
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Physiol., 12 April 2023
Sec. Invertebrate Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1182610
This article is part of the Research TopicInvertebrates as Model Organisms: Opportunities and Challenges in Physiology and Bioscience ResearchView all 6 articles
The Drosophila heart tube seems simple, yet it has notable anatomic complexity and contains highly specialized structures. In fact, the development of the fly heart tube much resembles that of the earliest stages of mammalian heart development, and the molecular-genetic mechanisms driving these processes are highly conserved between flies and humans. Combined with the fly’s unmatched genetic tools and a wide variety of techniques to assay both structure and function in the living fly heart, these attributes have made Drosophila a valuable model system for studying human heart development and disease. This perspective focuses on the functional and physiological similarities between fly and human hearts. Further, it discusses current limitations in using the fly, as well as promising prospects to expand the capabilities of Drosophila as a research model for studying human cardiac diseases.
At first sight flies, and by extension their physiology, seem far removed from humans, yet the fly’s simple heart tube harbors more complexity than initially assumed. The Drosophila heart has distinct morphological features, including an anterior aorta structure that is separated from the posterior heart chamber by an aortic valve to ensure posterior to anterior flow, and it has inflow tracts named ostia (Bodmer, 1995; Rotstein and Paululat, 2016) (Figure 1). One notable difference is that fly has an open circulatory system, i.e., all hemolymph is oxygenated (akin mammalian arterial blood) and flows via channel-like trajectories formed by the internal organs and by fibromuscular septa or diaphragms (Hillyer and Pass, 2020), with the whole body acting as trachea (Figure 1). Despite this difference, the earliest stages of heart development are extremely well conserved from flies to humans. These cover the migration of the bilateral rows of cardiac progenitor cells towards the midline to their fusion to form the heart tube (Bodmer, 1995; Ahmad, 2017). These early similarities go beyond structure to include molecular genetics (Olson, 2006; Souidi and Jagla, 2021). In fact, the first gene known to control heart development, tinman (tin), was discovered in flies (Azpiazu and Frasch, 1993; Bodmer, 1993). This then led to the identification of its homolog, Nkx2.5, as a key transcription factor for mammalian heart development (Komuro and Izumo, 1993; Lyons et al., 1995; Tanaka et al., 1999). Many of the major genetic pathways in human heart development, like WNT, TGFβ, and FGF, as well as key transcription factors, like NKX2.5, QRSL1 (a.k.a. GATA), TBX (T-box), MEF2, and the HAND family, are evolutionarily conserved and have homologs in flies, zebrafish, and mice (Bodmer, 1995; Olson, 2006; Cui et al., 2018). For example, Wnt4 is required for the development of the ostia in the fly heart (Chen et al., 2016); and Wnt signaling is a key regulator of mammalian heart development, during which Wnt mediates cardiac specification, proliferation, and patterning (Gessert and Kühl, 2010; Li D. et al., 2022). The shared developmental genetic, molecular, cellular, and functional mechanisms culminate in shared physiology. Therefore, despite the simpler heart structure in fly and the evolutionary distance between flies and humans, the fly heart’s structural and functional similarities to the human heart during early development combined with its genetic tools and resources have made Drosophila a valuable model system to study human cardiac diseases (to highlight a few: Qian et al., 2008; Ma et al., 2010; Neely et al., 2010; den Hoed et al., 2013; Haack et al., 2013; Zhu et al., 2017a; Iuso et al., 2018; Kronert et al., 2018; Palandri et al., 2018; Ekure et al., 2021) (Table 1).
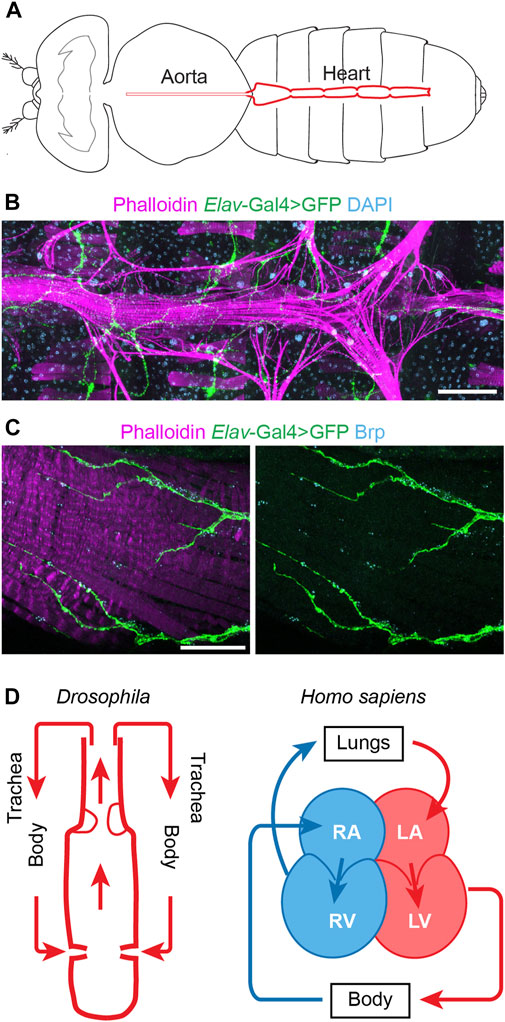
FIGURE 1. The Drosophila and human hearts. (A) Schematic illustration of an adult fly (dorsal view). The aorta and the heart are shown in red. (B) Representative image of a Drosophila adult heart (ventral view). Phalloidin stains filament actin in red. The elav-Gal4>GFP (elav, embryonic legal abnormal vision; GFP, green fluorescent protein) labels cardiac neuron fiber in green. DAPI stains DNA in blue. The scale bar represents 100 μm. Image was obtained using a ZEISS Apotome fluorescence microscope using a 20× objective and ZEISS Zen (blue edition) acquisition software. (C) Representative image of the posterior Drosophila adult heart (ventral view). Phalloidin stains filament actin in red. The elav-Gal4>GFP (elav, embryonic legal abnormal vision; GFP, green fluorescent protein) labels cardiac neuronal axons in green. Brp, Bruchpilot, is in blue and labels the neuromuscular junctions. The scale bar represents 20 μm. Images were obtained using a ZEISS LSM900 confocal microscope using a 63× objective and ZEISS Zen (blue edition) acquisition software. (D) Schematic representations of the fly (Drosophila) and human (Homo sapiens) hearts. Arrows indicate the direction of flow for the fly hemolymph and human blood (blue, oxygen poor; red, oxygen rich blood). Structure inside the fly heart depicts the aortic valve, and flow enters the heart at the inflow tracts with ostia (valves). Human heart chambers: RA, right atrium; RV, right ventricle; LA, left atrium; LV, left ventricle.
