- 1Jilin Engineering Research Center of Resource Insects Industrialization, Institute of Biological Control, Jilin Agricultural University, Changchun, China
- 2Institute of Plant Protection, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 3Key Laboratory of Green Pesticide and Agricultural Bioengineering of Ministry of Education, Guizhou University, Guiyang, China
The GATA transcription factor Pannier is identified as the major regulatory gene in color pattern formation in the Asian multi-colored ladybird beetle (Harmonia axyridis). however, the mechanisms of Pannier in regulating melanin synthesis and development in H. axyridis remain elusive. In this study, we identified and characterized Pannier in H. axyridis (HaPnr) and showed it to have two alternative spliced variants named HaPnr-α and HaPnr-β. Analyses of developmental stage expression patterns revealed that HaPnr, HaPnr-α and HaPnr-β were constitutively expressed throughout all developmental stages. To examine the role of HaPnr in H. axyridis development, RNA interference was performed in late larvae (the fourth instar) and early pupae (the first day of pupa stage). The transcript levels of HaPnr were effectively suppressed after the injection of double-stranded RNA of HaPnr (dsHaPnr). The fourth instar larvae injected with dsHaPnr reduced the pupation rates to only 61.50%, compared with 88.5% in the dsGFP-injected group. The un-pupated larvae gradually died after 1 week, and visually unaffected pupae emerged into abnormal adults with malformed hind wings and melanin absent from the cuticle. These abnormal adults gradually died 10 days after eclosion. However, when early pupae were injected with dsHaPnr, the normal eclosion rate was achieved at 88.41% on day 6 after the injection. In addition, these successful eclosion adults also showed an absence of melanin in the cuticle, but they could mate normally and have normal fecundity as compared with the control. We further demonstrated that the suppression of HaPnr-α or HaPnr-β individually did not affect the pupation and eclosion process. The suppression of HaPnr-α expression resulted in elytra melanin decreasing in both the conspicua and the succinea subgroup in H. axyridis. Even though the suppression of HaPnr-β expression only affected the melanin synthesis in the succinea subgroup, it significantly prolonged the time taken for melanin synthesis to occur in the conspicua subgroup in H. axyridis. These results indicate that HaPnr plays an essential role in insect development, especially during their metamorphosis, and also support our hypothesis that HaPnr could regulate melanin synthesis in H. axyridis under the combined action with its two splicing variants, HaPnr-α and HaPnr-β.
Introduction
Insects are among the species with the most abundant phenotypic diversity in the world (Moczek, 2010). Phenotypic diversity occurs with important biological and physiological significance in insect development and evolution (Chau and Goodisman, 2017). Phenotypic plasticity is a regulatory mechanism for organisms to respond to environmental changes. In insects, melanization is a significant and common phenomenon in phenotypic diversity on the cuticle. Since the first finding of a melanized individual of Bistom betularia in Manchester in 1864, many scholars have focused on and examined the melanization phenomenon, its creation mechanism, and its evolutionary significance (True, 2003). Many studies have demonstrated that melanization occurs with important physiological meaning in insects, such as melanic individuals protecting themselves through mimicry, such as the swallowtail butterfly and Papilio glaucus in Lepidoptera (Koch et al., 2000), while melanic individuals of Oncopeltus fasciatus have higher fitness in low temperature (Sharma et al., 2016). In addition, the intraspecific diversification of elytra melanin patterns in ladybird beetle showed obvious mating selection preference (Wang et al., 2009; Wang et al., 2013). As a result, a foundation for developing particular melanized strains needs to be established by studying the melanization mechanism in insects. Previous studies have demonstrated that there are significant differences in the melanization mechanism between insects and vertebrates because of pigment synthesis differences. Vertebrates use specialized cell types and cell migration to create body pigment patterns (Hoekstra, 2006), while insects typically produce their body color pigment (or precursor) in epidermal cells then secrete these substances into the developing cuticle (True, 2003). The synthesis and deposition of insect melanin pigments can be influenced by environmental factors and be regulated by pivotal genes (Wittkopp and Beldade, 2009; Van der Burg et al., 2020).
The Asian multi-colored ladybird beetle, Harmonia axyridis (Coleoptera: Coccinellidae), is known for its voracious predatory capacity on various key pests (Koch, 2003; Chen et al., 2019a). In addition, it also works as an ideal model organism to study insect phenotype variations because of more than 200 different elytral color patterns in H. axyridis (Gautier et al., 2018; Ando and Niimi, 2019). There are four common color patterns of elytra that have been widely studied in H. axyridis, which include: succinea, axyridis, spectabilis, and conspicua (Ando et al., 2018; Gautier et al., 2018; Xiao et al., 2020). This striking intraspecific variation has prompted the investigation of its genetic, biochemical, and evolutionary meaning (Bezzerides et al., 2007; Ando et al., 2018). A previous study demonstrated that the elytra color pattern variation in H. axyridis is a melanization phenomenon in the cuticle (True, 2003). In our previous studies, we have confirmed that dopamine melanin synthesis is the primary pathway in the elytra of H. axyridis (Chen et al., 2019b; Xiao et al., 2020). Meanwhile, the aspartate-β-alanine-NBAD pathway being able to regulate the number and size of melanin spots in elytra has been reported in H. axyridis (Zhang et al., 2020; Zhang et al., 2021). Recently, new insights into the effect of the transcription factor Pannier on the regulation of pigment synthesis were discovered in H. axyridis (Niimi and Ando, 2021). With the advancement of next-generation sequencing (NGS) technologies, the ortholog of the Drosophila melanogaster Pannier gene was identified as the regulatory gene of wing color pattern polymorphism at the h locus in H. axyridis. Duals regulate functions of the Pannier gene on color pattern polymorphism in H. axyridis. First, repeated inversions at the upstream region of the first intron of Pannier resulted in the four common subgroup formations from the evolutionary genetic aspect. Second, Pannier could regulate melanin synthesis, further influencing color pattern formation from the physiological aspect (Ando et al., 2018; Gautier et al., 2018).
The biological function of Pannier has been studied in the genetically tractable organism D. melanogaster. Pannier was first reported in D. melanogaster in 1993 (Ramain et al., 1993), and is one of five GATA transcription factors in insects (Minakhina et al., 2011). Previous studies demonstrated that Pannier plays important roles in several developmental progresses in D. melanogaster. In the embryo’s development, Pannier is essential for dorsal closure (Herranze and Morata, 2001) and hemocyte maturation and differentiation (Minakhina et al., 2011). During imaginal development, the function of Pannier is to control imaginal disc development, while it acts upstream of wingless (wg) to direct dorsal eye disc development (Maurel-Zaffran and Treisman, 2000). Otherwise, Pannier controls compound eye size by activating target genes during dorsal–ventral axis formation in eye–antennal disc development (Buchberger et al., 2021). Moreover, Pannier also plays an important role in establishing and maintaining proper heart function in D. melanogaster (Gajewski et al., 1999; Qian and Bodmer, 2009; Lovato et al., 2015). However, the detailed function of Pannier in H. axyridis is still unknown. Thus, it is necessary to investigate the physiological function of Pannier in H. axyridis to deepen the understanding of the role of Pannier in the development of insects.
Even though the role of Pannier in elytral pigmentation during pupal development was discovered in H. axyridis, the detailed regulatory mechanisms remain elusive. To better understand the regulation mechanisms of Pannier in H. axyridis and further its physiological function in development, we: 1) sequenced and characterized cDNA, putatively encoding Pannier from H. axyrids, an emerging model organism in multiple phenotype evolution studies; 2) examined the developmental stage-dependent expression profiles of HaPnr; and 3) investigated the roles of HaPnr in melanin synthesis and insect development by using RNA interference. Our results provide crucial evidence that Pannier regulates melanin synthesis through a special action mode with its splicing variants and is an essential gene during the developmental process in H. axyridis.
