
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol., 15 September 2022
Sec. Gastrointestinal Sciences
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.903302
This article is part of the Research TopicCell-To-Cell Communications In Alcohol-Associated, Metabolic-Related And Viral Liver DiseasesView all 5 articles
Neoplasms contain tumor-initiating stem-like cells (TICs) that are characterized by increased drug resistance. The incidence of many cancer types have trended downward except for few cancer types, including hepatocellular carcinoma (HCC). Therefore mechanism of HCC development and therapy resistance needs to be understood. These multiple hits by hepatitis C virus (HCV) eventually promotes transformation and TIC genesis, leading to HCC development. This review article describes links between HCV-associated HCC and TICs. This review discusses 1) how HCV promotes genesis of TICs and HCC development; 2) how this process avails itself as a novel therapeutic target for HCC treatment; and 3) ten hall marks of TIC oncogenesis and HCC development as targets for novel therapeutic modalities.
Hepatitis B and C virus (HBV/HCV) infection, alcoholism and obesity are major risk factors for hepatocellular carcinoma (HCC) (Okuda, 2000; Crippin et al., 2002). Among all the risk factors, HCV infection is a major and highest risk factor for developing HCC because it promotes fibrosis and cirrhosis (El-Serag and Rudolph, 2007). Approximately 90% of HCV-associated cancers present in advanced fibrosis or cirrhosis. Other nonviral factors (such as alcoholism and obesity) account for about 20% of HCC cases (El-Serag and Mason, 1999) since diabetes and obesity are the strongest metabolic factors associated with HCC (Hagstrom et al., 2018). The incidence of liver cancer is rising with an estimated 841,080 (4.7%) new cases and 781,631 deaths for 2018 (Bray et al., 2018; Gerbes et al., 2018; Kulik et al., 2018; Roche et al., 2018). HCV infection is present in around 50% of cases and the incidence of HCV-induced HCC is falling (Roche et al., 2018).
Treatment options for HCC are limited and not encouraging. The 3-years survival rate of HCC is 13%–21% without any curative treatment (Ebara et al., 1986; Barbara et al., 1992). The 5-years survival rate of HCC is less than 5% with or without therapeutic intervention (El-Serag and Mason, 1999), even in advanced countries such as the United States. (Okuda, 2000; Liang and Heller, 2004). Since the incidence rate of extrahepatic metastasis is 13% at 5 years (Kanda et al., 2008), liver resection is a viable option for HCC combined with cirrhosis (Nakamura et al., 2014). However, only 10%–23% of HCC patients are candidates for surgery (Sonnenday et al., 2007; Shah et al., 2011). Thus, HCV-associated HCC remains an incurable malignancy and an urgent unmet medical need. TIC-mediated HCC development are clinically important. The lifetime risk of HCC in chronically infected HCV individuals is 2%–7% (Di Bisceglie et al., 2003), although it may take 30–40 years for HCC to develop in these patients. Furthermore, HCV affects more than 75 million people worldwide (Okuda, 2000; Yao and Terrault, 2001; Okuda et al., 2002). The main risk factors for developing HCC are viral hepatitis infections such as HBV and HCV. However, the incidence of HCC is rising in non-alcoholic fatty liver disease (Holmes and Chung, 2016; Suresh et al., 2020; Huang et al., 2021).
Chronic liver damage caused by viral infection and environmental factors (such as alcohol or metabolic syndrome) can result in increased risk for HCC. The cirrhotic liver is a permissive factor for HCC due to the large regenerative activity repetitive damage-regeneration cycles or formation of dysplastic nodules (International Consensus Group for Hepatocellular Neoplasia, 2009). Thus, the cirrhotic liver may be considered a pre-neoplastic “cancer field” comprised of genetically abnormal but non-neoplastic tissue that is at high risk for malignant transformation (Figure 1) (Braakhuis et al., 2003). Understanding the molecular mechanisms of HCV-induced hepatocarcinogenesis will aid in the development of improved therapeutic modalities (Crippin et al., 2002).
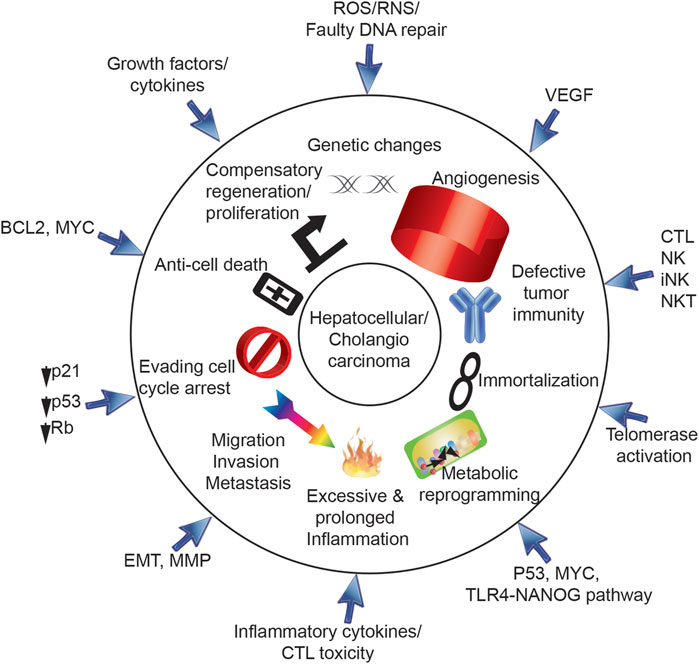
FIGURE 1. Ten hallmarks for cancer development via HCV. Ten hallmarks of cancer are triggered by HCV, including 1) genetic changes, 2) angiogenesis, 3) defective tumor immunity, 4) immortalization, 5) metabolic reprogramming, 6) excessive and prolonged inflammation, 7) migration/invasion/metastasis, 8) evading cell cycle arrest, 9) anti-cell death and 10) compensatory regeneration/proliferation. HCV core protein, E1, E2, NS3, NS5A, and NS5B are involved in progression of HCV-induced HCC.
HCV-associated cirrhosis can result in HCC by ultimately promoting tumor-initiating stem-like cells (TIC) formation since deposition of extracellular matrix (ECM), including collagen and laminin, promotes tumor-prone microenvironment. These TICs can develop into several different types of liver cancer, e.g., HCC and cholangiocarcinoma (CC). TICs are resistant to conventional chemotherapy and immunotherapy and persist as recurrent tumors or circulating tumor cells. TICs share key features with embryonic stem cells (ESCs) present in preimplantation blastocyst stage embryos, including the expression of a core pluripotency-associated transcription factor (TF) network (Kim et al., 2010; Ikushima et al., 2011). Liver progenitor cells have asymmetric cell division process. In untransformed stem cells, self-renewal occurs through asymmetric cell division, in which one daughter cell retains the multipotent progenitor status of its parent while the other cell commits to a specialized cell fate. In contrast to ESCs, TICs fail to control the self-renewing mode of cell division. As asymmetric cell division mechanism is disrupted in HCCs, TICs exhibit a loss of this intrinsic asymmetry without regulated differentiated daughter cells, leading to the ectopic implementation of stem cell gene expression programs in both progeny cells. This leads to subsequent unchecked expansion of the progenitor cell pool (Cicalese et al., 2009; Knoblich, 2010; Martin-Belmonte and Perez-Moreno, 2012). Thus, understanding the mechanism of genesis of TICs paves the way for novel therapeutic approaches, including cell fate-determinant molecule NUMB, which is p53-MDM2 associated proteins. This key cell fate determinant molecules are targeted by interacting protein TBC1D15 in TICs (Feldman et al., 2013).
Ten hallmarks of cancer (Hanahan and Weinberg, 2011) are triggered by HCV in hepatocytes (Figures 1, 2), including 1) genetic changes, 2) angiogenesis, 3) defective tumor immunity, 4) immortalization, 5) metabolic reprogramming, 6) excessive and prolonged inflammation, 7) migration/invasion/metastasis, 8) evasion of cell cycle arrest, 9) anti-cell death and 10) compensatory regeneration/proliferation. Following ten sections describe ten hall marks of genesis of TICs induced by HCV infection and/or comorbidity, including environmental factors, such as alcoholism, obesity, cirrhosis and xenobiotic agents.
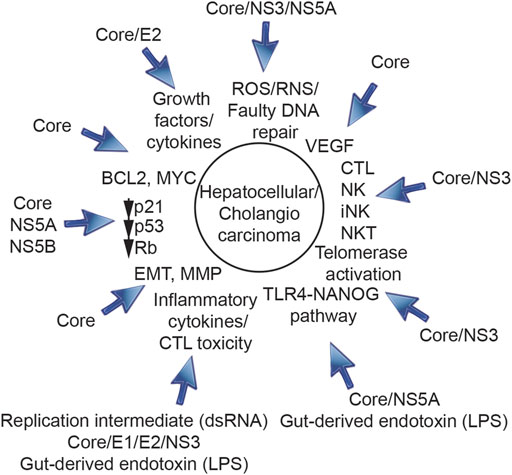
FIGURE 2. Roles of HCV structural proteins (core protein, E1, E2) and non-structural proteins (NS3, NS5A, and NS5B) in progression of HCV-induced HCC. HCV core protein, E1, E2, NS3, NS5A, and NS5B are involved in initiation, promotion and progression of HCV-induced HCC and genesis of TICs.
The following text enumerates HCV connections to a series of cellular “Cancer Hallmarsks.” This is important since multiple hits transforms hepatocytes to TIC and HCC. Understanding links between cancer hallmarks and TIC genesis makes conceptual advances to help advance cancer research and therapy by clarifying novel targeting therapy and innovative strategy to suppress theHCC recurrence problems and metastatic spread of HCC cells.
Defects in DNA repair genes cause genetic instability, gross chromosomal rearrangements and accumulation of mutations, leading ultimately to neoplastic transformation. Both homologous recombination and nonhomologous end joining (NHEJ) play a role in the repair of double-strand DNA breaks (DSBs) in mammalian cells (Hiom, 1999). The interaction of broken DNA with members of the Rad52 epistasis group, including Rad51, a mammalian homologue of bacterial RecA, initiates homologous recombination repair (Hiom, 1999). Following DNA damage, Rad51 is redistributed within the nucleus (Haaf et al., 1995; Baumann and West, 1998) and induces the ATP-dependent homologous strand pairing reaction that initiates recombination. In contrast, NHEJ works by non-homology-dependent ligation of broken DNA ends. DNA-dependent protein kinase (DNA-PK) and its associated proteins Ku70, Ku80, and Xrcc4 mediate NHEJ (Song et al., 2003).
Structural variations (STVs) of chromosomes include translocation, deletion, or inversion of chromosomes of gene APC, and tandem duplications (Fujimoto et al., 2016). HCCs contain broad genomic gains (1q, 5p, 6p, 8q, 17q, 20q, and Xq) and deletions (1p, 4p-q, 6q, 8p, 13p-q, 16p-q, 17p, 21p-q, and 22q) (Totoki et al., 2011; Guichard et al., 2012; Kan et al., 2013; Ahn et al., 2014; Schulze et al., 2015), suggesting that STVs increase the expression of oncogenes and/or decrease the expression of tumor suppressor genes to promote hepatocarcinogenesis.
The take-away message is that HCV-associated HCCs have frequent chromosomal aberrations. These frequent chromosomal aberrations can be targeted and translated into therapies. The status of current related therapeutic strategies is under development. HCV-associated HCCs have frequent MYC loci amplification. These constitutive MYC activation can be targeted and translated into therapies.
Mutations in tumor-suppressor genes or proto-oncogenes or the activity of growth factors during chronic HCV infection transforms hepatocytes, cholangiocytes, and liver progenitor cells (Simonetti et al., 1992). Whole genome and exome analysis demonstrated that TP53, CTNNB1, and chromatin modulators, including ARID1A and ARID2, are the most frequently mutated coding genes in HCC (Shibata and Aburatani, 2014; Totoki et al., 2014; Fujimoto et al., 2016). Loss-of-function ARID1A mutations are correlated with poor prognosis, sorafenib resistance, HCC invasion and metastasis (Nhieu et al., 1999). The most frequently mutated driver genes in human alcohol-associated HCCs, but not in dysplastic macronodules (Guichard et al., 2012), are in the chromatin remodeling complex (ARID1A) (8%–38%), β-Catenin/Wnt (activating mutations in exon 3 of CTNNB1), and TP53 (30%–65%). CTNNB1 point mutations occurred in serine residues (Ser 33, 37, and 45) and through the destruction complex (GSK3β and CSNK1A1). The more frequent mutations in non-coding regions are found in the TERT promoter and TFPI2. Long intergenic noncoding RNAs, including NEAT1 and MALAT1, are also frequently mutated (Fujimoto et al., 2016). Genomic gain of function causes focal amplifications in cancer-related genes such as VEGFA and FGF3/4/19/CCND1, which is associated with a good response to the multi-kinase inhibitor sorafenib (Arao et al., 2013; Llovet, 2014). Point mutations and also STV breakpoints in HCC tissues are detected in cancer-driver genes, including TERT, ARID1A, ARID2, and PTEN, (Fujimoto et al., 2016). Therefore, both nucleotide variants and STVs of chromosomes are detected in hepatitis virus-related HCCs. These diver mutations direct self0renewal ability of TICs.
The take-away message is that tumor driver gene mutations in TP53, CTNNB1, ARID1A make hepatocytes susceptible for HCC development. These driver mutation are targeted and translated into therapies, including ICG-001 for CTNNB1 mutation (Delgado et al., 2014; Lin et al., 2016) and Adenovirus expressing functional p53 (Anderson et al., 1998). The status of current related therapeutic strategies showed promising responses for ICG-001 therapy and Ad-p53 (Anderson et al., 1998).
HCV-associated HCC has a 3–5 times higher mutation rate than associated non-tumor liver tissues. Furthermore, the ratio of amino acid replacement and silent mutations in the tumors, but not in the neighboring non-tumor tissues, was significantly higher than the ratio expected in the absence of selection of growth advantage of premalignant cells (Machida et al., 2004a) since chronic hepatocellular turnover could select for cells with genetic or epigenetic changes that confer growth advantages allowing clonal expansion. For example, HCV infection induced a mutator phenotype, which involves enhanced mutations in many somatic genes, including immunoglobulin (Ig) genes, proto-oncogenes and tumor suppressor genes (Machida et al., 2004a). HCV infection promotes error-prone DNA polymerase expression and increases mutation frequencies by induction of ROS and reactive nitrogen species and by inhibition of DNA repair mechanisms (Machida et al., 2004a). Therefore, HCV-induced mutations contribute to the eventual selection and amplification of certain deleterious mutations of proto-oncogenes or tumor suppressor genes in tumors. Accordingly HCV-associated oncogenesis is characterized by a long latency period due to the need for multiple hits.
These mutator phenotype can be great target for immune checkpoint inhibitor since high genetic instability is good prognosis maker for immune checkpoint inhibitor. Indeed, immune checkpoint inhibitor Anti-PD-1 therapy got FDA approval after landmark clinical trial (El-Khoueiry et al., 2017).