CHD affects over 1% of all live births, making it the most frequent type of birth defect (Pierpont et al., 2018). Although the contribution of genetics to CHD has been well-established, identifying the causal genetic mutations for individual CHD patients has proven difficult. Tremendous effort has been invested to identify CHD-linked causal variants. For example, the Pediatric Cardiac Genomic Consortium (PCGC) (Zaidi et al., 2013; Homsy et al., 2015; Jin et al., 2017) incorporates CHD-related findings from many studies including Kids First, Center for Mendelian Genomics (CMG), Undiagnosed Disease Network (UDN) (Posey et al., 2019), and Deciphering Developmental Disorders (DDD) (Sifrim et al., 2016; Verheije et al., 2019). These large-scale genomic sequencing projects, together with studies led by cardiologists and medical geneticists, have identified thousands of novel candidate genes and variants in patients with CHD.
In the absence of independent patient families that each carry the same genetic variant, functional validation is essential to establish causality. Screening such a large number of candidate CHD genes is not feasible using conventional mammalian research models, which are too time-consuming and costly for these purposes. Drosophila could bridge this gap because most disease-causing genes have homologs in fly (Ugur et al., 2016). For example, the highly conserved NKX2.5 (tinman in fly) which plays a crucial role in heart development is also a hotspot for genetic variants that have been linked to CHD (McElhinney et al., 2004; Pashmforoush et al., 2004). In addition, the fly has an unprecedented arsenal of genetic tools that enable precise genetic manipulation to target specific developmental time points, or specific tissues and even cell-types within those tissues (Hales et al., 2015; Zhao et al., 2021). Over the years many techniques to assay heart structure and function in the fly have been developed; these have consistently shown the physiological similarities of Drosophila and human heart development and function, as well as dysfunction when studying disease-associated genes and variants (Wolf and Rockman, 2008; Manivannan et al., 2020). Altogether, these features make the fly a versatile model system, capable of rapid cost-effective screens of hundreds of candidate genetic variants for CHD. One such study used an RNAi-based functional screen of 134 genes associated with CHD, of which over 70 genes were shown to be involved in Drosophila heart development thus supporting their causality (Zhu et al., 2017a). One of the hits was WD repeat domain 5 (WDR5); silencing its homolog Wds in the fly heart caused complete developmental lethality and abnormal cardiac morphology in late larvae, including reduced cardiac myofibers and increased Pericardin deposition (Zhu et al., 2017a). Notably, overexpressing wildtype human WDR5 restored the cardiac phenotype in flies with heart-specific deficiency for Wds, whereas human WDR5 carrying a patient variant could not (Zhu et al., 2017a). These findings demonstrate the physiological (gene-structure) homology between human and fly key cardiac genes. Another study identified 19 deleted de novo copy number variants (CNVs) covering hitherto not associated candidate disease genes in a cohort of 167 patients with CHD (Schroeder et al., 2019). These were then tested using parallel screens in human induced pluripotent stem cell (iPSC)-derived multipotent cardiac progenitor cells and a Drosophila in vivo heart model. Flies with heart-specific deficiency for candidate genes showed phenotypes ranging from a completely absent heart to structural and functional defects that included reduced or abolished contractility (Schroeder et al., 2019).
Like CHD, cardiomyopathies are genetically and phenotypically diverse (Arad et al., 2002; Richard et al., 2006; Ware et al., 2021). Among the many cardiomyopathy-associated genes is Lamin A/C (LMNA), one of the most sequenced human genes. It has hundreds of variants associated with multiple cardiomyopathies including those with pediatric onset (Heller et al., 2017; Kervella et al., 2022). Cardiomyopathy clinically manifests as systolic and diastolic dysfunction, arrhythmia, and increased risk of heart failure. These functional manifestations have been linked to structural issues of increased ventricular size, thickening of the ventricles (i.e., hypertrophy), and stiffening of the ventricular walls (Lee et al., 2017; El Hadi et al., 2023). Multiple structural and functional readouts that have been established for the fly heart can capture these phenotypes. Brightfield microscopy of histological sections or micro computerized tomography, a 3D X-ray imaging technique, can be used to determine the thickness of the heart muscle wall (Migunova et al., 2021; Petersen et al., 2022). Whereas high-speed movies of semi intact Drosophila heart preparations (Ocorr et al., 2007), in vivo imaging of the heart in intact flies using high resolution optical coherence microscopy which yields imaging similar to ultrasound (Migunova et al., 2021), or optical coherence tomography which is similar to echocardiography in humans (Wolf et al., 2006), can be used to quantify muscle wall thickness and function, including diastolic diameter, end systolic diameter, and fractional shortening. Finally, the cardiac flow, a measure of contractile force, can be measured by a dye injection assay that times the flow from injection site to target site (Zhu et al., 2017a; 2017b), or with intravital imaging which enables life tracking of the heart wall, quantitation of the chamber diameter during contraction (systole) and relaxation (diastole), and fractional shortening, as well as estimates of cardiac output and stroke volume using segmentation algorithms (Klassen et al., 2017).
The above techniques to observe the heart in flies have been successfully used in fly models for diverse cardiomyopathies including modulators of the EGF receptor signaling pathway associated with dilated (Yu et al., 2010) or hypertrophic (Yu et al., 2013) cardiomyopathy; in which flies showed increased cardiac chamber size, and cardiac hypertrophy with reduced contraction, respectively. Tropomyosin II null (TM23) mutant flies showed cardiac arrhythmia reminiscent of the clinical observation in patients with Tropomyosin-associated dilated cardiomyopathy (Ma et al., 2010). Moreover, these techniques to study the fly heart have been applied to establish genetic causality for dilated cardiomyopathy using fly models deficient for Phosphopantothenoylcysteine synthetase (PPCS) and transgenic flies that caried PPCS with patient variants (Iuso et al., 2018). Affected flies showed reduced viability, increased heart rate, increased arrhythmia index, reduced systolic function, and increased heart wall shortening, reminiscent of the pathophysiology observed in the patients (Iuso et al., 2018). The methods to observe fly hearts have also been applied to establish causality for Zinc phosphodiesterase ELAC protein 2 (ELAC2) genetic variants in a rare form of severe infantile cardiomyopathy (Migunova et al., 2021). Transgenic flies expressing patient mutations in the homologous fly gene (RNaseZ) displayed cardiac hypertrophy and reduced contraction mimicking the clinical pathology in patients (Migunova et al., 2021). In a final example, the techniques to study the heart in fly were used to establish causality for variants in Myosin light chain 2 (MYL2) associated with hypertrophic cardiomyopathy of infantile onset and characterized by mitral valve dysplasia, resulting in infant death (Manivannan et al., 2020). Silencing the fly homologous gene Mlc2 in the fly heart resulted in developmental lethality and decreased fractional shortening similar to the patients that carried loss-of-function variants (Manivannan et al., 2020). These phenotypes could be rescued by expressing wildtype human MYL2, but not by MYL2 patient variant cDNA (Manivannan et al., 2020). These studies exemplify both the relevance and the potential of using fly models to study myocardial contractility defects.
