
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol., 06 October 2022
Sec. Cell Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.953880
Diabetes mellitus (DM) is a fast-growing chronic metabolic disorder that leads to significant health, social, and economic problems worldwide. Chronic hyperglycemia caused by DM leads to multiple devastating complications, including macrovascular complications and microvascular complications, such as diabetic cardiovascular disease, diabetic nephropathy, diabetic neuropathy, and diabetic retinopathy. Numerous studies provide growing evidence that aberrant expression of and mutations in RNA-binding proteins (RBPs) genes are linked to the pathogenesis of diabetes and associated complications. RBPs are involved in RNA processing and metabolism by directing a variety of post-transcriptional events, such as alternative splicing, stability, localization, and translation, all of which have a significant impact on RNA fate, altering their function. Here, we purposed to summarize the current progression and underlying regulatory mechanisms of RBPs in the progression of diabetes and its complications. We expected that this review will open the door for RBPs and their RNA networks as novel therapeutic targets for diabetes and its related complications.
Diabetes mellitus is one of the fastest-growing metabolic disorders characterized by chronic hyperglycemia. In recent decades, the global prevalence of diabetes in adults has been growing at an astonishing rate. It is estimated that in 2045 there will be 693 million adults who suffered from diabetes worldwide (Cho et al., 2018; Harding et al., 2019). Depending on the different mechanisms, diabetes can be divided into two main forms, type 1 diabetes mellitus (T1DM) and type 2 diabetes mellitus (T2DM). T1DM is considered an auto-immune disease defined by islets β-cell damage and absolute lack of insulin. T2DM is often accompanied by relatively insufficient insulin secretion and insulin resistance, preventing insulin from stimulating glucose uptake into target tissues, resulting in elevated blood glucose levels (Atkinson et al., 2014; Petersmann et al., 2019; Weir et al., 2020). Persistent hyperglycemia affects nearly every tissue of the body that causes severe macrovascular and microvascular complications, with retinopathy, cardiomyopathy, nephropathy, neuropathy, and peripheral vascular disease serving as key avenues of morbidity (Cole and Florez, 2020). Currently, a thorough knowledge of the molecular pathophysiology of diabetic complications remains elusive. Emerging evidence supports that the RNA-binding proteins are involved in the occurrence and development of diabetes and its complications (Nutter and Kuyumcu-Martinez, 2018; Salem et al., 2019; Good and Stoffers, 2020; Kelaini et al., 2021). The specific mechanisms will be described in detail in the present review.
RBPs are typically considered as proteins that are responsible for modulating post-transcriptional gene expression in the eukaryotic cells (Gerstberger et al., 2014). Thousands of such RBPs have been discovered and investigated over the years. RBPs can recognize and interact with their target RNAs to form ribonucleoprotein (RNP) complexes which control almost every aspect of post-transcriptional processing of target RNA substrates, including pre-mRNA splicing, translational control, cleavage and polyadenylation, RNA stability, RNA localization, nuclear export, and RNA editing (Van Nostrand et al., 2020). In recent years, numerous RBPs have been demonstrated to be involved in many human diseases, from cardiovascular diseases and endocrine dysfunction to cancer, and neurodegenerative disorders (Pereira et al., 2017; Lujan et al., 2018; Yang et al., 2018; Cao et al., 2021; Kelaini et al., 2021; Klim et al., 2021). It has been shown that post-transcriptional dysregulation is linked to diabetes mellitus, which serves as a reminder that RBPs may be crucial in the pathogenesis of diabetes and its related complications (Nutter and Kuyumcu-Martinez, 2018).
In this review, we will provide a brief overview of the mechanisms by which RBPs exercise their functions. Then, we will explain how these RBPs are dysregulated and their contribution to the pathological process of diabetes mellitus. In addition, we would like to focus on the relationships between RBPs and diabetic complications. We believe that a comprehensive understanding of the RBPs’ role in diabetes and its associated complications may aid in the development of innovative treatments in clinic.
The mRNA life cycle is a complex system that includes the process of transforming the newly transcribed mRNA molecules to fully functional mature mRNA transcripts. RBPs play an essential role in this process. Recent studies have revealed some RBPs do not have typical RNA binding domains (RBDs) but are replaced by at least one intrinsically disordered region (IDR) through which they can not only be involved in aggregation of RNPs, but also directly engage in RNA binding (Hentze et al., 2018). However, most RBPs are considered to interact with their target RNAs by a limited set of RBDs, such as the RNA recognition motif (RRM), hnRNP K homology domain (KH), zinc-finger, and DEAD/DEAH box helicase (Van Nostrand et al., 2020). The interaction of RBP-RNA occurs at RBD, which is mainly located within 5′and 3′untranslated regions (UTRs) of RNA, although it can also be found at the intronic and exonic regions. RBPs usually have a series of repeats RBDs that work together to improve specificity and affinity for their target mRNAs. Multiple target mRNAs can have their expression controlled by a single RBP. Multiple RBPs can interact with the same mRNA, playing a role in either cooperation or competition (Pope and Medzhitov, 2018; Van Nostrand et al., 2020).
The mechanisms of post-transcriptional regulation by RBPs are complex and elaborate, including 5′capping, alternative splicing of pre-mRNAs, polyadenylation, RNA degradation and stabilization, RNA localization and export, proteins translation (Figure 1) (Corley et al., 2020). RBPs can promote the maturation of RNA via pre-mRNA alternative splicing, polyadenylation, RNA editing, and the addition of the 5′ cap in the target RNAs. RBPs can regulate pre-mRNA alternative splicing (AS) through binding to the pre-mRNA and interacting with the spliceosome components, which generate variant protein isoforms from a single gene, resulting in transcriptome and proteome diversity (Sperling, 2017). Previous studies demonstrated that AS controlled by RBPs plays a crucial role in diabetes and its complications (Verma et al., 2013; Nutter et al., 2016; Gazzara et al., 2017; Belanger et al., 2019; Verma et al., 2022). The role of RBPs in AS will be discussed in detail in the next section.
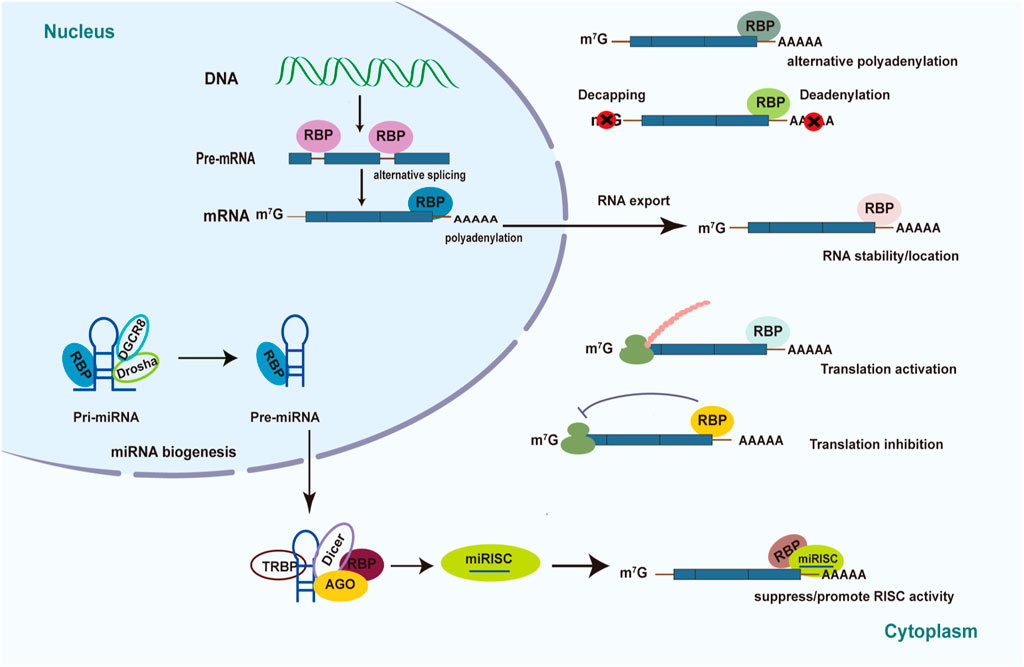
FIGURE 1. Mechanism of posttranscriptional regulation controlled by RBPs in diabetes. Schematic diagrams summarize the various roles of RBPs played in diabetes pathology and diabetic complications. RBPs have the ability to determine RNA’s fate through pre-mRNA splicing, translational control, polyadenylation, RNA stability, RNA localization, RNA export, and miRNA-mediated processing.
mRNA can be exported from the nucleus to the cytoplasm to perform the function of protein translation. RBPs such as eIF4E is essential for translation of majority of mRNAs. It has been reported that mRNA Cap-binding protein eIF4E can recognize the structure of the 5′-m7GTP cap of mRNA and assemble it into eIF4F complexes which can recruit ribosomes onto mRNA to perform translational functions (Lazaris-Karatzas et al., 1990; Osborne and Borden, 2015; Ho and Lee, 2016). However, under diabetic conditions, the formation of eIF4F complexes is inhibited and thus affects the translation rate, contributing to the development of diabetic complications (Schrufer et al., 2010; Dennis et al., 2015; Miller et al., 2016). Besides, RBPs can trigger the degradation and RNA decay process by binding to cis-regulatory RNA elements and recruiting mediators (Pérez-Ortín et al., 2013). For example, 5′ cap can be removed by decapping enzymes while 3′-poly A tail can be diminished by deadenylating enzymes. A well-known element that mediates degradation is the AU-rich element (ARE) located in the 3′ UTRs of mRNA (Mayr, 2019). Multiple studies showed that RBPs such as Tristetraprolin (TTP) and ELAV families can regulate the degradation of target mRNAs that contain the ARE element (Makita et al., 2021; Sidali et al., 2021). Transcription and degradation rates together regulate the content of intracellular mRNA.