Materials and Methods
Insect Culture
Adults of H. axyridis were collected from wheat fields (39°95′ N, 116°28′ E) on the experimental field of the Beijing Academy of Agriculture and Forestry Sciences (BAAFS), Beijing, China, in May 2014. The ladybird beetles were transported to an automated environmental management system controlled (Sunauto, Beijing, China) rearing room. They were maintained in aluminum frame cages (50 cm × 50 cm × 50 cm) with 100-mesh plastic gauze coverings at 25 ± 1°C, 60% relative humidity, and 16 h: 8 h (Light: Dark) photoperiod. Each cage housed 40 pairs of adults, which were fed daily on vicia aphids, Megoura japonica Matsumura (Hemiptera: Aphididae) on seedlings of broad bean, Vicia faba L., cv. ‘LinCan-5’).
Total RNA Isolation and Reverse Transcription
TRIzol reagent (Invitrogen, Carlsbad, CA, United States) was used to extract total RNA from pupa of H. axyridis samples, and the RNA concentration was determined at 260 nm using a NanoDrop ONE spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). After the total RNA (1.0 μg) was used for cDNA synthesis using the First Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) with oligo (dT)18 as the primer in a 20 μL reaction system. The first-strand cDNA was used in all subsequent analyses.
Subcloning and Sequencing of HaPnr Coding Sequences
One pair of gene specific primers were designed on the HaPnr gene prediction in NCBI (Accession number: LC269051.1) to amplify open reading frame (ORF) fragments by PCR for the full-length cDNA corresponding to the entire protein coding regions (Table 1). The PCR products were subjected to electrophoresis on 1% agarose gel. The PCR bands were excised and purified using QIAEX II Agarose Gel Extraction Kit (QIAGEN, Hilden, Germany). The purified PCR fragment was ligated into pcDNA3.1 (+) vector (Invitrogen, Carlsbad, CA, United States). The ligation mixtures were then used to transform Fast-T1 bacterial cells (Vazyme, Nanjing, China). Plasmids were isolated from the bacterial cells and sequenced by a DNA sequencing company (Sangon, Shanghai, China).
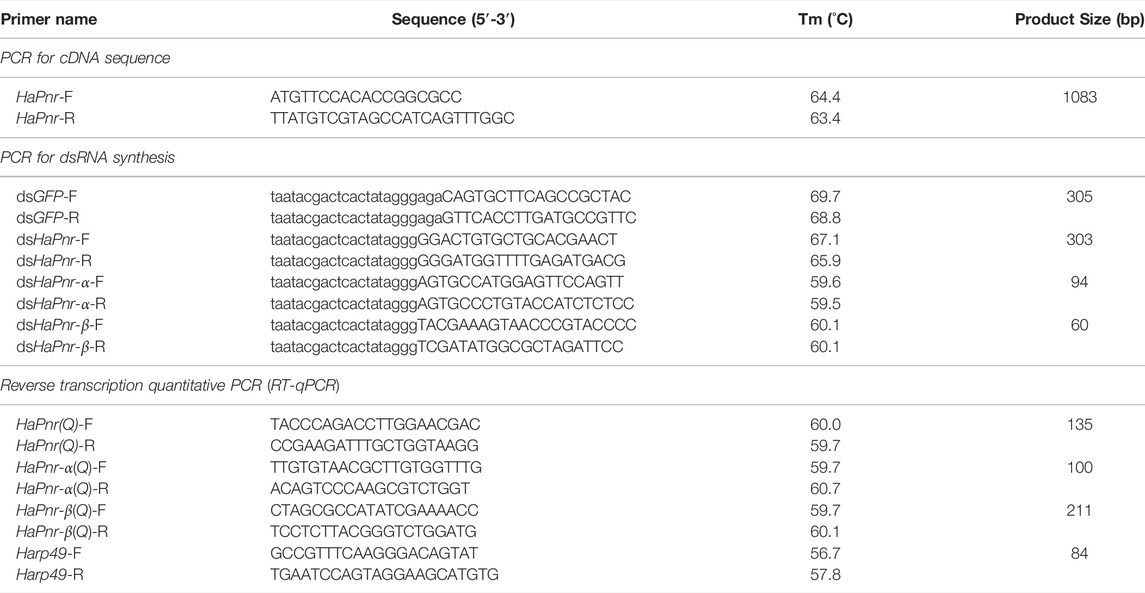
TABLE 1. Primers used to amplify HaPnr cDNA sequence, synthesize dsRNA and analyze transcript levels.
Analyses of HaPnr Coding Sequences, Deduced Amino Acid, and Gene Sequences
The amino acid sequence of a putative HaPnr protein was deduced from its cDNA, and molecular mass and isoelectric point of the deduced protein were calculated by using online tools of ExPASy website (http://www.expasy.org/tools/). Structure homology modeling was done using SWISS-MODEL (https://swissmodel.expasy.org). Multiple amino acid sequence alignment of all known insect Pnrs found in UniProt was carried out using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). We performed insect Pannier phylogenetic analysis using the neighbor-joining algorithm by using Mega X software. To evaluate the branch strength of the tree, a bootstrap analysis of 1000 replications was performed. The gene structure of HaPnr was revealed by comparing the full-length cDNA sequence with its corresponding gene sequence from genome data (http://v2.insect-genome.com/Organism/418).
Analysis of Expression Profiles by Reverse Transcription Quantitative PCR
The relative transcript levels of HaPnr including its two splicing variants, HaPnr-α and HaPnr-β, were analyzed by reverse transcription quantitative PCR (RT-qPCR) using SYBR Green with the Applied Biosystems® Real-time PCR Instrument (ABI Laboratories, Hercules, CA, United States). For developmental expression profiling, samples were prepared from the embryos of one-, two- and three-day-old eggs, larvae of all four instars, pupae ranging from one to 4 days in age, and initial eclosion adults, including males and females. Gene-specific primers (Table 1) were designed using the Primer3Plus (Untergasser et al., 2007) based on our obtained HaPnr, HaPnr-α and HaPnr-β sequence information. The ribosomal protein S49 (Harp49) in H. axyridis was used as an internal reference gene.
The optimized quantitative PCR program consisted of an initial denaturation at 95°C for 30 s, followed by 40 cycles of 95°C for 10 s, and 60°C for 30 s. After PCR, amplification specificity was verified by obtaining the dissociation curve, in which the PCR product were followed by one repeat of 95°C for 15 s, 60°C for 1 minute, and 95°C for 15 s. The specificity of each reaction was evaluated based on the melting temperatures of the PCR products. RT-qPCR was performed with three biological replicates, each with three technical replicates. The transcript levels of HaPnr, HaPnr-α and HaPnr-β were expressed as the normalized transcript abundance using Harp49 as an internal reference gene, individually. The relative HaPnr, HaPnr-α and HaPnr-β transcript levels were calculated according to the 2-△△Ct method.
Functional Analysis of HaPnr
The role of HaPnr in H. axyridis development was investigated using RNA interference (RNAi). Double stranded RNAs (dsRNAs) were synthesized using MEGAscript® RNAi Kit (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. Relevant information on the primers used for dsRNA synthesis is given in Table 1. The fourth instar larvae and one-day-old pupae were chosen for injections of HaPnr, HaPnr-α and HaPnr-β dsRNA (dsHaPnr, dsHaPnr-α and dsHaPnr-β) at doses of 300 ng/individual; To serve as controls, similar numbers of the same life stages were injected with dsRNA of the green fluorescent protein gene (dsGFP) at the same dose. The injection of dsGFP resulted in a mortality rate of less than 10%. Each RNAi experiment was carried out three times with at least 40 insects each time. The injected insects were reared in regular conditions, and phenotypes (elytral coloration) of the insects was recorded daily following injection. RT-qPCR was used to monitor the change in HaPnr, HaPnr-α and HaPnr-β transcript level after the injection. Three insects from each time point were then pooled as a sample for total RNA extraction. The analysis was performed with three biological samples per time point, with three technical replicates per sample. Phenotype images were taken using Zeiss Microscope SteREO Discovery V20 (Carl Zeiss, Germany), all using the same magnification, exposure time, and light intensity. Images were then selected for depiction of the most representative phenotypes.
