- 1Department of Entomology, College of Plant Protection, Nanjing Agricultural University, Nanjing, China
- 2Institute of Plant Protection, Suzhou Academy of Agricultural Sciences, Suzhou, China
- 3Anhui Province Key Laboratory of Pollutant Sensitive Materials and Environmental Remediation, Anhui Provincial Engineering Laboratory for Efficient Utilization of Featured Resource Plants, College of Life Sciences, Huaibei Normal University, Huaibei, China
- 4Phytotoxicity Research Department, Central Agricultural Pesticide Laboratory, Agricultural Research Center, Giza, Egypt
Insects have sensitive olfactory systems to interact with environment and respond to the change in host plant conditions. Key genes in the system can be potential targets for developing new and efficient pest behaviour control methods. Riptortus pedestris is an important soybean pest in East Asia and has caused serious damage to the soybean plants in Huang-Huai-Hai region of China. However, the current treatment of pests is dominated by chemical insecticides and lacks efficient sustainable prevention and control technologies. In this study, we identified 49 putative odorant-binding proteins (OBPs) (43 were new genes) and 25 chemosensory proteins (CSPs) (17 were new genes) in R. pedestris genome. These OBP and CSP genes are clustered in highly conserved groups from other hemipteran species in phylogenetic trees. Most RpedOBPs displayed antennal-biased expression. Among the 49 RpedOBPs, 33 were significantly highly expressed in the antennae, including three male-biased and nine female-biased. While many RpedCSPs were detected both in the antennae and in non-antennal tissues, only 11 RpedCSPs displayed antennal-biased expression, in which four RpedCSPs were male-biased and five RpedCSPs were female-biased. Some OBP and CSP genes showed sex-biased expression profiles. Our results not only provide a foundation for future exploration of the functions of RpedOBPs and RpedCSPs but also aid in developing environmentally friendly insecticides in the future.
Introduction
Insects have a sensitive olfactory system, which enhances their ability to adapt to the complex external environment to accurately complete behavioural reactions such as feeding, mating, and avoiding natural enemies (Leal, 2013; Robertson, 2019). A large amount of studies on the molecular mechanisms of insect olfactory systems have found that the accurate operation of the system is inseparable from the participation of olfactory genes such as odorant-binding proteins (OBPs), chemosensory proteins (CSPs), and olfactory receptors (ORs) (Zhang et al., 2013; Glaser et al., 2015; Li et al., 2015; Elfekih et al., 2016; Larter et al., 2016; Paula et al., 2016; Renou and Anton, 2020; Rihani et al., 2021).
OBPs and CSPs are located in the lymph of insect antennal sensilla, and can accurately bind to external odorants and transport them to the corresponding ORs, ionotropic receptors (IRs) or gustatory receptors (GRs) to initiate behavioural responses. Therefore, the action of OBPs and CSPs is the first step in activating insect olfactory perception (Zhou, 2010; Dani et al., 2011; Pelosi et al., 2018; Rihani et al., 2021), which can be used as potential target genes to develop new and efficient pest behaviour control agents. Since the discovery of the first OBP and CSP in Antheraea polyphemus (Vogt and Riddiford, 1981) and Drosophila melanogaster (McKenna et al., 1994), respectively, a large number of OBPs and CSPs have been confirmed and studied in different insects (Latorre-Estivalis et al., 2021; Li et al., 2021; He et al., 2022).
These two types of genes have been the subject of studies on evolution, molecular structure, tissue distribution, and functional analysis (Spinelli et al., 2012; Pelosi et al., 2018; Li et al., 2021). It is now clear that both OBPs and CSPs are soluble small-molecule proteins. Generally, OBPs use six positionally conserved cysteines to form three interlocking disulfide bridges that stabilise the three-dimensional structure of the proteins. OBPs can be divided into three distinct subfamilies: classic OBPs (six conserved cysteines), minus-C OBPs (four conserved cysteines), and plus-C OBPs (more than six conserved cysteines) (Zhou, 2010; Schultze et al., 2012; Spinelli et al., 2012; Li et al., 2013; He and He, 2014; Zhang et al., 2017b). Compared with OBPs, CSPs are smaller, display greater evolutionary conservation, and have only two disulfide bridges with four conserved cysteines (Maleszka and Stange, 1997; Bohbot et al., 1998; Pelosi et al., 2005; Zhang et al., 2014; Zhu J. et al., 2016; Yi et al., 2017; Pelosi et al., 2018). Additionally, OBPs are often specifically or highly expressed in the antennae and are mainly involved in odorant recognition (Krieger et al., 1996; Sengul and Tu, 2010; Missbach et al., 2015; Zhang et al., 2017a), whereas many CSPs are expressed in the antennal and other non-olfactory organs (Pelosi et al., 2005; Vogt, 2005; Zhang et al., 2013; Zhang et al., 2014; Zhang L.-W. et al., 2016; Chen G.-L. et al., 2018). This suggests that CSPs may play both olfactory and non-olfactory roles in insects.
The bean bug Riptortus pedestris (Fabricius) (Hemiptera: Alydidae) is an important soybean pest in East Asia (Xu et al., 2021) and has a wide range of hosts. In addition to soybean, it can also harm Cruciferae, Gramineae, and other crops (Huang et al., 2021). R. pedestris damages soybeans by sucking, which results in poor growth and development of plants and insufficient pods (Chen J. H. et al., 2018). In recent years, R. pedestris has caused serious damage to the soybean plants in Huang-Huai-Hai region of China and has greatly reduced the yield of soybean, or lost the harvest. It has now become an important pest in China’s summer soybean producing areas (Chen J. H. et al., 2018; Li et al., 2019). However, the current treatment of pests is still dominated by the use of chemical insecticides and lacks efficient green prevention and control technologies. This has become a growing consensus that the development of green and efficient behaviour disruptors is a popular research direction based on the exploration of insect olfactory systems (Sun et al., 2011; Cui and Zhu, 2016; Qin et al., 2020). In this study, we identified 49 OBPs and 25 CSPs in the R. pedestris genome and found that these genes were clustered in highly conserved groups comprising OBP and CSP genes from other hemipteran species, respectively. The gene expression profiles of OBPs and CSPs showed that most RpedOBPs displayed antennal-biased expression, while many RpedCSPs were highly expressed in the antennae and non-antennal tissues, and some genes showed sex-biased expression. These results will help us identify the functions of RpedOBPs and RpedCSPs and develop environmentally friendly insecticides against this pest in the future.
Materials and Methods
Insect Rearing and Tissue Extraction
R. pedestris were fed with bean sprouts and maintained at a temperature of 26 ± 1°C under a 14:10 h light:dark photoperiod. Female and male adults, as well as larvae, were placed in insect cages for reproduction. Fifth instar larvae were collected and raised separately to obtain three-day-old virgin adults. The heads, thoraxes, abdomens, legs, wings, and antennae of virgin adults were collected. All collected samples were immediately frozen in liquid nitrogen and stored at ˗80°C for future use.
Sequence Data Collection and Analyses
Genome data, gene, protein, RNA and annotation files of R. pedestris were obtained from the Genome Warehouse (https://ngdc.cncb.ac.cn/gwh/Assembly/18849/show). We used the protein data of R. pedestris and blasted with different database of Nr (Non-Redundant Protein Sequence), Nt (nucleotide sequence database), UniProt (The Universal Protein Resource), KOG (Clusters of orthologous groups for eukaryotic complete genomes), eggNOG (evolutionary genealogy of genes: No-supervised Orthologous Groups), Interpro (the integrative protein signature), GO (Gene Ontology), and KEGG (Kyoto Encyclopedia of Genes and Genomes) databases (Huang et al., 2021), so R. pedestris proteins were annotated based on homology (These data were derived from a previous genome paper). We acquired the genes and ORF sequences of OBPs and CSPs by corresponding protein ID using R. pedestris genomic database. To ensure the accuracy of gene sequences, we used OBP and CSP genes to blast with Nr (Non-Redundant Protein Sequence) database in NCBI BLAST (http://blast.ncbi.nlm.nih.gov/). We also selected some genes (RpedOBP8, RpedOBP37, RpedCSP5) to clone and sent to sequencing to verify the correctness of the sequences. A total of 49 OBPs and 25 CSPs were obtained based on the similarity analysis. Putative N-terminal signal peptides of all OBPs and CSPs were predicted using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/), (Petersen et al., 2011).