RBPs can also control the post-transcriptional regulatory process by regulating the biogenesis and function of non-coding RNAs (Ho et al., 2021; Yao et al., 2022). For instance, RBPs are required to generate miRNAs and miRNA-mediated gene expression (Figure 1) (Ciafrè and Galardi, 2013). Immature miRNAs are normally translated into long primary transcripts (pri-miRNAs) with a stem-loop structure in the canonical miRNA biosynthesis pathway (Velázquez-Cruz et al., 2021). RBPs can influence the processing of pri-miRNA and precursor (pre-miRNA) biogenesis by identifying and binding sequences or special structures of the hairpin RNA. For example, RBP Lin28 can inhibit pri- and pre-let-7 miRNA biogenesis by interacting with the terminal loop of these immature miRNA via Drosha and Dicer (Mayr and Heinemann, 2013). Furthermore, overexpression of Lin28 was reported to increase glucose utilization in different tissues as well as prevent weight gain by suppressing let-7 miRNA biogenesis (Zhu et al., 2011; Shinoda et al., 2013). The interaction between Lin28 and let-7 miRNA may affect the pancreatic β-cell functions (Sung et al., 2019). RBPs have the ability to influence the stability and turnover of mature miRNAs (Ciafrè and Galardi, 2013; Connerty et al., 2015; Fukao et al., 2015). The Argonaute (AGO) proteins bind to double-stranded miRNAs and combine with them to form the miRNA-induced silencing complex (miRISC), in which one strand of the RNA duplex becomes functional while the other is deleted. The mature single-stranded miRNA generally binds to the 3′UTR of their target mRNAs, directing the translational inhibition and RNA degradation activity of miRISC. RBPs can bind to the 3′UTR of the target mRNAs to suppress the functions of miRNA via competing for the same binding motif or restructuring the target RNAs. In addition, RBPs can also alter the structure of 3′UTRs to facilitate miRNA binding, thereby promoting gene post-transcriptional regulation mediated by miRNA (Connerty et al., 2015; Velázquez-Cruz et al., 2021).
Pancreatic islet β-cells are marked by their ability to synthesize and secrete large amounts of insulin, which maintain metabolic homeostasis via lowering glycemia (Campbell and Newgard, 2021). Although the pathogenesis of the two types of diabetes is not exactly consistent, T1DM and T2DM share common pathologies, such as decreased β-cell mass and loss of insulin secretory function (Eizirik et al., 2020). RBPs regulate a variety of processes in pancreatic β-cell, including insulin synthesis and secretion (Good and Stoffers, 2020; Demir et al., 2021). The abundant RNA-binding protein PTBP1 is the most well-studied regulator of insulin secretion (Magro and Solimena, 2013). In pancreatic β-cells, PTBP1 stabilizes preproinsulin mRNA by binding to the pyrimidine-rich region in its 3′UTR, thereby promoting the protein level of insulin. And it is regulated by glucose stimulation. In rat insulinoma INS-1 cells, suppression of PTBP1 by RNAi reduces insulin secretion (Knoch et al., 2004; Knoch et al., 2014). Besides, PTBP1 can also bind and stabilize 3′UTR of islet cell autoantigen (ICA512) mRNA, which is considered as an integral membrane protein of the insulin immature secretory granules (SGs). PTBP1 may stimulate the translation of insulin SG proteins via cap-independent mechanisms, which may be mediated by PTBP1 binding to the 5′UTR of the human preproinsulin (Ins2) mRNA (Knoch et al., 2004; Fred et al., 2011; Kulkarni et al., 2011; Knoch et al., 2014). In line with these finds, it was later discovered that the expression of PTBP1 in glucose-induced β-cells is mediated by the insulin receptor (IR) signaling pathway through Akt, and silencing Akt can significantly reduce the level of PTBP1 expression (Jeong et al., 2018). Accordingly, the level of RBP HuD decreased in β-cells of diabetes. HuD can bind to the Ins2 5′UTR to inhibit the translation of Ins2 and reduce insulin production. After glucose stimulation, Ins2 mRNA is promptly released from HuD, accompany by enabling translation of Ins2 mRNA. HuD knockout mice exhibit increased insulin levels in β-cells, while HUD overexpressed mice do the opposite (Lee et al., 2012). Furthermore, HuD enhances mitofusin 2 (Mfn2) expression level by binding to the 3′UTR of Mfn2 mRNA. In pancreatic β-cells, its decreased expression causes mitochondrial dysfunction (Hong et al., 2020). RBP hnRNPK, a member of the poly C-binding protein family, is phosphorylated and upregulated in islets under conditions associated with T2D. HnRNPK can bind to the poly C-rich fragments in JUND mRNA 3′UTR, thus influencing β-cell redox homeostasis and apoptosis. Post-transcriptional upregulation of JUND is blocked due to hnRNPK deletion during metabolic stress. Besides, DDX3X is essential for the efficient translation of JUND mRNA by interacting with hnRNPK (Good et al., 2019). In addition, overexpression of RBP Lin28a protects pancreatic β-cells from damage caused by streptozotocin (STZ) both in vitro and in vivo. Lin28a enhanced cell survival and proliferation through activating the PI3K-Akt signaling pathway, which is possibly regulated by let-7 (Sung et al., 2019). DDX1, an RNA-binding protein of the DEAD-box helicase superfamily, can promote translation activity of insulin mRNA by binding to its mRNA. Free Fatty Acids (FFAs) treatment causes DDX1 to be phosphorylated and dissociated from insulin mRNA, resulting in insulin translation inhibition (Li et al., 2018). RNA-binding protein CUGBP1 is upregulated in the islets of diabetic mice. CUGBP1 reduces insulin secretion in reply to glucose and GLP-1 stimulation by binding to the 3′UTR ATTTGTT sequence of PDE3B (Zhai et al., 2016).
RNA-binding proteins can not only directly bind to mRNA to regulate β-cell function and insulin secretion, but there have also been instances of RBPs and circular RNA interacting to exercise regulatory activities (Wu et al., 2022). For example, TDP-43 is a nuclear protein that acts as a regulator of gene expression as well as a DNA- and RNA-binding protein involved in RNA metabolism. It has recently been shown that knockout of TDP-43 in β-cells leads to defective insulin secretion (Araki et al., 2019). The intronic circRNA (ci-Ins2/ci-INS), generated by second intron excised from the primary insulin transcript, can interact with the RBP TDP-43 thereby controlling the expression of genes essential for insulin secretion in β-cells (Stoll et al., 2020). CircPPM1F competitively interacted with RBP HuR to suppress PPM1F translation, thus leading to pancreatic β-cell apoptosis through promoting M1 macrophage activation. Besides, two RBPs EIF4A3 and FUS might be oppositely regulated and maintained the expression of circPPM1F during the progression of T1DM (Zhang et al., 2020).
Tristetraprolin (TTP, also known as ZFP36) is an RBP that depresses post-transcriptional gene expression via interacting with AU-rich elements (AREs) in the 3′UTR of target mRNAs(Blackshear, 2002). TTP is induced by insulin stimulation in vitro and in vivo (Cao et al., 2008). The levels of TTP are decreased in the livers of diabetic mice and humans. TTP suppression may be due to insulin resistance and reduced AKT signal that regulates TTP at the promoter level under diabetic conditions. TTP binds to FGF21 mRNA 3′UTR leading to a degeneration of FGF21. TTP-KO mice may improve systemic glucose tolerance and insulin sensitivity by increasing liver-induced FGF21 (Sawicki et al., 2018). RBP hnRNP A1 interacts with glycogen synthase (gys1) and stabilizes its mRNA, thus facilitating glycogen synthesis in muscle tissue and preserving insulin sensitivity. Severe insulin resistance is caused by the absence of hnRNP A1 in mice fed a high-fat diet (HFD) (Zhao et al., 2020). In addition, it was also reported that myeloid-specific loss of TTP protects against glucose intolerance and improves insulin sensitivity in obesity (Caracciolo et al., 2018). Insulin-like growth factor binding protein (IGFBP1) modulates cellular responses independently of IGF binding through interaction of the Arg-Gly-Asp (RGD) sequence of IGFBP-1 with the cell surface integrin receptors. Previous studies have indicated that increasing the levels of circulating IGFBP1 improved insulin sensitivity in mice and humans (Gokulakrishnan et al., 2012; Rajwani et al., 2012). Furthermore, IGFBP1 increases insulin sensitivity by RGD integrin-binding domain and activation of focal adhesion kinase (FAK), thus improving glucose uptake in skeletal muscle cells. In response to glucose stimulation, RGD peptides can also increase insulin secretion of β-cells via FAK and integrin-linked kinase (ILK) activation (Haywood et al., 2017).
Diabetic neuropathy affects at least half of patients with the development of diabetes. Patients with diabetes are characterized by signs of axonal degeneration and incomplete regeneration, demyelinating, and microangiopathy (Feldman et al., 2019; Calcutt, 2020). It has been reported that decrease of RNA-binding protein ZBP1 fails axonal RNA localization into the injured axons after sciatic nerve injury in T1DM rodent model induced by streptozotocin. This failure of RNA mobilization links to a reduction in axonal regeneration. When over-expression of ZBP1, this RBP can rescue in vitro growth defects in injured dorsal root ganglion (DRGs) from diabetic rats (Jones et al., 2021). Thus it shows that ZBP1 is a crucial savior in regeneration after axonal injury in diabetic rats.
Elav-like gene encodes Hu proteins, which belong to the RBPs superfamily. The Hu proteins family has three neuronal-specific members HuB, HuC, and HuD (encoded by Elav-like 2,3,4 genes respectively), while the fourth is HuR or HuA (encoded by Elav-like 1 gene) is omnipresent (Ambrosio et al., 2021). In another study, the neuronal-specific Hu proteins expression level was not correlated with its own gene in the thermal hypoalgesia condition caused by the advanced diabetic neuropathy. Moreover, the levels of Elavl2 and Elavl3 are reduced, while HuB is upregulated and HuD is downregulated in diabetic mice, compared to control one. Compared to control mice, Elavl genes and Hu proteins levels are significantly downregulated on the premise that algesic profile is unchanged under exposure to thermal radiation in diabetic-resistance mice (Mustăciosu et al., 2019). It has been certificated HuD protein upregulation in thermal hyperalgesia, which is the early phases of diabetes while otherwise in the thermal hypoalgesia condition caused by the advanced phases of diabetes (Sanna et al., 2014; Sanna et al., 2015). Previous studies indicated HuD can promote nerve regeneration and axon repair through interacting with mRNA by regulating its location or stabilizing the target mRNA (Wang et al., 2015; Gomes et al., 2017; Sanna et al., 2017). Therefore, it is reasonable to believe that the regulation of thermal hypoalgesia due to advanced diabetic neuropathy is closely related to changes in the post-transcriptional regulation of RNA in which RBPs are involved. What is more, the expression level of HuC in DRG neurons of rats with diabetic neuropathy is increased and is closely related to diabetic colonic hypersensitivity according to our unpublished research. We believe that the crucial role of Hu protein family in diabetic neuropathy can be further comprehended along with emerging research.