To examine the effect of HaPnr suppression on fecundity in H. axyridis, the one-day-old pupae were injected with dsHaPnr (300 ng/pupa) or dsGFP as controls. After the injected pupae developed into adults, the males and females were separated and paired randomly due to the disappeared characteristic of color type in adults when HaPnr was suppressed in the pupa stage. Each treatment consisted of three pairs of a male and a female, and each treatment was repeated three times. Eggs were collected for 15 days after eclosion. Egg hatchability was examined 3 days after the eggs were collected.
Statistical Analysis
The level of HaPnr, HaPnr-α and HaPnr-β transcript in RT-qPCR analysis was expressed as a percentage of the level in the controls by dividing the relative expression value (REV) in the dsHaPnr-injected insects by REV in the dsGFP-injected insects and multiplying by 100. The REVs from developmental stages were arcsine square-root transformed before being subjected to ANOVA followed by Duncan’s test. Percent data from the RNAi experiments were subjected to ANOVA followed by Duncan’s test to separate means (IBM SPSS Statistics 22.0 software).
Results
Analysis of HaPnr Gene Sequences, cDNA and Amino Acid
The length of the HaPnr gene sequence is 23,604 base pairs (bp) from the transcription start site to the transcription end site, containing four exons and three introns. The HaPnr gene is located on chromosome 4. HaPnr has two splicing variants, HaPnr-α and HaPnr-β (Figure 1A). Sequencing analysis of the components of HaPnr-α and HaPnr-β found that alternative splicing occurred in second exon (Figure 1B). The open read frame (ORF) of HaPnr-α is 1083 bp, encoding 360 amino acids (aa). The predicted molecular mass and isoelectric point are 38.91 kDa and 9.19, respectively. The ORF of HaPnr-β is 996 bp, encoding 331 aa. The predicted molecular mass and isoelectric point are 35.96 kDa and 9.30, respectively. The zinc-finger domain is the representative characteristic of the transcription factor Pannier. Our results show that HaPnr-α has two zinc finger domains and HaPnr-β has one zinc finger domain based on the SWISS-MODEL analysis (Figure 1C). Detailed information regarding the zinc-finger domain located on HaPnr sequences is shown in Supplementary Figures S1, S2.
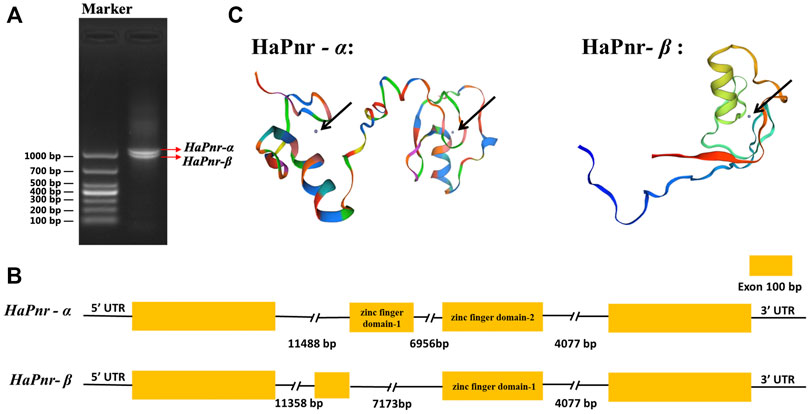
FIGURE 1. Electropherogram of HaPnr gene amplified, DL1000 (Takara Bio Inc., Japan) was used as DNA marker for agarose gel electrophoresis (A). Schematic diagram for the organization of HaPnr-α and HaPnr-β (B) and the location of zinc-finger domain in the predicted protein structure chart of HaPnr-α and HaPnr-β (black arrow) (C).
Phylogenetic Relationship of HaPnr With Other Insect Panniers
All amino acid sequences of Pannier for phylogenetic analysis were obtained from the UniProt database, forming five distinctive groups representing different orders: Coleoptera, including Coccinella septempunctata (CsPnr, amino acid sequence identity to HaPnr-α 92.47%, to HaPnr-β 76.15%), Tribolium castaneum (TcPnr, identity to HaPnr-α 78.80%, to HaPnr-β 70.31%), Onthophagus binodis (ObPnr, identity to HaPnr-α 72.73%, to HaPnr-β 59.62%), O. sagittarius (OsPnr, identity to HaPnr-α 72.73%, to HaPnr-β 60.09%); Diptera, including D. quadrilineata (DqPnr, identity to HaPnr-α 56.10%, to HaPnr-β 42.70%), D. melanogaster (DmPnr, identity to HaPnr-α 61.11%, to HaPnr-β 51.35%), Anopheles gambiae (AgPnr, identity to HaPnr-α 59.25%, to HaPnr-β 40.58%), Megaselia abdita (MaPnr, identity to HaPnr-α 66.33%, to HaPnr-β 48.78%), Calliphora vicina (CvPnr, identity to HaPnr-α 57.88%, to HaPnr-β 39.47%), Ceratitis capitata (CcPnr, identity to HaPnr-α 58.08%, to HaPnr-β 47.33%), Clogmia albipunctata (CaPnr, identity to HaPnr-α 75%, to HaPnr-β 40.62%); Hemiptera, including Oncopeltus fasciatus (OfPnr, identity to HaPnr-α 81.61%, to HaPnr-β 72.84%); Lepidoptera, including Danaus plexippus (DpPnr, identity to HaPnr-α 50.42%, to HaPnr-β 39.66%) and Lxodida, including Ornithodoros erraticus (OePnr, identity to HaPnr-α 44.68%, to HaPnr-β 37.53%). Based on the amino acid identity levels, HaPnr appears to be more related to those in Coleoptera than to those in other orders.
Developmental Stage-specific Expression Patterns of HaPnr
Analyses of the developmental stage-specific expression pattern of HaPnr by using RT-qPCR in the egg, larval, pupal, and adult stages showed constitutive expression (Figure 3A). The highest expression occurred in one-day-old eggs, and other expression peaks were found in the first day of larva and pupa stage. Based on the two splicing variants of HaPnr, two specifics primers of RT-qPCR were designed on the specific domain to analyze developmental stage expression patterns of HaPnr-α and HaPnr-β, respectively (Supplementary Figure S3). Our results show that these two splicing variants have similar expression tendency with HaPnr. Additionally, the highest expression also occurred in one-day-old eggs (Figure 3B,C).
RNAi of HaPnr in the Fourth Instar Larvae and Its Effect on Pupation, Eclosion and Elytral Melanization
Due to two splicing variants having occurred in HaPnr, the dsRNA corresponding to HaPnr was designed on the common region and the primers of RT-qPCR were also designed on the common region that has no overlap with dsRNA domain (Supplementary Figure S3). When the fourth instar larvae of H. axyridis were injected with dsHaPnr, the transcript levels of HaPnr were significantly suppressed on days 4, 6 and 8 after injection; the remaining HaPnr was only 40% that of the dsGFP-injected group on day 4 after injection (Figure 4A). Consequently, the injection of dsHaPnr in the fourth instar larvae led to steadily decreasing survival frequency. By day 16 after the injection, only a 16.4% survival ratio was observed in dsHaPnr-injected larvae (Figure 4B). When HaPnr was suppressed in the fourth instar larvae, a significantly decreased pupation rate was observed as compared with the control (Figure 4C). In the control group with dsGFP injection, 88.5% of larvae could successfully pupate. However, in the dsHaPnr-injected group, approximately 61.5% of larvae could pupate, but these pupae showed an abnormal phenotype in which melanin on the cuticle was decreased as compared with the control group. The remaining 38.5% of larvae in the dsHaPnr-injected group could not accomplish the pupate process and died in larva form. Furthermore, these dead individuals showed epidermis softening and an abnormal darkening in color (Figure 4E). Suppressing the expression level of HaPnr in the fourth instar larvae also affects the eclosion process. These successfully pupating individuals with abnormal melanization in the epidermis were examined to calculate the eclosion rate. However, 100% of insects with an overall abnormal eclosion in the dsHaPnr injected group showed a malformed hind wing and the exuvium was attached to the body. In addition, the melanin in the head, pronotum and elytra were totally absent as compared with the control. We also observed that the original melanin synthesis areas are transformed into a yellow color, leading to a loss of color pattern characteristic in the elytra (Figure 4F). All the abnormal adults died within 10 days after the eclosion.