RNA Isolation and cDNA Synthesis
Total RNA was extracted using a FastPure® Cell/Tissue Total RNA Isolation Kit (Vazyme, Nanjing, China) following the manufacturer’s instructions, and RNA quality was checked using a spectrophotometre (NanoDropTM 2000; Thermo Fisher Scientific, United States). Single-stranded cDNA templates were synthesised from 1 μg of total RNA from various tissue samples using the MonScript™ RTlll Super Mix with dsDNase (Two-Step) (Monad, Shanghai, China).
Quantitative Real Time-Polymerase Chain Reaction
The qRT-PCR primers for 49 OBPs and 25 CSPs (Supplementary Table S1) were designed using Beacon Designer 7.9 (PREMIER Biosoft International, CA, United States). Expression profilings of RpedOBPs and RpedCSPs were performed using qRT-PCR in a LightCycler® 96 (Roche, Switzerland) with a mixture of 5 μL MonAmp™ ChemoHS qPCR Mix (Monad, Shanghai, China), 0.2 μL of each primer (10 μM), 2.5 ng of sample cDNA, and 3.6 μL of sterilised ultrapure H2O. The reaction program was as follows: 10 min at 95°C, 40 cycles at 95°C for 10 s, 60°C for 10 s, and 72°C for 30 s. The results were analysed using LightCycler® 96 SW 1.1. Then, fluorescence was measured over a 55–95°C melting curve to detect a single gene-specific peak and to check the absence of primer dimer peaks; a single and discrete peak was detected for all primers tested. Negative controls consisted of non-template reactions in which the cDNA was replaced with H2O.
The expression levels of RpedOBPs and RpedCSPs were calculated relative to the reference genes RpedGAPDH (R. pedestris glyceraldehyde-3-phosphate dehydrogenase) and RpedEF (R. pedestris elongation factor) using the Q-Gene method in Microsoft Excel-based software Visual Basic (Muller et al., 2002; Simon, 2003). For each sample, three biological replicates were performed with three technical replicates per biological replicate.
Sequence Analyses
Based on sequence alignments, all phylogenetic trees in this study were constructed using the MEGA7 software (Kumar et al., 2016) using the neighbour-joining method, and each tree was tested by bootstrapping with 1,000 replicates. A phylogenetic tree was constructed based on the alignment results of cytochrome oxidase subunit I (COI) genes from different species (Aphis gossypii: KR017753.1, Nilaparvata lugens: AB325705.1, Sogatella furcifera: LC005703.1, Adelphocoris lineolatus: MZ608737.1, Adelphocoris suturalis: KY367052.1, Nysius ericae: KM022105.1, and R. pedestris: MG838422.1). The amino acid sequences of the RpedOBPs, RpedCSPs, and other hemipteran OBPs and CSPs were listed in Supplementary Table S2. The totals numbers of OBPs and CSPs in other insects have been reported in previous studies (Gu et al., 2011; Gu et al., 2013; Cao et al., 2014; He and He, 2014; Xue et al., 2014; Yang et al., 2014; Zhou et al., 2014; He et al., 2015; Zhou et al., 2015; Cui et al., 2017). Gene structure and exon/intron structure maps were generated using TBTools (version 1.098728) (Chen et al., 2020) based on R. pedestris annotated file (Gene Location Visualisation from GTF/GFF and Visualisation of Gene Structure). Pairwise similarity of sequences was also generated by TBTools based protein sequences (Protein Pairwise Similarity Matrix). Expression heat maps and bars were drawn using TBtools and GraphPad Prism 9.0, respectively, based on the qRT-PCR results.
Statistical Analysis
The qRT-PCR data (mean ± SE) of RpedOBPs and RpedCSPs from various samples were subjected to one-way nested analysis of variance (ANOVA) followed by a least significant difference test (LSD) to compare means using the SPSS Statistics software (version 22.0; SPSS Inc., Chicago, IL, United States).
Results and Discussion
Identification of OBP and CSP Genes in R. pedestris
Recent progress in the whole-genome sequencing provides insights into the molecular mechanisms of olfaction in insects (Cheng et al., 2017; Wan et al., 2019). Second- and third-generation sequencing methods have also been successfully used for R. pedestris genome assembly. Based on BUSCO completeness and contig length, the Wtdbg2 assembly was used for the draft assembly of the R. pedestris genome (Huang et al., 2021). We downloaded the R. pedestris genome, protein, RNA and annotation files using Genome Warehouse and further annotated them using the Nr, Nt, SwissProt, KOG, eggNOG, Interpro, GO, and KEGG databases. A total of 53 OBPs and 25 CSPs of R. pedestris were identified and corrected using the following correction through BLAST. Finally, 49 OBPs and 25 CSPs were identified and named RpedOBP1-49 (43 were new genes), RpedCSP1-25 (17 were new genes) (Table 1). The predicted results of the sequences analysis revealed that all OBPs and CSPs had full-length open reading frames (ORF), and 39 OBPs and 23 CSPs had a signal peptide, respectively. The 49 OBPs without signal peptides share 22.5–99.19% amino acid identities with each other, while the 25 CSPs share 22.5–99.07% amino acid identities with each other (Supplementary Table S3). Full-length sequences of the 49 RpedOBPs and 25 RpedCSPs were presented in the supplementary files.
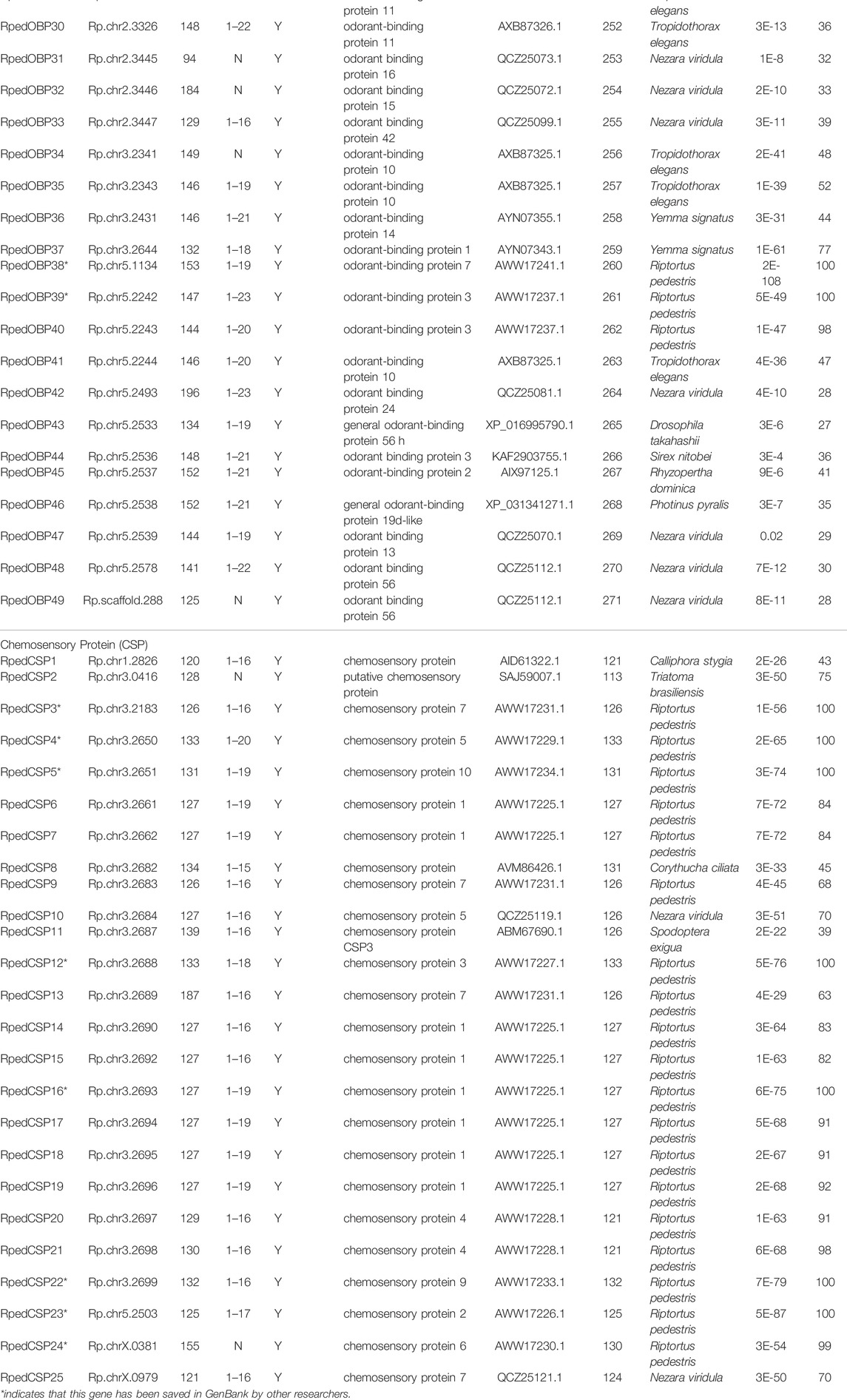
The number of RpedOBP and RpedCSP genes identified for R. pedestris is larger than those in other hemipterans (Figure 1). For example, 11 OBPs and 17 CSPs were identified in N. lugens (Xue et al., 2014; Yang et al., 2014; Zhang Y.-N. et al., 2016), 14 OBPs and 8 CSPs in A. lineolatus (Gu et al., 2011; Zhang Y.-N. et al., 2016), 16 OBPs and eight CSPs in A. suturalis (Cui et al., 2017), 28 OBPs and 16 CSPs in N. ericae (Zhang Y.-N. et al., 2016), nine OBPs and nine CSPs in A. gossypii (Gu et al., 2013; Zhang Y.-N. et al., 2016), and 12 OBPs and nine CSPs were identified in S. furcifera (He and He, 2014; Zhou et al., 2015). The differences in gene numbers may be explained by: 1) the different behaviours of different insects requiring distinct molecular mechanisms that have been developed over evolutionary time, and 2) the genomic data of R. pedestris that is more conducive to the comprehensive mining of OBP and CSP genes than other hemipteran species.