Diabetic nephropathy (DN) is one of the most common chronic complications of both type 1 and 2 diabetes and is considered as a main cause of end-stage renal disease (ESRD). Glomerular basement membrane thickening, mesangial growth and hypertrophy, and the accumulation of extracellular matrix (ECM) proteins are all hallmarks of DN (Kanwar et al., 2011; Zoja et al., 2020). In a type 1 diabetes model, the RNA-binding protein HuR rapidly upregulated NAPDH oxidase 4 (NOX4) expression levels by binding to AU-rich elements (Ares) in the NOX4 mRNA 3′UTR, which induced mesangial cell (MC) fibrotic injury and kidney damage, and a reno-protective role was shown by suppressing HuR expression in type 1 diabetic mouse models (Shi et al., 2020). HuR can also bind to the 3′UTR Ares of the NOD2, increasing NOD2 expression and mRNA stability, which leads to glomerular mesangial cells damage and proteinuria in diabetic rats (Shang et al., 2015). In addition, transforming growth factor-β1 (TGF-β1) can cause mesangial extracellular matrix (ECM) proteins like collagen type 1-α2 (Col1a2) and type 4-α1 (Col4a1) to accumulate. Let-7 family miRNAs protect mouse mesangial cells (MMC) from collagen accumulation by inhibiting the levels of Col1a2 and Col4a1. Under diabetic conditions, elevated TGF-β1 expressions cause an increase in RBP Lin28b level, which is considered as a crucial inhibitor of let-7 miRNA biogenesis, thereby leading to the decrease of let-7 miRNA and the accumulation of mesangial ECM proteins (Park et al., 2014). RBP IMP2 can regulate the translation of Laminin-β2 (LAMB2), which is a component of the glomerular basement membrane and is associated with actin during translation. Decreased expression of IMP2 and Lamb2 in the diabetic condition leads to impaired mesangial cell migration and proteinuria (Schaeffer et al., 2012).
As noted above, there is a multitude of RBPs involved in the pathogenesis of glomerular mesangial cells damage and kidney injury. But do RBPs have an effect on renal parenchymal cells in diabetic conditions? The answer is obvious. Heterogeneous nuclear ribonucleoproteins (hnRNPs) are pre-mRNA binding proteins that can regulate the processing of mRNA. In renal proximal tubular cells of Akita hnRNP F-Tg mice, selective overexpression of RBP hnRNP F lowers expressions of angiotensinogen (Agt) and TGF-β1 and reduces kidney hypertrophy and glomerulotubular fibrosis (Lo et al., 2012). HnRNP F can suppress the transcriptional activity of rat Agt gene promoter by binding to the insulin-responsive element (IRE) (Wei et al., 2005). RNA-binding protein TTP expression was significantly reduced, while HuR expression was elevated in glomerular podocytes of patients with DKD and db/db mice. The expression of Interleukin (IL)-17 and claudin-1 are enhanced in the glomeruli, which are considered as targets of TTP and HuR. Treating db/db mice with GSK-3β small molecule inhibitors Eliminates changes in TTP and HuR in the glomeruli and mitigates overexpression of their target genes, which in turn also alleviates proteinuria and DKD pathology (Guo et al., 2020). It was known in previous studies that TTP may negatively regulate the progression of DKD, whereas HuR does the opposite (Khalaj et al., 2017; Ross et al., 2017). The imbalance between them may play a significant role in the occurrence and development of DKD. Moreover, glucose in high concentration could upregulate miR-138 level and repress the expression of SIRT1 by binding to its 3′UTR, resulting in the TTP inhibition in cultured podocytes as well as db/db mice renal tissues. Lower TTP expression causes an increase in the expression of inflammatory factors, leaving podocytes in an inflammatory state for an extended period of time, which leads to loss of normal morphology and function (Liu et al., 2021). RBP IGFBP-1 expression is reduced and affects the function of podocytes via β1-integrin/FAK signaling in human type 2 diabetic glomeruli (Lay et al., 2021).
Diabetic cardiomyopathy is a type of heart disease characterized by insulin resistance in heart tissue, compensatory hyperinsulinemia, and hyperglycemia progression which can give rise to heart failure (HF). And it occurs in the absence of basic cardiac diseases such as hypertension, coronary artery disease, and heart valve disease (Jia et al., 2018; Dillmann, 2019). CELF1, also known as CUG-BP, is a highly conserved RNA binding protein that regulates alternative splicing, polyadenylation, mRNA stability, and translation of target transcripts. Previous studies showed that CELF1 is up-regulating in the hearts of T1DM mice, but diabetes-induced AS alterations are consistent with CELF1 depletion or decreased CELF1 splicing activity (Blech-Hermoni et al., 2016; Belanger et al., 2018). Interestingly, RBFox2, an RNA-binding protein belonging to the RBFOX family, that is involved in AS regulation in heart diseases, shows the same trend as CELF1 (Gazzara et al., 2017; Verma et al., 2022). RBFox2 regulates cardiac function-related genes associated with diabetic cardiomyopathy. Though levels of RBFox2 protein are increased in the heart of diabetics, RBFox2 AS activity is low. This is due to the production of a dominant negative isoform of RBFox2 that blocks RBFox2-mediated AS, thereby damaging cardiomyocytes. Dominant negative RBFox2 expression is exclusive to diabetes and appears in its early stages, therefore it might be served as a potential target for treating diabetic cardiomyopathy (Nutter et al., 2016). Recently, research showed a spliced variant of RNA-binding protein PTBP1 is expressed aberrantly in T1DM mouse hearts compared with normal newborn mouse hearts. This PTBP1 spliced variant induced by diabetes has a lower inhibitory splicing activity. Furthermore, PTBP1 and RBFox2 regulate AS of some of their targets antagonistically (Belanger et al., 2019). Besides, another study indicated that CUG-BP (also known as CELF1)/RBFox2 can be phosphorylated and up-regulated by activating PKC signaling in diabetic heart, which in turn alters the AS of gene and contribute to diabetic cardiomyopathy pathogenesis (Verma et al., 2013). In addition, RBFox2 may regulate the AS of genes associated with cGMP-PKG-Ca2+ signaling pathway and lead to cardiomyopathy and heart failure (Wan et al., 2020). Lin28, an RNA-binding protein that comes in two forms: Lin28a and Lin28b, is essential for glucose metabolism (Zhu et al., 2011). Lin28a levels were significantly reduced in the diabetic mice hearts. Over-expression of Lin28a protects against diabetic cardiomyopathy through improving left ventricular ejection fraction (LVEF), promoting autophagy, and decreasing apoptosis, which is regulated by inhibiting activation of PKA/ROCK2 pathway (Sun et al., 2016). Moreover, Lin28a′s protective effects, induced by activation of autophagy, were dependent on Mst1 inhibition in diabetic mouse cardiomyocytes (You et al., 2020). Another RBP Quaking 5 (QKI) level was deficient in diabetic ob/ob mice myocardium. QKI-5 overexpression undermines the stability of FoxO1 mRNA thus inhibiting FoxO1 overactivation, which diminishes nitrosative stress and endoplasmic reticulum stress in ob/ob myocardium (Guo et al., 2014).
Vascular endothelial cell (EC) dysfunction is largely acknowledged as a major contribution to the pathophysiology of cardiovascular disease in people with diabetes. RBP QKI is a member of the signal transduction and activation of RNA (STAR) family, and it is linked to diabetic cardiomyopathy and atherosclerosis (Yang et al., 2018). Quaking 5 (QKI-5), Quaking 6 (QKI-6) and Quaking 7 (QKI-7) are three primary QKI transcript isoforms that have been reported to have important roles in the vascular system. For example, QKI-5 and QKI-6 have been demonstrated to play a key role in cardiovascular health regulation and maintenance through their involvement in a variety of processes such as EC and vascular smooth muscle cell differentiation, apoptosis, and neovascularization (Caines et al., 2019). Recently studies have implicated that QKI-7 expression in diabetic EC is elevated, and QKI-7 can bind to its downstream targets to promote their mRNA degradation. Furthermore, two RBPs CUG-BP and hnRNPM are involved in the regulation of QKI-7. It has been shown that these two RBPs are acting as a vital upstream factor of QKI-7 and regulating the transcription network in diabetes. An imbalance of CUG-BP/hnRNPM regulation causes up-regulation of QKI-7, which increases their target mRNA degradation and finally leads to diabetic endothelial dysfunction (Yang et al., 2020). Lin28 is an RBP involved in kidney and cardiac complications of diabetes (Park et al., 2014). Lin28 levels decreased in the hearts of T1DM mice (Sun et al., 2016; You et al., 2020). Emerging evidence has shown that Lin28 can prevent endothelial oxidative stress in response to high glucose by stabilizing OGG1 mRNA (Tao et al., 2021).
In addition to endothelial cell dysfunction, diabetic vascular disease can further alter capillary density to affect coronary flow velocity reserve (CFVR), which in turn develops into coronary microvascular disease (CMD) (Si et al., 2021). Previous research showed that HuR overexpression promotes angiogenesis via stabilizing VEGF-A mRNA and modifying endothelial cell angiogenic activity (Chang et al., 2013). In addition, diabetes attenuates the expression of Cx40, a gap junction channel protein, in cardiac ECs and impairs coronary microvascular function via downregulating the level of RNA-binding protein HuR. Overexpression of CX40 increased the density of capillary and ameliorated CFVR in diabetic mice (Si et al., 2021).