RNAi of HaPnr in One-Day-Old Pupae and Its Effect on Eclosion and Elytral Melanization
When early (1 day old) pupae were injected with dsHaPnr, the transcript levels of HaPnr were also significantly suppressed on days 2 and 4 after the injection. The HaPnr transcript levels were suppressed by 44.5% on day 2 after the injection of dsHaPnr (Figure 5A). Unlike with dsHaPnr injection in the fourth instar larvae, the suppressed expression level of HaPnr in one-day-old pupae did not affect the metamorphosis process from pupae to the adult stage (Figure 5C). Even though the pupae could eclose, but these adults showed an abnormal phenotype in which melanin was missing from the head, pronotum and elytra in H. axyridis. As with the phenotype in which HaPnr was suppressed in the fourth instar larvae, this melanin-lacking adult also displayed an original melanin synthesis area which transformed into a yellow color and lost its color pattern characteristics in its elytra (Figure 5D). These melanin-lacking ladybird beetles showed a normal activity state and mated normally. Thus, the fecundity of melanin-lacking and control ladybird beetles were calculated 10 days after eclosion. Our results show that there is no significant difference in fecundity and egg hatching rate between the dsHaPnr- and dsGFP-injected groups. The cumulative egg number within 15 days is 740 ± 99 and 781 ± 60.47 in the dsGFP and dsHaPnr group, respectively (Figure 5E). In addition, the hatching rate of eggs is 95.13 ± 0.94% and 90.87 ± 2.26% in the dsGFP and dsHaPnr group, respectively (Figure 5F).
RNAi of HaPnr-α and HaPnr-β in the Fourth Instar Larvae and Its Effect on Elytral Melanization
To further understand the individual roles of the two splicing variant transcripts in the development and melanization of H. axyridis, we performed an experiment that injected HaPnr-α and HaPnr-β into the fourth instar larvae, respectively. The specific primer of dsRNA synthesis and RT-qPCR of each splicing variant was designed on a specific region (Supplementary Figure S3). Our results show that dsHaPnr-α dramatically reduced its transcript levels without significantly affecting the non-target mRNA levels on day 2 after injection (Figure 6A), but on day 4 after injection, the transcript levels of HaPnr-β was significantly increased as compared with control (Figure 6B). In addition, dsHaPnr-β dramatically reduced its transcript levels without significantly affecting the non-target mRNA levels on day 2 and 4 after injection (Figure 6C,D). The transcript levels of HaPnr-α and HaPnr-β were suppressed by 44% and 42% on day 2 after the injection of dsHaPnr-α and dsHaPnr-β as compared with their respective control (Figure 6A,C). Suppressing the expression level of HaPnr-α and HaPnr-β individually had no significant effect on the pupation and eclosion rate. The pupation rates were 83.3% and 85.7% on day 7 after injection with dsHaPnr-α and dsHaPnr-β as compared with their respective control, dsGFP injection (93.3%), respectively (Figure 7A,D). Furthermore, the eclosion rates were 85.33% and 82.4% on day 12 after injection with dsHaPnr-α and dsHaPnr-β as compared with their respective control, dsGFP injection (90.8%), respectively (Figure 7B,E). Suppressing the expression level of HaPnr-α in the fourth instar larvae resulted in decreased elytra melanin in both the conspicua and the succinea subgroup in H. axyridis (Figure 7C). In addition, suppressing the expression level of HaPnr-β in the fourth instar larvae resulted in decreased elytra melanin in the succinea subgroup only in H. axyridis. The conspicua subgroup of H. axyridis showed normal melanin levels when HaPnr-β was suppressed in the fourth instar larvae as compared with dsGFP injection (Figure 7F). However, we observed that the synthetic time of melanin was more than 12 h after eclosion, which was significantly longer than 2 h in the dsGFP-injected control.
Discussion
The transcription factor Pannier, as a member of the GATA family, is involved in various biological processes during development in insects (Calleja et al., 2000; Buchberger et al., 2021). Recently, it was discovered to have a novel regulatory role in melanin synthesis in H. axyridis (Ando et al., 2018; Gautier et al., 2018; Ando and Niimi, 2019; Niimi and Ando, 2021). However, the detailed biological functions and regulated mechanisms of melanin in H. axyridis are still elusive. In the present study, we identified a Pannier homology from H. axyridis and characterized its cDNA and deduced amino acid sequences. Furthermore, we took advantage of the robust RNAi response of the insect to demonstrate its important biological function in pupation, eclosion and melanin synthesis in H. axyridis.
The model insect D. melanogaster study demonstrated that Pannier encodes two structurally related isoforms that are differentially expressed during development (Fromrntal-Ramain et al., 2008). Our results are consistent with the finding in D. melanogaster that HaPnr also has two splicing variants, HaPnr-α and HaPnr-β (Figure 1A). One of the most important features of the transcription factor is the zinc-finger domain (Han et al., 2012). In our study, the deduced protein sequence of the HaPnr gene exhibits the common features of the transcription factor: the HaPnr-α coded protein has two zinc finger domains, while the HaPnr-β coded protein has one zinc finger domain (Figure 1C), which is different from D. melanogaster, which has two zinc finger domains in the two isoforms, respectively (Fromrntal-Ramain et al., 2008; Valadez-Graham et al., 2012). Phylogenetic analysis of Pannier protein sequences of 15 insect species obtained from UniProt showed that the Pannier protein is conserved in the Coleoptera intragroup (Figure 2).
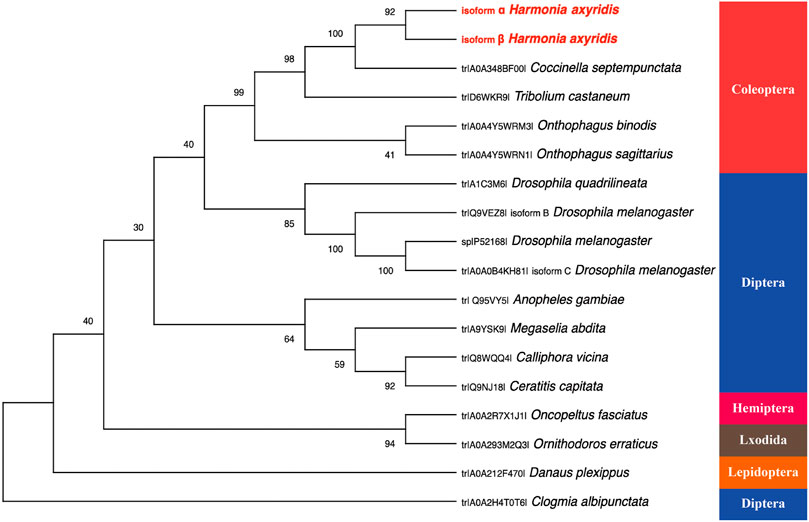
FIGURE 2. The rooted phylogenetic tree of deduced Pannier coding sequences from 15 insect species as constructed by the neighbor-jointing method. All the names used in the tree consist of the UniProt entry number and species name.