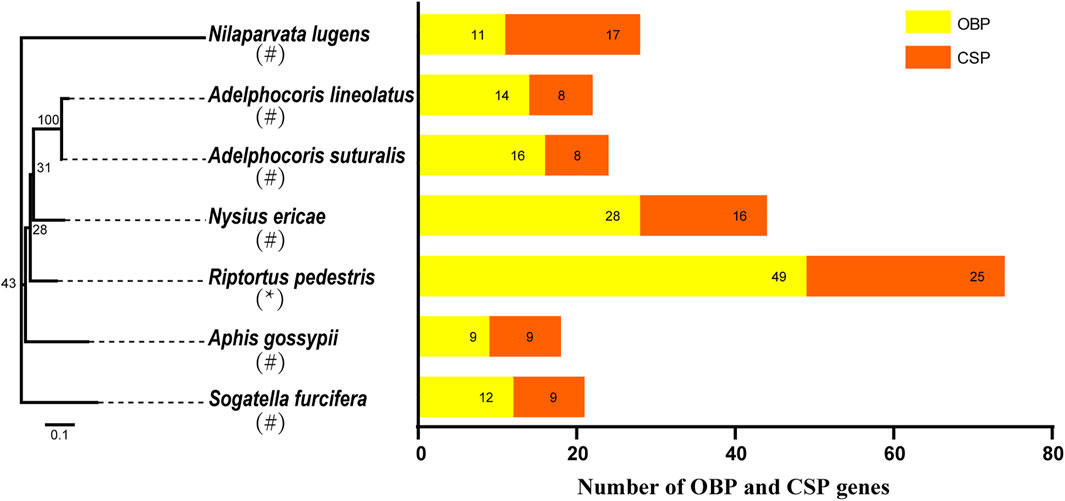
FIGURE 1. The number of OBP and CSP genes in different insect species, obtained from genome (*) or antennal transcriptome (#). The digits by the histogram bars represent number of OBP and CSP genes in different hemipteran species (Aphis gossypii, Nilaparvata lugens, Sogatella furcifera, Adelphocoris lineolatus, Adelphocoris suturalis, Nysius ericae, R. pedestris) and phylogenetic tree was built by these hemipteran species COI genes.
Localization of OBPs and CSPs in the R. pedestris Genome
To clarify the position of OBPs and CSPs in chromosomes, we carried out chromosome location analysis of all genes, and the results showed that 48 OBP genes were distributed across four chromosomes (Figure 2A); only RpedOBP49 was located on scaffold056, which could not be mapped to a chromosome based on the current genome assembly. Thirty OBPs clustered together on chromosome 2, followed by chromosome 5 (11 OBPs), 3 (four OBPs), and 1 (three OBPs). Twenty-five CSP genes were distributed across four chromosomes, with 21 CSPs clustered together on chromosome 3 and the others on chromosome X (two CSPs), 1 (one CSP), and 5 (one CSP) (Figure 2B). More than 60% OBP and 80% CSP genes are located within clusters, as seen in other insects (Gong et al., 2009; Gu et al., 2013), indicating a relatively recent expansion of the OBP and CSP families of R. pedestris and the diverse functions of genes have evolved in response to different odorants in the environment.
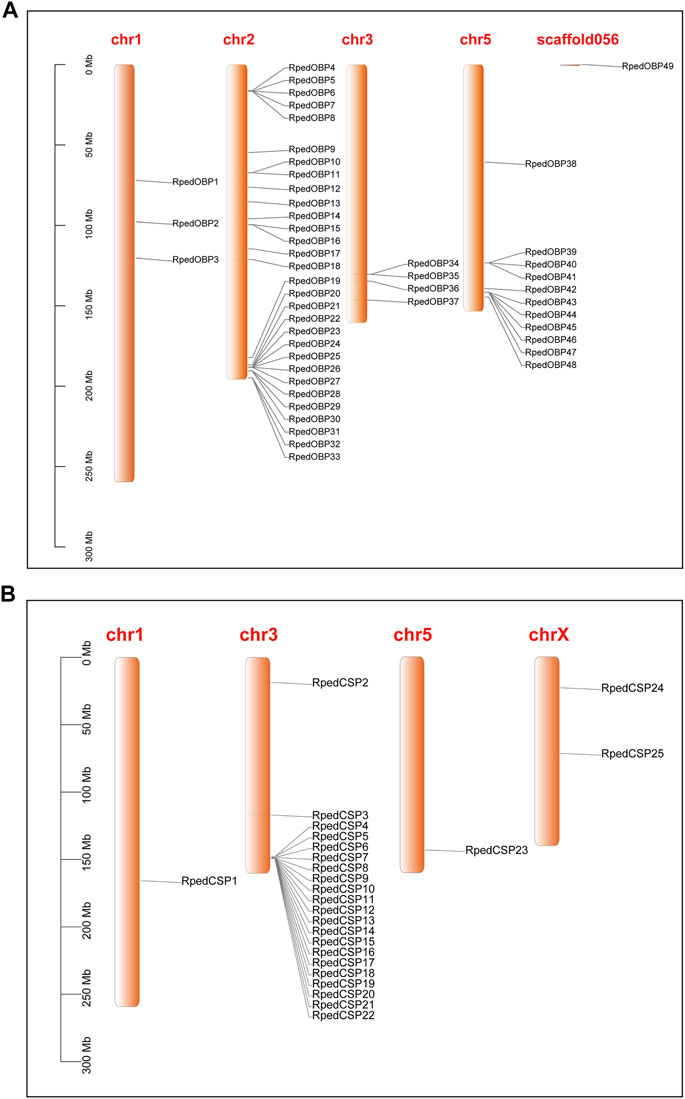
FIGURE 2. Localization of OBPs and CSPs in the R. pedestris genome. Based on the annotation file of R. pedestris, we acquired the localization of OBP and CSP genes in genome. (A) The RpedOBPs in the R. pedestris genome, they clustered together in 4 chromosomes and one scaffold. (B) The RpedCSPs in the R. pedestris genome, they clustered together in 4 chromosomes.
Genomic Structure of R. pedestris OBPs and CSPs
To further clarify the genomic structural characteristics of OBPs and CSPs, we obtained the gene lengths and intron numbers of OBPs and CSPs based on the genome annotation file of R. pedestris (Figure 3). The lengths of the OBP genes ranged from 3.065 to 46.888 kb, with 33 OBPs having six introns and the other 16 OBPs having four, five, seven, eight, nine, and 12 introns, respectively (Figure 3A). The CSP genes were much shorter, ranging from 2.114 to 34.628 kb, with one, two, or three introns (Figure 3B). The phylogenetic trees of OBPs and CSPs in R. pedestris showed that the genes clustered together tended to have similar genomic structures, which also implies that they may have similar functions. The sequences of RpedOBPs were longer and had more introns than those of RpedCSPs, indicating that they may have complex features of functional differentiation.
FIGURE 3. Genomic structure of R. pedestris OBPs and CSPs. Based on the annotation file of R. pedestris, we acquired genomic structures of OBP and CSP genes. (A) Numbers of exon/intron and exon/intron structures of RpedOBPs. (B) Numbers of exon/intron and exon/intron structures of RpedCSPs.