Diabetic retinopathy (DR) is a common complication of DM, which remains a leading cause of vision damage or loss among working-age adults worldwide. Neovascularization plays an indispensable role in DR (Wang and Lo, 2018; Kang and Yang, 2020). DR involves early changes in the retina, characterized by vascular endothelial growth factor (VEGF) signal enhancement in various dysfunctions. It has been proven that inhibiting VEGF-mediated pathological angiogenesis enhances vision in DR patients (Stitt et al., 2013). Under the diabetic condition, the RNA-binding protein HuR is upregulated and binds to VEGF mRNA to regulate its stability, thereby enhancing its protein expression which leads to an abnormal increase in VEGF in the retina of diabetic rats (Amadio et al., 2010; Amadio et al., 2012). Besides, increased HuR and VEGF were suppressed by HuR silencing via intravitreal injection of small interfering RNA (siRNA) nanoparticles, which protect rat retinal tissue from damage caused by DM (Amadio et al., 2016). Furthermore, recent studies have shown that the expression of VEGF-A164 is time-specific (Bucolo et al., 2021). And VP12/14 and VP12/11, two derivatives containing indole structures, can regulate HuR expression and reduce the levels of VEGF and TNF-α release by human retinal endothelial cells (HRECs) exposed to high glucose (HG) conditions (Platania et al., 2020). HuR may represent a new target to inhibit the increased expression of VEGF, thus improving diabetic retinal vascular hyperplasia and inflammation. RNA-binding protein hnRNPA2B1 was confirmed to be a downstream target of Transthyretin (TTR) in human retinal microvascular endothelial cells (HRMECs). TTR can interact with hnRNPA2B1 to form a TTR-hnRNPA2B1 complex, which plays a critical role in TTR’s anti-angiogenesis function in hyperglycemia via the STAT4/miR-223e3p/FBXW7 signaling pathway (Gu et al., 2021). The expression level of RBP ZFR is meaningfully elevated both in vitro and in vivo in HRMECs in response to high glucose. Furthermore, ZFR can also enhance proliferation and migration in HRMECs. Besides, ZFR expression stimulated by high glucose can be attenuated by suppressing O-GlcNAcylation activity (Xing et al., 2019). The stable expression of RBP Lin-28 homolog b (lin28b) can promote VEGF expression (Wu et al., 2013; Weiße et al., 2020). It has been revealed that miR-152 can specifically target lin28b 3′UTR. Under high glucose conditions, miR-152 expression was significantly repressed, whereas lin28b expression was meaningfully augmented. Overexpression of lin28b increased the angiogenesis and the protein levels of proangiogenesis factors while inhibiting the function of miR-152 overexpression in both hRECs and hRMECs (Fu and Ou, 2020).
Furthermore, retinal neurodegeneration also occurs in the etiology of DR, which is mainly characterized by apoptosis and glial changes (Simo et al., 2018). Glia cells are considered as the interface between the vasculature and neurons (Hammes, 2018). mRNA Cap-binding protein eIF4E can recognize the structure of the 5′-m7GTP cap of mRNA and assemble it into eIF4F complexes, which can recruit ribosomes onto mRNA to perform translational functions. The recruitment of ribosomes, which can occur via a cap-dependent or cap-independent mechanism, limits the rate of mRNA translation. The interaction between 4E-BP1 and eIF4E promotes the dissociation of eIF4E from eIF4F complexes, which inhibits cap-dependent and promotes cap-independent translation (Schrufer et al., 2010; Dennis et al., 2015; Miller et al., 2016). In Muller cells and retina of diabetic rats, high glucose conditions increase REDD1 levels and enhance the binding of 4E-BP1 to eIF4E. This reduces the overall rate of protein synthesis and cap-dependent mRNA translation accompanied by upregulated cap-independent VEGF mRNA translation, which is thought to be a key mechanism in the development of DR (Dennis et al., 2015). In addition, retinal protein O-GlcNAcylation promotes cap-independent Cd40 mRNA translation through a 4E-BP1 dependent mechanism under a diabetic condition in Muller glia cells. Elevated expression of the CD40 protein in Muller glial cells leads to chronic retinal inflammation correlates with DR (Dierschke et al., 2020).
Emerging evidence indicates that dysregulation of RBPs is linked to a variety of disorders and affects almost every stage of the disease progression. There is a lot of literature on RBPs and their implicates in diabetes, but it is fragmented and lacks systematic reviews. With a high incidence of diabetes and severe chronic complications of multiple systems, a thorough understanding of the post-transcriptional regulatory role of RBPs in diabetes and its complications is critical to the development of novel RNA-based therapies. Here, for the first time, we categorize and summarize some common and relatively mature RBPs according to different systemic complications (Figure 2) (Table 1). We hope that both new therapeutic developers and researchers working on RBPs in the diabetes field can find some convenient and useful information from this review.
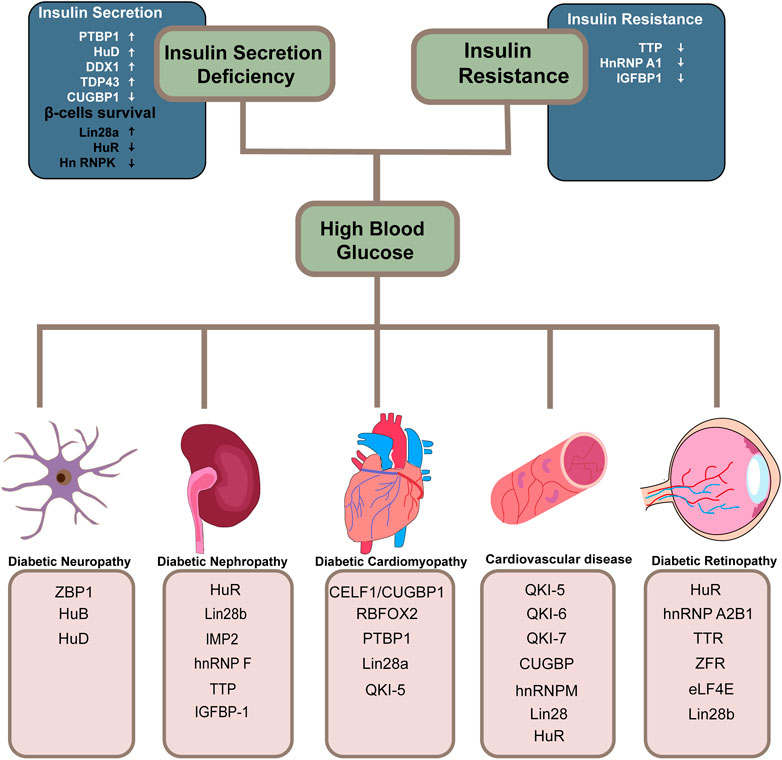
FIGURE 2. Overview of RBPs involved in diabetes and its related systematic complications. RNA-binding proteins implicated in the two decisive links in the progression of diabetes and its related systematic complications are summarized. The arrows in the diagram point to either protective effects or the opposite.
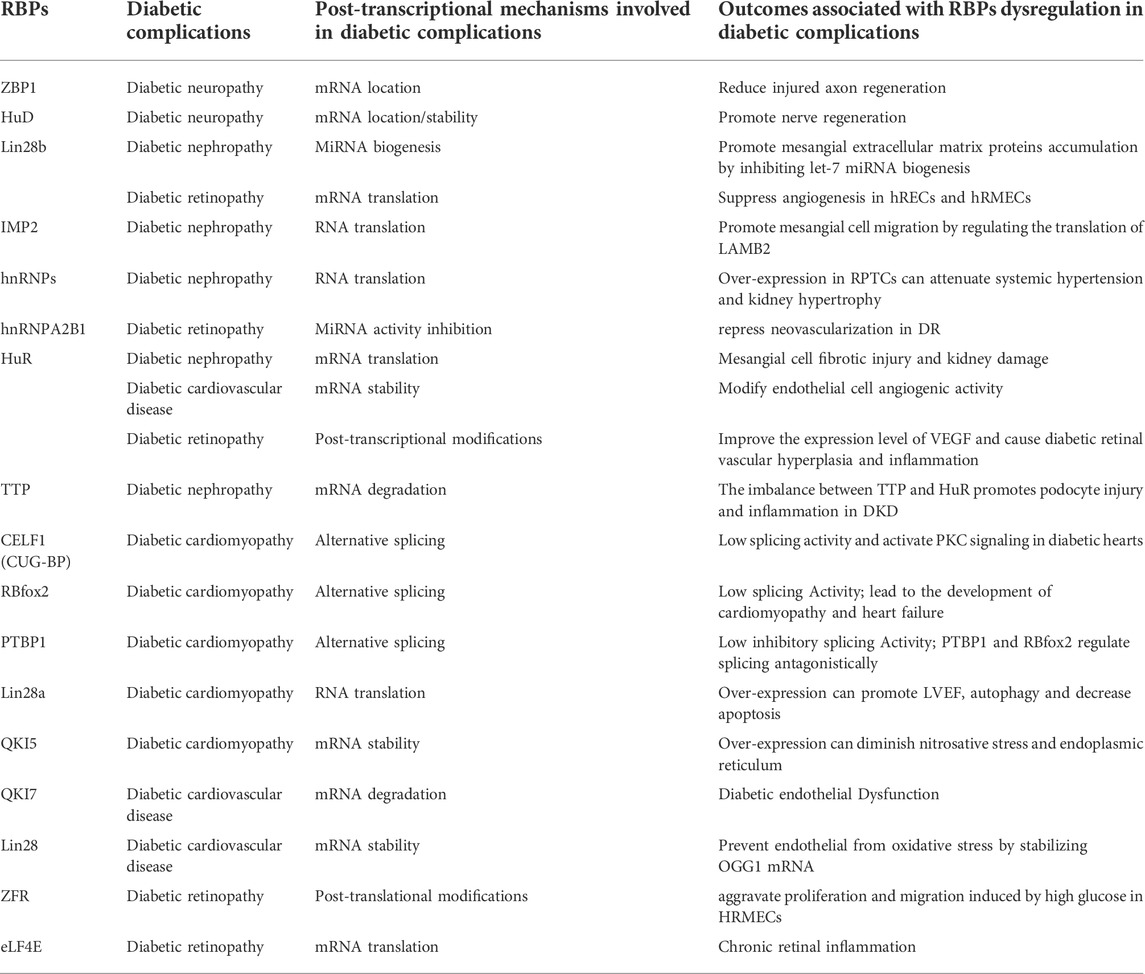
TABLE 1. The regulatory mechanism of RNA-Binding proteins and the outcomes of their dysregulation in diabetic complications.
