
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 10 April 2023
Sec. Invertebrate Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1177297
This article is part of the Research TopicApplied Bioinformatics in Insect PhysiologyView all 5 articles
Chemosensation of tarsi provides moths with the ability to detect chemical signals which are important for food recognition. However, molecular mechanisms underlying the chemosensory roles of tarsi are still unknown. The fall armyworm Spodoptera frugiperda is a serious moth pest that can damage many plants worldwide. In the current study, we conducted transcriptome sequencing with total RNA extracted from S. frugiperda tarsi. Through sequence assembly and gene annotation, 23 odorant receptors 10 gustatory receptors and 10 inotropic receptors (IRs) were identified. Further phylogenetic analysis with these genes and homologs from other insect species indicated specific genes, including ORco, carbon dioxide receptors, fructose receptor, IR co-receptors, and sugar receptors were expressed in the tarsi of S. frugiperda. Expression profiling with RT-qPCR in different tissues of adult S. frugiperda showed that most annotated SfruORs and SfruIRs were mainly expressed in the antennae, and most SfruGRs were mainly expressed in the proboscises. However, SfruOR30, SfruGR9, SfruIR60a, SfruIR64a, SfruIR75d, and SfruIR76b were also highly enriched in the tarsi of S. frugiperda. Especially SfruGR9, the putative fructose receptor, was predominantly expressed in the tarsi, and with its levels significantly higher in the female tarsi than in the male ones. Moreover, SfruIR60a was also found to be expressed with higher levels in the tarsi than in other tissues. This study not only improves our insight into the tarsal chemoreception systems of S. frugiperda but also provides useful information for further functional studies of chemosensory receptors in S. frugiperda tarsi.
Daily living and reproductive behaviors of insects, such as feeding, mating, host-seeking, oviposition, and avoiding predators, rely on the ability of chemoreception (Dahanukar et al., 2005). Chemoreception, mainly includes olfaction and gustation, refers to the sensing of chemical cues from the external environment. Of which, olfaction means the detection of airborne chemical molecules, such as host plant volatiles and sex pheromones (Carey and Carlson, 2011; Zhang et al., 2014; He et al., 2022), whereas gustation refers to the detection of contact chemicals, such as amino acids, bitter and sweet tastants (Chapman, 2003; Depetris-Chauvin et al., 2015).
When the insect has landed on plants, it subsequently taps the leaves with their tarsi, detects the chemicals on the surface of the plants, and thus directs its food seeking behaviors. Insect tarsal chemosensilla are proposed to be involved in this process through sensing of food chemicals (Blaney and Simmonds, 1990; Maher and Thiery, 2004; Gaaboub et al., 2005). The chemoreceptive function of tarsal chemosensilla in detecting various food chemicals had been demonstrated in many noctuid species. For example, specific chemosensilla on the tarsi of autumn gum moth Mnesampela privata had been found to be sensitive to salts, sugars, and amino acids (Calas et al., 2009). The fifth tarsomere of the forelegs of female Helicoverpa armigera possesses 14 gustatory chemosensilla which can be robustly provoked by sugars (i.e., maltose, sucrose, fructose, and glucose), lysine, and myo-inositol (Zhang et al., 2010).
Insect chemosensory receptors, including the odorant receptors (ORs), the gustatory receptors (GRs), and the ionotropic receptors (IRs), are expressed on the dendrite membrane of the chemosensory neurons (CSNs) that located within the chemosensilla. Chemosensory receptors could selectively recognize external ligands and are the primary determinants of the detection spectrum of chemosensilla (Lu et al., 2007; Leal, 2013; Li and Liberles, 2015). Researches on insect ORs, GRs, and IRs were all firstly launched in Drosophila melanogaster (Gao and Chess, 1999; Clyne et al., 2000; Benton et al., 2009). In the last 20 years, along with the great advance of molecular biological techniques, the study of chemosensory receptors has been unfolded in a variety of insect species (Benton et al., 2006; Wilson, 2013; Gadenne et al., 2016; Paoli and Galizia, 2021). Insect ORs are proposed to function as non-selective cation channels via heterodimerization with a subunit called ORco which is highly conserved among species (Sato, et al., 2008; Butterwick et al., 2018). Ligand spectra of OR repertoires have been studied in several species, such as D. melanogaster (Hallem and Carlson, 2006), Anopheles gambiae (Carey et al., 2010; Wang et al., 2010), Spodoptera littoralis (de Fouchier et al., 2017), and H. armigera (Guo et al., 2021). A great number of moth-ORs in the detection of sex pheromones have also been characterized, and these ORs were specially called pheromone receptors (PRs) (Zhang et al., 2014; Yang and Wang, 2020). Insect GRs are mainly located in taste organs, mediate the perception of non-volatile chemical cues and CO2 (Scott et al., 2001; Jones et al., 2007). Insect GRs can act independently or as heteromultimers, depending on the gene types (Sato et al., 2011; Xu et al., 2016). Insect IRs constitute an evolutionary distinct family of chemosensory receptors. Members in this family are three transmembrane proteins homologous to ionotropic glutamate receptors but have a divergent ligand-binding domain (Benton et al., 2009). Insect IRs can be mainly divided into two classes. “Antennal IRs” are usually expressed in the antennae and are relatively conserved among species. “Divergent IRs” appear to exist in different tissues, and their types are variable among species (Croset et al., 2010). Insect IRs are supposed to function as ion channels, and form heterodimers with one or more conserved co-receptors (i.e., IR8a, IR25a, IR76b, and IR93a) (Abuin et al., 2011; Silbering et al., 2011). IRs are demonstrated to be involved in food detection (Koh et al., 2014; Ganguly et al., 2017), acid or amine sensing (Ai et al., 2013; Hussain et al., 2018), and courtship promotion (Grosjean et al., 2011).
The function of specific chemosensory receptors expressed in the tarsal chemosensilla had been investigated in several insect species. For example, 28 GRs were identified in the tarsal sensilla of D. melanogaster and demonstrated to be involved in the recognition of sweet or bitter compounds (Ling et al., 2014). In vitro and in vivo approaches showed that PxutGR1 in tarsal sensilla responds specifically to the stimulant synephrine and represents a key receptor in host specialization in the butterfly Papilio xuthus (Ozaki et al., 2011). Combing ectopia expression as well as behavioral and electrophysiological investigation, PrapGr28 that shows high expression in the female tarsi of Pieris rapae is demonstrated to be the receptor that detects the bitter compound sinigrin (Yang et al., 2021). However, systematic identification of chemosensory receptors in the tarsi of Lepidoptera is relatively limited. In a recent study, 18 ORs, 9 GRs, and 7 IRs were annotated in the tarsal transcriptome of the Noctuidae species Helicoverpa zea (Dou et al., 2019). To provide more useful information for further research on molecular mechanisms underlying the food detection in moths, analyses of chemosensory receptors in the tarsi of more species are still needed.
Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae) is an important agricultural pest of many crops, such as maize, rice, cotton, sorghum, and sugar cane (Sparks, 1979; Meagher and Nagoshi, 2012). It is pandemic throughout the North and South America and making a significant threat in sub-Saharan Africa (Nagoshi et al., 2018) and Asia (Wu et al., 2019). In behavioural assays, adult S. frugiperda females of which tarsi stimulated with a range of sugars, amino acids, allelochemics, and plant extracts showed proboscis extension reflexes. Further electrophysiological recordings from tarsal chemosensilla identified different neurons responsive to sugars, amino acids, and allelochemicals. And three neurons were found to be responsive to the plant extracts (Blaney and Simmonds, 1990). However, chemosensory mechanisms concerning such electrophysiological and behavioral reactions are still unknown.