Viral or immune-mediated reactive oxygen species (ROS) induce oxidative stress. ROS-associated oxidative DNA damage (such as 8-oxo-dG) promotes DNA mutagenesis, leading to oncogenic transformation in chronic hepatitis C (Figure 2). For example, the HCV core protein promotes formation of intracellular ROS, both in vitro (Okuda et al., 2002) and in vivo (Korenaga et al., 2005), via its localization to the mitochondria and inhibition of electron transport (Korenaga et al., 2005). The electron flow that interacts with oxygen molecules results in formation ROS prior to reaching the cytochrome oxidase complex. HCV infection induces ROS (Choi and Ou, 2006), which leads to oxidative DNA damages and lipid peroxidation in HCV-infected cells (Machida et al., 2006). Consequently pretreatment of HCV-infected cells with ROS inhibitors prevented mitochondrial damage and the production of ROS (Machida et al., 2006). ROS with NO (Machida et al., 2004b) induces steatosis (through lipid peroxides) and oncogenesis (through DNA mutations and STAT3 activation), leading to acute hepatocyte damage (via production of STAT3) (Machida et al., 2006). Individual liver cells undergo disease evolution to reproduce HCV-associated HCC (Salk et al., 2010). Activated inflammatory cells (CTL or NK) also release ROS and nitrogen (RNS) species and induce lipid peroxidation (Bartsch and Nair, 2006), leading to a pro-carcinogenic microenvironment. Viral particle was consisted with structural proteins, nucleocapside Core, Envelop genes E1 and E2 and p7. Non-structural proteins are consisted of Serine protease NS2 and NS3, protease cofactor NS4A, NS4B, NS5A, and RNA-dependent RNA polymerase NS5B (Holmes and Chung, 2016). The expression of some HCV proteins, in particular structural nucleocapside protein core and non-structural viral protein NS5A, may also contribute directly to the induction of oxidative stress (Figure 2).
The take-away message is that HCV-associated ROS production promotes DNA damages and inhibits DNA repair, leading to mutator phenotypes. These HCV-associated mutaor phenotype can be targeted by vitamin treatment. However current clinical reports are not optimistic for HCC patients.
An HCV-induced oxidative environment may overwhelm cellular antioxidant and DNA-repair mchanisms, leading to accumulation of DSBs and chromosomal abnormalities in HCV-infected cells. As discussed above, HCV infection induces a mutator phenotype by causing DSBs (Machida et al., 2004a) through induction of iNOS mRNA and nitric oxide (NO) production by the viral core protein and the NS3 protein (Machida et al., 2004b). HCV non-structural protein NS3 blocks the cellular repair process. The ataxia-telangiectasia mutated kinase (ATM) is not only required for HCV replication (Ariumi et al., 2008) but also interacts with the HCV NS3-4A protein complex, resulting in impaired DNA damage responses and enhanced sensitivity to ionizing irradiation (Lai et al., 2008). The resulting increased rate of double-stranded DNA breaks is a possible direct viral causal role in tumorigenesis (Figure 3) (Lai et al., 2008).
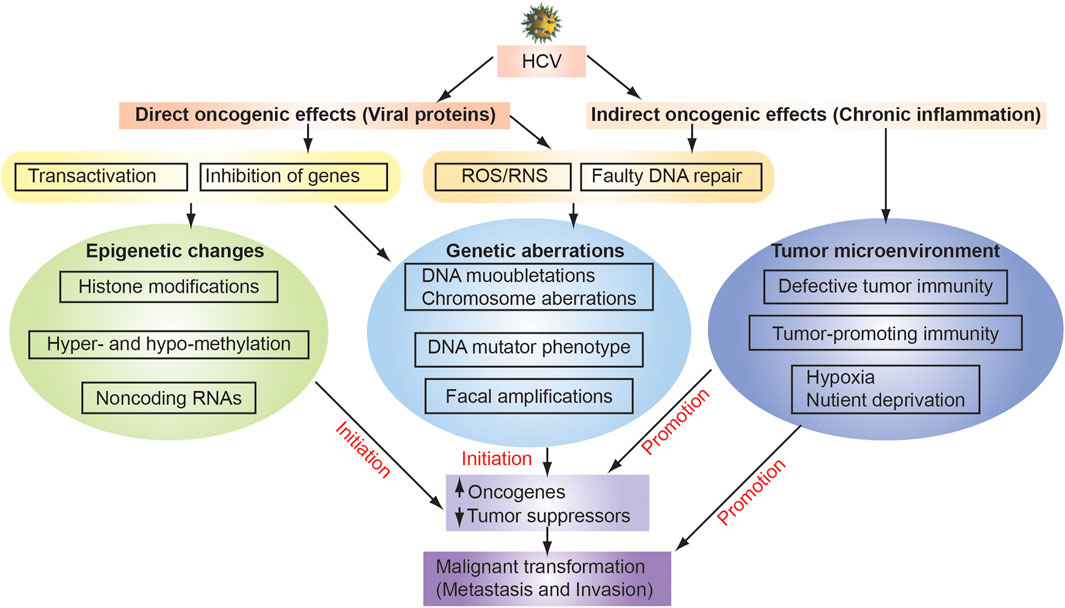
FIGURE 3. Mechanisms of HCV-associated hepatocarcinogenesis. HCV infection causes chronic inflammation in the liver as indirect oncogenic carcinogenic effects, leading to genetic alteration through ROS generation, elevated expression of DNA mutator phenotype, and dysfunction of DNA repair. As indirect oncogenic carcinogenic effects induce tumor microenvironment with defective tumor immunity, tumor-promoting immunity and hypocix and nutriet deprived environment. Environmental factors (e.g., Alcohol, obesity, and high fat diet) enhance sensitivity of livers to LPS. Direct effects of hepatitis virus, including the oncogenic effects of HBV genome integration and HBx protein expression, can also contribute to increased genomic instability. These multiple factors coordinately induce the accumulation of genetic and epigenetic alterations in liver tissue underlying chronic hepatitis or cirrhosis, leading to the development of HCC.
DNA repair proteins prevent DNA mutations caused by oxidative damage, but are vulnerable to nitric oxide (NO)-induced oxidative damage. This is because of sulfhydryl, tyrosyl, and/or phenolic side chains in their active sites (Starke, Chen, Bapna, Lesnefsky, Mieyal; Jaiswal et al., 2000; Jaiswal et al., 2001). Suppression of DNA repair, coupled with the induction of DNA breaks by viral proteins, increases the mutation frequency and chromosome rearrangements in virus-infected cells. Indeed, HCV core proteins generate a chronic oxidative stress causing chromosomal and mitochondrial DNA instability (Hoshida et al., 2014).
The take-away message is that inhibition of DNA repair mechanism promotes mutator phenotype in HCV-associated HCC and these mutator phenotype should be reat targets for immune checkpoint inhibitor treatment (refer to Section 3). In cancer treatment, blocking poly (ADP-ribose) polymerase (PARP) prevents cancer cells from repairing their DNA damage, causing them to die (Buisson et al., 2010). Therefore PARP inhibitor treatment may be another strategy to reduce HCCs. The status of current related therapeutic strategies showed that PARP inhibitor inhibit many types of cancer development, possible for HCC as well. These genetic alterations induce self-rewnal ability of TICs.
Angiogenesis is elevated in highly vascularized tumors, including HCCs. The multi-tyrosine kinase inhibitor sorafenib (FDA-approved) is used in HCC patients to inhibit angiogenesis-inducing cytokine VEGF and the MAP Kinase, Raf/Mek/Erk pathways. Rapamycin is also used to inhibit the PI3K/Akt/mTOR pathway (Llovet and Bruix, 2008; Newell et al., 2009). Constitutive expression of Myc oncogene, platelet derived growth factor (PDGF), or VEGFA all lead to HCC development. Transgenic mouse studies demonstrated that tumor angiogenesis and recurrence is linked to MYC, PDGF and VEGFA pathways (Llovet and Bruix, 2008; Newell et al., 2009). Indeed, the angiogenesis biomarkers VEGF and Ang2 (angiogenin, ribonuclease A family, member 2) were independent predictors of advanced HCC patient survival (Llovet et al., 2012).
The take-away message is that angiogenesis is one of the most effective targets for HCC treatment since multi-kinase inhibitors Sorafenib and Rigorafenib are two FDA-approved chemotherapeutic drugs for HCC treatment after multinational, randomized, placebo-controlled, phase III Sorafenib HCC Assessment Randomized Protocol (SHARP) trial (Llovet et al., 2008). In HCC patients positive for anti-HCV antibody, sorafenib treatment improved median overall survival (OS), time to progression (TTP), disease control rate (DCR) (Llovet et al., 2008), indicating that these kinase signaling pathways maintain HCV-associated HCCs. New multikinase inhibitor Rigorafenib is used for Sorafenib-failure HCC patients.
Defective tumor immunity allows unrestricted HCC grow without tumor surveillace immune protection. HCV infection is a predisposing condition for HCC as viral clearance by the immune system since immune system cannot remove HCV-infected cells in almost all cases. Virus-specific CD8+ cytotoxic T lymphocytes (CTL) (Weiner et al., 1995; Cooper et al., 1999) clears HCV only in a minority (∼30%–40%) of cases (Hajarizadeh et al., 2013), leading to persistent lifelong infection with continuous immune-mediated hepatic inflammation.
Although mixed lymphocyte infiltration (T lymphocytes and NK cells) occurs in HCC tissues of patients (Rehermann, 2013), these immune cells are not cytoxic to cancer cells. Some of HCCs are tumors that do not have infiltration of immune cells, so called “cold” tumorsthat do not frequently respond to immune checkpoint inhibitor therapies. Indeed, HCV infection and/or viral proteins inhibit a variety of fuctions in many immune cell types, including CTL, CD4+ T, dendritic cells, macrophages and B cells (Bowen and Walker, 2005), indicating that HCV infection is associated with “cold” tumor phenotype.
Nonetheless infected hepatocytes do activate innate immunity by sensing HCV RNA motifs through RIG-I and TLR3. This leads to activation of the NF-κB pathway and generation of interferons and other pro-inflammatory cytokines. Furthermore the viral polymerase NS5B directly activates the inflammatory cascade through NF-κB in a MAVS and TBK-1 dependent manner, resulting in secretion of IL-6 and type I IFN (Yu et al., 2012).
Immune defects were targeted and translated into immunotherapies. Immunotherapy by the transplantation of donor bone-marrow stem cells kills tumor cells in the recipient (Dean et al., 2005). Thus, isolated TICs from a patient could be lethally irradiated and used to autologously ‘immunize’ the patient or used ex vivo activate donor immune cells against the patient’s TICs (Dean et al., 2005).
Immune checkpoint inhibitor anti-PD-1 therapy got FDA approval after landmark clinical trial (El-Khoueiry et al., 2017). As high genetic instability is good prognosis maker for immune checkpoint inhibitor, HCV-associated mutator phenotype (Machida et al., 2004a) can be great target for immune checkpoint inhibitor.
Human TERT cis-activation and telomerase enzymatic activity are associated with hepatocyte immortalization. HCV infection stimulates continuous growth and upregulates telomerase expression resulting in immortalization (Ray and Meyer, 2000). Furthermore, TERT promoter mutations are involved in TERT transactivation (Fujimoto et al., 2016). This immortalization process is prerequisite for step-wise carcinogenesis, which ultimately contributing to TIC formation.
Viral oncogenesis is characterized by two category, direct effects by viral proteins and indirect effects by inflammation (Figure 3). Direct oncogenic effects occur by expression of viral proteins, whereas indirect oncogenic effects occur by inflammation that is elicited by viral replication. HCV RNA is detected both in HCC and in surrounding non-tumor tissues (Alam et al., 2002; Sobesky et al., 2007). Both HCV structural and non-structural proteins are implicated in pro- and anti-apoptotic effects of hepatocytes, in eliciting inflammation and cancer-promoting signaling pathways, including WNT and sonic hedgehog pathways (McGivern and Lemon, 2011). Direct effects include viral protein-mediated transactivation or transcriptional suppression of genes, especially proto-oncogenes or tumor suppressor genes. Multifunctional HCV nucleocapsid core protein inhibits apoptosis, signal transduction, reactive oxygen species (ROS) formation, lipid metabolism, transcriptional activation, transformation and immune modulation. These additional hallmarks are observed in genesis of tumor-generating TICs when inoculated into immunocompromised mice. Therefore, a detailed and complete understanding of these mechanisms will contribute greatly to the development of new therapeutic strategies.
The take-away message is that HCV infection and/or viral proteins immortalize hepatocytes and/or cholangiocytes and/or liver progenitor cells through constitutive Telomerase activation. Telomere and/or other immrtalization siugnals can be targeted and translated into therapies. The status of current related therapeutic strategies include telomerase inhibitors and other kinase inhibitors.
Compelling evidence identifies obesity/alcoholism and HCV as co-morbidity risk factors for HCC. The risk for HCC, as assessed by odds ratio, increases from 8 to 12 to 48–54, if HCV patients have concomitant obesity or alcoholism (Hassan et al., 2002; Yuan et al., 2004; Artinyan et al., 2010). Demographic data indicate increased HCC occurrence in african-americans, Hispanics/latinos, low income individuals and rural poor especially among alcoholics or obese patients in these populations. This synergism in HCC development is explained by gut microbiota changes to the TLR4-NANOG signaling pathways. As mentioned above the innate immune system induces the expression of pattern recognition molecular pattern Toll-like receptor 4 (TLR4) following HCV infection. This is observed in hepatocytes of transgenic mouse models, liver cell cultures and during natural infection. This induction of TLR4 sensitizes hepatocytes to endotoxemia induced by obesity and alcohol. Subsequent TLR4 signaling, induces the stem marker NANOG in the HCV nonstructural protein Ns5a transgenic (Tg) mice but not in wild type or Ns5a Tg mice deficient in TLR4. Only the combined effects of alcohol/obvesity and HCV infection lead to NANOG induction and liver tumors.
Obesity leads to persistent inflammation. This condition increases gut permeability allowing increased blood levels of endotoxins such as lipopolysaccharides (LPS), which in turn activate toll-like receptors (TLRs) (Hritz et al., 2008). TLR activation induces the production of cytokines and the inflammatory response, ultimately leading to liver injury and the development of obesity/alcohol-related liver disease (Figure 4). TLR4 activation in immune cells, however, does not induce stemness or pluripotency transcription factor, such as NANOG, without HCV infection or NS5A protein expression in transgenic mouse model, indicating that enhanced TLR4 expression induced by NS5A protein or other causal effects is required for excessive signaling in order to turn on NANOG. Furthermore, mature cells have an enhanced methylation on NANOG promoter while progenitor cells or embryonic stem cells have less methylation of NANOG promoter, indicating that hypomethylation status of NANOG promoter is also required for NANOG induction.
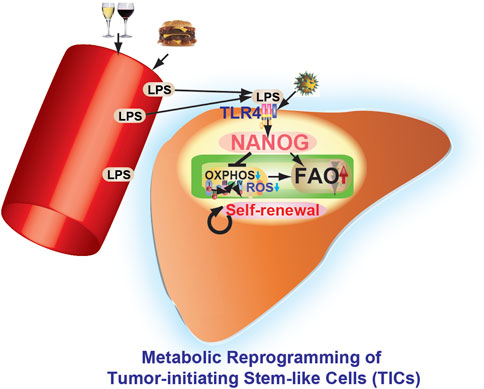
FIGURE 4. Pluripotency transcription factor NANOG contributes to cancer progression by mitochondrial reprogramming leading to the genesis of TICs. Environmental factors (alcohol and high-fat diet) and virus infection (i.e., hepatitis C virus) promote metabolic reprogramming and other characteristics of TICs. Obesity and alcoholism increase gut permeability leading to endotoxemia, which in turn activates Toll-like receptor 4 (TLR4) in the liver with induction of pluripotency transcription factor NANOG and an inflammatory response. This leads to subsequent development of obesity/alcohol-related liver cancer. NANOG ChIP-seq identified novel gene targets needed for oxidative phosphorylation (OXPHOS) and fatty acid oxidation (FAO). OXPHOS and fatty acid metabolism are identified as major pathways contributing to NANOG-mediated oncogenesis. NANOG-ChIP sequencing, gene profiling, proteomics, and metabolomics approaches were all combined to identify the altered pathway(s) in tumors. NANOG repressed OXPHOS and mitochondrial reactive oxygen species (ROS) in TICs. Restoration of OXPHOS and inhibition of FAO restored drug susceptibility of TICs. Identification of novel metabolic pathways provides potential drug targets for neutralizing the activity of highly malignant TICs found in cancer patients.