Nearly all metabolic pathways are shared between flies and humans (Bharucha, 2009), making fly a notable model system to study metabolic and diet-associated diseases, such as obesity (high-fat diet fly models) and diabetes (high-sugar diet fly models). Obesity caused by high-fat diet is a major contributor to diabetes and related cardiovascular complications. In patients, obesity-associated cardiomyopathy is marked by altered metabolism, ventricular hypertrophy and remodeling, diastolic and systolic dysfunction, abnormal conduction, atrial fibrillation (i.e., arrhythmia), and ultimately heart failure (Piché et al., 2020). Flies fed a high-fat-diet mimic the features of metabolic syndrome in patients, including elevated lipid levels and altered insulin and glucose homeostasis (Birse et al., 2010). Moreover, the diet has detrimental effects on the fly heart, including reduced contractility, conduction blocks (i.e., anterior and posterior heart beat at different rates), dysfunctional ostia (valves at the inflow tracts), and structural defects and disorganization of the myofibrils (Birse et al., 2010). Moreover, the cardiac dysfunction induced by high-fat-diet in Drosophila was shown to persist for two subsequent generations (Guida et al., 2019). This heredity was linked to lasting metabolic changes mediated by epigenetic modifications (Guida et al., 2019). Many gaps remain in our understanding of the pathomechanisms underlying obesity-induced cardiomyopathy and the inherited risks for future generations; Drosophila high-fat-diet models could provide a valuable contribution to this research.
Incentivized by the conserved metabolic pathways and the range of genetic tools available in fly, Drosophila models for type I and type II diabetes have been established. Glucose homeostasis is highly conserved and includes fly functional equivalents of mammalian insulin and glucagon (Liguori et al., 2021). Similar to patients, diabetic flies display signs of hyperglycemia, hypertrehalosemia, peripheral resistance to exogenous insulin, and accumulation of triglyceride (Na et al., 2013). Flies on a high-sucrose diet displayed increased cardiac arrhythmia, and increased diastole and systole (without fractional shortening) (Na et al., 2013) reminiscent of diabetes-associated dilated cardiomyopathy. The functional outcomes were accompanied by structural defects, including increased Pericardin deposition in fly heart tissue, a measure of cardiac fibrosis (Na et al., 2013) which is a common complication in patients with diabetes (Armstrong et al., 2017). These cardiac symptoms in fly were more severe at higher dietary sugar content and with age/prolonged exposure, ultimately resulting in a shortened lifespan (Na et al., 2013). Besides genetic factors, environmental factors greatly contribute to the risk of developing diabetes. Drosophila can be a powerful model in studying these gene-environment interactions. For example, maternal diabetes-induced fetal hyperglycemia is associated with a five-fold increased risk for CHD. Combining an assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) of a high-glucose in vitro model, and maternal diabetic mouse and Drosophila models, revealed a conserved interaction between Notch1 signaling (gene) and high-glucose (environment); controlled by a Jarid2 (repressor of Notch signaling)-mediated epigenetic mechanism (Basu et al., 2017). This study demonstrates that fly diabetic models can be valuable tools in unraveling the gene-environment interactions that underly diabetes-associated cardiomyopathy.
Aging is associated with increased risk for cardiovascular disease, including increased prevalence of atrial fibrillation (i.e., arrhythmia) and decreased diastolic function of the left ventricle, even in the absence of major cardiovascular risk factors (Dai et al., 2012; Gude et al., 2018; Go et al., 2023). Similarly, in Drosophila, aging leads to decreased cardiac output (Klassen et al., 2017) and increased cardiac arrhythmia (Ocorr et al., 2007; Taghli-Lamallem et al., 2008). Due to the flies’ relatively short lifespan, with a median lifetime of 63 days (Taghli-Lamallem et al., 2008), aging-related studies of cardiac morphology and function can be accomplished within practicable timeframes. Electrophysiology can be used to measure cardiac action potentials, like electrocardiography in humans (Johnson et al., 2001; Papaefthmiou and Theophilidis, 2001; Lalevée et al., 2006). Aging flies displayed a reduced resting heart rate and increased cardiac arrhythmia events, as well as cardiovascular stress-induced maximal heart rate and increased heart failure (Paternostro et al., 2001; Wessells et al., 2004). These findings are similar to the cardiac functional decline observed in the aging human heart (Dai et al., 2012; Gude et al., 2018; Go et al., 2023).
Fly models of aging have also been used to study the molecular-genetic pathways that underly age-related cardiac pathophysiology. The insulin and mTOR pathways play key regulatory roles in aging (Kim, 2007; Papadopoli et al., 2019), and their disruption leads to similar defects of the heart in aging flies (Wessells et al., 2004; Luong et al., 2006). A fly study identified several effectors that act at the interchange of insulin and mTOR signaling, and demonstrated their importance during age-related cardiac decline (Wessells et al., 2009). Another study of the aging fly heart found that cardiac decline and arrhythmias were accompanied by reduced expression of KCNQ (fly homolog of mammalian KCNQ1-encoded voltage-gated potassium channel alpha subunits) (Ocorr et al., 2007). Moreover, hearts of young flies deficient for KCNQ displayed prolonged contractions and fibrillation, reminiscent of the cardiac arrhythmic phenotypes in patients with mutations in KCNQ1 (torsade de pointes/congenital long QT syndrome) (Ocorr et al., 2007). A comparative study of age-related cardiac transcriptomic changes identified pathways involved in remodeling of the extra-cellular matrix, mitochondrial metabolism, protein handling, and contractile functions, that were conserved between Drosophila and rodents (Cannon et al., 2017). Besides, like individual aging rodent hearts, the gene expression changes between individual aging fly hearts showed little overlap. The findings suggest that different transcriptional paths can lead to similar age-related cardiac decline (Cannon et al., 2017). Taken together, these studies demonstrate the proficiency of the Drosophila system to model the physiology underlying the aging heart in the absence of major cardiovascular risks.