In this study, we conducted systematic transcriptome analysis of male and female S. frugiperda tarsi. A total of 43 chemoreceptor genes, including 23 ORs, 10 GRs, and 10 IRs were identified. Phylogenetic relationships of the candidate genes with chemoreceptors from other insect species were then analyzed to infer the putative functions of these genes. Finally, expression profiles of the candidate genes in different chemosensory tissues of adult S. frugiperda were investigated. Some chemosensory receptors that were highly expressed in the tarsi were detected. These findings give us useful information for further chemosensory mechanism investigation in the tarsi of S. frugiperda as well as in the tarsi of other moth species.
S. frugiperda larvae were originally collected from the corn field in Shidian county, Baoshan, Yunnan Province, China, and successive generations were maintained in the laboratory under constant conditions of 27°C ± 1°C, 70%–75% relative humidity, and a 16 h: 8 h light/dark photoperiod. The larvae were fed with artificial diets (Guo et al., 2022). Adults were provided unlimited access to 10% (v/v) honey solutions.
For transcriptome analysis, tarsi of 80 three-day-old virgin male and female moths were collected separately. For RT-qPCR measurements, tissues including male and female antennae, proboscises, tarsi, and female ovipositor (together with the pheromone gland) were collected from 50 to 80 three-day-old virginal moths. All collected samples were immersed in liquid nitrogen until the total RNA extraction.
Total RNA was extracted from the tarsi of S. frugiperda using Trizol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. The RNA concentration was determined with an ND-2000 spectrophotometer (Nanodrop, Wilmington, DE, United States). cDNA libraries were constructed following our previously described protocols (Sun et al., 2020), and then sequenced on an Illumina HiSeq2000 platform (Illumina, United States) at Sangon Biotech (Shanghai, China) Co., Ltd. De novo assembly and annotation of unigenes were performed as follows: the raw reads were firstly processed for removing the adapter sequences and low-quality bases so as to obtain the clean reads (Bolger et al., 2014). The GC-content and Q30 values were calculated to assess the sequence quality. The clean reads were then trimmed using Trimmomatic-0.30, and then merged and assembled using Trinity (. 2.4.0). The Trinity outputs were clustered by tgicl, and then capped using Cap3 to made a collection of unigenes. The unigenes were then searched (E-values < 1e-5) for homologous sequences using BLAST tool in NCBI Nr (non-redundant) database, and the COG (Clusters of Orthologous Groups of proteins), Swiss-Prot, PFAM, KEGG (Kyoto encyclopedia of genes and genomes), and GO (gene ontology) databases. The identified putative chemosensory receptor genes were manually verified by BLASTx searching against the NCBI Nr database. The open reading frames of the putative genes were predicted by translate (https://web.expasy.org/translate/). Expression levels of candidate genes in the tarsi of S. frugiperda were estimated by the TPM (transcripts per kilobase of exon per million mapped) values using RSEM (http://deweylab.github.io/RSEM/).
Phylogenetic analyses were caried out based on the amino sequences of the candidate genes and homologs of other insect species. The OR and GR sequences from S. frugiperda (this study), Bombyx mori, and H. armigera; the IR sequences from S. frugiperda (this study), H. armigera, and D. melanogaster were applied in the phylogenetic analysis, respectively. Detailed methods for constructing the neighbor-joining trees were similar as we previously descried (Sun et al., 2020). The amino acid sequences used for tree building are listed in Supplementary Table S1. Node support was assessed using a bootstrap procedure based on 1,000 replicates. Constructed phylogenetic trees were colored with Figtree software (v1.4.2).
Total RNA was extracted from different tissue samples of S. frugiperda using Trizol reagent according to the manufacturer’s protocols. cDNA was then synthesized with M-MLV reverse transcriptase (Promega, Madison, WI, United States), following the manufacturer’s instructions. RT-qPCR reactions were carried out on Agilent qPCR System (Mx3005P, Agilent Technologies, CA, United States). The primer sequences which were designed with Primer Premier 5.0 software are listed in Supplementary Table S2. All reactions were performed (for three times) in a total volume of 20 µL containing 10 µL SYBR Premix Ex TaqⅡ (Takara, Dalian, China), 0.4 mM of each primer, and 2.5 ng of sample cDNA under the following conditions: 1 cycle of 94°C for 30 s; 40 cycles of 94°C for 5 s, 58°C for 30 s, and 72°C for 25 s; followed by 1 cycle of 94°C for 15 s, 60°C for 1 min, 94°C for 15 s. The 2−ΔΔCT method was applied to calculate the expression levels of candidate genes (Schmittgen and Livak, 2008). β-actin gene of S. frugiperda was chosen as the internal control to normalize expression levels. Amplification curves (S-shaped) and CT values (ranging from 17.2 to 18.3) for the reactions of the β-actin gene were carefully checked to make sure it is consistent across different tissues. The primer efficiency was evaluated by standard amplification curves constructed with 10-fold dilutions of cDNA samples. The efficiency percentage (90–110%) was validated within the acceptable range. All experiments were repeated three times using three independent RNA samples. Figures were made using GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA). The data were analyzed by ANOVA followed by Tukey’s multiple comparison test, and all figures were made in GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA, United States). The level of significance was set at p < 0.05.
A total of 48.25 million and 57.31 million clean reads (mean length 141 bp) were produced from the male and female tarsi samples, respectively. The Q30 bases ratio in each sample was ≥93.23%. All clean reads from both samples were assembled and generated a total of 100,048 unigenes. According to the size distribution analysis result, 14.55% of the unigenes were ≥1,000 bp. The N50 length of the unigenes was 968 bp and the mean length of the unigenes was 670.79 bp (Table 1). All the obtained unigenes were then aligned to Nr, KEGG, Swiss-Prot, PFAM, COG, and GO databases. Base on the BLASTx results, 56.01% (56037) of the unigenes had matches in at least one database. Among the unigenes (27,918) that annotated to the Nr database, 84.34% (23,547) had best hits to sequences from lepidopteran species (Supplementary Figure S1).
Blast2GO was then used to classify the genes into functional categories. Among the 100,048 unigenes, 31,635 (31.62%) were assigned to at least one GO group. Among which, 30,284 were allocated to the biological process group, 17,798 to the cellular component group, and 15,597 to the molecular function group. The most enriched terms were “binding” in the molecular function, “cell” in the biological process, and “metabolic process” in the biological process (Supplementary Figure S2).
Based on the Blastx results, a total of 23 candidate OR genes (SfruORs) were identified in the transcriptome of S. frugiperda tarsi. Twenty-one of these genes have putative full-length ORFs. Details for the 23 SfruORs, including BLASTx best hits, lengths, and sequences are listed in Table 2 and Supplementary Table S3. Among the 23 annotated SfruORs in S. frugiperda tarsi, 12 (SfruOR1/12b/17/25/27/34/38/40/49b/57/62/64, accession numbers QQ442940−QQ442951) have not been reported in previous genome or transcriptome studies of this species (Gouin et al., 2017; Qiu et al., 2020; Sun et al., 2022). To keep consistency, candidate genes in our study were numbered following the names of previously reported sequences in S. frugiperda (if possible) or following the names of the best hit homologs in other lepidopteran species.
To infer the putative functions of the candidate SfruORs in this study, phylogenetic relationships were analyzed based on the alignment with ORs from B. mori and H. armigera. According to the neighbor-joining tree, ORcos were highly conserved and clustered in the ORco branch. Other 22 SfruORs were scatter in different “ordinary ORs” branches, and we did not find the putative “PR” genes in the transcriptome of S. frugiperda tarsi (Figure 1).