We found that HaPnr, including its splicing variants HaPnr-α and HaPnr-β, were constitutively expressed in all developmental stages. The highest expression of HaPnr occurred in the 1-day-old egg stage, and other expression peaks occurred in 1-day-old larvae and 1-day-old pupae (Figure 3A). Our results show that the HaPnr transcript peaked in early embryogenesis, then rapidly decreased during later embryogenesis, increased before the transition phase from embryo to the first instar larva and was maintained at an intermediate level during larva stage, then rapidly increased before the transition phase from larva to pupa stage and was also maintained at an intermediate level during the pupal stage. The high expression level of HaPnr in the initial stage of each metamorphosis process indicates that it plays a predominant role in the morphogenesis process in H. axyridis.
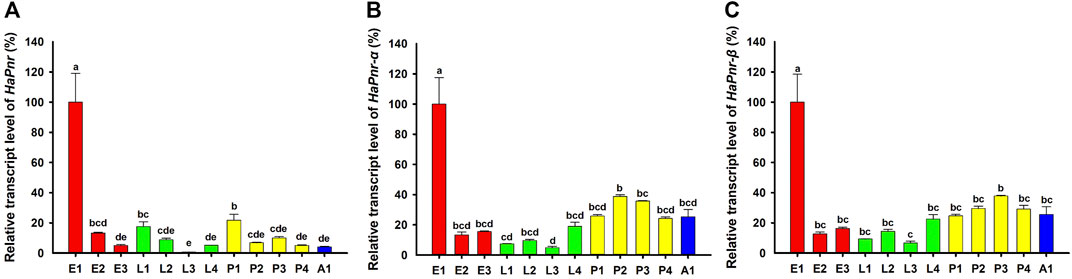
FIGURE 3. Relative transcript levels of HaPnr (A), HaPnr-α (B), and HaPnr-β (C) at different development stages of H. axyridis as determined by RT—qPCR. E1, E2, and E3 represent 1-, 2-, 3-day-old eggs; L1, L2, L3 and L4 represent first, second, third, and fourth instar larvae; P1, P2, P3 and P4 represent 1-, 2-, 3-, 4-day-old pupae; and A1 represents 1-day-old adults, respectively. Different letters above the standard error bars indicate significant differences based on ANOVA followed by Duncan’s multiple comparison test (p < 0.05). H. axyridis ribosomal protein 49 (Harp49) was used as an internal reference gene to normalize the differences among the samples. Relative expression levels for HaPnr were calculated based on the highest expressions of HaPnr in the one-day-old egg sample in developmental stage.
To determine the detailed biological function of HaPnr during metamorphosis (pupation and eclosion), we performed a functional assay to suppress the expression level of HaPnr in the fourth instar larvae. Specifically, in the fourth instar larvae, injection of dsHaPnr correnponding to a common region of the HaPnr-α and HaPnr-β transcript resulted in the significant decrease of its transcript level (Figure 4A). The results indicated that HaPnr plays an important role in the development process. Such suppression resulted in high mortality within 16 days after the injection (Figure 4B). The results of HaPnr was suppressed in the fourth instar larvae resulted in two major lines of evidence to support its essential role in pupation and eclosion process. First, suppressing the expression level of HaPnr led to a significantly reduced pupation rate (Figure 4C), and the un-pupated larvae eventually died after 1 week (Figure 4E). Second, even when a small proportion of the dsHaPnr-injected larvae were able to pupate, these pupae were unable to emerge into normal adults, and these abnormal adults died within 10 days after eclosion (Figure 4D,F). Our results are in agreement with a previous study in D. melanogaster demonstrating that the first exon of Pannier is inserted into a transposable element carrying the Gal4 gene, resulting in higher mortality in homozygote Pannier-Gal4 flies (Heitzler et al., 1996; Valadez-Graham et al., 2012).
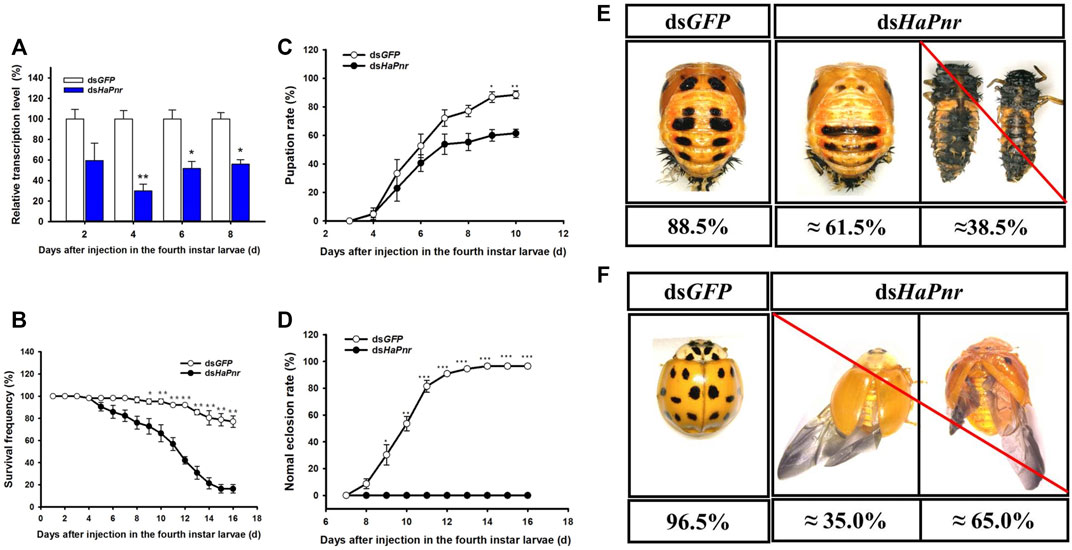
FIGURE 4. Suppression of HaPnr transcript in the fourth instar larvae of H. axyridis injected with dsHaPnr and dsGFP at 300 ng/larva as determined by RT-qPCR (A). The time-dependent larval survival frequency (B), pupation rate (C), and normal eclosion rate (D) in dsHaPnr and dsGFP-treated larvae. The phenotype of pupae (E) and adults (F) that injected with dsGFP and dsHaPnr with 300 ng/larva in the fourth instar larvae. The results are presented as the mean and standard errors of three replicates; each was performed with an RNA sample prepared from three insects. Asterisks above the standard error bars indicate significantly differences (Duncan tests, p < 0.05).
To further confirm the role of HaPnr in the metamorphosis process, we continue to perform detailed functional analyses by using RNAi to suppress the expression level of HaPnr in one-day-old pupae. Specifically, the injection of dsHaPnr corresponding to a common region of its two splicing variants transcript resulted in a dramatic suppression of its transcript level in one-day-old pupae (Figure 5A). However, suppression of HaPnr expression in the early pupa stage did not affect the eclosion process (Figure 5C). dsHaPnr-injected pupae were able to pupate into adults, but with an abnormal phenotype in which melanin disappeared from the head, pronotum and elytra (Figure 5D). We found that ladybird beetles missing melanin could mate normally and perform oviposition. Additionally, there were no significant differences in fecundity and egg hatching ratio between the dsHaPnr- and dsGFP-injection groups (Figure 5E,F). Previous research reported that Pannier could regulate imaginal disc development in insects (Maurel-Zaffran and Treisman, 2000; Orose et al., 2010; Buchberger et al., 2021). Imaginal discs are defined as clusters of undifferentiated embryonic cells in holometabolous insects that proliferate during larval stages, then differentiate during the pupal stage (Blair, 2009). In the present study, severe lethality occurred when the expression level of HaPnr was suppressed in the fourth instar larvae; however, no mortality was observed when early pupae (1 day old) were injected with dsHaPnr. Through comprehensive analysis of the difference in lethality resulting from HaPnr suppression in the fourth instar larvae and early pupa (1 day old) stage, our results demonstrate that the crucial regulation time of HaPnr in development is before the pupa stage (the fourth instar larvae or pre-pupae stage). Furthermore, we speculate that HaPnr may regulate development of the wing disc in the final larval stage due to the malformed hind wing which was obtained in the absence of HaPnr expression (Figure 4F).
