
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol., 29 June 2023
Sec. Gastrointestinal Sciences
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1129889
C2H2 zinc finger (C2H2-ZF) proteins are the majority group of human transcription factors and they have many different molecular functions through different combinations of zinc finger domains. Hepatocellular carcinoma (HCC) is one of the most prevalent malignant tumors and the main reason for cancer-related deaths worldwide. More and more findings support the abnormal expression of C2H2-ZF protein in the onset and progression of HCC. The C2H2-ZF proteins are involved in various biological functions in HCC, such as EMT, stemness maintenance, metabolic reprogramming, cell proliferation and growth, apoptosis, and genomic integrity. The study of anti-tumor drug resistance also highlights the pivotal roles of C2H2-ZF proteins at the intersection of biological functions (EMT, stemness maintenance, autophagy)and chemoresistance in HCC. The involvement of C2H2-ZF protein found recently in regulating different molecules, signal pathways and pathophysiological activities indicate these proteins as the possible therapeutic targets, and diagnostic or prognostic biomarkers for HCC.
Hepatocellular carcinoma (HCC) is a frequent malignant tumor with high morbidity and mortality and has been the third leading cause of cancer-related death worldwide (Sung et al., 2021). Treatment strategies for HCC require interdisciplinary and multidisciplinary approaches, including surgery, liver transplantation, tumor ablation, transcatheter arterial chemical embolization (TACE), interventional radiology, chemotherapy, tumor-targeted therapy, and immunotherapy. The clinical presentation of HCC has improved over the past 10 years with the emergence of adjuvant medications following surgical resection, ablation or chemoprevention (Villanueva, 2019). Although interventional therapy can dramatically improve patient prognosis when HCC is detected early, early detection of HCC is crucial for patients and highly related to patient prognosis, most patients with HCC are unfortunately diagnosed in the advanced phase, which reduces the likelihood of curable resection or liver transplantation. Systemic treatment is the most often used option for individuals with unresectable advanced HCC. Sorafenib, lenvatinib, regorafenib, cabozantinib, nivolumab, and ramucirumab are different drugs of targeted therapies that target cancer-promoting mechanisms at the level of gene expression (Llovet et al., 2018; Raoul et al., 2018). At present, there are no biomarkers that can satisfactorily predict the track of HCC, and no drugs can effectively target chemotherapy resistance or metastatic cells (Huang et al., 2020). Thus, understanding the potential progress of onset and development of HCC is very important in order to find more biomarkers and develop individualized treatment, which is beneficial to improve the prognosis of patients with HCC.
Transcription factors (TFs) play an important role in gene expression control by attaching to target DNA in a sequence-specific way. The Cys2-His2 (C2H2) zinc finger proteins represent the majority group of human TFs (Lander et al., 2001; Urrutia, 2003), almost half of which mainly bind to endogenous retroelements (EREs) (Najafabadi et al., 2015). C2H2 zinc finger (C2H2-ZF) protein is a transcription factor protein through a variety of structural shapes and motifs, with a variety of different molecular functions, including protein-protein or protein-RNA interaction, RNA binding and sequence-specific binding to DNA involved in transcriptional regulation (Stubbs et al., 2011; Weirauch and Hughes, 2011). The structure of the C2H2-ZF protein consists of a C-terminal DNA-binding C2H2-ZF domain and an N-terminal effect domain. These effect domains include POZ domain, also known as BTB domain, KRAB (Krüppel-Associated Box) domain, SCAN (SRE-ZBP, CTfin51, AW-1, and Number 18 cDNA) domain and SET {suppressor of variegation 3 to 9 [Su(var)3–9], enhancer of zeste [E(z)], and trithorax (Tx)} (Laity et al., 2001) domain. C2H2-ZF protein plays a key role in the binding of DNA sequences and conservative connectors between fingers through amino acid on the finger a-helix (Figure 1) (Lee et al., 1989). The BTB/POZ domain is a protein-protein interaction motif that has been conserved throughout evolution. The BTB (BroadComplex, Tamarack, and Bric-a-brac) domain improves the particularity and affinity of DNA binding by interacting with dimerization or its functional contact with genetic code areas. It also inhibits transcription by interacting with other families of proteins (BCL-6/PLZF) (Melnick et al., 2002). The KRAB domain, as well as BTB/POZ domain, plays a role in the transcriptional inhibition module, which recruits its cofactor TRIM28 to participate in silencing foreign retroviruses and ERE (Rowe and Trono, 2011). SCAN domain is a leucine-rich domain. Like the BTB/POZ domain, the SCAN box can mediate homologous and heterologous dimerization between certain SCAN domain family members (Collins et al., 2001). Some of these domains are related to the transcriptional control of genes involved in lipid metabolism, growth factors, and other genes essential for cell maintenance and development (Sander et al., 2003). The proteins containing the SET domain are part of a class of enzymes with the same common domain that methylate their substrate by using the cofactor S-adenosine-L-methionine (SAM). Regulation of signal pathways, transcription factors, and tumor suppressors are formed by the methylation of histones or the targeting of non-histone substrates, and tissue homeostasis is maintained (Herz et al., 2013). Therefore, through different combinations of zinc finger domains, C2H2-ZF proteins could enhance different roles in regulation through diverse cellular environments. In this review, we have briefly introduced the network regulation and anti-tumor drug resistance mechanisms of C2H2-ZF proteins in HCC (Tables 1, 2), which not only shows that C2H2-ZF proteins play significant roles in the development of HCC, but also suggests them as potential therapeutic targets and HCC biomarkers with diagnostic or prognostic value.
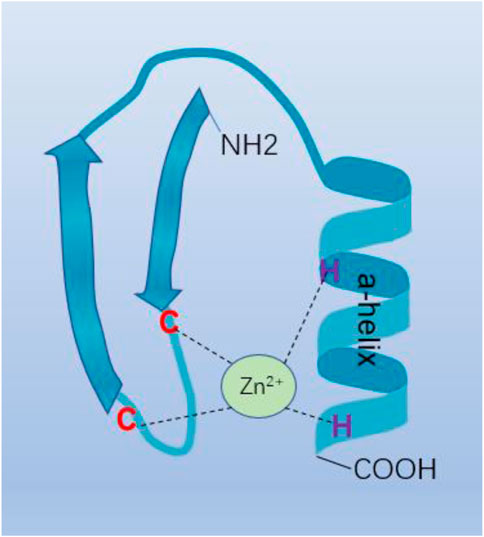
FIGURE 1. C2H2-type zinc finger motif. C2H2-ZF protein plays an important role in the binding of DNA sequences and conservative connectors between fingers through amino acid on the finger a-helix.
A change from the epithelial to the mesenchymal state known as the epithelial-mesenchymal transition (EMT) changes the adhesion molecules that the cell expresses, enabling it to take on a migratory and invasive activity (Nieto et al., 2016). EMT endows cells with the characteristics to migrate and invade, induces stem cells, inhibits cell death and senescence, and relates to autoimmunity. EMT occurs as a result of multiple signaling pathways that trigger the production of particular transcription factors known as EMT-TF (such as Snail, ZEB1, ZEB2, Slug, etc.).
Zinc finger E-box-binding homeobox 1 (ZEB1) is an EMT-TF of the ZFH (zinc-finger-homeodomain) family. It contains two C2H2-ZF domains that interact with E2-box-like CACCT(G) motifs in the target gene regulatory regions and a central POU-like homologous domain that does not interact with DNA (Figure 2), which could mainly participate in protein-protein interactivity (Vandewalle et al., 2008). ZEB1 promotes EMT by inhibiting the expression of E-cadherin, which is a kind of cell adhesion molecule and inhibits tumorigenesis and invasion through the process of malignant transformation in HCC (Vandewalle et al., 2008). The downregulation of E-cadherin eventually leads to EMT, which is a key biological event in tumor cell metastasis. ZEB1 is highly expressed in HCC and its high expression is associated with clinical stage, morphological and pathological features of the tumor (including tumor size, intrahepatic metastasis, and vascular invasion) (Zhou et al., 2012). Insulin-like growth factor binding protein 2 (IGFBP2) promotes p65 nuclear translocation, which interacts with the promoter of ZEB1 and stimulates nuclear factor kappa B (NF-B) and ZEB1 expression in HCC (Guo et al., 2020). Moreover, thioredoxin domain-containing protein 12 (TXNDC12) increases the downstream ZEB1-mediated EMT process via activating Wnt/β-Catenin signal pathway to promote intimal metastasis and HCC metastasis (Yuan et al., 2020). ZEB1 participates in different axis reactions by interacting with different molecules (Figure 3).
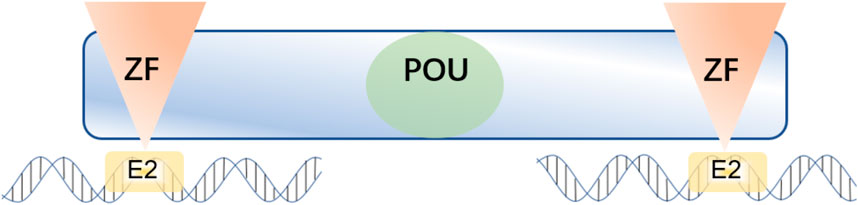
FIGURE 2. ZEB1 contains two C2H2-ZF domains at the C-terminal and N-terminal regions that bind to E2-box-like CACCT(G) sequences in the target gene promoter region, and a central POU-like homologous domain.
