Nat. Genet. 49, 1025–1034. doi:10.1038/ng.3871
Ma, X., Zhu, P., Ding, Y., Zhang, H., Qiu, Q., Dvornikov, A. V., et al. (2020). Retinoid X receptor alpha is a spatiotemporally predominant therapeutic target for anthracycline-induced cardiotoxicity. Sci. Adv. 6, eaay2939. doi:10.1126/sciadv.aay2939
Macri, C., Wang, F., Tasset, I., Schall, N., Page, N., Briand, J. P., et al. (2015). Modulation of deregulated chaperone-mediated autophagy by a phosphopeptide. Autophagy 11, 472–486. doi:10.1080/15548627.2015.1017179
Mangos, S., Lam, P. Y., Zhao, A., Liu, Y., Mudumana, S., Vasilyev, A., et al. (2010). The ADPKD genes pkd1a/b and pkd2 regulate extracellular matrix formation. Dis. Model Mech. 3, 354–365. doi:10.1242/dmm.003194
Mccampbell, K. K., Springer, K. N., and Wingert, R. A. (2014). Analysis of nephron composition and function in the adult zebrafish kidney. J. Vis. Exp. 90, e51644. doi:10.3791/51644
Mcconnachie, D. J., Stow, J. L., and Mallett, A. J. (2021). Ciliopathies and the kidney: A review. Am. J. Kidney Dis. 77, 410–419. doi:10.1053/j.ajkd.2020.08.012
Menezes, L. F., Lin, C. C., Zhou, F., and Germino, G. G. (2016). Fatty acid oxidation is impaired in an orthologous mouse model of autosomal dominant polycystic kidney disease. EBioMedicine 5, 183–192. doi:10.1016/j.ebiom.2016.01.027
Nagao, S., Kusaka, M., Nishii, K., Marunouchi, T., Kurahashi, H., Takahashi, H., et al. (2005). Androgen receptor pathway in rats with autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 16, 2052–2062. doi:10.1681/ASN.2004070595
Neugarten, J., Acharya, A., and Silbiger, S. R. (2000). Effect of gender on the progression of nondiabetic renal disease: A meta-analysis. J. Am. Soc. Nephrol. 11, 319–329. doi:10.1681/ASN.V112319
Neugarten, J., and Golestaneh, L. (2022). Sex differences in acute kidney injury. Semin. Nephrol. 42, 208–218. doi:10.1016/j.semnephrol.2022.04.010
Porath, B., Gainullin, V. G., Cornec-Le Gall, E., Dillinger, E. K., Heyer, C. M., Hopp, K., et al. (2016). Mutations in GANAB, encoding the glucosidase IIα subunit, cause autosomal-dominant polycystic kidney and liver disease. Am. J. Hum. Genet. 98, 1193–1207. doi:10.1016/j.ajhg.2016.05.004
Reed, B. Y., Masoumi, A., Elhassan, E., Mcfann, K., Cadnapaphornchai, M. A., Maahs, D. M., et al. (2011). Angiogenic growth factors correlate with disease severity in young patients with autosomal dominant polycystic kidney disease. Kidney Int. 79, 128–134. doi:10.1038/ki.2010.355
Reed, B. Y., Mcfann, K., Bekheirnia, M. R., Nobakhthaghighi, N., Masoumi, A., Johnson, A. M., et al. (2008). Variation in age at ESRD in autosomal dominant polycystic kidney disease. Am. J. Kidney Dis. 51, 173–183. doi:10.1053/j.ajkd.2007.10.037
Reiter, J. F., and Leroux, M. R. (2017). Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18, 533–547. doi:10.1038/nrm.2017.60
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi:10.1093/bioinformatics/btp616
Sardiello, M., Palmieri, M., Ronza, A., Medina, D. L., Valenza, M., Gennarino, V. A., et al. (2009). A gene network regulating lysosomal biogenesis and function. Science 325, 473–477. doi:10.1126/science.1174447
Senum, S. R., Li, Y. S. M., Benson, K. A., Joli, G., Olinger, E., Lavu, S., et al. (2022). Monoallelic IFT140 pathogenic variants are an important cause of the autosomal dominant polycystic kidney-spectrum phenotype. Am. J. Hum. Genet. 109, 136–156. doi:10.1016/j.ajhg.2021.11.016
Shaheen, R., Almoisheer, A., Faqeih, E., Babay, Z., Monies, D., Tassan, N., et al. (2015). Identification of a novel MKS locus defined by TMEM107 mutation. Hum. Mol. Genet. 24, 5211–5218. doi:10.1093/hmg/ddv242
Shih, Y. H., Zhang, Y., Ding, Y., Ross, C. A., Li, H., Olson, T. M., et al. (2015). Cardiac transcriptome and dilated cardiomyopathy genes in zebrafish. Circ. Cardiovasc Genet. 8, 261–269. doi:10.1161/CIRCGENETICS.114.000702
Shillingford, J. M., and Shayman, J. A. (2023). Functional TFEB activation characterizes multiple models of renal cystic disease and loss of polycystin-1. Am. J. Physiol. Ren. Physiol. 324, F404–F422. doi:10.1152/ajprenal.00237.2022
Smith, U. M., Consugar, M., Tee, L. J., Mckee, B. M., Maina, E. N., Whelan, S., et al. (2006). The transmembrane protein meckelin (MKS3) is mutated in Meckel-Gruber syndrome and the wpk rat. Nat. Genet. 38, 191–196. doi:10.1038/ng1713