The TLR4-NANOG axis promotes metabolic reprogramming in hepatocytes via activation of fatty acid oxidation and inhibition of oxidative phosphorylation (OXPHOS) (Chen et al., 2016). Liver TICs are additionally sensitized to leptin and leptin exposure increases the expression and activity of an intrinsic pluripotency-associated transcriptional network comprised of STAT3, SOX2, OCT4, and NANOG. This axis is important but as previously mentioned, HCV infection is also required. This indicate that there are other predisposing factors for oncogenesis and tiC genesis beside TLR4 signaling activation.
Therefore, metabolic inhibitors and/or stemness inhibitors may prevent HCC self-renewal ability to kill HCC cells. Specific metabolic pathways may be novel therapeutic targets in order to selectively kill HCC cancer cells.
Chronic immune-mediated inflammation leads to repeated hepatocyte destruction and regeneration. As a consequence, these events lead to fibrogenic wound-healing responses which drive HCC or CC in chronic hepatitis C cases (Figures 1, 2). The prolonged liver inflammation coupled with the repeated liver regeneration process is conducive to multi-step hepatocarcinogenesis (Figure 3) (Simonetti et al., 1992).
Host pathogen-associated molecular pattern (PAMP) receptors sense double-stranded viral RNA replication intermediates to activate IFN Regulatory Factors IRF3/7 and NF-κB, leading to the induction of IFNs and related IFN-stimulated genes (ISGs). Persistent HCV infection induces ISG (Wieland et al., 2014) although HCV antagonizes RNA-sensor mechanisms for viral persistence (Li and Lemon, 2013). For negative feedback pathway, the HCV RNA replicase complex downregulates viral RNA synthesis, to maintain replication at low levels and minimizing oxidative damage (Yamane et al., 2014). HCV persistence induces hepatic oxidative DNA damage in chronic hepatitis C (Shimoda et al., 1994; Fujita et al., 2008).
Direct-acting antiviral agents might promote tumour occurrence in patients with cirrhosis, or recurrence in patients with presumed cure of hepatocellular carcinoma while DAA significantly reduces viral load. In view of the potential clinical implications, this controversy calls for a thorough and expeditious consideration of the hypothetical oncogenic activity of novel HCV drugs (Llovet and Villanueva, 2016).
Due to the “new” antiviral drugs available for HCV treatment, the incidence of HCC has changed. However, some patients who achieved sustained viral response develop HCC.
Mouse hepatocytes that express HCV-NS5A in liver upregulate the expression of Toll-like receptor 4 (TLR4) and develop liver tumors containing NANOG positive, tumor-initiating stem-like cells (TICs). The TLR signaling pathway is often upregulated in chronic liver diseases, especially since many different liver cell types express TLRs (Testro and Visvanathan, 2009). Hepatocytes express TLR1 through TLR9. Stellate cells express TLR2, 3, and 4. Bile duct epithelium expresses TLR2, 3, 4, and 5. Kupffer cells express TLR2, 3, and 4. Chronic alcohol consumption is associated with activation of TLR1, 2, and 6–9, which further increases the TNF-α response to LPS in mice (Testro and Visvanathan, 2009). Human monocytes exposed to ethanol for a week develop hypersensitivity to LPS through decreased IRAK-M expression, which activates mitogen-activated protein kinase (MAPK) and NF-κB following TLR4 signaling. This leads to activation of NF-κB, AP-1, and ERK (Mandrekar et al., 2009) and associated inflammatory response.
Nanog is one of the core transcription factors found in pluripotent embryonic stem cells (ESCs) (Martin, 1981). It is essential for maintaining self-renewal and pluripotency of both human and mouse embryonic stem cells (Loh et al., 2006; Wang et al., 2006; Rao and Orkin, 2006; Pan and Thomson, 2007). Overexpression of Nanog induces and maintains the pluripotency and self-renewing characteristics of ESCs under what normally would be differentiation-inducing culture conditions (Chambers et al., 2003). Recently, Nanog expression has been reported in human neoplasms, including germ cell tumors (Ezeh et al., 2005; Hart et al., 2005; Hoei-Hansen et al., 2005; Santagata et al., 2007), breast carcinomas (Ezeh et al., 2005), osteosarcoma (Gibbs et al., 2005), and HCC (Ma et al., 2008). Ectopic expression of Nanog induces an oncogenic potential in NIH3T3 (Zhang et al., 2005).
Nanog expression alone is not as effective as TLR4 activation in liver tumorigenesis, as shown by our cell transplantation experiments (Machida et al., 2009). TLR4 activation induces other tumor-driver genes which cooperatively work with Nanog to initiate liver oncogenesis. Thus, Nanog is essential for TLR4-dependent oncogenesis, but it alone is poorly oncogenic. This highlights the importance of alcohol and HCV NS5A synergism for liver tumor induction, especially in mice The importance of Nanog as a direct downstream gene of TLR4 in liver oncogenesis is summarized in Figure 4 (Feldman et al., 2012). Thus, pharmacologic inhibition of TLR4 signaling, including TLR4 antagonist (Eritoran or FP7) (Perrin-Cocon et al., 2017), may become a viable therapeutic strategy for HCV-associated liver tumors.
The take-away message is that TLR4-NANOG pathway is novel therapeutic targets.
The status of current related therapeutic strategies is under development.
Epithelial-mesenchymal transition promotes cell migration and invasion, ultimately leading to metastasis. This transition also occurs in development of HCC. As discussed earlier, the combined effect of TLR4-NANOG signaling promotes the development of TICs and tumorigenesis in transgenic mice expressing NS5A. These mice, when placed on a Western diet high in cholesterol and saturated fat (HCFD) activate the TLR4-NANOG axis in combination with the leptin receptor (OB-R)-pSTAT3 signaling pathways. The net resul is the occurrence of liver tumorigenesis through an exaggerated mesenchymal phenotype with prominent Twist1-expressing TICs (Uthaya Kumar et al., 2016).
Therefore, TWIST1 targetting therapy may prevent HCC cell spread into distal organs. Preventing metastasis spread would be effective strategy to keep HCC in primary organ site.
Epigenetics is stable alterations in gene expression without genetic modifications in the sequence (Herceg, 2007). Epigenetic and genetic mechanisms silences cellular genes leading to transformation in human cancers, including HCC (Vaissiere et al., 2008). Contribution of different epigenetic factors, including genomic DNA methylation, histone modifications, and miRNA regulation, contribute to HCC dissemination, invasion, and metastasis. The reversal of deregulated epigenetic changes is emerging treatment of HCC (Nishida and Goel, 2011). High-throughput screening provides targeting inflammation-epigenome cross-talk in HCC to discover novel epigenetic targets (Herceg and Paliwal, 2011). Epigenetic Mechanism promotes the HBV/HCV-Related HCC tumorigenesis (Rongrui et al., 2014)
Non-coding RNAs [LncRNA and microRNA (miRNA)] are dysregulated in HCV-induced liver carcinogenesis via regulation of gene expression. LncRNAs expression (LINC01419, AK021443, UCA1, and WRAP53) are increased in HCV-related HCCs compared to non-cancerous tissues (dysplasia) while lncRNA AF070632 is decreased in advanced HCC samples compared with early HCC. These lncRNAs are associated with Child-Pugh score. LINC01419 and AK021443 were mostly involved in cell cycle progression, whereas AF070632 regulates cofactor binding, oxidation-reduction and carboxylic acid catabolic process (Zhang et al., 2015). Two lncRNAs, including urothelial carcinoma associated-1 (UCA1) and WD repeat containing, antisense to TP53 (WRAP53) are upregulated in serum. UCA1 and WRAP53 (+) HCC patients had a decreased recurrence-free survival (RFS) and increased cumulative hazards. WRAP53 was an independent prognostic factor of RFS (Kamel et al., 2016). Some of these lncRNAs were dysregulated predominantly in one specific hepatitis virus-related HCC, including PCAT-29 in HBV-related HCC, aHIF and PAR5 in HCV-related HCC, and Y3 in HDV-related HCC. DBH-AS1, hDREH and hPVT1 were differentially expressed in HCC of different viral etiology (Zhang et al., 2016).
LncRNA and miRNAs have been associated with HCC (Wong et al., 2018). Due to this review being focused on TICs, more details about TICs and non-coding RNAs are described (such as (Huang et al., 2018; Machida, 2020; Rojas et al., 2022): More details about TICs and non-coding RNAs are included such as: (Huang et al., 2018; Machida, 2020; Rojas et al., 2022). In Table 1 HCV-associated HCC are involved in alterations of lncRNAs [(Hou and Bonkovsky, 2013; Lange et al., 2013; Zhang et al., 2015; Zhang et al., 2016; Fu et al., 2016; Shi et al., 2016; Hai et al., 2017; Wong et al., 2018; Sur et al., 2018; Toraih et al., 2018; Wang et al., 2018; Zhang et al., 2018; Cheng et al., 2019; Refai et al., 2019; Wang et al., 2019; Wu et al., 2019; Yang et al., 2019; Zhao et al., 2019; Zheng et al., 2019; Zhong et al., 2019; El-Khazragy et al., 2020; Ferrasi et al., 2020; Lorini et al., 2020; Machida, 2020; Mohyeldeen et al., 2020; Morishita et al., 2020; Oura et al., 2020; Unfried and Fortes, 2020; Jing et al., 2021; Sabry et al., 2021; Wong and Wong, 2021; Yao et al., 2021; Wang et al., 2022; Wei et al., 2022)]. Upregulated lncRNAs include NORAD (LINC00657), HCP5, lnc-HOTAIR (HOX antisense intergenic RNA), CASC11, HEIM, eosinophil granule ontogeny transcript (EGOT), lncRNA SEMA3B-AS1 [SEMA3B Antisense RNA 1 (Head To Head)], TPT-1S, LINC01189. In contrast, TPT1-AS1, LINC01152, aHIF and PWAR5 (PAR5) are downregulated.
miRNA targets hundred mRNAs, miRs are diagnostic markers and therapeutic target for personalized therapy. miRNAs are differentially expressed in liver cancer and are related to different stages of liver carcinogenesis, supporting the diagnosis and prognosis tools of miRNAs in HCC patient. miRNAs promote or inhibits carcinogenesis via activation of oncogenes and/or suppression of tumor suppressors (Chen, 2005). HCV requires liver-specific miR-122 for replication (Jopling et al., 2005). Sequestering miR-122 in patients leads to a dose-dependent decrease in HCV viremia in phase 2a trial (Janssen et al., 2013). Mice lacking miR-122 have high tumor incidence (Tsai et al., 2012). The miR-122 abundance is reduced in human advanced fibrosis (Trebicka et al., 2013) and in therapresistant HCV patients (Sarasin-Filipowicz et al., 2009). Recruitment of miR-122 to the HCV genome depletes this important liver-specific miR-122. HCV sequesters anti-tumorigenic miR-122 to promote HCC development.
miR-21, miR-17, miR-222, miR-224, and miR-221 are increased in liver cancer (Borel et al., 2012) (Ladeiro et al., 2008) while miR-200, let-7, miR-29, miR-123, miR-122, miR-199a, and miR-199 b are decreased in HCCs (Hou et al., 2011; Huang and He, 2011; Anwar and Lehmann, 2015). miR-199a/b-3p prevents the p21-stimulated kinase 4/Raf/MEK/ERL pathway and suppresses HCC.
Down-regulation of miR-199a/b is associated with poor prognosis and low survival rate (Li et al., 2011). Increased miR-224 is associated with malignancy aggression, deteriorated liver function, and poor prognosis (Wang et al., 2008; Zhuang and Meng, 2015).
Induction of HCV proteins or the infection of HCC cells with HCV cell culture (HCVcc) suppresses histone H4 methylation/acetylation and histone H2AX phosphorylation for HCC development, indicating that HCV-induced overexpression of PP2Ac are associated with HCC via deregulation of epigenetic histone modifications (Duong et al., 2010). HCV infection upregulates histone deacetylation (HDAC) activity through affecting hepcidin expression, a key suppressor of iron availability (Miura et al., 2008). The induced HCV oxidative stress leads to suppression of hepcidin expression by increased HDAC function. HCV increases histone deacetylation (HDAC) activity through negative regulator of iron availability (hepcidin expression) (Miura et al., 2008). Furthermore, antiviral agents IFN with epigenetic drugs (such as DNMT inhibitors or HDAC inhibitors) counteract epigenome changes with cytokines (Muller, 2006).
HCV caused epigenetic alteration mainly occurred on DNA repair-related genes. Induction of HCV proteins or the infection of HCC cells with HCVcc inhibits histone H4 methylation/acetylation and histone H2AX phosphorylation and inhibited DNA damage repair, indicating that HCV-induced overexpression of PP2Ac promotes hepatocarcinogenesis via dysregulation of epigenetic histone modifications (Duong et al., 2010).
HCV infection accelerates or inhibits the methylation process. DNA methyltransferases (DNMTs) methylates DNA. HCV upregulates DNA methyl transferases, which further block tumor suppressor genes leading to HCC (Tian et al., 2013). HCV core protein upregulates both mRNA and protein expression levels of DNMT1 and DNMT3b, which promotes DNA methylation in HCV-infected hepatocytes (Benegiamo et al., 2012). HCV core protein increases the mRNA and protein levels of DNMT1 and DNMT3b, leading to epigenetic alteration of HCV patients (Benegiamo et al., 2012). HCV tissues have seven hypermethylated markers (COX2, MINT1, CACNA1G, RASSF2, MINT2, Reprimo, and DCC) in comparison to both HBV and normal liver tissues (Nishida et al., 2008). Different HCV proteins NS5A (Kasprzak and Adamek, 2008) promotes hepatocarcinogenesis.
Epigenetic event Contribution in HCC development.
• DNA hypermethylation
○ CDH1: Cell adhesion and metastasis
○ RASSF1A: Cell cycle dysregulation
○ P21WAF1/CIP1: Cell cycle dysregulation
○ Gadd45: Response to genotoxic stress
○ MGMT: Dysfunction of DNA repair
○ APC: Cell cycle dysregulation/Dysfunction of DNA repair
• DNA hypomethylation (demethylation)
○ STAT1: Upregulation of JAK/STAT pro-tumorigenic signaling
○ COX-2: Inflammation
• Histone modification
○ PP2Ac: Inflammation
The adenomatous polyposis coli (APC) tumor suppressor gene encodes a large protein with multiple cellular functions and interactions, including signal transduction in the WNT-signaling pathway (Lee et al., 2006). The APC promoter is methylated in up to 81% of patients with viral hepatitis-induced HCC (Lee et al., 2003). A next generation sequencing of CpG methylation site demonstrates that APC was hypermethylated in HCC tissues to their corresponding non-tumorous tissues (Archer et al., 2010). In contrast, NOTCH4, EMR3, HDAC9, DCL1, HLA-DOA, HLA-DPA1, and ERN1 were hypomethylated in HCC (Archer et al., 2010).
The take-away message is that epigenetic regulation promotes envation, migration and metastatic characteristics thorugh aberrant expression of non-coding RNA, histone modification and DNA methylation. These aberrant epigenetic regulation can be targeted and translated into therapies. Indeed, miR-122 restoration strategy is being tested in clinical trials. The status of current related therapeutic strategies includes new miR and/or ncRNA targeted therapies.