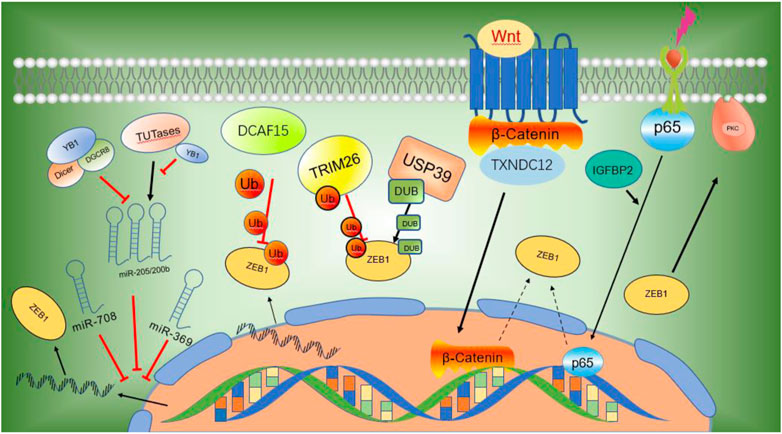
FIGURE 3. ZEB1 participates in different axis reactions through interacting with different molecules.
MicroRNAs (miRNAs) have been demonstrated to suppress the expression of critical cancer-related genes and are implicated in the development and progression of human cancer (Calin and Croce, 2006). MicroRNAs exert inhibitory activity by interacting with ZEB1 mRNA on its 3′untranslated regions (UTRs), repressing ZEB1 expression post-transcriptionally or inducing mRNA degradation (Esquela-Kerscher and Slack, 2006). Y-box binding protein 1 (YB1) inhibits the maturation of miR-205 and miR-200b through binding to microprocessors or uridylyl transferases. The downregulation of miR-205 and miR-200b enhances ZEB1 transcription, resulting in enhanced cell movement and malignant transformation (Liu et al., 2021). Other studies have shown that miR-369 inhibits the expression of ZEB1 mRNA and protein by interacting with its 3′-UTR in HCC cells. MiR-369 highly expresses in HCC and inhibits EMT by promoting the expression of E-cadherin while decreasing the expression of the mesenchymal biomarkers vimentin and N-cadherin (Dong Y. et al., 2021). Li and colleagues reported that miR-708 could decrease the expression of ZEB1 by specifically interacting with ZEB1 mRNA on its 3′-UTR, and therefore inhibit the growth, proliferation, and metastasis of HCC through mediating Wnt/β-Catenin signal (Li LY. et al., 2021).
ZEB1 is used as the substrate of E3 ubiquitin ligase. Tumor suppressor genes DDB1 and CUL4 are the core components of CRL4-DCAF15 (DDB1 and CUL4 associated factor 15), E3 ubiquitin ligase, which specifically recognizes zinc finger domain on the N-terminal of ZEB1 and promotes ZEB1 ubiquitination and degradation, thereby suppressing cell growth, proliferation, and metastasis as well as EMT in HCC cells (Dong X. et al., 2021). Other studies have shown that Ubiquitin-specific peptidase 39 (USP39) deubiquitinates and suppresses ZEB1 degradation, thus promoting the development of EMT and the onset of hepatocellular carcinoma. E3 ligase TRIM26 also degrades ZEB1 through ubiquitination and inhibits cell growth, development, and metastasis of HCC. USP39 and TRIM26 bind to each other directly and maintain the ubiquitin level of ZEB1, thus determining the proliferation and migration of HCC (Li X. et al., 2021).
KLF17, SP/KLF zinc finger protein family, inhibits EMT and metastasis of cancer. The DNA binding domain (DBD) or C2H2-ZF domain on its C-terminal of KLF17 binds to the relevant G/C box and CACCC box of the gene and inhibits transcription (Sun et al., 2013; Lea et al., 2021). Ali and colleagues have found a new mechanism of interaction between TGF-β/Smad-KLF17 axis in the progress of tumor metastasis. KLF17 promotes TGF/Smad-dependent signaling to prevent tumor development. At the same time, it was also found that TGF/Smad3 signaling enhances the expression of KLF17, resulting in a positive feedback loop (Ali et al., 2015). Sun found that miR-9 downregulated the expression of KLF17 protein through directly binding to KLF17 on its 3′-UTR region, resulting in up-regulating EMT-related genes (ZO-1, Vimentin, and Fibronectin (FN)) and promoting HCC migration and invasion (Sun et al., 2013).
Cancer stem cells (CSCs) are a category of cancer cells. The ability of CSCs to escape cell death and metastasis is of great significance to tumorigenesis (Plaks et al., 2015; Batlle and Clevers, 2017). The increased transcription of pluripotency-associated factors (including SOX2, c-myc, BMI1, Nanog, etc.) in CSCs is related to tumor progression, chemotherapy resistance, and recurrence (Wang et al., 2010; Sekiya and Suzuki, 2011; Shan et al., 2012; Ladewig et al., 2013; Zhang et al., 2016; Novak et al., 2020). C2H2-ZF protein could bind to pluripotency-associated factors. As a result, numerous transcription factor binding loci (MTL) are formed. In addition, most of the surface markers (such as CD13, CD24, CD44, CD90, CD133, and EpCAM) are connected to the incidence and progression of malignancies in HCC (Yang et al., 2008; Yamashita et al., 2009; Yang et al., 2009; Haraguchi et al., 2010; Lee et al., 2011; Tang et al., 2012; Kim and Park, 2014; Yan et al., 2015; Batlle and Clevers, 2017). C2H2-ZF protein regulates the mRNA or protein expression of these markers and promotes growth and proliferation through its regulatory actions on liver CSCs. Therefore, investigating the transcription regulatory mechanisms of the self-renewal and pluripotency of CSC is helpful to HCC patients.
ZNF687 directly binds the promoter and increases the transcriptional expression of pluripotency-related factors (BMI1, Oct4, and Nanog), promotes the tumorigenesis of HCC cells in vivo, and increases the HCC SP+/CD133+ populations in vitro. CD133 regulates the incidence and growth of CSCs in HCC via NTS, IL-8, CXCL1, and MAPK signals (Tang et al., 2012). This study also found that the interaction between ZNF687 and the activity enhancer MBD3/NuRD complex promotes Nanog-induced pluripotency (Kloet et al., 2015), suggesting that ZNF687 has been linked to transcription activation and cell reprogramming regulation. Nanog-positive cells show intensive self-renewal, clone formation, tumor initiation, resistance to therapeutic drugs such as sorafenib and cisplatin, and a strong ability for tumor migration (Shan et al., 2012). At the same time, it was found that ZNF687 may induce the upregulation of pluripotency-related factors through the Wnt/β-catenin signal pathway (Zhang et al., 2017).
Tsai and colleagues reported that the miR-200b-ZEB1 circuits are linked to the initiation and development of various CSCs. ZEB1 binds to the promoters of surface markers, causing CD13 and CD24 to be upregulated and EpCAM to be downregulated. CD13 can reduce DNA damage induced by ROS and protect cells from apoptosis (Haraguchi et al., 2010). Through STAT3-mediated Nanog overexpression, CD24+ hepatoma cells activate tumor development and self-renewal (Yamashita et al., 2009; Lee et al., 2011). EpCAM is a biomarker of hepatic stem cells (HPSCs), which activates the Wnt/β-catenin signals and maintains the characteristics of HSPCs and also can activate c-Myc, Oct3/4, SOX2, and KLF4 to induce fibroblasts to differentiate into pluripotent stem cells in HCC (Yamashita et al., 2009; Kim and Park, 2014). MicroRNA-200b directly inhibits the expression of BMI1 and ZEB1. BMI1 gives cells the ability to renew themselves and promotes cell growth and development, colony formation, invasion, and metastasis (Zhang et al., 2016). Downregulation of miR-200b combined with overexpression of ZEB1 promotes the creation and maintenance of CD13+/CD24+/EpCAM-CSCs. In contrast, upregulation of miR-200b combined with downregulation of ZEB1 leads HCC cells to EpCAM + CSCs. Downregulation of miR-200b was linked to tumor recurrence and prognosis, suggesting that the miR-200b/ZEB1 axis is critical in the formation and maintenance of CSCs for tumorigenesis, relapse, and chemotherapy resistance. (Tsai et al., 2017).
Zinc finger protein X-linked (ZFX) encodes on the mammalian X chromosome and contains a DBD with 13 C2H2-ZF at the 3′-end (Schneider-Gädicke et al., 1989). Lai and colleagues found that ZFX binds to Nanog and SOX2 and therefore activates their transcription in HCC and promotes tumor growth. SOX2 contributes to cancer progression and augments therapy resistance in HCC (Novak et al., 2020). On the contrary, the knockout of ZFX can cause the G0/G1 cell cycle to halt, decrease the ability of cell proliferation and colony formation of liver cancer cells, and increase the sensitivity of HCC cells to chemotherapeutic drug cisplatin (Lai et al., 2014). Wang found that ZFX activates and maintains the cell stem of EpCAM + CSCs through the Wnt/β-catenin signal. ZFX promotes the nuclear transcription of β-catenin, enhances its transcriptional activity, and induces the expression of downstream genes such as CylinD1, thus promoting the growth and proliferation of liver stem cells (Wang et al., 2017).
ZBP-89 (also called ZNF148), as a tumor suppressor, can suppress the development of liver cancer in a variety of ways. In patients with HCC, the overexpression of ZBP-89 may indicate improved survival and decreased recurrence (Wang et al., 2018). Wang and colleagues have found that ZBP-89 downregulates the stem cell-like characteristics in HCC by inhibiting Notch1 signals. ZBP-89 locates and binds to Notch1 intracellular domain (NICD) in the nucleus, competitively blocks the NICD transcriptional activator complex, and inhibits the expression of EpCAM and CD44 in a dosage-dependent manner, which has a negative regulatory effect on liver cancer stem cells. CD44 is also found to have a critical player that is a key regulator of CSC features such as self-renewal, tumorigenesis, invasion, and drug resistance (Yan et al., 2015). At the same time, they also found that NOTCH1 may suppress ZBP-89 and form a negative feedback loop to sustain cancer cell existence (Wang et al., 2020).