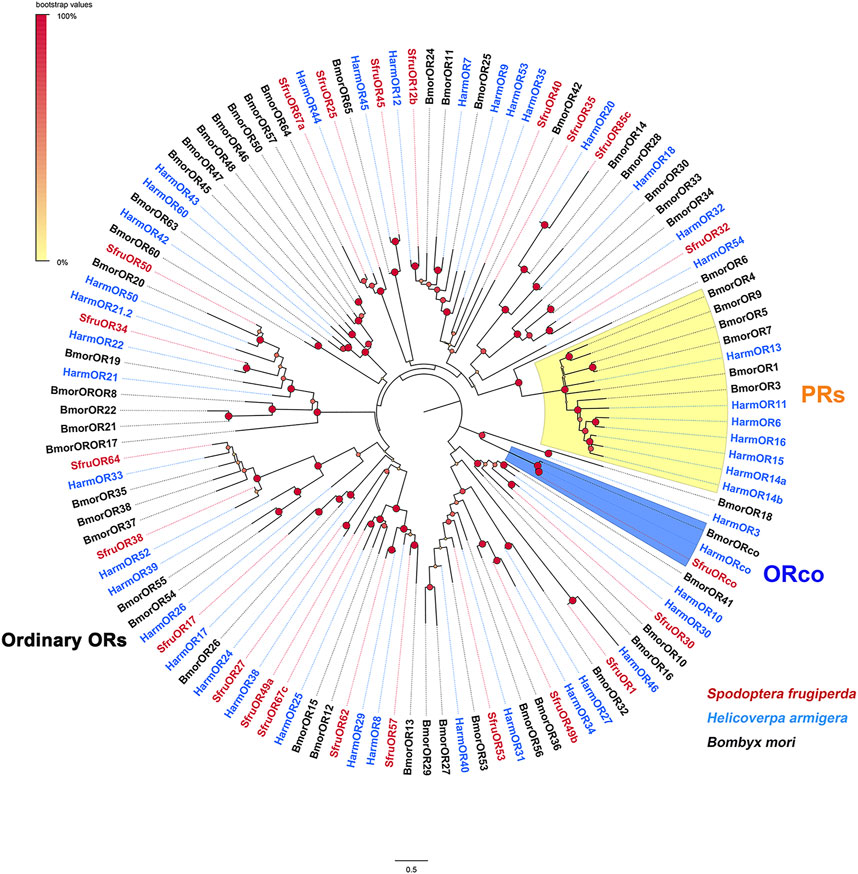
FIGURE 1. Phylogenetic analysis of the candidate SfruORs from the transcriptome of S. frugiperda tarsi and ORs from H. armigera and B. mori.
The expression of the 43 (23 SfruORs,10 SfruGRs, and 10 SfruIRs) candidate receptor genes was then normalized across the transcriptome sequences using the TPM calculation method. For SfruORs, despite the inconsistency of TPM values between females and males, SfruOR30 was the highest transcribed gene (5.03/4.20 TPM, females/males, similarly hereinafter) among all the annotated SfruORs in the tarsi of S. frugiperda. Other two candidate SfruORs, SfruOR17 and SfruOR27, showed high expression levels as indicated by the TPM values (2.35/1.65 for SfruOR17, 0.56/3.05 for SfruOR27). Significantly, transcripts of SfruORco were also detected in the tarsi, although its TPM values were quite low (0.16/0.61) (Figure 2).
RT-qPCR was further performed to study the expression profiles of candidate receptor genes in various chemosensory tissues of adult S. frugiperda, including the antennae, tarsi, proboscises, and ovipositors. As shown in Figure 3, all of the SfruOR genes were mainly expressed in the antennae, and the levels of SfruOR34/45/53/57/64/67c/85c were higher in female antennae than in male ones. The results also showed that SfruOR30 was highly expressed in the tarsi (both sexes), with its levels lower than in the antennae, but higher than in other tested tissues (i.e., proboscises and ovipositors).
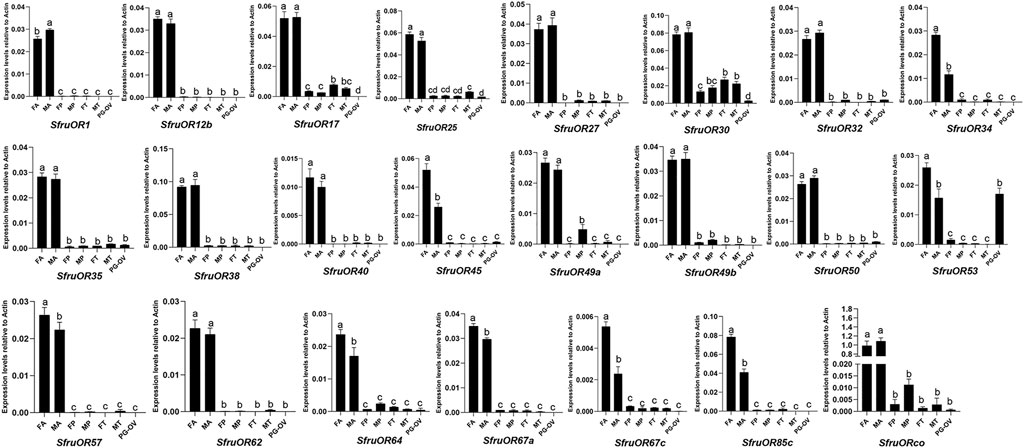
FIGURE 3. Expression patterns of candidate SfruORs in different chemosensory tissues of adult S. frugiperda. FA: female antennae, MA: male antennae, FP: female proboscises, MP: male proboscises, FT: female tarsi, MT: male tarsi, PG-OV: pheromone gland-ovipositor. Different letters indicate significant difference based on a one-way ANOVA followed by Tukey’s multiple comparison test. Error bars show the standard errors of the means (+SE), p < 0.05, n = 3.
Ten putative SfruGRs were identified based on the analysis of the transcriptome of S. frugiperda tarsi. Except for SfruGR6, all the other annotated SfruGRs possess complete ORFs (Table 2). Among the 10 SfruGRs, SfruGR1/2/3/9 have been reported in previous larval antennae and maxilla transcriptome studies of this species (Sun et al., 2022). SfruGR4/5/6//7/8/10, which have best hits to the putative gustatory receptor for sugar taste 64a/e/f-like in genome sequences, have not yet been annotated in other transcriptome data of this species.
A phylogenetic tree was then built with GRs from S. frugiperda (this study), B. mori, and H. armigera. The results showed that SfruGR1, SfruGR2, and SfruGR3, which grouped with BmorGR1/HarmGR1, BmorGR2/HarmGR2, and BmorGR3/HarmGR3 linages, were putative CO2 receptors. SfruGR9, clustered with the BmorGR9, was putative fructose receptor. Other six SfruGRs (SfruGR4/5/6/7/8/10) clustered in the “sugar-taste receptors” groups (Figure 4).
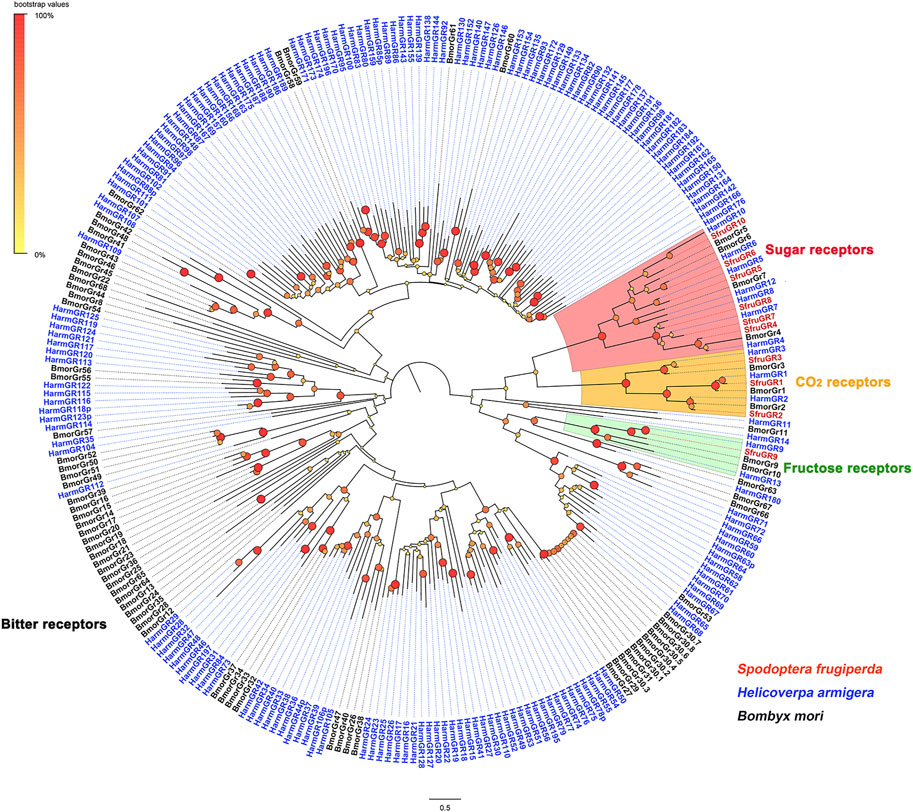
FIGURE 4. Phylogenetic analysis of the candidate SfruGRs from the transcriptome of S. frugiperda tarsi and GRs from H. armigera and B. mori.