Phylogenetic Analyses of Hemipteran OBPs and CSPs
Two phylogenetic trees were constructed for the OBPs and CSPs using protein sequences from R. pedestris, A. lineolatus, Apolygus lucorum, Aphis gossypii, and other hemipteran species (Gu et al., 2011; Gu et al., 2013; Cao et al., 2014; He and He, 2014; Xue et al., 2014; Yang et al., 2014; Zhou et al., 2014; He et al., 2015; Zhou et al., 2015; Cui et al., 2017). Similar to that in other studies (Gu et al., 2011; Zhang Y.-N. et al., 2016; Cui et al., 2017), the OBP tree in this study showed that eight RpedOBPs (OBP1, 5–9, 13, and 42) could be divided into the Plus-C OBP subfamily, and the other 41 RpedOBPs clustered into the classic OBP subfamily (Figure 4). In the constructed CSP tree, our results indicated that all 25 RpedCSPs were distributed along various branches, and each clustered with at least one other moth orthologue (Figure 5). The diversity of the RpedOBPs and RpedCSPs families suggests a role for positive selection in the rapid evolution and functional diversification of these genes. We speculate that both RpedOBP and RpedCSP genes had some gene expansions, such as OBP1/5/6/7/8/13, OBP23/10/48/16/33/32/31/49/46/45/44/47/43, OBP25/26/27/28, OBP40/39/41/34/15/36/35/11, CSP13/8/19/22, and CSP12/15/14/16/17/19/18/6/7, indicating that these genes may be involved in the recognition of important odorants related to R. pedestris behaviour (Pelosi et al., 2005; Matsuo et al., 2007; Gu et al., 2012; Poivet et al., 2013; Martin-Blazquez et al., 2017; He et al., 2019).
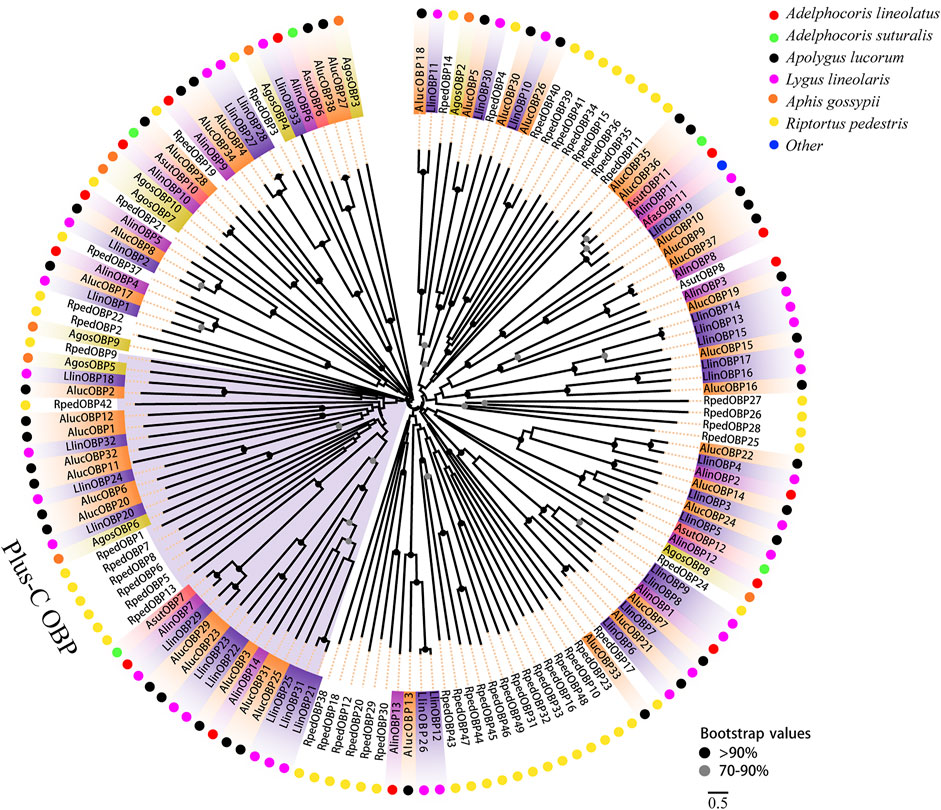
FIGURE 4. Phylogenetic tree of OBPs. Phylogenetic neighbor-joining tree was constructed using MEGA7 based on alignment results of MEGA7. A phylogenetic neighbor-joining tree was constructed using the OBPs protein that removed signal peptide. Bootstrap values higher than 70 were displayed.
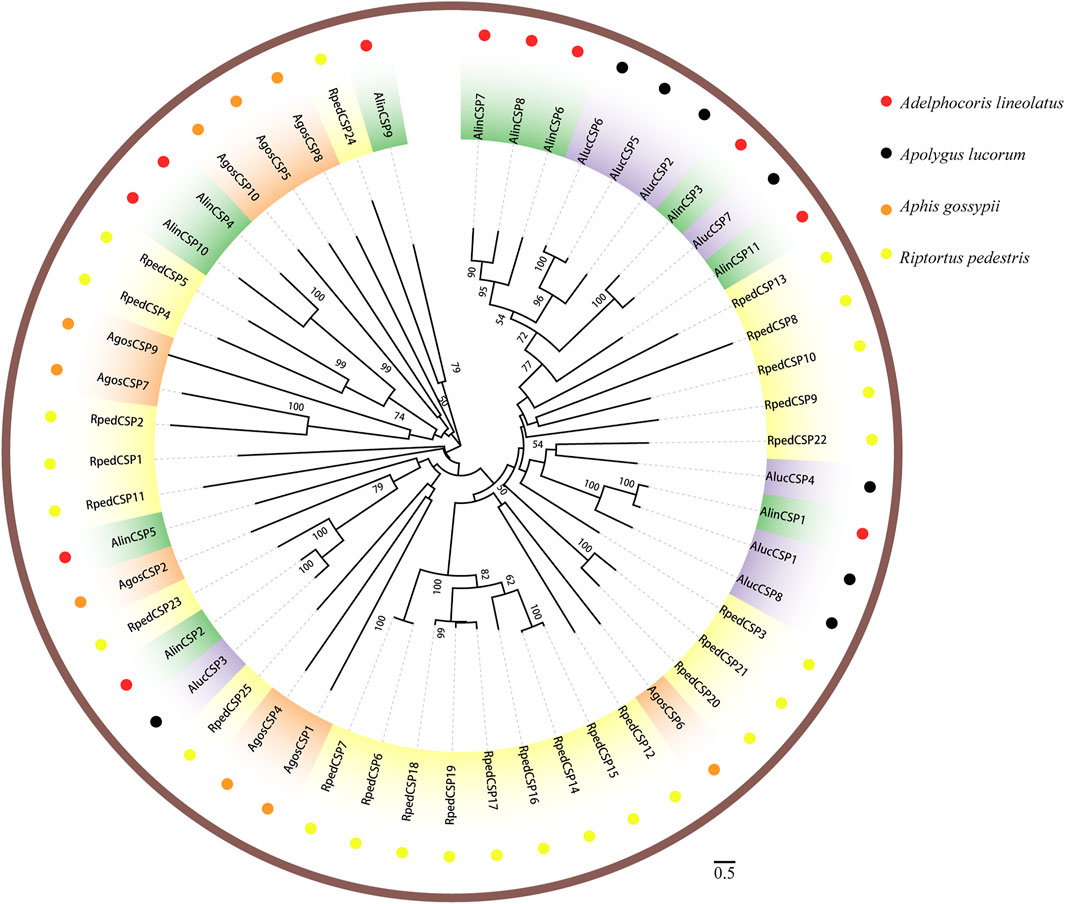
FIGURE 5. Phylogenetic tree of CSPs. Phylogenetic neighbor-joining tree was constructed using MEGA7 based on alignment results of MEGA7. A phylogenetic neighbor-joining tree was constructed using the CSPs protein that removed signal peptide. Bootstrap values higher than 50 were displayed.