In this review, we focus on the molecular mechanisms of RBPs and mRNA interactions, which have an either positive or negative impact on diabetes. The functional interactions between RBPs and non-coding RNAs, including microRNAs and circular RNAs, are another essential aspect that is briefly explored in this study. We have spent a lot of sections discussing that dysregulation of RBP leads to abnormal function of its interacting nucleic acids or proteins in diabetes. However, RBP’s own activity is profoundly controlled by post-translational modifications (PTMs), Which is also an important mechanism that determines the occurrence and development of diseases (Lovci et al., 2016). PTMs generally refers to enzymatic reactions that occur after protein synthesis. PTMs follow a variety of signaling transductions that induce proteins to form covalent bonds with new functional chemical groups such as phosphate, methyl, acetyl, and ubiquitin (Deribe et al., 2010). PTMs can significantly alter the activity and properties of RBPs, resulting in changes in regulating protein activity, stability, localization, turnover and degradation (Velázquez-Cruz et al., 2021). Phosphorylation is the most common and widely explored among various types of PTMs. For example, several specific phosphorylations of hnRNPK by specific kinases can alter hnRNPK protein subcellular localization, stability, or affinity for binding targets (Xu et al., 2019). PTMs can also alter the Subcellular localization of HuR, and most phosphorylation of HuR occurs in its hinged region (Grammatikakis et al., 2017). In addition, the nuclear import of serine/arginine-rich (SR) protein family requires phosphorylation by the SR protein kinases 1 and 2 (SRPK1/2) (Long et al., 2019). These post-translational modifications also play an important role in diabetes. O-linked N-acetylglucosamine (O-GlcNAc) glycosylation is involved in the pathogenesis of diabetes and its related complications by O-linked addition of GlcNAc (O-GlcNAcylation) to Ser/Thr residues of proteins (Issad et al., 2010; Zhu and Hart, 2021). There are multiple studies reported that O-GlcN acylation enhancement of retinal proteins in rodent models of type 1 and type 2 diabetes (Mellor et al., 2015; Peterson and Hart, 2016; Masaki et al., 2020). The cap-binding protein eIF4E is more readily sequestered in the mice with DR, due to the repressor of mRNA translation 4E-BP1 being O-GlcNAcylated (Dierschke et al., 2019). O-GlcNAc signaling activation also increases the level of RBP ZFR under high glucose condition, which aggravates proliferation and migration induced in HRMECs (Xing et al., 2019). It is not difficult to grasp that PTM as a major element governs the properties and function of RBPs with highly dynamic and largely reversible. We do not describe in detail the regulation of PTMs to RBPs and the complex signaling pathways it orchestrates. However, a thorough understanding of the molecular underpinnings of disease-associated PTMs dysregulation on RBPs is necessary for fully comprehending the pathophysiological process of diseases.
In summary, we expect to fully understand the dynamic RBPs-mediated regulatory network in diabetes. Correcting gene expression abnormalities in diabetic patients by targeting the interaction between RBPs and their target RNAs could be an effective approach. RNA-based therapies are primarily designed drugs to imitate or antagonize specific RNA processes by mimicking the action of protective RBPs or inhibiting the action of pathogenic RBPs. Many candidate strategies are being applied to target RBPs for therapeutics in pre-clinical or clinical trials, such as small-molecule inhibitors, therapeutic small peptides, anti-sense oligonucleotides (ASOs), and siRNA (Chi et al., 2017; Mohibi et al., 2019). Besides, circular RNAs are also considered as a potential strategy that can be designed to bind RBDs of RBPs and compete with target RNAs (Mohibi et al., 2019). However, how to improve the target specificity is also a tough problem that needs to be overcome. Although many issues and connections of RBPs remains to be explored and solved, existing knowledge and growing evidence show that we have an opportunity to enter a new era in the therapies of diabetes and its related complications.
SZ conceived and drafted the manuscript, XY, MJ and LM revised the draft version of the manuscript. JH reviewed and edited the article. H-HZ designed and supervised the study and edited the manuscript. H-HZ or JH is the guarantor of this study. All authors contributed to the article and approved the submitted version.
This work was supported by grant from the National Natural Science Foundation of China (82071234 to H-HZ, 82170836 to JH) and from the Jiangsu Youth Medical Talents Project (QNRC2016874 to H-HZ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Amadio M., Bucolo C., Leggio G. M., Drago F., Govoni S., Pascale A. (2010). The PKCβ/HuR/VEGF pathway in diabetic retinopathy. Biochem. Pharmacol. 80 (8), 1230–1237. doi:10.1016/j.bcp.2010.06.033
Amadio M., Osera C., Lupo G., Motta C., Drago F., Govoni S., (2012). Protein kinase C activation affects, via the mRNA-binding Hu-antigen R/ELAV protein, vascular endothelial growth factor expression in a pericytic/endothelial coculture model. Mol. Vis. 18, 2153–2164.
Amadio M., Pascale A., Cupri S., Pignatello R., Osera C., (2016). Nanosystems based on siRNA silencing HuR expression counteract diabetic retinopathy in rat. Pharmacol. Res. 111, 713–720. doi:10.1016/j.phrs.2016.07.042
Ambrosio F. A., Coricello A., Costa G., Lupia A., Micaelli M., Marchesi N., et al. (2021). Identification of compounds targeting HuD. Another brick in the wall of neurodegenerative disease treatment. J. Med. Chem. 64 (14), 9989–10000. doi:10.1021/acs.jmedchem.1c00191
Araki K., Araki A., Honda D., Izumoto T., Hashizume A., Hijikata Y., et al. (2019). TDP-43 regulates early-phase insulin secretion via CaV1.2-mediated exocytosis in islets. J. Clin. Invest 129 (9), 3578–3593. doi:10.1172/jci124481
Atkinson M. A., Eisenbarth G. S., Michels A. W. (2014). Type 1 diabetes. Lancet 383 (9911), 69–82. doi:10.1016/s0140-6736(13)60591-7
Belanger K., Nutter C. A., Li J., Tasnim S., Liu P., Yu P., et al. (2018). CELF1 contributes to aberrant alternative splicing patterns in the type 1 diabetic heart. Biochem. Biophys. Res. Commun. 503 (4), 3205–3211. doi:10.1016/j.bbrc.2018.08.126
Belanger K., Nutter C. A., Li J., Yu P., Kuyumcu-Martinez M. N. (2019). A developmentally regulated spliced variant of PTBP1 is upregulated in type 1 diabetic hearts. Biochem. Biophys. Res. Commun. 509 (2), 384–389. doi:10.1016/j.bbrc.2018.12.150
Blackshear P. J. (2002). Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30 (6), 945–952. doi:10.1042/bst0300945
Blech-Hermoni Y., Dasgupta T., Coram R. J., Ladd A. N. (2016). Identification of targets of CUG-BP, elav-like family member 1 (CELF1) regulation in embryonic heart muscle. PLoS One 11 (2), e0149061. doi:10.1371/journal.pone.0149061
Bucolo C., Barbieri A., Vigano I., Marchesi N., Bandello F., Drago F., et al. (2021). Short-and long-term expression of vegf: A temporal regulation of a key factor in diabetic retinopathy. Front. Pharmacol. 12, 707909. doi:10.3389/fphar.2021.707909
Caines R., Cochrane A., Kelaini S., Vila-Gonzalez M., Yang C., Eleftheriadou M., et al. (2019). The RNA-binding protein QKI controls alternative splicing in vascular cells, producing an effective model for therapy. J. Cell Sci. 132 (16), jcs230276. doi:10.1242/jcs.230276
Calcutt N. A. (2020). Diabetic neuropathy and neuropathic pain: A (con)fusion of pathogenic mechanisms? Pain 161 (1), S65–s86. doi:10.1097/j.pain.0000000000001922
Campbell J. E., Newgard C. B. (2021). Mechanisms controlling pancreatic islet cell function in insulin secretion. Nat. Rev. Mol. Cell Biol. 22 (2), 142–158. doi:10.1038/s41580-020-00317-7
Cao H., Urban J. F., Anderson R. A. (2008). Insulin increases tristetraprolin and decreases VEGF gene expression in mouse 3T3-L1 adipocytes. Obes. (Silver Spring) 16 (6), 1208–1218. doi:10.1038/oby.2008.65
Cao J., Yan W., Ma X., Huang H., Yan H. (2021). Insulin-like growth factor 2 mRNA-binding protein 2-a potential link between type 2 diabetes mellitus and cancer. J. Clin. Endocrinol. Metab. 106 (10), 2807–2818. doi:10.1210/clinem/dgab391
Caracciolo V., Young J., Gonzales D., Ni Y., Flowers S. J., Summer R., et al. (2018). Myeloid-specific deletion of Zfp36 protects against insulin resistance and fatty liver in diet-induced obese mice. Am. J. Physiol. Endocrinol. Metab. 315 (4), E676–e693. doi:10.1152/ajpendo.00224.2017
Chang S. H., Lu Y. C., Li X., Hsieh W. Y., Xiong Y., Ghosh M., et al. (2013). Antagonistic function of the RNA-binding protein HuR and miR-200b in post-transcriptional regulation of vascular endothelial growth factor-A expression and angiogenesis. J. Biol. Chem. 288 (7), 4908–4921. doi:10.1074/jbc.M112.423871