Zinc finger and homeboxes 2 (ZHX2) was initially identified to regulate the oncofetal gene AFP, H19, and GPC3 in mice, showing that it is involved in HCC. Lin and colleagues have proved that ZHX2 plays a tumor-inhibitory role in liver cancer. ZHX2 inhibits the demethylation of histone H3 lysine36 (H3K36) in the promoter region of stemness-related transcription factors (including SOX2 and Oct4) mediated by lysine demethylase 2A (KDM2A), and suppresses liver CSCs activity and sorafenib resistance. The overexpression of Oct4 induces the activation of TCL1 and AKT to mediate chemotherapy resistance. The Oct4-TCL4-AKT axis affects the proliferation of embryonic stem cells and CSCs by inhibiting apoptosis (Wang et al., 2010). In addition, there is a negative association between the expression of ZHX2 and KDM2A in human HCC, and high ZHX2 expression and low KDM2A expression are associated with the prognosis in HCC (Lin et al., 2020).
Metabolic reprogramming is a marker of malignant tumors, affecting the growth, development, and tumor microenvironment in HCC (Sun et al., 2018). In the occurrence and development of HCC, liver cancer cells could obtain energy and biomass synthesis to gain support for cell survival, and escape immune surveillance and proliferative growth by reprogramming their catabolism and anabolism (Guo et al., 2014). Metabolic changes are accompanied by tumor progression and metastasis and diffusion of cancer cells, which require nutritional absorption and biosynthesis in the early stage of tumor growth and depend on new ways to promote tumor metastasis, invasion, and chemical drug resistance in the advanced stage of HCC (Faubert et al., 2020). The metabolic interaction between tumor cells and the tumor ecosystem will promote therapeutic resistance (Schulze and Harris, 2012), so the metabolic changes of cancer cells and the mutual degradation of the microenvironment become another key mechanism to regulate cancer progression.
In cancer cells, when glycolysis flux is upregulated, pyruvate can produce lactate under oxygen-rich environments, a process called the Warburg’s effect or aerobic glycolysis (Koppenol et al., 2011; Bensaad and Harris, 2014). Many cancers, including liver cancer, are marked by enhanced glucose absorption and glycolysis, suggesting that metabolic changes provide a growth advantage for tumor cells (Schulze and Harris, 2012). Cancer cells promote the production of ATP and lactic acid buildup, regulate the pH value of the tumor ecosystem, and affect the signal pathway via Warburg’s effect, which offers cancer cells selective benefits in terms of growth, continued existence, migration, and metastasis (Lunt and Vander Heiden, 2011; Teoh and Lunt, 2018).
Zinc finger protein acts on the key enzyme regulating the rate-limiting stage in glycolysis, and promotes or inhibits the aerobic glycolysis of tumor cells by up-regulating or inhibiting the rate-limiting enzyme. For example, there are two isoforms of pyruvate kinase M(PKM). PKM1 is abundant in mature cells, which increases ATP production, while PKM2 is commonly abundant in embryos and cancer tissue, which boosts the Warburg’s effect of HCC (Yang and Lu, 2013). The expression of PKM2 offers cancer a selective survival benefit. PKM2 participates in the metabolism of cancer reprogramming, and promotes tumor growth and development, which prompt people to study the metabolic and regulatory process behind the role of PKM2 in HCC (Benjamin Daniel et al., 2012).
Past research has discovered that E3 ligase ZFP91 interacts with hnRNP A1, a splicing regulator, to boost proteasome degradation of hnRNP A1. As a result, hnRNP A1-dependent PKM mRNA pre-mRNA splicing is inhibited, causing PKM1 isoform formation while inhibiting PKM2 isoform formation. The results show that ZEP91 inhibits liver cancer cells growth and proliferation, which participates in ubiquitination and catabolism of substrates, and suppresses metabolic programming, cell growth, and development of liver cancer (Chen et al., 2020).
The latest study found that silencing ZEB1 in different HCC cell lines substantially reduced levels of protein and aerobic glycolysis of the muscle isoform of phosphofructokinase-1 (PFKM), another rate-limiting enzyme in glycolysis, which was characterized by a decrease in glucose uptake and lactic acid production. The exogenous expression of PFKM significantly reduced this decline. Further study found that ZEB1 interacts with the non-classical ZEB1 binding motif in the PFKM promoter region, thus activating its transcription (Zhou et al., 2021). It is suggested that a new mechanism of direct relationship between zinc finger protein and promoting glycolysis and Warburg effect in the development of HCC.
In addition to glucose, the increase in lipid synthesis has been considered a component of cancer cell metabolic reprogramming (Schulze and Harris, 2012). Lipids synthesized by tumor cells are important components of biofilm lipids and important substrates of energy metabolism and produce lipid signal molecules to promote tumor proliferation and metastasis (Menendez and Lupu, 2007). Non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH) could accelerate liver fibrosis, cirrhosis, end-stage liver disease, and eventually develop into liver cancer, indicating that lipid metabolism may be a determining factor in the development of NAFLD to HCC (NAFLD- NASH-HCC) (Schuppan and Schattenberg, 2013; Jiang et al., 2014). In recent years, some zinc finger proteins have been shown to transcribe key enzymes or lipid metabolism-related genes and proteins and affect the progression of NAFLD-NASH-HCC (Wu et al., 2020; Yu et al., 2020).
Lipoprotein lipase (LPL) is a key enzyme in lipid metabolism, which encourages the development and proliferation of liver cancer through the uptake of exogenous lipids. It has been found that LPL is the target of ZHX2. In NAFLD, ZHX2 inhibits NAFLD-HCC progression and cell growth retardation through transcriptional inhibition of LPL expression. In HCC cells, ZHX2 inhibits LPL and further suppresses the proliferation, lipid deposition, growth, and formation of exogenic and spontaneous tumors (Wu et al., 2020). Further studies found that ZHX2 boosted the transcription of miR-24–3p, which aimed at SREBP1c and caused its destruction. Moreover, ZHX2 inhibited de novo lipogenesis (DNL) and the development and transfer of HCC (Wu et al., 2020).
Sterol-responsive element-binding protein 1c (SREBP1c) is an important TF for DHL and is an important link between tumor signal transduction and tumor metabolism (Lee et al., 2019). SREBP1c participates in the production of cholesterol and fatty acids and has been proven to be overexpressed in HCC (Guo et al., 2014). Tumor cells reactivate DNL to provide nutrition for tumor cells, which is conducive to tumor cell growth and tumor symbiosis (Schulze and Harris, 2012). Therefore, zinc finger protein can delay the progression from NAFLD to HCC and the growth and invasion of HCC via transcriptional control of key regulatory factors in DNL.
Cell cycle disorder can contribute to excessive cell division, and sustaining proliferative signaling is one of the markers of various malignant tumors (Hanahan and Weinberg, 2011). Cell cycle proteins and cell division cycle proteins are required for cell cycle precise control.
In the G1 phase, cyclin-dependent kinases (CDK) 4 and 6 bind to one of the three D-type cyclins (D1, D2, D3) in different environments and promote cell cycle transition after transcriptional induction (Malumbres, 2014; Ingham and Schwartz, 2017). C2H2-ZF protein 384 (ZNF384) upregulates the expression of CyclinD1 by recognizing the adenine-rich region of the promoter of CyclinD1. The interaction between CyclinD1 and CDK4 or CDK6 promotes the G1/S stage transformation and the multiplication in liver cancer (He et al., 2019). Cell division cycle 6 (CDC6) regulates DNA replication in mammalian cells, which regulates DNA replication or checkpoint mechanism (Karanika et al., 2017).
CDC6 participates in the assembly of the complex before replication and promotes the G1/S phase transition (Borlado and Mendez, 2008). The DNA over-replication found in cancer cells following the ectopic expression of CDC6 (Vaziri et al., 2003). C2H2-ZF protein 143 (ZNF143) promoted the expression of CDC6 by directly activating the transcription of histone demethylase mineral dust-induced gene (MDIG), which reduced H3K9me3 enrichment in the CDC6 promoter region and promoted the cell cycle progression of HCC (Zhang et al., 2020).
In addition, C2H2-ZF protein 233 (ZNF233), the Krüppel C2H2-ZF family, accelerates growth and proliferation by promoting the G1/S phase transition in HCC cells. Clinical studies have shown that the level of expressiveness of ZNF233 is associated with the tumor grade, stage, and clinical presentation of patients with HCC (Xie et al., 2018).
Anoikis, a type of apoptosis that occurs when epithelial or endothelial cells detach from the extracellular matrix, is also another form of “loss-of-signal"-induced cell death (ECM). Unligated ECM proteins of the integrin family give up to stimulate pro-survival signaling under this circumstance, ultimately causing cell death (Green and Llambi, 2015). Focal adhesion kinase (FAK) is a common and important tyrosine kinase that transduces signals from integrins, growth, and endocrine elements and is involved in many essential biological functions and activities (Franchini, 2012). Src family kinases, a player in promoting metastasis in many tumor types, are critical components of the anoikis resistance pathway (Dai et al., 2016). Src recruits FAK and promotes Src phosphorylation (Acebron et al., 2020). The activation of Src activates downstream target protein kinase AKT and regulates cell resistance to apoptosis (Dai et al., 2016). As a receptor of intracellular ROS level, C2H2-ZF protein 32 (ZNF32) maintains redox homeostasis and promotes the resistance of HCC to anoikis through inhibiting excessive ROS accumulation of HCC and activating Src/FAK signal pathway (Li et al., 2019).
Because genomic dysfunction affects many other human diseases in addition to cancer and aging (Sarek et al., 2015). It is essential to understand the regulation and maintenance of the genome, including DNA damage response (DDR) and telomere maintenance (Jackson and Bartek, 2009). Zinc finger domains are found in some DNA damage factors and telomere-associated proteins. Zinc finger domains have exceptional binding flexibility and may attach to Nucleotides, molecules, and protein post-translational modifications (PTMs), which are involved in genome integrity (Gamsjaeger et al., 2007).