Expression Profiles of R. pedestris OBP and CSP Genes
We used qRT-PCR to assess expression profiles of all R. pedestris OBPs and CSPs in the heads, thoraxes, abdomens, legs, wings, and antennae of the adults. The results showed that all OBPs and CSPs were expressed in the adult antennae of R. pedestris. Among the 49 RpedOBPs, 33 (approximately 67%) were significantly highly expressed in the antennae, including three male-biased (RpedOBP19, RpedOBP21, and RpedOBP32) and nine female-biased (RpedOBP2, RpedOBP6, RpedOBP9, RpedOBP17, RpedOBP24, RpedOBP34, RpedOBP36, RpedOBP48, and RpedOBP49). Among the 49 RpedOBPs, RpedOBP37 exhibited the highest expression level in male and female antennae (Figure 6). Compared to RpedOBPs, RpedCSPs were highly expressed in adult antennae as well as in non-antennal tissues. Of the 25 identified RpedCSP genes, only 11 RpedCSPs (approximately 44%) displayed antennal-biased expression; four RpedCSPs (RpedCSP3, RpedCSP12, RpedCSP20, and RpedCSP21) were male-biased and five RpedCSPs (RpedCSP4, RpedCSP9, RpedCSP11, RpedCSP13, and RpedCSP24) were female-biased in their expression (Figure 7). Several studies have shown that OBPs and CSPs are required for the correct recognition of some odorants from the external environment (Zhang et al., 2014; Liu et al., 2015; Chen G.-L. et al., 2018; Pelosi et al., 2018; Zhang et al., 2020a; Zhang et al., 2020b), therefore, we infer that the 33 RpedOBPs and 11 RpedCSPs highly expressed in adult antennae are likely to be involved in the crucial odorant reorganisation of R. pedestris (Krieger et al., 1996; Bohbot and Vogt, 2005; Zhang et al., 2014; Missbach et al., 2015; Chen G.-L. et al., 2018). The sex-biased RpedOBPs and RpedCSPs may be involved in the reorganisation of plant volatiles from oviposition sites or other sex-related odorants (He et al., 2010; Zhou et al., 2013; Zhang et al., 2019). Further analysis is needed to explore their exact roles, such as through fluorescence competitive binding assays (Liu et al., 2015; Ingham et al., 2020; Li et al., 2022), CRISPR/Cas9 mediated genome editing (Zhu G.-H. et al., 2016; Han et al., 2022), and gene mutations (Stowers and Logan, 2008; Zhang et al., 2020b).
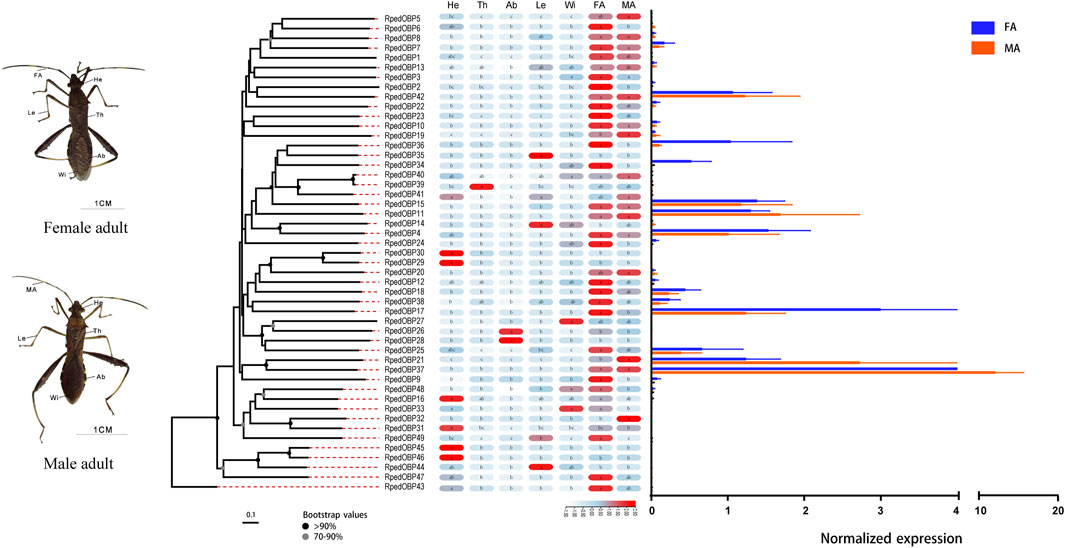
FIGURE 6. Tissue expression profiles of R. pedestris OBPs by qRT-PCR. The relative expression level is presented as mean ± SE (n = 3). The heatmap use Log2 and row scale based the relative expression level data. Different capital letters mean a significant difference between tissues (p < 0.05, ANOVA, LSD). He, heads; Th, thoraxes; Ab, abdomens; Le, legs; Wi, wings; FA, female antennae; MA, male antennae.
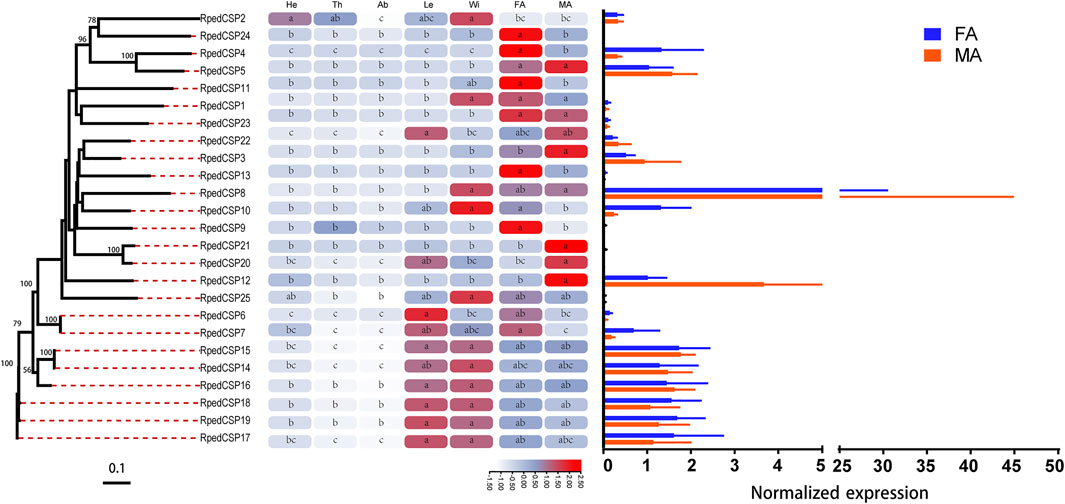
FIGURE 7. Tissue expression profiles of R. pedestris CSPs by qRT-PCR. The relative expression level is presented as mean ± SE (n = 3). The heatmap use Log2 and row scale based the relative expression level data. Different capital letters mean a significant difference between tissues (p < 0.05, ANOVA, LSD). He, heads; Th, thoraxes; Ab, abdomens; Le, legs; Wi, wings; FA, female antennae; MA, male antennae.
Similar to the findings of previous studies (Zhang et al., 2013; McKenzie et al., 2014; Gu et al., 2015; Zhang L.-W. et al., 2016), we also found that there were 12 RpedOBP and six RpedCSP genes highly expressed in non-antennal tissues, including four leg-biased genes (RpedOBP14, RpedOBP35, RpedOBP44, and RpedCSP6), six head-biased genes (RpedOBP16, RpedOBP29, RpedOBP30, RpedOBP31, RpedOBP45, and RpedOBP46), one thorax-biased gene (RpedOBP39), two abdomen-biased genes (RpedOBP26 and RpedOBP28), and five wing-biased genes (RpedCSP1, RpedCSP2, RpedCSP8, RpedCSP10, and RpedCSP25), indicating that these genes may have other non-olfactory functions.
Conclusion
In conclusion, we identified 49 OBPs and 25 CSPs in the R. pedestris genome and found that these genes were clustered in highly conserved groups comprising OBP and CSP genes from other hemipteran species. To further understand the functions of these genes, we conducted comprehensive and comparative phylogenetic analyses and studied the gene expression profiles of OBPs and CSPs. We found that most RpedOBPs displayed antennal-biased expression, but many RpedCSPs were detected in the antennae and were highly expressed in non-antennal tissues, and some genes showed characteristics of sex-biased expression. Tissue- and sex-biased expression patterns will help us identify the functions of RpedOBPs and RpedCSPs, which will also aid in understanding the olfactory mechanism of R. pedestris and the development of environmentally friendly insecticides against this pest in the future.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
JL, BL, and XZ conceived and designed the experimental plan. MY, WY, and SM performed the experiment. JL, MY, YD, CW, and XZ processed and analyzed the experiment data. YD, XL, and YW provided important suggestions to help modify the manuscript. JL, MY, YD, and XZ wrote the manuscript.