Decades of research contributions from Drosophila models have taught us much about the developing and aging heart in health and disease. However, several challenges remain, which when overcome would greatly expand the opportunities to use fly models in studies of cardiac disease. Whereas single-cell RNA sequencing technology has provided detailed transcriptomic profiles of the developing heart in humans (Asp et al., 2019; Cui et al., 2019; Litvinukova et al., 2020; Tucker et al., 2020) and several model systems, including mouse (DeLaughter et al., 2016; Li et al., 2016; Li et al., 2019b.; Li et al., 2019a.; Gladka et al., 2018; Hu et al., 2018; Jia et al., 2018; Lescroart et al., 2018; Skelly et al., 2018; Farbehi et al., 2019; Goodyer et al., 2019), zebrafish (Danio rerio) (Burkhard and Bakkers, 2018; Yuan et al., 2018; Honkoop et al., 2019; Weinberger et al., 2020), and sea squirt (Ciona robusta) (Wang et al., 2019); for Drosophila single-cell RNA sequencing data is only available for the adult heart (Li et al., 2022a). Knowing the cell types that make up the fly heart at crucial developmental stages would facilitate determining the extent of evolutionary conservation of heart development and its underlying molecular pathways. In addition, these data would enable more direct comparisons between different cardiac cell types in flies and human. This knowledge could be used to better assign disease subtypes, especially those with genotypic and phenotypic overlap; for example, by targeting RNAi and/or human cDNA carrying patient variants to the fly equivalent of the cells affected in patients. Together, the better understanding would aid translation of the findings in fly to applications in patients.
It is worth noting that to accommodate its open circulatory system, the fly carries additional pulsatile organs, known as antennal/frontal accessory pulsatile organ and wing hearts, that ensure circulation of hemolymph (fly blood) throughout its antennae and wings, respectively (Tögel et al., 2013; Hillyer and Pass, 2020; Kay et al., 2021). The muscle cells that make up the wing hearts originate from a select group of pericardial progenitors, marked by the expression of Even skipped (Eve) and early loss of Tinman (Tögel et al., 2008). Pathological conditions that affect the equivalent cell in the human heart—single-cell RNA sequencing data of the fly, mammalian, and human hearts could shed light on this—could potentially benefit from studying the fly wing hearts specifically.
Arrhythmia can present as either a primary or a secondary cause. For example, as a complication in diverse CHD and cardiomyopathies, as well as in the aging heart in the absence of major cardiovascular risk factors. A clear pathophysiological understanding is crucial to distinguish between arrhythmia as a primary or secondary feature. In humans, the heart’s rhythm is regulated by the sinoatrial node (SA node), the atrioventricular node (AV node), the bundle of His, and Purkinje fibers which make up the cardiac conduction system (Chloe Li et al., 2022). The SA node acts as the biological pacemaker, its excitation signals the start of a heartbeat (Hanna et al., 2021). The SA node is controlled by the vagus nerve which innervates both the heart muscle cells and the conduction system (Capilupi et al., 2020). However, the exact neural circuitry and the molecular mechanisms that coordinate the parasympathetic (relaxes heart rate) and sympathetic (increases heart rate) actions of the vagus nerve are not fully understood. Likewise, the Drosophila heart is innervated by peripheral neurons (Dulcis and Levine, 2005) (Figure 1). Although, many questions remain regards the fly heart conduction system. A study into myotonic dystrophy type 1 (DM1) demonstrates the potential of Drosophila to model human cardiac conduction defects. The study simulated the DM1-associated misbalance between two RNA binding factors, Muscleblind like splicing regulator (MBNL1) and CUGBP Elav-like family member 1 (CELF1), in the fly heart; this led to dysregulated calcium signaling genes including straightjacket (stj)/α2δ3, which encodes a voltage-gated calcium channel subunit (Auxerre-Plantié et al., 2019). In the flies, dysregulated stj resulted in an asynchronous heartbeat, indicative of abnormal conduction. Moreover, the study showed altered expression of α2δ3 in heart tissue from patients with DM1-associated conduction defects (Auxerre-Plantié et al., 2019). Better understanding the similarities and differences between the human and fly cardiac conduction systems would aid the development of additional fly models for arrhythmia. A variety of imaging tools to assay the heart cycle and rhythm (Wolf et al., 2006; Ocorr et al., 2007; Zhu et al., 2017a; Klassen et al., 2017; Migunova et al., 2021) and electrophysiology to measure the energy fluxes (Johnson et al., 2001; Papaefthmiou and Theophilidis, 2001; Lalevée et al., 2006) are already available and will aid this research direction.
In addition to accommodating studies of gene-environment interactions, like fly models for diabetes, Drosophila provides an excellent opportunity to study polygenic causation, which has likewise been difficult to study using conventional animal models. However, Genomic studies have found many patients with CHD or cardiomyopathy that likely have a polygenic cause (Zaidi et al., 2013; Homsy et al., 2015; Jin et al., 2017; Ware et al., 2021). For these studies too, the fly could be immensely valuable. In fact, several polygenic fly models for cardiac disease have been generated (Qian and Bodmer, 2012), thus demonstrating feasibility. The numerous readily available transgenic fly lines, unmatched genetic tools, and rapid crosses with large progenies make for relatively straightforward development of polygenic fly models that carry the genetic variant combinations identified in patients. Moreover, the compact Drosophila genome carries little redundancy which facilitates data interpretation. These studies could reveal new disease mechanisms relevant to the human heart.
The flies’ established track record combined with the latest technology and assays for fly cardiac function, and the many new avenues about to be explored make Drosophila a very exciting model system to study a wide variety of aspects of human cardiac diseases.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
YZ, JL, and ZH constructed the outline and main ideas. YZ generated the data for figure. JL and YZ wrote the detailed sections. JL and ZH edited the manuscript.