Song, Z., Zhang, X., Jia, S., Yelick, P. C., and Zhao, C. (2016). Zebrafish as a model for human ciliopathies. J. Genet. Genomics 43, 107–120. doi:10.1016/j.jgg.2016.02.001
Srivastava, A., Sarkar, H., Gupta, N., and Patro, R. (2016). RapMap: A rapid, sensitive and accurate tool for mapping RNA-seq reads to transcriptomes. Bioinformatics 32, i192–i200. doi:10.1093/bioinformatics/btw277
Stewart, J. H. (1994). End-stage renal failure appears earlier in men than in women with polycystic kidney disease. Am. J. Kidney Dis. 24, 181–183. doi:10.1016/s0272-6386(12)80179-x
Stringer, K. D., Komers, R., Osman, S. A., Oyama, T. T., Lindsley, J. N., and Anderson, S. (2005). Gender hormones and the progression of experimental polycystic kidney disease. Kidney Int. 68, 1729–1739. doi:10.1111/j.1523-1755.2005.00589.x
Sullivan-Brown, J., Schottenfeld, J., Okabe, N., Hostetter, C. L., Serluca, F. C., Thiberge, S. Y., et al. (2008). Zebrafish mutations affecting cilia motility share similar cystic phenotypes and suggest a mechanism of cyst formation that differs from pkd2 morphants. Dev. Biol. 314, 261–275. doi:10.1016/j.ydbio.2007.11.025
Sun, Z., Amsterdam, A., Pazour, G. J., Cole, D. G., Miller, M. S., and Hopkins, N. (2004). A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 131, 4085–4093. doi:10.1242/dev.01240
Sun, Z., and Hopkins, N. (2001). vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 15, 3217–3229. doi:10.1101/gad946701
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612. doi:10.1093/nar/gkaa1074
Terabayashi, T., Germino, G. G., and Menezes, L. F. (2020). Pathway identification through transcriptome analysis. Cell Signal 74, 109701. doi:10.1016/j.cellsig.2020.109701
Torres, V. E., and Harris, P. C. (2014). Strategies targeting cAMP signaling in the treatment of polycystic kidney disease. J. Am. Soc. Nephrol. 25, 18–32. doi:10.1681/ASN.2013040398
Veiras, L. C., Girardi, A. C. C., Curry, J., Pei, L., Ralph, D. L., Tran, A., et al. (2017). Sexual dimorphic pattern of renal transporters and electrolyte homeostasis. J. Am. Soc. Nephrol. 28, 3504–3517. doi:10.1681/ASN.2017030295
Volff, J. N. (2005). Genome evolution and biodiversity in teleost fish. Hered. (Edinb) 94, 280–294. doi:10.1038/sj.hdy.6800635
Ward, C. J., Hogan, M. C., Rossetti, S., Walker, D., Sneddon, T., Wang, X., et al. (2002). The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat. Genet. 30, 259–269. doi:10.1038/ng833
Wingert, R. A., Selleck, R., Yu, J., Song, H. D., Chen, Z., Song, A., et al. (2007). The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 3, 1922–1938. doi:10.1371/journal.pgen.0030189
Woods, I. G., Kelly, P. D., Chu, F., Ngo-Hazelett, P., Yan, Y. L., Huang, H., et al. (2000). A comparative map of the zebrafish genome. Genome Res. 10, 1903–1914. doi:10.1101/gr.10.12.1903
Zhou, J. X., and Torres, V. E. (2023). Drug repurposing in autosomal dominant polycystic kidney disease. Kidney Int. 103, 859–871. doi:10.1016/j.kint.2023.02.010
Zhou, X., Fan, L. X., Sweeney, W. E., Denu, J. M., Avner, E. D., and Li, X. (2013). Sirtuin 1 inhibition delays cyst formation in autosomal-dominant polycystic kidney disease. J. Clin. Invest. 123, 3084–3098. doi:10.1172/JCI64401
Zhu, P., Qiu, Q., Harris, P. C., Xu, X., and Lin, X. (2021). Mtor haploinsufficiency ameliorates renal cysts and cilia abnormality in adult zebrafish tmem67 mutants. J. Am. Soc. Nephrol. 32, 822–836. doi:10.1681/ASN.2020070991
Keywords: zebrafish, cystic kidney disease, sexual diamorphism, transcriptome, ciliopathy
Citation: Koslow M, Zhu P, McCabe C, Xu X and Lin X (2023) Kidney transcriptome and cystic kidney disease genes in zebrafish. Front. Physiol. 14:1184025. doi: 10.3389/fphys.2023.1184025
Received: 10 March 2023; Accepted: 20 April 2023;
Published: 04 May 2023.
Edited by:
Chi-Wen Lung, Asia University, TaiwanReviewed by:
Yori Pusparani, Universitas Budi Luhur, IndonesiaCopyright © 2023 Koslow, Zhu, McCabe, Xu and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolei Xu, eHUueGlhb2xlaUBtYXlvLmVkdQ==; Xueying Lin, bGluLnh1ZXlpbmdAbWF5by5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.