Funding
This work was supported by the Anhui Provincial Key Research and Development Program (202104a06020035), Natural Science Fund of Education Department of Anhui Province, China (KJ 2021B10 and KJ2021ZD0059), Anhui Provincial Natural Science Foundation (2008085MC63), Overseas Visiting Program of Outstanding Young Talents Program of Anhui Province, China (gxgwfx2019023), National College Students’ Innovation and Entrepreneurship Training Program (202110373017 and 202113620004), Anhui College Students’ Innovation and Entrepreneurship Training Program (202110373108, 202010373096, and S202013620012).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Bachelor students Hui-Ru Zhang, Dong Zhang, Hui Zhang, Xiao-Ya Zhang (Huaibei Normal University, China) for their help in rearing insects.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.949607/full#supplementary-material
References
Bohbot J., Sobrio F., Lucas P., Nagnan-Le Meillour P. (1998). Functional Characterization of a New Class of Odorant-Binding Proteins in the MothMamestra Brassicae. Biochem. Biophysical Res. Commun. 253 (2), 489–494. doi:10.1006/bbrc.1998.9806
Bohbot J., Vogt R. G. (2005). Antennal Expressed Genes of the Yellow Fever Mosquito (Aedes aegypti L.); Characterization of Odorant-Binding Protein 10 and Takeout. Insect Biochem. Mol. Biol. 35 (9), 961–979. doi:10.1016/j.ibmb.2005.03.010
Cao D., Liu Y., Walker W. B., Li J., Wang G. (2014). Molecular Characterization of the Aphis Gossypii Olfactory Receptor Gene Families. PLoS One 9 (6), e101187. doi:10.1371/journal.pone.0101187
Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: an Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen G.-L., Pan Y.-F., Ma Y.-F., Wang J., He M., He P. (2018). Binding Affinity Characterization of an Antennae-Enriched Chemosensory Protein from the White-Backed Planthopper, Sogatella Furcifera (Horváth), with Host Plant Volatiles. Pesticide Biochem. Physiology 152, 1–7. doi:10.1016/j.pestbp.2018.09.006
Chen J. H., Cui J., Tang J. W., Bi R., Zhang J. P., Shi S. S. (2018). Effects of Temperature on the Growth and Development of Riptortus Pedestris Fabricius. Chi J. Oil Crop Sci. 40 (4), 579–584. doi:10.7505/j.issn.1007-9084.2018.04.016
Cheng T., Wu J., Wu Y., Chilukuri R. V., Huang L., Yamamoto K., et al. (2017). Genomic Adaptation to Polyphagy and Insecticides in a Major East Asian Noctuid Pest. Nat. Ecol. Evol. 1 (11), 1747–1756. doi:10.1038/s41559-017-0314-4
Cui G. Z., Zhu J. J. (2016). Pheromone-Based Pest Management in China: Past, Present, and Future Prospects. J. Chem. Ecol. 42 (7), 557–570. doi:10.1007/s10886-016-0731-x
Cui H.-H., Gu S.-H., Zhu X.-Q., Wei Y., Liu H.-W., Khalid H. D., et al. (2017). Odorant-binding and Chemosensory Proteins Identified in the Antennal Transcriptome of Adelphocoris Suturalis Jakovlev. Comp. Biochem. Physiology Part D Genomics Proteomics 24, 139–145. doi:10.1016/j.cbd.2016.03.001
Dani F. R., Michelucci E., Francese S., Mastrobuoni G., Cappellozza S., La Marca G., et al. (2011). Odorant-binding Proteins and Chemosensory Proteins in Pheromone Detection and Release in the Silkmoth Bombyx mori. Chem. Senses 36 (4), 335–344. doi:10.1093/chemse/bjq137
Elfekih S., Chen C.-Y., Hsu J.-C., Belcaid M., Haymer D. (2016). Identification and Preliminary Characterization of Chemosensory Perception-Associated Proteins in the Melon Fly Bactrocera Cucurbitae Using RNA-Seq. Sci. Rep. 6, 19112. doi:10.1038/srep19112
Glaser N., Gallot A., Legeai F., Harry M., Kaiser L., Le Ru B., et al. (2015). Differential Expression of the Chemosensory Transcriptome in Two Populations of the Stemborer Sesamia Nonagrioides. Insect Biochem. Mol. Biol. 65, 28–34. doi:10.1016/j.ibmb.2015.07.008
Gong D.-P., Zhang H.-J., Zhao P., Xia Q.-Y., Xiang Z.-H. (2009). The Odorant Binding Protein Gene Family from the Genome of Silkworm, Bombyx mori. BMC Genomics 10, 332. doi:10.1186/1471-2164-10-332
Gu S.-H., Wang S.-P., Zhang X.-Y., Wu K.-M., Guo Y.-Y., Zhou J.-J., et al. (2011). Identification and Tissue Distribution of Odorant Binding Protein Genes in the Lucerne Plant Bug Adelphocoris Lineolatus (Goeze). Insect Biochem. Mol. Biol. 41 (4), 254–263. doi:10.1016/j.ibmb.2011.01.002
Gu S.-H., Wang S.-Y., Zhang X.-Y., Ji P., Liu J.-T., Wang G.-R., et al. (2012). Functional Characterizations of Chemosensory Proteins of the Alfalfa Plant Bug Adelphocoris Lineolatus Indicate Their Involvement in Host Recognition. PLoS One 7 (8), e42871. doi:10.1371/journal.pone.0042871
Gu S.-H., Wu K.-M., Guo Y.-Y., Field L. M., Pickett J. A., Zhang Y.-J., et al. (2013). Identification and Expression Profiling of Odorant Binding Proteins and Chemosensory Proteins between Two Wingless Morphs and a Winged Morph of the Cotton Aphid Aphis Gossypii Glover. PLoS One 8 (9), e73524. doi:10.1371/journal.pone.0073524
Gu S.-H., Zhou J.-J., Gao S., Wang D.-H., Li X.-C., Guo Y.-Y., et al. (2015). Identification and Comparative Expression Analysis of Odorant Binding Protein Genes in the Tobacco Cutworm Spodoptera Litura. Sci. Rep. 5, 13800. doi:10.1038/srep13800
Han W.-K., Yang Y.-L., Si Y.-X., Wei Z.-Q., Liu S.-R., Liu X.-L., et al. (2022). Involvement of GOBP2 in the Perception of a Sex Pheromone Component in Both Larval and Adult Spodoptera Litura Revealed Using CRISPR/Cas9 Mutagenesis. Insect Biochem. Mol. Biol. 141, 103719. doi:10.1016/j.ibmb.2022.103719
He M., He P. (2014). Molecular Characterization, Expression Profiling, and Binding Properties of Odorant Binding Protein Genes in the Whitebacked Planthopper, Sogatella Furcifera. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 174, 1–8. doi:10.1016/j.cbpb.2014.04.008
He M., Ma Y.-F., Guo H., Liu X.-Z., Long G.-J., Wang Q., et al. (2022). Genome-wide Identification and Expression Pattern Analysis of Novel Chemosensory Genes in the German Cockroach Blattella germanica. Genomics 114 (2), 110310. doi:10.1016/j.ygeno.2022.110310
He M., Zhang Y.-N., He P. (2015). Molecular Characterization and Differential Expression of an Olfactory Receptor Gene Family in the White-Backed Planthopper Sogatella Furcifera Based on Transcriptome Analysis. PLoS One 10 (11), e0140605. doi:10.1371/journal.pone.0140605
He P., Chen G. L., Li S., Wang J., Ma Y. F., Pan Y. F., et al. (2019). Evolution and Functional Analysis of Odorant‐binding Proteins in Three Rice Planthoppers: Nilaparvata Lugens Sogatella Furcifera and Laodelphax Striatellus. Pest. Manag. Sci. 75 (6), 1606–1620. doi:10.1002/ps.5277
He X., Tzotzos G., Woodcock C., Pickett J. A., Hooper T., Field L. M., et al. (2010). Binding of the General Odorant Binding Protein of Bombyx mori BmorGOBP2 to the Moth Sex Pheromone Components. J. Chem. Ecol. 36 (12), 1293–1305. doi:10.1007/s10886-010-9870-7