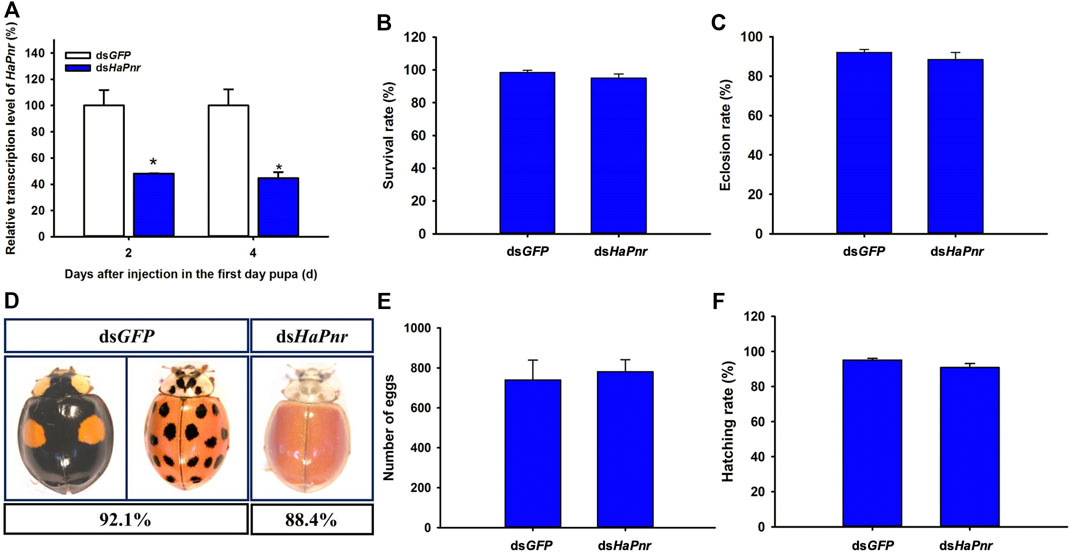
FIGURE 5. Suppression of HaPnr transcript in the one-day-old pupa of H. axyridis injected with dsHaPnr and dsGFP at 300 ng/pupa as determined by RT-qPCR (A). The survival rate (B) and eclosion rate (C) of the one-day-old pupae injected with dsHaPnr and dsGFP, respectively. The phenotype of adults after the one-day-old pupae were injected with dsHaPnr and dsGFP, respectively (D). The cumulative egg number of three pairs of ladybirds within 15 days (E) and the hatching rate of eggs (F). The results are presented as the mean and standard errors of three replicates; each was performed with an RNA sample prepared from three insects. Asterisks above the standard error bars indicate significantly different (Duncan tests, p < 0.05).
Beyond the regulatory role of HaPnr in pupation and eclosion process, it also plays a predominant role in regulating melanin synthesis in H. axyridis. The suppression of HaPnr in the fourth instar larvae resulted in melanin disorder in pupae and melanin disappearing from the head, pronotum and elytra in adults (Figure 4E,F). Furthermore, melanin-lacking H. axyridis were also obtained when HaPnr was suppressed in one-day-old pupae (Figure 5D). Our results are consistent with recent studies focusing on color pattern polymorphism research, which also demonstrated that HaPnr could regulate melanin synthesis in H. axyridis (Gautier et al., 2018; Ando et al., 2018). In our previous study, suppression of the expression level of dopa decarboxylase (DDC) also resulted in a melanin-absent phenotype, which is the crucial gene in dopamine melanin synthesis. Even though the melanin disappeared in the elytra in H. axyridis, we can easily discern the characteristic of color pattern because of the original melanin area transforming into a light-yellow color when HaDDC was suppressed (Xiao et al., 2020; Chen et al., 2019b). However, we did not discern the color pattern in H. axyridis when HaPnr was suppressed in the fourth instar larvae or one-day-old pupae due to the whole elytra represent the same yellow color (Figures 4F, 5D). We speculated that HaPnr has additional functions which could regulate pigment cell differentiation. In a previous study, the researchers also presented a similar viewpoint that Pannier may be expressed in specific cells in the forewing in H. axyridis, which promotes differentiation in black pigment cells and suppresses differentiation into red pigment cell (Niimi and Ando, 2021).
Indeed, the mode of action of Pannier splicing variants was reported in D. melanogaster, displaying antagonistic activities in the regulation of wingless expression during imaginal disc development (Fromrntal-Ramain et al., 2008). In order to analysis the sole role of HaPnr splicing variants in pupation and eclosion process in H. axyridis, special dsRNA corresponding to the specific region of HaPnr-α and HaPnr-β was designed and injected into the fourth instar larvae in H. axyridis (Supplementary Figure S3). Specifically, the injection of dsRNA corresponding to a specific region of HaPnr-α and HaPnr-β transcript resulted in a dramatic suppression of their transcript level in the fourth instar larvae, respectively (Figure 6A–D). Unlike with the suppression of the expression level of HaPnr in the fourth instar larvae, suppressing the expression level of its splicing variant individually has no significant effect on pupation and eclosion (Figure 7A,B,D,E), indicating that any of the individual splice variants are able to perform the HaPnr genetic functions in development. We continue to analyze the phenotype induced by HaPnr-α or HaPnr-β being suppressed in the fourth instar larvae individually. Our results show that the melanin was significantly decreased in the succinea subgroup in H. axyridis when both HaPnr-α and HaPnr-β were suppressed in the fourth instar larvae, individually. Even though the melanin was synthesized normally in the conspicua pattern in H. axyridis, the coloration time was significantly longer as compared with the control (Figure 7C,F). As compared with the phenotype when HaPnr was suppressed in the fourth instar larvae and one-day-old pupae, melanin was totally absent, and the color pattern characteristic was lost. Our present data highlight that HaPnr function in regulated melanin synthesis is achieved by two structurally related isoforms. In addition, through comprehensive analysis of the distribution of zinc-finger domain in HaPnr-α and HaPnr-β, we speculate that HaPnr-α may play a predominant role in regulating melanin synthesis with two zinc finger domain and HaPnr-β may play an auxiliary role in regulating melanin synthesis with one zinc finger domain.
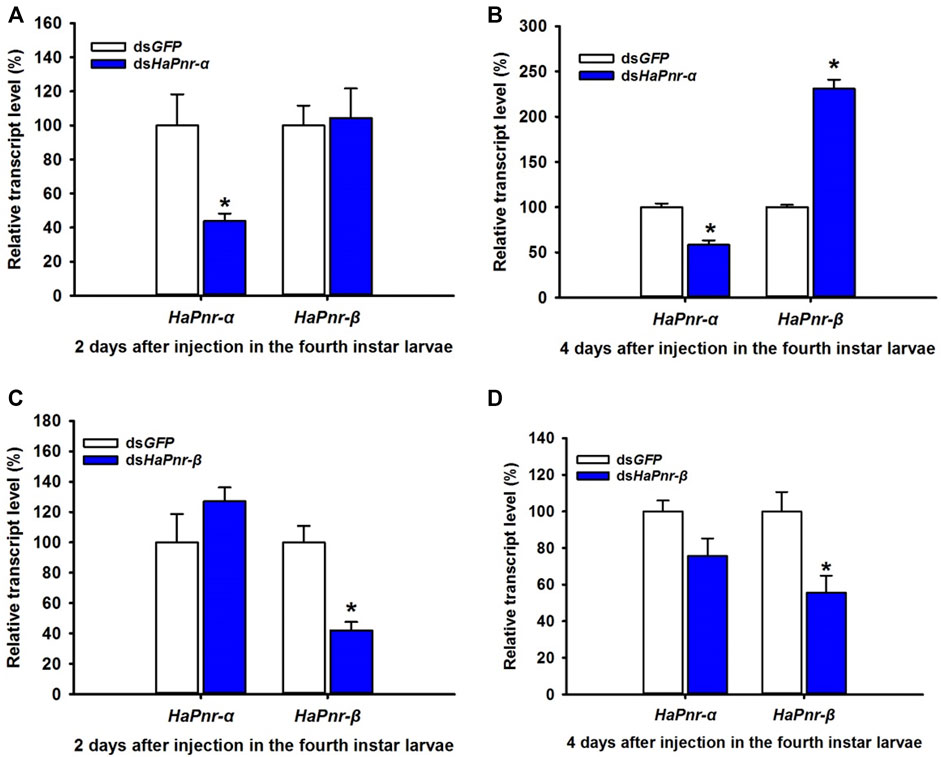
FIGURE 6. Suppression of HaPnr-α (A,B) and HaPnr-β (C,D) transcript in the fourth instar larvae of H. axyridis injected with dsHaPnr-α, dsHaPnr-β, and dsGFP at 300 ng/larva as determined by RT-qPCR.