According to the TPM values, transcript levels of SfruGR9 in the tarsi were more abundant than that of the other annotated SfruGRs, and with its TPM value higher in the female tarsi (6.49 TPM) than in the male ones (3.83 TPM). Other two sugar-taste receptors, SfruGR6 and SfruGR7, were also highly enriched in the tarsi, and with their transcript levels similar between females and males. In comparison, the two putative CO2 receptors, SfruGR2 (0/0.02) and SfruGR3 (0.16/0) have the lowest TPM values in the S. frugiperda tarsi (Figure 5).
Based on RT-qPCR analyses, SfruGR9 showed significantly higher expression levels in the tarsi than in other tissues, and its expression in female tarsi was more abundant than in the male ones. This is accordant with the expression profiles of SfruGR9 as that reported by Sun et al., 2022. Eight SfruGR genes, SfruGR1/2/3/5/6/7/8/10, were mainly expressed in the proboscises of both sexes, among which, SfruGR10 was also highly expressed in the tarsi of both sexes. On contrast, SfruGR4 was mainly expressed in the antennae, and with levels higher in the female moths than in the male ones (Figure 6).
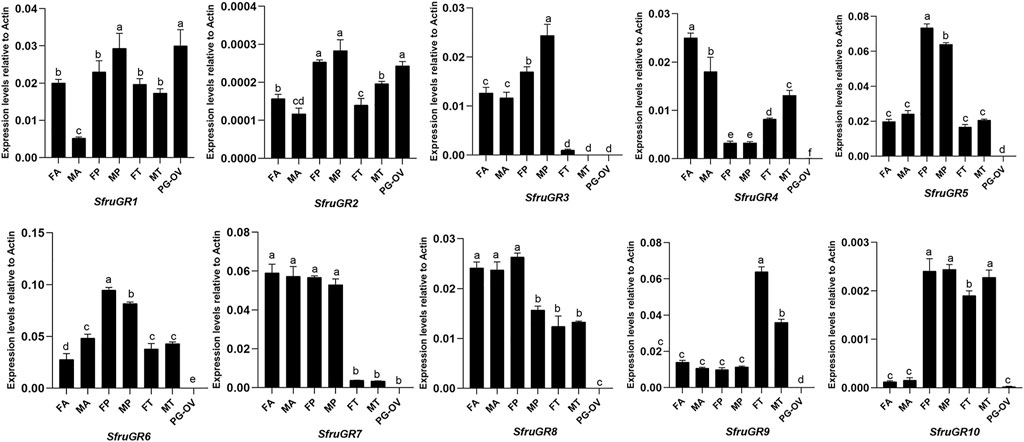
FIGURE 6. Expression patterns of candidate SfruGRs in different chemosensory tissues of adult S. frugiperda. FA: female antennae, MA: male antennae, FP: female proboscises, MP: male proboscises, FT: female tarsi, MT: male tarsi, PG-OV: female pheromone gland-ovipositor. Different letters indicate significant difference based on a one-way ANOVA followed by Tukey’s multiple comparison test. Error bars show the standard errors of the means (+SE), p < 0.05, n = 3.
We annotated 10 SfruIRs in the transcriptome of S. frugiperda tarsi. All of these candidate IRs were identified possessing complete ORFs (Table 2, Supplementary Table S3). Based on the Blastx results, sequences of 9 SfruIRs have been uploaded to the NCBI database previously, except for SfruIR8a (accession number OQ442939), which has not been reported in previous genome or transcriptome studies of this species (Gouin et al., 2017; Qiu et al., 2020; Sun et al., 2022).
A neighbor-joining tree was then constructed to predict evolutionary relationships between SfruIRs (this study) and IRs from D. melanogaster and H. armigera. Predictably, all of the four putative SfruIR co-receptors, SfruIR8a, SfruIR25a, SfruIR76b, and SfruIR93a, clustered in the co-receptor lineages of IR8a, IR25a, IR76b, and IR93a, respectively. The other 6 SfruIRs (SfruIR21a/60a/64a/75a/75d/75p) were assigned to the “antennal IR” clade (Figure 7).
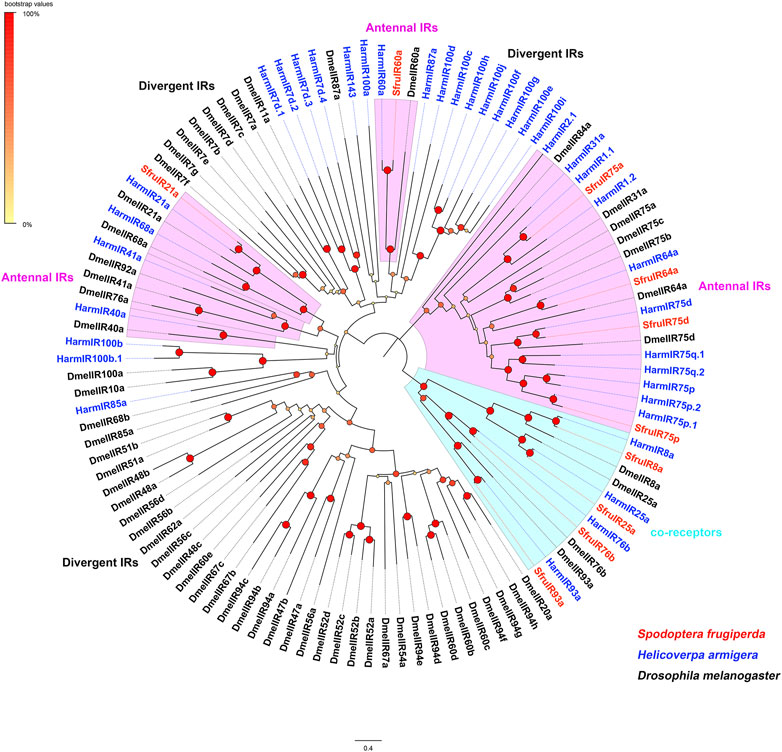
FIGURE 7. Phylogenetic analysis of the candidate SfruIRs from the transcriptome of S. frugiperda tarsi and IRs from H. armigera and D. melanogaster.
TPM value analysis indicated that the “antennal IR”, SfruIR60a, had the highest transcript levels (13.25/19.21 TPM) in the S. frugiperda tarsi among all the annotated SfruIRs. Another “antennal IR”, SfruIR75d, was also abundantly transcribed in the tarsi (4.3/3.76 TPM). Among the 4 annotated co-receptors, SfruIR76b was the most enriched one in the tarsi (8.5/10.43 TPM). Other two co-receptors, SfruIR8a and SfruIR93a, showed quite low values of 0.02/0 TPM and 0.03/0.19 TPM, respectively (Figure 8).