Thel tumor suppressor p53 protein coordinates cell-cycle arrest, senescence, and apoptosis in response to DNA damage and cellular stresses (Bieging et al., 2014). Mutations in p53-DNAb-binding domains that disrupt DNA binding ability of p53 are associated with many human cancers, including HCC (Hussain et al., 2007; Guichard et al., 2012). HCV proteins target tumor suppressor genes and proto-oncogenes. For example, three HCV proteins, including core, NS3, and NS5A, interact with tumor suppressor p53 when overexpressed in cell culture (McGivern and Lemon, 2011). There is conflicting data on whether core interaction with p53 results in activation or inhibition of p53 target genes, but thismay reflect differences in the level of core expression (high level core expression is needed) (Kao et al., 2004). The NS3-p53 interaction blocks apoptosis in vitro (Deng et al., 2006) and NS5A interaction with p53 results in p53 redistribution to the peri-nuclear membrane (Majumder et al., 2001). The impact of HCV proteins on p53 activity and interactions between HCV proteins and p53 are controversial since the cell-lines that are most permissive for HCV (Huh-7 hepatoma cells and their derivatives: Huh7.5 and Huh7.5.1) express a mutated, inactive form of p53 (Bressac et al., 1990; Hsu et al., 1993). Additionally, the retinoblastoma tumor suppressor protein (Rb) interacts with the HCV NS5B protein, leading to its poly-ubiquitination and degradation to promote S phase entry (Munakata et al., 2005). The take-away message is that HCV infection and/or viral proteins inactivates tumor suressor pathways. Therefore, restoration of p53 function, by use of Adenovirus expressing functional p53, may restore tumor suppressor function and selectively kill HCC cells.
HCV infection modulates several genes, including SOCS-1 Suppressor of cytokine signaling (SOCS) 1 (negative regulator of the JAK/STAT pathway). Expression of SOCS1 leads to reduced JAK /STAT phosphorylation, reduced STAT dimerization and imports to the nucleus and reduced transcription of target genes (Chim et al., 2004). JAK/STAT pathway is pro-tumor signaling (Calvisi et al., 2006). The take-away message is that HCV infection and/or viral proteins constitutively activate proto-oncogenes and/or inhibits tumor suppressors. Therefore, STAT3 is targeted and translated into STAT3 inhibitor therapies.
Stem cells have three major characteristics: self-renewal, asymmetric and multiple cell division (clonality), and plasticity. Hepatic small oval progenitor cells around the peripheral branches of the bile ducts, the canals of Hering, differentiates into biliary epithelial cells and hepatocytes (Roskams et al., 2004). These oval liver progenitor cells share molecular markers with adult hepatocytes [albumin, cytokeratin 7 (CK7), CK19, oval cell markers (OV-6, A6, and OV-1), chromogranin-A, NCAM (neural cell adhesion molecule)] and fetal hepatocytes (α-fetoprotein) (Roskams et al., 2004; Roskams, 2006). They are also positive for more common stem cell markers such as CD34+, Thy-1+ (Burke et al., 2007). A CD117+/CD133+ hepatic precursors are detected in regenerating liver tissue (Craig et al., 2004) while a CD45–/CD90+ tumor subpopulation are detected in HCC (Yang et al., 2008). The CD90+ cells are not present in the normal liver and, when injected into immunodeficient mice, create tumors repeatedly. In human HCC and HCC cell lines, specifically CD133(+) cells, not CD133(−)cells, had the ability to self-renew, create differentiated progenies, and form tumors (Ma et al., 2007).
Forty percent of HCC have clonality, and thus are considered to originate from progenitor/stem cells (Alison, 2005; Roskams, 2006; Zender et al., 2006; Tang et al., 2008). TICs express stemness genes, including CD133 (Prominin in mice), Wnt/β-catenin, Nanog (Feldman et al., 2012), Notch, Hedgehog/SMO, Bmi, Oct3/4 (Beachy et al., 2004; Chambers and Smith, 2004; Valk-Lingbeek et al., 2004), NOTCH, BMI, OCT3/4, CD44 (cell adhesion molecule) and CD34. The CD133 subpopulations displayed similar expression for CD29 (integrin β1), CD49f (integrin α6), CD90, and CD117 (c-kit: gastrointestinal stroma tumor), indicating these makers are still not definitive TIC markers (Ma et al., 2007). CD133+/CD49f + HCC TICs confer resistance to chemotherapy, which hampers efficacy of therapy in HCC (Rountree et al., 2008). Thus, HCV infection is associated with TICs and HCC development.
Dysregulated signaling and gene expression promotes the plasticity of TICs to resist current FDA-approved therapies, such as sorafenib or regorafenib or anti-PD1 immune checkpoint inhibitor treatment. TICs of HCC are observed to have several elevated oncogenic and anti-apoptotic signaling pathways such as PI3K/AKT (Ma et al., 2008), signal transducer and activator of transcription 3 (STAT3) (Wurmbach et al., 2007; Yeoh et al., 2007), Notch (Dando et al., 2005), hedgehog (Sicklick et al., 2006a; Sicklick et al., 2006b) and transforming growth factor-beta (TGF-β) (Kitisin et al., 2007; Nguyen et al., 2007).
Normal stem cells and TICs express high levels of ATP-binding cassette (ABC) transporters, such as ABCB1, and the half-transporter ABCG2 identified in mitoxantrone-resistant cells (Shepard et al., 2003; Schneiderman et al., 2010). Cancer cells in culture become resistant to cytotoxic anticancer drugs through multiple pathways, such as increased active efflux at the plasma membrane (MDR1, MRP family members, and MXR), reduced drug uptake, expression of one or more energy-dependent transporters that specifically detect and eject intracellular anticancer drugs, insensitivity to drug-induced apoptosis and induction of drug-detoxifying mechanisms (Gottesman, 2002). The drug-transporting property of TICs conferred by ABC transporters is the basis for the observed ‘side-population’ phenotype that is identified by exclusion of the fluorescent dye Hoechst 33,342. Therefore, tumors may have an intrinsic population of drug-resistant pluripotent cells that survive chemotherapy and thus repopulate the tumor (Gottesman, 2002). The take-away message is that drug-transporting property of TICs are rationale target to suppress drug-resisstance phenotype of TICs. Therefore, ABCG2 inhibitors (GF120918 and tariquidar) will inhibit both ABCG2 and ABCB1 to overcome drug resistance by inhibition of the Hedgehog-Patched receptor signaling protein, Smoothened.
Chronic liver inflammation causes liver injury with compensatory hepatocellular proliferation which indirectly promotes HCC. Regenerative pathways promote dedifferentiation and proliferation to replace damaged tissue in inflammatory liver disease and hepatocellular necrosis associated with chronic hepatitis C infection.
For example, the Wnt/β-catenin pathway is directly altered by HCV. Initially, NS5A promotes β-catenin stabilization through inactivation of GSK3-β, which normally promotes β-catenin degradation (Street et al., 2005; Park et al., 2009). Addiotional study shows that NS5A directly interacts with β-catenin and stabilizes β-catenin (Milward et al., 2010). Similarly GSK3-β is also a target of hepatitis B virus X-protein which results in stabilization of β-catenin. HCV interacts with the TGF-β pathway leading to cytostatic response or fibrogenic responses. HCV NS5A blocks TGF-β signaling through direct interaction with its receptor, TGF-β receptor I (TβR-I) (Choi and Hwang, 2006). Antagonism of TGF-β signalingpromotes liver damage, fibrosis and cancer. Furthemore, HCV core variant isolated from HCC, but not from surrounding liver tissue blocks TGF-β signaling through interactions with SMAD3 (Pavio et al., 2005).
Cells within such a precancerous field contain mutations that predispose progression to a cancerous phenotype. What is the impact of chronic hepatocyte turnover on tumorigenesis? Perhaps apoptosis of infected hepatocytes,either by immune- or virus-mediated mechanisms with compensatory hepatocellular proliferation, promotes carcinogenesis during decades of chronic HCV infection.
The association of liver injury with HCV infection is depicted in Figure 4. Core protein activates cellular oncoproteins and NF-κB cell signaling pathways and causes p53 and pRb inactivation to initiate genomic instability and uncontrollable cellular proliferations (Smirnova et al., 2006). HCV nonstructural proteins NS5A and NS3 alter host expression and promote liver cell proliferation, leading to HCC development (Jeong et al., 2012). NS5A suppresses immune responses, inactivates tumor suppressors, inhibits apoptosis, and disrupts liver homeostas, thus leading a to a primary liver.
The take-away message is that HCV promotes HCC growth by compensatory regeneration and proliferation. Compensatory regeneration and proliferation were targeted and translated into therapies. the status of current related therapeutic strategies showed sorafenib and regorafenib inhibits compensatory regeneration and proliferation.
The TIC population is a prime origin of cancer recurrence in drug-treated patients with HCC. TICs are generated in HCV carcinogenesis (Figure 5). TICs acuired tumor-initiation property. The molecular pathways of HCV-induced carcinogenesis may involve indirect, non-virological factors such as the induction of chronic liver inflammation and regeneration that lead to the emergence of mutated cells with high proliferation rates. In addition, HCV infection may also involve viral gene products that stimulate the production of ROS with the expression of error-prone DNA polymerases. These diverse pathways highlight the complicated interplays between the virus and its host in HCV associated carcinogenesis. Given the latter pathway, potential use of ROS and or iNOS inhibitors may be useful for treating HCC patients with HCV co-morbidity. Transformed cells are further altered and progressed to TICs (Figure 5). Expansion of TICs requires self-renewal property (Figure 5). Environmental factors (alcoholism, obesity and carcinogen exposure) and HCV infection are invoved in initiation and promotion (Figure 5). Chronic HCV infection results in a high frequency of HCC that displays non-metastatic and multicentric characteristics.
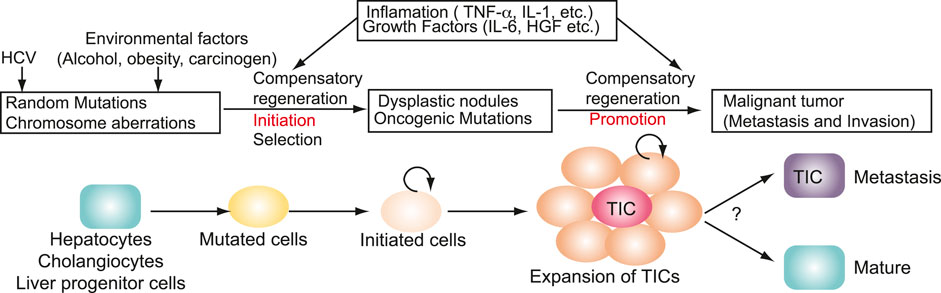
FIGURE 5. Genesis of TICs in HCV carcinogenesis. Environmental factors (alcoholism, obesity and carcinogen exposure) and HCV infection are invoved in initiation and promotion. Transformed cells are further altered and progressed to TICs. Expansion of TICs requires self-renewal property. TICs acuired tumor-initiation property.
Additional therapies are desperately needed since current treatments (sorafenib, regorafenib or anti-PD1 immune checkpoint inhibitors) have limitationsbecause eventual treatment failures leads to cancer metastasis. Future investigative projects need to address the specific treatment needs of patients with HCC. Current FDA-approved immunotherapy, such as anti-PD-1 or anti-CTLA4 therapy has a limited efficacy for a small fraction of HCC patients (10%–25% range undergoing monotherapy). The remaining HCC patients did not respond to this monotherapy, indicating that an immune-checkpoint inhibitor approach has a limited efficacy and other immune mechanisms may be needed to have synergism with tumor-killing cells, such as antigen presenting cells, including dendritic cells and B cells. However inclusion of immune checkpoint inhibitors with combination therapy may break immune tolerance and improve the therapeutic efficacy of this approach.
Simply extending patient life spans by several months is not suffiecient. The goal should be to expand the completely cured patient population. This is the biggest challenge of researchers to transform a “previously “incurable” malignancy into a curable illness. Discovery of new immune checkpoint inhibitors and incorporation into combination therapies may be a new therapeutic avenue to drastically improve HCC treatment. HCC should not be a death sentence and may become a curable malignancy providing foundational research findings can be translated into game-changing innovative treatments (Refer to the outstanding questions box).
Finally, personalized medicine approaches will stratify the HCC patient population into distinct subpopulations that may be responsive to HCC-type specific treatments. As presented in this review, there are several avenues of liver morbidities leading to HCC. Thus, treatment options may likely reflect the original triggering event leading to HCC.
Keigo Machida, Writing manuscript.
This research was supported by NIH grants 1R01AA018857-01 and 1R21AA025470 - 01A1, pilot project funding (5P30DK048522-13). P50AA011999 (Research Project, Animal Core, Morphology Core, and Pilot Project Program), R24AA012885 (Non-Parenchymal Liver Cell Core), Zumberge Foundation, AI83025U19, U19 AI 83025, the Cell and Tissue Imaging Core of the USC Research Center for Liver Diseases (P30 DK048522), CA123328, and CA108302. This research is also supported by a Research Scholar Grant, RSG MPC122545 and pilot funding (IRG-58-007-48) from American Cancer Society. Animal imaging was performed by the USC Molecular Imaging Center supported by NIH/NVRR S10. Tissue pathological slide preparation was performed by Ms. Moli Chen in Translational Pathology Core of Norris Comprehensive Cancer Center. Liver tissues were obtained from The Liver Tissue Cell Distribution System (LTCDS) in University of Minnesota.
We thank Stanley M Tahara (USC) for critical comments and editorial assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahn S. M., Jang S. J., Shim J. H., Kim D., Hong S. M., Sung C. O., et al. (2014). Genomic portrait of resectable hepatocellular carcinomas: Implications of RB1 and FGF19 aberrations for patient stratification. Hepatology 60 (6), 1972–1982. doi:10.1002/hep.27198
Alam S. S., Nakamura T., Naganuma A., Nozaki A., Nouso K., Shimomura H., et al. (2002). Hepatitis C virus quasispecies in cancerous and noncancerous hepatic lesions: The core protein-encoding region. Acta Med. Okayama 56 (3), 141–147. doi:10.18926/AMO/31716
Alison M. R. (2005). Liver stem cells: Implications for hepatocarcinogenesis. Stem Cell Rev. 1 (3), 253–260. doi:10.1385/SCR:1:3:253
Anderson S. C., Johnson D. E., Harris M. P., Engler H., Hancock W., Huang W. M., et al. (1998). p53 gene therapy in a rat model of hepatocellular carcinoma: intra-arterial delivery of a recombinant adenovirus. Clin. Cancer Res. 4 (7), 1649–1659.