This work was supported by National Institutes of Health grant R01-HL134940 to ZH.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmad, S. M. (2017). Conserved signaling mechanisms in Drosophila heart development. Dev. Dyn. 246, 641–656. doi:10.1002/dvdy.24530
Arad, M., Seidman, J. G., and Seidman, C. E. (2002). Phenotypic diversity in hypertrophic cardiomyopathy. Hum. Mol. Genet. 11, 2499–2506. doi:10.1093/hmg/11.20.2499
Armstrong, A. C., Ambale-Venkatesh, B., Turkbey, E., Donekal, S., Chamera, E., Backlund, J.-Y., et al. (2017). Association of cardiovascular risk factors and myocardial fibrosis with early cardiac dysfunction in type 1 diabetes: The diabetes control and complications trial/epidemiology of diabetes interventions and complications study. Diabetes Care 40, 405–411. doi:10.2337/dc16-1889
Asp, M., Giacomello, S., Larsson, L., Wu, C., Furth, D., Qian, X., et al. (2019). A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell 179, 1647–1660. e19. doi:10.1016/j.cell.2019.11.025
Auxerre-Plantié, E., Nakamori, M., Renaud, Y., Huguet, A., Choquet, C., Dondi, C., et al. (2019). Straightjacket/α2δ3 deregulation is associated with cardiac conduction defects in myotonic dystrophy type 1. Elife 8, e51114. doi:10.7554/eLife.51114
Azpiazu, N., and Frasch, M. (1993). Tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 7, 1325–1340. doi:10.1101/gad.7.7b.1325
Basu, M., Zhu, J.-Y., LaHaye, S., Majumdar, U., Jiao, K., Han, Z., et al. (2017). Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease. JCI Insight 2, e95085. doi:10.1172/jci.insight.95085
Bharucha, K. N. (2009). The epicurean fly: Using Drosophila melanogaster to study metabolism. Pediatr. Res. 65, 132–137. doi:10.1203/PDR.0b013e318191fc68
Birse, R. T., Choi, J., Reardon, K., Rodriguez, J., Graham, S., Diop, S., et al. (2010). High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab. 12, 533–544. doi:10.1016/j.cmet.2010.09.014
Bodmer, R. (1995). Heart development in Drosophila and its relationship to vertebrates. Trends cardiovasc. Med. 5, 21–28. doi:10.1016/1050-1738(94)00032-Q
Bodmer, R. (1993). The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 118, 719–729. doi:10.1242/dev.118.3.719
Burkhard, S. B., and Bakkers, J. (2018). Spatially resolved RNA-sequencing of the embryonic heart identifies a role for Wnt/β-catenin signaling in autonomic control of heart rate. Elife 7, e31515. doi:10.7554/eLife.31515
Cannon, L., Zambon, A. C., Cammarato, A., Zhang, Z., Vogler, G., Munoz, M., et al. (2017). Expression patterns of cardiac aging in Drosophila. Aging Cell 16, 82–92. doi:10.1111/acel.12559
Capilupi, M. J., Kerath, S. M., and Becker, L. B. (2020). Vagus nerve stimulation and the cardiovascular system. Cold Spring Harb. Perspect. Med. 10, a034173. doi:10.1101/cshperspect.a034173
Chen, Z., Zhu, J.-Y., Fu, Y., Richman, A., and Han, Z. (2016). Wnt4 is required for ostia development in the Drosophila heart. Dev. Biol. 413, 188–198. doi:10.1016/j.ydbio.2016.03.008
Chloe Li, K. Y., Cook, A. C., and Lovering, R. C. (2022). GOing forward with the cardiac conduction system using gene ontology. Front. Genet. 13, 802393. doi:10.3389/fgene.2022.802393
Cui, M., Wang, Z., Bassel-Duby, R., and Olson, E. N. (2018). Genetic and epigenetic regulation of cardiomyocytes in development, regeneration and disease. Development 145, dev171983. doi:10.1242/dev.171983
Cui, Y., Zheng, Y., Liu, X., Yan, L., Fan, X., Yong, J., et al. (2019). Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep. 26, 1934–1950. e5. doi:10.1016/j.celrep.2019.01.079
Dai, D.-F., Chen, T., Johnson, S. C., Szeto, H., and Rabinovitch, P. S. (2012). Cardiac aging: From molecular mechanisms to significance in human health and disease. Antioxid. Redox Signal. 16, 1492–1526. doi:10.1089/ars.2011.4179
DeLaughter, D. M., Bick, A. G., Wakimoto, H., McKean, D., Gorham, J. M., Kathiriya, I. S., et al. (2016). Single-cell resolution of temporal gene expression during heart development. Dev. Cell 39, 480–490.
den Hoed, M., Eijgelsheim, M., Esko, T., Brundel, B. J. J. M., Peal, D. S., Evans, D. M., et al. (2013). Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat. Genet. 45, 621–631.
Dulcis, D., and Levine, R. B. (2005). Glutamatergic innervation of the heart initiates retrograde contractions in adult Drosophila melanogaster. J. Neurosci. 25, 271–280. doi:10.1523/JNEUROSCI.2906-04.2005
Ekure, E. N., Adeyemo, A., Liu, H., Sokunbi, O., Kalu, N., Martinez, A. F., et al. (2021). Exome sequencing and congenital heart disease in sub-saharan africa. Circ. Genom Precis. Med. 14, e003108. doi:10.1161/CIRCGEN.120.003108
El Hadi, H., Freund, A., Desch, S., Thiele, H., and Majunke, N. (2023). Hypertrophic, dilated, and arrhythmogenic cardiomyopathy: Where are we? Biomedicines 11, 524. doi:10.3390/biomedicines11020524
Farbehi, N., Patrick, R., Dorison, A., Xaymardan, M., Janbandhu, V., Wystub-Lis, K., et al. (2019). Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife 8, e43882. doi:10.7554/eLife.43882
Gessert, S., and Kühl, M. (2010). The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ. Res. 107, 186–199. doi:10.1161/CIRCRESAHA.110.221531
Gladka, M. M., Molenaar, B., de Ruiter, H., van der Elst, S., Tsui, H., Versteeg, D., et al. (2018). Single-cell sequencing of the healthy and diseased heart reveals cytoskeleton-associated protein 4 as a new modulator of fibroblasts activation. Circulation 138, 166–180. doi:10.1161/CIRCULATIONAHA.117.030742
Go, A. S., Al-Khatib, S. M., Desvigne-Nickens, P., Bansal, N., Bushnell, C. D., Fang, M. C., et al. (2023). Research opportunities in stroke prevention for atrial fibrillation: A report from a national heart, lung, and blood institute virtual workshop. Stroke 54, e75–e85. doi:10.1161/STROKEAHA.121.038273