ZNF451 is a class of SUMO E3 ligases and a multifunctional DDR factor, containing two N-terminal SIM domains and a ubiquitin-interacting motif. Extensive biochemical analysis shows that the C2H2-ZF domains and tandem SIM domains are the keys to its E3 ligase activity (Cappadocia et al., 2015; Eisenhardt et al., 2015; Schellenberg et al., 2017). ZNF451 and Topoisomerase 2 (TOP2) are involved in DDR and break the DNA double helix and topological changes (Deweese and Osheroff, 2009; Schellenberg et al., 2017), which could affect DNA supercoil formation during transcription and replication, and lead to genomic instability (Ju et al., 2006; Pommier, 2013; Tammaro et al., 2013; Madabhushi et al., 2015). Shanbhag found that DNA double-strand breaks (DSB) cause extensive local and global changes in chromatin structure, many of which depend on ATM kinases, leading to transcriptional silencing (Shanbhag et al., 2010). DNA double-strand break (DSB), one of the main types of DNA damage, promotes cell cycle halt and DDR to maintain the integrity of the genome via activating a series of cell activities.
CDC6 can protect the integrity of the genome via activating DDR (Yoshida et al., 2010), but overexpression of CDC6 may lead to repetitive replication and genomic instability, which is essential for tumor progression (Liontos et al., 2007). Zhang and colleagues proposed a new pathway of ZNF143-MDIG-CDC6 regulating in liver cells (Zhang et al., 2020). In addition, Understanding the interplay between transcription and DNA repair mechanisms will give critical insights and, eventually, novel therapeutic options for a variety of disorders linked with genome maintenance abnormalities (Sebastian and Oberdoerffer, 2017).
In mammalian cells, the maintenance of the Chromatin structure promotes the self-renewal of human embryonic stem cells (HESCs) and induced pluripotent stem cells (HiPSCs) (Blasco, 2007; Huang et al., 2011; Pucci et al., 2013; Dan et al., 2014; Rivera et al., 2017). Telomere erosion and telomere lengthening mechanisms that are two opposite forces determine the length of the telomere.
C2H2-ZF protein has become a key controller of telomere length. Dan has shown that zinc finger and SCAN domain containing 4 (Zscan4) inhibits telomere extension linked with the expression of Zscan4 by recruiting Uhrf1 and Dnmt1 (the main component of DNA methylation mechanism) and promoting their degradation, blocking DNA demethylation (Dan et al., 2017).
ZNF827 is abundant in telomeres and uses the ALT pathway to recruit DNA repair factors, which encourage homologous recombination and telomere lengthening. Conomos believe that ZNF827 attachment to human telomeres induces telomeric chromatin modification and the formation of an atmosphere that encourages telomere-telomere recombination, as well as the integration and regulation of numerous molecular components of ALT function (Conomos et al., 2014).
ZBTB48 (also known as HKR3 or TZAP) has 11 C2H2-ZF domains, three of which interact with TTAGGG motifs specifically. It was found that ZBTB48 binds to double-stranded TTAGGG repeats and acts as a negative regulator on telomeres (Li J. et al., 2017; Jahn et al., 2017; Kappei et al., 2017). When ZBTB48 is located at telomeres, a process called telomere trimming is triggered, resulting in the rapid deletion of telomere repeat sequences (Li J. et al., 2017). Due to the phenomenon that telomere maintenance is commonly and initially changed in HCC, telomerase targeting is a promising anti-cancer strategy.
HCC is distinguished by a high level of inherent chemoresistance, resulting in a restricted effect of chemotherapy and recurrence since therapy. Multidrug resistance is one of the causes of poor cure rates in advanced HCC (Khaled et al., 2022). Zinc finger proteins are not only taking part in the initiation, growth, progression, and metastasis of HCC but also in drug resistance to chemotherapy. Zinc finger proteins regulate the resistance of chemotherapeutic drugs and targeted drugs through various mechanisms and pathways. According to the research in recent years, we will elaborate on the regulation of C2H2-ZF protein in chemotherapeutic resistance from three aspects of EMT, stemness maintenance, and autophagy (Figure 4).
The role of EMT in anti-tumor drug resistance has been more and more recognized. C2H2-ZF proteins such as ZEB1, ZEB2, Slug, and GLI1 participate in the progression of EMT can also promote anti-tumor drug resistance.
ZEB1-induced EMT can lead to resistance to conventional chemotherapy. ZEB1 can classify hepatocellular carcinoma into epithelial type and mesenchymal type in vivo and in vitro. Mesenchymal/metastatic cancer cells need to activate the protein kinase C (PKC) pathway to survive (Singh et al., 2009). Sreekumar and colleagues have demonstrated that the PKC signal is stimulated in mesenchymal HCC cells, which helps them proliferate. Mesenchymal cells and chemotherapy-resistant cells that express ZEB1 are dependent on PKCa, which can be eliminated selectively by PKC inhibitor UCN-01. The inactivation of PKCa also reduces the activity of HCC, showing that UCN-01 in conjunction with chemotherapy can improve the therapeutic effect of ZEB1-positive HCC (Sreekumar et al., 2019). LONG studies have shown that the upregulation of ZEB1 expression is associated with the EMT characteristics of HCC, while inhibition of ZEB1 expression makes cells sensitive to chemotherapy (Long et al., 2019). Targeted inhibition of the ZEB1 gene may contribute to clinical chemotherapy resistance.
ZEB2 has a significant impact on the progression of HCC by regulating EMT. The interaction between Linc-ROR and miR-145 upregulates the transcription of ZEB2, which promote the proliferation and metastasis of HCC cells (Li C. et al., 2017). Recent research found that miR-212–3p reduces the transcription of ZEB2 and reduces the EMT, migration, and metastasis of HCC by targeting ZEB2, thus reducing paclitaxel resistance, which provides a new target for liver cancer chemotherapy (Yang et al., 2020).
Snail family transcriptional repressor 2 (SNAI2/Slug), belongs to the part of the Snail family of C2H2-ZF TFs and is a transcriptional repressor that interacts with E-box domains (Hajra et al., 2002). Slug is the key regulator of EMT, inducing EMT and inhibiting E-cadherin in various epithelial cells (Shih et al., 2005; Uchikado et al., 2005). Previous studies have shown that Slug inhibits multidrug resistance by inhibiting ABCB1 in vivo and in vitro, thus exerting tumor inhibitory effect (Hoshida et al., 2016). In contrast, recent research has shown that ABCB1 is downregulated in HCC cells undergoing EMT, while chemotherapy resistance to cisplatin has more than doubled. It is concluded that the chemotherapy resistance of HCC is sometimes influenced by changing Slug. The expression of Slug could affect chemotherapy resistance, depending on the cell environment (Karaosmanoglu et al., 2018).
Hedgehog (HH) signaling pathway is closely related to cancer growth and differentiation. Abnormal HH signal has been considered to be an important signal pathway and therapeutic target for human tumors (Lum and Beachy, 2004). HH signal is closely related to many basic processes, and the imbalance of this signal axis may lead to tumor occurrence, metastasis, and drug resistance. HH signaling has been found to promote tumor development at the molecular level by influencing tumor growth, EMT, CSC development, and acquired drug resistance (Ochoa et al., 2010; Sari et al., 2018). C2H2-ZF protein GLI1, the terminal effector of HH signaling, is a potential marker of signaling activation because it promotes gene expression. Chen and colleagues have found that EMT and elevated HH signaling activation may be causes of chemical resistance and invasion in poorly differentiated HCC cells (Chen et al., 2011). Further study showed that the protein level of GLI1 in poorly differentiated HCC cells was significantly increased. After the decrease of GLI1 expression, the responsiveness of HCC cells to doxorubicin and cisplatin increased significantly. Targeted HH signaling pathway gives a fresh viewpoint on drug resistance in the treatment of HCC (Zhou et al., 2020).
Stemness maintenance is related to tumor recurrence and chemotherapy resistance (vide supra), in which the high expression of Nanog and Oct4 in CSCs promotes chemotherapy resistance, and the surface markers CD24 and CD90 are related to chemotherapy resistance in HCC.
ZNF687 upregulates pluripotency-related factors. Overexpression of ZNF687 made cisplatin-treated liver cells resistant to apoptosis, and silencing ZNF687 significantly enhanced the sensitivity of HCC to cisplatin. These results indicate that ZNF687 has a significant impact on the chemical resistance of HCC (Zhang et al., 2017).
Zinc finger protein X-linked (ZFX) binds to the promoters of Nanog and SOX2 to promote tumor growth. In contrast, the knockout of ZFX increases the chemotherapeutic responsiveness to HCC (Wang et al., 2017). ZHX2 inhibited the demethylation of SOX2, Nanog, and Oct4, and inhibited the chemical resistance of liver cancer (Lin et al., 2020).
Autophagy is a critical biological function that influences cell metabolism, proteome, quantity, and quality of organelles, and changes cell function in different ways (Kimmelman and White, 2017). In cancer, it can inhibit the tumor or increase survival under the pressure of treatment (chemotherapy and targeted drugs), thereby increasing treatment resistance (Thorburn et al., 2014; Singh et al., 2018). Continuous exposure to sorafenib can activate endoplasmic reticulum stress (ERS)-related proteins, as well as induce autophagy and sorafenib resistance. Inhibition of ERS can reduce autophagy and reverse sorafenib resistance (Cheng et al., 2020). ZNF263 is an ERS-specific transcription factor and the downregulation of its expression reduces chemotherapy resistance and increases apoptosis rate. Knockout of ZNF263 effectively alleviates the chemotherapy resistance of ERS-related autophagy-induced HCC cells (Cui et al., 2020). The study of ERS-specific transcription factors provides a different strategy to deal with drug resistance: inhibition of autophagy to avoid resistance to kinase inhibitors such as sorafenib (Amaravadi et al., 2016).