Anwar S. L., Lehmann U. (2015). MicroRNAs: Emerging novel clinical biomarkers for hepatocellular carcinomas. J. Clin. Med. 4 (8), 1631–1650. doi:10.3390/jcm4081631
Arao T., Ueshima K., Matsumoto K., Nagai T., Kimura H., Hagiwara S., et al. (2013). FGF3/FGF4 amplification and multiple lung metastases in responders to sorafenib in hepatocellular carcinoma. Hepatology 57 (4), 1407–1415. doi:10.1002/hep.25956
Archer K. J., Mas V. R., Maluf D. G., Fisher R. A. (2010). High-throughput assessment of CpG site methylation for distinguishing between HCV-cirrhosis and HCV-associated hepatocellular carcinoma. Mol. Genet. Genomics 283 (4), 341–349. doi:10.1007/s00438-010-0522-y
Ariumi Y., Kuroki M., Dansako H., Abe K. I., Ikeda M., Wakita T., et al. (2008). The DNA damage sensors ataxia-telangiectasia mutated kinase and checkpoint kinase 2 are required for hepatitis C virus RNA replication. J. Virol. 82 (19), 9639–9646. doi:10.1128/JVI.00351-08
Artinyan A., Mailey B., Sanchez-Luege N., Khalili J., Sun C. L., Bhatia S., et al. (2010). Race, ethnicity, and socioeconomic status influence the survival of patients with hepatocellular carcinoma in the United States. Cancer 116 (5), 1367–1377. doi:10.1002/cncr.24817
Barbara L., Benzi G., Gaiani S., FusconiF. , Zironi G., Siringo S., et al. (1992). Natural history of small untreated hepatocellular carcinoma in cirrhosis: A multivariate analysis of prognostic factors of tumor growth rate and patient survival. Hepatology 16 (1), 132–137. doi:10.1002/hep.1840160122
Bartsch H., Nair J. (2006). Chronic inflammation and oxidative stress in the Genesis and perpetuation of cancer: Role of lipid peroxidation, DNA damage, and repair. Langenbecks Arch. Surg. 391 (5), 499–510. doi:10.1007/s00423-006-0073-1
Baumann P., West S. C. (1998). DNA end-joining catalyzed by human cell-free extracts. Proc. Natl. Acad. Sci. U. S. A. 95 (24), 14066–14070. doi:10.1073/pnas.95.24.14066
Beachy P. A., Karhadkar S. S., Berman D. M. (2004). Tissue repair and stem cell renewal in carcinogenesis. Nature 432 (7015), 324–331. doi:10.1038/nature03100
Benegiamo G., Vinciguerra M., Mazzoccoli G., Piepoli A., Andriulli A., Pazienza V. (2012). DNA methyltransferases 1 and 3b expression in Huh-7 cells expressing HCV core protein of different genotypes. Dig. Dis. Sci. 57 (6), 1598–1603. doi:10.1007/s10620-012-2160-1
Bieging K. T., Mello S. S., Attardi L. D. (2014). Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 14 (5), 359–370. doi:10.1038/nrc3711
Borel F., Konstantinova P., Jansen P. L. M. (2012). Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J. Hepatol. 56 (6), 1371–1383. doi:10.1016/j.jhep.2011.11.026
Bowen D. G., Walker C. M. (2005). Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 436 (7053), 946–952. doi:10.1038/nature04079
Braakhuis B. J., Tabor M. P., Kummer J. A., Leemans C. R., Brakenhoff R. H. (2003). A genetic explanation of slaughter's concept of field cancerization: Evidence and clinical implications. Cancer Res. 63 (8), 1727–1730.
Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 68 (6), 394–424. doi:10.3322/caac.21492
Bressac B., Galvin K. M., Liang T. J., Isselbacher K. J., Wands J. R., OzturkM. (1990). Abnormal structure and expression of p53 gene in human hepatocellular carcinoma. Proc. Natl. Acad. Sci. U. S. A. 87 (5), 1973–1977. doi:10.1073/pnas.87.5.1973
Buisson R., Dion-Cote A. M., Coulombe Y., Launay H., Cai H., Stasiak A. Z., et al. (2010). Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 17 (10), 1247–1254. doi:10.1038/nsmb.1915
Burke Z. D., Thowfeequ S., Peran M., Tosh D. (2007). Stem cells in the adult pancreas and liver. Biochem. J. 404 (2), 169–178. doi:10.1042/BJ20070167
Calvisi D. F., Ladu S., Gorden A., Farina M., Conner E. A., Lee J. S., et al. (2006). Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology 130 (4), 1117–1128. doi:10.1053/j.gastro.2006.01.006
Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., et al. (2003). Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113 (5), 643–655. doi:10.1016/s0092-8674(03)00392-1
Chambers I., Smith A. (2004). Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene 23 (43), 7150–7160. doi:10.1038/sj.onc.1207930
Chen C. L., Uthaya Kumar D. B., Punj V., Xu J., Sher L., Tahara S. M., et al. (2016). NANOG metabolically reprograms tumor-initiating stem-like cells through tumorigenic changes in oxidative phosphorylation and fatty acid metabolism. Cell Metab. 23 (1), 206–219. doi:10.1016/j.cmet.2015.12.004
Chen C. Z. (2005). MicroRNAs as oncogenes and tumor suppressors. N. Engl. J. Med. 353 (17), 1768–1771. doi:10.1056/NEJMp058190
Cheng N., Wu J., Yin M., Xu J., Wang Y., Chen X., et al. (2019). LncRNA CASC11 promotes cancer cell proliferation in hepatocellular carcinoma by inhibiting miRNA-188-5p. Biosci. Rep. 39 (4), 1–7. doi:10.1042/BSR20190251
Chim C. S., Fung T. K., Cheung W. C., Liang R., Kwong Y. L. (2004). SOCS1 and SHP1 hypermethylation in multiple myeloma: Implications for epigenetic activation of the jak/STAT pathway. Blood 103 (12), 4630–4635. doi:10.1182/blood-2003-06-2007
Choi J., Ou J. H. (2006). Mechanisms of liver injury. III. Oxidative stress in the pathogenesis of hepatitis C virus. Am. J. Physiol. Gastrointest. Liver Physiol. 290 (5), G847–G851. doi:10.1152/ajpgi.00522.2005
Choi S. H., Hwang S. B. (2006). Modulation of the transforming growth factor-beta signal transduction pathway by hepatitis C virus nonstructural 5A protein. J. Biol. Chem. 281 (11), 7468–7478. doi:10.1074/jbc.M512438200
Cicalese A., Bonizzi G., Pasi C. E., Faretta M., Ronzoni S., Giulini B., et al. (2009). The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138 (6), 1083–1095. doi:10.1016/j.cell.2009.06.048
Cooper S., Erickson A. L., Adams E. J., Kansopon J., Weiner A. J., Chien D. Y., et al. (1999). Analysis of a successful immune response against hepatitis C virus. Immunity 10 (4), 439–449. doi:10.1016/s1074-7613(00)80044-8
Craig C. E., Quaglia A., Selden C., Lowdell M., Hodgson H., Dhillon A. P. (2004). The histopathology of regeneration in massive hepatic necrosis. Semin. Liver Dis. 24 (1), 49–64. doi:10.1055/s-2004-823101
Crippin J. S., McCashland T., Terrault N., Sheiner P., Charlton M. R. (2002). A pilot study of the tolerability and efficacy of antiviral therapy in hepatitis C virus-infected patients awaiting liver transplantation. Liver Transpl. 8 (4), 350–355. doi:10.1053/jlts.2002.31748
Dando J. S., Tavian M., Catelain C., Poirault S., Bennaceur-Griscelli A., Sainteny F., et al. (2005). Notch/Delta4 interaction in human embryonic liver CD34+ CD38- cells: Positive influence on BFU-E production and LTC-IC potential maintenance. Stem Cells 23 (4), 550–560. doi:10.1634/stemcells.2004-0205
Dean M., Fojo T., Bates S. (2005). Tumour stem cells and drug resistance. Nat. Rev. Cancer 5 (4), 275–284. doi:10.1038/nrc1590
Delgado E. R., Yang J., So J., Leimgruber S., Kahn M., Ishitani T., et al. (2014). Identification and characterization of a novel small-molecule inhibitor of beta-catenin signaling. Am. J. Pathol. 184 (7), 2111–2122. doi:10.1016/j.ajpath.2014.04.002
Deng L., Nagano-Fujii M., Tanaka M., Nomura-Takigawa Y., Ikeda M., Kato N., et al. (2006). NS3 protein of Hepatitis C virus associates with the tumour suppressor p53 and inhibits its function in an NS3 sequence-dependent manner. J. Gen. Virol. 87 (6), 1703–1713. doi:10.1099/vir.0.81735-0
Di Bisceglie A. M., Lyra A. C., Schwartz M., Reddy R. K., Martin P., Gores G., et al. (2003). Hepatitis C-related hepatocellular carcinoma in the United States: Influence of ethnic status. Am. J. Gastroenterol. 98 (9), 2060–2063. doi:10.1111/j.1572-0241.2003.t01-1-07641.x
Duong F. H., Christen V., Lin S., Heim M. H. (2010). Hepatitis C virus-induced up-regulation of protein phosphatase 2A inhibits histone modification and DNA damage repair. Hepatology 51 (3), 741–751. doi:10.1002/hep.23388
Ebara M., OhtoM. , Shinagawa T., SugiuraN. , Kimura K., MatSutani S., et al. (1986). Natural history of minute hepatocellular carcinoma smaller than three centimeters complicating cirrhosis. A study in 22 patients. Gastroenterology 90 (2), 289–298. doi:10.1016/0016-5085(86)90923-6
El-Khazragy N., Elshimy A. A., Hassan S. S., Shaaban M. H., Bayoumi A. H., El Magdoub H. M., et al. (2020). lnc-HOTAIR predicts hepatocellular carcinoma in chronic hepatitis C genotype 4 following direct-acting antivirals therapy. Mol. Carcinog. 59 (12), 1382–1391. doi:10.1002/mc.23263
El-Khoueiry A. B., Sangro B., Yau T., Crocenzi T. S., Kudo M., Hsu C., et al. (2017). Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 389 (10088), 2492–2502. doi:10.1016/S0140-6736(17)31046-2
El-Serag H. B., Mason A. C. (1999). Rising incidence of hepatocellular carcinoma in the United States. N. Engl. J. Med. 340 (10), 745–750. doi:10.1056/NEJM199903113401001
El-Serag H. B., Rudolph K. L. (2007). Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 132 (7), 2557–2576. doi:10.1053/j.gastro.2007.04.061
Ezeh U. I., Turek P. J., Reijo R. A., Clark A. T. (2005). Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are expressed in both seminoma and breast carcinoma. Cancer 104 (10), 2255–2265. doi:10.1002/cncr.21432
Feldman D. E., Chen C., Punj V., Tsukamoto H., Machida K. (2012). Pluripotency factor-mediated expression of the leptin receptor (OB-R) links obesity to oncogenesis through tumor-initiating stem cells. Proc. Natl. Acad. Sci. U. S. A. 109 (3), 829–834. doi:10.1073/pnas.1114438109
Feldman D. E., Chen C., Punj V., Machida K. (2013). The TBC1D15 oncoprotein controls stem cell self-renewal through destabilization of the Numb-p53 complex. PLoS One 8 (2), e57312. doi:10.1371/journal.pone.0057312
Ferrasi A. C., Fernandez G. J., Grotto R. M. T., Silva G. F., Goncalves J., Costa M. C., et al. (2020). New LncRNAs in chronic hepatitis C progression: From fibrosis to hepatocellular carcinoma. Sci. Rep. 10 (1), 9886. doi:10.1038/s41598-020-66881-2
Fu N., Niu X., Wang Y., Du H., Wang B., Du J., et al. (2016). Role of LncRNA-activated by transforming growth factor beta in the progression of hepatitis C virus-related liver fibrosis. Discov. Med. 22 (119), 29–42.
Fujimoto A., Furuta M., Totoki Y., Tsunoda T., Kato M., Shiraishi Y., et al. (2016). Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 48 (5), 500–509. doi:10.1038/ng.3547
Fujita N., Sugimoto R., MaN. , Tanaka H., IwasaM. , KobaYashi Y., et al. (2008). Comparison of hepatic oxidative DNA damage in patients with chronic Hepatitis B and C. J. Viral Hepat. 15 (7), 498–507. doi:10.1111/j.1365-2893.2008.00972.x
Gerbes A., Zoulim F., Tilg H., Dufour J. F., Bruix J., Paradis V., et al. (2018). Gut roundtable meeting paper: Selected recent advances in hepatocellular carcinoma. Gut 67 (2), 380–388. doi:10.1136/gutjnl-2017-315068
Gibbs C. P., Kukekov V. G., Reith J. D., Tchigrinova O., Suslov O. N., Scott E. W., et al. (2005). Stem-like cells in bone sarcomas: Implications for tumorigenesis. Neoplasia 7 (11), 967–976. doi:10.1593/neo.05394
Gottesman M. M. (2002). Mechanisms of cancer drug resistance. Annu. Rev. Med. 53, 615–627. doi:10.1146/annurev.med.53.082901.103929
Guichard C., Amaddeo G., Imbeaud S., Ladeiro Y., Pelletier L., Maad I. B., et al. (2012). Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 44 (6), 694–698. doi:10.1038/ng.2256
Haaf T., Golub E. I., Reddy G., Radding C. M., Ward D. C. (1995). Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. U. S. A. 92 (6), 2298–2302. doi:10.1073/pnas.92.6.2298
Hagstrom H., Tynelius P., Rasmussen F. (2018). High BMI in late adolescence predicts future severe liver disease and hepatocellular carcinoma: A national, population-based cohort study in 1.2 million men. Gut 67 (8), 1536–1542. doi:10.1136/gutjnl-2016-313622
Hai H., Tamori A., Thuy L. T. T., Yoshida K., Hagihara A., Kawamura E., et al. (2017). Polymorphisms in MICA, but not in DEPDC5, HCP5 or PNPLA3, are associated with chronic hepatitis C-related hepatocellular carcinoma. Sci. Rep. 7 (1), 11912. doi:10.1038/s41598-017-10363-5
Hajarizadeh B., Grebely J., Dore G. J. (2013). Epidemiology and natural history of HCV infection. Nat. Rev. Gastroenterol. Hepatol. 10 (9), 553–562. doi:10.1038/nrgastro.2013.107
Hanahan D., Weinberg R. A. (2011). Hallmarks of cancer: The next generation. Cell 144 (5), 646–674. doi:10.1016/j.cell.2011.02.013
Hart A. H., Hartley L., Parker K., Ibrahim M., Looijenga L. H. J., Pauchnik M., et al. (2005). The pluripotency homeobox gene NANOG is expressed in human germ cell tumors. Cancer 104 (10), 2092–2098. doi:10.1002/cncr.21435
Hassan M. M., Hwang L. Y., Hatten C. J., Swaim M., Li D., Abbruzzese J. L., et al. (2002). Risk factors for hepatocellular carcinoma: Synergism of alcohol with viral hepatitis and diabetes mellitus. Hepatology 36 (5), 1206–1213. doi:10.1053/jhep.2002.36780
Herceg Z. (2007). Epigenetics and cancer: Towards an evaluation of the impact of environmental and dietary factors. Mutagenesis 22 (2), 91–103. doi:10.1093/mutage/gel068
Herceg Z., Paliwal A. (2011). Epigenetic mechanisms in hepatocellular carcinoma: How environmental factors influence the epigenome. Mutat. Res. 727 (3), 55–61. doi:10.1016/j.mrrev.2011.04.001
Hiom K. (1999). Dna repair: Rad52 - the means to an end. Curr. Biol. 9 (12), R446–R448. doi:10.1016/s0960-9822(99)80278-4
Hoei-Hansen C. E., Almstrup K., Nielsen J. E., BraSk Sonne S., GraemN. , Skakkebaek N. E., et al. (2005). Stem cell pluripotency factor NANOG is expressed in human fetal gonocytes, testicular carcinoma in situ and germ cell tumours. Histopathology 47 (1), 48–56. doi:10.1111/j.1365-2559.2005.02182.x
Holmes J. A., Chung R. T. (2016). Viral hepatitis: HCV compartmentalization in HCC: Driver, passenger or both? Nat. Rev. Gastroenterol. Hepatol. 13 (5), 254–256. doi:10.1038/nrgastro.2016.46