Goodyer, W. R., Beyersdorf, B. M., Paik, D. T., Tian, L., Li, G., Buikema, J. W., et al. (2019). Transcriptomic profiling of the developing cardiac conduction system at single-cell resolution. Circ. Res. 125, 379–397. doi:10.1161/CIRCRESAHA.118.314578
Gude, N. A., Broughton, K. M., Firouzi, F., and Sussman, M. A. (2018). Cardiac ageing: Extrinsic and intrinsic factors in cellular renewal and senescence. Nat. Rev. Cardiol. 15, 523–542. doi:10.1038/s41569-018-0061-5
Guida, M. C., Birse, R. T., Dall’Agnese, A., Toto, P. C., Diop, S. B., Mai, A., et al. (2019). Intergenerational inheritance of high fat diet-induced cardiac lipotoxicity in Drosophila. Nat. Commun. 10, 193. doi:10.1038/s41467-018-08128-3
Haack, T. B., Kopajtich, R., Freisinger, P., Wieland, T., Rorbach, J., Nicholls, T. J., et al. (2013). ELAC2 mutations cause a mitochondrial RNA processing defect associated with hypertrophic cardiomyopathy. Am. J. Hum. Genet. 93, 211–223. doi:10.1016/j.ajhg.2013.06.006
Hales, K. G., Korey, C. A., Larracuente, A. M., and Roberts, D. M. (2015). Genetics on the fly: A primer on the Drosophila model system. Genetics 201, 815–842. doi:10.1534/genetics.115.183392
Hanna, P., Dacey, M. J., Brennan, J., Moss, A., Robbins, S., Achanta, S., et al. (2021). Innervation and neuronal control of the mammalian sinoatrial node a comprehensive atlas. Circ. Res. 128, 1279–1296. doi:10.1161/CIRCRESAHA.120.318458
Heller, F., Dabaj, I., Mah, J. K., Bergounioux, J., Essid, A., Bönnemann, C. G., et al. (2017). Cardiac manifestations of congenital LMNA-related muscular dystrophy in children: Three case reports and recommendations for care. Cardiol. Young 27, 1076–1082. doi:10.1017/S1047951116002079
Hillyer, J. F., and Pass, G. (2020). The insect circulatory system: Structure, function, and evolution. Annu. Rev. Entomol. 65, 121–143. doi:10.1146/annurev-ento-011019-025003
Homsy, J., Zaidi, S., Shen, Y., Ware, J. S., Samocha, K. E., Karczewski, K. J., et al. (2015). De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350, 1262–1266. doi:10.1126/science.aac9396
Honkoop, H., de Bakker, D. E., Aharonov, A., Kruse, F., Shakked, A., Nguyen, P. D., et al. (2019). Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. Elife 8, e50163. doi:10.7554/eLife.50163
Hu, P., Liu, J., Zhao, J., Wilkins, B. J., Lupino, K., Wu, H., et al. (2018). Single-nucleus transcriptomic survey of cell diversity and functional maturation in postnatal mammalian hearts. Genes Dev. 32, 1344–1357. doi:10.1101/gad.316802.118
Iuso, A., Wiersma, M., Schüller, H.-J., Pode-Shakked, B., Marek-Yagel, D., Grigat, M., et al. (2018). Mutations in PPCS, encoding phosphopantothenoylcysteine synthetase, cause autosomal-recessive dilated cardiomyopathy. Am. J. Hum. Genet. 102, 1018–1030. doi:10.1016/j.ajhg.2018.03.022
Jia, G., Preussner, J., Chen, X., Guenther, S., Yuan, X., Yekelchyk, M., et al. (2018). Single cell RNA-seq and ATAC-seq analysis of cardiac progenitor cell transition states and lineage settlement. Nat. Commun. 9, 4877. doi:10.1038/s41467-018-07307-6
Jin, S. C., Homsy, J., Zaidi, S., Lu, Q., Morton, S., DePalma, S. R., et al. (2017). Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet. 49, 1593–1601. doi:10.1038/ng.3970
Johnson, E., Ringo, J., and Dowse, H. (2001). Dynamin, encoded by shibire, is central to cardiac function. J. Exp. Zool. 289, 81–89. doi:10.1002/1097-010x(20010201)289:2<81::aid-jez1>3.0.co;2-t
Kay, A. R., Eberl, D. F., and Wang, J. W. (2021). Myogenic contraction of a somatic muscle powers rhythmic flow of hemolymph through Drosophila antennae and generates brain pulsations. J. Exp. Biol. 224, jeb242699. doi:10.1242/jeb.242699
Kervella, M., Jahier, M., Meli, A. C., and Muchir, A. (2022). Genome organization in cardiomyocytes expressing mutated A-type lamins. Front. Cell Dev. Biol. 10, 1030950. doi:10.3389/fcell.2022.1030950
Kim, S. K. (2007). Common aging pathways in worms, flies, mice and humans. J. Exp. Biol. 210, 1607–1612. doi:10.1242/jeb.004887
Klassen, M. P., Peters, C. J., Zhou, S., Williams, H. H., Jan, L. Y., and Jan, Y. N. (2017). Age-dependent diastolic heart failure in an in vivo Drosophila model. Elife 6, e20851. doi:10.7554/eLife.20851
Komuro, I., and Izumo, S. (1993). Csx: A murine homeobox-containing gene specifically expressed in the developing heart. Proc. Natl. Acad. Sci. U. S. A. 90, 8145–8149. doi:10.1073/pnas.90.17.8145
Kronert, W. A., Bell, K. M., Viswanathan, M. C., Melkani, G. C., Trujillo, A. S., Huang, A., et al. (2018). Prolonged cross-bridge binding triggers muscle dysfunction in a Drosophila model of myosin-based hypertrophic cardiomyopathy. Elife 7, e38064. doi:10.7554/eLife.38064
Lalevée, N., Monier, B., Sénatore, S., Perrin, L., and Sémériva, M. (2006). Control of cardiac rhythm by ORK1, a Drosophila two-pore domain potassium channel. Curr. Biol. 16, 1502–1508. doi:10.1016/j.cub.2006.05.064
Lee, T. M., Hsu, D. T., Kantor, P., Towbin, J. A., Ware, S. M., Colan, S. D., et al. (2017). Pediatric cardiomyopathies. Circ. Res. 121, 855–873. doi:10.1161/circresaha.116.309386
Lescroart, F., Wang, X., Lin, X., Swedlund, B., Gargouri, S., Sanchez-Danes, A., et al. (2018). Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science 359, 1177–1181. doi:10.1126/science.aao4174
Li, D., Sun, J., and Zhong, T. P. (2022a). Wnt signaling in heart development and regeneration. Curr. Cardiol. Rep. 24, 1425–1438. doi:10.1007/s11886-022-01756-8
Li, G., Tian, L., Goodyer, W., Kort, E. J., Buikema, J. W., Xu, A., et al. (2019a). Single cell expression analysis reveals anatomical and cell cycle-dependent transcriptional shifts during heart development. Development 146, dev173476. doi:10.1242/dev.173476
Li, G., Xu, A., Sim, S., Priest, J. R., Tian, X., Khan, T., et al. (2016). Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Dev. Cell 39, 491–507. doi:10.1016/j.devcel.2016.10.014