Chi X., Gatti P., Papoian T. (2017). Safety of antisense oligonucleotide and siRNA-based therapeutics. Drug Discov. Today 22 (5), 823–833. doi:10.1016/j.drudis.2017.01.013
Cho N. H., Shaw J. E., Karuranga S., Huang Y., da Rocha Fernandes J. D., Ohlrogge A. W., et al. (2018). IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 138, 271–281. doi:10.1016/j.diabres.2018.02.023
Ciafrè S. A., Galardi S. (2013). microRNAs and RNA-binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA Biol. 10 (6), 935–942. doi:10.4161/rna.24641
Cole J. B., Florez J. C. (2020). Genetics of diabetes mellitus and diabetes complications. Nat. Rev. Nephrol. 16 (7), 377–390. doi:10.1038/s41581-020-0278-5
Connerty P., Ahadi A., Hutvagner G. (2015). RNA binding proteins in the miRNA pathway. Int. J. Mol. Sci. 17 (1), 31. doi:10.3390/ijms17010031
Corley M., Burns M. C., Yeo G. W. (2020). How RNA-binding proteins interact with RNA: Molecules and mechanisms. Mol. cell 78 (1), 9–29. doi:10.1016/j.molcel.2020.03.011
Demir S., Nawroth P. P., Herzig S., Ekim Ustunel B. (2021). Emerging targets in type 2 diabetes and diabetic complications. Adv. Sci. (Weinheim, Baden-Wurttemberg, Ger. 8 (18), e2100275. doi:10.1002/advs.202100275
Dennis M. D., Kimball S. R., Fort P. E., Jefferson L. S. (2015). Regulated in development and DNA damage 1 is necessary for hyperglycemia-induced vascular endothelial growth factor expression in the retina of diabetic rodents. J. Biol. Chem. 290 (6), 3865–3874. doi:10.1074/jbc.M114.623058
Deribe Y. L., Pawson T., Dikic I. (2010). Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17 (6), 666–672. doi:10.1038/nsmb.1842
Dierschke S. K., Miller W. P., Favate J. S., Shah P., Imamura Kawasawa Y., Salzberg A. C., et al. (2019). O-GlcNAcylation alters the selection of mRNAs for translation and promotes 4E-BP1-dependent mitochondrial dysfunction in the retina. J. Biol. Chem. 294 (14), 5508–5520. doi:10.1074/jbc.RA119.007494
Dierschke S. K., Toro A. L., Miller W. P., Sunilkumar S., Dennis M. D. (2020). Diabetes enhances translation of Cd40 mRNA in murine retinal Muller glia via a 4E-BP1/2-dependent mechanism. J. Biol. Chem. 295 (31), 10831–10841. doi:10.1074/jbc.RA120.013711
Dillmann W. H. (2019). Diabetic cardiomyopathy. Circ. Res. 124 (8), 1160–1162. doi:10.1161/circresaha.118.314665
Eizirik D. L., Pasquali L., Cnop M. (2020). Pancreatic β-cells in type 1 and type 2 diabetes mellitus: Different pathways to failure. Nat. Rev. Endocrinol. 16 (7), 349–362. doi:10.1038/s41574-020-0355-7
Feldman E. L., Callaghan B. C., Pop-Busui R., Zochodne D. W., Wright D. E., Bennett D. L., et al. (2019). Diabetic neuropathy. Nat. Rev. Dis. Prim. 5 (1), 42. doi:10.1038/s41572-019-0097-9
Fred R. G., Sandberg M., Pelletier J., Welsh N. (2011). The human insulin mRNA is partly translated via a cap- and eIF4A-independent mechanism. Biochem. Biophys. Res. Commun. 412 (4), 693–698. doi:10.1016/j.bbrc.2011.08.030
Fu X., Ou B. (2020). miR-152/LIN28B axis modulates high-glucose-induced angiogenesis in human retinal endothelial cells via VEGF signaling. J. Cell Biochem. 121 (2), 954–962. doi:10.1002/jcb.28978
Fukao A., Aoyama T., Fujiwara T. (2015). The molecular mechanism of translational control via the communication between the microRNA pathway and RNA-binding proteins. RNA Biol. 12 (9), 922–926. doi:10.1080/15476286.2015.1073436
Gazzara M. R., Mallory M. J., Roytenberg R., Lindberg J. P., Jha A., Lynch K. W., et al. (2017). Ancient antagonism between CELF and RBFOX families tunes mRNA splicing outcomes. Genome Res. 27 (8), 1360–1370. doi:10.1101/gr.220517.117
Gerstberger S., Hafner M., Tuschl T. (2014). A census of human RNA-binding proteins. Nat. Rev. Genet. 15 (12), 829–845. doi:10.1038/nrg3813
Gokulakrishnan K., Velmurugan K., Ganesan S., Mohan V. (2012). Circulating levels of insulin-like growth factor binding protein-1 in relation to insulin resistance, type 2 diabetes mellitus, and metabolic syndrome (Chennai Urban Rural Epidemiology Study 118). Metabolism 61 (1), 43–46. doi:10.1016/j.metabol.2011.05.014
Gomes C., Lee S. J., Gardiner A. S., Smith T., Sahoo P. K., Patel P., et al. (2017). Axonal localization of neuritin/CPG15 mRNA is limited by competition for HuD binding. J. cell Sci. 130 (21), 3650–3662. doi:10.1242/jcs.201244
Good A. L., Haemmerle M. W., Oguh A. U., Doliba N. M., Stoffers D. A. (2019). Metabolic stress activates an ERK/hnRNPK/DDX3X pathway in pancreatic β cells. Mol. Metab. 26, 45–56. doi:10.1016/j.molmet.2019.05.009
Good A. L., Stoffers D. A. (2020). Stress-induced translational regulation mediated by RNA binding proteins: Key links to β-cell failure in diabetes. Diabetes 69 (4), 499–507. doi:10.2337/dbi18-0068
Grammatikakis I., Abdelmohsen K., Gorospe M. (2017). Posttranslational control of HuR function. Wiley Interdiscip. Rev. RNA 8 (1), 1. doi:10.1002/wrna.1372
Gu Y., Hu D., Xin Y., Shao J. (2021). Transthyretin affects the proliferation and migration of human retinal microvascular endothelial cells in hyperglycemia via hnRNPA2B1. Biochem. Biophys. Res. Commun. 557, 280–287. doi:10.1016/j.bbrc.2021.04.035
Guo J., Lei M., Cheng F., Liu Y., Zhou M., Zheng W., et al. (2020). RNA-binding proteins tristetraprolin and human antigen R are novel modulators of podocyte injury in diabetic kidney disease. Cell death Dis. 11 (6), 413. doi:10.1038/s41419-020-2630-x
Guo W., Jiang T., Lian C., Wang H., Zheng Q., Ma H. (2014). QKI deficiency promotes FoxO1 mediated nitrosative stress and endoplasmic reticulum stress contributing to increased vulnerability to ischemic injury in diabetic heart. J. Mol. Cell Cardiol. 75, 131–140. doi:10.1016/j.yjmcc.2014.07.010
Hammes H. P. (2018). Diabetic retinopathy: Hyperglycaemia, oxidative stress and beyond. Diabetologia 61 (1), 29–38. doi:10.1007/s00125-017-4435-8
Harding J. L., Pavkov M. E., Magliano D. J., Shaw J. E., Gregg E. W. (2019). Global trends in diabetes complications: A review of current evidence. Diabetologia 62 (1), 3–16. doi:10.1007/s00125-018-4711-2
Haywood N. J., Cordell P. A., Tang K. Y., Makova N., Yuldasheva N. Y., Imrie H., et al. (2017). Insulin-like growth factor binding protein 1 could improve glucose regulation and insulin sensitivity through its RGD domain. Diabetes 66 (2), 287–299. doi:10.2337/db16-0997
Hentze M. W., Castello A., Schwarzl T., Preiss T. (2018). A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19 (5), 327–341. doi:10.1038/nrm.2017.130
Ho J. J. D., Lee S. (2016). A cap for every occasion: Alternative eIF4F complexes. Trends Biochem. Sci. 41 (10), 821–823. doi:10.1016/j.tibs.2016.05.009
Ho J. J. D., Man J. H. S., Schatz J. H., Marsden P. A. (2021). Translational remodeling by RNA-binding proteins and noncoding RNAs. Wiley Interdiscip. Rev. RNA 12 (5), e1647. doi:10.1002/wrna.1647
Hong Y., Tak H., Kim C., Kang H., Ji E., Ahn S., et al. (2020). RNA binding protein HuD contributes to β-cell dysfunction by impairing mitochondria dynamics. Cell Death Differ. 27 (5), 1633–1643. doi:10.1038/s41418-019-0447-x
Issad T., Masson E., Pagesy P. (2010). O-GlcNAc modification, insulin signaling and diabetic complications. Diabetes Metab. 36 (1), 423–435. doi:10.1016/j.diabet.2010.09.001
Jeong D. E., Heo S., Han J. H., Lee E. Y., Kulkarni R. N., Kim W. (2018). Glucose controls the expression of polypyrimidine tract-binding protein 1 via the insulin receptor signaling pathway in pancreatic β cells. Mol. Cells 41 (10), 909–916. doi:10.14348/molcells.2018.0147
Jia G., Whaley-Connell A., Sowers J. R. (2018). Diabetic cardiomyopathy: A hyperglycaemia- and insulin-resistance-induced heart disease. Diabetologia 61 (1), 21–28. doi:10.1007/s00125-017-4390-4
Jones J. I., Costa C. J., Cooney C., Goldberg D. C., Ponticiello M., Cohen M. W., et al. (2021). Failure to upregulate the RNA binding protein ZBP after injury leads to impaired regeneration in a rodent model of diabetic peripheral neuropathy. Front. Mol. Neurosci. 14, 728163. doi:10.3389/fnmol.2021.728163
Kang Q., Yang C. (2020). Oxidative stress and diabetic retinopathy: Molecular mechanisms, pathogenetic role and therapeutic implications. Redox Biol. 37, 101799. doi:10.1016/j.redox.2020.101799
Kanwar Y. S., Sun L., Xie P., Liu F. Y., Chen S. (2011). A glimpse of various pathogenetic mechanisms of diabetic nephropathy. Annu. Rev. Pathol. 6, 395–423. doi:10.1146/annurev.pathol.4.110807.092150
Kelaini S., Chan C., Cornelius V. A., Margariti A. (2021). RNA-binding proteins hold key roles in function, dysfunction, and disease. Biology 10 (5), 366. doi:10.3390/biology10050366
Khalaj K., Ahn S. H., Bidarimath M., Nasirzadeh Y., Singh S. S., Fazleabas A. T., et al. (2017). A balancing act: RNA binding protein HuR/TTP axis in endometriosis patients. Sci. Rep. 7 (1), 5883. doi:10.1038/s41598-017-06081-7
Klim J. R., Pintacuda G., Nash L. A., Guerra San Juan I., Eggan K. (2021). Connecting TDP-43 pathology with neuropathy. Trends Neurosci. 44 (6), 424–440. doi:10.1016/j.tins.2021.02.008