Hoshida Y., Fuchs B. C., Bardeesy N., Baumert T. F., Chung R. T. (2014). Pathogenesis and prevention of hepatitis C virus-induced hepatocellular carcinoma. J. Hepatol. 61 (1), S79–S90. doi:10.1016/j.jhep.2014.07.010
Hou J., Lin L., Zhou W., Wang Z., Ding G., Dong Q., et al. (2011). Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 19 (2), 232–243. doi:10.1016/j.ccr.2011.01.001
Hou W., Bonkovsky H. L. (2013). Non-coding RNAs in hepatitis C-induced hepatocellular carcinoma: Dysregulation and implications for early detection, diagnosis and therapy. World J. Gastroenterol. 19 (44), 7836–7845. doi:10.3748/wjg.v19.i44.7836
Hritz I., Mandrekar P., Velayudham A., Catalano D., Dolganiuc A., Kodys K., et al. (2008). The critical role of toll-like receptor (TLR) 4 in alcoholic liver disease is independent of the common TLR adapter MyD88. Hepatology 48 (4), 1224–1231. doi:10.1002/hep.22470
Hsu I. C., Tokiwa T., Bennett W., Metcalf R. A., Welsh J. A., Sun T., et al. (1993). p53 gene mutation and integrated Hepatitis B viral DNA sequences in human liver cancer cell lines. Carcinogenesis 14 (5), 987–992. doi:10.1093/carcin/14.5.987
Huang D. Q., El-Serag H. B., Loomba R. (2021). Global epidemiology of NAFLD-related HCC: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 18 (4), 223–238. doi:10.1038/s41575-020-00381-6
Huang G., Jiang H., He Y., Lin Y., Xia W., Luo Y., et al. (2018). LncMAPK6 drives MAPK6 expression and liver TIC self-renewal. J. Exp. Clin. Cancer Res. 37 (1), 105. doi:10.1186/s13046-018-0770-y
Huang S., He X. (2011). The role of microRNAs in liver cancer progression. Br. J. Cancer 104 (2), 235–240. doi:10.1038/sj.bjc.6606010
Hussain S. P., Schwank J., StaibF. , Wang X. W., Harris C. C. (2007). TP53 mutations and hepatocellular carcinoma: Insights into the etiology and pathogenesis of liver cancer. Oncogene 26 (15), 2166–2176. doi:10.1038/sj.onc.1210279
Ikushima H., Todo T., Ino Y., Takahashi M., Saito N., Miyazawa K., et al. (2011). Glioma-initiating cells retain their tumorigenicity through integration of the Sox axis and Oct4 protein. J. Biol. Chem. 286 (48), 41434–41441. doi:10.1074/jbc.M111.300863
International Consensus Group for Hepatocellular Neoplasia (2009). Pathologic diagnosis of early hepatocellular carcinoma: A report of the international consensus group for hepatocellular neoplasia. Hepatology 49 (2), 658–664. doi:10.1002/hep.22709
Jaiswal M., LaRusso N. F., Burgart L. J., Gores G. J. (2000). Inflammatory cytokines induce DNA damage and inhibit DNA repair in cholangiocarcinoma cells by a nitric oxide-dependent mechanism. Cancer Res. 60 (1), 184–190. doi:10.1053/gast.2001.20875
Jaiswal M., LaRusso N. F., Shapiro R. A., Billiar T. R., Gores G. J. (2001). Nitric oxide-mediated inhibition of DNA repair potentiates oxidative DNA damage in cholangiocytes. Gastroenterology 120 (1), 190–199. doi:10.1053/gast.2001.20875
Janssen H. L., Reesink H. W., Lawitz E. J., Zeuzem S., Rodriguez-Torres M., Patel K., et al. (2013). Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368 (18), 1685–1694. doi:10.1056/NEJMoa1209026
Jeong S. W., Jang J. Y., Chung R. T. (2012). Hepatitis C virus and hepatocarcinogenesis. Clin. Mol. Hepatol. 18 (4), 347–356. doi:10.3350/cmh.2012.18.4.347
Jing G. Y., Zheng X. Z., Ji X. X. (2021). lncRNA HAND2-AS1 overexpression inhibits cancer cell proliferation in hepatocellular carcinoma by downregulating RUNX2 expression. J. Clin. Lab. Anal. 35 (4), e23717. doi:10.1002/jcla.23717
Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., Sarnow P. (2005). Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309 (5740), 1577–1581. doi:10.1126/science.1113329
Kamegaya Y., Hiasa Y., Zukerberg L., Fowler N., Blackard J. T., Lin W., et al. (2005). Hepatitis C virus acts as a tumor accelerator by blocking apoptosis in a mouse model of hepatocarcinogenesis. Hepatology 41 (3), 660–667. doi:10.1002/hep.20621
Kamel M. M., Matboli M., Sallam M., Montasser I. F., Saad A. S., El-Tawdi A. H. F. (2016). Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma. Transl. Res. 168, 134–145. doi:10.1016/j.trsl.2015.10.002
Kan Z., Zheng H., Liu X., Li S., Barber T. D., Gong Z., et al. (2013). Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 23 (9), 1422–1433. doi:10.1101/gr.154492.113
Kanda M., Tateishi R., Yoshida H., Sato T., Masuzaki R., Ohki T., et al. (2008). Extrahepatic metastasis of hepatocellular carcinoma: Incidence and risk factors. Liver Int. 28 (9), 1256–1263. doi:10.1111/j.1478-3231.2008.01864.x
Kang Y., Zhu X., Xu Y., Tang Q., Huang Z., Zhao Z., et al. (2018). Energy stress-induced lncRNA HAND2-AS1 represses HIF1α-mediated energy metabolism and inhibits osteosarcoma progression. Am. J. Cancer Res. 8 (3), 526–537.
Kao C. F., Chen S. Y., Chen J. Y., Wu Lee Y. H. (2004). Modulation of p53 transcription regulatory activity and post-translational modification by hepatitis C virus core protein. Oncogene 23 (14), 2472–2483. doi:10.1038/sj.onc.1207368
Kasprzak A., Adamek A. (2008). Role of hepatitis C virus proteins (C, NS3, NS5A) in hepatic oncogenesis. Hepatol. Res. 38 (1), 1–26. doi:10.1111/j.1872-034X.2007.00261.x
Kim J., Woo A. J., Chu J., Snow J. W., Fujiwara Y., Kim C. G., et al. (2010). A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143 (2), 313–324. doi:10.1016/j.cell.2010.09.010
Kitisin K., GaNesaNN. , Tang Y., Jogunoori W., Volpe E. A., Kim S. S., et al. (2007). Disruption of transforming growth factor-beta signaling through beta-spectrin ELF leads to hepatocellular cancer through cyclin D1 activation. Oncogene 26 (50), 7103–7110. doi:10.1038/sj.onc.1210513
Knoblich J. A. (2010). Asymmetric cell division: Recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11 (12), 849–860. doi:10.1038/nrm3010
Korenaga M., Wang T., Li Y., Showalter L. A., Chan T., Sun J., et al. (2005). Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J. Biol. Chem. 280 (45), 37481–37488. doi:10.1074/jbc.M506412200
Kulik L., Heimbach J. K., Zaiem F., Almasri J., Prokop L. J., Wang Z., et al. (2018). Therapies for patients with hepatocellular carcinoma awaiting liver transplantation: A systematic review and meta-analysis. Hepatology 67 (1), 381–400. doi:10.1002/hep.29485
Ladeiro Y., Couchy G., Balabaud C., Bioulac-Sage P., Pelletier L., Rebouissou S., et al. (2008). MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology 47 (6), 1955–1963. doi:10.1002/hep.22256
Lai C. K., Jeng K. S., Machida K., Cheng Y. S., Lai M. M. C. (2008). Hepatitis C virus NS3/4A protein interacts with ATM, impairs DNA repair and enhances sensitivity to ionizing radiation. Virology 370 (2), 295–309. doi:10.1016/j.virol.2007.08.037
Lange C. M., Bibert S., Dufour J. F., Cellerai C., Cerny A., Heim M. H., et al. (2013). Comparative genetic analyses point to HCP5 as susceptibility locus for HCV-associated hepatocellular carcinoma. J. Hepatol. 59 (3), 504–509. doi:10.1016/j.jhep.2013.04.032
Lee H. C., Kim M., Wands J. R. (2006). Wnt/Frizzled signaling in hepatocellular carcinoma. Front. Biosci. 11, 1901–1915. doi:10.2741/1933
Lee S., Lee H. J., Kim J. H., Lee H. S., Jang J. J., Kang G. H. (2003). Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am. J. Pathol. 163 (4), 1371–1378. doi:10.1016/S0002-9440(10)63495-5
Lerat H., Honda M., Beard M. R., Loesch K., Sun J., Yang Y., et al. (2002). Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology 122 (2), 352–365. doi:10.1053/gast.2002.31001
Li D., Liu X., Lin L., Hou J., Li N., Wang C., et al. (2011). MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J. Biol. Chem. 286 (42), 36677–36685. doi:10.1074/jbc.M111.270561
Li K., Lemon S. M. (2013). Innate immune responses in hepatitis C virus infection. Semin. Immunopathol. 35 (1), 53–72. doi:10.1007/s00281-012-0332-x
Liang T. J., Heller T. (2004). Pathogenesis of hepatitis C-associated hepatocellular carcinoma. Gastroenterology 127 (5), S62–S71. doi:10.1053/j.gastro.2004.09.017
Lim L. J., Wong S. Y. S., Huang F., Lim S., Chong S. S., Ooi L. L., et al. (2019). Roles and regulation of long noncoding RNAs in hepatocellular carcinoma. Cancer Res. 79 (20), 5131–5139. doi:10.1158/0008-5472.CAN-19-0255
Lin H. H., Feng W. C., Lu L. C., Shao Y. Y., Hsu C. H., Cheng A. L. (2016). Inhibition of the Wnt/β-catenin signaling pathway improves the anti-tumor effects of sorafenib against hepatocellular carcinoma. Cancer Lett. 381 (1), 58–66. doi:10.1016/j.canlet.2016.07.013
Llovet J. M., Bruix J. (2008). Molecular targeted therapies in hepatocellular carcinoma. Hepatology 48 (4), 1312–1327. doi:10.1002/hep.22506
Llovet J. M. (2014). Focal gains of VEGFA: Candidate predictors of sorafenib response in hepatocellular carcinoma. Cancer Cell 25 (5), 560–562. doi:10.1016/j.ccr.2014.04.019
Llovet J. M., Pena C. E. A., Lathia C. D., Shan M., Meinhardt G., Bruix J., et al. (2012). Plasma biomarkers as predictors of outcome in patients with advanced hepatocellular carcinoma. Clin. Cancer Res. 18 (8), 2290–2300. doi:10.1158/1078-0432.CCR-11-2175
Llovet J. M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J. F., et al. (2008). Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359 (4), 378–390. doi:10.1056/NEJMoa0708857
Llovet J. M., Villanueva A. (2016). Liver cancer: Effect of HCV clearance with direct-acting antiviral agents on HCC. Nat. Rev. Gastroenterol. Hepatol. 13 (10), 561–562. doi:10.1038/nrgastro.2016.140
Loh Y. H., Wu Q., Chew J. L., Vega V. B., Zhang W., Chen X., et al. (2006). The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38 (4), 431–440. doi:10.1038/ng1760
Lorini S., Gragnani L., Zignego A. L. (2020). The relevance of MicroRNAs in the pathogenesis and prognosis of HCV-disease: The emergent role of miR-17-92 in cryoglobulinemic vasculitis. Viruses 12 (12), E1364. doi:10.3390/v12121364
Ma S., Chan K. W., Hu L., Lee T. K. W., Wo J. Y. H., Ng I. O. L., et al. (2007). Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 132 (7), 2542–2556. doi:10.1053/j.gastro.2007.04.025
Ma S., Lee T. K., Zheng B. J., Chan K. W., Guan X. Y. (2008). CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene 27 (12), 1749–1758. doi:10.1038/sj.onc.1210811
Machida K. (2020). Cell fate, metabolic reprogramming and lncRNA of tumor-initiating stem-like cells induced by alcohol. Chem. Biol. Interact. 323, 109055. doi:10.1016/j.cbi.2020.109055
Machida K., Cheng K. T. N., Sung V. M. H., Shimodaira S., Lindsay K. L., Levine A. M., et al. (2004a). Hepatitis C virus induces a mutator phenotype: Enhanced mutations of immunoglobulin and protooncogenes. Proc. Natl. Acad. Sci. U. S. A. 101 (12), 4262–4267. doi:10.1073/pnas.0303971101
Machida K., Cheng K. T. H., Lai C. K., Jeng K. S., Sung V. M. H., Lai M. M. C. (2006). Hepatitis C virus triggers mitochondrial permeability transition with production of reactive oxygen species, leading to DNA damage and STAT3 activation. J. Virol. 80 (14), 7199–7207. doi:10.1128/JVI.00321-06
Machida K., Cheng K. T. H., Sung V. M. H., Lee K. J., Levine A. M., Lai M. M. C. (2004b). Hepatitis C virus infection activates the immunologic (type II) isoform of nitric oxide synthase and thereby enhances DNA damage and mutations of cellular genes. J. Virol. 78 (16), 8835–8843. doi:10.1128/JVI.78.16.8835-8843.2004
Machida K., Tsukamoto H., Mkrtchyan H., Duan L., Dynnyk A., Liu H. M., et al. (2009). Toll-like receptor 4 mediates synergism between alcohol and HCV in hepatic oncogenesis involving stem cell marker Nanog. Proc. Natl. Acad. Sci. U. S. A. 106 (5), 1548–1553. doi:10.1073/pnas.0807390106
Majumder M., Ghosh A. K., Steele R., Ray R., Ray R. B. (2001). Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J. Virol. 75 (3), 1401–1407. doi:10.1128/JVI.75.3.1401-1407.2001
Mandrekar P., Bala S., Catalano D., Kodys K., Szabo G. (2009). The opposite effects of acute and chronic alcohol on lipopolysaccharide-induced inflammation are linked to IRAK-M in human monocytes. J. Immunol. 183 (2), 1320–1327. doi:10.4049/jimmunol.0803206
Martin G. R. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U. S. A. 78 (12), 7634–7638. doi:10.1073/pnas.78.12.7634
Martin-Belmonte F., Perez-Moreno M. (2012). Epithelial cell polarity, stem cells and cancer. Nat. Rev. Cancer 12 (1), 23–38. doi:10.1038/nrc3169
McGivern D. R., Lemon S. M. (2011). Virus-specific mechanisms of carcinogenesis in hepatitis C virus associated liver cancer. Oncogene 30 (17), 1969–1983. doi:10.1038/onc.2010.594
Milward A., Mankouri J., Harris M. (2010). Hepatitis C virus NS5A protein interacts with beta-catenin and stimulates its transcriptional activity in a phosphoinositide-3 kinase-dependent fashion. J. Gen. Virol. 91 (2), 373–381. doi:10.1099/vir.0.015305-0
Miura K., Taura K., Kodama Y., Schnabl B., Brenner D. A. (2008). Hepatitis C virus-induced oxidative stress suppresses hepcidin expression through increased histone deacetylase activity. Hepatology 48 (5), 1420–1429. doi:10.1002/hep.22486
Mohyeldeen M., Ibrahim S., Shaker O., Helmy H. (2020). Serum expression and diagnostic potential of long non-coding RNAs NEAT1 and TUG1 in viral hepatitis C and viral hepatitis C-associated hepatocellular carcinoma. Clin. Biochem. 84, 38–44. doi:10.1016/j.clinbiochem.2020.06.005
Morishita A., Fujita K., Iwama H., Chiyo T., Fujihara S., Oura K., et al. (2020). Role of microRNA-210-3p in Hepatitis B virus-related hepatocellular carcinoma. Am. J. Physiol. Gastrointest. Liver Physiol. 318 (3), G401–G409. doi:10.1152/ajpgi.00269.2019
Moriya K., Fujie H., Shintani Y., Yotsuyanagi H., TsuTsumi T., Ishibashi K., et al. (1998). The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 4 (9), 1065–1067. doi:10.1038/2053