Huang H. J., Ye Y. X., Ye Z. X., Yan X. T., Wang X., Wei Z. Y., et al. (2021). Chromosome‐level Genome Assembly of the Bean Bug Riptortus Pedestris. Mol. Ecol. Resour. 21 (7), 2423–2436. doi:10.1111/1755-0998.13434
Ingham V. A., Anthousi A., Douris V., Harding N. J., Lycett G., Morris M., et al. (2020). A Sensory Appendage Protein Protects Malaria Vectors from Pyrethroids. Nature 577 (7790), 376–380. doi:10.1038/s41586-019-1864-1
Krieger J., von Nickisch-Rosenegk E., Mameli M., Pelosi P., Breer H. (1996). Binding Proteins from the Antennae of Bombyx mori. Insect Biochem. Mol. Biol. 26 (3), 297–307. doi:10.1016/0965-1748(95)00096-8
Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Larter N. K., Sun J. S., Carlson J. R. (2016). Organization and Function of Drosophila Odorant Binding Proteins. Elife 5, e20242. doi:10.7554/eLife.20242
Latorre-Estivalis J. M., Grosse-Wilde E., da Rocha Fernandes G., Hansson B. S., Lorenzo M. G. (2022). Changes in Antennal Gene Expression Underlying Sensory System Maturation in Rhodnius prolixus. Insect Biochem. Mol. Biol. 140, 103704. doi:10.1016/j.ibmb.2021.103704
Leal W. S. (2013). Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu. Rev. Entomol. 58 (1), 373–391. doi:10.1146/annurev-ento-120811-153635
Li K., Zhang X., Guo J., Penn H., Wu T., Li L., et al. (2019). Feeding of Riptortus Pedestris on Soybean Plants, the Primary Cause of Soybean Staygreen Syndrome in the Huang-Huai-Hai River Basin. Crop J. 7, 360–367. doi:10.1016/j.cj.2018.07.008
Li L.-L., Xu J.-W., Yao W.-C., Yang H.-H., Dewer Y., Zhang F., et al. (2021). Chemosensory Genes in the Head of Spodoptera Litura Larvae. Bull. Entomol. Res. 111 (4), 454–463. doi:10.1017/S0007485321000109
Li L. L., Huang J. R., Xu J. W., Yao W. C., Yang H. H., Shao L., et al. (2022). Ligand‐binding Properties of Odorant‐binding Protein 6 in Athetis Lepigone to Sex Pheromones and Maize Volatiles. Pest Manag. Sci. 78 (1), 52–62. doi:10.1002/ps.6606
Li X.-M., Zhu X.-Y., Wang Z.-Q., Wang Y., He P., Chen G., et al. (2015). Candidate Chemosensory Genes Identified in Colaphellus Bowringi by Antennal Transcriptome Analysis. BMC Genomics 16 (1), 1028. doi:10.1186/s12864-015-2236-3
Li Z.-Q., Zhang S., Luo J.-Y., Cui J.-J., Ma Y., Dong S.-L. (2013). Two Minus-C Odorant Binding Proteins from Helicoverpa Armigera Display Higher Ligand Binding Affinity at Acidic pH Than Neutral pH. J. Insect Physiology 59 (3), 263–272. doi:10.1016/j.jinsphys.2012.12.004
Liu N.-Y., Yang F., Yang K., He P., Niu X.-H., Xu W., et al. (2015). Two Subclasses of Odorant-Binding Proteins inSpodoptera Exiguadisplay Structural Conservation and Functional Divergence. Insect Mol. Biol. 24 (2), 167–182. doi:10.1111/imb.12143
Maleszka R., Stange G. (1997). Molecular Cloning, by a Novel Approach, of a cDNA Encoding a Putative Olfactory Protein in the Labial Palps of the Moth Cactoblastis cactorum. Gene 202 (1-2), 39–43. doi:10.1016/s0378-1119(97)00448-4
Martín-Blázquez R., Chen B., Kang L., Bakkali M. (2017). Evolution, Expression and Association of the Chemosensory Protein Genes with the Outbreak Phase of the Two Main Pest Locusts. Sci. Rep. 7 (1), 6653. doi:10.1038/s41598-017-07068-0
Matsuo T., Sugaya S., Yasukawa J., Aigaki T., Fuyama Y. (2007). Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 5 (5), e118. doi:10.1371/journal.pbio.0050118
McKenna M. P., Hekmat-Scafe D. S., Gaines P., Carlson J. R. (1994). Putative Drosophila Pheromone-Binding Proteins Expressed in a Subregion of the Olfactory System. J. Biol. Chem. 269 (23), 16340–16347. doi:10.1016/s0021-9258(17)34013-9
McKenzie S. K., Oxley P. R., Kronauer D. J. (2014). Comparative Genomics and Transcriptomics in Ants Provide New Insights into the Evolution and Function of Odorant Binding and Chemosensory Proteins. BMC Genomics 15, 718. doi:10.1186/1471-2164-15-718
Missbach C., Vogel H., Hansson B. S., Groβe-Wilde E. (2015). Identification of Odorant Binding Proteins and Chemosensory Proteins in Antennal Transcriptomes of the Jumping BristletailLepismachilis Y-Signataand the FirebratThermobia domestica:Evidence for an Independent OBP-OR Origin. Chemse 40 (9), 615–626. doi:10.1093/chemse/bjv050
Muller P. Y., Janovjak H., Miserez A. R., Dobbie Z. (2002). Processing of Gene Expression Data Generated by Quantitative Real-Time RT-PCR. Biotechniques 32 (6), 1372–1379.
Paula D. P., Togawa R. C., Costa M. M. C., Grynberg P., Martins N. F., Andow D. A. (2016). Identification and Expression Profile of Odorant‐binding Proteins in Halyomorpha Halys (Hemiptera: Pentatomidae). Insect Mol. Biol. 25 (5), 580–594. doi:10.1111/imb.12243
Pelosi P., Calvello M., Ban L. (2005). Diversity of Odorant-Binding Proteins and Chemosensory Proteins in Insects. Chem. Senses 30 (Suppl. 1), i291–i292. doi:10.1093/chemse/bjh229
Pelosi P., Iovinella I., Zhu J., Wang G., Dani F. R. (2018). Beyond Chemoreception: Diverse Tasks of Soluble Olfactory Proteins in Insects. Biol. Rev. 93 (1), 184–200. doi:10.1111/brv.12339
Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: Discriminating Signal Peptides from Transmembrane Regions. Nat. Methods 8 (10), 785–786. doi:10.1038/nmeth.1701
Poivet E., Gallot A., Montagné N., Glaser N., Legeai F., Jacquin-Joly E. (2013). A Comparison of the Olfactory Gene Repertoires of Adults and Larvae in the Noctuid Moth Spodoptera Littoralis. PLoS One 8 (4), e60263. doi:10.1371/journal.pone.0060263
Qin Y. G., Yang Z. K., Song D. L., Wang Q., Gu S. H., Li W. H., et al. (2020). Bioactivities of Synthetic Salicylate‐substituted Carboxyl ( E )‐β‐Farnesene Derivatives as Ecofriendly Agrochemicals and Their Binding Mechanism with Potential Targets in Aphid Olfactory System. Pest Manag. Sci. 76 (7), 2465–2472. doi:10.1002/ps.5787
Renou M., Anton S. (2020). Insect Olfactory Communication in a Complex and Changing World. Curr. Opin. Insect Sci. 42, 1–7. doi:10.1016/j.cois.2020.04.004
Rihani K., Ferveur J.-F., Briand L. (2021). The 40-Year Mystery of Insect Odorant-Binding Proteins. Biomolecules 11 (4), 509. doi:10.3390/biom11040509
Robertson H. M. (2019). Molecular Evolution of the Major Arthropod Chemoreceptor Gene Families. Annu. Rev. Entomol. 64, 227–242. doi:10.1146/annurev-ento-020117-043322
Schultze A., Schymura D., Forstner M., Krieger J. (2012). Expression Pattern of a 'Plus-C' Class Odorant Binding Protein in the Antenna of the Malaria Vector Anopheles gambiae. Insect Mol. Biol. 21 (2), 187–195. doi:10.1111/j.1365-2583.2011.01125.x
Sengul M. S., Tu Z. (2010). Expression Analysis and Knockdown of Two Antennal Odorant-Binding Protein Genes inAedes Aegypti. J. Insect Sci. 10, 1–18. doi:10.1673/031.010.14131
Simon P. (2003). Q-Gene: Processing Quantitative Real-Time RT-PCR Data. Bioinformatics 19 (11), 1439–1440. doi:10.1093/bioinformatics/btg157