According to the expression profiles of SfruIRs in different chemosensory tissues, 9 of the annotated SfruIR genes were mainly expressed in the antennae, especially SfruIR8a, SfruIR75a, SfruIR75p, and SfruIR93a, were almost exclusively expressed in the antennae of both sexes. In comparison, the expression of SfruIR60a, SfruIR64a and SfruIR75d was detected in all of the tested tissues. Notably, the expression level of SfruIR60a was higher in the tarsi than in other tissues. Moreover, although the expression levels of SfruIR64a and SfruIR75d were much lower in the tarsi than in the antennae, they were similarly (SfruIR64a) or even higher (SfruIR75d) expressed in the tarsi comparing to that in the other tissues (Figure 9).
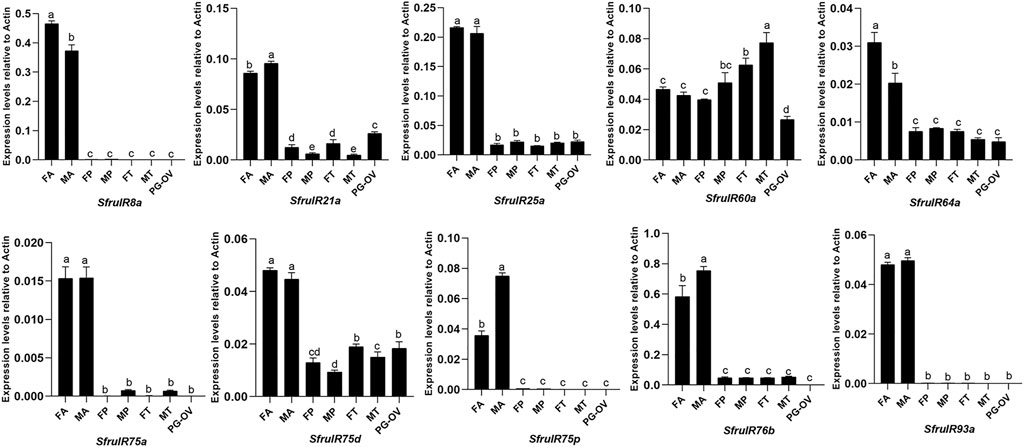
FIGURE 9. Expression patterns of candidate SfruIRs in different chemosensory tissues of adult S. frugiperda. FA: female antennae, MA: male antennae, FP: female proboscises, MP: male proboscises, FT: female tarsi, MT: male tarsi, PG-OV: female pheromone gland-ovipositor. Different letters indicate significant difference based on a one-way ANOVA followed by Tukey’s multiple comparison test. Error bars show the standard errors of the means (+SE), p < 0.05, n = 3.
Insect tarsi are important for the feeding behaviors. S. frugiperda is a worldwide pest which has potential to cause severe damage to more than 300 crop species. Molecular mechanisms underlying the chemosensation of S. frugiperda tarsi are still unknown. In the current study, we identified 23 ORs, 10 GRs, and 10 IRs in the tarsal transcriptome of S. frugiperda. This is more than that reported in the tarsi of H. zea (18 ORs, 9 GRs, and 7 IRs) (Dou et al., 2019), which provides high confidence in the quality of the transcriptome sequencing.
ORs are the vital factor in peripheral olfactory reception in insects (Leal, 2013). A total of 23 SfruORs were identified in our research. Notably, a trace of ORco was documented in the tarsi of S. frugiperda. Such expression profile may reflect the olfaction roles of the S. frugiperda tarsi. Furthermore, SfruORco was demonstrated to have the highest expression level in the antennae among all the annotated SfruORs. This is consistent with the previous suggestion that ORco is the most highly expressed ORs in antennae (Jones et al., 2005; Sun et al., 2020). The SfruORs that mainly expressed in the antennae may be involved in the olfaction of S. frugiperda. For example, HarmOR29, an ortholog of SfruOR62 (identity: 85.44%), mainly expressed in the antennae of H. armigera, was found to be triggered by methyl 4-hydroxybenzoate when heterologously expressed in Drosophila empty neurons (Guo et al., 2021). Although most of the candidate SfruORs are extensively expressed in the antennae, SfruOR30 and SfruOR53 was also found to be expressed in the tarsi or ovipositor (with lower level than in the antennae). We suggested that these 2 ORs may participate in the detection of feeding- or oviposition-related chemicals (Yactayo-Chang et al., 2021). According to the expression profile analyses, we did not observe obvious expression of other 21 SfruORs in the tarsi, this could be explained by the fact that OR genes are mainly expressed in the olfactory organ (i.e., antennae), rather than in the gustatory organs (e.g., tarsi).
In this study, we identified 10 GRs in the tarsi of S. frugiperda. By sequence alignment and phylogenetic analysis, these GRs were determined in the “CO2-receptors” sub-family (SfruGR1/2/3) and “sugar-receptors” sub-family (SfruGR4/5/6/7/8/9/10). We did not annotate putative members in the “bitter-taste receptors” clade. Similarly, only 1 bitter-taste receptor was reported in the tarsal transcriptome of H. zea. This may be due to the low transcript levels of most bitter-taste receptors (Xu et al., 2016; Yang et al., 2021). And the third-generation sequencing technique (full-length transcriptome sequencing) could help us to identify more receptors of which transcript levels are relatively low in the tested tissues. Furthermore, as shown in the RT-qPCR, in addition to be expressed in the tarsi, most of the annotated SfruGRs were also highly expressed in the proboscises. It has long been reported that many chemosensilla are distributed on the mouthpart and that the mouthpart functions in chemosensation in moths (Krenn, 2010; Liu et al., 2014; Hu et al., 2021). Therefore, systematic analysis of GRs in other taste organs such as proboscis are still needed in the future.
Phytophagous insects could detect and measure the CO2 gradients so as to direct their oviposition and feeding behaviors. Fresh flowers which produce more CO2 than old flowers indicate more nectars than the old ones (Guerenstein and Hildebrand, 2008). It is demonstrated that specialized CSNs that sense CO2 are distributed on the labial palps in Lepidoptera (Bogner et al., 1986). And 3 GRs (GR1, GR2, and GR3) are responsible for the detection of CO2 (Xu and Anderson, 2015; Ning et al., 2016). In this study, we annotated three CO2 GRs (SfruGR1/2/3) in the tarsi of S. frugiperda. Although moth GRs detecting CO2 have been only characterized in antennae and labial palps (Xu and Anderson, 2015; Ning et al., 2016). The identification of three candidate CO2 receptors in S. frugiperda tarsi indicate that moths may also sense CO2 via their tarsi.
The group of “sugar receptors” in moth GR family is mainly responsible for the detection of various sweet substances. In the transcriptome of S. frugiperda tarsi, we determined 7 GRs in the clade of sugar-taste receptors. This number is close to that reported in the genome (10 putative sugar-taste receptors) of this species (Gouin et al., 2017). And more than that reported (5) in the tarsal transcriptome of H. zea (Dou et al., 2019). Although a lot of studies have been made in unrevealing the function of insect sugar-taste GRs in taste perception, most of them have been carried out in the model insect D. melanogaster (Dahanukar et al., 2007; Moon et al., 2009; Lee et al., 2010; Miyamoto et al., 2012; Dweck and Carlson, 2020). An exception, GR9 homologs in the fructose-receptor clade have been well studied in moths. For example, through heterologous expression, GR9 in B. mori (BmorGR9) has been demonstrated to be sensitive to D-fructose (Sato et al., 2011); GR9 in H. armigera (HarmGR9) shows responses to D-fructose, D-galactose, and D-maltose (Xu et al., 2016); While in another study, HarmGR9 responds specifically to D-fructose (Jiang et al., 2015). In Spodoptera litura, SlitGR8 (ortholog of BmorGR9 and HarmGR9) has a specific response to D-fructose (Liu et al., 2019). In our study, according to the TPM values and RT-qPCR results, SfruGR9 was predominantly expressed in the tarsi (with higher levels than the other SfruGRs), and the level in female tarsi was significantly higher than that in the male ones, suggesting that SfruGR9 is responsible for the fructose detection during feeding and ovipostion in S. frugiperda.