Muller C. (2006). Chronic Hepatitis B and C--current treatment and future therapeutic prospects. Wien. Med. Wochenschr. 156 (13-14), 391–396. doi:10.1007/s10354-006-0314-5
Munakata T., Nakamura M., Liang Y., Li K., Lemon S. M. (2005). Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 102 (50), 18159–18164. doi:10.1073/pnas.0505605102
Naas T., Ghorbani M., Alvarez-Maya I., Lapner M., Kothary R., De Repentigny Y., et al. (2005). Characterization of liver histopathology in a transgenic mouse model expressing genotype 1a hepatitis C virus core and envelope proteins 1 and 2. J. Gen. Virol. 86 (8), 2185–2196. doi:10.1099/vir.0.80969-0
Nakamura Y., Mizuguchi T., Tanimizu N., Ichinohe N., Ooe H., Kawamoto M., et al. (2014). Preoperative hepatocyte transplantation improves the survival of rats with nonalcoholic steatohepatitis-related cirrhosis after partial hepatectomy. Cell Transpl. 23 (10), 1243–1254. doi:10.3727/096368913X668645
Newell P., Toffanin S., Villanueva A., Chiang D. Y., Minguez B., Cabellos L., et al. (2009). Ras pathway activation in hepatocellular carcinoma and anti-tumoral effect of combined sorafenib and rapamycin in vivo. J. Hepatol. 51 (4), 725–733. doi:10.1016/j.jhep.2009.03.028
Nguyen L. N., Furuya M. H., Wolfraim L. A., Nguyen A. P., Holdren M. S., Campbell J. S., et al. (2007). Transforming growth factor-beta differentially regulates oval cell and hepatocyte proliferation. Hepatology 45 (1), 31–41. doi:10.1002/hep.21466
Nhieu J. T., Renard C. A., Wei Y., Cherqui D., Zafrani E. S., Buendia M. A. (1999). Nuclear accumulation of mutated beta-catenin in hepatocellular carcinoma is associated with increased cell proliferation. Am. J. Pathol. 155 (3), 703–710. doi:10.1016/s0002-9440(10)65168-1
Nishida N., Goel A. (2011). Genetic and epigenetic signatures in human hepatocellular carcinoma: A systematic review. Curr. Genomics 12 (2), 130–137. doi:10.2174/138920211795564359
Nishida N., Nagasaka T., Nishimura T., Ikai I., Boland C. R., Goel A. (2008). Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology 47 (3), 908–918. doi:10.1002/hep.22110
Okuda K. (2000). Hepatocellular carcinoma. J. Hepatol. 32 (1), 225–237. doi:10.1016/s0168-8278(00)80428-6
Okuda M., Li K., Beard M. R., Showalter L. A., Scholle F., Lemon S. M., et al. (2002). Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology 122 (2), 366–375. doi:10.1053/gast.2002.30983
Oura K., Morishita A., Masaki T. (2020). Molecular and functional roles of MicroRNAs in the progression of hepatocellular carcinoma-A review. Int. J. Mol. Sci. 21 (21), E8362. doi:10.3390/ijms21218362
Pan G., Thomson J. A. (2007). Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 17 (1), 42–49. doi:10.1038/sj.cr.7310125
Park C. Y., Choi S. H., Kang S. M., Kang J. I., Ahn B. Y., Kim H., et al. (2009). Nonstructural 5A protein activates beta-catenin signaling cascades: Implication of hepatitis C virus-induced liver pathogenesis. J. Hepatol. 51 (5), 853–864. doi:10.1016/j.jhep.2009.06.026
Pavio N., Battaglia S., Boucreux D., Arnulf B., Sobesky R., Hermine O., et al. (2005). Hepatitis C virus core variants isolated from liver tumor but not from adjacent non-tumor tissue interact with Smad3 and inhibit the TGF-beta pathway. Oncogene 24 (40), 6119–6132. doi:10.1038/sj.onc.1208749
Perrin-Cocon L., Aublin-Gex A., Sestito S. E., Shirey K. A., Patel M. C., Andre P., et al. (2017). TLR4 antagonist FP7 inhibits LPS-induced cytokine production and glycolytic reprogramming in dendritic cells, and protects mice from lethal influenza infection. Sci. Rep. 7, 40791. doi:10.1038/srep40791
Rao S., Orkin S. H. (2006). Unraveling the transcriptional network controlling ES cell pluripotency. Genome Biol. 7 (8), 230. doi:10.1186/gb-2006-7-8-230
Ray R. B., Meyer K. (2000). Hepatitis C virus core protein promotes immortalization of primary human hepatocytes. Virology 271 (1), 197–204. doi:10.1006/viro.2000.0295
Refai N. S., Louka M. L., Halim H. Y., Montasser I. (2019). Long non-coding RNAs (CASC2 and TUG1) in hepatocellular carcinoma: Clinical significance. J. Gene Med. 21 (9), e3112. doi:10.1002/jgm.3112
Rehermann B. (2013). Pathogenesis of chronic viral hepatitis: Differential roles of T cells and NK cells. Nat. Med. 19 (7), 859–868. doi:10.1038/nm.3251
Roche B., Coilly A., Duclos-Vallee J. C., Samuel D. (2018). The impact of treatment of hepatitis C with DAAs on the occurrence of HCC. Liver Int. 38 (1), 139–145. doi:10.1111/liv.13659
Rojas A., Gil-Gomez A., de la Cruz-Ojeda P., Munoz-Hernandez R., Sanchez-Torrijos Y., Gallego-Duran R., et al. (2022). Long non-coding RNA H19 as a biomarker for hepatocellular carcinoma. Liver Int. 42, 1410–1422. doi:10.1111/liv.15230
Rongrui L., Na H., Zongfang L., Fanpu J., Shiwen J. (2014). Epigenetic mechanism involved in the HBV/HCV-related hepatocellular carcinoma tumorigenesis. Curr. Pharm. Des. 20 (11), 1715–1725. doi:10.2174/13816128113199990533
Roskams T. A., Theise N. D., Balabaud C., Bhagat G., Bhathal P. S., Bioulac-Sage P., et al. (2004). Nomenclature of the finer branches of the biliary tree: Canals, ductules, and ductular reactions in human livers. Hepatology 39 (6), 1739–1745. doi:10.1002/hep.20130
Roskams T. (2006). Liver stem cells and their implication in hepatocellular and cholangiocarcinoma. Oncogene 25 (27), 3818–3822. doi:10.1038/sj.onc.1209558
Rountree C. B., Senadheera S., Mato J. M., Crooks G. M., Lu S. C. (2008). Expansion of liver cancer stem cells during aging in methionine adenosyltransferase 1A-deficient mice. Hepatology 47 (4), 1288–1297. doi:10.1002/hep.22141
Sabry H. S., Tayel S. I., Enar M. E., Elabd N. S. (2021). Differential expression of long noncoding RNA in hepatocellular carcinoma on top of chronic HCV and HBV infections. Clin. Exp. Hepatol. 7 (4), 337–350. doi:10.5114/ceh.2021.111060
Salk J. J., Fox E. J., Loeb L. A. (2010). Mutational heterogeneity in human cancers: Origin and consequences. Annu. Rev. Pathol. 5, 51–75. doi:10.1146/annurev-pathol-121808-102113
Santagata S., Ligon K. L., Hornick J. L. (2007). Embryonic stem cell transcription factor signatures in the diagnosis of primary and metastatic germ cell tumors. Am. J. Surg. Pathol. 31 (6), 836–845. doi:10.1097/PAS.0b013e31802e708a
Sarasin-Filipowicz M., Krol J., Markiewicz I., Heim M. H., Filipowicz W. (2009). Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat. Med. 15 (1), 31–33. doi:10.1038/nm.1902
Schneiderman R. S., Shmueli E., Kirson E. D., Palti Y. (2010). TTFields alone and in combination with chemotherapeutic agents effectively reduce the viability of MDR cell sub-lines that over-express ABC transporters. BMC Cancer 10, 229. doi:10.1186/1471-2407-10-229
Schulze K., Imbeaud S., Letouze E., Alexandrov L. B., Calderaro J., Rebouissou S., et al. (2015). Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 47 (5), 505–511. doi:10.1038/ng.3252
Shah S. A., Smith J. K., Li Y., Ng S. C., Carroll J. E., Tseng J. F. (2011). Underutilization of therapy for hepatocellular carcinoma in the medicare population. Cancer 117 (5), 1019–1026. doi:10.1002/cncr.25683
Shepard R. L., Cao J., Starling J. J., Dantzig A. H. (2003). Modulation of P-glycoprotein but not MRP1- or BCRP-mediated drug resistance by LY335979. Int. J. Cancer 103 (1), 121–125. doi:10.1002/ijc.10792
Shi L., Peng F., Tao Y., Fan X., Li N. (2016). Roles of long noncoding RNAs in hepatocellular carcinoma. Virus Res. 223, 131–139. doi:10.1016/j.virusres.2016.06.008
Shibata T., Aburatani H. (2014). Exploration of liver cancer genomes. Nat. Rev. Gastroenterol. Hepatol. 11 (6), 340–349. doi:10.1038/nrgastro.2014.6
Shimoda R., NagashiMaM. , SakaMotoM. , YamaguchiN. , HirohaShi S., Yokota J., et al. (1994). Increased formation of oxidative DNA damage, 8-hydroxydeoxyguanosine, in human livers with chronic hepatitis. Cancer Res. 54 (12), 3171–3172.
Sicklick J. K., Li Y. X., Jayaraman A., Kannangai R., Qi Y., Vivekanandan P., et al. (2006a). Dysregulation of the Hedgehog pathway in human hepatocarcinogenesis. Carcinogenesis 27 (4), 748–757. doi:10.1093/carcin/bgi292
Sicklick J. K., Li Y. X., Melhem A., Schmelzer E., Zdanowicz M., Huang J., et al. (2006b). Hedgehog signaling maintains resident hepatic progenitors throughout life. Am. J. Physiol. Gastrointest. Liver Physiol. 290 (5), G859–G870. doi:10.1152/ajpgi.00456.2005
Simonetti R. G., Camma C., FiorelloF. , CottoneM. , RapicettaM. , Marino L., et al. (1992). Hepatitis C virus infection as a risk factor for hepatocellular carcinoma in patients with cirrhosis. A case-control study. Ann. Intern. Med. 116 (2), 97–102. doi:10.7326/0003-4819-116-2-97
Smirnova I. S., Aksenov N. D., Kashuba E. V., Payakurel P., Grabovetsky V. V., Zaberezhny A. D., et al. (2006). Hepatitis C virus core protein transforms murine fibroblasts by promoting genomic instability. Cell. Oncol. 28 (4), 177–190. doi:10.1155/2006/864648
Sobesky R., Feray C., Rimlinger F., Derian N., Dos Santos A., Roque-Afonso A. M., et al. (2007). Distinct hepatitis C virus core and F protein quasispecies in tumoral and nontumoral hepatocytes isolated via microdissection. Hepatology 46 (6), 1704–1712. doi:10.1002/hep.21898
Song E., Lee S. K., Dykxhoorn D. M., Novina C., Zhang D., Crawford K., et al. (2003). Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J. Virol. 77 (13), 7174–7181. doi:10.1128/jvi.77.13.7174-7181.2003
Sonnenday C. J., Dimick J. B., Schulick R. D., Choti M. A. (2007). Racial and geographic disparities in the utilization of surgical therapy for hepatocellular carcinoma. J. Gastrointest. Surg. 11 (12), 1636–1646. doi:10.1007/s11605-007-0315-8
Starke D. W., Chen Y., Bapna C. P., Lesnefsky E. J., Mieyal J. J. (1997). Sensitivity of protein sulfhydryl repair enzymes to oxidative stress. Free Radic. Biol. Med. 23 (3), 373–384. doi:10.1016/s0891-5849(97)00009-9
Street A., Macdonald A., McCormick C., Harris M. (2005). Hepatitis C virus NS5A-mediated activation of phosphoinositide 3-kinase results in stabilization of cellular beta-catenin and stimulation of beta-catenin-responsive transcription. J. Virol. 79 (8), 5006–5016. doi:10.1128/JVI.79.8.5006-5016.2005
Sur S., Sasaki R., Devhare P., Steele R., Ray R., Ray R. B. (2018). Association between MicroRNA-373 and long noncoding RNA NORAD in hepatitis C virus-infected hepatocytes impairs Wee1 expression for growth promotion. J. Virol. 92 (20), e01215–18. doi:10.1128/JVI.01215-18
Suresh D., Srinivas A. N., Kumar D. P. (2020). Etiology of hepatocellular carcinoma: Special focus on fatty liver disease. Front. Oncol. 10, 601710. doi:10.3389/fonc.2020.601710
Tanaka N., Moriya K., Kiyosawa K., Koike K., Aoyama T. (2008). Hepatitis C virus core protein induces spontaneous and persistent activation of peroxisome proliferator-activated receptor alpha in transgenic mice: Implications for HCV-associated hepatocarcinogenesis. Int. J. Cancer 122 (1), 124–131. doi:10.1002/ijc.23056
Tang Y., Kitisin K., Jogunoori W., Li C., Deng C. X., Mueller S. C., et al. (2008). Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc. Natl. Acad. Sci. U. S. A. 105 (7), 2445–2450. doi:10.1073/pnas.0705395105
Testro A. G., Visvanathan K. (2009). Toll-like receptors and their role in gastrointestinal disease. J. Gastroenterol. Hepatol. 24 (6), 943–954. doi:10.1111/j.1440-1746.2009.05854.x
Tian Y., Yang W., Song J., Wu Y., Ni B. (2013). Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Mol. Cell. Biol. 33 (15), 2810–2816. doi:10.1128/MCB.00205-13
Toraih E. A., Ellawindy A., Fala S. Y., Al Ageeli E., Gouda N. S., Fawzy M. S., et al. (2018). Oncogenic long noncoding RNA MALAT1 and HCV-related hepatocellular carcinoma. Biomed. Pharmacother. 102, 653–669. doi:10.1016/j.biopha.2018.03.105
Totoki Y., Tatsuno K., Covington K. R., Ueda H., Creighton C. J., Kato M., et al. (2014). Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 46 (12), 1267–1273. doi:10.1038/ng.3126
Totoki Y., Tatsuno K., Yamamoto S., Arai Y., Hosoda F., Ishikawa S., et al. (2011). High-resolution characterization of a hepatocellular carcinoma genome. Nat. Genet. 43 (5), 464–469. doi:10.1038/ng.804
Trebicka J., Anadol E., Elfimova N., Strack I., Roggendorf M., Viazov S., et al. (2013). Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J. Hepatol. 58 (2), 234–239. doi:10.1016/j.jhep.2012.10.015
Tsai W. C., Hsu S. D., Hsu C. S., Lai T. C., Chen S. J., Shen R., et al. (2012). MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. . 122 (8), 2884–2897. doi:10.1172/JCI63455
Unfried J. P., Fortes P. (2020). LncRNAs in HCV infection and HCV-related liver disease. Int. J. Mol. Sci. 21 (6), E2255. doi:10.3390/ijms21062255
Uthaya Kumar D. B., Chen C. L., Liu J. C., Feldman D. E., Sher L. S., French S., et al. (2016). TLR4 signaling via NANOG cooperates with STAT3 to activate Twist1 and promote formation of tumor-initiating stem-like cells in livers of mice. Gastroenterology 150 (3), 707–719. doi:10.1053/j.gastro.2015.11.002
Vaissiere T., Sawan C., Herceg Z. (2008). Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat. Res. 659 (1-2), 40–48. doi:10.1016/j.mrrev.2008.02.004
Valk-Lingbeek M. E., Bruggeman S. W. M., van Lohuizen M. (2004). Stem cells and cancer; the polycomb connection. Cell 118 (4), 409–418. doi:10.1016/j.cell.2004.08.005
Wakita T., Taya C., KAtsume A., Kato J., Yonekawa H., Kanegae Y., et al. (1998). Efficient conditional transgene expression in hepatitis C virus cDNA transgenic mice mediated by the Cre/loxP system. J. Biol. Chem. 273 (15), 9001–9006. doi:10.1074/jbc.273.15.9001
Wang H., Zhang X., Chen X., Zhang S., Yun Z., Gao Q., et al. (2022). Long non-coding RNA placentaspecific protein 2 regulates the chemosensitivity of cancer cells to cisplatin in hepatocellular carcinoma (HCC) by sponging microRNA-96 to upregulate X-linked inhibitor of apoptosis protein. Bioengineered 13 (4), 10765–10773. doi:10.1080/21655979.2022.2056815