C2H2-ZF proteins are abnormally expressed in HCC and involve in numerous HCC features like EMT, stemness maintenance, metabolic reprogramming, cell proliferation and growth, apoptosis, and genomic integrity. These proteins are also correlated with and regulate chemoresistance through targeting and modulating the activities of key molecules involved in these processes, such as EMT, stemness maintenance, and autophagy. However, the regulatory mechanisms of C2H2-ZF proteins in HCC are still unclear and the related studies are also still in the initial stage. This paper summarizes the regulatory role of C2H2-ZF proteins in hepatocarcinogenesis and progression as follows: 1. C2H2-ZF proteins play an important role in regulating the transcription of downstream genes that are involved in the proliferation, apoptosis, migration, and invasion of HCC (Liang et al., 2017). 2. The differential expression levels of C2H2-ZF proteins in HCC are regulated by tumor-associated miRNAs (Li C. et al., 2017). 3. Involvement in post-translational modifications through ubiquitination, a modality that can increase zinc finger protein transcriptional activation or inhibit post-translational regulation (Dong X. et al., 2021). 4. Zinc finger proteins show different sequence specificity in their ability to bind DNA, and the diversity of zinc finger proteins is demonstrated after different combinations of zinc finger motifs (Dan et al., 2017). 5. Transcriptional regulation is an extremely complex biological process, and the microenvironment within tumor cells may be an important factor in addition to the structural diversity of the same C2H2-ZF protein, which results in multiple transcriptional regulation patterns (Zhou et al., 2021). Therefore, the role of C2H2-ZF protein in regulating different molecules, signal pathways, and pathophysiological activities shows that C2H2-ZF protein could be used as diagnostic or prognostic markers and could be considered as potential therapeutic targets or sensitizers in HCC. We believe that C2H2-ZF proteins would have a broad application prospect in the diagnosis and drug treatment of HCC with the continuous improvement of the detection, verification, and functional analysis of them. However, more studies demand to pave the way for the clinical application of these findings.
QL: Writing—original draft. GT: Writing—review and editing. FW: Conceptualization, Funding acquisition, Supervision, Writing—review and editing. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acebron, I., Righetto, R. D., Schoenherr, C., de Buhr, S., Redondo, P., Culley, J., et al. (2020). Structural basis of Focal Adhesion Kinase activation on lipid membranes. EMBO J. 39 (19), e104743. PMID:32779739. doi:10.15252/embj.2020104743
Ali, A., Zhang, P., Liangfang, Y., Wenshe, S., Wang, H., Lin, X., et al. (2015). KLF17 empowers TGF-β/Smad signaling by targeting Smad3-dependent pathway to suppress tumor growth and metastasis during cancer progression. Cell Death Dis. 6, e1681. doi:10.1038/cddis.2015.48
Amaravadi, R., Kimmelman, A., White, E. J. G., and development, (2016). Recent insights into the function of autophagy in cancer. Genes Dev. 30, 1913–1930. PMID:27664235. doi:10.1101/gad.287524.116
Batlle, E., and Clevers, H. (2017). Cancer stem cells revisited. Nat. Med. 23, 1124–1134. PMID:28985214. doi:10.1038/nm.4409
Benjamin Daniel, I., Cravatt Benjamin, F., and Nomura Daniel, K. (2012). Global profiling strategies for mapping dysregulated metabolic pathways in cancer. Cell Metab. 16, 565–577. doi:10.1016/j.cmet.2012.09.013
Bensaad, K., and Harris, A. L. (2014). Hypoxia and metabolism in cancer. Adv. Exp. Med. Biol. 772, 1–39. PMID:24272352. doi:10.1007/978-1-4614-5915-6_1
Blasco, M. A. (2007). The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 8, 299–309. PMID:17363977. doi:10.1038/nrg2047
Borlado, L. R., and Mendez, J. (2008). CDC6: From DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis 29, 237–243. PMID:18048387. doi:10.1093/carcin/bgm268
Calin, G. A., and Croce, C. M. (2006). MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866. 857. doi:10.1038/nrc1997
Cappadocia, L., Pichler, A., and Lima, C. D. (2015). Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nat. Struct. Mol. Biol. 22, 968–975. PMID:26524494. doi:10.1038/nsmb.3116
Chen, D., Wang, Y., Lu, R., Jiang, X., Chen, X., Meng, N., et al. (2020). E3 ligase ZFP91 inhibits Hepatocellular Carcinoma Metabolism Reprogramming by regulating PKM splicing. Theranostics 10, 8558–8572. PMID:32754263. doi:10.7150/thno.44873
Chen, X., Lingala, S., Khoobyari, S., Nolta, J., Zern, M. A., and Wu, J. (2011). Epithelial mesenchymal transition and hedgehog signaling activation are associated with chemoresistance and invasion of hepatoma subpopulations. J. Hepatol. 55, 838–845. PMID:21334406. doi:10.1016/j.jhep.2010.12.043
Cheng, Z., Wei-Qi, J., and Jin, D. (2020). New insights on sorafenib resistance in liver cancer with correlation of individualized therapy. Biochim. Biophys. Acta Rev. Cancer 1874, 188382. PMID:32522600. doi:10.1016/j.bbcan.2020.188382
Collins, T., Stone, J. R., and Williams, A. J. (2001). All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol. Cell Biol. 21, 3609–3615. PMID:11340155. doi:10.1128/MCB.21.11.3609-3615.2001
Conomos, D., Reddel, R. R., and Pickett, H. A. (2014). NuRD-ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat. Struct. Mol. Biol. 21, 760–770. PMID:25150861. doi:10.1038/nsmb.2877
Cui, J., Liu, J., Fan, L., Zhu, Y., Zhou, B., Wang, Y., et al. (2020). A zinc finger family protein, ZNF263, promotes hepatocellular carcinoma resistance to apoptosis via activation of ER stress-dependent autophagy. Transl. Oncol. 13, 100851. PMID:32898766. doi:10.1016/j.tranon.2020.100851
Dai, H., Lv, Y. F., Yan, G. N., Meng, G., Zhang, X., and Guo, Q. N. (2016). RanBP9/TSSC3 complex cooperates to suppress anoikis resistance and metastasis via inhibiting Src-mediated Akt signaling in osteosarcoma. Cell Death Dis. 7, e2572. PMID:28032865. doi:10.1038/cddis.2016.436
Dan, J., Liu, Y., Liu, N., Chiourea, M., Okuka, M., Wu, T., et al. (2014). Rif1 maintains telomere length homeostasis of ESCs by mediating heterochromatin silencing. Dev. Cell 29, 7–19. PMID:24735877. doi:10.1016/j.devcel.2014.03.004
Dan, J., Rousseau, P., Hardikar, S., Veland, N., Wong, J., Autexier, C., et al. (2017). Zscan4 inhibits maintenance DNA methylation to facilitate telomere elongation in mouse embryonic stem cells. Cell Rep. 20, 1936–1949. PMID:28834755. doi:10.1016/j.celrep.2017.07.070
Deweese, J. E., and Osheroff, N. (2009). The DNA cleavage reaction of topoisomerase II: Wolf in sheep's clothing. Nucleic Acids Res. 37, 738–748. PMID:19042970. doi:10.1093/nar/gkn937
Dong, X., Han, Y., Zhang, E., Wang, Y., Zhang, P., Wang, C., et al. (2021b). Tumor suppressor DCAF15 inhibits epithelial-mesenchymal transition by targeting ZEB1 for proteasomal degradation in hepatocellular carcinoma. Aging (Albany NY) 13:10603–10618. PMID:33833131. doi:10.18632/aging.202823
Dong, Y., Li, F., Wang, J., Hu, J., Li, Z., Gu, Y., et al. (2021a). miR-369 inhibits Liver Cancer progression by targeting ZEB1 pathway and predicts the prognosis of HCC patients. J. Cancer 12, 3067–3076. PMID:33854606. doi:10.7150/jca.54759
Eisenhardt, N., Chaugule, V. K., Koidl, S., Droescher, M., Dogan, E., Rettich, J., et al. (2015). A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly. Nat. Struct. Mol. Biol. 22, 959–967. PMID:26524493. doi:10.1038/nsmb.3114
Esquela-Kerscher, A., and Slack, F. J. (2006). Oncovirus — microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269. doi:10.1038/nrc1840
Faubert, B., Solmonson, A., and DeBerardinis, R. J. (2020). Metabolic reprogramming and cancer progression. Science 368, eaaw5473. PMID:32273439. doi:10.1126/science.aaw5473
Franchini, K. G. (2012). Focal adhesion kinase — the basis of local hypertrophic signaling domains. J. Mol. Cell. Cardiol. 52, 485–492. doi:10.1016/j.yjmcc.2011.06.021
Gamsjaeger, R., Liew, C. K., Loughlin, F. E., Crossley, M., and Mackay, J. P. (2007). Sticky fingers: Zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 32, 63–70. PMID:17210253. doi:10.1016/j.tibs.2006.12.007
Green, D. R., and Llambi, F. (2015). Cell death signaling. Cold Spring Harb. Perspect. Biol. 7, a006080. PMID:26626938. doi:10.1101/cshperspect.a006080
Guo, D., Bell, E., Mischel, P., and Chakravarti, A. J. (2014). Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr. Pharm. Des. 20, 2619–2626. PMID:23859617. doi:10.2174/13816128113199990486
Guo, Q., Yu, D. Y., Yang, Z. F., Liu, D. Y., Cao, H. Q., and Liao, X. W. (2020). IGFBP2 upregulates ZEB1 expression and promotes hepatocellular carcinoma progression through NF-κB signaling pathway. Dig. Liver Dis. 52, 573–581. PMID:31818638. doi:10.1016/j.dld.2019.10.008
Hajra, K., Chen, D., and Fearon, E. (2002). The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 62, 1613–1618. PMID:11912130.