Li, H., Janssens, J., De Waegeneer, M., Kolluru, S. S., Davie, K., Gardeux, V., et al. (2022b). Fly cell atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Science 375, eabk2432. doi:10.1126/science.abk2432
Li, Z., Solomonidis, E. G., Meloni, M., Taylor, R. S., Duffin, R., Dobie, R., et al. (2019b). Single-cell transcriptome analyses reveal novel targets modulating cardiac neovascularization by resident endothelial cells following myocardial infarction. Eur. Heart J. 40, 2507–2520. doi:10.1093/eurheartj/ehz305
Liguori, F., Mascolo, E., and Vernì, F. (2021). The genetics of diabetes: What we can learn from Drosophila. Int. J. Mol. Sci. 22, 11295. doi:10.3390/ijms222011295
Litvinukova, M., Talavera-Lopez, C., Maatz, H., Reichart, D., Worth, C. L., Lindberg, E. L., et al. (2020). Cells of the adult human heart. Nature 588, 466–472. doi:10.1038/s41586-020-2797-4
Luong, N., Davies, C. R., Wessells, R. J., Graham, S. M., King, M. T., Veech, R., et al. (2006). Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 4, 133–142. doi:10.1016/j.cmet.2006.05.013
Lyons, I., Parsons, L. M., Hartley, L., Li, R., Andrews, J. E., Robb, L., et al. (1995). Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 9, 1654–1666. doi:10.1101/gad.9.13.1654
Ma, L., Bradu, A., Podoleanu, A. G., and Bloor, J. W. (2010). Arrhythmia caused by a Drosophila tropomyosin mutation is revealed using a novel optical coherence tomography instrument. PLoS One 5, e14348. doi:10.1371/journal.pone.0014348
Manivannan, S. N., Darouich, S., Masmoudi, A., Gordon, D., Zender, G., Han, Z., et al. (2020). Novel frameshift variant in MYL2 reveals molecular differences between dominant and recessive forms of hypertrophic cardiomyopathy. PLoS Genet. 16, e1008639. doi:10.1371/journal.pgen.1008639
McElhinney, D. B., Geiger, E., Blinder, J., Benson, D. W., and Goldmuntz, E. (2004). NKX2.5 mutations in patients with congenital heart disease. Acc. Curr. J. Rev. 13, 62–63. doi:10.1016/j.accreview.2003.12.072
Migunova, E., Theophilopoulos, J., Mercadante, M., Men, J., Zhou, C., and Dubrovsky, E. B. (2021). ELAC2/RNaseZ-linked cardiac hypertrophy in Drosophila melanogaster. Dis. Model. Mech. 14, dmm048931. doi:10.1242/dmm.048931
Na, J., Musselman, L. P., Pendse, J., Baranski, T. J., Bodmer, R., Ocorr, K., et al. (2013). A Drosophila model of high sugar diet-induced cardiomyopathy. PLoS Genet. 9, e1003175. doi:10.1371/journal.pgen.1003175
Neely, G. G., Kuba, K., Cammarato, A., Isobe, K., Amann, S., Zhang, L., et al. (2010). A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell 141, 142–153. doi:10.1016/j.cell.2010.02.023
Ocorr, K., Reeves, N. L., Wessells, R. J., Fink, M., Chen, H.-S. V., Akasaka, T., et al. (2007). KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc. Natl. Acad. Sci. U. S. A. 104, 3943–3948. doi:10.1073/pnas.0609278104
Olson, E. N. (2006). Gene regulatory networks in the evolution and development of the heart. Science 313, 1922–1927. doi:10.1126/science.1132292
Palandri, A., Martin, E., Russi, M., Rera, M., Tricoire, H., and Monnier, V. (2018). Identification of cardioprotective drugs by medium-scale in vivo pharmacological screening on a Drosophila cardiac model of Friedreich’s ataxia. Dis. Model. Mech. 11, dmm033811. doi:10.1242/dmm.033811
Papadopoli, D., Boulay, K., Kazak, L., Pollak, M., Mallette, F., Topisirovic, I., et al. (2019). mTOR as a central regulator of lifespan and aging. F1000Res 8. doi:10.12688/f1000research.17196.1
Papaefthmiou, C., and Theophilidis, G. (2001). An in vitro method for recording the electrical activity of the isolated heart of the adult Drosophila melanogaster. Vitro Cell. Dev. Biol. Anim. 37, 445–449. doi:10.1290/1071-2690(2001)037<0445:aivmfr>2.0.co;2
Pashmforoush, M., Lu, J. T., Chen, H., Amand, T. S., Kondo, R., Pradervand, S., et al. (2004). Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 117, 373–386. doi:10.1016/s0092-8674(04)00405-2
Paternostro, G., Vignola, C., Bartsch, D.-U., Omens, J. H., McCulloch, A. D., and Reed, J. C. (2001). Age-associated cardiac dysfunction in Drosophila melanogaster. Circ. Res. 88, 1053–1058. doi:10.1161/hh1001.090857
Petersen, C. E., Tripoli, B. A., Schoborg, T. A., and Smyth, J. T. (2022). Analysis of Drosophila cardiac hypertrophy by microcomputerized tomography for genetic dissection of heart growth mechanisms. Am. J. Physiology-Heart Circulatory Physiology 322, H296–H309. doi:10.1152/ajpheart.00387.2021
Piché, M.-E., Tchernof, A., and Després, J.-P. (2020). Obesity phenotypes, diabetes, and cardiovascular diseases. Circ. Res. 126, 1477–1500. doi:10.1161/CIRCRESAHA.120.316101
Pierpont, M. E., Brueckner, M., Chung, W. K., Garg, V., Lacro, R. V., McGuire, A. L., et al. (2018). Genetic basis for congenital heart disease: Revisited: A scientific statement from the American heart association. Circulation 138, e653–e711. doi:10.1161/CIR.0000000000000606
Posey, J. E., O’Donnell-Luria, A. H., Chong, J. X., Harel, T., Jhangiani, S. N., Coban Akdemir, Z. H., et al. Centers for Mendelian Genomics (2019). Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet. Med. 21, 798–812. doi:10.1038/s41436-018-0408-7
Qian, L., and Bodmer, R. (2012). Probing the polygenic basis of cardiomyopathies in Drosophila. J. Cell. Mol. Med. 16, 972–977. doi:10.1111/j.1582-4934.2012.01529.x
Qian, L., Mohapatra, B., Akasaka, T., Liu, J., Ocorr, K., Towbin, J. A., et al. (2008). Transcription factor neuromancer/TBX20 is required for cardiac function in Drosophila with implications for human heart disease. Proc. Natl. Acad. Sci. U. S. A. 105, 19833–19838. doi:10.1073/pnas.0808705105
Richard, P., Villard, E., Charron, P., and Isnard, R. (2006). The genetic bases of cardiomyopathies. J. Am. Coll. Cardiol. 48, A79–A89. doi:10.1016/j.jacc.2006.09.014
Rotstein, B., and Paululat, A. (2016). On the morphology of the Drosophila heart. J. Cardiovasc. Dev. Dis. 3, 15. doi:10.3390/jcdd3020015