Spinelli S., Lagarde A., Iovinella I., Legrand P., Tegoni M., Pelosi P., et al. (2012). Crystal Structure of Apis mellifera OBP14, a C-Minus Odorant-Binding Protein, and its Complexes with Odorant Molecules. Insect Biochem. Mol. Biol. 42 (1), 41–50. doi:10.1016/j.ibmb.2011.10.005
Stowers L., Logan D. W. (2008). LUSH Shapes up for a Starring Role in Olfaction. Cell 133 (7), 1137–1139. doi:10.1016/j.cell.2008.06.010
Sun Y., Qiao H., Ling Y., Yang S., Rui C., Pelosi P., et al. (2011). New Analogues of (E)-β-Farnesene with Insecticidal Activity and Binding Affinity to Aphid Odorant-Binding Proteins. J. Agric. Food Chem. 59 (6), 2456–2461. doi:10.1021/jf104712c
Vogt R. G. (2005). “Molecular Basis of Pheromone Detection in Insects,” in Comprehensive Insect Physiology, Biochemistry, Pharmacology and Molecular Biology Endocrinology. Editors K. Iatro LI Gilbert, and S. S. Gill (London: Elsevier), 753–803. doi:10.1016/b0-44-451924-6/00047-8
Vogt R. G., Riddiford L. M. (1981). Pheromone Binding and Inactivation by Moth Antennae. Nature 293 (5828), 161–163. doi:10.1038/293161a0
Wan F., Yin C., Tang R., Chen M., Wu Q., Huang C., et al. (2019). A Chromosome-Level Genome Assembly of Cydia Pomonella Provides Insights into Chemical Ecology and Insecticide Resistance. Nat. Commun. 10 (1), 4237. doi:10.1038/s41467-019-12175-9
Xu H., Zhao J., Li F., Yan Q., Meng L., Li B. (2021). Chemical Polymorphism Regulates the Attractiveness to Nymphs in the Bean Bug Riptortus Pedestris. J. Pest Sci. 94, 463–472. doi:10.1007/s10340-020-01268-w
Xue J., Zhou X., Zhang C.-X., Yu L.-L., Fan H.-W., Wang Z., et al. (2014). Genomes of the Rice Pest Brown Planthopper and its Endosymbionts Reveal Complex Complementary Contributions for Host Adaptation. Genome Biol. 15 (12), 521. doi:10.1186/s13059-014-0521-0
Yang K., He P., Dong S.-L. (2014). Different Expression Profiles Suggest Functional Differentiation Among Chemosensory Proteins in Nilaparvata Lugens (Hemiptera: Delphacidae). J. Insect Sci. 14 (1), 270. doi:10.1093/jisesa/ieu132
Yi X., Qi J., Zhou X., Hu M. Y., Zhong G. H. (2017). Differential Expression of Chemosensory-Protein Genes in Midguts in Response to Diet of Spodoptera Litura. Sci. Rep. 7 (1), 296. doi:10.1038/s41598-017-00403-5
Zhang L.-W., Kang K., Jiang S.-C., Zhang Y.-N., Wang T.-T., Zhang J., et al. (2016). Analysis of the Antennal Transcriptome and Insights into Olfactory Genes in Hyphantria cunea (Drury). PLoS One 11 (10), e0164729. doi:10.1371/journal.pone.0164729
Zhang L., Guo M., Zhuo F., Xu H., Zheng N., Zhang L. (2019). An Odorant-Binding Protein Mediates Sexually Dimorphic Behaviors via Binding Male-specific 2-heptanone in Migratory Locust. J. Insect Physiology 118, 103933. doi:10.1016/j.jinsphys.2019.103933
Zhang Y.-N., Jin J.-Y., Jin R., Xia Y.-H., Zhou J.-J., Deng J.-Y., et al. (2013). Differential Expression Patterns in Chemosensory and Non-chemosensory Tissues of Putative Chemosensory Genes Identified by Transcriptome Analysis of Insect Pest the Purple Stem Borer Sesamia Inferens (Walker). PLoS One 8 (7), e69715. doi:10.1371/journal.pone.0069715
Zhang Y.-N., Kang K., Xu L., Zhu X.-Y., Qian J.-L., Zhang Z.-J., et al. (2017a). Deep Sequencing of Antennal Transcriptome from Callosobruchus Chinensis to Characterize Odorant Binding Protein and Chemosensory Protein Genes. J. Stored Prod. Res. 74 (74), 13–21. doi:10.1016/j.jspr.2017.08.006
Zhang Y.-N., Xu J.-W., Zhang X.-C., Zhang X.-Q., Li L.-L., Yuan X., et al. (2020a). Organophosphorus Insecticide Interacts with the Pheromone-Binding Proteins of Athetis Lepigone: Implication for Olfactory Dysfunction. J. Hazard. Mater. 397, 122777. doi:10.1016/j.jhazmat.2020.122777
Zhang Y.-N., Ye Z.-F., Yang K., Dong S.-L. (2014). Antenna-predominant and Male-Biased CSP19 of Sesamia Inferens Is Able to Bind the Female Sex Pheromones and Host Plant Volatiles. Gene 536 (2), 279–286. doi:10.1016/j.gene.2013.12.011
Zhang Y.-N., Zhang X.-Q., Zhang X.-C., Xu J.-W., Li L.-L., Zhu X.-Y., et al. (2020b). Key Amino Acid Residues Influencing Binding Affinities of Pheromone-Binding Protein from Athetis Lepigone to Two Sex Pheromones. J. Agric. Food Chem. 68 (22), 6092–6103. doi:10.1021/acs.jafc.0c01572
Zhang Y.-N., Zhu X.-Y., Ma J.-F., Dong Z.-P., Xu J.-W., Kang K., et al. (2017b). Molecular Identification and Expression Patterns of Odorant Binding Protein and Chemosensory Protein Genes inAthetis lepigone(Lepidoptera: Noctuidae). PeerJ 5, e3157. doi:10.7717/peerj.3157
Zhang Y.-N., Zhu X.-Y., Zhang Q., Yin C.-Y., Dong Z.-P., Zuo L.-H., et al. (2016). De Novo assembly and Characterization of Antennal Transcriptome Reveal Chemosensory System in Nysius Ericae. J. Asia-Pacific Entomology 19, 1077–1087. doi:10.1016/j.aspen.2016.09.013
Zhou J.-J. (2010). Odorant-binding Proteins in Insects. Vitam. Horm. 83, 241–272. doi:10.1016/S0083-6729(10)83010-9
Zhou S.-S., Sun Z., Ma W., Chen W., Wang M.-Q. (2014). De Novo analysis of the Nilaparvata Lugens (Stål) Antenna Transcriptome and Expression Patterns of Olfactory Genes. Comp. Biochem. Physiology Part D Genomics Proteomics 9, 31–39. doi:10.1016/j.cbd.2013.12.002
Zhou W., Yuan X., Qian P., Cheng J., Zhang C., Gurr G., et al. (2015). Identification and Expression Profiling of Putative Chemosensory Protein Genes in Two Rice Planthoppers, Laodelphax Striatellus (Fallén) and Sogatella Furcifera (Horváth). J. Asia-Pacific Entomology 18 (4), 771–778. doi:10.1016/j.aspen.2015.09.006
Zhou X.-H., Ban L.-P., Iovinella I., Zhao L.-J., Gao Q., Felicioli A., et al. (2013). Diversity, Abundance, and Sex-specific Expression of Chemosensory Proteins in the Reproductive Organs of the Locust Locusta migratoria Manilensis. Biol. Chem. 394 (1), 43–54. doi:10.1515/hsz-2012-0114
Zhu G.-H., Xu J., Cui Z., Dong X.-T., Ye Z.-F., Niu D.-J., et al. (2016). Functional Characterization of SlitPBP3 in Spodoptera Litura by CRISPR/Cas9 Mediated Genome Editing. Insect Biochem. Mol. Biol. 75, 1–9. doi:10.1016/j.ibmb.2016.05.006
Keywords: Riptortus pedestris, genome analysis, odorant-binding proteins, chemosensory proteins, olfactory
Citation: Li J-B, Yin M-Z, Yao W-C, Ma S, Dewer Y, Liu X-Z, Wang Y-Y, Wang C-W, Li B-P and Zhu X-Y (2022) Genome-Wide Analysis of Odorant-Binding Proteins and Chemosensory Proteins in the Bean bug Riptortus pedestris. Front. Physiol. 13:949607. doi: 10.3389/fphys.2022.949607
Received: 21 May 2022; Accepted: 20 June 2022;
Published: 14 July 2022.
Edited by:
Wei Xu, Murdoch University, AustraliaReviewed by:
Nai-Yong Liu, Southwest Forestry University, ChinaXiaolong Liu, Hubei University, China
Da-Song Chen, Guangdong Academy of Science (CAS), China
Copyright © 2022 Li, Yin, Yao, Ma, Dewer, Liu, Wang, Wang, Li and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao-Ping Li, bGJwQG5qYXUuZWR1LmNu; Xiu-Yun Zhu, eHl6aHVoYm51QDE2My5jb20=
 Jin-Bu Li1,2
Jin-Bu Li1,2 Mao-Zhu Yin
Mao-Zhu Yin Bao-Ping Li
Bao-Ping Li