Knoch K. P., Bergert H., Borgonovo B., Saeger H. D., Altkruger A., Verkade P., et al. (2004). Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat. Cell Biol. 6 (3), 207–214. doi:10.1038/ncb1099
Knoch K. P., Nath-Sain S., Petzold A., Schneider H., Beck M., Wegbrod C., et al. (2014). PTBP1 is required for glucose-stimulated cap-independent translation of insulin granule proteins and Coxsackieviruses in beta cells. Mol. Metab. 3 (5), 518–530. doi:10.1016/j.molmet.2014.05.002
Kulkarni S. D., Muralidharan B., Panda A. C., Bakthavachalu B., Vindu A., Seshadri V. (2011). Glucose-stimulated translation regulation of insulin by the 5' UTR-binding proteins. J. Biol. Chem. 286 (16), 14146–14156. doi:10.1074/jbc.M110.190553
Lay A. C., Hale L. J., Stowell-Connolly H., Pope R. J. P., Nair V., Ju W., et al. (2021). IGFBP-1 expression is reduced in human type 2 diabetic glomeruli and modulates β1-integrin/FAK signalling in human podocytes. Diabetologia 64 (7), 1690–1702. doi:10.1007/s00125-021-05427-1
Lazaris-Karatzas A., Montine K. S., Sonenberg N. (1990). Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5' cap. Nature 345 (6275), 544–547. doi:10.1038/345544a0
Lee E. K., Kim W., Tominaga K., Martindale J. L., Yang X., Subaran S. S., et al. (2012). RNA-binding protein HuD controls insulin translation. Mol. cell 45 (6), 826–835. doi:10.1016/j.molcel.2012.01.016
Li Z., Zhou M., Cai Z., Liu H., Zhong W., Hao Q., et al. (2018). RNA-binding protein DDX1 is responsible for fatty acid-mediated repression of insulin translation. Nucleic Acids Res. 46 (22), 12052–12066. doi:10.1093/nar/gky867
Liu F., Guo J., Qiao Y., Pan S., Duan J., Liu D., et al. (2021). MiR-138 plays an important role in diabetic nephropathy through SIRT1-p38-TTP regulatory axis. J. Cell Physiol. 236 (9), 6607–6618. doi:10.1002/jcp.30238
Lo C.-S., Chang S.-Y., Chenier I., Filep J. G., Ingelfinger J. R., Zhang S. L., et al. (2012). Heterogeneous nuclear ribonucleoprotein F suppresses angiotensinogen gene expression and attenuates hypertension and kidney injury in diabetic mice. Diabetes 61 (10), 2597–2608. doi:10.2337/db11-1349
Long Y., Sou W. H., Yung K. W. Y., Liu H., Wan S. W. C., Li Q., et al. (2019). Distinct mechanisms govern the phosphorylation of different SR protein splicing factors. J. Biol. Chem. 294 (4), 1312–1327. doi:10.1074/jbc.RA118.003392
Lovci M. T., Bengtson M. H., Massirer K. B. (2016). Post-translational modifications and RNA-binding proteins. Adv. Exp. Med. Biol. 907, 297–317. doi:10.1007/978-3-319-29073-7_12
Lujan D. A., Ochoa J. L., Hartley R. S. (2018). Cold-inducible RNA binding protein in cancer and inflammation. Wiley Interdiscip. Rev. RNA 9 (2). doi:10.1002/wrna.1462
Magro M. G., Solimena M. (2013). Regulation of β-cell function by RNA-binding proteins. Mol. Metab. 2 (4), 348–355. doi:10.1016/j.molmet.2013.09.003
Makita S., Takatori H., Nakajima H. (2021). Post-transcriptional regulation of immune responses and inflammatory diseases by RNA-binding ZFP36 family proteins. Front. Immunol. 12, 711633. doi:10.3389/fimmu.2021.711633
Masaki N., Feng B., Breton-Romero R., Inagaki E., Weisbrod R. M., Fetterman J. L., et al. (2020). O-GlcNAcylation mediates glucose-induced alterations in endothelial cell phenotype in human diabetes mellitus. J. Am. Heart Assoc. 9 (12), e014046. doi:10.1161/jaha.119.014046
Mayr C. (2019). What are 3' UTRs doing? Cold Spring Harb. Perspect. Biol. 11 (10), a034728. doi:10.1101/cshperspect.a034728
Mayr F., Heinemann U. (2013). Mechanisms of Lin28-mediated miRNA and mRNA regulation a structural and functional perspective. Int. J. Mol. Sci. 14 (8), 16532–16553. doi:10.3390/ijms140816532
Mellor K. M., Brimble M. A., Delbridge L. M. (2015). Glucose as an agent of post-translational modification in diabetes New cardiac epigenetic insights. Life Sci. 129, 48–53. doi:10.1016/j.lfs.2014.03.020
Miller W. P., Mihailescu M. L., Yang C., Barber A. J., Kimball S. R., Jefferson L. S., et al. (2016). The translational repressor 4E-BP1 contributes to diabetes-induced visual dysfunction. Invest Ophthalmol. Vis. Sci. 57 (3), 1327–1337. doi:10.1167/iovs.15-18719
Mohibi S., Chen X., Zhang J. (2019). Cancer the'RBP'eutics-RNA-binding proteins as therapeutic targets for cancer. Pharmacol. Ther. 203, 107390. doi:10.1016/j.pharmthera.2019.07.001
Mustăciosu C. C., Banciu A., Rusu C. M., Banciu D. D., Savu D., Radu M., et al. (2019). RNA-binding proteins HuB, HuC, and HuD are distinctly regulated in dorsal root ganglia neurons from STZ-sensitive compared to STZ-resistant diabetic mice. Int. J. Mol. Sci. 20 (8), 1965. doi:10.3390/ijms20081965
Nutter C. A., Jaworski E. A., Verma S. K., Deshmukh V., Wang Q., Botvinnik O. B., et al. (2016). Dysregulation of RBFOX2 is an early event in cardiac pathogenesis of diabetes. Cell Rep. 15 (10), 2200–2213. doi:10.1016/j.celrep.2016.05.002
Nutter C. A., Kuyumcu-Martinez M. N. (2018). Emerging roles of RNA-binding proteins in diabetes and their therapeutic potential in diabetic complications. Wiley Interdiscip. Rev. RNA 9 (2), 1. doi:10.1002/wrna.1459
Osborne M. J., Borden K. L. (2015). The eukaryotic translation initiation factor eIF4E in the nucleus: Taking the road less traveled. Immunol. Rev. 263 (1), 210–223. doi:10.1111/imr.12240
Park J. T., Kato M., Lanting L., Castro N., Nam B. Y., Wang M., et al. (2014). Repression of let-7 by transforming growth factor-β1-induced Lin28 upregulates collagen expression in glomerular mesangial cells under diabetic conditions. Am. J. Physiol. Ren. Physiol. 307 (12), F1390–F1403. doi:10.1152/ajprenal.00458.2014
Pereira B., Billaud M., Almeida R. (2017). RNA-binding proteins in cancer: Old players and new actors. Trends Cancer 3 (7), 506–528. doi:10.1016/j.trecan.2017.05.003
Pérez-Ortín J. E., Alepuz P., Chávez S., Choder M. (2013). Eukaryotic mRNA decay: Methodologies, pathways, and links to other stages of gene expression. J. Mol. Biol. 425 (20), 3750–3775. doi:10.1016/j.jmb.2013.02.029
Petersmann A., Muller-Wieland D., Muller U. A., Landgraf R., Nauck M., Freckmann G., et al. (2019). Definition, classification and diagnosis of diabetes mellitus. Exp. Clin. Endocrinol. Diabetes 127 (1), S1–s7. doi:10.1055/a-1018-9078
Peterson S. B., Hart G. W. (2016). New insights: A role for O-GlcNAcylation in diabetic complications. Crit. Rev. Biochem. Mol. Biol. 51 (3), 150–161. doi:10.3109/10409238.2015.1135102
Platania C. B. M., Pittalà V., Pascale A., Marchesi N., Anfuso C. D., Lupo G., et al. (2020). Novel indole derivatives targeting HuR-mRNA complex to counteract high glucose damage in retinal endothelial cells. Biochem. Pharmacol. 175, 113908. doi:10.1016/j.bcp.2020.113908
Pope S. D., Medzhitov R. (2018). Emerging principles of gene expression programs and their regulation. Mol. Cell 71 (3), 389–397. doi:10.1016/j.molcel.2018.07.017
Rajwani A., Ezzat V., Smith J., Yuldasheva N. Y., Duncan E. R., Gage M., et al. (2012). Increasing circulating IGFBP1 levels improves insulin sensitivity, promotes nitric oxide production, lowers blood pressure, and protects against atherosclerosis. Diabetes 61 (4), 915–924. doi:10.2337/db11-0963
Ross E. A., Naylor A. J., O'Neil J. D., Crowley T., Ridley M. L., Crowe J., et al. (2017). Treatment of inflammatory arthritis via targeting of tristetraprolin, a master regulator of pro-inflammatory gene expression. Ann. Rheum. Dis. 76 (3), 612–619. doi:10.1136/annrheumdis-2016-209424
Salem E. S. B., Vonberg A. D., Borra V. J., Gill R. K., Nakamura T. (2019). RNAs and RNA-binding proteins in immuno-metabolic homeostasis and diseases. Front. Cardiovasc Med. 6, 106. doi:10.3389/fcvm.2019.00106
Sanna M. D., Ghelardini C., Galeotti N. (2017). HuD-mediated distinct BDNF regulatory pathways promote regeneration after nerve injury. Brain Res. 1659, 55–63. doi:10.1016/j.brainres.2017.01.019
Sanna M. D., Peroni D., Quattrone A., Ghelardini C., Galeotti N. (2015). Spinal RyR2 pathway regulated by the RNA-binding protein HuD induces pain hypersensitivity in antiretroviral neuropathy. Exp. Neurol. 267, 53–63. doi:10.1016/j.expneurol.2015.02.036
Sanna M. D., Quattrone A., Ghelardini C., Galeotti N. (2014). PKC-mediated HuD-GAP43 pathway activation in a mouse model of antiretroviral painful neuropathy. Pharmacol. Res. 81, 44–53. doi:10.1016/j.phrs.2014.02.004
Sawicki K. T., Chang H. C., Shapiro J. S., Bayeva M., De Jesus A., Finck B. N., et al. (2018). Hepatic tristetraprolin promotes insulin resistance through RNA destabilization of FGF21. JCI Insight 3 (13), e95948. doi:10.1172/jci.insight.95948
Schaeffer V., Hansen K. M., Morris D. R., LeBoeuf R. C., Abrass C. K. (2012). RNA-binding protein IGF2BP2/IMP2 is required for laminin-β2 mRNA translation and is modulated by glucose concentration. Am. J. physiology. Ren. physiology 303 (1), F75–F82. doi:10.1152/ajprenal.00185.2012