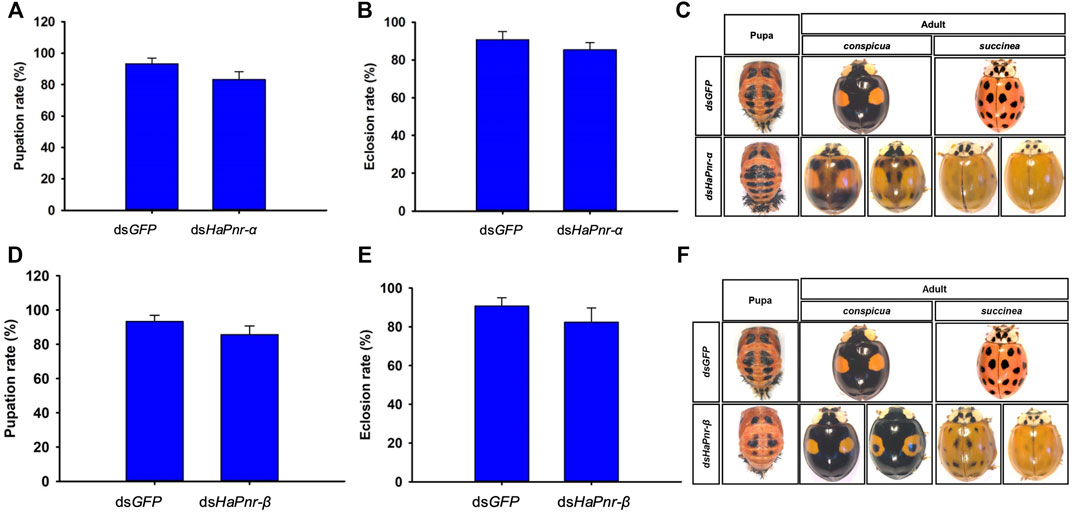
FIGURE 7. The pupation rate in dsHaPnr-α (A) and dsHaPnr-β (D) injected groups compared with dsGFP-treated larvae, respectively. The eclosion rate in dsHaPnr-α (B) and dsHaPnr-β (E) injected group compared with dsGFP-treated larvae, respectively. The pupae and adult’s phenotype after the fourth instar larvae injected with dsHaPnr-α (C) and dsHaPnr-β (F). The results are presented as the mean and standard errors of three replicates; each was performed with an RNA sample prepared from three insects. Asterisks above the standard error bars indicate significantly different (Duncan tests, p < 0.05).
In summary, previous research revealed evolutionally conserved mechanisms of Pannier in regulating imaginal disc development, dorsal closure, and the formation of ommateum and bristle. However, much of the evidence for such behaviors in insects comes from studies of the Pannier mutants of D. melanogaster. Thus, our results provide, for the first time, crucial evidence that HaPnr plays an essential role in insect metamorphosis in H. axyridis by using RNAi. In addition, HaPnr also regulates melanin synthesis under the combined action of its two splicing variants, HaPnr-α and HaPnr-β.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/, GenBank: ON186775, ON186776.
Author Contributions
RT, SW, LZ, and DX conceived and designed the experiments. RT, XC, and MW Performed the experiments. RT, QX, and DX Analyzed the data. QX, SW, and DX Contributed reagents/materials/analysis tools. RT, XC, SW, and DX Wrote the paper. RT, XC, SW, and DX Contributed with revisions. All authors read and approved the final manuscript.
Funding
The Technical Innovation Program of Beijing Academy of Agriculture and Forestry Sciences (KJCX20200110), the National Natural Science Foundation of China (32072479), and the Beijing Technology Program (Z201100008020014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank researchers in the Ministry of Agriculture Key Lab of Molecular Biology of Crop Pathogens and Insects in Zhejiang University for providing assistance in analyzing the genomic structure of Pannier in H. axyridis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.909258/full#supplementary-material
References
Ando T., Niimi T. (2019). Development and Evolution of Color Patterns in Ladybird Beetles: A Case Study in Harmonia axyridis. Develop. Growth Differ. 61, 73–84. doi:10.1111/dgd.12592
Ando T., Matsuda T., Goto K., Hara K., Ito A., Hirata J., et al. (2018). Repeated Inversions within a Pannier Intron Drive Diversification of Intraspecific Colour Patterns of Ladybird Beetles. Nat. Commun. 9, 3843. doi:10.1038/s41467-018-06116-1
Bezzerides A. L., McGraw K. J., Parker R. S., Husseini J. (2007). Elytra Color as a Signal of Chemical Defense in the Asian Ladybird Beetle Harmonia axyridis. Behav. Ecol. Sociobiol. 61, 1401–1408. doi:10.1007/s00265-007-0371-9
Blair S. S. (2009). “Imaginal Discs,” in Encyclopedia of Insects. 2nd Edn, Editors V. H. Resh, and R. T. Carde (San Diego, CA: Academic Press, Elsevier Science Publisher), 489–492.