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: The next generation. Cell 144, 646–674. PMID:21376230. doi:10.1016/j.cell.2011.02.013
Hanoun, N., Bureau, C., Diab, T., Gayet, O., Dusetti, N., Selves, J., et al. (2010). The SV2 variant of KLF6 is down-regulated in hepatocellular carcinoma and displays anti-proliferative and pro-apoptotic functions. J. Hepatol. 53, 880–888. PMID:20801538. doi:10.1016/j.jhep.2010.04.038
Haraguchi, N., Ishii, H., Mimori, K., Tanaka, F., Ohkuma, M., Kim, H. M., et al. (2010). CD13 is a therapeutic target in human liver cancer stem cells. J. Clin. Invest. 120, 3326–3339. PMID:20697159. doi:10.1172/JCI42550
He, L., Fan, X., Li, Y., Chen, M., Cui, B., Chen, G., et al. (2019). Overexpression of zinc finger protein 384 (ZNF 384), a poor prognostic predictor, promotes cell growth by upregulating the expression of Cyclin D1 in Hepatocellular carcinoma. Cell Death Dis. 10, 444. PMID:31168049. doi:10.1038/s41419-019-1681-3
Herz, H. M., Garruss, A., and Shilatifard, A. (2013). SET for life: Biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem. Sci. 38, 621–639. PMID:24148750. doi:10.1016/j.tibs.2013.09.004
Hoshida, Y., Zhao, X-Y., Li, L., Wang, X-B., Fu, R-J., Lv, Y-P., et al. (2016). Inhibition of Snail family transcriptional repressor 2 (SNAI2) enhances multidrug resistance of hepatocellular carcinoma cells. Plos One 11, e0164752. doi:10.1371/journal.pone.0164752
Huang, A., Yang, X. R., Chung, W. Y., Dennison, A. R., and Zhou, J. (2020). Targeted therapy for hepatocellular carcinoma. Signal Transduct. Target Ther. 5, 146. PMID:32782275. doi:10.1038/s41392-020-00264-x
Huang, J., Wang, F., Okuka, M., Liu, N., Ji, G., Ye, X., et al. (2011). Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 21, 779–792. PMID:21283131. doi:10.1038/cr.2011.16
Ingham, M., and Schwartz, G. K. (2017). Cell-cycle therapeutics come of age. J. Clin. Oncol. 35, 2949–2959. PMID:28580868. doi:10.1200/jco.2016.69.0032
Jackson, S. P., and Bartek, J. (2009). The DNA-damage response in human biology and disease. Nature 461, 1071–1078. PMID:19847258. doi:10.1038/nature08467
Jahn, A., Rane, G., Paszkowski-Rogacz, M., Sayols, S., Bluhm, A., Han, C. T., et al. (2017). ZBTB48 is both a vertebrate telomere-binding protein and a transcriptional activator. EMBO Rep. 18, 929–946. PMID:28500257. doi:10.15252/embr.201744095
Jiang, C. M., Pu, C. W., Hou, Y. H., Chen, Z., Alanazy, M., and Hebbard, L. (2014). Non alcoholic steatohepatitis a precursor for hepatocellular carcinoma development. World J. Gastroenterol. 20, 16464–16473. PMID:25469014. doi:10.3748/wjg.v20.i44.16464
Ju, B. G., Lunyak, V. V., Perissi, V., Garcia-Bassets, I., Rose, D. W., Glass, C. K., et al. (2006). A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802. PMID:16794079. doi:10.1126/science.1127196
Kappei, D., Scheibe, M., Paszkowski-Rogacz, M., Bluhm, A., Gossmann, T. I., Dietz, S., et al. (2017). Phylointeractomics reconstructs functional evolution of protein binding. Nat. Commun. 8, 14334. PMID:28176777. doi:10.1038/ncomms14334
Karanika, S., Karantanos, T., Li, L., Wang, J., Park, S., Yang, G., et al. (2017). Targeting DNA damage response in prostate cancer by inhibiting androgen receptor-CDC6-ATR-chk1 signaling. Cell Rep. 18, 1970–1981. doi:10.1016/j.celrep.2017.01.072
Karaosmanoglu, O., Banerjee, S., and Sivas, H. (2018). Identification of biomarkers associated with partial epithelial to mesenchymal transition in the secretome of slug over-expressing hepatocellular carcinoma cells. Cell Oncol. (Dordr) 41, 439–453. PMID:29858962. doi:10.1007/s13402-018-0384-6
Khaled, J., Kopsida, M., Lennernas, H., and Heindryckx, F. (2022). Drug resistance and endoplasmic reticulum stress in hepatocellular carcinoma. Cells 11, 632. PMID:35203283. doi:10.3390/cells11040632
Kim, H., and Park, Y. N. (2014). Hepatocellular carcinomas expressing 'stemness'-related markers: Clinicopathological characteristics. Dig. Dis. 32, 778–785. PMID:25376296. doi:10.1159/000368021
Kimmelman, A. C., and White, E. (2017). Autophagy and tumor metabolism. Cell Metab. 25, 1037–1043. doi:10.1016/j.cmet.2017.04.004
Kloet, S. L., Baymaz, H. I., Makowski, M., Groenewold, V., Jansen, P. W., Berendsen, M., et al. (2015). Towards elucidating the stability, dynamics and architecture of the nucleosome remodeling and deacetylase complex by using quantitative interaction proteomics. FEBS J. 282, 1774–1785. PMID:25123934. doi:10.1111/febs.12972
Koppenol, W. H., Bounds, P. L., and Dang, C. V. (2011). Otto Warburg's contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337. PMID:21508971. doi:10.1038/nrc3038
Ladewig, J., Koch, P., and Brustle, O. (2013). Leveling Waddington: The emergence of direct programming and the loss of cell fate hierarchies. Nat. Rev. Mol. Cell Biol. 14, 225–236. PMID:23486282. doi:10.1038/nrm3543
Lai, K. P., Chen, J., He, M., Ching, A. K., Lau, C., Lai, P. B., et al. (2014). Overexpression of ZFX confers self-renewal and chemoresistance properties in hepatocellular carcinoma. Int. J. Cancer 135, 1790–1799. PMID:24585547. doi:10.1002/ijc.28819
Laity, J., Lee, B., and Wright, P. J. (2001). Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46. PMID:11179890. doi:10.1016/s0959-440x(00)00167-6
Lander, E., Linton, L., Birren, B., Nusbaum, C., Zody, M., Baldwin, J., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. PMID:11237011. doi:10.1038/35057062
Lea, R., McCarthy, A., Boeing, S., Fallesen, T., Elder, K., Snell, P., et al. (2021). KLF17 promotes human naïve pluripotency but is not required for its establishment. Development 148. dev199378. PMID:34661235. doi:10.1242/dev.199378
Lee, G., Jang, H., Kim, Y., Choe, S., Kong, J., Hwang, I., et al. (2019). SREBP1c-PAX4 Axis mediates pancreatic β-cell compensatory responses upon metabolic stress. Diabetes 68, 81–94. PMID:30352876. doi:10.2337/db18-0556
Lee, M. S., Gippert, G. P., Soman, K. V., Case, D. A., and Wright, P. E. (1989). Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science 245, 635–637. doi:10.1126/science.2503871
Lee, T., Castilho, A., Cheung, V., Tang, K., Ma, S., and Ng, I. J. C. (2011). CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell 9, 50–63. PMID:21726833. doi:10.1016/j.stem.2011.06.005
Li, C., Lu, L., Feng, B., Zhang, K., Han, S., Hou, D., et al. (2017b). The lincRNA-ROR/miR-145 axis promotes invasion and metastasis in hepatocellular carcinoma via induction of epithelial-mesenchymal transition by targeting ZEB2. Sci. Rep. 7, 4637. PMID:28680141. doi:10.1038/s41598-017-04113-w
Li, J., Miralles Fusté, J., Simavorian, T., Bartocci, C., Tsai, J., Karlseder, J., et al. (2017a). Tap: A telomere-associated protein involved in telomere length control. Science 355, 638–641. PMID:28082411. doi:10.1126/science.aah6752
Li, K., Zhao, G., Ao, J., Gong, D., Zhang, J., Chen, Y., et al. (2019). ZNF32 induces anoikis resistance through maintaining redox homeostasis and activating Src/FAK signaling in hepatocellular carcinoma. Cancer Lett. 442, 271–278. doi:10.1016/j.canlet.2018.09.033
Li, L. Y., Yang, J. F., Rong, F., Luo, Z. P., Hu, S., Fang, H., et al. (2021a). ZEB1 serves an oncogenic role in the tumorigenesis of HCC by promoting cell proliferation, migration, and inhibiting apoptosis via Wnt/β-catenin signaling pathway. Acta Pharmacol. Sin. 42, 1676–1689. PMID:33514855. doi:10.1038/s41401-020-00575-3
Li, X., Yuan, J., Song, C., Lei, Y., Xu, J., Zhang, G., et al. (2021b). Deubiquitinase USP39 and E3 ligase TRIM26 balance the level of ZEB1 ubiquitination and thereby determine the progression of hepatocellular carcinoma. Cell Death Differ. 28, 2315–2332. PMID:33649471. doi:10.1038/s41418-021-00754-7
Liang, Y., Li, Q., Chen, K., Ni, W., Zhan, Z., Ye, F., et al. (2017). Zinc finger protein 307 functions as a tumor-suppressor and inhibits cell proliferation by inducing apoptosis in hepatocellular carcinoma. Oncol. Rep. 38, 2229–2236. PMID:28765950. doi:10.3892/or.2017.5868