Schrufer T. L., Antonetti D. A., Sonenberg N., Kimball S. R., Gardner T. W., Jefferson L. S. (2010). Ablation of 4E-BP1/2 prevents hyperglycemia-mediated induction of VEGF expression in the rodent retina and in Muller cells in culture. Diabetes 59 (9), 2107–2116. doi:10.2337/db10-0148
Shang J., Wan Q., Wang X., Duan Y., Wang Z., Wei X., et al. (2015). Identification of NOD2 as a novel target of RNA-binding protein HuR: Evidence from NADPH oxidase-mediated HuR signaling in diabetic nephropathy. Free Radic. Biol. Med. 79, 217–227. doi:10.1016/j.freeradbiomed.2014.12.013
Shi Q., Lee D.-Y., Féliers D., Abboud H. E., Bhat M. A., Gorin Y. (2020). Interplay between RNA-binding protein HuR and Nox4 as a novel therapeutic target in diabetic kidney disease. Mol. Metab. 36, 100968. doi:10.1016/j.molmet.2020.02.011
Shinoda G., Shyh-Chang N., Soysa T. Y., Zhu H., Seligson M. T., Shah S. P., et al. (2013). Fetal deficiency of lin28 programs life-long aberrations in growth and glucose metabolism. Stem Cells 31 (8), 1563–1573. doi:10.1002/stem.1423
Si R., Cabrera J. T. O., Tsuji-Hosokawa A., Guo R., Watanabe M., Gao L., et al. (2021). HuR/Cx40 downregulation causes coronary microvascular dysfunction in type 2 diabetes. JCI Insight 6 (21), e147982. doi:10.1172/jci.insight.147982
Sidali A., Teotia V., Solaiman N. S., Bashir N., Kanagaraj R., Murphy J. J., et al. (2021). AU-rich element RNA binding proteins: At the crossroads of post-transcriptional regulation and genome integrity. Int. J. Mol. Sci. 23 (1), 96. doi:10.3390/ijms23010096
Simo R., Stitt A. W., Gardner T. W. (2018). Neurodegeneration in diabetic retinopathy: Does it really matter? Diabetologia 61 (9), 1902–1912. doi:10.1007/s00125-018-4692-1
Sperling R. (2017). The nuts and bolts of the endogenous spliceosome. Wiley Interdiscip. Rev. RNA 8 (1), 1. doi:10.1002/wrna.1377
Stitt A. W., Lois N., Medina R. J., Adamson P., Curtis T. M. (2013). Advances in our understanding of diabetic retinopathy. Clin. Sci. (Lond) 125 (1), 1–17. doi:10.1042/cs20120588
Stoll L., Rodríguez-Trejo A., Guay C., Brozzi F., Bayazit M. B., Gattesco S., et al. (2020). A circular RNA generated from an intron of the insulin gene controls insulin secretion. Nat. Commun. 11 (1), 5611. doi:10.1038/s41467-020-19381-w
Sun S., Zhang M., Lin J., Hu J., Zhang R., Li C., et al. (2016). Lin28a protects against diabetic cardiomyopathy via the PKA/ROCK2 pathway. Biochem. Biophysical Res. Commun. 469 (1), 29–36. doi:10.1016/j.bbrc.2015.11.065
Sung Y., Jeong J., Kang R. J., Choi M., Park S., Kwon W., et al. (2019). Lin28a expression protects against streptozotocin-induced β-cell destruction and prevents diabetes in mice. Cell Biochem. Funct. 37 (3), 139–147. doi:10.1002/cbf.3376
Tao L., Fan X., Sun J., Zhang Z. (2021). Metformin prevented high glucose-induced endothelial reactive oxygen species via OGG1 in an AMPKα-Lin-28 dependent pathway. Life Sci. 268, 119015. doi:10.1016/j.lfs.2020.119015
Van Nostrand E. L., Freese P., Pratt G. A., Wang X., Wei X., Xiao R., et al. (2020). A large-scale binding and functional map of human RNA-binding proteins. Nature 583 (7818), 711–719. doi:10.1038/s41586-020-2077-3
Velázquez-Cruz A., Banos-Jaime B., Díaz-Quintana A., De la Rosa M. A., Díaz-Moreno I. (2021). Post-translational control of RNA-binding proteins and disease-related dysregulation. Front. Mol. Biosci. 8, 658852. doi:10.3389/fmolb.2021.658852
Verma S. K., Deshmukh V., Liu P., Nutter C. A., Espejo R., Hung M.-L., et al. (2013). Reactivation of fetal splicing programs in diabetic hearts is mediated by protein kinase C signaling. J. Biol. Chem. 288 (49), 35372–35386. doi:10.1074/jbc.M113.507426
Verma S. K., Deshmukh V., Thatcher K., Belanger K. K., Rhyner A. M., Meng S., et al. (2022). RBFOX2 is required for establishing RNA regulatory networks essential for heart development. Nucleic Acids Res. 50 (4), 2270–2286. doi:10.1093/nar/gkac055
Wan X., Belanger K., Widen S. G., Kuyumcu-Martinez M. N., Garg N. J. (2020). Genes of the cGMP-PKG-Ca(2+) signaling pathway are alternatively spliced in cardiomyopathy: Role of RBFOX2. Biochim. Biophys. Acta Mol. Basis Dis. 1866 (3), 165620. doi:10.1016/j.bbadis.2019.165620
Wang F., Tidei J. J., Polich E. D., Gao Y., Zhao H., Perrone-Bizzozero N. I., et al. (2015). Positive feedback between RNA-binding protein HuD and transcription factor SATB1 promotes neurogenesis. Proc. Natl. Acad. Sci. U. S. A. 112 (36), E4995–E5004. doi:10.1073/pnas.1513780112
Wang W., Lo A. C. Y. (2018). Diabetic retinopathy: Pathophysiology and treatments. Int. J. Mol. Sci. 19 (6), 1816. doi:10.3390/ijms19061816
Wei C. C., Guo D. F., Zhang S. L., Ingelfinger J. R., Chan J. S. (2005). Heterogenous nuclear ribonucleoprotein F modulates angiotensinogen gene expression in rat kidney proximal tubular cells. J. Am. Soc. Nephrol. 16 (3), 616–628. doi:10.1681/asn.2004080715
Weir G. C., Gaglia J., Bonner-Weir S. (2020). Inadequate β-cell mass is essential for the pathogenesis of type 2 diabetes. Lancet Diabetes Endocrinol. 8 (3), 249–256. doi:10.1016/s2213-8587(20)30022-x
Weiße J., Rosemann J., Krauspe V., Kappler M., Eckert A. W., Haemmerle M., et al. (2020). RNA-binding proteins as regulators of migration, invasion and metastasis in oral squamous cell carcinoma. Int. J. Mol. Sci. 21 (18), 6835. doi:10.3390/ijms21186835
Wu T., Jia J., Xiong X., He H., Bu L., Zhao Z., et al. (2013). Increased expression of Lin28B associates with poor prognosis in patients with oral squamous cell carcinoma. PLoS One 8 (12), e83869. doi:10.1371/journal.pone.0083869
Wu Y. L., Li H. F., Chen H. H., Lin H. (2022). Emergent roles of circular RNAs in metabolism and metabolic disorders. Int. J. Mol. Sci. 23 (3), 1032. doi:10.3390/ijms23031032
Xing X., Wang H., Zhang Y., Niu T., Jiang Y., Shi X., et al. (2019). O- glycosylation can regulate the proliferation and migration of human retinal microvascular endothelial cells through ZFR in high glucose condition. Biochem. Biophys. Res. Commun. 512 (3), 552–557. doi:10.1016/j.bbrc.2019.03.135
Xu Y., Wu W., Han Q., Wang Y., Li C., Zhang P., et al. (2019). Post-translational modification control of RNA-binding protein hnRNPK function. Open Biol. 9 (3), 180239. doi:10.1098/rsob.180239
Yang C., Eleftheriadou M., Kelaini S., Morrison T., González M. V., Caines R., et al. (2020). Targeting QKI-7 in vivo restores endothelial cell function in diabetes. Nat. Commun. 11 (1), 3812. doi:10.1038/s41467-020-17468-y
Yang C., Kelaini S., Caines R., Margariti A. (2018). RBPs play important roles in vascular endothelial dysfunction under diabetic conditions. Front. Physiol. 9, 1310. doi:10.3389/fphys.2018.01310
Yao Z. T., Yang Y. M., Sun M. M., He Y., Liao L., Chen K. S., et al. (2022). New insights into the interplay between long non-coding RNAs and RNA-binding proteins in cancer. Cancer Commun. (Lond) 42 (2), 117–140. doi:10.1002/cac2.12254
You P., Cheng Z., He X., Deng J., Diao J., Chen H., et al. (2020). Lin28a protects against diabetic cardiomyopathy through Mst1 inhibition. J. Cell Physiol. 235 (5), 4455–4465. doi:10.1002/jcp.29321
Zhai K., Gu L., Yang Z., Mao Y., Jin M., Chang Y., et al. (2016). RNA-binding protein CUGBP1 regulates insulin secretion via activation of phosphodiesterase 3B in mice. Diabetologia 59 (9), 1959–1967. doi:10.1007/s00125-016-4005-5
Zhang C., Han X., Yang L., Fu J., Sun C., Huang S., et al. (2020). Circular RNA circPPM1F modulates M1 macrophage activation and pancreatic islet inflammation in type 1 diabetes mellitus. Theranostics 10 (24), 10908–10924. doi:10.7150/thno.48264
Zhao M., Shen L., Ouyang Z., Li M., Deng G., Yang C., et al. (2020). Loss of hnRNP A1 in murine skeletal muscle exacerbates high-fat diet-induced onset of insulin resistance and hepatic steatosis. J. Mol. Cell Biol. 12 (4), 277–290. doi:10.1093/jmcb/mjz050
Zhu H., Shyh-Chang N., Segrè A. V., Shinoda G., Shah S. P., Einhorn W. S., et al. (2011). The Lin28/let-7 axis regulates glucose metabolism. Cell 147 (1), 81–94. doi:10.1016/j.cell.2011.08.033
Zhu Y., Hart G. W. (2021). Targeting O-GlcNAcylation to develop novel therapeutics. Mol. Asp. Med. 79, 100885. doi:10.1016/j.mam.2020.100885
Keywords: RNA-binding proteins, RNA-protein interaction, chronic complications, post-transcriptional gene regulation, diabetes mellitus
Citation: Zhang S, Yang X, Jiang M, Ma L, Hu J and Zhang H-H (2022) Post-transcriptional control by RNA-binding proteins in diabetes and its related complications. Front. Physiol. 13:953880. doi: 10.3389/fphys.2022.953880
Received: 27 May 2022; Accepted: 20 September 2022;
Published: 06 October 2022.
Edited by:
Rai Ajit K. Srivastava, Gemphire Therapeutics, United StatesReviewed by:
Yaseswini Neelamraju, University of Virginia, United StatesCopyright © 2022 Zhang, Yang, Jiang, Ma, Hu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ji Hu, aHVqaUBzdWRhLmVkdS5jbg==; Hong-Hong Zhang, emhoNTUwMjJAc2luYS5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.