Wang J., Rao S., Chu J., Shen X., Levasseur D. N., Theunissen T. W., et al. (2006). A protein interaction network for pluripotency of embryonic stem cells. Nature 444 (7117), 364–368. doi:10.1038/nature05284
Wang S., Jiang J., Zhang C., Zhang X., Wang C. (2019). Serum lincRNA-p21 expression in primary liver diseases and liver metastatic diseases. Pathol. Res. Pract. 215 (4), 779–783. doi:10.1016/j.prp.2019.01.014
Wang X., Peng J., Wang J., Li M., Wu D., Wu S., et al. (2018). Hepatitis C virus core impacts expression of miR122 and miR204 involved in carcinogenic progression via regulation of TGFBRAP1 and HOTTIP expression. Onco. Targets. Ther. 11, 1173–1182. doi:10.2147/OTT.S149254
Wang Y., Lee A. T. C., Ma J. Z. I., Wang J., Ren J., Yang Y., et al. (2008). Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J. Biol. Chem. 283 (19), 13205–13215. doi:10.1074/jbc.M707629200
Wei W., Huang X., Shen X., Lian J., Chen Y., Wang W., et al. (2022). Overexpression of IncRNA TPT1-AS1 suppresses hepatocellular carcinoma cell proliferation by downregulating CDK2. Crit. Rev. Eukaryot. Gene Expr. 32 (1), 1–9. doi:10.1615/CritRevEukaryotGeneExpr.2021039224
Weiner A., Erickson A. L., Kansopon J., Crawford K., Muchmor E., Hughes A. L., et al. (1995). Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc. Natl. Acad. Sci. U. S. A. 92 (7), 2755–2759. doi:10.1073/pnas.92.7.2755
Wieland S., Makowska Z., Campana B., Calabrese D., Dill M. T., Chung J., et al. (2014). Simultaneous detection of hepatitis C virus and interferon stimulated gene expression in infected human liver. Hepatology 59 (6), 2121–2130. doi:10.1002/hep.26770
Wong C. M., Tsang F. H. C., Ng I. O. L. (2018). Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 15 (3), 137–151. doi:10.1038/nrgastro.2017.169
Wong L. S., Wong C. M. (2021). Decoding the roles of long noncoding RNAs in hepatocellular carcinoma. Int. J. Mol. Sci. 22 (6), 3137. doi:10.3390/ijms22063137
Wu S., Ai H., Zhang K., Yun H., Xie F. (2019). Long non-coding RNA EGOT promotes the malignant phenotypes of hepatocellular carcinoma cells and increases the expression of HMGA2 via down-regulating miR-33a-5p. Onco. Targets. Ther. 12, 11623–11635. doi:10.2147/OTT.S218308
Wurmbach E., Chen Y. b., Khitrov G., Zhang W., Roayaie S., Schwartz M., et al. (2007). Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 45 (4), 938–947. doi:10.1002/hep.21622
Yamane D., McGivern D. R., Wauthier E., Yi M., Madden V. J., Welsch C., et al. (2014). Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat. Med. 20 (8), 927–935. doi:10.1038/nm.3610
Yang S., Cai H., Hu B., Tu J. (2019). LncRNA SAMMSON negatively regulates miR-9-3p in hepatocellular carcinoma cells and has prognostic values. Biosci. Rep. 39 (7). BSR20190615. doi:10.1042/BSR20190615
Yang Z. F., Ho D. W., Ng M. N., Lau C. K., Yu W. C., Ngai P., et al. (2008). Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell 13 (2), 153–166. doi:10.1016/j.ccr.2008.01.013
Yao F., Terrault N. (2001). Hepatitis C and hepatocellular carcinoma. Curr. Treat. Options Oncol. 2 (6), 473–483. doi:10.1007/s11864-001-0069-6
Yao Y., Shu F., Wang F., Wang X., Guo Z., Wang H., et al. (2021). Long noncoding RNA LINC01189 is associated with HCV-hepatocellular carcinoma and regulates cancer cell proliferation and chemoresistance through hsa-miR-155-5p. Ann. Hepatol. 22, 100269. doi:10.1016/j.aohep.2020.09.013
Yeoh G. C., Ernst M., Rose-John S., Akhurst B., Payne C., Long S., et al. (2007). Opposing roles of gp130-mediated STAT-3 and ERK-1/2 signaling in liver progenitor cell migration and proliferation. Hepatology 45 (2), 486–494. doi:10.1002/hep.21535
Yu G. Y., He G., Li C. Y., Tang M., Grivennikov S., Tsai W. T., et al. (2012). Hepatic expression of HCV RNA-dependent RNA polymerase triggers innate immune signaling and cytokine production. Mol. Cell 48 (2), 313–321. doi:10.1016/j.molcel.2012.07.032
Yuan J. M., Govindarajan S., Arakawa K., Yu M. C. (2004). Synergism of alcohol, diabetes, and viral hepatitis on the risk of hepatocellular carcinoma in blacks and whites in the U.S. Cancer 101 (5), 1009–1017. doi:10.1002/cncr.20427
Zender L., Spector M. S., Xue W., Flemming P., Cordon-Cardo C., Silke J., et al. (2006). Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125 (7), 1253–1267. doi:10.1016/j.cell.2006.05.030
Zhang C., Yang X., Qi Q., Gao Y., Wei Q., Han S. (2018). lncRNA-HEIH in serum and exosomes as a potential biomarker in the HCV-related hepatocellular carcinoma. Cancer Biomark. 21 (3), 651–659. doi:10.3233/CBM-170727
Zhang H., Zhu C., Zhao Y., Li M., Wu L., Yang X., et al. (2015). Long non-coding RNA expression profiles of hepatitis C virus-related dysplasia and hepatocellular carcinoma. Oncotarget 6 (41), 43770–43778. doi:10.18632/oncotarget.6087
Zhang J., Wang X., Chen B., Suo G., Zhao Y., Duan Z., et al. (2005). Expression of Nanog gene promotes NIH3T3 cell proliferation. Biochem. Biophys. Res. Commun. 338 (2), 1098–1102. doi:10.1016/j.bbrc.2005.10.071
Zhang Q., Matsuura K., Kleiner D. E., Zamboni F., Alter H. J., Farci P. (2016). Analysis of long noncoding RNA expression in hepatocellular carcinoma of different viral etiology. J. Transl. Med. 14 (1), 328. doi:10.1186/s12967-016-1085-4
Zhao B., Lu Y., Cao X., Zhu W., Kong L., Ji H., et al. (2019). MiRNA-124 inhibits the proliferation, migration and invasion of cancer cell in hepatocellular carcinoma by downregulating lncRNA-UCA1. Onco. Targets. Ther. 12, 4509–4516. doi:10.2147/OTT.S205169
Zheng Y., Lv P., Wang S., Cai Q., Zhang B., Huo F. (2019). LncRNA PLAC2 upregulates p53 to induce hepatocellular carcinoma cell apoptosis. Gene 712, 143944. doi:10.1016/j.gene.2019.143944
Zhong Y., Li Y., Song T., Zhang D. (2019). MiR-718 mediates the indirect interaction between lncRNA SEMA3B-AS1 and PTEN to regulate the proliferation of hepatocellular carcinoma cells. Physiol. Genomics 51 (10), 500–505. doi:10.1152/physiolgenomics.00019.2019
Genetically modified mouse models identify molecular and histological stages during the process of multistep hepatocarcinogenesis. Because of the large number of transgenic mice, only a small selection of representative models for HCC research are discussed in this review (Table A1).
Epigenetics is stable alterations in gene expression without genetic modifications in the DNA sequence (Herceg, 2007). Epigenetic and genetic mechanisms silences cellular genes leading to transformation in human cancers, including HCC (Vaissiere et al., 2008). Contribution of different epigenetic factors, including genomic DNA methylation, histone modifications, and miRNA regulation, contribute to HCC dissemination, invasion, and metastasis. The reversal of deregulated epigenetic changes is emerging treatment of HCC (Nishida and Goel, 2011). High-throughput screening provides targeting inflammation-epigenome cross-talk in HCC to discover novel epigenetic targets (Herceg and Paliwal, 2011). Epigenetic Mechanism promotes the HBV/HCV-Related HCC tumorigenesis (Rongrui et al., 2014). Several LncRNAs regulate cancer biology, including MALAT and etc. LncRNAs have pleiotropic effects on miRNAs, mRNAs, and proteins and alter miRNA and mRNA expression and stability, affect protein expression, degradation, structure, or interactions with transcriptional regulators. Therefore, novel lncRNAs serve as biomarkers to achieve precision therapy for HCC (Lim et al., 2019). The function of the 98% of non-coding sequences in the human genome is elusive (Wong et al., 2018).
Antiviral agents INF with epigenetic drugs (such as DNMT inhibitors or HDAC inhibitors) counteract epigenome changeseract epigenome changes with cytokines (Muller, 2006). Induction of HCV proteins or the infection of HCC cells with HCV cell culture (HCVcc) suppresses histone H4 methylation/acetylation and histone H2AX phosphorylation for HCC development, indicating that HCV-induced overexpression of PP2Ac are associated with HCC via deregulation of epigenetic histone modifications (Duong et al., 2010). HCV infection upregulates histone deacetylation (HDAC) activity through affecting hepcidin expression, a key suppressor of iron availability (Miura et al., 2008). The induced HCV oxidative stress leads to suppression of hepcidin expression by increased HDAC function. Non-coding RNAs [microRNA (miRNA)] are dysregulated in HCV-induced liver carcinogenesis via regulation of gene expression. miRNA targets hundred mRNAs, miRs are diagnostic markers and therapeutic target for personalized therapy. miRNAs promote or inhibits carcinogenesis via activation of oncogenes and/or suppression of tumor suppressors (Chen, 2005). miRNAs are differentially expressed in liver cancer and are related to different stages of liver carcinogenesis, supporting the diagnosis and prognosis tools of miRNAs in HCC patient. HCV requires liver-specific miR-122 for replication (Jopling et al., 2005). Sequestering miR-122 in patients leads to a dose-dependent decrease in HCV viremia in phase 2a trial (Janssen et al., 2013). Mice lacking miR-122 have high tumor incidence (Tsai et al., 2012). The miR-122 abundance is reduced in human advanced fibrosis (Trebicka et al., 2013) and in therapresistant HCV patients (Sarasin-Filipowicz et al., 2009). Recruitment of miR-122 to the HCV genome depletes this important liver-specific miR-122. HCV sequesters anti-tumorigenic miR-122 to promote HCC development. miR-21, miR-17, miR-222, miR-224, and miR-221 are increased in liver cancer (Borel et al., 2012) (Ladeiro et al., 2008) while miR-200, let-7, miR-29, miR-123, miR-122, miR-199a and miR-199 b are decreased in HCCs (Hou et al., 2011; Huang and He, 2011; Anwar and Lehmann, 2015). miR-199a/b-3p prevents the p21-stimulated kinase 4/Raf/MEK/ERL pathway and suppresses HCC. Downregulation of miR-199a/b is associated with poor prognosis and low survival rate (Li et al., 2011). Increased miR-224 is associated with malignancy aggression, deteriorated liver function, and poor prognosis (Wang et al., 2008; Zhuang and Meng, 2015).
HCV caused epigenetic alteration mainly occurred on DNA repair-related genes. Induction of HCV proteins or the infection of HCC cells with HCVcc inhibits histone H4 methylation/acetylation and histone H2AX phosphorylation and inhibited DNA damage repair, indicating that HCV-induced overexpression of PP2Ac promotes hepatocarcinogenesis via dysregulation of epigenetic histone modifications (Duong et al., 2010). HCV increases histone deacetylation (HDAC) activity through negative regulator of iron availability (hepcidin expression) (Miura et al., 2008).
HCV infection accelerates or inhibits the methylation process. DNA methyltransferases (DNMTs) methylates DNA. HCV upregulates DNA methyl transferases, which further block tumor suppressor genes leading to HCC (Tian et al., 2013). HCV core protein upregulates both mRNA and protein expression levels of DNMT1 and DNMT3b, which promotes DNA methylation in HCV-infected hepatocytes (Benegiamo et al., 2012). HCV core protein increases the mRNA and protein levels of DNMT1 and DNMT3b, leading to epigenetic alteration of HCV patients (Benegiamo et al., 2012). HCV tissues have seven hypermethylated markers (COX2, MINT1, CACNA1G, RASSF2, MINT2, Reprimo, and DCC) in comparison to both HBV and normal liver tissues (Nishida et al., 2008). Different HCV proteins NS5A (Kasprzak and Adamek, 2008) promotes hepatocarcinogenesis. Epigenetic event Contribution in HCC development.
• DNA hypermethylation
○ CDH1: Cell adhesion and metastasis
○ RASSF1A: Cell cycle dysregulation
○ P21WAF1/CIP1: Cell cycle dysregulation
○ Gadd45: Response to genotoxic stress
○ MGMT: Dysfunction of DNA repair
○ APC: Cell cycle dysregulation/Dysfunction of DNA repair
• DNA hypomethylation (demethylation)
○ STAT1: Upregulation of JAK/STAT pro-tumorigenic signaling
○ COX-2: Inflammation
• Histone modification
○ PP2Ac: Inflammation
HCV infection modulates several genes, including SOCS-1 Suppressor of cytokine signaling (SOCS) 1 (negative regulator of the JAK/STAT pathway). Expression of SOCS1 leads to reduced JAK /STAT phosphorylation, reduced STAT dimerization and imports to the nucleus and reduced transcription of target genes (Chim et al., 2004). JAK/STAT pathway is pro-tumor signaling (Calvisi et al., 2006).
The adenomatous polyposis coli (APC) tumor suppressor gene encodes a large protein with multiple cellular functions and interactions, including signal transduction in the WNT-signaling pathway (Lee et al., 2006). The APC promoter is methylated in up to 81% of patients with viral hepatitis-induced HCC (Lee et al., 2003). A next generation sequencing of CpG methylation site demonstrates that APC was hypermethylated in HCC tissues to their corresponding non-tumorous tissues (Archer et al., 2010). In contrast, NOTCH4, EMR3, HDAC9, DCL1, HLA-DOA, HLA-DPA1, and ERN1 were hypomethylated in HCC (Archer et al., 2010).
Keywords: cancer stem cell, tumor-initiating stem-like cells (TICs), drug resistance, HCV, hepatocellular carcinoma (HCC), obesity
Citation: Machida K (2022) HCV and tumor-initiating stem-like cells. Front. Physiol. 13:903302. doi: 10.3389/fphys.2022.903302
Received: 24 March 2022; Accepted: 11 July 2022;
Published: 15 September 2022.
Edited by:
Kusum K. Kharbanda, University of Nebraska Medical Center, United StatesReviewed by:
Manuel Romero-Gómez, Sevilla University, SpainCopyright © 2022 Machida. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keigo Machida, a2VpZ28ubWFjaGlkYUBtZWQudXNjLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.