Insects IRs not only contribute to olfaction and gustation, but are also involved in the sensing of temperature, humidity, and sound (Rytz et al., 2013; Knecht et al., 2016; Ni et al., 2016; Pitts et al., 2017; Hou et al., 2022). In this study, we annotated a total of 10 IRs in the tarsi of S. frugiperda. And four of them (SfruIR8a, SfruIR25a, SfruIR76b, and SfruIR93a) were determined in the IR co-receptors clade. RT-qPCR results showed that SfruIR76b was expressed with higher levels in the tarsi than that of the other 3 annotated IR co-receptors. It had been demonstrated that IR76b can function as the co-receptor of specific IRs tuned to amino acids in D. melanogaster (Ganguly et al., 2017). The participation of SfruIR76b in the sensing of amino acids in S. frugiperda tarsi remains to be elucidated. According to the phylogenetic analysis, the other 6 SfruIRs (SfruIR21a/60a/64a/75a/75d/75p) were grouped in the “antennal IRs”. Consistent with that reported in other insect species (Liu et al., 2018; Zhu et al., 2018), most of the annotated “antennal IRs” were mainly expressed in the antennae of S. frugiperda. However, SfruIR60a was highly expressed in all the tested tissues, and with its levels even higher in the tarsi than in other tissues. Similarly, expression investigation of HarmIR60a in olfactory and taste tissues of H. armigera demonstrated that HarmIR60a is expressed in antennae, proboscises, and legs (Liu et al., 2018). It had been found that IR60a group shared higher sequence similarities with divergent-IRs compared to antennal-IRs (Olivier et al., 2011). In our study, SfruIR60a is present between antennal-IRs and divergent-IRs, which is consistent with that reported in other moths (Liu et al., 2018; Hou et al., 2022). Thus, we speculated that SfruIR60a may bear dual roles of taste and smell in the tarsi of S. frugiperda. Although “divergent IRs” were found to be the largest group in D. melanogaster (Croset et al., 2010), In the current study, we did not identify members that belonged to the “divergent IRs” in the tarsal transcriptome of S. frugiperda. This may be caused by low expression levels of “divergent IRs” in the tarsi of S. frugiperda which need to be experimentally verified in the future.
By analyzing the transcriptome of the tarsi of S. frugiperda, we annotated 43 putative chemosensory receptor genes. We then used RT-qPCR to compare the expression of these genes in different chemosensory organs. The high expression of several chemosensory receptors in the tarsi suggests that these genes could be important in the tarsal chemosensation of food chemicals in S. frugiperda. The results should facilitate the study of the molecular mechanisms of chemosensation in the tarsi of S. frugiperda and of other moth species.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
J-FD and C-HT conceived and designed the study. D-XL and H-BY reared the insect and collected the biological material. J-FD extracted the RNA. H-QY performed the transcriptome data analysis. J-FD, H-BY and D-XL constructed the phylogenetic trees. J-FD, C-HT, and H-QY wrote the manuscript. All authors read and approved the final version of the manuscript.
This work was funded by National Key R&D Programs of China (Grant Nos. 2021YFD1400701) and Major Special Science and Technology Project of Henan Province (Grant Nos. 201300111500).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1177297/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Homologous species distribution in Nr database.
SUPPLEMENTARY FIGURE S2 | Functional annotation of the transcripts based on GO categorization.
Abuin, L., Bargeton, B., Ulbrich, M. H., Isacoff, E. Y., Kellenberger, S., and Benton, R. (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60. doi:10.1016/j.neuron.2010.11.042
Ai, M., Blais, S., Park, J. Y., Min, S., Neubert, T. A., and Suh, G. S. B. (2013). Ionotropic glutamate receptors IR64a and IR8a form a functional odorant receptor complex in vivo in Drosophila. J. Neurosci. 33, 10741–10749. doi:10.1523/JNEUROSCI.5419-12.2013
Benton, R., Sachse, S., Michnick, S. W., and Vosshall, L. B. (2006). Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4, e20. doi:10.1371/journal.pbio.0040020
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi:10.1016/j.cell.2008.12.001
Blaney, W. M., and Simmonds, M. S. J. (1990). A behavioural and electrophysiological study of the role of tarsal chemoreceptors in feeding by adults of Spodoptera, Heliothis virescens and Helicoverpa armigera. J. Insect Physiol. 36, 743–756. doi:10.1016/0022-1910(90)90048-K
Bogner, F., Boppré, M., Ernst, K. D., and Boeckh, J. (1986). CO2 sensitive receptors on labial palps of rhodogastria moths (Lepidoptera: Arctiidae): Physiology, fine structure and central projection. J. Comp. Physiol. A 158, 741–749. doi:10.1007/bf01324818
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi:10.1093/bioinformatics/btu170
Butterwick, J. A., Del Mármol, J., Kim, K. H., Kahlson, M. A., Rogow, J. A., Walz, T., et al. (2018). Cryo-EM structure of the insect olfactory receptor Orco. Nature 560, 447–452. doi:10.1038/s41586-018-0420-8
Calas, D., Marion-Poll, F., and Steinbauer, M. J. (2009). Tarsal taste sensilla of the autumn gum moth, Mnesampela privata: Morphology and electrophysiological activity. Entomol. Exp. Appl. 133, 186–192. doi:10.1111/j.1570-7458.2009.00921.x
Carey, A. F., and Carlson, J. R. (2011). Insect olfaction from model systems to disease control. Proc. Natl. Acad. Sci. U. S. A. 108, 12987–12995. doi:10.1073/pnas.1103472108
Carey, A. F., Wang, G., Su, C. Y., Zwiebel, L. J., and Carlson, J. R. (2010). Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464, 66–71. doi:10.1038/nature08834
Chapman, R. F. (2003). Contact chemoreception in feeding by phytophagous insects. Annu. Rev. Entomol. 48, 455–484. doi:10.1146/annurev.ento.48.091801.112629
Clyne, P. J., Warr, C. G., and Carlson, J. R. (2000). Candidate taste receptors in Drosophila. Science 287, 1830–1834. doi:10.1126/science.287.5459.1830
Croset, V., Rytz, R., Cummins, S. F., Budd, A., Brawand, D., Kaessmann, H., et al. (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6, e1001064. doi:10.1371/journal.pgen.1001064
Dahanukar, A., Hallem, E. A., and Carlson, J. R. (2005). Insect chemoreception. Curr. Opin. Neurobiol. 15, 423–430. doi:10.1016/j.conb.2005.06.001
Dahanukar, A., Lei, Y.-T., Kwon, J. Y., and Carlson, J. R. (2007). Two Gr genes underlie sugar reception in Drosophila. Neuron 56, 503–516. doi:10.1016/j.neuron.2007.10.024
de Fouchier, A., Walker, W. B., Montagné, N., Steiner, C., Binyameen, M., Schlyter, F., et al. (2017). Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat. Commun. 8, 15709. doi:10.1038/ncomms15709