Schroeder, A. M., Allahyari, M., Vogler, G., Missinato, M. A., Nielsen, T., Yu, M. S., et al. (2019). Model system identification of novel congenital heart disease gene candidates: Focus on RPL13. Hum. Mol. Genet. 28, 3954–3969. doi:10.1093/hmg/ddz213
Sifrim, A., Hitz, M.-P., Wilsdon, A., Breckpot, J., Turki, S. H. A., Thienpont, B., et al. (2016). Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat. Genet. 48, 1060–1065. doi:10.1038/ng.3627
Skelly, D. A., Squiers, G. T., McLellan, M. A., Bolisetty, M. T., Robson, P., Rosenthal, N. A., et al. (2018). Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 22, 600–610. doi:10.1016/j.celrep.2017.12.072
Souidi, A., and Jagla, K. (2021). Drosophila heart as a model for cardiac development and diseases. Cells 10. doi:10.3390/cells10113078
Taghli-Lamallem, O., Akasaka, T., Hogg, G., Nudel, U., Yaffe, D., Chamberlain, J. S., et al. (2008). Dystrophin deficiency in Drosophila reduces lifespan and causes a dilated cardiomyopathy phenotype. Aging Cell 7, 237–249. doi:10.1111/j.1474-9726.2008.00367.x
Tanaka, M., Chen, Z., Bartunkova, S., Yamasaki, N., and Izumo, S. (1999). The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development 126, 1269–1280. doi:10.1242/dev.126.6.1269
Tögel, M., Pass, G., and Paululat, A. (2013). In vivo imaging of Drosophila wing heart development during pupal stages. Int. J. Dev. Biol. 57, 13–24. doi:10.1387/ijdb.120111ap
Tögel, M., Pass, G., and Paululat, A. (2008). The Drosophila wing hearts originate from pericardial cells and are essential for wing maturation. Dev. Biol. 318, 29–37. doi:10.1016/j.ydbio.2008.02.043
Tucker, N. R., Chaffin, M., Fleming, S. J., Hall, A. W., Parsons, V. A., Bedi, K. C., et al. (2020). Transcriptional and cellular diversity of the human heart. Circulation 2020. doi:10.1161/CIRCULATIONAHA.119.045401
Ugur, B., Chen, K., and Bellen, H. J. (2016). Drosophila tools and assays for the study of human diseases. Dis. Model. Mech. 9, 235–244. doi:10.1242/dmm.023762
Verheije, R., Kupchik, G. S., Isidor, B., Kroes, H. Y., Lynch, S. A., Hawkes, L., et al. (2019). Heterozygous loss-of-function variants of MEIS2 cause a triad of palatal defects, congenital heart defects, and intellectual disability. Eur. J. Hum. Genet. 27, 278–290. doi:10.1038/s41431-018-0281-5
Wang, W., Niu, X., Stuart, T., Jullian, E., Mauck, W. M., Kelly, R. G., et al. (2019). A single-cell transcriptional roadmap for cardiopharyngeal fate diversification. Nat. Cell Biol. 21, 674–686. doi:10.1038/s41556-019-0336-z
Ware, S. M., Wilkinson, J. D., Tariq, M., Schubert, J. A., Sridhar, A., Colan, S. D., et al. (2021). Genetic causes of cardiomyopathy in children: First results from the pediatric cardiomyopathy genes study. J. Am. Heart Assoc. 10, e017731. doi:10.1161/JAHA.120.017731
Weinberger, M., Simoes, F. C., Patient, R., Sauka-Spengler, T., and Riley, P. R. (2020). Functional heterogeneity within the developing zebrafish epicardium. Dev. Cell 52, 574–590. e6. doi:10.1016/j.devcel.2020.01.023
Wessells, R., Fitzgerald, E., Piazza, N., Ocorr, K., Morley, S., Davies, C., et al. (2009). d4eBP acts downstream of both dTOR and dFoxo to modulate cardiac functional aging in Drosophila. Aging Cell 8, 542–552. doi:10.1111/j.1474-9726.2009.00504.x
Wessells, R. J., Fitzgerald, E., Cypser, J. R., Tatar, M., and Bodmer, R. (2004). Insulin regulation of heart function in aging fruit flies. Nat. Genet. 36, 1275–1281. doi:10.1038/ng1476
Wolf, M. J., Amrein, H., Izatt, J. A., Choma, M. A., Reedy, M. C., and Rockman, H. A. (2006). Drosophila as a model for the identification of genes causing adult human heart disease. Proc. Natl. Acad. Sci. U. S. A. 103, 1394–1399. doi:10.1073/pnas.0507359103
Wolf, M. J., and Rockman, H. A. (2008). Drosophila melanogaster as a model system for genetics of postnatal cardiac function. Drug Discov. Today Dis. Models 5, 117–123. doi:10.1016/j.ddmod.2009.02.002
Yu, L., Daniels, J., Glaser, A. E., and Wolf, M. J. (2013). Raf-mediated cardiac hypertrophy in adult Drosophila. Dis. Model. Mech. 6, 964–976. doi:10.1242/dmm.011361
Yu, L., Lee, T., Lin, N., and Wolf, M. J. (2010). Affecting Rhomboid-3 function causes a dilated heart in adult Drosophila. PLoS Genet. 6, e1000969. doi:10.1371/journal.pgen.1000969
Yuan, X., Song, M., Devine, P., Bruneau, B. G., Scott, I. C., and Wilson, M. D. (2018). Heart enhancers with deeply conserved regulatory activity are established early in zebrafish development. Nat. Commun. 9, 4977. doi:10.1038/s41467-018-07451-z
Zaidi, S., Choi, M., Wakimoto, H., Ma, L., Jiang, J., Overton, J. D., et al. (2013). De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220–223. doi:10.1038/nature12141
Zhao, Y., Lindberg, B. G., Esfahani, S. S., Tang, X., Piazza, S., and Engström, Y. (2021). Stop codon readthrough alters the activity of a POU/Oct transcription factor during Drosophila development. BMC Biol. 19, 185. doi:10.1186/s12915-021-01106-0
Zhu, J.-Y., Fu, Y., Nettleton, M., Richman, A., and Han, Z. (2017a). High throughput in vivo functional validation of candidate congenital heart disease genes in Drosophila. Elife 6, e22617. doi:10.7554/eLife.22617
Keywords: congenital heart disease, hypertrophic cardiomyopathy, cardiac arrhythmia, Drosophila, cardiac conduction, cardiac physiology
Citation: Zhao Y, van de Leemput J and Han Z (2023) The opportunities and challenges of using Drosophila to model human cardiac diseases. Front. Physiol. 14:1182610. doi: 10.3389/fphys.2023.1182610
Received: 09 March 2023; Accepted: 05 April 2023;
Published: 12 April 2023.
Edited by:
Jian Xu, East China Normal University, ChinaReviewed by:
Daniel F. Eberl, The University of Iowa, United StatesCopyright © 2023 Zhao, van de Leemput and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhe Han, emhhbkBzb20udW1hcnlsYW5kLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.