Lin, Q., Wu, Z., Yue, X., Yu, X., Wang, Z., Song, X., et al. (2020). ZHX2 restricts hepatocellular carcinoma by suppressing stem cell-like traits through KDM2A-mediated H3K36 demethylation. EBioMedicine 53, 102676. PMID:32114388. doi:10.1016/j.ebiom.2020.102676
Liontos, M., Koutsami, M., Sideridou, M., Evangelou, K., Kletsas, D., Levy, B., et al. (2007). Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 67, 10899–10909. PMID:18006835. doi:10.1158/0008-5472.CAN-07-2837
Liu, X., Chen, D., Chen, H., Wang, W., Liu, Y., Wang, Y., et al. (2021). YB1 regulates miR-205/200b-ZEB1 axis by inhibiting microRNA maturation in hepatocellular carcinoma. Cancer Commun. (Lond) 41, 576–595. PMID:34110104. doi:10.1002/cac2.12164
Llovet, J. M., Montal, R., Sia, D., and Finn, R. S. (2018). Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 15, 599–616. PMID:30061739. doi:10.1038/s41571-018-0073-4
Long, L., Xiang, H., Liu, J., Zhang, Z., and Sun, L. (2019). ZEB1 mediates doxorubicin (Dox) resistance and mesenchymal characteristics of hepatocarcinoma cells. Exp. Mol. Pathol. 106, 116–122. PMID:30615851. doi:10.1016/j.yexmp.2019.01.001
Lum, L., and Beachy, P. A. (2004). The hedgehog response network: Sensors, switches, and routers. Science 304, 1755–1759. PMID:15205520. doi:10.1126/science.1098020
Lunt, S. Y., and Vander Heiden, M. G. (2011). Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27, 441–464. PMID:21985671. doi:10.1146/annurev-cellbio-092910-154237
Madabhushi, R., Gao, F., Pfenning, A. R., Pan, L., Yamakawa, S., Seo, J., et al. (2015). Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161, 1592–1605. PMID:26052046. doi:10.1016/j.cell.2015.05.032
Malumbres, M. (2014). Cyclin-dependent kinases. Genome Biol. 15, 122. PMID:25180339. doi:10.1186/gb4184
Melnick, A., Carlile, G., Ahmad, K. F., Kiang, C. L., Corcoran, C., Bardwell, V., et al. (2002). Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol. Cell Biol. 22, 1804–1818. PMID:11865059. doi:10.1128/MCB.22.6.1804-1818.2002
Menendez, J. A., and Lupu, R. (2007). Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 7, 763–777. PMID:17882277. doi:10.1038/nrc2222
Najafabadi, H. S., Mnaimneh, S., Schmitges, F. W., Garton, M., Lam, K. N., Yang, A., et al. (2015). C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 33, 555–562. PMID:25690854. doi:10.1038/nbt.3128
Nieto, M. A., Huang, R. Y., Jackson, R. A., and Thiery, J. P. (2016). Emt: 2016. Cell 166, 21–45. PMID:27368099. doi:10.1016/j.cell.2016.06.028
Novak, D., Huser, L., Elton, J. J., Umansky, V., Altevogt, P., and Utikal, J. (2020). SOX2 in development and cancer biology. Semin. Cancer Biol. 67, 74–82. PMID:31412296. doi:10.1016/j.semcancer.2019.08.007
Ochoa, B., Syn, W. K., Delgado, I., Karaca, G. F., Jung, Y., Wang, J., et al. (2010). Hedgehog signaling is critical for normal liver regeneration after partial hepatectomy in mice. Hepatology 51, 1712–1723. PMID:20432255. doi:10.1002/hep.23525
Plaks, V., Kong, N., and Werb, Z. (2015). The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 16, 225–238. PMID:25748930. doi:10.1016/j.stem.2015.02.015
Pommier, Y. (2013). Drugging topoisomerases: Lessons and challenges. ACS Chem. Biol. 8, 82–95. PMID:23259582. doi:10.1021/cb300648v
Pucci, F., Gardano, L., and Harrington, L. (2013). Short telomeres in ESCs lead to unstable differentiation. Cell Stem Cell 12, 479–486. PMID:23561444. doi:10.1016/j.stem.2013.01.018
Raoul, J. L., Kudo, M., Finn, R. S., Edeline, J., Reig, M., and Galle, P. R. (2018). Systemic therapy for intermediate and advanced hepatocellular carcinoma: Sorafenib and beyond. Cancer Treat. Rev. 68, 16–24. PMID:29783126. doi:10.1016/j.ctrv.2018.05.006
Rivera, T., Haggblom, C., Cosconati, S., and Karlseder, J. (2017). A balance between elongation and trimming regulates telomere stability in stem cells. Nat. Struct. Mol. Biol. 24, 30–39. PMID:27918544. doi:10.1038/nsmb.3335
Rowe, H. M., and Trono, D. (2011). Dynamic control of endogenous retroviruses during development. Virology 411, 273–287. PMID:21251689. doi:10.1016/j.virol.2010.12.007
Sander, T. L., Stringer, K. F., Maki, J. L., Szauter, P., Stone, J. R., and Collins, T. (2003). The SCAN domain defines a large family of zinc finger transcription factors. Gene 310, 29–38. doi:10.1016/s0378-1119(03)00509-2
Sarek, G., Marzec, P., Margalef, P., and Boulton, S. J. (2015). Molecular basis of telomere dysfunction in human genetic diseases. Nat. Struct. Mol. Biol. 22, 867–874. PMID:26581521. doi:10.1038/nsmb.3093
Sari, I. N., Phi, L. T. H., Jun, N., Wijaya, Y. T., Lee, S., and Kwon, H. Y. (2018). Hedgehog signaling in cancer: A prospective therapeutic target for eradicating cancer stem cells. Cells 7, 208. PMID:30423843. doi:10.3390/cells7110208
Schellenberg, M., Lieberman, J., Herrero-Ruiz, A., Butler, L., Williams, J., Muñoz-Cabello, A., et al. (2017). ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links. Science 357:1412–1416. PMID:28912134. doi:10.1126/science.aam6468
Schneider-Gädicke, A., Beer-Romero, P., Brown, L., Mardon, G., Luoh, S., and Page, D. J. N. (1989). Putative transcription activator with alternative isoforms encoded by human ZFX gene. Nature 342:708–711. PMID:2512506. doi:10.1038/342708a0
Schulze, A., and Harris, A. L. (2012). How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 491, 364–373. doi:10.1038/nature11706
Schuppan, D., and Schattenberg, J. M. (2013). Non-alcoholic steatohepatitis: Pathogenesis and novel therapeutic approaches. J. Gastroenterol. Hepatol. 28 (1), 68–76. PMID:23855299. doi:10.1111/jgh.12212
Sebastian, R., and Oberdoerffer, P. (2017). Transcription-associated events affecting genomic integrity. Philos. Trans. R. Soc. Lond B Biol. Sci. 372, 20160288. PMID:28847825. doi:10.1098/rstb.2016.0288
Sekiya, S., and Suzuki, A. (2011). Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393. PMID:21716291. doi:10.1038/nature10263
Shan, J., Shen, J., Liu, L., Xia, F., Xu, C., Duan, G., et al. (2012). Nanog regulates self-renewal of cancer stem cells through the insulin-like growth factor pathway in human hepatocellular carcinoma. Hepatology 56, 1004–1014. PMID:22473773. doi:10.1002/hep.25745
Shanbhag, N. M., Rafalska-Metcalf, I. U., Balane-Bolivar, C., Janicki, S. M., and Greenberg, R. A. (2010). ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981. PMID:20550933. doi:10.1016/j.cell.2010.04.038
Shih, J-Y., Tsai, M-F., Chang, T-H., Chang, Y-L., Yuan, A., Yu, C-J., et al. (2005). Transcription repressor slug promotes carcinoma invasion and predicts outcome of patients with lung adenocarcinoma. Clin. Cancer Res. 11, 8070–8078. doi:10.1158/1078-0432.Ccr-05-0687
Singh, A., Greninger, P., Rhodes, D., Koopman, L., Violette, S., Bardeesy, N., et al. (2009). A gene expression signature associated with "K-Ras addiction" reveals regulators of EMT and tumor cell survival. Cancer Cell 15, 489–500. PMID:19477428. doi:10.1016/j.ccr.2009.03.022
Singh, S. S., Vats, S., Chia, A. Y., Tan, T. Z., Deng, S., Ong, M. S., et al. (2018). Dual role of autophagy in hallmarks of cancer. Oncogene 37, 1142–1158. PMID:29255248. doi:10.1038/s41388-017-0046-6
Sreekumar, R., Emaduddin, M., Al-Saihati, H., Moutasim, K., Chan, J., Spampinato, M., et al. (2019). Protein kinase C inhibitors override ZEB1-induced chemoresistance in HCC. Cell Death Dis. 10, 703. PMID:31543517. doi:10.1038/s41419-019-1885-6
Stubbs, L., Sun, Y., and Caetano-Anolles, D. (2011). Function and evolution of C2H2 zinc finger arrays. Subcell. Biochem. 52, 75–94. PMID:21557079. doi:10.1007/978-90-481-9069-0_4
Sun, J., Wang, B., Liu, Y., Zhang, L., Ma, A., Yang, Z., et al. (2014). Transcription factor KLF9 suppresses the growth of hepatocellular carcinoma cells in vivo and positively regulates p53 expression. Cancer Lett. 355, 25–33. doi:10.1016/j.canlet.2014.09.022
Sun, L., Suo, C., Li, S. T., Zhang, H., and Gao, P. (2018). Metabolic reprogramming for cancer cells and their microenvironment: Beyond the Warburg Effect. Biochim. Biophys. Acta Rev. Cancer 1870, 51–66. PMID:29959989. doi:10.1016/j.bbcan.2018.06.005