Depetris-Chauvin, A., Galagovsky, D., and Grosjean, Y. (2015). Chemicals and chemoreceptors: Ecologically relevant signals driving behavior in Drosophila. Front. Ecol. Evol. 3, 41. doi:10.3389/fevo.2015.00041
Dou, X., Liu, S., Ahn, S.-J., Choi, M.-Y., and Jurenka, R. (2019). Transcriptional comparison between pheromone gland-ovipositor and tarsi in the corn earworm moth Helicoverpa zea. Comp. Biochem. Physiol. Part D. Genomics Proteomics 31, 100604. doi:10.1016/j.cbd.2019.100604
Dweck, H. K. M., and Carlson, J. R. (2020). Molecular logic and evolution of bitter taste in Drosophila. Curr. Biol. 30, 17–30.e3. doi:10.1016/j.cub.2019.11.005
Gaaboub, I., Schuppe, H., and Newland, P. L. (2005). Position-dependent sensitivity and density of taste receptors on the locust leg underlies behavioural effectiveness of chemosensory stimulation. J. Comp. Physiol. A 191, 281–289. doi:10.1007/s00359-004-0582-8
Gadenne, C., Barrozo, R. B., and Anton, S. (2016). Plasticity in insect olfaction: To smell or not to smell. Annu. Rev. Entomol. 61, 317–333. doi:10.1146/annurev-ento-010715-023523
Ganguly, A., Pang, L., Duong, V. K., Lee, A., Schoniger, H., Varady, E., et al. (2017). A molecular and cellular context-dependent role for Ir76b in detection of amino acid taste. Cell Rep. 18, 737–750. doi:10.1016/j.celrep.2016.12.071
Gao, Q., and Chess, A. (1999). Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics 60, 31–39. doi:10.1006/geno.1999.5894
Gouin, A., Bretaudeau, A., Nam, K., Gimenez, S., Aury, J. M., Duvic, B., et al. (2017). Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci. Rep. 7, 11816. doi:10.1038/s41598-017-10461-4
Grosjean, Y., Rytz, R., Farine, J. P., Abuin, L., Cortot, J., Jefferis, G. S. X. E., et al. (2011). An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature 478, 236–240. doi:10.1038/nature10428
Guerenstein, P. G., and Hildebrand, J. G. (2008). Roles and effects of environmental carbon dioxide in insect life. Annu. Rev. Entomol. 53, 161–178. doi:10.1146/annurev.ento.53.103106.093402
Guo, H., Gong, X. L., Li, G. C., Mo, B. T., Jiang, N. J., Huang, L. Q., et al. (2022). Functional analysis of pheromone receptor repertoire in the fall armyworm, Spodoptera frugiperda. Pest Manag. Sci. 78, 2052–2064. doi:10.1002/ps.6831
Guo, M. B., Du, L. X., Chen, Q. Y., Feng, Y. L., Zhang, J., Zhang, X. X., et al. (2021). Odorant receptors for detecting flowering plant cues are functionally conserved across moths and butterflies. Mol. Biol. Evol. 38, 1413–1427. doi:10.1093/molbev/msaa300
Hallem, E. A., and Carlson, J. R. (2006). Coding of odors by a receptor repertoire. Cell 125, 143–160. doi:10.1016/j.cell.2006.01.050
He, P., Liu, Y., Hull, J. J., Zhang, Y. N., Zhang, J., Guo, X. J., et al. (2022). Editorial: Insect olfactory proteins (from gene identification to functional characterization), volume II. Front. Physiol. 13, 858728. doi:10.3389/fphys.2022.858728
Hou, X. Q., Zhang, D. D., Powell, D., Wang, H. L., Andersson, M. N., and Löfstedt, C. (2022). Ionotropic receptors in the turnip moth Agrotis segetum respond to repellent medium-chain fatty acids. BMC Biol. 20, 34. doi:10.1186/s12915-022-01235-0
Hu, G. L., Zhang, C. M., Wang, Z. Q., Chen, Q. X., and Lu, J. Q. (2021). Sensilla of the antenna and proboscis of Athetis lepigone (Möschler) (Lepidoptera: Noctuidae). J. Morphol. 282, 733–745. doi:10.1002/jmor.21342
Hussain, A., Zhang, M., Üçpunar, H. K., Svensson, T., Quillery, E., Gompel, N., et al. (2018). Ionotropic chemosensory receptors mediate the taste and smell of polyamines. PLoS Biol. 14, e1002454. doi:10.1371/journal.pbio.1002454
Jiang, X. J., Ning, C., Guo, H., Jia, Y. Y., Huang, L. Q., Qu, M. J., et al. (2015). Gustatory receptor tuned to D-fructose in antennal sensilla chaetica of Helicoverpa armigera. Insect biochem. Mol. Biol. 60, 39–46. doi:10.1016/j.ibmb.2015.03.002
Jones, W. D., Cayirlioglu, P., Kadow, I. G., and Vosshall, L. B. (2007). Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445, 86–90. doi:10.1038/nature05466
Jones, W. D., Nguyen, T. A., Kloss, B., Lee, K. J., and Vosshall, L. B. (2005). Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Curr. Biol. 15, R119–R121. doi:10.1016/j.cub.2005.02.007
Knecht, Z. A., Silbering, A. F., Ni, L., Klein, M., Budelli, G., Bell, R., et al. (2016). Distinct combinations of variant ionotropic glutamate receptors mediate thermosensation and hygrosensation in Drosophila. eLife 5, e17879. doi:10.7554/eLife.17879
Koh, T. W., He, Z., Gorur-Shandilya, S., Menuz, K., Larter, N. K., Stewart, S., et al. (2014). The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83, 850–865. doi:10.1016/j.neuron.2014.07.012
Krenn, H. W. (2010). Feeding mechanisms of adult Lepidoptera: Structure, function, and evolution of the mouthparts. Annu. Rev. Entomol. 55, 307–327. doi:10.1146/annurev-ento-112408-085338
Leal, W. S. (2013). Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi:10.1146/annurev-ento-120811-153635
Lee, Y., Kim, S. H., and Montell, C. (2010). Avoiding DEET through insect gustatory receptors. Neuron 67, 555–561. doi:10.1016/j.neuron.2010.07.006
Li, Q., and Liberles, S. D. (2015). Aversion and attraction through olfaction. Curr. Biol. 25, R120–R129. doi:10.1016/j.cub.2014.11.044
Ling, F., Dahanukar, A., Weiss, L. A., Kwon, J. Y., and Carlson, J. R. (2014). The molecular and cellular basis of taste coding in the legs of Drosophila. J. Neurosci. 34, 7148–7164. doi:10.1523/JNEUROSCI.0649-14.2014
Liu, N. Y., Xu, W., Dong, S. L., Zhu, J. Y., Xu, Y. X., and Anderson, A. (2018). Genome-wide analysis of ionotropic receptor gene repertoire in Lepidoptera with an emphasis on its functions of Helicoverpa armigera. Insect biochem. Mol. Biol. 99, 37–53. doi:10.1016/j.ibmb.2018.05.005
Liu, X. L., Yan, Q., Yang, Y. L., Hou, W., Miao, C. L., Peng, Y. C., et al. (2019). A gustatory receptor GR8 tunes specifically to D-fructose in the common cutworm Spodoptera litura. Insects 10, 272. doi:10.3390/insects10090272
Liu, Y. L., Guo, H., Huang, L. Q., Pelosi, P., and Wang, C. Z. (2014). Unique function of a chemosensory protein in the proboscis of two Helicoverpa species. J. Exp. Biol. 217, 1821–1826. doi:10.1242/jeb.102020
Lu, T., Qiu, Y. T., Wang, G., Kwon, J. Y., Rutzler, M., Kwon, H.-W., et al. (2007). Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr. Biol. 17, 1533–1544. doi:10.1016/j.cub.2007.07.062
Maher, N., and Thiery, D. (2004). Distribution of chemo- and mechanoreceptors on the tarsi and ovipositor of female European grapevine moth, Lobesia botrana. Entomol. Exp. Appl. 110, 135–143. doi:10.1111/j.0013-8703.2004.00131.x
Meagher, R. L., and Nagoshi, R. N. (2012). Differential feeding of fall armyworm Lepidoptera (Lepidoptera: Noctuidae) host strains on meridic and natural diets. Ann. Entomol. Soc. Am. 105, 462–470. doi:10.1603/AN11158