Buchberger E., Bilen A., Ayaz S., Salamanca D., Matas de las Heras C., Niksic A., et al. (2021). Variation in Pleiotropic Hub Gene Expression is Associated with Interspecific Differences in Head Shape and Eye Size in Drosophila. Mol. Biol. Evol. 38, 1924–1942. doi:10.1093/molbev/msaa335
Calleja M., Herranz H., Estella C., Casal J., Lawrence P., Simpson P., et al. (2000). Generation of Medial and Lateral Dorsal Body Domains by the Pannier Gene of Drosophila. Development 127, 3971–3980. doi:10.1242/dev.127.18.3971
Chau L. M., Goodisman M. A. D. (2017). Gene Duplication and the Evolution of Phenotypic Diversity in Insect Societies. Evolution 71, 2871–2884. doi:10.1111/evo.13356
Chen X., Xiao D., Du X., Zhang F., Zang L., Wang S. (2019a). Impact of Polymorphism and Abiotic Conditions on Prey Consumption by Harmonia axyridis. Entomol. Gen. 39, 251–258. doi:10.1127/entomologia/2019/0874
Chen X., Xiao D., Du X., Guo X., Zhang F., Desneux N., et al. (2019b). The Role of the Dopamine Melanin Pathway in the Ontogeny of Elytral Melanization in Harmonia axyridis. Front. Physiol. 10, 1066. doi:10.3389/fphys.2019.01066
Fromental-Ramain C., Vanolst L., Delaporte C., Ramain P. (2008). Pannier Encodes Two Structurally Related Isoforms that are Differentially Expressed during Drosophila Development and Display Distinct Functions during Thorax Patterning. Mech. Dev. 125, 43–57. doi:10.1016/j.mod.2007.10.008
Gajewski K., Fossett N., Molkentin J. D., Schulz R. A. (1999). The Zinc finger Proteins Pannier and GATA4 Function as Cardiogenic Factors in Drosophila. Development 126, 5679–5688. doi:10.1242/dev.126.24.5679
Gautier M., Yamaguchi J., Foucaud J., Loiseau A., Ausset A., Facon B., et al. (2018). The Genomic Basis of Color Pattern Polymorphism in the Harlequin Ladybird. Curr. Biol. 28, 3296–3302. doi:10.1016/j.cub.2018.08.023
Han Y.-W., Matsumoto T., Yokota H., Kashiwazaki G., Morinaga H., Hashiya K., et al. (2012). Binding of Hairpin Pyrrole and Imidazole Polyamides to DNA: Relationship between Torsion Angle and Association Rate Constants. Nucleic Acids Res. 40, 11510–11517. doi:10.1093/nar/gks897
Heitzler P., Haenlin M., Ramain P., Calleja M., Simpson P. (1996). A Genetic Analysis of Pannier, a Gene Necessary for Viability of Dorsal Tissues and Bristle Positioning in Drosophila. Genetics 143, 1271–1286. doi:10.1093/genetics/143.3.1271
Herranz H., Morata G. (2001). The Functions of Pannier during Drosophila Embryogenesis. Development 128, 4837–4846. doi:10.1242/dev.128.23.4837
Hoekstra H. E. (2006). Genetics, Development and Evolution of Adaptive Pigmentation in Vertebrates. Heredity 97, 222–234. doi:10.1038/sj.hdy.6800861
Koch P. B., Behnecke B., ffrench-Constant R. H. (2000). The Molecular Basis of Melanism and Mimicry in a Swallowtail Butterfly. Curr. Biol. 10, 591–594. doi:10.1016/s0960-9822(00)00494-2
Koch R. L. (2003). The Multicolored Asian Lady Beetle, Harmonia axyridis: A Review of its Biology, Uses in Biological Control, and Non-target Impacts. J. Insect Sci. 3, 1–16. doi:10.1673/031.003.3201
Lovato T. L., Sensibaugh C. A., Swingle K. L., Martinez M. M., Cripps R. M. (2015). The Drosophila Transcription Factors Tinma and Pannier Activate and Collaborate with Myocyte Enhancer Factor-2 to Promote Heart Cell Fate. Plos One 10, e0132965. doi:10.1371/journal.pone.0132965
Maurel-Zaffran C., Treisman J. E. (2000). Pannier Acts Upstream of Wingless to Direct Dorsal Eye Disc Development in Drosophila. Development 127, 1007–1016. doi:10.1242/dev.127.5.1007
Minakhina S., Tan W., Steward R. (2011). JAK/STAT and the GATA Factor Pannier Control Hemocyte Maturation and Differentiation in Drosophila. Develop. Biol. 352, 308–316. doi:10.1016/j.ydbio.2011.01.035
Moczek A. P. (2010). Phenotypic Plasticity and Diversity in Insects. Phil. Trans. R. Soc. B 365, 593–603. doi:10.1098/rstb.2009.0263
Niimi T., Ando T. (2021). Evo-devo of Wing Colour Patterns in Beetles. Curr. Opin. Genet. Develop. 69, 97–102. doi:10.1016/j.gde.2021.02.007
Oros S. M., Tare M., Kango-Singh M., Singh A. (2010). Dorsal Eye Selector Pannier (Pnr) Suppresses the Eye Fate to Define Dorsal Margin of the Drosophila Eye. Develop. Biol. 346, 258–271. doi:10.1016/j.ydbio.2010.07.030
Qian L., Bodmer R. (2009). Partial Loss of GATA Factor Pannier Impairs Adult Heart Function in Drosophila. Hum. Mol. Genet. 18, 3153–3163. doi:10.1093/hmg/ddp254
Ramain P., Heitzler P., Haenlin M., Simpson P. (1993). Pannier, a Negative Regulator of Achaete and Scute in Drosophila, Encodes a Zinc finger Protein with Homology to the Vertebrate Transcription Factor GATA-1. Development 119, 1277–1291. doi:10.1242/dev.119.4.1277
Sharma A. I., Yanes K. O., Jin L., Garvey S. L., Taha S. M., Suzuki Y. (2016). The Phenotypic Plasticity of Developmental Modules. Evodevo 7, 15. doi:10.1186/s13227-016-0053-7
True J. R. (2003). Insect Melanism: The Molecules Matter. Trends Ecol. Evol. 18, 640–647. doi:10.1016/j.tree.2003.09.006
Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J. A. M. (2007). Primer3Plus, an Enhanced Web Interface to Primer3. Nucleic Acids Res. 35, W71–W74. doi:10.1093/nar/gkm306
Valadez-Graham V., Yoshioka Y., Velazquez O., Kawamori A., Vázquez M., Neumann A., et al. (2012). XNP/dATRX Interacts with DREF in the Chromatin to Regulate Gene Expression. Nucleic Acids Res. 40, 1460–1474. doi:10.1093/nar/gkr865
Van der Burg K. R. L., Lewis J. J., Brack B. J., Fandino R. A., Mazo-Vargas A., Reed R. D. (2020). Genomic Architecture of a Genetically Assimilated Seasonal Color Pattern. Science 370 (370), 721–725. doi:10.1126/science.aaz3017
Wang S., Michaud J. P., Zhang R. Z., Zhang F., Liu S. (2009). Seasonal Cycles of Assortative Mating and Reproductive Behaviour in Polymorphic Populations of Harmonia axyridis in China. Ecol. Entomol. 34, 483–494. doi:10.1111/j.1365-2311.2008.01075.x
Wang S., Michaud J. P., Tan X. L., Murray L., Zhang F. (2013). Melanism in a Chinese Population of Harmonia axyridis (Coleoptera: Coccinellidae): A Criterion for Male Investment with Pleiotropic Effects on Behavior and Fertility. J. Insect Behav. 26, 679–689. doi:10.1007/s10905-013-9384-6
Wittkopp P. J., Beldade P. (2009). Development and Evolution of Insect Pigmentation: Genetic Mechanisms and the Potential Consequences of Pleiotropy. Semin. Cel Develop. Biol. 20, 65–71. doi:10.1016/j.semcdb.2008.10.002
Xiao D., Chen X., Tian R., Wu M., Zhang F., Zang L., et al. (2020). Molecular and Potential Regulatory Mechanisms of Melanin Synthesis in Harmonia axyridis. Int. J. Mol. Sci. 21, 2088. doi:10.3390/ijms21062088
Zhang Y., Wang X.-X., Feng Z.-J., Cong H.-S., Chen Z.-S., Li Y.-D., et al. (2020). Superficially Similar Adaptation within One Species Exhibits Similar Morphological Specialization but Different Physiological Regulations and Origins. Front. Cel Dev. Biol. 8, 300. doi:10.3389/fcell.2020.00300
Keywords: pannier, melanin, development, Harmonia axyridis, metamorphosis
Citation: Tian R, Chen X, Wu M, Xu Q, Wang S, Zang L and Xiao D (2022) The Molecular Properties and Roles of Pannier in Harmonia axyridis’s Metamorphosis and Melanin Synthesis. Front. Physiol. 13:909258. doi: 10.3389/fphys.2022.909258
Received: 31 March 2022; Accepted: 18 April 2022;
Published: 03 May 2022.
Edited by:
Ya-Nan Zhang, Huaibei Normal University, ChinaReviewed by:
Wen Liu, Huazhong Agricultural University, ChinaHuipeng Pan, South China Agricultural University, China
Copyright © 2022 Tian, Chen, Wu, Xu, Wang, Zang and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liansheng Zang, bHN6YW5nQGd6dS5lZHUuY25sc3phbmdAZ3p1LmVkdS5jbg==; Da Xiao, eGlhb2RhQGlwZXBiYWFmcy5jbg==
 Renbin Tian1,2
Renbin Tian1,2 Su Wang
Su Wang Da Xiao
Da Xiao