Sun, Z., Han, Q., Zhou, N., Wang, S., Lu, S., Bai, C., et al. (2013). MicroRNA-9 enhances migration and invasion through KLF17 in hepatocellular carcinoma. Mol. Oncol. 7, 884–894. 884. doi:10.1016/j.molonc.2013.04.007
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. PMID:33538338. doi:10.3322/caac.21660
Tammaro, M., Barr, P., Ricci, B., and Yan, H. (2013). Replication-dependent and transcription-dependent mechanisms of DNA double-strand break induction by the topoisomerase 2-targeting drug etoposide. PLoS One 8, e79202. PMID:24244448. doi:10.1371/journal.pone.0079202
Tang, K. H., Ma, S., Lee, T. K., Chan, Y. P., Kwan, P. S., Tong, C. M., et al. (2012). CD133(+) liver tumor-initiating cells promote tumor angiogenesis, growth, and self-renewal through neurotensin/interleukin-8/CXCL1 signaling. Hepatology 55, 807–820. PMID:21994122. doi:10.1002/hep.24739
Teoh, S. T., and Lunt, S. Y. (2018). Metabolism in cancer metastasis: Bioenergetics, biosynthesis, and beyond. Wiley Interdiscip. Rev. Syst. Biol. Med. 10. PMID:28990735. doi:10.1002/wsbm.1406
Thorburn, A., Thamm, D. H., and Gustafson, D. L. (2014). Autophagy and cancer therapy. Mol. Pharmacol. 85, 830–838. PMID:24574520. doi:10.1124/mol.114.091850
Tsai, S. C., Lin, C. C., Shih, T. C., Tseng, R. J., Yu, M. C., Lin, Y. J., et al. (2017). The miR-200b-ZEB1 circuit regulates diverse stemness of human hepatocellular carcinoma. Mol. Carcinog. 56, 2035–2047. PMID:28383782. doi:10.1002/mc.22657
Uchikado, Y., Natsugoe, S., Okumura, H., Setoyama, T., Matsumoto, M., Ishigami, S., et al. (2005). Slug Expression in the E-cadherin preserved tumors is related to prognosis in patients with esophageal squamous cell carcinoma. Clin. Cancer Res. 11, 1174–1180. PMID:15709186. doi:10.1158/1078-0432.1174.11.3
Urrutia, R. (2003). KRAB-containing zinc-finger repressor proteins. Genome Biol. 4, 231. PMID:14519192. doi:10.1186/gb-2003-4-10-231
Vandewalle, C., Van Roy, F., and Berx, G. (2008). The role of the ZEB family of transcription factors in development and disease. Cell. Mol. Life Sci. 66, 773–787. doi:10.1007/s00018-008-8465-8
Vaziri, C., Saxena, S., Jeon, Y., Lee, C., Murata, K., Machida, Y., et al. (2003). A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11, 997–1008. PMID:12718885. doi:10.1016/s1097-2765(03)00099-6
Villanueva, A. (2019). Hepatocellular carcinoma. N. Engl. J. Med. 380, 1450–1462. PMID:30970190. doi:10.1056/NEJMra1713263
Wang, C., Fu, S. Y., Wang, M. D., Yu, W. B., Cui, Q. S., Wang, H. R., et al. (2017). Zinc finger protein X-linked promotes expansion of EpCAM(+) cancer stem-like cells in hepatocellular carcinoma. Mol. Oncol. 11, 455–469. PMID:28156061. doi:10.1002/1878-0261.12036
Wang, N., Li, M. Y., Liu, Y., Yu, J., Ren, J., Zheng, Z., et al. (2020). ZBP-89 negatively regulates self-renewal of liver cancer stem cells via suppression of Notch1 signaling pathway. Cancer Lett. 472, 70–80. PMID:31874246. doi:10.1016/j.canlet.2019.12.026
Wang, N., Wang, S., Yang, S. L., Liu, L. P., Li, M. Y., Lai, P. B. S., et al. (2018). Targeting ZBP-89 for the treatment of hepatocellular carcinoma. Expert Opin. Ther. Targets 22, 817–822. PMID:30142986. doi:10.1080/14728222.2018.1516753
Wang, X. Q., Ongkeko, W. M., Chen, L., Yang, Z. F., Lu, P., Chen, K. K., et al. (2010). Octamer 4 (Oct4) mediates chemotherapeutic drug resistance in liver cancer cells through a potential Oct4-AKT-ATP-binding cassette G2 pathway. Hepatology 52, 528–539. PMID:20683952. doi:10.1002/hep.23692
Weirauch, M. T., and Hughes, T. R. (2011). A catalogue of eukaryotic transcription factor types, their evolutionary origin, and species distribution. Subcell. Biochem. 52, 25–73. PMID:21557078. doi:10.1007/978-90-481-9069-0_3
Wu, Z., Ma, H., Wang, L., Song, X., Zhang, J., Liu, W., et al. (2020). Tumor suppressor ZHX2 inhibits NAFLD-HCC progression via blocking LPL-mediated lipid uptake. Cell Death Differ. 27, 1693–1708. PMID:31740790. doi:10.1038/s41418-019-0453-z
Xie, W., Qiao, X., Shang, L., Dou, J., Yang, X., Qiao, S., et al. (2018). Knockdown of ZNF233 suppresses hepatocellular carcinoma cell proliferation and tumorigenesis. Gene 679, 179–185. PMID:30179682. doi:10.1016/j.gene.2018.08.070
Yamashita, T., Ji, J., Budhu, A., Forgues, M., Yang, W., Wang, H. Y., et al. (2009). EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 136, 1012–1024. PMID:19150350. doi:10.1053/j.gastro.2008.12.004
Yan, Y., Zuo, X., and Wei, D. (2015). Concise review: Emerging role of CD44 in cancer stem cells: A promising biomarker and therapeutic target. Stem Cells Transl. Med. 4, 1033–1043. PMID:26136504. doi:10.5966/sctm.2015-0048
Yang, J., Cui, R., and Liu, Y. (2020). MicroRNA-212-3p inhibits paclitaxel resistance through regulating epithelial-mesenchymal transition, migration and invasion by targeting ZEB2 in human hepatocellular carcinoma. Oncol. Lett. 20, 23. PMID:32774496. doi:10.3892/ol.2020.11884
Yang, W., and Lu, Z. (2013). Regulation and function of pyruvate kinase M2 in cancer. Cancer Lett. 339, 153–158. PMID:23791887. doi:10.1016/j.canlet.2013.06.008
Yang, X. R., Xu, Y., Yu, B., Zhou, J., Li, J. C., Qiu, S. J., et al. (2009). CD24 is a novel predictor for poor prognosis of hepatocellular carcinoma after surgery. Clin. Cancer Res. 15, 5518–5527. PMID:19706825. doi:10.1158/1078-0432.CCR-09-0151
Yang, Z. F., Ho, D. W., Ng, M. N., Lau, C. K., Yu, W. C., Ngai, P., et al. (2008). Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell 13, 153–166. PMID:18242515. doi:10.1016/j.ccr.2008.01.013
Yoshida, K., Sugimoto, N., Iwahori, S., Yugawa, T., Narisawa-Saito, M., Kiyono, T., et al. (2010). CDC6 interaction with ATR regulates activation of a replication checkpoint in higher eukaryotic cells. J. Cell Sci. 123, 225–235. PMID:20048340. doi:10.1242/jcs.058693
Yu, X., Lin, Q., Wu, Z., Zhang, Y., Wang, T., Zhao, S., et al. (2020). ZHX2 inhibits SREBP1c-mediated de novo lipogenesis in hepatocellular carcinoma via miR-24-3p. J. Pathol. 252, 358–370. PMID:32770671. doi:10.1002/path.5530
Yuan, K., Xie, K., Lan, T., Xu, L., Chen, X., Li, X., et al. (2020). TXNDC12 promotes EMT and metastasis of hepatocellular carcinoma cells via activation of beta-catenin. Cell Death Differ. 27, 1355–1368. PMID:31570854. doi:10.1038/s41418-019-0421-7
Zhang, L., Huo, Q., Ge, C., Zhao, F., Zhou, Q., Chen, X., et al. (2020). ZNF143-Mediated H3K9 trimethylation upregulates CDC6 by activating MDIG in hepatocellular carcinoma. Cancer Res. 80, 2599–2611. PMID:32312832. doi:10.1158/0008-5472.CAN-19-3226
Zhang, R., Wu, W. R., Shi, X. D., Xu, L. B., Zhu, M. S., Zeng, H., et al. (2016). Dysregulation of Bmi1 promotes malignant transformation of hepatic progenitor cells. Oncogenesis 5, e203. PMID:26926789. doi:10.1038/oncsis.2016.6
Zhang, T., Huang, Y., Liu, W., Meng, W., Zhao, H., Yang, Q., et al. (2017). Overexpression of zinc finger protein 687 enhances tumorigenic capability and promotes recurrence of hepatocellular carcinoma. Oncogenesis 6, e363. PMID:28737756. doi:10.1038/oncsis.2017.63
Zhou, X. T., Ding, J., Li, H. Y., Zuo, J. L., Ge, S. Y., Jia, H. L., et al. (2020). Hedgehog signalling mediates drug resistance through targeting TAP1 in hepatocellular carcinoma. J. Cell Mol. Med. 24, 4298–4311. PMID:32108992. doi:10.1111/jcmm.15090
Zhou, Y., Lin, F., Wan, T., Chen, A., Wang, H., Jiang, B., et al. (2021). ZEB1 enhances Warburg effect to facilitate tumorigenesis and metastasis of HCC by transcriptionally activating PFKM. Theranostics 11, 5926–5938. PMID:33897890. doi:10.7150/thno.56490
Keywords: C2H2 zinc finger proteins, hepatocellular carcinoma, EMT, stemness maintenance, metabolic reprogramming, anti-tumor drug resistance
Citation: Li Q, Tan G and Wu F (2023) The functions and roles of C2H2 zinc finger proteins in hepatocellular carcinoma. Front. Physiol. 14:1129889. doi: 10.3389/fphys.2023.1129889
Received: 22 December 2022; Accepted: 20 June 2023;
Published: 29 June 2023.
Edited by:
Stephen J. Pandol, Cedars Sinai Medical Center, United StatesCopyright © 2023 Li, Tan and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fan Wu, d3VmYW43OTExQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.