Miyamoto, T., Slone, J., Song, X., and Amrein, H. (2012). A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell 151, 1113–1125. doi:10.1016/j.cell.2012.10.024
Moon, S. J., Lee, Y., Jiao, Y., and Montell, C. (2009). A Drosophila gustatory receptor essential for aversive taste and inhibiting male-to-male courtship. Curr. Biol. 19, 1623–1627. doi:10.1016/j.cub.2009.07.061
Nagoshi, R. N., Goergen, G., Tounou, K. A., Agboka, K., Koffi, D., and Meagher, R. L. (2018). Analysis of strain distribution, migratory potential, and invasion history of fall armyworm populations in northern Sub-Saharan Africa. Sci. Rep. 8, 3710. doi:10.1038/s41598-018-21954-1
Ni, L., Klein, M., Svec, K. V., Budelli, G., Chang, E. C., Ferrer, A. J., et al. (2016). The ionotropic receptors IR21a and IR25a mediate cool sensing in Drosophila. eLife 5, e13254. doi:10.7554/eLife.13254
Ning, C., Yang, K., Xu, M., Huang, L. Q., and Wang, C. Z. (2016). Functional validation of the carbon dioxide receptor in labial palps of Helicoverpa armigera moths. Insect biochem. Mol. Biol. 73, 12–19. doi:10.1016/j.ibmb.2016.04.002
Olivier, V., Monsempes, C., Francois, M. C., Poivet, E., and Jacquin-Joly, E. (2011). Candidate chemosensory ionotropic receptors in a Lepidoptera. Insect Mol. Biol. 20, 189–199. doi:10.1111/j.1365-2583.2010.01057.x
Ozaki, K., Ryuda, M., Yamada, A., Utoguchi, A., Ishimoto, H., Calas, D., et al. (2011). A gustatory receptor involved in host plant recognition for oviposition of a swallowtail butterfly. Nat. Commun. 2, 542. doi:10.1038/ncomms1548
Paoli, M., and Galizia, G. C. (2021). Olfactory coding in honeybees. Cell Tissue Res. 383, 35–58. doi:10.1007/s00441-020-03385-5
Pitts, R. J., Derryberry, S. L., Zhang, Z., and Zwiebel, L. J. (2017). Variant ionotropic receptors in the malaria vector mosquito Anopheles gambiae tuned to amines and carboxylic acids. Sci. Rep. 7, 40297. doi:10.1038/srep40297
Qiu, L., He, L., Tan, X., Zhang, Z., Wang, Y., Li, X., et al. (2020). Identification and phylogenetics of Spodoptera frugiperda chemosensory proteins based on antennal transcriptome data. Comp. Biochem. Physiol. Part D. Genomics Proteomics 34, 100680. doi:10.1016/j.cbd.2020.100680
Rytz, R., Croset, V., and Benton, R. (2013). Ionotropic receptors (IRs): Chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect biochem. Mol. Biol. 43, 888–897. doi:10.1016/j.ibmb.2013.02.007
Sato, K., Pellegrino, M., Nakagawa, T., Vosshall, L. B., and Touhara, K. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006. doi:10.1038/nature06850
Sato, K., Tanaka, K., and Touhara, K. (2011). Sugar-regulated cation channel formed by an insect gustatory receptor. Proc. Natl. Acad. Sci. U. S. A. 108, 11680–11685. doi:10.1073/pnas.1019622108
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi:10.1038/nprot.2008.73
Scott, K., Brady, R., Cravchik, A., Morozov, P., Rzhetsky, A., Zuker, C., et al. (2001). A chemosensory gene family encoding candidate gustatory and olfactory receptor in Drosophila. Cell 104, 661–673. doi:10.1016/s0092-8674(01)00263-x
Silbering, A. F., Rytz, R., Grosjean, Y., Abuin, L., Ramdya, P., Jefferis, G. S., et al. (2011). Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 31, 13357–13375. doi:10.1523/JNEUROSCI.2360-11.2011
Sparks, A. N. (1979). A review of the biology of the fall armyworm. Flo. Entomol. 62, 82–87. doi:10.2307/3494083
Sun, Y. L., Dong, J. F., Gu, N., and Wang, S. L. (2020). Identification of candidate chemosensory receptors in the antennae of the variegated cutworm, Peridroma saucia Hübner, based on a transcriptomic analysis. Front. Physiol. 11, 39. doi:10.3389/fphys.2020.00039
Sun, Y. L., Jiang, P. S., Dong, B. X., Tian, C. H., and Dong, J. F. (2022). Candidate chemosensory receptors in the antennae and maxillae of Spodoptera frugiperda (J.E. Smith) larvae. Front. Physiol. 2022 13, 970915. doi:10.3389/fphys.2022.970915
Wang, G., Carey, A. F., Carlson, J. R., and Zwiebel, L. J. (2010). Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. U. S. A. 107, 4418–4423. doi:10.1073/pnas.0913392107
Wilson, R. I. (2013). Early olfactory processing in Drosophila: Mechanisms and principles. Annu. Rev. Neurosci. 36, 217–241. doi:10.1146/annurev-neuro-062111-150533
Wu, Q. L., Jiang, Y. Y., and Wu, K. M. (2019). Analysis of migration routes of the fall armyworm Spodoptera frugiperda (J.E. Smith) from Myanmar to China. Plant Prot. 45 (1–6), 18. doi:10.16688/j.zwbh.2019047
Xu, W., and Anderson, A. (2015). Carbon dioxide receptor genes in cotton bollworm Helicoverpa armigera. Naturwissenschaften 102, 11. doi:10.1007/s00114-015-1260-0
Xu, W., Papanicolaou, A., Zhang, H.-J., and Anderson, A. (2016). Expansion of a bitter taste receptor family in a polyphagous insect herbivore. Sci. Rep. 6, 23666. doi:10.1038/srep23666
Yactayo-Chang, J. P., Mendoza, J., Willms, S., Rering, C. C., Beck, J. J., and Block, A. K. (2021). Zea mays volatiles that influence oviposition and feeding behaviors of Spodoptera frugiperda. J. Chem. Ecol. 47, 799–809. doi:10.1007/s10886-021-01302-w
Yang, J., Guo, H., Jiang, N. J., Tang, R., Li, G. C., Huang, L. Q., et al. (2021). Identification of a gustatory receptor tuned to sinigrin in the cabbage butterfly Pieris rapae. PLoS Genet. 17, e1009527. doi:10.1371/journal.pgen.1009527
Yang, K., and Wang, C. Z. (2020). Review of pheromone receptors in heliothine species: Expression, function, and evolution. Entomol. Exp. Appl. 169, 156–171. doi:10.1111/eea.12982
Zhang, J., Walker, W. B., and Wang, G. R. (2014). Pheromone reception in moths: From molecules to behaviors. Prog. Mol. Biol. Transl. 130, 109–128. doi:10.1016/bs.pmbts.2014.11.005
Zhang, Y. F., van Loon, J. J. A., and Wang, C. Z. (2010). Tarsal taste neuron activity and proboscis extension reflex in response to sugars and amino acids in Helicoverpa armigera (Hübner). J. Exp. Biol. 213, 2889–2895. doi:10.1242/jeb.042705
Keywords: tarsal transcriptome, gustatory receptor, odorant receptor, ionotropic receptor, expression profile, Spodoptera frugiperda
Citation: Dong J-F, Yang H-B, Li D-X, Yu H-Q and Tian C-H (2023) Identification and expression analysis of chemosensory receptors in the tarsi of fall armyworm, Spodoptera frugiperda (J. E. Smith). Front. Physiol. 14:1177297. doi: 10.3389/fphys.2023.1177297
Received: 01 March 2023; Accepted: 29 March 2023;
Published: 10 April 2023.
Edited by:
Ya-Nan Zhang, Huaibei Normal University, ChinaReviewed by:
Fengqi Li, Guizhou University, ChinaCopyright © 2023 Dong, Yang, Li, Yu and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Qi Yu, aHF5dUBpbmZvbWFpbC5tbnIuZ292LmNu; Cai-Hong Tian, Y2FpaG9uZ3RpYW5AaG5hZ3JpLm9yZy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.